Abstract
Cyclical activation and inactivation of Rho family small G proteins, such as Rho, Rac, and Cdc42, are needed for moving cells to form leading edge structures in response to chemoattractants. However, the mechanisms underlying the dynamic regulation of their activities are not fully understood. We recently showed that another small G protein, Rap1, plays a crucial role in the platelet-derived growth factor (PDGF)-induced formation of leading edge structures and activation of Rac1 in NIH3T3 cells. We showed here that knockdown of afadin, an actin-binding protein, in NIH3T3 cells resulted in a failure to develop leading edge structures in association with an impairment of the activation of Rap1 and Rac1 and inactivation of RhoA in response to PDGF. Overexpression of a constitutively active mutant of Rap1 (Rap1-CA) and knockdown of SPA-1, a Rap1 GTPase-activating protein that was negatively regulated by afadin by virtue of binding to it, in afadin-knockdown NIH3T3 cells restored the formation of leading edge structures and the reduction of the PDGF-induced activation of Rac1 and inactivation of RhoA, suggesting that the inactivation of Rap1 by SPA-1 is responsible for inhibition of the formation of leading edge structures. The effect of Rap1-CA on the restoration of the formation of leading edge structures and RhoA inactivation was diminished by additional knockdown of ARAP1, a Rap-activated Rho GAP, which localized at the leading edges of moving NIH3T3 cells. These results indicate that afadin regulates the cyclical activation and inactivation of Rap1, Rac1, and RhoA through SPA-1 and ARAP1.
Cell migration is a spatiotemporally regulated process involving the formation and disassembly of protrusions, such as filopodia and lamellipodia, ruffles, focal complexes, and focal adhesions. At the leading edges of moving cells, the continuous formation and disassembly of these protrusive structures are tightly regulated by the actions of the Rho family small G proteins, including RhoA, Rac1, and Cdc42. RhoA regulates the formation of stress fibers and focal adhesions, whereas Rac1 and Cdc42 regulate the formation of lamellipodia and filopodia, respectively (1, 2). In addition, both Rac1 and Cdc42 regulate the formation of focal complexes (3, 4). In order to have cells keep moving, each member of the Rho family small G proteins should cyclically be active and inactive as these leading edge structures are dynamically formed and disassembled. Rac1 and Cdc42 must be activated and RhoA must be inactivated at focal complexes, and vice versa at focal adhesions. Thus, the cyclical activation and inactivation of the Rho family small G proteins are critical for turnover of the transformation of focal complexes into focal adhesions during cell movement. The activities of these small G proteins are tightly regulated by guanine nucleotide exchange factors and GTPase-activating proteins (GAPs).2 It is likely that signals from receptors and integrins cooperatively regulate the dynamics of this spatial and temporal activation and inactivation of the Rho family small G proteins. However, the molecular mechanisms of their cyclical activation and inactivation through the regulation of guanine nucleotide exchange factors and GAPs at the leading edges remain largely unknown.
We recently showed that platelet-derived growth factor (PDGF) receptor (PDGFR), integrin αvβ3, and Necl-5 associate with each other and form a complex and that this complex is clustered at the leading edges of directionally moving NIH3T3 cells in response to PDGF (5, 6). We also demonstrated that PDGF induces the activation of Rap1, which then induces the activation of Rac1 (7). Overexpression of Rap1GAP to inactivate Rap1 inhibits the PDGF-induced formation of leading edge structures, cell movement, and activation of Rac1, suggesting that, in addition to the activation of Rap1, the subsequent activation of Rac1 and presumably the inactivation of RhoA may be critical for the PDGF-induced migration of NIH3T3 cells.
Afadin is a nectin- and F-actin-binding protein that is involved in the formation of adherens junctions in cooperation with nectin and cadherin (8). Afadin has multiple domains: two Ras association (RA) domains, a forkhead-associated domain, a dilute domain, a PSD-95-Dlg-1-ZO-1 domain, three proline-rich domains, and an F-actin-binding domain at the C terminus and localizes to adherens junctions in epithelial cells (9). Afadin-knock-out mice showed impaired formation of the cell-cell junction during embryogenesis (10, 11). Although Ras small G protein was initially identified as an interacting molecule with the RA domain of afadin (12), other studies demonstrate that afadin binds GTP-bound Rap1 with a higher affinity than GTP-bound Ras or GTP-bound Rap2 (13, 14). In addition to the functional role of afadin in the organization of cell-cell adhesion, we recently found that, in NIH3T3 cells that do not form cell-cell junctions, afadin did not associate with nectin, localized at the leading edges during cell movement, and was involved in their directional, but not random, movement. The interaction of afadin with Rap1 at the leading edge was necessary for the PDGF-induced directional movement of NIH3T3 cells. Thus, in addition to that in the formation of adherens junctions, afadin plays another role in directional cell movement in NIH3T3 cells.
In a series of studies using afadin-knockdown NIH3T3 cells, we found that neither lamellipodia, ruffles, nor focal complexes are formed, suggesting that Rap1 may be inactivated and, conversely, RhoA may be activated in the reduced state of afadin. Here we first examined this possibility and found that Rap1 is indeed inactivated, whereas RhoA is activated in afadin-knockdown NIH3T3 cells. To understand the mechanisms of how the activities of Rap1 and RhoA are regulated in afadin-knockdown NIH3T3 cells, we searched for afadin-interacting proteins that could potentially regulate Rap1 activity and sought Rap1 targets that might regulate RhoA activity. We focused on SPA-1 and ARAP1 and found that these proteins coordinately regulate the activities of these small G proteins. SPA-1 is a GAP for Rap1 that interacts with afadin (15), whereas ARAP1 is a Rho GAP that binds Rap1 and could be activated by virtue of this binding (16). We describe here how afadin regulates the cyclical activation and inactivation of Rap1, Rac1, and RhoA through SPA-1 and ARAP1 at the leading edges of moving NIH3T3 cells. We conclude that afadin is critical for the coordinated regulation of the activation of Rap1 and Rac1 and subsequent inactivation of RhoA necessary for cell movement.
EXPERIMENTAL PROCEDURES
Plasmid Construction
Expression vectors for GFP-tagged Rap1 (pEGFP-Rap1), FLAG-tagged Rap1-CA (pIRM21-FLAG-Rap1-CA), and GFP-tagged Rap1-CA (pEGFP-Rap1-CA) were prepared as described (7, 17, 18). Expression vectors for Myc-tagged dominant negative Rac1 and RhoA (pEF-BOS-Myc Rac1-DN and pEF-BOS-Myc RhoA-DN, respectively) and FLAG-tagged constitutively active Rac1 (pEF-BOS-FLAG Rac1-CA) were prepared as described previously (19, 20). A plasmid expressing EGFP-tagged afadin (pEGFP-C1M-afadin) was prepared as described elsewhere (21). Plasmids expressing FLAG-tagged SPA-1 and FLAG-tagged SPA-1 lacking the GAP-related domain (SPA-1-ΔGAP) were kindly provided by Prof. N. Minato (Kyoto University Graduate School of Medicine, Kyoto, Japan). A plasmid expressing FLAG-tagged ARAP1 was a kind gift from Dr. K. Miura (Osaka Bioscience Institute, Suita, Japan). Expression vectors for HA-tagged ARAP1 and mutants of ARAP1, in which the RA domain and the Rho GAP domain were deleted (ARAP1-ΔRBD and ARAP1-ΔRGD, respectively), were constructed by standard molecular biology methods. The constructs of SPA-1 contained the following amino acids (aa): pSRα-FLAG-SPA-1, aa 1–1038; pSRα-FLAG-SPA-1-ΔGAP, aa 212–532 deletion. The constructs of ARAP1 contained the following aa: pCMV-HA-ARAP1, aa 75–1210; pCMV-HA-ARAP1-ΔRBD, aa 932–1021 deletion; pCMV-HA-ARAP1-ΔRGD, aa 714–899 deletion. Plasmids expressing siRNA-resistant mutants were generated using the QuikChange site-directed mutagenesis kit (Stratagene).
