Abstract
Fibulin-4 and -5 are extracellular glycoproteins with essential non-compensatory roles in elastic fiber assembly. We have determined how they interact with tropoelastin, lysyl oxidase, and fibrillin-1, thereby revealing how they differentially regulate assembly. Strong binding between fibulin-4 and lysyl oxidase enhanced the interaction of fibulin-4 with tropoelastin, forming ternary complexes that may direct elastin cross-linking. In contrast, fibulin-5 did not bind lysyl oxidase strongly but bound tropoelastin in terminal and central regions and could concurrently bind fibulin-4. Both fibulins differentially bound N-terminal fibrillin-1, which strongly inhibited their binding to lysyl oxidase and tropoelastin. Knockdown experiments revealed that fibulin-5 controlled elastin deposition on microfibrils, although fibulin-4 can also bind fibrillin-1. These experiments provide a molecular account of the distinct roles of fibulin-4 and -5 in elastic fiber assembly and how they act in concert to chaperone cross-linked elastin onto microfibrils.
Fibulins are a family of extracellular glycoproteins containing contiguous calcium-binding epidermal growth factor-like domains (cbEGFs)3 and a characteristic C-terminal fibulin (FC) domain (1–3). Recent studies have revealed that fibulin-4 and -5 are both essential for elastic fiber formation (4–7). Fibulin-4 is widely expressed from early embryogenesis and is necessary for normal vascular, lung, and skin development, since mice that lack fibulin-4 do not form elastic fibers and die perinatally (5). Furthermore, mice with reduced fibulin-4 expression develop aneurysms (8). Fibulin-5 is abundant in the aorta and large arteries during embryogenesis and following vascular injury (9, 10). Lack of fibulin-5 causes a less severe phenotype, with viable homozygous mice, but the elastic fibers in skin, lungs, and aorta are irregular and fragmented (6, 7), and there is altered vascular remodeling (11). These mice models also highlight that fibulin-4 and -5 have non-compensatory roles in elastic fiber formation. Mutations in both molecules can cause cutis laxa, a heritable disorder associated with elastic fiber degeneration leading to sagging skin, vascular tortuosity, and emphysematous lungs (12–15). A third isoform, fibulin-3, may play a minor role in elastic fiber formation, since its deficiency disrupts elastic fibers in Bruch's membrane of the eye (16) and vaginal tissues (17).
Elastic fiber formation is a complex multistep process (18–20). Initial pericellular microassembly of tropoelastin, which may involve the 67-kDa elastin-binding protein receptor, generates elastin globules that are stabilized by desmosine cross-links catalyzed mainly by lysyl oxidase (LOX) but also by LOXL1 (LOX-like 1). These globules are deposited on a fibrillin microfibril template, where they coalesce and undergo further cross-linking to form the elastin core of mature fibers. The ability of fibulin-4 and -5 to bind tropoelastin and fibrillin-1, the major structural component of microfibrils, supports a model in which these fibulins direct elastin deposition on microfibrils (4–7, 21–25). This model does not delineate the unique molecular contributions of fibulin-4 and -5 to elastic fiber formation, but some molecular differences have emerged. Tropoelastin was bound more strongly by fibulin-5 than by fibulin-4, whereas fibulin-5 was at the microfibril-elastin interface, but perichondrial fibulin-4 localized mainly to microfibrils (4).
Fibulin-4 null mice offer tantalizing clues to how fibulin-4 contributes to elastic fiber formation (5). They had dramatically reduced (94%) desmosine cross-links despite no change in elastin or LOX expression levels, and electron-dense rodlike structures were prominent within elastin aggregates. Morphologically similar structures seen after chemically inhibiting LOX were previously identified as glycosaminoglycans, which can bind charged free ϵ-amino groups on lysines in tropoelastin (26). However, fibulin-4+/− mice showed ∼20% increase in desmosine (5). LOX-null mice have phenotypic features similar to those of fibulin-4 null mice, dying perinatally with 60% reduced desmosine cross-links and major abnormalities in vascular and other elastic tissues (27, 28). In contrast, LOXL1-null mice are viable but have reduced desmosine (29), whereas fibulin-5 null mice have a 16% reduction in desmosine cross-links and survive well into adulthood (7). Detection of the LOXL1 pro-domain in fibulin-5 null mice skin but not wild-type skin implicates fibulin-5 in activation of LOXL1 (30).
We and others have shown that fibrillin-1 and the microfibrillar protein MAGP-1 can both directly bind tropoelastin (31–34). However, the fibulin-null mice show that the fibrillin-1 interaction with tropoelastin is insufficient to support elastic fiber formation in vivo. Fibulin-5 has been reported to facilitate tropoelastin binding to the N-terminal half of fibrillin-1 (21). A study of elastin polypeptide self-assembly through coacervation and maturation phases showed that, although the N-terminal half of fibrillin-1 increased maturation velocity and droplet clustering, fibulin-4 and -5 both slowed maturation and limited globule growth (35). These studies imply that fibulins and fibrillin-1 act together to regulate elastin accretion on microfibrils.
To gain further insights into the contributions of fibulin-4 and -5 to elastic fiber formation, we have delineated how they interact with tropoelastin, LOX, and fibrillin-1. Novel findings are that fibulin-4 directly binds LOX, and this interaction enhances fibulin-4 binding to tropoelastin, thus forming a ternary complex that may be critical for elastin cross-linking. Fibulin-5 can concurrently bind fibulin-4 and tropoelastin, but the interaction of both fibulins with fibrillin-1 strongly inhibits their binding to tropoelastin. These interactions indicate the molecular basis of how fibulins act as chaperones for deposition of elastin onto microfibrils. Our study thus provides a molecular account of the differential roles of fibulins-4 and -5 in elastic fiber formation.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant Proteins
All human elastic fiber proteins were expressed with an N-terminal His6 tag, using the mammalian expression vector pCEP-His and 293-EBNA cells, and purified using nickel affinity chromatography as described (22, 33, 36–41) (Fig. 1 and supplemental Fig. S1).
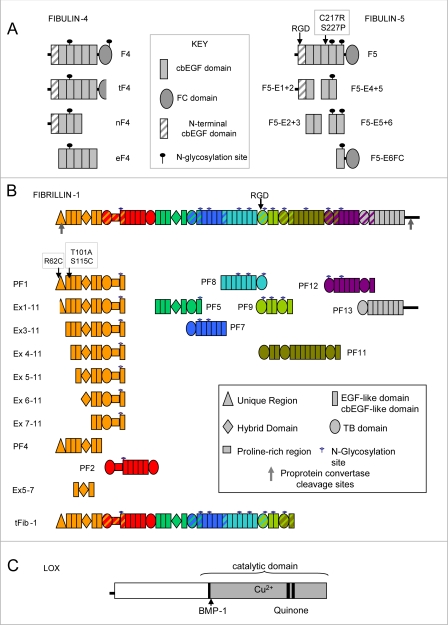
FIGURE 1.
A, recombinant human fibulin-4 and -5. Domain structures of full-length fibulin-4 (F4), C-terminally truncated fibulin-4 (tF4), the N-terminal half of fibulin-4 (nF4), and the five central cbEGFs (eF4) are shown, with a key of the domains and N-glycosylation sites. Also shown are the domain structures of full-length fibulin-5 (F5) with the sites of the two cutis laxa mutations (F5C217R and F5S227P) and domain pair fragments. Details of the domains are given (see key). For SDS-PAGE analysis of recombinant fibulins, see supplemental Fig. S1A. B, recombinant human fibrillin-1. Domain structures of full-length fibrillin-1, a C-terminally truncated two-third fragment of fibrillin-1 (tFib-1), overlapping fragments, and N-terminal deletion fragments are shown, with a key of the domains and N-glycosylation sites. All fragments have been described (39, 55) except for tFib-1 (for mass spectrometry of tFib-1, see supplemental Fig. S2B). Details of the domains are given (see key), and the colors indicate overlapping recombinant fragments. For SDS-PAGE analysis of recombinant tFib-1, see supplemental Fig. S1B. C, recombinant human LOX. The molecular arrangement of full-length human LOX is shown. BMP-1, position of the bone morphogenetic protein-1 cleavage site. For SDS-PAGE analysis of recombinant LOX, see supplemental Fig. S1C. EGF, epidermal growth factor.
Full-length human fibulin-4 (residues 26–418) was designated F4 (Fig. 1A). A truncated version of full-length fibulin-4 (designated tF4) was expressed by one culture of transfected cells; analysis by mass spectrometry revealed the absence of the C-terminal sequence (53 residues, comprising the FC domain and 6 residues of the preceding linker region) (supplemental Fig. S2A). We also expressed the N-terminal four domains of fibulin-4 (residues 26–217; designated nF4) and the central cbEGFs 2–6 (residues 98–303; designated eF4). The C-terminal four domains did not express efficiently.
Full-length human fibulin-5 (residues 42–448) was designated F5 (Fig. 1A). Full-length fibulin-5 incorporating two cutis laxa mutations, C217R and S227P (F5C217R and F5S227P) (13), were expressed together with five overlapping domain pairs of fibulin-5 (residues 42–167 (F5-E1+2), 127–206 (F5-E2+3), 207–287 (F5-E4+5), 247–333 (F5-E5+6), and 288–448 (F5-E6FC)) (Fig. 1A). A sixth domain pair (F5-E3+4) did not express efficiently.
Human fibrillin-1 fragments generated for this study were PF1, PF2, PF4, PF5, PF7, PF8, PF9, PF11, PF12, and PF13 (33, 37–41) (Fig. 1B). We also generated PF1 variants with the Marfan mutation R62C, T101A, or S115C. In addition, we used our panel of N-terminal deletion mutants: PF1 encoded by exons 1–11 (residues 31–489), Ex1–11 encoded by residues 45–489), Ex3–11 encoded by exons 3–11 (residues 81–489), Ex4–11 encoded by exons 4–11 (residues 115–489), Ex5–11 encoded by exons 5–11 (residues 147–489), Ex6–11 encoded by exons 6–11 (residues 179–489), Ex7–11 encoded by exons 7–11 (residues 246–489), and a three-domain fragment Ex5–7 encoded by exons 5–7 (residues 147–287) (39). Cells transfected with full-length fibrillin-1 expressed a large fragment comprising the N-terminal two-thirds of the molecule (250 kDa; designated tFib-1) (Fig. 1B and supplemental Fig. S1B); mass spectrometry indicated that the cleavage site was after the domain encoded by exon 43 (supplemental Fig. S2B). Human full-length LOX (Fig. 1C) was expressed and purified, as outlined above (supplementary Fig. S1C). Recombinant human tropoelastin lacking domain 26A was a generous gift from Dr. A. S. Weiss (Sydney, Australia) (42, 43).
