Abstract
Although glucocorticoids suppress proliferation of many cell types and are used in the treatment of certain cancers, trials of glucocorticoid therapy in breast cancer have been a disappointment. Another suggestion that estrogens may affect glucocorticoid action is that the course of some inflammatory diseases tends to be more severe and less responsive to corticosteroid treatment in females. To date, the molecular mechanism of cross-talk between estrogens and glucocorticoids is poorly understood. Here we show that, in both MCF-7 and T47D breast cancer cells, estrogen inhibits glucocorticoid induction of the MKP-1 (mitogen-activated protein kinase phosphatase-1) and serum/glucocorticoid-regulated kinase genes. Estrogen did not affect glucocorticoid-induced glucocorticoid receptor (GR) nuclear translocation but reduced ligand-induced GR phosphorylation at Ser-211, which is associated with the active form of GR. We show that estrogen increases expression of protein phosphatase 5 (PP5), which mediates the dephosphorylation of GR at Ser-211. Gene knockdown of PP5 abolished the estrogen-mediated suppression of GR phosphorylation and induction of MKP-1 and serum/glucocorticoid-regulated kinase. More importantly, after PP5 knockdown estrogen-promoted cell proliferation was significantly suppressed by glucocorticoids. This study demonstrates cross-talk between estrogen-induced PP5 and GR action. It also reveals that PP5 inhibition may antagonize estrogen-promoted events in response to corticosteroid therapy.
Breast cancer is a leading cause of cancer mortality among women. In 2004, 186,772 women were diagnosed with breast cancer and 40,954 women died from breast cancer in United States (1). The female hormone, estrogen, promotes breast cancer cell growth via the estrogen receptor (ER),2 which is expressed in ∼60% of breast cancers (2). Another consequence of estrogen is suggested by observations that the course of some allergic, autoimmune, and malignant diseases is more severe and less responsive to corticosteroid treatment in females (3–5), implicating a role for estrogen in glucocorticoid resistance.
There are two forms of ER, ERα and ERβ, that reside in the cell membrane, cytoplasm, and nucleus (6, 7). Nuclear ER regulates gene transcription by binding to DNA directly at estrogen-response elements or indirectly through interactions with transcriptional factors (7). Membrane-bound ER participates in cell signal transduction by activating G protein subunits and subsequently augments downstream kinase activities, such as p38 and ERK, in endothelial and breast cancer cells (8, 9). Binding to estrogen causes a conformational change in the ER that promotes the assembly of an active transcription complex at estrogen-induced genes such as c-myc and cyclin D1, which mediate the promotion of cell proliferation (10, 11).
Glucocorticoids are well known for their anti-inflammatory, immunosuppressive, and anti-proliferative actions (12–14). They bind to the glucocorticoid receptor (GR) and regulate gene expression by mechanisms similar to ER. However, direct binding to DNA is accomplished through distinct DNA sequence motifs or glucocorticoid-responsive elements (GRE) to regulate the expression of specific genes, such as mitogen-activated protein kinase phosphatase-1 (MKP-1) and serum/glucocorticoid-regulated kinase (SGK) (15–18).
Three pathways have been reported to affect GR phosphorylation and activity. First, MAPK family members p38, JNK, and ERK regulate GR activity differentially. Activation of JNK and ERK inhibits GR transcriptional enhancement, and inhibition of JNK and ERK by inhibitors enhances GR function (19–21). The role of p38 in modulation of GR activity remains controversial (22, 23). Second, cyclin-dependent kinases (CDK) phosphorylate GR and regulate its activity. CDK2 phosphorylate rat GR at Ser-224 and Ser-232 (24, 25), and CDK5 suppresses GR transcriptional activity by attenuating binding of transcriptional cofactors to glucocorticoid-responsive promoters (26). Third, serine/threonine protein phosphatases (PP) negatively regulate GR phosphorylation. Inhibition of PP1, PP2A, PP2B, and PP5 by protein phosphatase inhibitors okadaic acid and calyculin A potentiates GR activity and increases GR phosphorylation (27, 28). Unlike PP1 and PP2A, PP5 acts predominantly in protein complexes because the N-terminal domain of PP5 folds over the catalytic site blocking access to substrates in the absence of other proteins (29, 30). PP5 has been identified in complexes containing GR and heat shock protein 90 (hsp90) (31, 32), suggesting that PP5 may regulate GR activity.
Glucocorticoids have been used in breast cancer therapy to antagonize the growth-promoting effect of estrogen. Nonetheless, clinical trials of glucocorticoid monotherapy in breast cancer have shown only a modest response (33). In advanced breast cancer meta-analyses, the addition of glucocorticoids to either chemotherapy or other endocrine therapy has resulted in increased response rates, but not increased survival (33, 34). To date, the mechanism of glucocorticoid resistance in breast cancer has not been elucidated but would be important to understand if estrogen-driven corticosteroid resistance is to be circumvented. In this study, we investigated the three GR-regulating pathways discussed above, and we identified PP5 to be involved in the inhibition of GR activity by estrogen providing a novel mechanism of cross-talk between estrogen and glucocorticoids.
EXPERIMENTAL PROCEDURES
Materials
17β-Estradiol (E2), dexamethasone (DEX), ICI 182,780, nonimmune rabbit serum, and monoclonal anti-β-actin antibody were purchased from Sigma. PD98059 and roscovitine were purchased from Calbiochem. Purified mouse anti-glucocorticoid receptor antibody was purchased from BD Biosciences. Rabbit polyclonal antibody to glucocorticoid receptor, rabbit polyclonal antibody to phospho-glucocorticoid receptor (Ser-226) antibody, PP5 antibody, and mouse monoclonal antibody to TATA-binding protein (TBP) were purchased from Abcam Inc. (Cambridge, MA). Phospho-glucocorticoid receptor (Ser-211) antibody was purchased from Cell Signaling (Danvers, MA). Rabbit IgG was purchased from Southern Biotechnology Association, Inc. (Birmingham, AL). Normal mouse IgG1 and protein A/G PLUS-agarose were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). FuGENE 6 transfection reagent was purchased from Roche Applied Science. NE-PER nuclear and cytoplasmic extraction reagents were purchased from Pierce. SureSilencing shRNA plasmids were purchased from SuperArray Bioscience Corp. (Frederick, MD). ON-TARGETplus SMARTpool siRNA against PP5 and ON-TARGETplus nontargeting pool siRNA were purchased from Dharmacon (Lafayette, CO). CellQuanti-MTTTM cell viability assay kit was purchased from BioAssay Systems (Hayward, CA). SuperBlock was purchased from Skytec (Logan, UT). Nonimmune donkey serum was purchased from Jackson ImmunoResearch (West Grove, PA). Anti-mouse or anti-rabbit horseradish peroxidase-labeled IgG was purchased from Amersham Biosciences. Chemiluminescent reagent was purchased from PerkinElmer Life Sciences.
Cell Culture and Treatment
MCF-7, T47D, and MDA-MB-231 cell lines were purchased from American Type Culture Collection. For routine proliferation, MCF-7 and MDA-MB-231 cell lines were cultured in minimum Eagle's medium; T47D was cultured in RPMI 1640 medium, supplemented with 10% fetal calf serum, 50 μg/ml streptomycin, and 50 units/ml penicillin. Cells were cultured in hormone-free medium (phenol red-free minimum Eagle's medium containing 2.5% charcoal-stripped serum) at least 2 days before they were treated with 10 nm E2 and 100 nm DEX for the time length as indicated below. An equal volume of ethanol was used as vehicle control.