Cell Culture, Transfection, and siRNA Experiment
NIH3T3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) calf serum. Afadin-knockdown stable NIH3T3 cell lines (afadin-knockdown NIH3T3 cells) were established as described (21) and cultured in DMEM supplemented with 10% calf serum and G418 (Nacalai Tesque). For transient expression experiments, cells were transfected with various expression vectors and siRNAs by the use of Lipofectamine 2000 reagent or Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Stable NIH3T3 and afadin-knockdown stable NIH3T3 cell lines in which GFP or GFP-Rap1-CA were stably expressed (GFP-NIH3T3, GFP-Rap1-CA-NIH3T3, afadin-KD-GFP-NIH3T3, and afadin-KD-GFP-Rap1-CA-NIH3T3 cells) were obtained by infection of pMSCVpuro-EGFP-C1M and pMSCVpuro-EGFP-C1M-bV12Rap1B recombinant retroviruses, respectively, and selection with puromycin.
Antibodies and Reagents
Rat anti-Necl-5 monoclonal antibody (mAb) was prepared as described (22). Hamster anti-integrin αV and anti-integrin β3 mAbs (BD Biosciences), rabbit anti-PDGFRβ mAb (Abcam), goat anti-ARAP1 polyclonal Ab (pAb) (Abcam), rabbit anti-Rap1 pAb (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), mouse anti-Rac1 mAb (BD Biosciences), rabbit anti-RhoA pAb (Santa Cruz Biotechnology), rabbit anti-Rap1GAP mAB (EPITOMICS), mouse anti-afadin mAb (BD Biosciences), rabbit anti-GFP pAb (Medical and Biological Laboratories), mouse anti-FLAG mAb (Sigma), rabbit anti-HA pAb (Abcam), rabbit anti-Myc pAb (Cell Signaling Technology), mouse anti-HA mAb (Covance), and rhodamine-phalloidin (Molecular Probes) were purchased from commercial sources. Horseradish peroxidase-conjugated secondary Abs were purchased from GE Healthcare. Fluorophore (fluorescein isothiocyanate, Cy3, and Cy5)-conjugated secondary Abs were purchased from Jackson ImmunoResearch. Rabbit anti-SPA-1 and anti-ARAP1 pAbs were kindly provided by Prof. N. Minato and Dr. K. Miura, respectively. Human recombinant PDGF-BB was purchased from PEPROTECH. Vitronectin was purified from human plasma (Kohjinbio) as described (23). Stealth RNAis for SPA-1 and ARAP1 were purchased from Invitrogen.
Directional Stimulation with PDGF
To generate a concentration gradient of PDGF, a μ-Slide VI flow (uncoated; Ibidi) was used (5). In brief, the μ-Slide VI flow has six parallel channels, which were coated with 5 μg/ml vitronectin, according to the manufacturer's protocol. Cells were plated at a density of 5 × 103 cells/cm2, cultured for 16 h, and starved of serum with DMEM containing 0.5% bovine serum albumin (BSA) for 1 h. The concentration gradient of PDGF was applied using DMEM containing 0.5% BSA and 30 ng/ml PDGF according to the manufacturer's protocol. After 30 min, cells were fixed with acetone/methanol (1:1) or, alternatively, 4% paraformaldehyde/phosphate-buffered saline (PBS) and then incubated with 100% methanol at −20 °C. Cells were incubated with 1% BSA in PBS and then incubated with 20% BlockAce in PBS, followed by immunofluorescence microscopy (24). To analyze the leading edge formation quantitatively, the signal for F-actin, a major component of peripheral ruffles, was observed. Two investigators blindly counted at least 100 cells of each type, and the results were averaged. The cell morphology was classified into four categories: “lamellipodia with ruffles,” “lamellipodia without ruffles,” “thin and angular shapes,” and “round shapes.”
Pull-down Assays for Small G Proteins
Pull-down assays were performed as described (25). In brief, cells were plated on vitronectin-coated dishes and cultured overnight. After 1 h of serum starvation, cells were treated with PDGF-BB and then washed twice with 5 ml of ice-cold PBS, lysed in Buffer A (50 mm Tris/HCl, pH 7.4, 150 mm NaCl, 5 mm MgCl2, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm Na3VO4, 10 μg/ml leupeptin, 2 μg/ml aprotinin, and 10 μm 4-amidinophenylmethanesulfonyl fluoride) containing 30 μg of GST fusion proteins, GST-RalGDS-RBD for Rap1, GST-PAK-CRIB for Rac1, or GST-Rhotekin for RhoA, and incubated at 2 °C for 30 min. The cell extract was obtained by centrifugation at 20,000 × g at 0 °C for 10 min and incubated with 50 μl of glutathione-Sepharose beads (GE Healthcare) at 2 °C for 1 h. After the beads were washed with Buffer A, proteins bound to the beads were eluted with SDS sample buffer and subjected to SDS-PAGE followed by Western blotting.
Reverse Transcription (RT)-PCR
Total mRNA was isolated from NIH3T3 cells and J774.1 cells by the use of TRIzol (Invitrogen). Equal amounts of mRNAs were subjected to RT-PCR using Transcriptor First Strand cDNA Synthesis kit (Roche Applied Science) with specific primers for ARAP1 (5′-ttgacgactctgactatgatgatgt-3′ and 5′-ggcgtccttgttactgtcaaagtat-3′), ARAP2 (5′-tttggagcagtacctcttacatttc-3′ and 5′-cagtaatggaatcggaacactctat-3′), ARAP3 (5′-atgatgcctaatgccatctactttg-3′ and 5′-gtcactgccaaagtacattagactc-3′), or glyceraldehyde-3-phosphate dehydrogenase (5′-acggatttggtcgtattgggc-3′ and 5′-ttgacggtgccatggaatttg-3′).
Western Blotting
Cells were untransfected or transfected with the indicated siRNA, cultured for 48 h, washed with ice-cold PBS, and lysed with lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm Na2EDTA, 1 mm EGTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm Na3VO4, 1 μg/ml leupeptin, and 1 mm phenylmethylsulfonyl fluoride). The lysates were subjected to centrifugation at 12,000 × g for 10 min. The supernatant was mixed with 5× Laemmli buffer and boiled. The samples were subjected to SDS-PAGE and transferred to polyvinylidene difluoride membranes. Membranes were blocked in 5% nonfat dry milk and then incubated with primary Abs, followed by incubation with horseradish peroxidase-conjugated secondary Abs. After incubation with SuperSignal West Pico chemiluminescent substrate or SuperSignal West Femto maximum sensitivity substrate (Pierce), signals were detected using the LAS-3000mini imaging system (Fujifilm).