Purification and Biophysical Characterization of Recombinant Proteins
After nickel affinity chromatography, monomeric recombinant proteins were isolated by Superdex S200 10/300 GL size fractionation (GE Healthcare) in 10 mm Hepes-buffered saline, pH 7.4, containing 150 mm NaCl (HBS), using an AKTA purifier system. cDNA sequencing, SDS-PAGE, Western blotting with anti-His antibody (Sigma), and mass spectrometry confirmed the correct products (Fig. 1 and supplemental Figs. S1 and S2; data not shown). All molecules with NX(S/T) motifs were N-glycosylated, as determined by treatment with peptide:N-glycanase F (New England Biolabs). In all cases, the monomer fractions from S200 chromatography were equilibrated in HBS containing 0.5 mm CaCl2 (HBS+Ca) for subsequent binding assays.
Monomers of fibulin-4 (F4, nF4, and tF4) and fibulin-5 (F5, F5S227P, and domain pairs F5-E1+2, F5-E2+3, F5-E4+5, F5-E5+6, and F5-E6FC) that had been isolated by Superdex 200 gel filtration in HBS, were analyzed by multiangle laser light scattering (MALLS) in HBS or HBS+Ca. Samples eluting from the column passed through a Wyatt EOS 18-angle light scattering detector fitted with a 688-nm laser and an Optilab r-EX refractometer. The solute molecular mass was determined using in-line MALLS attached to quasielastic light scattering and a differential refractometer (Wyatt Technology Corp.) (Table 1 and supplemental Fig. S3). The monomers had molecular masses that corresponded well with SDS-PAGE analysis (see supplemental Fig. S1).
TABLE 1.
MALLS analysis of fibulin-4 and -5
Fibulin-4 and -5 were analyzed by MALLS to obtain details of their molecular masses in solution (for domain structures, see Fig. 1A). Without added CaCl2 (HBS buffer), F4 and nF4 were monomeric. In the presence of 0.5 mm CaCl2 (HBS+Ca buffer), apparent dimers predominated, and some larger aggregates were also detected (data not shown). Similarly, F5 was monomeric in HBS (below) but dimerized in HBS+Ca (data not shown).4 Fibulin-5 mutant F5S227P and fibulin-5 domain pairs were also monomeric in HBS. Calculated molecular masses from primary sequences (without carbohydrate) and SDS-PAGE-determined molecular masses are also shown.
| Molecule (buffer) | MALLS-measured molecular mass | SDS-PAGE-measured molecular mass | Calculated molecular mass |
|---|---|---|---|
| kDa | kDa | kDa | |
| F4 (HBS) monomer | 52 | 52 | 49 |
| nF4 (HBS) monomer | 30 | 40 | 27 |
| F4 (HBS+Ca) “dimer” | 94 | ||
| F4 (HBS+Ca) monomer | 56 | ||
| nF4 (HBS+Ca) “dimer” | 60 | ||
| nF4 (HBS+Ca) monomer | 31 | ||
| F5 (HBS) monomer | 51 | 53 | 50 |
| F5-S227P (HBS) monomer | 52 | ||
| F5-E1+2 (HBS) monomer | 20 | 25 | 14 |
| F5-E2+3 (HBS) monomer | 13 | 15 | 9 |
| F5-E4+5 (HBS) monomer | 13 | 15 | 9 |
| F5-E5+6 (HBS) monomer | 16 | 18 | 10 |
| F5-E6FC (HBS) monomer | 24 | 30 | 19 |
LOX monomers were isolated on a Superdex 200 gel filtration column in 10 mm Tris, pH 7.4, with 150 mm NaCl containing 1 mm CaCl2 (TBS+Ca). MALLS analysis revealed that LOX was monomeric with a molecular mass of 47.5 kDa, compared with 50 kDa by SDS-PAGE (supplemental Fig. S1). Analysis of LOX by circular dichroism revealed 5% α-helix, 31% β-strand, and 13% β-turn. Analysis of LOX by analytical ultracentrifugation revealed a molecular mass of 48.9 kDa, measured S20,w of 2.2, calculated hydrodynamic radius of 5.41, and frictional ratio of 2.25.
Single Particle Averaging of Fibulin-4
Purified F4 monomers in 10 mm Tris, pH 7.4, containing 500 mm NaCl (TBS) were adsorbed onto glow-discharged carbon-coated 400-mesh copper grids, negatively stained with 2% uranyl acetate, and analyzed by single particle averaging, as described (Fig. 2) (37). Briefly, samples were observed at ×52,000 magnification using an FEI Tecnai 12 transmission electron microscope operating at 120 kV with a LaB6 filament, and images were recorded by a CCD camera (TVIPS Tem Cam, 2048 × 2048 resolution) using low dose (<10e−/Å2) at defocus values of 2 μm. The pixel size was 1.9 Å. Images were analyzed using electron microscopy analysis (EMAN) (44) for all data processing. The data set contained 3538 particles, which were aligned and classified by reference-free methods. A preliminary three-dimensional structure was calculated by averaging classes that represented distinct views of the sample. Fourier common lines were applied to determine the relative orientation of these classes. The preliminary structure was subsequently refined by iterative (eight times) projection matching. Each refinement was examined by the convergence of the Fourier shell correlation coefficient. Final resolution of the F4 reconstruction was ∼20 Å using the Fourier shell correlation = 0.5 criterion.
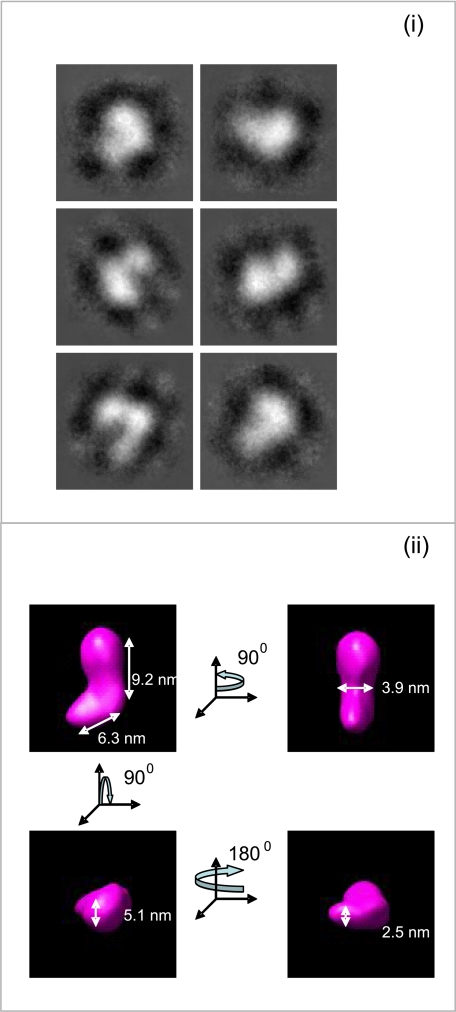
FIGURE 2.
Electron microscopy and single particle averaging of full-length fibulin-4. F4 was equilibrated in TBS prior to adsorption onto carbon-coated grids. i, representative class averages from a data set of 3538 fibulin-4 molecules. ii, a three-dimensional reconstruction of fibulin-4 was generated using angular reconstitution. For i and ii, the box size is 19 × 19 nm. The data indicate that the length of fibulin-4 is 13–14 nm with a central bend and a globular end-domain. This result extends previous rotary shadowing studies of fibulin-4 molecules, which indicated short rods of 10–20 nm in length and an end globule (4, 51).
BIAcore Analysis of Interactions with Tropoelastin and LOX
For kinetic binding studies by surface plasmon resonance, a BIAcore biosensor was used (BIAcore 3000; GE Healthcare). Tropoelastin or LOX was immobilized onto CM5 sensor chips by amine coupling using 1-ethyl-3(3-dimethylaminopropyl)-carbodiimide hydrochloride, N-hydroxysuccinimide, and ethanolamine-HCl, as described (33, 39). All binding experiments were performed in HBS+Ca. LOX, fibulin-4, or fibulin-5 was injected at concentration ranges of 2–10 μg/ml for F4 or 0.4–10 μg/ml for F5 or LOX for 3–5 min at a flow rate of 30 μl/min. Curves were fitted using a 1:1 Langmuir association/dissociation model (BIAevaluation 4.1; GE Healthcare). This model was found to fit the tropoelastin-fibulin-5 and LOX-fibulin-4 data, with low χ2 values. χ2 values are a standard statistical measure of the closeness of fit (mean square of the signal noise). Each binding interaction was performed at least twice, with one concentration in duplicate each time. Some BIAcore experiments investigated potential inhibitory effects of a given elastic fiber protein on binding of a second protein to immobilized tropoelastin or LOX. Kinetic analysis was performed using ligand protein (F5 at 100 nm; F4 at 200 nm; LOX at 200 nm) preincubated with a 0.01 nm to 3 μm concentration of the potential inhibitor protein or with a non-binding control fibrillin-1 fragment PF12, in HBS+Ca, as described (38). Inhibition curves were plotted using the response value of each normalized curve at the end of the association period.