Proliferation Assay
6 × 103 MCF-7 cells were plated in flat bottom 96-well plates and cultured in hormone-free medium. Two days later, 10 nm E2 was added. The following day, 100 nm DEX was added alone or in combination with E2, and the cells were allowed to grow for an additional 2 days. In experiments that examined the effect of PP5 knockdown on MCF-7 cell line proliferation, the cells were transfected with 0.05 μg of SureSilencing shRNA plasmid per well 24 h prior E2 treatment. The number of viable cells was determined with CellQuanti-MTTTM cell viability assay kit according to the manufacturer's instructions.
Real Time PCR
105 cells per well were cultured in hormone-free medium in 24-well plates and treated with hormones and inhibitors as indicated. Total RNA was prepared using RNeasy mini kit (Qiagen, Valencia, CA). After reverse transcription, 500 ng of cDNA from each sample were analyzed by real time PCR using the dual-labeled fluorigenic probe method on an ABI Prism 7300 real time PCR system (Applied Biosystems). All primers were purchased from Applied Biosystems (Foster City, CA). The ΔΔCt method was utilized to calculate the relative change in target gene expression as an approximation of transcription based on the change in threshold values for control versus treated cells (the cycle number at which the fluorescent signaling crosses the “threshold” or logarithmic increases in cDNA concentration). This method assumes that both reference gene (internal control, i.e. β-actin used in this study) and target genes have similar amplification efficiencies.
Immunofluorescence Assay
GR nuclear translocation and its phosphorylation in response to DEX was analyzed according to Ref. 20 with modifications. In brief, 105 MCF-7 cells were cultured on 18-mm round coverslips in 12-well plates. Cells were fixed with 4% paraformaldehyde at room temperature for 10 min, permeabilized 15 min in Permeabilization Buffer (PBS containing 0.1% Tween 20, 0.1% bovine serum albumin, 0.01% saponin), and blocked at 37 °C for 1 h in Blocking Buffer (2.25% bovine serum albumin, 45% SuperBlock, 10% nonimmune donkey serum). Total GR and phospho-GR (Ser-211) antibodies were diluted 1:50 in Permeabilization Buffer and incubated with the cells at 4 °C overnight. Corresponding amounts of mouse IgG1 and rabbit IgG were used as negative controls, respectively. The cells were washed in PBS containing 0.1% Tween 20 for 20 min, followed by incubation with Cy3-conjugated secondary antibody (donkey anti-mouse or donkey anti-rabbit, diluted 1:500 in Permeabilization Buffer containing 300 nm 4′,6-diamidino-2-phenylindole) at room temperature for 1 h. Cells were washed again and mounted on slides. All slides were analyzed by fluorescence microscopy (Leica Microsystems, Wetzlar, Germany) with the imaging software Slidebook (Intelligent Imaging Innovations, Denver, CO). Mean fluorescence intensity in the cell nuclei defined by 4′,6-diamidino-2-phenylindole staining was assessed. Fifty to 100 cells were analyzed per slide.
Western Blot
Protein samples were resolved on 4–12% BisTris gel (Invitrogen) and transferred to polyvinylidene difluoride membranes. The membranes were incubated in PBS containing specific antibodies, 5% dry milk, and 0.05% Tween 20 at 4 °C overnight. Subsequently, membranes were washed in PBS, 0.05% Tween 20 and incubated for 1 h at room temperature with anti-mouse or anti-rabbit horseradish peroxidase-labeled IgG (1:10,000), washed, incubated with chemiluminescent reagent, and processed for autoradiography.
Knockdown of PP5
105 MCF-7 cells were plated in each well of a 24-well plate and cultured in hormone-free medium. 24 h later, cells were transfected with 100 nm siRNA (or 0.1 μg of shRNA) in 1 ml of medium containing 1 μl of FuGENE 6 transfection reagent. Corresponding amounts of control siRNA or shRNA plasmids were used.
Chromatin Immunoprecipitation Assay
GR binding to GRE was assessed by chromatin immunoprecipitation assay as described previously (35) with modifications. Briefly, 2.5 × 106 cells were used in each precipitation. After sonication, chromatin solution was pre-cleared with 60 μl of protein A/G PLUS-agarose beads and 20 μl of nonspecific serum, followed by precipitation with 60 μl of protein A/G PLUS-agarose beads and specific antibody. Precipitated chromatin complexes were removed from the beads through incubation at 65 °C for 30 min with 550 μl of Elution Buffer (50 mm Tris, pH 8.0, 1 mm EDTA, 1% SDS). 500 μl of eluates were mixed with 25 μl of 5 m NaCl, 1 μl of RNase A (10 mg/ml, DNase-free) and incubated at 65 °C overnight. Samples were then digested with proteinase K, and DNA was purified with QIAquick columns (Qiagen, Valencia, CA) as indicated by the manufacturer, except that the sample was first mixed with PBI buffer (supplied by the manufacturer) for 30 min with agitation (36). Precipitated DNA was quantified by quantitative real time PCR using SYBR green (Applied Biosystems). Primers used to amplify GRE in SGK gene promoter were as follows: 5′-CTTGTTACCTCCTCACGTG-3′ (forward); 5′-GTCGTCTCTGCACTAAAGG-3′ (reverse).
Statistical Analyses
Results are expressed as the mean ± S.E. Statistical analysis was conducted using GraphPad Prism, version 5 (GraphPad Software, La Jolla, CA). Responses within an experiment were expressed as fold change over the control setting. These data were analyzed by the paired Student's t test, pairing by experiment. Before testing, paired difference distributions were examined for outliers, which can indicate violation to the normality assumption of the t test. No outliers were apparent. Tests were performed only for specific pre-planned treatment comparisons. Differences were considered significant at p < 0.05. A minimum of three independent experiments were conducted to allow for statistical comparisons.
RESULTS
Estrogen-promoted MCF-7 Cell Proliferation Is Not Affected by Glucocorticoids
In this study, we chose the MCF-7 breast adenocarcinoma cell line as a model for our experiments. This cell line is known to proliferate in response to estrogen stimulation (37). To study whether glucocorticoids can inhibit estrogen-mediated cell growth, a proliferation assay was carried out. MCF-7 cells were first cultured in hormone-free medium for 2 days to deplete hormones in the cells and then pretreated with 10 nm E2 (the only form of estrogen used in this study) or an equal volume of vehicle (ethanol) for 24 h followed by 100 nm DEX treatment. The purpose of pretreating the cells with estrogen was to mimic the in vivo state, because the breast cancer cells are under estrogen influence before glucocorticoid treatment. Estrogen promoted cell growth by 2.09 ± 0.06-fold as compared with mock control. No change in cell proliferation was noted when cells were cultured in the presence of both estrogen and DEX (1.97 ± 0.04-fold as compared with mock control), indicating that estrogen-promoted cell growth was not inhibited by glucocorticoid treatment.