Boyden Chamber Assays
Cell migration was investigated using Boyden chamber assays as described (26). In brief, cell culture inserts with PET membranes (8.0-μm pores; BD Biosciences) were coated with 5 μg/ml vitronectin for 1 h and then blocked with 1% BSA at 37 °C for 30 min. NIH3T3 cells, which had been serum-starved with DMEM supplemented with 0.5% BSA for 1 h, were detached with 0.05% trypsin and 0.53 mm EDTA and then treated with a trypsin inhibitor (Sigma). The cells were then resuspended in DMEM supplemented with 0.5% BSA and plated at a density of 5 × 104 cells/insert. The cells were incubated at 37 °C for 4 h in the presence or absence of 30 ng/ml PDGF-BB. PDGF-BB was added only to the bottom well to generate a concentration gradient. After incubation, the inserts were washed with PBS, and the cells were fixed with 3.7% formaldehyde and stained with crystal violet. The cells that had not migrated were removed by wiping the top of the membrane with a cotton swab. The number of migrated cells in five randomly chosen fields per filter was counted by microscopic examination.
RESULTS
Reduced Activation of Rap1 and Rac1 and Reduced Inactivation of RhoA by Knockdown of Afadin
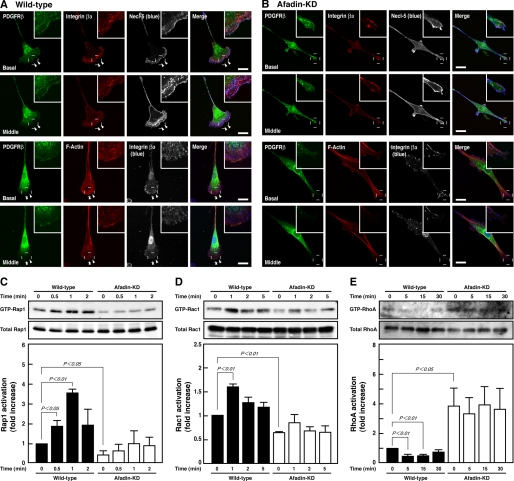
We first examined the effect of knockdown of afadin on the activities of Rap1, Rac1, and RhoA in NIH3T3 cells that were stimulated by PDGF. Wild-type and afadin-knockdown NIH3T3 cells were sparsely plated on μ-slide VI flow dishes precoated with vitronectin, an extracellular matrix protein that binds integrin αVβ3 (27), starved of serum, and directionally stimulated by PDGF. Most NIH3T3 cells became polarized and formed protrusive lamellipodia and ruffles over the lamellipodia at the leading edges in the direction of higher concentrations of PDGF, whereas afadin-knockdown NIH3T3 cells morphologically showed elongated shapes with no marked lamellipodia or ruffles and developed more prominent stress fibers than in wild-type cells (Fig. 1, A and B) (21). In wild-type cells, the immunofluorescence signals for Necl-5, PDGFR, and integrin β3 were concentrated and co-localized at the ruffles. The signal for Necl-5 was observed at focal complexes under the ruffles, whereas the signal for integrin β3 was apparent at both focal complexes and focal adhesions. These results were consistent with our earlier observations (5). Pull-down assays showed that PDGF increased the amounts of GTP-bound Rap1 and Rac1 in wild-type NIH3T3 cells, whereas in afadin-knockdown cells, the amounts of GTP-bound Rap1 and Rac1 were decreased even in the absence of PDGF as compared with those in wild-type cells, and PDGF failed to increase them (Fig. 1, C and D). PDGF decreased the amount of GTP-bound RhoA in wild-type cells, whereas it was markedly increased even in the absence of PDGF, and PDGF failed to decrease it in afadin-knockdown cells as compared with that in wild-type cells (Fig. 1E). The total amounts of these small G proteins were not different between these two types of cells. These results indicate that afadin regulates the PDGF-induced activation of Rap1 and Rac1 and inactivation of RhoA.
FIGURE 1.
Reduced activation of Rap1 and Rac1 and reduced inactivation of RhoA by knockdown of afadin. A and B, immunofluorescence images of wild-type (A) and afadin-knockdown (afadin-KD) NIH3T3 cells (B) cultured on vitronectin-coated μ-slide dishes. Cells were stimulated by PDGF for 30 min and then stained with various combinations of the anti-PDGFRβ mAb, the anti-integrin β3 mAb, the anti-Necl-5 mAb, and phalloidin. Arrowheads, leading edges; insets, higher magnification images of the leading edges; scale bars, 20 μm. The results shown are representative of three independent experiments. C–E, inhibition of the PDGF-induced activation of Rap1 and Rac1 and inactivation of RhoA in afadin-knockdown NIH3T3 cells. Wild-type and afadin-knockdown NIH3T3 cells were plated, starved of serum, and cultured in the presence of 15 ng/ml PDGF for the indicated periods of time. The cell lysates were subjected to pull-down assays followed by Western blotting using the anti-Rap1 pAb (C), the anti-Rac1 mAb (D), and the anti-RhoA pAb (E). Bars in the graphs represent the relative intensity of GTP-Rap1, GTP-Rac1, and GTP-RhoA normalized for the total amount of Rap1 (n = 3), Rac1 (n = 10), and RhoA (n = 5), respectively, as compared with a value in the absence of PDGF in wild-type NIH3T3 cells, which is expressed as 1.