Solid Phase Binding and Inhibition Assays
Because fibrillin-1 and the two fibulins when immobilized to BIAcore CM5 chips do not interact sufficiently with other proteins and to investigate further the interactions between LOX and tropoelastin and between fibrillin-1 and LOX, solid phase assays were utilized, as described (22, 33, 39). Soluble ligands were biotinylated using sulfosuccinimidyl 2-(biotinamido)-ethyl-1,3-dithiopropionate (Pierce). Flat-bottomed microtiter plates were coated with tropoelastin, LOX, fibulin-4 or -5, or N-terminal fibrillin-1 fragments, at 100 nm in 10 mm Tris, pH 7.4, containing 150 mm NaCl and 1 mm CaCl2 (TBS+Ca) buffer, overnight at 4 °C. Nonspecific binding sites were blocked with TBS+Ca containing 5% bovine serum albumin at room temperature for at least 2 h. The plates were washed three times with TBS+Ca, 0.1% bovine serum albumin and incubated with 100 nm biotinylated ligand. Control wells with only soluble biotinylated ligand were included in all experiments. After a further three washes, plates were incubated with a 1:500 dilution of extravidin peroxidase conjugate at room temperature for 20 min. Bound protein was quantified after four more washes by a colorimetric assay using 40 mm 2,2′-azino-bis(3-ethyl-benzthiazoline-6-sulfonic acid) solution (Sigma) for 10–20 min at room temperature. Plates were read at a wavelength of 405 nm. Wells were washed four times, and the color was developed. Any nonspecific binding was detected by blocking wells before incubation with bovine serum albumin only. All experiments were performed in triplicate, and all assays were repeated at least twice. In some experiments, the effects of inhibition of binding by other elastic fiber molecules or by heparin (3 kDa; Sigma) were conducted with increasing concentrations of potential inhibitors preincubated with soluble ligand. For kinetic and statistical analysis, GraphPad Prism was used to obtain dissociation constants (Kd) (33, 40), and unpaired t tests were used to calculate significance. Results were statistically significant when the p value is <0.05 (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Knockdown and Induced Expression of Elastic Fiber Proteins
Reverse transcription-PCR analysis of the expression of fibulin-4 and -5 was conducted on ARPE-19 cells (supplemental Fig. S5A). Validated siRNAs (pools of three target-specific siRNAs; Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); sc-75017 and sc-43121) were used to knock down human fibulin-4 in the ARPE-19 retinal pigmented epithelial cell line, with scrambled siRNA and no-siRNA controls. Cells (5 × 105 cells) were transfected with 200 nm siRNAs for fibulin-4 or with scrambled siRNAs (200 nm) by electroporation using cell-specific Nucleofector reagent (Amaxa). Parallel siRNA experiments were conducted on ARPE-19 cells that had been transduced with the pQCXIP retroviral vector (Clontech) for constitutive expression of human tropoelastin (supplemental Fig. S5B) and on transduced control ARPE-19 cells. Control and siRNA-transfected cells were plated at confluence and cultured for up to 7 days in Dulbecco's minimum essential medium supplemented with 10% fetal calf serum, penicillin, and streptomycin. Knockdown efficacy was confirmed to be 70% at the mRNA level by quantitative PCR (supplemental Fig. S5A) and was also reduced at the protein level by Western blotting, using an anti-fibulin-4-specific polyclonal antiserum raised to nF4 (1:1000 dilution). Anti-fibulin-5 antibody (1 μg/ml monoclonal antibody 3095; R&D Systems) was also used (not shown). Using the pSuper.retro.neo+gfp (pSR) vector (Oligoengine), stable knockdown of fibulin-5 was achieved in RFL-6 rat fetal lung fibroblasts (13) and confirmed at mRNA (supplemental Fig. S5C) and protein levels (not shown).
Light and Electron Microscopy of Cell Layers
Cell layers from fibulin-5 shRNA and control cultures were imaged by light microscopy after Miller's staining. Cells (8 × 104 cells/coverslip in 24-well plates) were cultured for 8–12 days before being fixed with 4% paraformaldehyde for 20 min. Coverslips were washed in phosphate-buffered saline before being oxidized with acidified potassium permanganate. Samples were washed in distilled H2O, decolorized with 1% oxalic acid, washed in 95% ethanol and H2O, and stained with Miller's stain (GCC Diagnostics) for 3 h. Samples were washed in 95% ethanol and distilled H2O before being counterstained with Van Gieson stain (GCC Diagnostics). Prior to mounting, samples were washed in 95% ethanol and distilled H2O.
Cells grown on Aclar film were fixed with 2.5% glutaraldehyde in 0.1 m cacodylate buffer, postfixed with 1% osmium tetroxide for 1 h, and treated with 1% tannic acid for 1 h and with 1% uranyl acetate for 1 h. They were dehydrated with alcohol series and embedded in TAAB LV resin. Ultrathin en face sections were cut at a Reichert Ultracut S Ultramicrotome and contrasted with lead citrate. Sections were observed with an FEI Tecnai Biotwin 12 microscope at 100 kV accelerating velocity.
Desmosine Cross-link Assay
Experiments were conducted to analyze desmosine cross-link formation, using ARPE-19 cells. We used untreated control ARPE-19 cells, ARPE-19 cells stably expressing transfected tropoelastin (supplemental Fig. S5), and ARPE-19 cells cultured in the presence of supplemented exogenous human tropoelastin (10 μg/ml) (45). Cells were subjected to siRNA knockdown of fibulin-4, and after culture for up to 7 days, cell layers were scraped into tubes in 100 μl of distilled H2O. ARPE-19 cell layers were kindly analyzed for elastin desmosine cross-links by Dr. Barry Starcher.
RESULTS
We generated and characterized recombinant human fibulin-4 and -5, fibrillin-1, and LOX (Fig. 1, Table 1, and supplemental Figs. S1–S3). These molecules were used to analyze the interactions of both fibulins with tropoelastin, LOX, and fibrillin-1 and to determine how fibulin-4 and -5 differentially regulate elastin cross-linking and deposition.
After nickel chromatography, monomers of full-length human fibulin-4 (F4), the N-terminal half of fibulin-4 (nF4), the central five cbEGF-like domains (eF4), and a C-terminally truncated fragment (tF4) were isolated by Superdex S200 size fractionation in HBS. SDS-PAGE analysis indicated molecular masses for F4 of 52 kDa, for nF4 of 40 kDa, and for eF4 of 36 kDa (supplemental Fig. S1A). The truncated molecule tF4 formed disulfide-bonded dimers of 75 kDa. MALLS analysis in HBS confirmed that F4 in solution was monomeric in the absence of CaCl2, with a molecular mass of 52 kDa (Table 1 and supplemental Fig. S3). In the presence of CaCl2, F4 contained some monomers but was predominantly dimeric. Electron microscopy and single particle averaging revealed that F4 has an elongated shape (13–14-nm length) with 6–7-nm diameter and a small globular end (Fig. 2).
After nickel chromatography, monomers of full-length fibulin-5 (F5) had a molecular mass of 53 kDa by SDS-PAGE (supplemental Fig. S1A). MALLS analysis in HBS indicated a solution molecular mass for F5 of 51 kDa and for the mutant F5S227P of 52 kDa (Table 1 and supplemental Fig. S3). Solution molecular masses for each monomeric fibulin-5 domain pair fragment in HBS by MALLS range from 13 to 24 kDa (Table 1). Recombinant fibulin-5 had a similar structure to fibulin-4 by single particle averaging.4
Defining Molecular Interactions of Fibulin-4 and -5
Monomeric molecules and fragments were equilibrated in HBS+Ca for interaction studies.
Tropoelastin Binds Fibulin-5 More Strongly than Fibulin-4
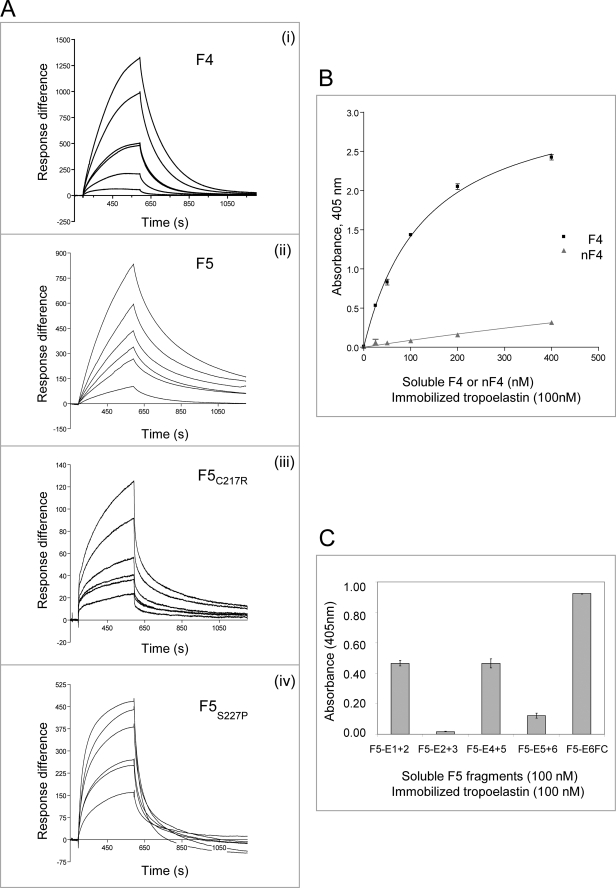
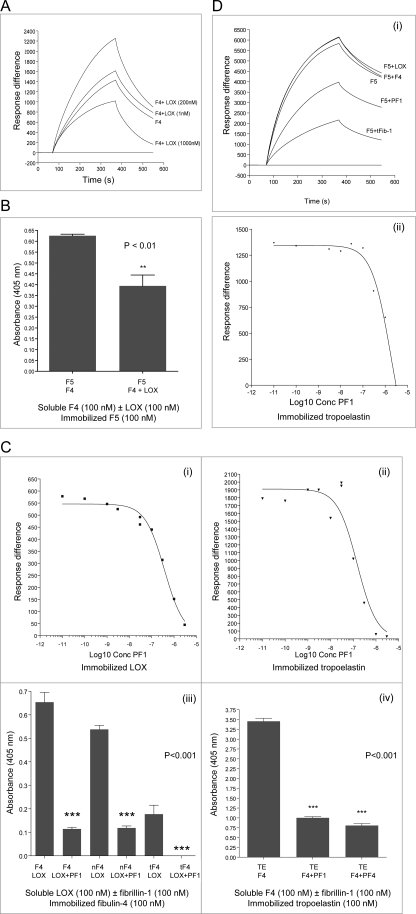
Tropoelastin was reported to bind to murine fibulin-4, using solid phase binding and co-immunoprecipitation (5). Here, BIAcore analysis confirmed that F4 bound tropoelastin (Fig. 3A). The F4 interaction with tropoelastin indicated more rapid dissociation kinetics than F5 (see below), but binding affinities could not be determined using the 1:1 Langmuir association/dissociation model (BIAevaluation version 4.1; GE Healthcare). Solid phase binding assays indicated that soluble F4 bound immobilized tropoelastin with a KD of 131 ± 8 nm but showed virtually no binding to the fibulin-4 fragments nF4, eF4, or tF4 (Fig. 3B) (data not shown). Thus, fibulin-4 binds tropoelastin with moderate affinity, with the binding site at or adjacent to the fibulin-4 FC domain.
FIGURE 3.