Estrogen Inhibits Glucocorticoid Action
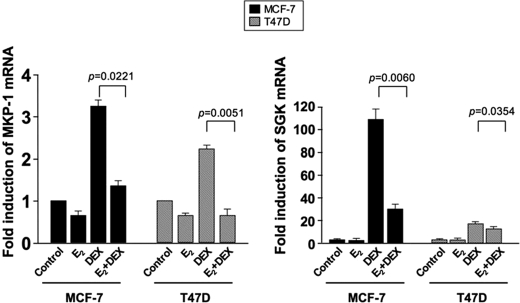
To further explore how estrogen affects the action of glucocorticoids, we assessed the effect of estrogen on DEX induction of MKP-1 and SGK, two well known glucocorticoid-responsive genes (17, 18). DEX alone induced MKP-1 and SGK by 3.23 ± 0.16- and 108.30 ± 9.69-fold, respectively, for 3 h in the MCF-7 cell line. Preincubating MCF-7 cells with estrogen for 24 h prior to DEX treatment significantly inhibited MKP-1 and SGK induction by a mean of 85% (n = 3) and 74% (n = 3), respectively (Fig. 1). Similar estrogen effects were observed in another breast cancer cell line, T47D (Fig. 1). Significantly lower GR expression was found in the T47D cell line as compared with MCF-7 cell line (data not shown). 8 h of stimulation with DEX was determined as an optimal time point for MKP-1 and SGK induction in this cell line.
FIGURE 1.
Estrogen inhibits glucocorticoid induction of MKP-1 and SGK mRNA in MCF-7 and T47D cells. MCF-7 and T47D cells were cultured with 10 nm E2 or vehicle control for 24 h, with or without 100 nm DEX added during the last 3 and 8 h, respectively. MKP-1 and SGK mRNA levels were detected by real time PCR and were normalized to actin mRNA. Fold induction in mRNA expression was calculated as compared with the corresponding vehicle control conditions. Values represent mean ± S.E. (n = 3 experiments).
Estrogen Exerts Its Effect on Glucocorticoids through ER
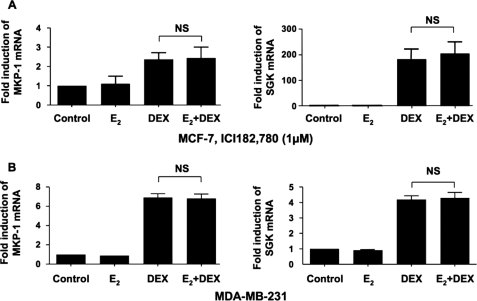
To investigate whether estrogen inhibited DEX induction of MKP-1 and SGK through ER, we employed a selective ER inhibitor ICI 182,780 (inhibitory concentration of 50% (IC50) = 0.29 nm) to antagonize ER in MCF-7 cells. The results demonstrated that with the presence of 1 μm ICI 182,780, DEX induced both MKP-1 and SGK by 2.37 ± 0.36- and 182.90 ± 38.98-fold, respectively, and the DEX-mediated induction of these genes was no longer inhibited by preincubating the cells with estrogen (Fig. 2A). These data indicate that estrogen exerts its inhibitory effect on glucocorticoid action through the ER. In all experiments using inhibitors, MCF-7 cells were treated in parallel with estrogen and DEX as mentioned above without inhibitors. The results showed that the cells were responding to hormones the same way as described in Fig. 1 (data not shown). In the ERα-negative MDA-MB-231 cell line, which expresses only ERβ (38, 39), DEX induction of MKP-1 and SGK was not affected by estrogen (Fig. 2B). These data indicate that estrogen exerts its inhibitory effect on glucocorticoid action through ERα.
FIGURE 2.
Estrogen exerts its effect on glucocorticoids through ER in MCF-7 cells. A, MCF-7 cells were treated with E2 and DEX as described in Fig. 1. 1 μm ICI 182,780 was added to all culture conditions at the time when E2 was added. B, MDA-MB-231 cells were cultured with 10 nm E2 for 24 h, with or without 100 nm DEX added during the last 3 h. Fold inductions in mRNA expression were calculated as compared with the corresponding vehicle control conditions. Values represent mean ± S.E. (n = 3 experiments). NS, not significant.
Estrogen Has No Effect on GR Nuclear Translocation but Inhibits GR Phosphorylation at Ser-211
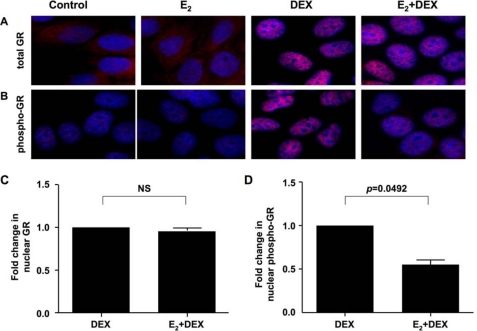
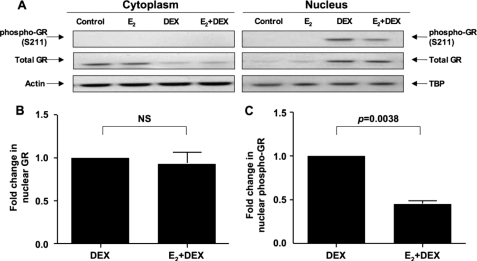
Because DEX induces gene expression through GR, which accumulates in the nucleus after ligand binding, and Ser-211 phosphorylation is associated with the transcriptionally active form of GR (40), we tested GR nuclear translocation and GR phosphorylation at Ser-211 by immunofluorescence (Fig. 3) and Western blot (Fig. 4). DEX alone increased GR nuclear localization with concomitant loss of cytoplasmic GR. This was not affected by preincubation of the cells with estrogen (Fig. 3, A and C). DEX-induced GR phosphorylation at Ser-211 was observed only in the nucleus and was significantly inhibited by estrogen by a mean of 39% (n = 3) (Fig. 3, B and D). Consistent with the immunofluorescence assay results, Western blot of cytoplasmic and nuclear fractions showed that estrogen significantly inhibited DEX-mediated Ser-211 GR phosphorylation by a mean of 55% (n = 3) (Fig. 4, A and C) without affecting GR nuclear localization (Fig. 4, A and B). We also tested GR phosphorylation at Ser-226. No phosphorylation at this site was observed in either the absence or presence of glucocorticoid and/or estrogen (data not shown). The other GR phosphorylation sites were not tested because there were no commercially available antibodies.
FIGURE 3.
DEX-induced GR nuclear translocation and its phosphorylation at Ser-211 in MCF-7 cells treated with estrogen as detected by immunofluorescence assay. Original magnification, ×400; blue, 4′,6-diamidino-2-phenylindole nuclear staining; red, Cy3-GR. MCF-7 cells were cultured on coverslips with 10 nm E2 or vehicle control for 24 h, with or without 100 nm DEX added during the last 1 h. A, total GR staining. B, Ser-211-phosphorylated GR staining. Mean fluorescence intensity in the nuclear area of the cells was measured to calculate fold changes in the nuclear total GR (C) and Ser-211-phosphorylated nuclear GR (D) in the cells treated with DEX alone (set as 1) versus cells cultured with E2 and DEX. Values represent mean ± S.E. (n = 3 experiments). 50–100 cells were analyzed in each experiment. NS, not significant.
FIGURE 4.
DEX-induced GR nuclear translocation and its phosphorylation at Ser-211 in MCF-7 cells treated with estrogen as detected by Western blot. A, cells were treated with hormones as described in Fig. 3. Nuclear and cytoplasmic protein samples were prepared using NE-PER nuclear and cytoplasmic extraction reagents and blotted with antibodies against Ser-211-phosphorylated GR. The membranes were stripped and reprobed with antibodies against total GR. Actin and TBP were used as loading controls for cytoplasmic and nuclear proteins, respectively. Images are representative of three independent experiments. Fold changes in the densitometry readings of nuclear total GR normalized to TBP (B) and Ser-211-phosphorylated nuclear GR normalized to total GR (C) in the cells treated with DEX alone (set as 1) versus cells cultured with E2 and DEX are provided. NS, not significant.