Restoration of the Reduced Activation of Rac1 and Inactivation of RhoA and the Impaired Formation of Leading Edge Structures by Expression of a Constitutively Active Mutant of Rap1 in Afadin-knockdown NIH3T3 Cells
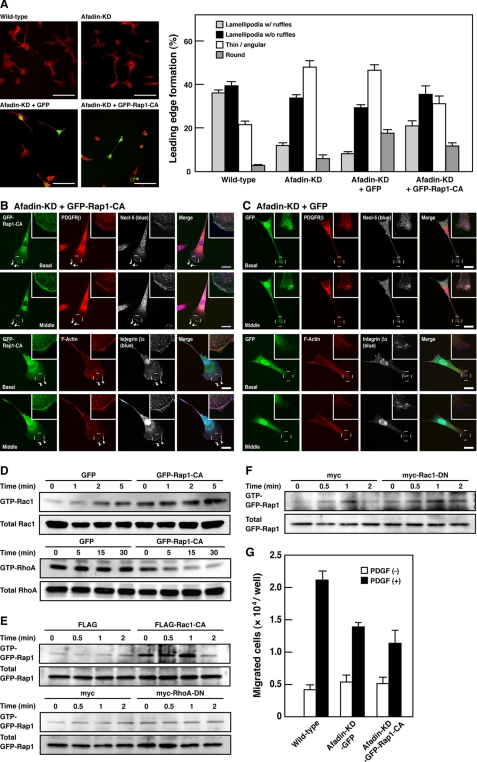
To examine whether a decrease in Rap1 activity is responsible for the impaired formation of leading edge structures in afadin-knockdown NIH3T3 cells, we tested a possibility that Rap1-CA could restore the impaired formation of leading edge structures in afadin-knockdown NIH3T3 cells. Analysis of cell morphology revealed that most wild-type NIH3T3 cells formed the polarized structures with lamellipodia (Fig. 2A). On the other hand, the number of cells forming the polarized structures with lamellipodia was decreased, whereas the number of cells with thin and angular shapes was increased in afadin-knockdown NIH3T3 cells. An expression of GFP-Rap1-CA, but not GFP, in afadin-knockdown NIH3T3 cells increased the percentage of the cells to form the polarized structures with lamellipodia and decreased the percentage of the cells with thin and angular shapes. An expression of GFP-Rap1-CA reduced stress fibers and reformed lamellipodia, ruffles, and focal complexes at the leading edges, similar to those in wild-type NIH3T3 cells as the immunofluorescence signals for Necl-5, PDGFR, and integrin β3 were concentrated and co-localized at the ruffles (Fig. 2B). The signal for Necl-5 was observed at focal complexes under the ruffles, whereas the signal for integrin β3 was apparent at both focal complexes and focal adhesions. On the other hand, GFP-transfected cells showed elongated shapes with no marked lamellipodia or ruffles and displayed prominent stress fibers, as did afadin-knockdown cells (Fig. 2C). These results indicate that the reduced activation of Rap1 is responsible for the impaired formation of leading edge structures in afadin-knockdown NIH3T3 cells.
FIGURE 2.
Restoration of the reduced activation of Rac1 and inactivation of RhoA and the impaired formation of leading edge structures by expression of a constitutively active mutant of Rap1 in afadin-knockdown NIH3T3 cells. A, morphology of wild-type and afadin-knockdown NIH3T3 cells transiently expressing GFP or GFP-Rap1-CA. Top, representative images. Scale bars, 100 μm. Bottom, quantitative analysis of the formation of leading edge structures. The results shown are the means ± S.E. of three independent experiments. B and C, immunofluorescence images of afadin-knockdown NIH3T3 cells transiently expressing GFP-Rap1-CA (B) or GFP (C) cultured on vitronectin-coated μ-slide dishes. Cells were stained with various combinations of the anti-integrin β3 mAb, the anti-Necl-5 mAb, the anti-PDGFR pAb, and phalloidin. Arrowheads, leading edges; insets, higher magnification images of the leading edges; scale bars, 20 μm. The results shown are representative of three independent experiments. D, restoration of the reduced activation of Rac1 and inactivation of RhoA by Rap1-CA. Rac1 and RhoA activities were analyzed in afadin-knockdown NIH3T3 cells stably expressing GFP-tagged Rap1-CA (afadin-KD-GFP-Rap1-CA-NIH3T3 cells) or GFP (afadin-KD-GFP-NIH3T3 cells). Cells were starved of serum and cultured in the presence of 15 ng/ml PDGF for the indicated periods of time. The cell lysates were subjected to the pull-down assays followed by Western blotting using the anti-Rac1 mAb and the anti-RhoA pAb. The results shown are representative of three independent experiments. E, effects of Rac1-CA and RhoA-DN on the PDGF-induced activation of Rap1. Afadin-knockdown NIH3T3 cells, co-transfected with GFP-tagged Rap1 and either constitutively active Rac1, dominant-negative RhoA, or vector alone were plated on vitronectin-coated dishes, starved of serum, and then cultured in the presence of 15 ng/ml PDGF for the indicated periods of time. The cell lysates were subjected to the pull-down assays followed by Western blotting using the anti-GFP pAb. The results shown are representative of three independent experiments. F, no effects of Rac1-DN on the PDGF-induced activation of Rap1. Wild-type NIH3T3 cells, co-transfected with GFP-tagged Rap1 and dominant-negative mutant Rac1 or vector alone, were plated on vitronectin-coated dishes, starved of serum, and then cultured in the presence of 15 ng/ml PDGF for the indicated periods of time. The cell lysates were subjected to the pull-down assays followed by Western blotting using the anti-GFP pAb. The results shown are representative of three independent experiments. G, inhibition of directional cell movement by Rap1-CA. Afadin-KD-GFP-Rap1-CA-NIH3T3 and afadin-KD-GFP-NIH3T3 cells were starved of serum and incubated on the cell culture inserts coated with vitronectin in the presence or absence of 30 ng/ml PDGF in the bottom wells for 4 h. *, p < 0.05. The results shown are the means ± S.E. of the three independent experiments.
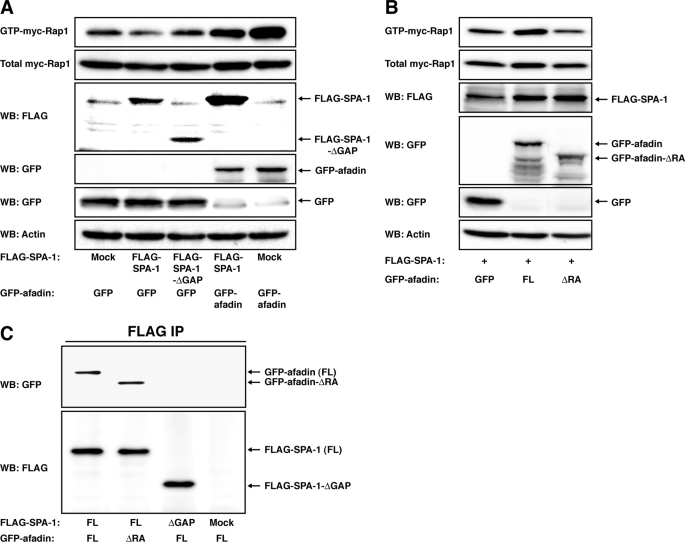
To examine whether suppression of Rap1 activity is responsible for the reduced activation of Rac1 and the reduced inactivation of RhoA in afadin-knockdown NIH3T3 cells, we tested the possibility that Rap1-CA could restore the activation levels of these small G proteins in afadin-knockdown NIH3T3 cells. For this purpose, we established and used afadin-knockdown NIH3T3 cells stably expressing GFP (afadin-KD-GFP-NIH3T3 cells) or GFP-tagged Rap1-CA (afadin-KD-GFP-Rap1-CA-NIH3T3 cells). Treatment of afadin-KD-GFP-NIH3T3 cells with PDGF induced the activation of Rac1 in a time-dependent manner, but the PDGF-induced activation of Rac1 was augmented in afadin-KD-GFP-Rap1-CA-NIH3T3 cells as compared with that in afadin-KD-GFP-NIH3T3 cells (Fig. 2D). In addition, the level of the amount of GTP-bound RhoA was significantly higher in afadin-KD-GFP-NIH3T3 cells than that in afadin-KD-GFP-Rap1-CA-NIH3T3 cells (Fig. 2D). Notably, the PDGF-induced inactivation of RhoA was observed in afadin-KD-GFP-Rap1-CA-NIH3T3 cells (Fig. 2D). These results indicate that Rap1-CA can restore the reduced activation of Rac1 and inactivation of RhoA in afadin-knockdown cells.