Molecular interactions of tropoelastin with fibulin-4 and -5. A, BIAcore analysis of interactions of immobilized tropoelastin with full-length fibulin-4 (F4) (i) or fibulin-5 (F5, F5C217R, or F5S227P) (ii–iv). These soluble ligands were injected over tropoelastin immobilized on a CM5 chip (see Table 2). Each sensorgram shows analyte concentrations ranging from 2 to 10 μg/ml (for F4) or from 0.4 to 10 μg/ml (for F5), with duplicate concentrations included in every run. One representative experiment is shown. Response difference is the difference between experimental and control flow cells, in response units. Time is shown in seconds. F4, F5C217R, and F5S227P showed faster dissociation than F5 from tropoelastin. B, solid phase binding curves showing that soluble biotinylated F4 bound strongly to immobilized tropoelastin but nF4 binding was very weak. One representative experiment is shown. Data are shown with the negative (biotinylated F4 only) control subtracted. Results are shown as the mean ± S.E. of triplicate values. C, solid phase binding assays localizing tropoelastin binding sites on biotinylated fibulin-5. Three of the domain pair fragments, F5-E1+2, F5-E4+5, and F5-E6FC, bound well to immobilized tropoelastin (KD values of 332, 452, and 965 nm, respectively; binding curves are shown in supplemental Fig. S4A), but fragments F5-E2+3 and F5-E5+6 interacted only very weakly. One representative experiment is shown. Data are shown with the negative (biotinylated F5 fragments only) control subtracted. Results are shown as the mean ± S.E. of triplicate values.
Tropoelastin was reported to bind fibulin-5 in N- and C-terminal regions (24, 25). Here, BIAcore analysis revealed that F5 bound tropoelastin strongly (KD = 64 ± 40 nm) (Table 2 and Fig. 3A). Full-length cutis laxa mutants F5C217R and F5S227P (mutations in cbEGF4) both showed markedly reduced binding affinities for tropoelastin, with rapid dissociation kinetics (Table 2 and Fig. 3A). In solid phase binding assays, soluble fibulin-5 domain pair fragments (F5-E1+2, F5-E4+5, and F5-E6FC) bound immobilized tropoelastin, with the strongest binding by F5-E6FC (Fig. 3C; for binding curves, see supplemental Fig. S4A). The tropoelastin interaction with F5-E6FC was partially calcium-dependent (not shown). Binding of the full-length mutants and fragment F5-E4+5 to tropoelastin indicate that cbEGF4 contributes to F5-tropoelastin interactions. Thus, fibulin-5 binds tropoelastin strongly, with binding sites in N- and C-terminal and central cbEGF array regions.
TABLE 2.
BIAcore surface plasmon resonance analysis of fibulin-4 interactions with LOX and fibulin-5 interactions with tropoelastin
Surface plasmon resonance binding was carried out as described under “Experimental Procedures.” The kinetics of the evaluation of the interactions was performed according to a 1:1 binding model. Values are mean ± S.E. of at least two separate experiments.
| ka | kd | KD | χ2 | |
|---|---|---|---|---|
| ×103m−1s−1 | ×10e−3s−1 | nm | ||
| Tropoelastin-F5 | 44 ± 20 | 1.3 ± 0.1 | 64 ± 40 | 14.8 ± 9.2 |
| Tropoelastin-F5C217R | 8.4 ± 2.7 | 4.6 ± 0.4 | 865 ± 492 | 3.3 ± 1.2 |
| Tropoelastin-F5S227P | 20 ± 13 | 9.3 ± 2.3 | 3423 ± 1439 | 22.4 ± 13.6 |
| LOX-F4 | 21.0 ± 0.7 | 0.7 ± 0.1 | 33 ± 3 | 4.6 ± 0.9 |
| LOX-nF4 | 24.5 ± 8.5 | 0.4 ± 0.2 | 14 ± 6 | 1.1 ± 0.1 |
LOX Binds Strongly to Fibulin-4 but Weakly to Fibulin-5
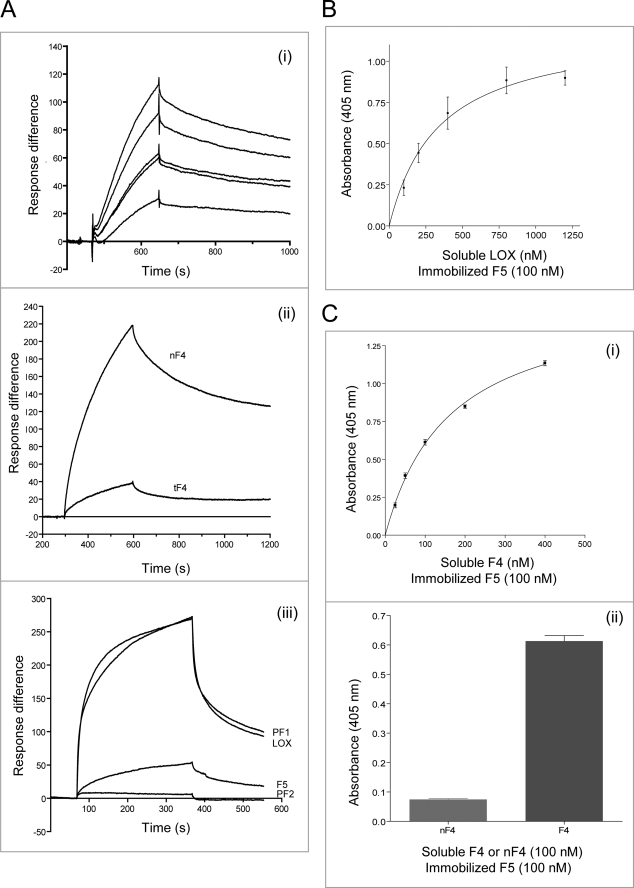
LOX, the major cross-linking enzyme of elastic fibers, is targeted to elastic fibers through its pro-domain (46). Using BIAcore, we examined whether full-length human LOX (see Fig. 1C) interacts with fibulin-4 and -5. F4 bound to immobilized LOX with high affinity (KD = 33 ± 3 nm) (Table 2 and Fig. 4A). nF4 also bound LOX strongly with a KD of 14 ± 6 nm (Table 2 and Fig. 4A), but tF4 bound only weakly (Fig. 4A), and the central eF4 fragment did not bind LOX (not shown). Thus, we have identified a strong binding site for LOX within the N-terminal half of fibulin-4. Using BIAcore, we also confirmed binding of LOX to immobilized F4 (Fig. 4A).
FIGURE 4.
Molecular interactions of LOX with fibulin-4 and -5. A, BIAcore analysis of interactions of immobilized LOX with soluble F4 (i) or nF4 or tF4 (ii) injected over LOX immobilized on a CM5 chip (see Table 2). In iii, BIAcore interactions of immobilized F4 with soluble LOX, PF1, and F5 are shown (virtually no interaction was detected with PF2). In i, the sensorgram shows analyte concentrations ranging from 2 to 10 μg/ml, with duplicate concentrations included in every run. One representative experiment in each case is shown. Response difference is the difference between experimental and control flow cells, in response units. Time is shown in seconds. F4 and nF4 bound strongly to LOX, but tF4 bound weakly. In parallel solid phase assays, F4 was confirmed to bind strongly to LOX (not shown). B, solid phase binding assays showing soluble biotinylated LOX binding to immobilized F5 (KD = 338 nm). One representative experiment is shown in each case. Data are shown with the negative (biotinylated LOX only) control subtracted. Results are shown as the mean ± S.E. of triplicate values. Soluble biotinylated LOX also bound strongly to four of the fibulin-5 domain pair fragments (F5-E1+2, F5-E2+3, F5-E4+5, and F5-E6FC), but in parallel BIAcore experiments, there was no detectable binding of full-length fibulin-5 (F5) to LOX (not shown). C, solid phase binding assays showing soluble biotinylated F4 binding to immobilized F5. i and ii, F4 bound strongly to F5, but nF4 shows virtually no binding to F5 (ii). Thus, the binding site for fibulin-5 is in the C-terminal half of fibulin-4. One representative experiment is shown. Data are shown with the negative (biotinylated F4 or nF4 only) control subtracted. Results are shown as the mean ± S.E. of triplicate values.
BIAcore analysis did not detect any F5 binding to LOX. However, solid phase binding assays did detect relatively weak interactions between soluble LOX and immobilized F5 (KD = 304 ± 69 nm) (Fig. 4B) and between soluble LOX and immobilized fibulin-5 domain pairs F5-E1+2, F5-E2+3, F5-E4+5, and F5-E6FC (not shown). Thus, fibulin-5 does not contain a strong specific binding site for LOX.
Fibulin-4 Binds Fibulin-5
Solid phase binding assays revealed that soluble F4 can bind immobilized F5 with moderate affinity (KD = 161 ± 10 nm) (Fig. 4C). This novel interaction was with the C-terminal half of fibulin-4, since soluble F4 (100 nm) bound, but soluble nF4 (100 nm) showed virtually no binding to immobilized F5 (Fig. 4C). Similarly, BIAcore revealed that soluble F5 binds immobilized F4, with a KD of 182 nm (χ2 = 1.27) (Fig. 4A). However, none of the fibulin-5 domain pair fragments showed strong binding to immobilized F4 (not shown). This novel interaction between fibulins may contribute to regulating elastin globule formation and deposition.
N-terminal Fibrillin-1 Binds Differentially to Fibulin-4 and -5
We and others have reported that the N-terminal half of fibrillin-1 binds to fibulin-4 and -5 (21, 22), and the third EGF-like domain and first hybrid domain of fibrillin-1 are implicated (21). Here, solid phase and BIAcore binding assays provided detailed analysis of the binding specificities of fibulin-4 and -5 to a panel of fibrillin-1 fragments (39). Our data show that these fibulins differentially interact with N-terminal fibrillin-1.