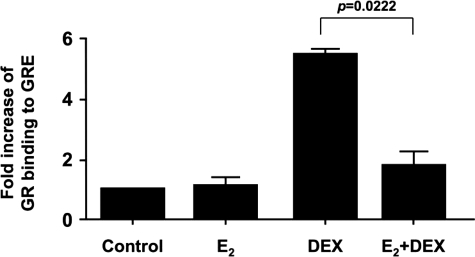
Estrogen Inhibits GR Binding to GRE of SGK Promoter
Chromatin immunoprecipitation was then performed to test GR binding to a well characterized GRE in the promoter region of the SGK gene (18, 41). Within 1 h, DEX induced GR binding to the GRE by 5.49 ± 0.15-fold, and this was significantly inhibited by estrogen by a mean of 82% (n = 3) (Fig. 5). This result suggests that estrogen inhibits DEX induction of SGK by reducing GR recruitment to the SGK promoter.
FIGURE 5.
E2 inhibits GR binding to GRE of SGK promoter. MCF-7 cells were cultured with 10 nm E2 or vehicle control for 24 h, with or without 100 nm DEX added during the last 1 h. The recruitment of GR to GRE of SGK promoter was determined by chromatin immunoprecipitation assay. Quantity of precipitated DNA was normalized to input DNA, and fold increase in GR binding to the GRE was calculated based on the vehicle control. Values represent mean ± S.E. (n = 3 experiments).
Estrogen Suppresses GR Activity through PP5
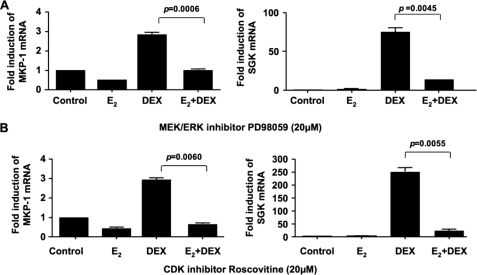
Because MAPK, CDK, and protein phosphatase can regulate GR activity, we examined the effect of estrogens on these three pathways (19–32). First, we screened MAPK phosphorylation using human phospho-MAPK array kit from R & D Systems and found that estrogen treatment increased ERK phosphorylation in MCF-7 cells (data not shown) confirming published data (8, 42). Inhibition of ERK phosphorylation with 20 μm PD98059 (MEK/ERK inhibitor, IC50 = 2 μm), as confirmed by Western blot (data not shown), did not diminish estrogen inhibition of the DEX-mediated induction of either MKP-1 or SGK (Fig. 6A). Similar results were seen when the cells were treated with 20 μm roscovitine (26, 43), a selective CDK2 (IC50 = 700 nm) and CDK5 (IC50 = 200 nm) inhibitor (Fig. 6B) indicating that neither ERK nor CDK pathways were involved in the estrogen-mediated inhibition of glucocorticoid action.
FIGURE 6.
Estrogen inhibition of glucocorticoid function in MCF-7 cells is not affected by ERK and CDK inhibitors. MCF-7 cells were treated with E2 and DEX as described in Fig. 1, and 20 μm MEK/ERK inhibitor PD98059 (A) or 20 μm CDK inhibitor roscovitine (B) were added to the medium 1 h before DEX. Fold inductions in MKP-1 and SGK mRNA expression were calculated as compared with the corresponding vehicle control conditions. Values represent mean ± S.E. (n = 3 experiments).
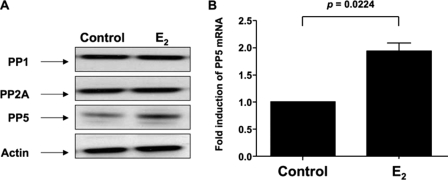
To examine a possible role of protein phosphatase in ER-GR cross-talk, we assessed PP1, PP2A, and PP5 expression in MCF-7 cells following estrogen stimulation. We found that PP5 expression was significantly induced by estrogen both at the protein level (Fig. 7A) and the mRNA level (Fig. 7B). No difference was observed in the expression of PP1 and PP2A.
FIGURE 7.
E2 increases PP5 expression in MCF-7 cells. A, MCF-7 cells were incubated with 10 nm E2 or vehicle control for 24 h, and whole cell extracts were prepared, and PP1, PP2A, and PP5 expression in the cells were analyzed by Western blot. B, PP5 mRNA level was detected by real time PCR after 12 h of incubation with E2 or vehicle control and was normalized to actin mRNA. Fold inductions in PP5 mRNA expression were calculated as compared with vehicle control. Values represent mean ± S.E. (n = 3 experiments).
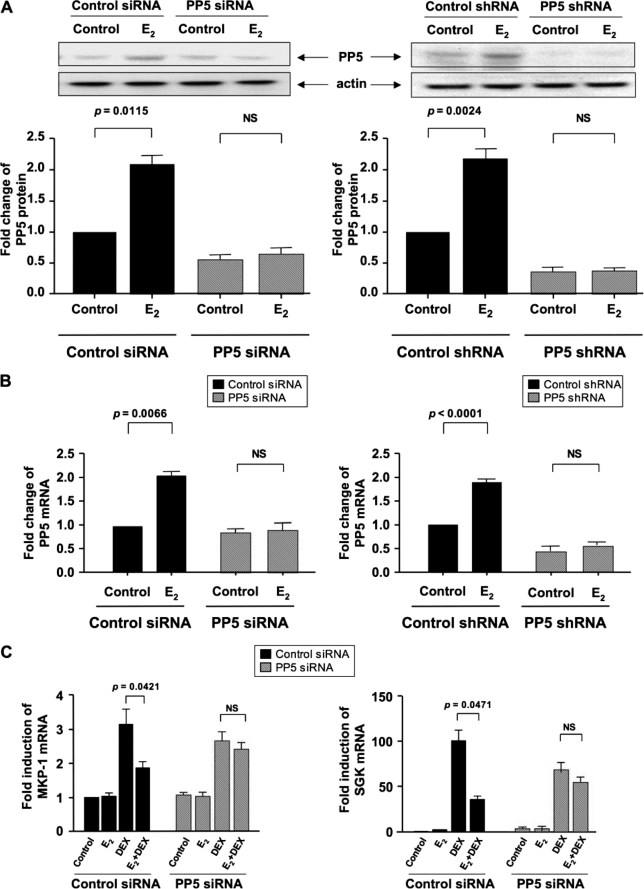
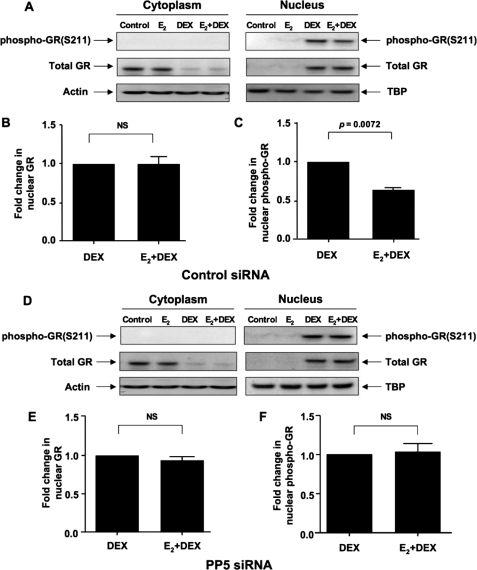
To determine whether estrogen exerts its inhibitory effect on glucocorticoid action through PP5, we specifically knocked down PP5 by RNA interference, and then assessed DEX induction of MKP-1 and SGK. Estrogen-induced PP5 expression was completely abolished by either siRNA or shRNA (Fig. 8, A and B); shRNA more effectively knocked down the base-line expression of PP5 (Fig. 8A). As before, DEX induction of MKP-1 and SGK was inhibited by estrogen if the cells were transfected with control siRNA (Fig. 8C). In contrast, when the cells were transfected with PP5 siRNA, estrogen-mediated inhibition was almost entirely abrogated (Fig. 8C). These data indicate that estrogen suppresses GR activity through increased expression of PP5. Following PP5 knockdown, DEX-mediated GR phosphorylation at Ser-211 was assessed (Fig. 9). Upon PP5 knockdown, DEX-induced GR phosphorylation at Ser-211 was no longer inhibited by estrogen, as compared with control siRNA treatment.