We further examined the possibility that activation of Rac1 or inactivation of RhoA could restore Rap1 activation levels. To detect clearly the effects of Rap1-CA and RhoA-DN, GFP-tagged Rap1 was co-expressed in afadin-knockdown NIH3T3 cells, and GFP-Rap1 from transfected cells was then selectively detected by subsequent anti-GFP blotting of precipitated GTP-bound Rap1. Introducing Rac1-CA, but not RhoA-DN, restored the amount of GTP-bound Rap1 (Fig. 2E), indicating that restoring Rac1 activity, but not RhoA activity, reversed Rap1 activity in afadin-knockdown NIH3T3 cells. Conversely, introducing Rac1-DN in wild-type NIH3T3 cells did not affect the PDGF-induced Rap1 activation (Fig. 2F). Thus, once Rap1 is activated in response to PDGF, activated Rap1 stimulates activation of Rac1, and activated Rac1 then potentiates the PDGF-induced activation of Rap1 in a positive feedback manner.
Boyden chamber assays revealed that knockdown of afadin reduced a motile activity in NIH3T3 cells and that overexpression of Rap1-CA did not restore the reduced cell motile activity despite the restoration of morphology and the activation levels of Rac1 and RhoA (Fig. 2G). These results indicate that cyclical, but not persistent, activation and inactivation of Rap1, Rac1, and RhoA are needed for efficient cell movement.
Restoration of the Reduced Activation of Rap1 and the Impaired Formation of Leading Edge Structures by Knockdown of SPA-1 in Afadin-knockdown NIH3T3 Cells
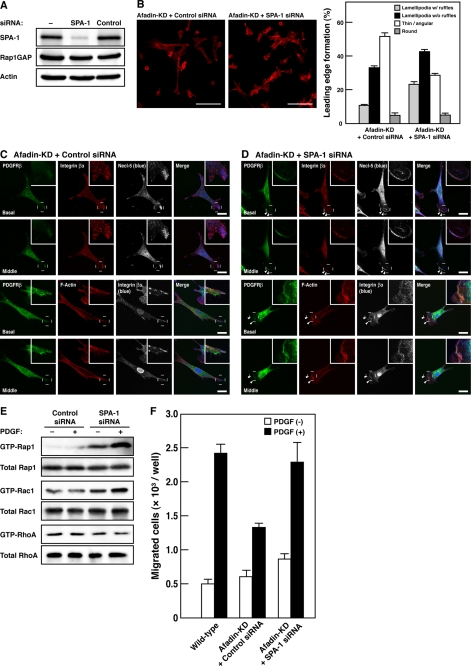
To understand the mechanism by which afadin regulates the Rap1 activity, we examined the possibility that SPA-1, a Rap1 GAP known to bind afadin (15), is involved in this regulation. When SPA-1 was additionally silenced by means of SPA-1 siRNA in afadin-knockdown NIH3T3 cells, the expression of SPA-1 was indeed reduced, whereas the expression of other proteins, such as Rap1GAP and actin, remained unchanged (Fig. 3A). Analysis of cell morphology revealed that afadin- and SPA-1-double knockdown NIH3T3 cells showed morphological phenotypes similar to those of wild-type NIH3T3 cells (compare Fig. 3, B and D, with Figs. 2A and 1A, respectively). Knockdown of SPA-1 restored the number of cells to form the polarized structures with lamellipodia in afadin-knockdown NIH3T3 cells (Fig. 3B). Afadin- and SPA-1-double knockdown NIH3T3 cells formed lamellipodia, ruffles, and focal complexes at the leading edges in response to PDGF (Fig. 3, C and D). Knockdown of SPA-1 restored the amounts of GTP-bound Rap1, Rac1, and RhoA and the cell motile activity in afadin-knockdown NIH3T3 cells, as estimated by the pull-down assays and Boyden chamber assays, respectively (Fig. 3, E and F). These results indicate that SPA-1 is critical for the reduced activation of Rap1, which is responsible for the impaired formation of leading edge structures, the reduced activation of Rac1 and inactivation of RhoA, and the impaired cell movement.
FIGURE 3.
Restoration of the reduced activation of Rap1 and Rac1, the reduced inactivation of RhoA, and the impaired formation of leading edge structures by knockdown of SPA-1 in afadin-knockdown NIH3T3 cells. A, knockdown of SPA-1 in afadin-knockdown NIH3T3 cells. The cell lysates of NIH3T3 cells untransfected or transfected with SPA-1 siRNA or non-silencing control siRNA were subjected to Western blotting using the anti-SPA-1 pAb, the anti-Rap1GAP mAb, and the anti-actin mAb. B, effect of knockdown of SPA-1 on morphology of afadin-knockdown NIH3T3 cells. Top, representative images. Scale bars, 100 μm. Bottom, quantitative analysis of the formation of leading edge structures. The results shown are the means ± S.E. of three independent experiments. C and D, immunofluorescence images of afadin-knockdown NIH3T3 cells transiently transfected with non-silencing control siRNA (C) or SPA-1 siRNA (D) cultured on vitronectin-coated μ-slide dishes. Cells were stained with various combinations of the anti-PDGFRβ mAb, the anti-integrin β3 mAb, the anti-Necl-5 mAb, and phalloidin. Arrowheads, leading edges; insets, higher magnification images of the leading edges; scale bars, 20 μm. E, reactivation of Rap1 and Rac1 and reinactivation of RhoA by knockdown of SPA-1. The activities of Rap1, Rac1, and RhoA were measured by the pull-down assays. Afadin-knockdown NIH3T3 cells transiently transfected with SPA-1 siRNA or non-silencing control siRNA were plated on vitronectin-coated μ-slide dishes, starved of serum, and cultured in the presence of 15 ng/ml PDGF for 1 min (Rap1), 2 min (Rac1), and 15 min (RhoA). The cell lysates were subjected to the pull-down assays followed by Western blotting using the anti-Rap1 pAb, the anti-Rac1 mAb, and the anti-RhoA pAb. F, the effect of knockdown of SPA-1 on directional cell movement. Wild-type and afadin-knockdown NIH3T3 cells transiently transfected with SPA-1 siRNA or non-silencing control siRNA were starved of serum and incubated on the cell culture inserts coated with vitronectin in the presence of 30 ng/ml PDGF in the bottom wells for 4 h. The number of migrated cells was counted. The results shown are the means ± S.E. of the three independent experiments.