BIAcore analysis showed that soluble PF1 bound strongly to immobilized F4 with steady-state affinity of 74 ± 5 nm (Fig. 4A), but virtually no binding to overlapping PF2 was detected. However, using solid phase assays, soluble F4 bound strongly to the N-terminal fibrillin-1 fragment PF1 (KD = 134 ± 17 nm) and to PF2 (KD = 194 ± 26 nm), which does not contain the first hybrid motif (Fig. 5A); both fragments contain the first TB module, proline-rich region, and following EGF-like domain (see Fig. 1B) (39). F4 also bound strongly to three PF1 deletion fragments (Ex1–11, Ex3–11, and Ex5–11) and moderately to Ex6–11 (Fig. 5A). However, there was only low F4 binding to fibrillin-1 fragment PF4, which contains the first hybrid motif, and virtually no binding to fibrillin-1 fragment Ex5–7, which comprises the first hybrid motif and flanking domains, or to fibrillin-1 fragment Ex7–11, which comprises five domains downstream of the first hybrid motif (Fig. 5A). Since fibulin-4 can bind PF2, the first hybrid motif is probably not essential for binding to N-terminal fibrillin-1 (Fig. 5C), although this motif probably influences N-terminal fibrillin-1 conformation. Both F4 and nF4 bound PF1 with similar kinetics, implicating the N-terminal half of fibulin-4 in binding fibrillin-1 (Fig. 5A).
FIGURE 5.
Molecular interactions of fibrillin-1 with fibulin-4 and -5. A, solid phase binding assays showing soluble biotinylated F4 binding to immobilized overlapping fragments encompassing full-length fibrillin-1 (i) and to N-terminal (PF1) deletion and short fragments (ii). F4 bound well to the N-terminal fragments PF1 and PF2 and to several N-terminal deletion fragments but very weakly to PF4, Ex5–7, and Ex7–11. In iii, binding curves show that soluble biotinylated F4 bound to immobilized PF1 and PF2. In iv, binding curves are shown for soluble biotinylated F4 and nF4 binding to immobilized PF1, indicating that the N-terminal half of fibulin-4 binds N-terminal fibrillin-1. One representative experiment is shown in each case. Data are shown with the negative (biotinylated F4 or nF4 only) control subtracted. Results are shown as the mean ± S.E. of triplicate values. B, solid phase binding assays show soluble biotinylated fibrillin1 (PF1 fragment) binding to immobilized F5 (i) and soluble biotinylated F5 binding to immobilized N-terminal fibrillin-1 fragments (ii). F5 bound well to the N-terminal fragments PF1 and PF4 and to several N-terminal deletion fragments but weakly to PF2 and Ex7–11. One representative experiment is shown. Data are shown with the negative (biotinylated F5 only) control subtracted. Results are shown as the mean ± S.E. of triplicate values. C, domain diagram of fibrillin-1 (see Fig. 1 for key), highlighting potential binding sites for F4 and F5, as well as reported binding sites for tropoelastin (33, 39). D, solid phase assays showing soluble biotinylated fibulin-5 fragments (F5-E1+2 (i) or F5-E6FC (ii)) binding to immobilized PF1 or to three different PF1 (Marfan syndrome) mutants (Fig. 1B). Fibulin-5 fragments bound well to PF1, but binding was significantly reduced to all of these PF1 mutants, especially PF1S115C. Data are shown with the negative (biotinylated protein only) control subtracted. Results are shown as the mean ± S.E. of triplicate values. Significant differences in binding of each fibulin-5 fragment to PF1 or to the PF1 mutants are shown as follows: ***, p < 0.001 (unpaired t test). In each case, a representative experiment is shown.
Solid phase binding assays showed that the soluble fibrillin-1 N-terminal fragment PF1 bound strongly to F5 (KD = 63 ± 14 nm), and soluble F5 also bound strongly to PF4 and Ex1–11 (Fig. 5B). There was also F5 binding to the deletion fragments Ex3–11, Ex4–11, Ex5–11, Ex6–11, and Ex5–7 but virtually no F5 binding to Ex7–11 or PF2, neither of which contains the first hybrid motif (Fig. 5B). Thus, F5 binds to N-terminal fragments that contain this hybrid motif (Fig. 5C), and there may be a second binding site N-terminal to Ex5–7. In terms of PF1 binding site(s) on fibulin-5, four of the fibulin-5 domain pairs (F5-E1+2, F5-E4+5, F5-E5+6, and F5-E6FC) bound PF1, with the strongest binding by F5-E6FC and F5-E1+2 (KD values of 187 ± 31 and 344 ± 54 nm, respectively) (supplemental Fig. S4B). The interaction with F5-E6FC was partially calcium-dependent (data not shown). Thus, both fibulin-5 N- and C-terminal regions bind N-terminal fibrillin-1. Marfan mutations (R62C, T101A, and S115C) in the N-terminal region of PF1 significantly reduced binding to F5-E1+2 and F5-E6FC (Fig. 5D), possibly due to long range conformation effects.
LOX Interacts Strongly with N-terminal Fibrillin-1 and with Tropoelastin
Using BIAcore, no binding was detected between immobilized LOX and soluble N-terminal fibrillin-1 (PF1 ligand). However, solid phase assays detected a strong interaction between soluble LOX and immobilized PF1, with a KD of 26 ± 11 nm, and at reduced levels to N-terminal deletion fragments Ex1–11 and Ex3–11 (Fig. 6A). However, LOX did not bind strongly to any other fibrillin-1 fragments. Thus, we have demonstrated that LOX interacts strongly and specifically with N-terminal fibrillin-1, although this interaction is ablated by LOX immobilization on BIAcore chips.
FIGURE 6.
Molecular interactions of LOX with fibrillin-1 and tropoelastin. A, solid phase binding assays showing soluble biotinylated LOX binding to immobilized fibrillin-1. i and ii, LOX binds to the N-terminal fibrillin-1 fragment PF1 and shows reduced binding to PF1 deletion fragments but no binding to Ex5–7. iii, binding curve showing soluble biotinylated LOX binding to immobilized PF1. One representative experiment is shown. Data are shown with the negative (biotinylated LOX only) control subtracted. Results are shown as the mean ± S.E. of triplicate values. B, solid phase binding curve showing soluble biotinylated LOX binding to immobilized tropoelastin. One representative experiment is shown. Data are shown with the negative (biotinylated LOX only) control subtracted. Results are shown as the mean ± S.E. of triplicate values.
Using BIAcore, there was no detectable interaction between soluble LOX and immobilized tropoelastin. However, solid phase assays detected a strong interaction between LOX and immobilized tropoelastin, with a KD of 49 ± 11 nm (Fig. 6B). Thus, LOX binds tropoelastin strongly, although this interaction is ablated by immobilization of tropoelastin on BIAcore chips.
Defining Fibulin Complexes Using Competitive Inhibition Binding Assays
Having defined binding interactions of fibulin-4 and -5 with tropoelastin, LOX, and fibrillin-1 and identified novel interactions between fibulin-4 and -5 and between LOX and N-terminal fibrillin-1, we investigated competition between these interactions in order to gain insights into how these complexes contribute to elastic fiber formation (for a model, see Fig. 9).
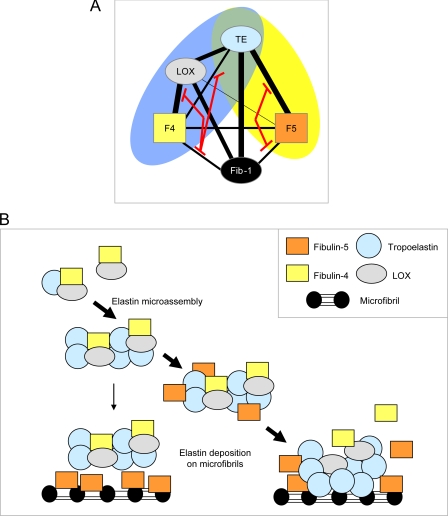
FIGURE 9.
Modeling elastic fiber interactions of fibulin-4 and -5. A, schematic diagram showing the elastic fiber interactions of fibulin-4 and -5. Thick black lines represent strong interactions, and thinner black lines show weaker interactions. Red lines indicate how fibrillin-1 binding to fibulins inhibits their interactions with LOX and tropoelastin. The blue oval highlights the ternary complex between fibulin-4, LOX, and tropoelastin, and the yellow oval highlights the complex between fibulin-5 and tropoelastin. TE, tropoelastin; Fib-1, fibrillin-1; F4, fibulin-4; F5, fibulin-5. B, schematic model indicating how fibulin interactions may contribute to elastic fiber assembly. Our binding data indicate that fibulin-4-LOX interactions facilitate formation of ternary complexes with tropoelastin; these complexes may regulate LOX activation and elastin cross-linking. Following fibulin-5 depletion, elastin appears aggregated and distinct from microfibrils, so fibulin-5 must normally regulate elastin globules and direct their interaction with microfibrils, either by first associating with microfibrils and then attracting elastin globules to bind microfibrils or by first associating with elastin globules and then facilitating their deposition on microfibrils. Our data support the latter model. Once localized at microfibrils, elastin probably interacts directly with fibrillin-1, since these interactions can be high affinity (39), and fibulin-5 interactions with fibrillin-1 and tropoelastin are mutually inhibitory. Active LOX may remain associated with and cross-link coalescing elastin globules on microfibrils. Fibulin-4 and -5 may remain associated with microfibrils or may “recycle” for further elastin deposition.
Fibulin-4 Forms a Ternary Complex with LOX and Tropoelastin
Since the N-terminal half of fibulin-4 binds LOX strongly (see Table 2 and Fig. 4A), we investigated whether this interaction regulates binding of fibulin-4 to tropoelastin (Fig. 7, A and B).
FIGURE 7.