FIGURE 8.
Estrogen inhibition of the glucocorticoid function in MCF-7 cells is abolished by PP5 knockdown. MCF-7 cells were transfected with 100 nm PP5 siRNA (or 0.1 μg of shRNA plasmids). Corresponding amounts of control siRNA or control shRNA plasmids were used for the control treatment groups. 24 h post-transfection the cells were treated with 10 nm E2 or vehicle control for an additional 24 h (A) or 12 h (B) to analyze PP5 protein and mRNA expression, respectively. Whole cell extracts were prepared, and PP5 was detected by Western blot. Band densitometry readings were normalized to actin control. PP5 mRNA expression was analyzed by real time PCR. Fold changes in PP5 protein and mRNA expression were calculated based on vehicle-treated control siRNA condition. MCF-7 cells were transfected with 100 nm of the control siRNA (C), and 24 h post-transfection the cells were treated with E2 and DEX as described in Fig. 1. MKP-1 and SGK mRNA fold inductions were calculated as compared with the corresponding vehicle control conditions. Values represent mean ± S.E. (n = 3 experiments).
FIGURE 9.
Estrogen has no effect on GR phosphorylation after PP5 knockdown in MCF-7 cells. MCF-7 cells were transfected with 100 nm of the control siRNA (A) or PP5 siRNA (D), and 24 h post-transfection the cells were stimulated with 10 nm E2 or vehicle control for 24 h, with or without 100 nm DEX added during the last 1 h. Total GR and Ser-211-phosphorylated GR were detected by Western blot as described in Fig. 4. Fold changes in the densitometry readings of nuclear total GR normalized to TBP (B and E) and Ser-211-phosphorylated nuclear GR normalized to total GR (C and F) in the cells treated with DEX alone (set as 1) versus cells cultured with E2 and DEX were calculated. Values represent mean ± S.E. (n = 3 experiments).
Glucocorticoids Suppress Estrogen-induced Proliferation in the Absence of PP5
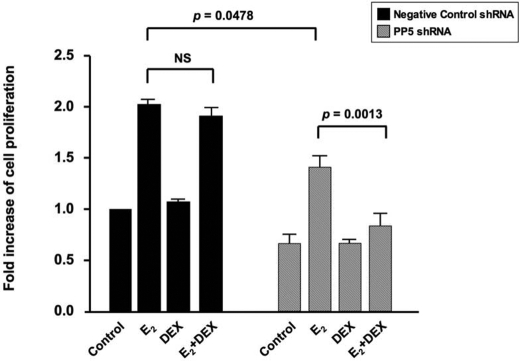
As described above glucocorticoids have no effect on estrogen-promoted cell growth, and the subsequent studies suggest that this may be because GR activity is inhibited by estrogen through PP5. To definitively test our proposed mechanism, we knocked down PP5 and assessed MCF-7 proliferation in response to estrogen or to estrogen plus DEX. In cells transfected with the control shRNA plasmid, estrogen alone promoted cell growth by 2.02 ± 0.04-fold, and this increase was unaffected when DEX was also added (Fig. 10). The knockdown of PP5 itself inhibited estrogen-induced cell proliferation by a mean of 65% (n = 3) as compared with estrogen-induced proliferation in cells transfected with the control shRNA (p = 0.0478) possibly because of activation of p53-mediated growth arrest (44). However, the addition of DEX significantly inhibited estrogen-mediated cell proliferation even further, resulting in a mean of 79% inhibition (n = 3) as compared with estrogen-induced proliferation of MCF-7 cells transfected with the PP5 shRNA plasmid (Fig. 10), substantiating the key role of PP5 in estrogen-mediated antagonism of the anti-proliferative effects of glucocorticoids.
FIGURE 10.
DEX significantly inhibited estrogen-promoted MCF-7 cell proliferation after PP5 knockdown. MCF-7 cells were transfected with 0.05 μg of the negative control shRNA plasmid or SureSilencing PP5 shRNA plasmid, and 24 h post-transfection the cells were stimulated with 10 nm E2 or vehicle control for 3 days with or without 100 nm DEX added during the last 2 days. Fold increase in viable cells was calculated by normalizing all values to the vehicle-treated negative control shRNA condition. Values represent mean ± S.E. (n = 3 experiments).
DISCUSSION
This study demonstrates for the first time that estrogen-induced PP5 in breast cancer cells ablates GR function via reduction in ligand-mediated Ser-211 GR phosphorylation. Furthermore, inhibition of PP5 induction by estrogen restores DEX-induced GR phosphorylation and allows GR-mediated growth arrest in the presence of estrogen. These findings have important implications for breast cancer therapy because they provide an explanation for the limited benefit observed in clinical trials utilizing corticosteroids as monotherapy (33, 34). Our study also suggests a potential antagonistic role of estrogen-induced PP5 in cellular glucocorticoid-mediated events in females, and it may provide an explanation of why the course of some allergic, autoimmune, and malignant diseases tends to be more severe and less responsive to corticosteroid treatment in females (3–5).
MKP-1 and SGK were chosen as well characterized glucocorticoid-inducible genes to study the inhibitory effects of estrogen on glucocorticoid-regulated targets. MKP-1 was originally identified as an ERK-specific phosphatase (45, 46). However, MKP-1 can also dephosphorylate and inactivate both the stress-activated protein kinase/JNK and p38 (47–49). The rapid induction of MKP-1 by DEX and the presence of at least three putative GREs in its promoter region suggest that it is transcriptionally regulated by the GR (17). It has been reported that induction of MKP-1 in MCF-7 cells results in cell growth suppression (50). SGK was first described in rat mammary epithelia as an immediate early response gene that is rapidly induced in vivo by glucocorticoids (51). The primary role of SGK is thought to involve the regulation of epithelial ion transport (52). It was also reported that glucocorticoid-induced SGK protected breast cancer cell lines from apoptosis (53). However, only a subset of breast cancer cell lines can be protected by glucocorticoid from apoptosis, and this does not occur in MCF-7 and T47D cell lines used in this study (53), which makes these cell lines a good model to study the ER-GR cross-talk. Thus, the role of SGK in cell growth regulation may be cell type-specific and deserves further investigation. Furthermore, we find that the GR fails to load at the SGK GRE upon ligand binding. This provides a mechanistic basis for the estrogen-mediated suppression of GR action.