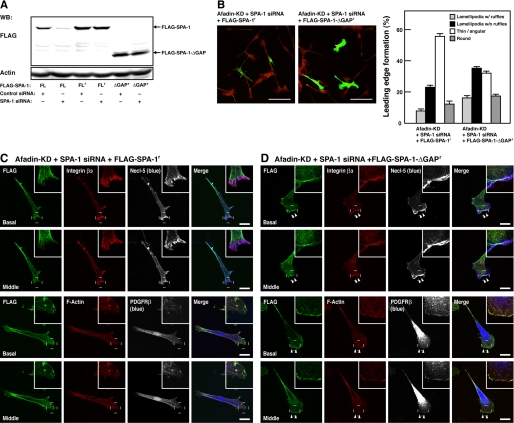
To assess whether the effect of SPA-1 siRNA indeed resulted from knockdown of SPA-1, we developed a mutant of FLAG-tagged SPA-1 that was not silenced by SPA-1 siRNA (FLAG-SPA-1r) (Fig. 4A). Re-expression of SPA-1 insensitive to siRNA in afadin- and SPA-1-double knockdown NIH3T3 cells showed phenotypes similar to those of afadin-knockdown NIH3T3 cells (compare Fig. 4, B and C, with Figs. 2A and 1B, respectively). However, an expression of an siRNA-insensitive deletion mutant of SPA-1 that lacks the Rap GAP domain (SPA-1-ΔGAPr) failed to restore the phenotypes (Fig. 4, B and D). These results indicate that SPA-1 inactivates Rap1 in the absence of afadin.
FIGURE 4.
Restoration of the effect of knockdown of SPA-1 by re-expression of SPA-1. A, construction of siRNA-resistant mutants of SPA-1. HEK293 cells were transiently co-transfected with various mutants of SPA-1 and either non-silencing control siRNA or SPA-1 siRNA. The cell lysates were subjected to the Western blotting using the anti-FLAG mAb and the anti-actin mAb. FL, full-length SPA-1; FLr, full-length SPA-1 resistant to SPA-1 siRNA; ΔGAPr, a GAP domain deletion mutant resistant to SPA-1 siRNA. B, effects of SPA-1 mutants on morphology of SPA-1- and afadin-double knockdown NIH3T3 cells. Afadin-knockdown NIH3T3 cells co-transfected with SPA-1 siRNA and either the SPA-1 siRNA-resistant SPA-1 (FLAG-SPA-1r) or SPA-1-ΔGAP (FLAG-SPA-1-ΔGAPr) mutants were stained with the anti-FLAG mAb (green) and rhodamine-phalloidin (red). Top, representative images. Scale bars, 100 μm. Bottom, quantitative analysis of the formation of leading edge structures. The results shown are the means ± S.E. of three independent experiments. C and D, immunofluorescence images of afadin-knockdown NIH3T3 cells transiently co-transfected with SPA-1 siRNA and either FLAG-SPA-1r (C) or FLAG-SPA-1-ΔGAPr (D). Cells were stained with various combinations of the anti-FLAG mAb, the anti-integrin β3 mAb, the anti-Necl-5 mAb, the anti-PDGFRβ mAb, and phalloidin. Arrowheads, leading edges; insets, higher magnification images of the leading edges; scale bars, 20 μm. The results shown are representative of three independent experiments.
Mechanisms of the Regulation of Rap1 Activation by Afadin
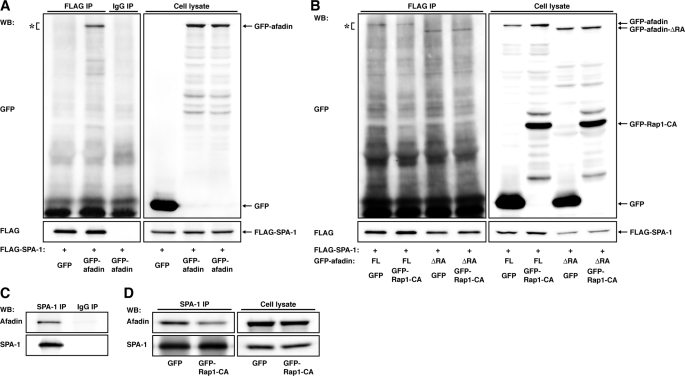
Our results suggest that afadin prevents SPA-1 from inactivating Rap1 by virtue of its binding to SPA-1 or Rap1. We examined whether Rap1 regulates the interaction between afadin and SPA-1 and tested the possibility that afadin binds SPA-1 and thereby inhibits its Rap GAP activity. Co-imunoprecipitation assays revealed that GFP-afadin bound FLAG-SPA-1 but that this binding was inhibited by GFP-Rap1-CA (Fig. 5, A and B). The effect of GFP-Rap1-CA was absent when afadin-ΔRA (GFP-afadin-ΔRA), a deletion mutant of afadin lacking the RA domain, was transfected instead of full-length afadin. Consistently, endogenous afadin was co-immunoprecipitated with SPA-1, and the amount of co-immunoprecipitated afadin was decreased by an expression of GFP-Rap1-CA in NIH3T3 cells (Fig. 5, C and D). These results indicate that Rap1-CA reduces the association between afadin and SPA-1.
FIGURE 5.
Regulation of the interaction between afadin and SPA-1 by Rap1. A, co-immunoprecipitation of afadin with SPA-1. HEK293 cells were transfected with FLAG-SPA-1 and GFP-afadin. FLAG-SPA-1 was immunoprecipitated (IP) using anti-FLAG mAb, and samples were assessed by Western blotting (WB) using the anti-FLAG mAb and the anti-GFP pAb. B, inhibition of binding of SPA-1 to afadin by Rap1-CA. HEK293 cells were transfected with various combinations of FLAG-SPA-1, GFP-afadin, GFP-afadin-ΔRA, GFP-Rap1-CA, and GFP, as indicated. FLAG-SPA-1 was immunoprecipitated using the anti-FLAG mAb, and samples were assessed by Western blotting using the anti-FLAG mAb and the anti-GFP pAb. C, co-immunoprecipitation of afadin with SPA-1 in NIH3T3 cells. SPA-1 was immunoprecipitated using the anti-SPA-1 pAb, and samples were assessed by Western blotting using the anti-afadin mAb and the anti-SPA-1 pAb. D, inhibition of binding of SPA-1 to afadin by Rap1-CA. SPA-1 was immunoprecipitated using the anti-SPA-1 pAb in GFP-NIH3T3 and GFP-Rap1-CA-NIH3T3 cells, and samples were assessed by Western blotting using the anti-afadin mAb and the anti-SPA-1 pAb.
We then examined the effect of afadin on the Rap GAP activity of SPA-1. An expression of FLAG-SPA-1 decreased the amount of GTP-bound Myc-Rap1, and this decrease was reversed by an expression of GFP-afadin (Fig. 6A). These results suggest the possibility that the interaction of afadin with SPA-1, which is negatively regulated by Rap1, inhibits the Rap GAP activity of SPA-1. However, GFP-afadin increased GTP-bound Myc-Rap1 in the absence of SPA-1, indicating that afadin positively regulates the Rap1 activity irrespective of SPA-1 (Fig. 6A). Notably, GFP-afadin-ΔRA, which was able to bind FLAG-SPA-1 (Fig. 6C), failed to increase the amount of GTP-bound Myc-Rap1 (Fig. 6B). Taken together, these results indicate that the interaction of afadin with Rap1 rather than with SPA-1 is probably responsible for the prevention of SPA-1 from inactivating Rap1.
FIGURE 6.