Molecular interactions and competition assays. A, analysis of the effect of LOX on soluble F4 interacting with immobilized tropoelastin. In BIAcore experiments, F4 was soluble ligand alone or preincubated with LOX. F4 binding to tropoelastin was enhanced following preincubation of F4 with LOX (both at 200 nm) but reduced following F4 preincubation with 1000 nm LOX. (LOX does not bind immobilized tropoelastin; not shown.) One representative experiment is shown. Response difference is the difference between experimental and control flow cells, in response units. Time is shown in seconds. B, solid phase binding assay showing that preincubation of soluble biotinylated F4 with LOX (both at 100 nm) partially inhibited the interaction of F4 with immobilized F5. A significant difference in binding of F4 preincubated with LOX to F5 was shown as follows: **, p < 0.01 (unpaired t test). One representative experiment is shown. Data are shown with the negative (biotinylated protein only) control subtracted. Results are shown as the mean ± S.E. of triplicate values. C, analysis of the effect of PF1 on soluble F4 interacting with immobilized LOX or tropoelastin. In BIAcore experiments (i and ii), F4 was a soluble ligand, alone or preincubated with increasing concentrations of PF1 (Log10 Conc PF1). F4 binding to LOX (i) and to tropoelastin (ii) was inhibited by PF1, with EC50 values of 365 and 115 nm, respectively. In each case, one representative experiment is shown. Response difference is the difference between experimental and control flow cells, in response units. iii, solid phase binding assays confirmed both that soluble biotinylated LOX (100 nm) interacted with immobilized F4 and nF4 but more weakly with tF4 and that preincubation of LOX with PF1 (100 nm) strongly inhibited all of these interactions. Significant differences in binding of LOX preincubated with PF1 to F4, nF4, or tF4 were shown as follows: ***, p < 0.001 (unpaired t test). iv, solid phase binding assays confirmed both that soluble biotinylated F4 (100 nm) interacted with immobilized tropoelastin and that preincubation of soluble F4 with PF1 or PF4 (all at 100 nm) strongly inhibited this interaction. Significant differences in binding of F4 or F4 preincubated with PF1 to tropoelastin were shown as follow: ***, p < 0.001 (unpaired t test). In each case, one representative experiment is shown. Data are shown with the negative (biotinylated protein only) control subtracted. Results are shown as the mean ± S.E. of triplicate values. BIAcore analysis of soluble F4 interacting with immobilized tropoelastin confirmed that preincubation of F4 with tFib-1 (a C-terminally truncated two-thirds fragment of fibrillin-1) markedly inhibits F4 binding to tropoelastin (see supplemental Fig. S4D). D, BIAcore analysis of soluble F5 interacting with immobilized tropoelastin. In i, F5 was soluble ligand alone, or preincubated with either LOX or F4 or with the fibrillin-1 fragments PF1 or tFib-1 (all at 200 nm). The ability of F5 to bind tropoelastin was unaffected by preincubation with LOX or F4 but was inhibited by PF1 (which does not contain tropoelastin binding sites) and inhibited more strongly by tFib-1 (which contains tropoelastin binding sites; see Fig. 4D). ii, increasing concentrations of soluble PF1 (Log10 Conc PF1) inhibited binding of F5 to tropoelastin. In each case, one representative experiment is shown. Response difference is the difference between experimental and control flow cells, in response units.
Using BIAcore (see above), F4 bound immobilized tropoelastin with relatively rapid dissociation kinetics, but LOX did not bind immobilized tropoelastin (Fig. 3A) (data not shown). Here, BIAcore experiments revealed that F4 binding to immobilized tropoelastin was markedly enhanced when preincubated with LOX (equimolar at 200 nm), although binding was partially inhibited when 200 nm fibulin-4 was preincubated with 1000 nm LOX (Fig. 7A). nF4 did not bind tropoelastin (see Fig. 3B) even in the presence of LOX (which binds nF4; see Fig. 4A), so the enhanced binding to tropoelastin in the presence of equimolar LOX was not directly LOX-mediated. Fibulin-4 may, by binding LOX, act as a template for tropoelastin, thereby facilitating its cross-linking by LOX.
Fibulin-5 Does Not Inhibit Fibulin-4 Binding to Tropoelastin
Although fibulin-4 binds fibulin-5 with moderate affinity (see Fig. 4C), preincubation of F4 with F5 (both at 100 nm) did not significantly affect F4 binding to immobilized tropoelastin (not shown). However, preincubation of F4 with LOX (both at 100 nm) partially inhibited binding of F4 to immobilized F5 (Fig. 7B), so F4-LOX complexes limit association of F4 with immobilized F5. F4-F5 complexes may contribute to elastic fiber formation after elastin cross-linking.
Fibrillin-1 Inhibits Fibulin-4 Binding to LOX and Tropoelastin
Fibulin-4 (F4 and nF4) binds N-terminal fibrillin-1 (PF1) with moderate affinity (see Fig. 5A). In BIAcore experiments, preincubation of F4 (200 nm) with PF1 (0–3000 nm) inhibited the binding of F4 to immobilized LOX (EC50 365 nm) and to immobilized tropoelastin (EC50 115 nm) (Fig. 7C). A downstream fibrillin-1 fragment (PF12) that does not bind F4 did not inhibit binding of LOX or tropoelastin to F4 (not shown). In solid phase assays, preincubation of LOX with PF1 (both at 100 nm; these ligands bind each other strongly; see Fig. 6A) strongly inhibited LOX binding to immobilized F4, nF4, or tF4 (Fig. 7C). Similarly, solid phase and BIAcore assays showed that preincubation of F4 and fibrillin-1 (PF1, PF4, or tFib-1, the C-terminally truncated two-thirds fibrillin-1 fragment) (all at 100 nm) strongly inhibited F4 binding to tropoelastin (Fig. 7C and supplemental Fig. S4D).
Thus, fibrillin-1 disrupts formation of fibulin-4 interactions with LOX and with tropoelastin. This competition may enhance deposition of cross-linked elastin globules onto microfibrils.
Fibrillin-1 Inhibits Fibulin-5 Binding to Tropoelastin
As shown in binding assays (see Figs. 3A, 4 (B and C), and 5B), F5 strongly binds to tropoelastin and to N-terminal fibrillin-1 (PF1) and moderately binds to F4 but binds weakly to LOX. Here, BIAcore experiments showed that preincubation of F5 with LOX or F4 did not affect the interaction between F5 and immobilized tropoelastin (Fig. 7D). However, preincubation of F5 with PF1 or tFib-1 inhibited F5 binding to tropoelastin (Fig. 7D).
Although PF1 does not bind tropoelastin, we previously reported two downstream sites within the N-terminal half of fibrillin-1 and a third C-terminal fibrillin-1 region that all bind tropoelastin strongly (see Fig. 5C) (33, 39). El-Hallous et al. (21) reported that the N-terminal half of fibrillin-1 did not bind tropoelastin but that fibulin-5 (more than fibulin-4) facilitated tropoelastin binding to this region of fibrillin-1. Here, we confirmed by BIAcore and solid phase assays that a C-terminally truncated two-thirds fibrillin-1 fragment (tFib-1) binds immobilized tropoelastin (supplemental Fig. S4C) (not shown) and found that fibulin-5 neither inhibited nor stimulated tropoelastin binding to this large fibrillin-1 fragment (not shown).
Hence, fibulin-5 can either bind tropoelastin or fibrillin-1, probably at the same or an overlapping site. This competition may facilitate fibulin-5 targeting of tropoelastin onto microfibrils.
Investigating the Contribution of Fibulin-4 to Elastin Cross-linking
Recently, Nonaka et al. (45) showed that ARPE-19 cultures supplemented with tropoelastin form desmosine cross-links, confirming LOX activity in these cultures and that transfection with fibulin-5 enhanced cross-links. Here, this culture model was used, with fibulin-4 siRNA knockdown, to investigate contributions of fibulin-4 to elastin cross-linking.
First, we showed that control ARPE-19 cells expressed moderate levels of fibulin-4 but only low levels of fibulin-5 and that siRNA of fibulin-4 achieved 70% knockdown (supplemental Fig. S5A), but fibulin-5 levels remained low (not shown). We ensured the presence of tropoelastin by transfecting cells with tropoelastin (supplemental Fig. S5B). Biological duplicate experiments showed that scrambled tropoelastin-transfected ARPE-19 cells contained 12.5 ± 3.8 pmol of desmosine/mg of protein, whereas tropoelastin-transfected fibulin-4 siRNA knockdown ARPE-19 cells had 15.4 ± 4.3 pmol of desmosine/mg of protein. Increased desmosine was also found in the knockdown cultures supplemented with 10 μg/ml exogenous human tropoelastin. These data are consistent with increased desmosine in fibulin-4+/− mice (5).
Defining the Contribution of Fibulin-5 to Ordered Elastin Deposition
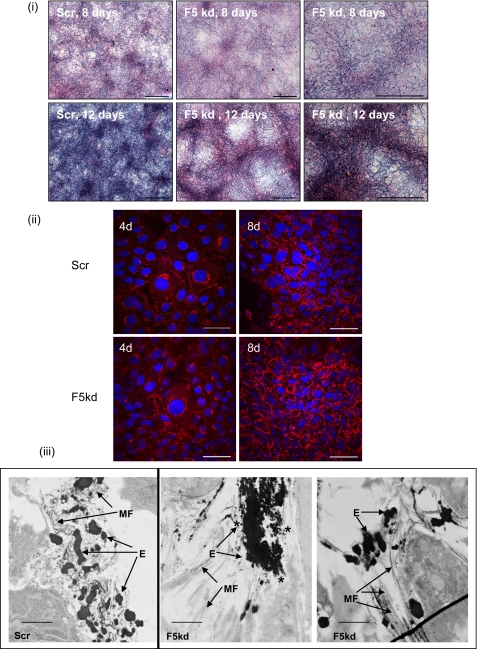
The rat fetal lung fibroblast line, RFL6, expresses abundant elastin and deposits elastic fibers (13). In RFL6 cells with fibulin-5 stably knocked down by shRNA (supplemental Fig. S5C) but expressing fibulin-4, numerous thick elastin-containing arrays were clearly apparent by light microscopy using Miller's elastin stain (Fig. 8). Transmission electron microscopy revealed the presence of large elastin globules and of numerous small, relatively uniform sized elastin globules with limited association with microfibrils; these images show similarities with data reported by Choi et al. (30). Hence, fibulin-5 is necessary for deposition of uniform elastin globules on microfibrils.
FIGURE 8.
shRNA knockdown of fibulin-5 in RFL6 cells. i, Miller's elastin staining of RFL6 scrambled control (Scr) and fibulin-5 shRNA knockdown (F5 kd) cultures (8 and 12 days); this stain highlights elastin as dark deposits. The scrambled control cultures contained a dense network of elastin, whereas the F5 knockdown cells formed prominent thick elastin arrays. Scale bars, 250 μm. ii, immunofluorescence microscopy of RFL6 scrambled control and fibulin-5 shRNA knockdown cultures (4 and 8 days; 4d and 8d, respectively), using elastin RA75 polyclonal antibody (red) and cell nuclear staining (4′,6-diamidino-2-phenylindole; blue). By 8 days, the fibulin-5 knockdown cultures showed more prominent staining of elastin than the scrambled control cultures. Scale bars, 50 μm. iii, transmission electron microscopy of RFL6 scrambled control and fibulin-5 shRNA knockdown cultures at 10 days. In the scrambled control cultures, elastin was accreting within microfibril bundles, whereas in the fibulin-5 knockdown cells, small elastin globules (indicated by an asterisk) formed large aggregates that were mainly distinct from microfibrils. MF, microfibrils; E, elastin. Scale bars, 1.4 μm.