The phosphorylation status of all three major families of MAPKs, the p38, JNK, and ERK, is essential in understanding the roles these signaling molecules play in cell function and disease. Our primary screening of MAPK phosphorylation found that only ERK phosphorylation is induced by estrogen. Further study using MEK/ERK inhibitor PD98059 excluded ERK to be the mediator of the effect of estrogen on glucocorticoid function in the MCF-7 cell line. Similarly, we excluded CDK2 and CDK5 using the CDK-specific inhibitor roscovitine. We then tested the protein phosphatase pathway in estrogen-treated MCF-7 cells and found expression of PP5 to be significantly increased by estrogen both at the protein and the mRNA level. This is consistent with previous reports that PP5 can be induced by estrogen (37), but it had not been previously established whether PP5 mediates the cross-talk between estrogen and glucocorticoids. To address this issue, we knocked down PP5 expression in the MCF-7 cell line and found that the inhibitory effect estrogen of on glucocorticoid induction of both MKP-1 and SGK was abolished. This novel discovery provides direct proof for the first time that PP5 bridges the cross-talk between estrogen and glucocorticoids.
The work by Honkanen and co-workers (37, 54, 55) that first explored a role of PP5 in breast cancer demonstrated the presence of the estrogen-response element in PP5 promoter and showed that PP5 can be induced by estrogen (37). It was found that the PP5 overexpression in MCF-7 cells allows rapid cell proliferation. When PP5 expression was inhibited by the synthetic oligonucleotide ISIS 15534 then cell proliferation was suppressed (37). It was suggested that PP5 provides a growth advantage to estrogen-responsive tumors (54, 55). In an MCF-7 mouse xenograph model of tumor development, the constitutive overexpression of PP5 was associated with accelerated tumor growth in a high estrogen environment. However, PP5 overexpression alone failed to produce spontaneous tumors in a low estrogen environment (55). PP5 has been shown to associate with estrogen receptors, resulting in suppression of ER-dependent transcription, as a feedback control mechanism (56). No experiments have been performed to determine whether the inhibition of estrogen-induced PP5 in breast cancer cell lines would allow DEX-mediated growth arrest.
Selective estrogen receptor modulators, such as tamoxifen (57), and aromatase inhibitors that inhibit estrogen synthesis (58) are used currently for the clinical management of ER-positive breast cancer. However, there are great variations among patients in both the therapeutic efficacy and side effects of these drugs (59). It was reported that long term selective estrogen receptor modulators therapy causes the development of acquired resistance (60); serious systemic side effects had been noted for the aromatase inhibitors (61). Our study suggests an alternative therapeutic approach in managing such cases. It demonstrates that suppression of estrogen-induced PP5 enhances the efficacy of corticosteroids and allows GR-mediated tumor growth arrest in the presence of estrogen.
In another cell line, A549, which is a lung epithelial cell line and is responsive to DEX-mediated growth arrest, it was shown that inhibition of endogenous PP5 by ISIS 15534 induces inhibition of cell growth via the p53 pathway and enhances GR transcriptional activity (44). It was determined that PP5 inhibits p53 phosphorylation at Ser-15 and suppresses p53 activity in the cells (44, 62). In contrast, glucocorticoids induce the same Ser-15 p53 phosphorylation, and this induces p21 expression, which mediates G1 growth arrest (62). In this study we tested whether PP5 suppression would relieve estrogen-mediated suppression of the GR function in the estrogen-responsive breast cancer model. We found that PP5 knockdown enhances GR function in breast cancer cells because of restitution of ligand-mediated Ser-211 GR phosphorylation.
GR has been well characterized to include three major functional domains as follows: N-terminal domain, DNA binding domain, and ligand binding domain. The N-terminal domain contains a transcriptional activation region (AF1) required for maximal transcriptional activity of GR. The AF1 of human GR has three residues (Ser-203, Ser-211, and Ser-226) that can be phosphorylated and affect GR function (for review see Ref. 63). It was reported that the Ser-203-phosphorylated GR is confined to the cytoplasm and to the perinuclear region; the Ser-226-phosphorylated GR inhibits transcription; and the Ser-211-phosphorylated GR is strictly agonist-dependent, localized to the nucleus, and strongly correlates with GR transcriptional activation (for review see Ref. 40). Inhibition of GR phosphorylation at Ser-211 is associated with decreased nuclear retention of GR and decreased gene transcription. Some GR-regulated gene promoters were found to be extremely sensitive to GR phosphorylation at Ser-211 as shown by inhibition of DEX-mediated gene transcription when S211A GR mutants were overexpressed in the cells. It was suggested that Ser-211 phosphorylation promotes GR conformational change that facilitates GR interaction with the coactivator MED14. MED14-dependent GR-regulated targets were found to be the most reliant on GR phosphorylation at Ser-211. Overexpression of the S226A mutant mainly enhanced DEX-mediated gene transcription as compared with wild type GR (64).
In our study, we observed that glucocorticoids strongly induced GR phosphorylation at Ser-211 within 1 h in MCF-7 cells, and this phosphorylation of GR at Ser-211 was significantly inhibited by estrogen. This is a critical result that supports our observation that estrogen inhibits glucocorticoid-induced GR binding to GRE and induction of MKP-1 and SGK. Furthermore, our PP5 knockdown experiments proved that estrogen-induced PP5 dephosphorylated the GR Ser-211 phosphorylation that was induced by glucocorticoids, resulting in reduced DEX-induced Ser-211 GR phosphorylation in the cells treated with estrogen. Data from the osteosarcoma cell line U2OS provided evidence that glucocorticoids induced both Ser-211 and Ser-226 phosphorylation as well as higher phosphorylation at Ser-211 relative to Ser-226. This correlated with GR nuclear localization and greater transcriptional activity (64). However, we did not see any Ser-226 phosphorylation in the MCF-7 cell line, which indicated GR phosphorylation could be cell type-specific. Because of the lack of commercially available chromatin immunoprecipitation grade Ser-211-phosphorylated GR antibody, we were unable to estimate Ser-211-phosphorylated GR binding to the SGK promoter. The data presented here with respect to Ser-211 phosphorylation mainly serve as an indicator that the changes in GR phosphorylation are important in estrogen-glucocorticoid cross-talk. However, the data do not exclude the possible contribution of other phosphorylation sites in GR in this process.
In addition to the breast cancer literature that described the role of PP5 in estrogen-mediated cell growth, basic molecular biology studies demonstrated the association of PP5 with the ligand binding domain of GR (interaction with hsp90 via the tetratricopeptide domain of PP5) (65). Furthermore, it was demonstrated that upon ligand binding PP5 dissociates from the GR (65). Garabedian and co-workers (27) have shown that ligand-bound PP5 in the absence of ligand dephosphorylates GR in a U2OS osteosarcoma cell line that was designed to overexpress wild type human GR. PP5 siRNA experiments in these cells demonstrated a somewhat enhanced DEX-induced Ser-203, Ser-211, and Ser-226 GR phosphorylation (27). The induction of several GR targets via transactivation (IRF8, ladinin, IGFBP-1, but not GILZ) was inhibited upon PP5 silencing. This suggested that PP5 modification of GR phosphorylation has selective effects on GR target gene induction (27). In this study we demonstrate that estrogen-induced PP5 dephosphorylates DEX-induced GR phosphorylation at Ser-211 and reduces GR transcriptional activity. Inhibition of estrogen-induced PP5 restores GR function. Previous publications indicate that estrogen can also inhibit glucocorticoid action by lowering the GR level (66, 67). In our study we did not see GR level change in 24 h (data not shown), although GR phosphorylation is affected. This suggests that estrogen inhibits GR action through not only one pathway.