Inhibition of the Rap GAP activity of SPA-1 by afadin. A and B, HEK293 cells were transfected with various combinations of FLAG (Mock), FLAG-SPA-1, FLAG-SPA-1-ΔGAP, GFP, GFP-afadin, GFP-afadin-ΔRA, and Myc-Rap1, as indicated. Rap1 activity was assessed by a pull-down assay using the anti-Myc pAb. The expression of FLAG-SPA-1 and GFP-tagged proteins was assessed by Western blotting (WB) using the anti-FLAG mAb and the anti-GFP pAb, respectively. C, co-immunoprecipitation of afadin with SPA-1 but not SPA-1-ΔGAP. HEK293 cells were transfected with various combinations of FLAG (Mock), FLAG-SPA-1, FLAG-SPA-1-ΔGAP, GFP-afadin, and GFP-afadin-ΔRA, as indicated. FLAG-SPA-1 or FLAG-SPA-1-ΔGAP was immunoprecipitated using the anti-FLAG mAb, and samples were assessed by Western blotting using the anti-afadin mAb and the anti-FLAG mAb. The results shown are representative of two independent experiments, and identical results were obtained.
Expression and Localization of ARAP1 at the Leading Edges in NIH3T3 Cells
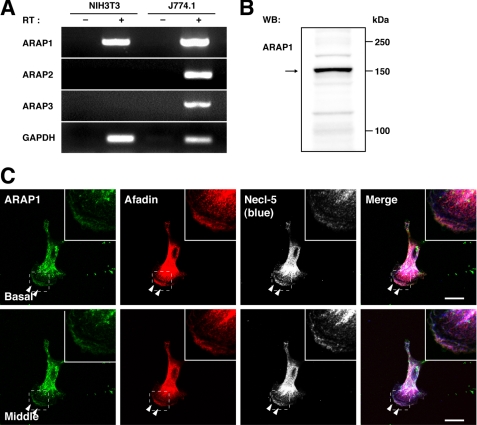
It was reported that Rap1 regulates the Rac1 activity through Vav2 and Tiam1, guanine nucleotide exchange factors for Rac1 (28). In this paper, an expression of a constitutively active mutant of Rac1 induces the inactivation of RhoA. This result provides the possibility that Rap1 may induce the inactivation of RhoA through the activation of Rac1. However, transfection of a constitutively active mutant of Rac1, V12Rac1, to afadin-knockdown NIH3T3 cells did not restore the phenotypes (data not shown), implying that another mechanism by which Rap1 regulates the activity of RhoA may be involved. Therefore, we next studied how the PDGF-mediated activation of Rap1 induces the inactivation of RhoA. ARAP is a Rho GAP that binds Rap1 and is activated by this binding (29), suggesting that this molecule may be involved in the Rap1-induced inactivation of RhoA. To test this possibility, we initially analyzed the expression of ARAP in NIH3T3 cells. ARAP constitutes a family consisting of three members, ARAP1, ARAP2, and ARAP3 (16, 30). RT-PCR analysis demonstrated that ARAP1 mRNA was abundantly expressed, whereas ARAP2 and ARAP3 mRNAs were hardly detected (Fig. 7A), indicating that ARAP1 is selectively expressed in NIH3T3 cells. Western blotting showed that ARAP1 was indeed expressed in this cell line (Fig. 7B). We then examined the intracellular localization of ARAP1 in moving NIH3T3 cells. When ARAP1 was stained in directionally moving NIH3T3 cells under the same conditions as described above, the immunofluorescence signal for ARAP1 was concentrated at the ruffles over lamellipodia and focal complexes but not at focal adhesions (Fig. 7C). These results indicate that ARAP1 is expressed and concentrated at the leading edges of directionally moving NIH3T3 cells.
FIGURE 7.
The expression and localization of ARAP1 at the leading edges in moving NIH3T3 cells. A, the expression of ARAP mRNAs. Total mRNA was extracted from NIH3T3 and J774.1 cells and subjected to the RT-PCR analyses using specific primers for ARAP1, ARAP2, and ARAP3. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. B, the expression of ARAP1. The cell lysate of NIH3T3 cells was subjected to Western blotting (WB) using the anti-ARAP1 pAb. C, immunofluorescence images of NIH3T3 cells cultured on vitronectin-coated μ-slide dishes. The cells were triple-stained with the anti-ARAP1 pAb, the anti-afadin mAb, and the anti-Necl-5 mAb. Arrowheads, leading edges; insets, higher magnification images of the leading edges; scale bars, 20 μm. The results shown are representative of three independent experiments.
Inactivation of RhoA by Rap1 through ARAP1 in Afadin-knockdown NIH3T3 Cells
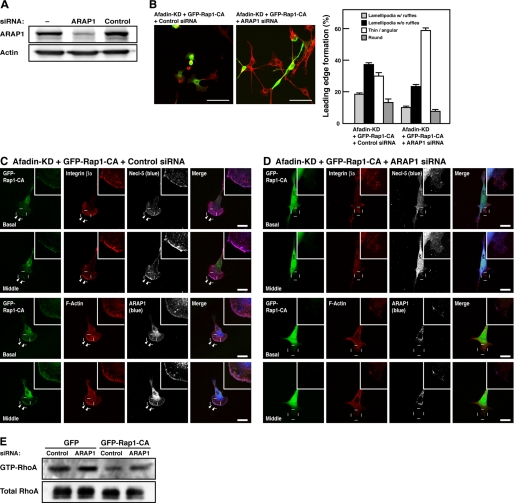
To examine whether ARAP1 mediates the Rap1-CA-induced inactivation of RhoA in afadin-knockdown NIH3T3 cells, we tested the effect of knockdown of ARAP1. Western blotting showed that ARAP1 was effectively knocked down by ARAP1 siRNA in afadin-knockdown NIH3T3 cells (Fig. 8A). Knockdown of ARAP1 cancelled the restoration of cell morphology by Rap1-CA in afadin-knockdown NIH3T3 cells (Fig. 8B). In ARAP1- and afadin-double knockdown NIH3T3 cells, an expression of Rap1-CA failed to induce the inactivation of RhoA; Rap1-CA could not diminish stress fibers or form lamellipodia, ruffles, or focal complexes (Fig. 8, C and D). Although knockdown of ARAP1 showed no significant effect on the amount of GTP-bound RhoA in afadin-KD-GFP-NIH3T3 cells in which the activity of Rap1 was suppressed, ARAP1 partially restored it in afadin-KD-GFP-Rap1-CA-NIH3T3 cells (Fig. 8E), indicating that ARAP1 acts as a Rho GAP downstream of Rap1. Thus, ARAP1 mediates the Rap1-induced inactivation of RhoA.
FIGURE 8.