Investigating Whether Heparin Influences Fibulin-mediated Elastic Fiber Formation
Heparan sulfate is abundant at the cell surface as a component of syndecan and glypican receptors, and heparan sulfate interacts with tropoelastin and enhances its coacervation (47–50). Preincubation of tropoelastin with heparin (10 and 100 nm) caused significant but limited reduction in binding of fibulin-4 and -5 to tropoelastin (supplemental Fig. S6A). Preincubation of fibulin-5 domain pair fragments F5-E1+2, F5-E4+5, and F5-E6FC (which bind tropoelastin; see Fig. 3C; supplemental Fig. 4A) with heparin also significantly inhibited their ability to interact with tropoelastin (supplemental Fig. S6B). Heparin also partially inhibited F5-E6FC binding to PF1 but enhanced the interaction of F5-E1+2 with PF1 (not shown). Thus, cell surface heparan sulfate subtly influences fibulin-mediated elastic fiber interactions.
DISCUSSION
Elastic fiber formation is a complex multistage process that proceeds through the formation of cross-linked elastin globules and their deposition on microfibrils, where they coalesce to form fibers (19, 20). Knock-out mice revealed that fibulin-4 and -5 have essential non-compensatory roles in this process (5–7), but their roles in this process remain unresolved. To gain insights into their distinct contributions, we delineated how they interact with major elastic fiber molecules (with tropoelastin; with LOX, which cross-links tropoelastin; and with fibrillin-1, which forms the microfibril template) and have identified novel fibulin-fibulin interactions and fibulin interactions with LOX and fibrillin-1. The first stage of elastin deposition is envisioned to involve the pericellular assembly of elastin into globules and their stabilization through cross-linking by LOX; our data show that fibulin-4 strongly binds LOX, and this interaction enhances formation of a ternary complex that can regulate elastin cross-linking (Fig. 9). The second stage of elastin deposition encompasses deposition of elastin globules on microfibrils and their accretion to form a fiber; our data show that fibulin-5 can interact with tropoelastin or with fibrillin-1, implying a chaperone role for fibulin-5 in directing elastin onto microfibrils (Fig. 9).
MALLS analysis of fibulin-4 revealed that it can form dimers in the presence of calcium. This property may reside within the N-terminal half of the molecule, since an N-terminal fragment (nF4) also underwent calcium-dependent dimerization, which suggests parallel dimers. Within the extracellular matrix, fibulin-4 is exposed to low millimolar calcium and may be dimeric; all binding assays were therefore conducted in the presence of calcium. The analysis of fibulin-4 by rotary shadowing electron microscopy has indicated a short rodlike structure with a globular end-domain (4, 51). Here, single-particle averaging electron microscopy of negatively stained fibulin-4, which provides enhanced molecular details, reveals a bent rodlike structure. It is unclear whether the globular end-domain is the N or C terminus.
The absence of fibulin-4 causes a much more severe perinatal-lethal elastic fiber phenotype than loss of fibulin-5, and the drastic reduction in desmosine and concomitant appearance of rodlike structures likely to contain glycosaminoglycans (26) indicates a critical regulatory role in elastin cross-linking (5). We have identified a high affinity LOX binding site within the N-terminal half of fibulin-4 and shown that fibulin-4 binding to LOX enhances tropoelastin binding and ternary complex formation. Reduction of fibulin-4 on a background of low fibulin-5 enhanced LOX-mediated elastin cross-linking, which is similar to that in fibulin-4+/− mice, which showed ∼20% increase in desmosine (5). The fibulin-4/fibulin-5 ratio may be critical, since Nonaka et al. (45), using the same cell system, showed increased desmosines when cells were transfected with fibulin-5. The novel interaction between fibulin-4 and -5 that we have described here may contribute after initial elastin cross-linking but is unlikely to be essential for cross-linking, since LOX partially inhibited this interaction, whereas mice with no fibulin-5 have only 16% reduction in desmosine cross-links (7). With low levels of fibulin-4 and -5, elastin probably forms larger globules that may be rapidly cross-linked. Overall, these data confirm that fibulin-4 plays a central regulator role in cross-linking, probably by acting as a template to control LOX activation and to juxtapose it with tropoelastin. Consequent lysine-derived cross-link formation will displace glycosaminoglycans and allow normal elastin assembly. Although LOX and tropoelastin can both bind fibrillin-1, microfibrils are insufficient to support elastin cross-linking in fibulin-4 null mice. Depletion of fibulin-4 may enhance free LOX, which could alter cell behavior by oxidizing cell surface proteins, such as platelet-derived growth factor receptors, (52).
We and others have reported that fibulin-4 and -5 bind N-terminal fibrillin-1 (21, 22, 53) and microfibrils (22). Here, we show exquisite differential features to the binding of both fibulins to fibrillin-1, since fibulin-4 can bind fibrillin-1 in the presence or absence of the first hybrid motif, whereas fibulin-5 binds fragments that contain this motif. These Marfan mutations in N-terminal fibrillin-1 that reduce binding to F5-E1+2 and F5-E6FC imply long range conformation effects. Fibrillin-1 competes with LOX and tropoelastin to bind fibulin-4, perhaps competing for the same site on fibulin-4, so the fibulin-4·LOX·tropoelastin complex may be important before elastin deposition on microfibrils. Although N-terminal fibrillin-1 is known to be a “sticky” sequence, its specific interactions shown here with LOX, fibulin-5, and tropoelastin highlight its central role in regulating elastin deposition on microfibrils.
Tissue-purified LOX was reported to cross-link recombinant tropoelastin in the absence of other molecules (54) and bound to the C-terminal half of tropoelastin by ligand blot overlay assay (46). Here, using BIAcore, repeated attempts failed to detect any direct binding of LOX to immobilized tropoelastin or of fibrillin-1 to immobilized LOX. However, using solid phase assays, strong interactions were detected between LOX and immobilized tropoelastin and between LOX and immobilized fibrillin-1. Clearly, immobilization of tropoelastin and LOX onto CM5 chips impedes these interactions. Molecules are generally covalently coupled to these chips via amines (often lysyl), as well as thiol, aldehyde, or carboxyl groups. It is thus important to confirm BIAcore data by solid phase assays.
Elastic fiber formation may proceed by the deposition and ordered accretion of elastin globules on microfibrils (18–20). Here, depletion of fibulin-5 caused the appearance of elastin-rich thick fibrous structures, as seen by light microscopy, and of numerous small elastin globules accreted into large aggregates adjacent to microfibril bundles, as seen by electron microscopy. Thus, fibulin-5 must normally facilitate deposition of uniform elastin globules onto microfibrils and prevent uncontrolled aggregation. These findings are consistent with a study of fibulin-5 null mice dermis in which elastin globules were adjacent to but not integrated within microfibril arrays (30). Although fibulin-4 and LOX can both bind fibrillin-1, they do not efficiently target elastin onto microfibrils or prevent aberrant elastin aggregation when fibulin-5 is depleted. Our data showing that fibrillin-1 and tropoelastin compete to bind fibulin-5 are consistent with fibulin-5 chaperoning cross-linked elastin globules onto microfibrils, where they may then be stabilized by elastin interactions with fibrillin-1 (Fig. 9). It seems unlikely that fibulin-5 facilitates deposition of individual tropoelastin monomers onto microfibrils, since oxidation by LOX precedes elastin deposition on microfibrils (20), whereas we have shown that fibrillin-1 inhibits the ternary complexes of fibulin-4, LOX, and tropoelastin that may facilitate cross-linking. In summary, our study provides molecular explanations for the non-overlapping essential roles of fibulin-4 and -5 in elastic fiber formation.
Supplementary Material
Acknowledgments
We thank L. Ward and D. Ng for assistance in preparing lysyl oxidase (LOX) and acknowledge the support of Dr. T. Jowitt and colleagues in the Faculty Biomolecules Core Facility and Electron Microscopy Facility.
This work was supported by Wellcome Trust Grants 081930 and 072291, Medical Research Council Grant G0200246, and European Union Grant LSHM-CT-2005-018960.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
Jones, R. P. O., Wang, M.-C., Jowitt, T. A., Ridley, C., Mellody, K. T., Howard, M., Wang, T., Bishop, P. N., Lotery, A. J., Kielty, C. M., Baldock, C., and Trump, D. (July 17, 2009) J. Biol. Chem. 10.1074/jbc.M109.011627.
- cbEGF
- calcium-binding epidermal growth factor-like domain
- FC
- C-terminal fibulin
- LOX
- lysyl oxidase
- MALLS
- multiangle laser light scattering
- siRNA
- small interfering RNA
- shRNA
- short hairpin RNA.