The accumulated body of literature from several fields suggests that PP5 and GR actions are naturally in fine balance, and different scenarios can unfold when this balance is disturbed. This study demonstrates for the first time the following: 1) when PP5 is overproduced due to estrogen stimulation in breast cancer cells, it decreases DEX-induced Ser-211 phosphorylation of the endogenous GR. This inhibits DEX-mediated induction of MKP-1 and SGK. 2) Inhibition of PP5 induction by estrogen restores DEX-induced Ser-211 GR phosphorylation and MKP/SGK induction. 3) Suppression of estrogen-induced PP5 in breast cancer cell lines restores DEX-mediated growth arrest. Our study therefore demonstrates a cross-talk between estrogen-induced PP5 and GR action (depicted in Fig. 11) that highlights a potential relevance to human disease not only for treatment of breast cancer but also potentially opening up new directions in exploring gender differences in response to corticosteroid therapy.
FIGURE 11.
Proposed mechanism of cross-talk between estrogen and glucocorticoids. Upon ligand binding GR accumulates in the cell nucleus and is highly phosphorylated at Ser-211. The phosphorylated GR is transcriptionally active and binds as a homodimer to a specific palindromic DNA sequence, termed a GRE, located in the regulatory regions of target genes, such as MKP-1 and SGK. The induction of the target genes mediates the cell growth arrest. Estrogen induces the expression of PP5 that binds to the Ser-211-phosphorylated GR and dephosphorylates it, dampening its ability to bind GRE. Thus, the expression of glucocorticoid-inducible genes is inhibited. This supports estrogen-mediated cell proliferation.
Acknowledgment
We thank Maureen Sandoval for assistance with preparation of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01AI070140 from NIAID. This work was also supported by the University of Colorado Cancer Center.
- ER
- estrogen receptor
- MAPK
- mitogen-activated protein kinase
- JNK
- c-Jun N-terminal kinase
- ERK
- extracellular signal-regulated kinase
- GR
- glucocorticoid receptor
- GRE
- glucocorticoid-responsive element
- shRNA
- short hairpin RNA
- TBP
- TATA-binding protein
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PBS
- phosphate-buffered saline
- DEX
- dexamethasone
- E2
- 17β-estradiol
- PP
- protein phosphatase
- SGK
- serum/glucocorticoid-regulated kinase
- CDK
- cyclin-dependent kinase
- MEK
- mitogen-activated protein kinase kinase
- siRNA
- small interfering RNA.
REFERENCES
- 1.United States Cancer Statistics Working Group (2007) United States Cancer Statistics, pp. 1–684, Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 2.McGuire W. L. (1980) Recent Prog. Horm. Res. 36, 135–156 [DOI] [PubMed] [Google Scholar]
- 3.Tantisira K. G., Colvin R., Tonascia J., Strunk R. C., Weiss S. T., Fuhlbrigge A. L.Childhood Asthma Management Program Research Group (2008) Am. J. Respir. Crit. Care Med. 178, 325–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fagan J. K., Scheff P. A., Hryhorczuk D., Ramakrishnan V., Ross M., Persky V. (2001) Ann. Allergy Asthma Immunol. 86, 177–184 [DOI] [PubMed] [Google Scholar]
- 5.Rohleder N., Schommer N. C., Hellhammer D. H., Engel R., Kirschbaum C. (2001) Psychosom. Med. 63, 966–972 [DOI] [PubMed] [Google Scholar]
- 6.Levin E. R. (2001) J. Appl. Physiol. 91, 1860–1867 [DOI] [PubMed] [Google Scholar]
- 7.Kushner P. J., Agard D., Feng W. J., Lopez G., Schiau A., Uht R., Webb P., Greene G. (2000) Novartis Found. Symp. 230, 20–40 [DOI] [PubMed] [Google Scholar]
- 8.Razandi M., Pedram A., Levin E. R. (2000) J. Biol. Chem. 275, 38540–38546 [DOI] [PubMed] [Google Scholar]
- 9.Razandi M., Pedram A., Merchenthaler I., Greene G. L., Levin E. R. (2004) Mol. Endocrinol. 18, 2854–2865 [DOI] [PubMed] [Google Scholar]
- 10.Hermeking H., Rago C., Schuhmacher M., Li Q., Barrett J. F., Obaya A. J., O'Connell B. C., Mateyak M. K., Tam W., Kohlhuber F., Dang C. V., Sedivy J. M., Eick D., Vogelstein B., Kinzler K. W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2229–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altucci L., Addeo R., Cicatiello L., Dauvois S., Parker M. G., Truss M., Beato M., Sica V., Bresciani F., Weisz A. (1996) Oncogene 12, 2315–2324 [PubMed] [Google Scholar]
- 12.Barnes P. J. (1998) Clin. Sci. 94, 557–572 [DOI] [PubMed] [Google Scholar]
- 13.Mouhieddine O. B., Cazals V., Kuto E., Le Bouc Y., Clement A. (1996) Endocrinology 137, 287–295 [DOI] [PubMed] [Google Scholar]
- 14.Ito K., Chung K. F., Adcock I. M. (2006) J. Allergy Clin. Immunol. 117, 522–543 [DOI] [PubMed] [Google Scholar]
- 15.Beato M., Herrlich P., Schütz G. (1995) Cell 83, 851–857 [DOI] [PubMed] [Google Scholar]
- 16.Jenkins B. D., Pullen C. B., Darimont B. D. (2001) Trends Endocrinol. Metab. 12, 122–126 [DOI] [PubMed] [Google Scholar]
- 17.Kassel O., Sancono A., Krätzschmar J., Kreft B., Stassen M., Cato A. C. (2001) EMBO J. 20, 7108–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itani O. A., Liu K. Z., Cornish K. L., Campbell J. R., Thomas C. P. (2002) Am. J. Physiol. Endocrinol. Metab. 283, E971–E979 [DOI] [PubMed] [Google Scholar]
- 19.Wang X., Wu H., Lakdawala V. S., Hu F., Hanson N. D., Miller A. H. (2005) Neuropsychopharmacology 30, 242–249 [DOI] [PubMed] [Google Scholar]
- 20.Li L. B., Goleva E., Hall C. F., Ou L. S., Leung D. Y. (2004) J. Allergy Clin. Immunol. 114, 1059–1069 [DOI] [PubMed] [Google Scholar]
- 21.Rogatsky I., Logan S. K., Garabedian M. J. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2050–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller A. L., Webb M. S., Copik A. J., Wang Y., Johnson B. H., Kumar R., Thompson E. B. (2005) Mol. Endocrinol. 19, 1569–1583 [DOI] [PubMed] [Google Scholar]
- 23.Wang X., Wu H., Miller A. H. (2004) Mol. Psychiatry 9, 65–75 [DOI] [PubMed] [Google Scholar]
- 24.Wang Z., Garabedian M. J. (2003) J. Biol. Chem. 278, 50897–50901 [DOI] [PubMed] [Google Scholar]
- 25.Krstic M. D., Rogatsky I., Yamamoto K. R., Garabedian M. J. (1997) Mol. Cell. Biol. 17, 3947–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kino T., Ichijo T., Amin N. D., Kesavapany S., Wang Y., Kim N., Rao S., Player A., Zheng Y. L., Garabedian M. J., Kawasaki E., Pant H. C., Chrousos G. P. (2007) Mol. Endocrinol. 21, 1552–1568 [DOI] [PubMed] [Google Scholar]
- 27.Wang Z., Chen W., Kono E., Dang T., Garabedian M. J. (2007) Mol. Endocrinol. 21, 625–634 [DOI] [PubMed] [Google Scholar]
- 28.Nordeen S. K., Moyer M. L., Bona B. J. (1994) Endocrinology 134, 1723–1732 [DOI] [PubMed] [Google Scholar]
- 29.Swingle M. R., Honkanen R. E., Ciszak E. M. (2004) J. Biol. Chem. 279, 33992–33999 [DOI] [PubMed] [Google Scholar]
- 30.Yang J., Roe S. M., Cliff M. J., Williams M. A., Ladbury J. E., Cohen P. T., Barford D. (2005) EMBO J. 24, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silverstein A. M., Galigniana M. D., Chen M. S., Owens-Grillo J. K., Chinkers M., Pratt W. B. (1997) J. Biol. Chem. 272, 16224–16230 [DOI] [PubMed] [Google Scholar]
- 32.Chen M. S., Silverstein A. M., Pratt W. B., Chinkers M. (1996) J. Biol. Chem. 271, 32315–32320 [DOI] [PubMed] [Google Scholar]
- 33.Keith B. D. (2008) BMC Cancer 8, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludwig Breast Cancer Study Group (1985) Cancer Res. 45, 4454–4459 [PubMed] [Google Scholar]
- 35.Zhang Y., Wang J. S., Chen L. L., Zhang Y., Cheng X. K., Heng F. Y., Wu N. H., Shen Y. F. (2004) J. Biol. Chem. 279, 42545–42551 [DOI] [PubMed] [Google Scholar]
- 36.Métivier R., Penot G., Hübner M. R., Reid G., Brand H., Kos M., Gannon F. (2003) Cell 115, 751–763 [DOI] [PubMed] [Google Scholar]
- 37.Urban G., Golden T., Aragon I. V., Scammell J. G., Dean N. M., Honkanen R. E. (2001) J. Biol. Chem. 276, 27638–27646 [DOI] [PubMed] [Google Scholar]
- 38.Im J. Y., Park H., Kang K. W., Choi W. S., Kim H. S. (2008) Chem. Biol. Interact. 172, 235–244 [DOI] [PubMed] [Google Scholar]
- 39.Chen B., Gajdos C., Dardes R., Kidwai N., Johnston S. R., Dowsett M., Jordan V. C. (2005) Int. J. Oncol. 27, 327–335 [PubMed] [Google Scholar]
- 40.Ismaili N., Garabedian M. J. (2004) Ann. N.Y. Acad. Sci. 1024, 86–101 [DOI] [PubMed] [Google Scholar]
- 41.Boonyaratanakornkit V., Bi Y., Rudd M., Edwards D. P. (2008) Steroids 73, 922–928 [DOI] [PubMed] [Google Scholar]
- 42.Lu Q., Pallas D. C., Surks H. K., Baur W. E., Mendelsohn M. E., Karas R. H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17126–17131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prall O. W., Rogan E. M., Musgrove E. A., Watts C. K., Sutherland R. L. (1998) Mol. Cell. Biol. 18, 4499–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuo Z., Urban G., Scammell J. G., Dean N. M., McLean T. K., Aragon I., Honkanen R. E. (1999) Biochemistry 38, 8849–8857 [DOI] [PubMed] [Google Scholar]
- 45.Sun H., Charles C. H., Lau L. F., Tonks N. K. (1993) Cell 75, 487–493 [DOI] [PubMed] [Google Scholar]
- 46.Noguchi T., Metz R., Chen L., Mattéi M. G., Carrasco D., Bravo R. (1993) Mol. Cell. Biol. 13, 5195–5205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y., Gorospe M., Yang C., Holbrook N. J. (1995) J. Biol. Chem. 270, 8377–8380 [DOI] [PubMed] [Google Scholar]
- 48.Imasato A., Desbois-Mouthon C., Han J., Kai H., Cato A. C., Akira S., Li J. D. (2002) J. Biol. Chem. 277, 47444–47450 [DOI] [PubMed] [Google Scholar]
- 49.Lasa M., Abraham S. M., Boucheron C., Saklatvala J., Clark A. R. (2002) Mol. Cell. Biol. 22, 7802–7811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y. W., Huang S. C., Lin-Shiau S. Y., Lin J. K. (2005) Carcinogenesis 26, 1296–1306 [DOI] [PubMed] [Google Scholar]
- 51.Webster M. K., Goya L., Firestone G. L. (1993) J. Biol. Chem. 268, 11482–11485 [PubMed] [Google Scholar]
- 52.Loffing J., Flores S. Y., Staub O. (2006) Annu. Rev. Physiol. 68, 461–490 [DOI] [PubMed] [Google Scholar]
- 53.Mikosz C. A., Brickley D. R., Sharkey M. S., Moran T. W., Conzen S. D. (2001) J. Biol. Chem. 276, 16649–16654 [DOI] [PubMed] [Google Scholar]
- 54.Golden T., Aragon I. V., Rutland B., Tucker J. A., Shevde L. A., Samant R. S., Zhou G., Amable L., Skarra D., Honkanen R. E. (2008) Biochim. Biophys. Acta 1782, 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Golden T., Aragon I. V., Zhou G., Cooper S. R., Dean N. M., Honkanen R. E. (2004) Cancer Lett. 215, 95–100 [DOI] [PubMed] [Google Scholar]
- 56.Ikeda K., Ogawa S., Tsukui T., Horie-Inoue K., Ouchi Y., Kato S., Muramatsu M., Inoue S. (2004) Mol. Endocrinol. 18, 1131–1143 [DOI] [PubMed] [Google Scholar]
- 57.Lee W. L., Cheng M. H., Chao H. T., Wang P. H. (2008) Taiwan J. Obstet. Gynecol. 47, 24–31 [DOI] [PubMed] [Google Scholar]
- 58.Herold C. I., Blackwell K. L. (2008) Clin. Breast Cancer 8, 50–64 [DOI] [PubMed] [Google Scholar]
- 59.Weinshilboum R. (2008) Adv. Exp. Med. Biol. 630, 220–231 [DOI] [PubMed] [Google Scholar]
- 60.Sengupta S., Jordan V. C. (2008) Adv. Exp. Med. Biol. 630, 206–219 [DOI] [PubMed] [Google Scholar]
- 61.Nabholtz J. M. (2008) Ther. Clin. Risk Manag. 4, 189–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Urban G., Golden T., Aragon I. V., Cowsert L., Cooper S. R., Dean N. M., Honkanen R. E. (2003) J. Biol. Chem. 278, 9747–9753 [DOI] [PubMed] [Google Scholar]
- 63.Kumar R., Thompson E. B. (2005) J. Steroid Biochem. Mol. Biol. 94, 383–394 [DOI] [PubMed] [Google Scholar]
- 64.Chen W., Dang T., Blind R. D., Wang Z., Cavasotto C. N., Hittelman A. B., Rogatsky I., Logan S. K., Garabedian M. J. (2008) Mol. Endocrinol. 22, 1754–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hinds T. D., Jr., Sánchez E. R. (2008) Int. J. Biochem. Cell Biol. 40, 2358–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krishnan A. V., Swami S., Feldman D. (2001) J. Steroid Biochem. Mol. Biol. 77, 29–37 [DOI] [PubMed] [Google Scholar]
- 67.Kinyamu H. K., Archer T. K. (2003) Mol. Cell. Biol. 23, 5867–5881 [DOI] [PMC free article] [PubMed] [Google Scholar]