REFERENCES
- 1.Chu M. L., Tsuda T. (2004) Birth Defects Res. C Embryo Today 72, 25–36 [DOI] [PubMed] [Google Scholar]
- 2.de Vega S., Iwamoto T., Yamada Y. (2009) Cell Mol. Life Sci. 66, 1890–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Timpl R., Sasaki T., Kostka G., Chu M. L. (2003) Nat. Rev. Mol. Cell Biol. 4, 479–489 [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi N., Kostka G., Garbe J. H., Keene D. R., Bächinger H. P., Hanisch F. G., Markova D., Tsuda T., Timpl R., Chu M. L., Sasaki T. (2007) J. Biol. Chem. 282, 11805–11816 [DOI] [PubMed] [Google Scholar]
- 5.McLaughlin P. J., Chen Q., Horiguchi M., Starcher B. C., Stanton J. B., Broekelmann T. J., Marmorstein A. D., McKay B., Mecham R., Nakamura T., Marmorstein L. Y. (2006) Mol. Cell Biol. 26, 1700–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura T., Lozano P. R., Ikeda Y., Iwanaga Y., Hinek A., Minamisawa S., Cheng C. F., Kobuke K., Dalton N., Takada Y., Tashiro K., Ross J., Jr., Honjo T., Chien K. R. (2002) Nature 415, 171–175 [DOI] [PubMed] [Google Scholar]
- 7.Yanagisawa H., Davis E. C., Starcher B. C., Ouchi T., Yanagisawa M., Richardson J. A., Olson E. N. (2002) Nature 415, 168–171 [DOI] [PubMed] [Google Scholar]
- 8.Hanada K., Vermeij M., Garinis G. A., de Waard M. C., Kunen M. G., Myers L., Maas A., Duncker D. J., Meijers C., Dietz H. C., Kanaar R., Essers J. (2007) Circ. Res. 100, 738–746 [DOI] [PubMed] [Google Scholar]
- 9.Nakamura T., Ruiz-Lozano P., Lindner V., Yabe D., Taniwaki M., Furukawa Y., Kobuke K., Tashiro K., Lu Z., Andon N. L., Schaub R., Matsumori A., Sasayama S., Chien K. R., Honjo T. (1999) J. Biol. Chem. 274, 22476–22483 [DOI] [PubMed] [Google Scholar]
- 10.Kowal R. C., Richardson J. A., Miano J. M., Olson E. N. (1999) Circ. Res. 84, 1166–1176 [DOI] [PubMed] [Google Scholar]
- 11.Spencer J. A., Hacker S. L., Davis E. C., Mecham R. P., Knutsen R. H., Li D. Y., Gerard R. D., Richardson J. A., Olson E. N., Yanagisawa H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2946–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dasouki M., Markova D., Garola R., Sasaki T., Charbonneau N. L., Sakai L. Y., Chu M. L. (2007) Am. J. Hum. Genet. A 143A, 2635–2641 [DOI] [PubMed] [Google Scholar]
- 13.Hu Q., Loeys B. L., Coucke P. J., De Paepe A., Mecham R. P., Choi J., Davis E. C., Urban Z. (2006) Hum. Mol. Genet. 15, 3379–3386 [DOI] [PubMed] [Google Scholar]
- 14.Hu Q., Reymond J. L., Pinel N., Zabot M. T., Urban Z. (2006) J. Invest. Dermatol. 126, 283–290 [DOI] [PubMed] [Google Scholar]
- 15.Hucthagowder V., Sausgruber N., Kim K. H., Angle B., Marmorstein L. Y., Urban Z. (2006) Am. J. Hum. Genet. 78, 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu L., Garland D., Yang Z., Shukla D., Rajendran A., Pearson E., Stone E. M., Zhang K., Pierce E. A. (2007) Hum. Mol. Genet. 16, 2411–2422 [DOI] [PubMed] [Google Scholar]
- 17.Rahn D. D., Acevedo J. F., Roshanravan S., Keller P. W., Davis E. C., Marmorstein L. Y., Word R. A. (2009) Am. J. Pathol. 174, 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kielty C. M. (2006) Expert Rev. Mol. Med. 8, 1–23 [DOI] [PubMed] [Google Scholar]
- 19.Wagenseil J. E., Mecham R. P. (2007) Birth Defects Res. C Embryo Today 81, 229–240 [DOI] [PubMed] [Google Scholar]
- 20.Sato F., Wachi H., Ishida M., Nonaka R., Onoue S., Urban Z., Starcher B. C., Seyama Y. (2007) J. Mol. Biol. 369, 841–851 [DOI] [PubMed] [Google Scholar]
- 21.El-Hallous E., Sasaki T., Hubmacher D., Getie M., Tiedemann K., Brinckmann J., Bätge B., Davis E. C., Reinhardt D. P. (2007) J. Biol. Chem. 282, 8935–8946 [DOI] [PubMed] [Google Scholar]
- 22.Freeman L. J., Lomas A., Hodson N., Sherratt M. J., Mellody K. T., Weiss A. S., Shuttleworth A., Kielty C. M. (2005) Biochem. J. 388, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirai M., Ohbayashi T., Horiguchi M., Okawa K., Hagiwara A., Chien K. R., Kita T., Nakamura T. (2007) J. Cell Biol. 176, 1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wachi H., Nonaka R., Sato F., Shibata-Sato K., Ishida M., Iketani S., Maeda I., Okamoto K., Urban Z., Onoue S., Seyama Y. (2008) J. Biochem. 143, 633–639 [DOI] [PubMed] [Google Scholar]
- 25.Zheng Q., Davis E. C., Richardson J. A., Starcher B. C., Li T., Gerard R. D., Yanagisawa H. (2007) Mol. Cell Biol. 27, 1083–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fornieri C., Baccarani-Contri M., Quaglino D., Jr., Pasquali-Ronchetti I. (1987) J. Cell Biol. 105, 1463–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornstra I. K., Birge S., Starcher B., Bailey A. J., Mecham R. P., Shapiro S. D. (2003) J. Biol. Chem. 278, 14387–14393 [DOI] [PubMed] [Google Scholar]
- 28.Mäki J. M., Sormunen R., Lippo S., Kaarteenaho-Wiik R., Soininen R., Myllyharju J. (2005) Am. J. Pathol. 167, 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X., Zhao Y., Gao J., Pawlyk B., Starcher B., Spencer J. A., Yanagisawa H., Zuo J., Li T. (2004) Nat. Genet. 36, 178–182 [DOI] [PubMed] [Google Scholar]
- 30.Choi J., Bergdahl A., Zheng Q., Starcher B., Yanagisawa H., Davis E. C. (2009) Matrix Biol. 28, 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarke A. W., Wise S. G., Cain S. A., Kielty C. M., Weiss A. S. (2005) Biochemistry 44, 10271–10281 [DOI] [PubMed] [Google Scholar]
- 32.Jensen S. A., Reinhardt D. P., Gibson M. A., Weiss A. S. (2001) J. Biol. Chem. 276, 39661–39666 [DOI] [PubMed] [Google Scholar]
- 33.Rock M. J., Cain S. A., Freeman L. J., Morgan A., Mellody K., Marson A., Shuttleworth C. A., Weiss A. S., Kielty C. M. (2004) J. Biol. Chem. 279, 23748–23758 [DOI] [PubMed] [Google Scholar]
- 34.Trask T. M., Trask B. C., Ritty T. M., Abrams W. R., Rosenbloom J., Mecham R. P. (2000) J. Biol. Chem. 275, 24400–24406 [DOI] [PubMed] [Google Scholar]
- 35.Cirulis J. T., Bellingham C. M., Davis E. C., Hubmacher D., Reinhardt D. P., Mecham R. P., Keeley F. W. (2008) Biochemistry 47, 12601–12613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lomas A. C., Mellody K. T., Freeman L. J., Bax D. V., Shuttleworth C. A., Kielty C. M. (2007) Biochem. J. 405, 417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baldock C., Siegler V., Bax D. V., Cain S. A., Mellody K. T., Marson A., Haston J. L., Berry R., Wang M. C., Grossmann J. G., Roessle M., Kielty C. M., Wess T. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11922–11927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cain S. A., Baldock C., Gallagher J., Morgan A., Bax D. V., Weiss A. S., Shuttleworth C. A., Kielty C. M. (2005) J. Biol. Chem. 280, 30526–30537 [DOI] [PubMed] [Google Scholar]
- 39.Cain S. A., Baldwin A. K., Mahalingam Y., Raynal B., Jowitt T. A., Shuttleworth C. A., Couchman J. R., Kielty C. M. (2008) J. Biol. Chem. 283, 27017–27027 [DOI] [PubMed] [Google Scholar]
- 40.Marson A., Rock M. J., Cain S. A., Freeman L. J., Morgan A., Mellody K., Shuttleworth C. A., Baldock C., Kielty C. M. (2005) J. Biol. Chem. 280, 5013–5021 [DOI] [PubMed] [Google Scholar]
- 41.Mellody K. T., Freeman L. J., Baldock C., Jowitt T. A., Siegler V., Raynal B. D., Cain S. A., Wess T. J., Shuttleworth C. A., Kielty C. M. (2006) J. Biol. Chem. 281, 31854–31862 [DOI] [PubMed] [Google Scholar]
- 42.Martin S. L., Vrhovski B., Weiss A. S. (1995) Gene 154, 159–166 [DOI] [PubMed] [Google Scholar]
- 43.Toonkool P., Jensen S. A., Maxwell A. L., Weiss A. S. (2001) J. Biol. Chem. 276, 44575–44580 [DOI] [PubMed] [Google Scholar]
- 44.Ludtke S. J., Baldwin P. R., Chiu W. (1999) J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 45.Nonaka R., Onoue S., Wachi H., Sato F., Urban Z., Starcher B. C., Seyama Y. (2009) Clin. Biochem. 42, 713–721 [DOI] [PubMed] [Google Scholar]
- 46.Thomassin L., Werneck C. C., Broekelmann T. J., Gleyzal C., Hornstra I. K., Mecham R. P., Sommer P. (2005) J. Biol. Chem. 280, 42848–42855 [DOI] [PubMed] [Google Scholar]
- 47.Gheduzzi D., Guerra D., Bochicchio B., Pepe A., Tamburro A. M., Quaglino D., Mithieux S., Weiss A. S., Pasquali Ronchetti I. (2005) Matrix Biol. 24, 15–25 [DOI] [PubMed] [Google Scholar]
- 48.Tu Y., Weiss A. S. (2008) Biomacromolecules 9, 1739–1744 [DOI] [PubMed] [Google Scholar]
- 49.Wu W. J., Vrhovski B., Weiss A. S. (1999) J. Biol. Chem. 274, 21719–21724 [DOI] [PubMed] [Google Scholar]
- 50.Broekelmann T. J., Kozel B. A., Ishibashi H., Werneck C. C., Keeley F. W., Zhang L., Mecham R. P. (2005) J. Biol. Chem. 280, 40939–40947 [DOI] [PubMed] [Google Scholar]
- 51.Giltay R., Timpl R., Kostka G. (1999) Matrix Biol. 18, 469–480 [DOI] [PubMed] [Google Scholar]
- 52.Lucero H. A., Ravid K., Grimsby J. L., Rich C. B., DiCamillo S. J., Mäki J. M., Myllyharju J., Kagan H. M. (2008) J. Biol. Chem. 283, 24103–24117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ono R. N., Sengle G., Charbonneau N. L., Carlberg V., Bächinger H. P., Sasaki T., Lee-Arteaga S., Zilberberg L., Rifkin D. B., Ramirez F., Chu M. L., Sakai L. Y. (2009) J. Biol. Chem. 284, 16872–16881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bedell-Hogan D., Trackman P., Abrams W., Rosenbloom J., Kagan H. (1993) J. Biol. Chem. 268, 10345–10350 [PubMed] [Google Scholar]
- 55.Bax D. V., Mahalingam Y., Cain S., Mellody K., Freeman L., Younger K., Shuttleworth C. A., Humphries M. J., Couchman J. R., Kielty C. M. (2007) J. Cell Sci. 120, 1383–1392 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.