Inactivation of RhoA by Rap1-CA through ARAP1 in afadin-knockdown NIH3T3 cells. A, knockdown of ARAP1 in afadin-knockdown NIH3T3 cells. The cell lysates of NIH3T3 cells untransfected or transfected with ARAP1 siRNA or non-silencing control siRNA were subjected to Western blotting using the anti-ARAP1 pAb and the anti-actin mAb. B, restoration of morphology of Rap1-CA-transfected afadin-knockdown NIH3T3 cells by knockdown of ARAP1. Afadin-knockdown NIH3T3 cells co-transfected with GFP-tagged Rap1-CA and either ARAP1 siRNA or non-silencing control siRNA were stained with rhodamine-phalloidin (red). Top, representative images. Scale bars, 100 μm. Bottom, quantitative analysis of the formation of leading edge structures. The results shown are the means ± S.E. of three independent experiments. C and D, immunofluorescence images of afadin-knockdown NIH3T3 cells co-transfected with GFP-tagged Rap1-CA and either non-silencing control siRNA (C) or ARAP1 siRNA (D) cultured on vitronectin-coated μ-slide dishes. The cells were stained with various combinations of the anti-integrin β3 mAb, the anti-Necl-5 mAb, the anti-ARAP1 pAb, and phalloidin. Arrowheads, leading edges; insets, higher magnification images of the leading edges; scale bars, 20 μm. E, restoration of the Rap1-induced inactivation of RhoA by knockdown of ARAP1. Afadin-knockdown NIH3T3 cells stably expressing GFP-tagged Rap1-CA (afadin-KD-GFP-Rap1-CA-NIH3T3 cells) or GFP (afadin-KD-GFP-NIH3T3 cells) were transfected with ARAP1 siRNA or non-silencing control siRNA. The cell lysates were subjected to the pull-down assay followed by Western blotting using the anti-RhoA pAb. The results shown are representative of three independent experiments.
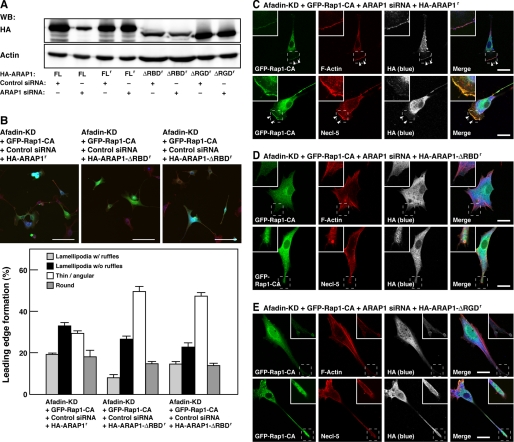
To further confirm that Rap1 activates ARAP1 and then activated ARAP1 induces the inactivation of RhoA, siRNA-insensitive full-length ARAP1 (HA-ARAP1r) or a siRNA-insensitive mutant of ARAP1, of which the Rap-binding domain or the Rho GAP domain was deleted (HA-ARAP1-ΔRBDr and HA-ARAP1-ΔRGDr, respectively) (Fig. 9A), was co-expressed with Rap1-CA in ARAP1- and afadin-double knockdown cells. Co-expression of HA-ARAP1r with Rap1-CA restored the morphological phenotypes of ARAP1- and afadin-double knockdown cells, whereas co-expression of HA-ARAP1-ΔRBDr or HA-ARAP1-ΔRGDr with Rap1-CA did not show this effect (Fig. 9, B–E). These results indicate that the effect of ARAP1 siRNA is indeed mediated by knockdown of ARAP1 and that both the Rap binding activity and the Rho GAP activity are needed for the ability of ARAP1 to inactivate RhoA. Collectively, Rap1 inactivates RhoA through ARAP1 in afadin-knockdown NIH3T3 cells.
FIGURE 9.
Requirement of the Rap-binding domain and the Rho GAP domain for inactivating RhoA by ARAP1. A, construction of siRNA-resistant mutants of ARAP1. HEK293 cells were transiently co-transfected with various HA-tagged mutants of ARAP1 and either non-silencing control siRNA or ARAP1 siRNA. The cell lysates were subjected to the Western blotting using the anti-HA pAb and the anti-actin mAb. FL, full-length ARAP1; FLr, full-length ARAP1 resistant to ARAP1 siRNA; ΔRBDr, a Rap-binding domain deletion mutant resistant to ARAP1 siRNA; ΔRGDr, a Rho GAP domain deletion mutant resistant to ARAP1 siRNA. B, the effects of ARAP1 mutants on morphology of Rap1-CA-transfected ARAP1- and afadin-double knockdown NIH3T3 cells. Afadin-knockdown NIH3T3 cells co-transfected with GFP-Rap1-CA, ARAP1 siRNA, and either ARAP1 siRNA-resistant HA-tagged full-length ARAP1 (HA-ARAP1r), ARAP1-ΔRBD (HA-ARAP1-ΔRBDr), or ARAP1-ΔRGD (HA-ARAP1-ΔRGDr) mutants were stained with anti-HA mAb (blue) and rhodamine-phalloidin (red). Top, the representative images. Scale bars, 100 μm. Bottom, quantitative analysis of the formation of leading edge structures. The results shown are the means ± S.E. of three independent experiments. C–E, immunofluorescence images of afadin-knockdown NIH3T3 cells co-transfected with GFP-Rap1-CA, ARAP1 siRNA, and either HA-ARAP1r (C), HA-ARAP1-ΔRBDr (D), or HA-ARAP1-ΔRGDr (E) cultured on vitronectin-coated μ-slide dishes. Cells were stained with phalloidin and the anti-HA mAb. Arrowheads, leading edges; scale bars, 20 μm. The results shown are representative of three independent experiments.
DISCUSSION
In the present study, we demonstrated that, in comparison with wild-type NIH3T3 cells, afadin-knockdown NIH3T3 cells show the reduced activation of Rap1 and Rac1 and the reduced inactivation of RhoA induced by PDGF. Although the difference was statistically significant, the amplitude of Rac1 activation by PDGF was less than that of Rap1 activation. This apparently small increase may be caused by the activation locally at the leading edge or the high background. We previously reported that Rap1 is critical for the formation of leading edge structures, such as lamellipodia, ruffles, and focal complexes, and cell movement in response to PDGF in NIH3T3 cells (7). Inhibition of Rap1 by overexpression of Rap1GAP resulted in an impairment of the formation of these leading edge structures in association with the reduced Rac1 activity and enhanced RhoA activity. Overall phenotypes of Rap1GAP-expressing cells were similar to those of afadin-knockdown cells. Consistently, an expression of Rap1-CA restored the reduced activation of Rap1 and Rac1 and the reduced inactivation of RhoA induced by PDGF. Therefore, the reduced activation of Rap1 is critical for the reduced activation of Rac1 and the reduced inactivation of RhoA in afadin-knockdown cells. In addition, introducing Rac1-CA, but not RhoA-DN, restored the Rap1 activation in response to PDGF in afadin-knockdown NIH3T3 cells (Fig. 2E). These results account for a cross-talk between Rap1 and Rac1. We reported that inhibiting Rap1 decreases Rac1 activation in response to PDGF in NIH3T3 cells (7). Introducing Rac1-DN did not affect the PDGF-induced Rap1 activation in wild-type NIH3T3 cells (Fig. 2F). We therefore conclude that PDGF induces Rap1 activation, which in turn leads to Rac1 activation. The PDGF-induced activation of Rac1 is necessary for clustering of the Necl-5-integrin αVβ3 complex, which then enhances the PDGF-induced activation of Rac1 (5, 6). Collectively, afadin plays an important role in the regulation of Rap1 activation, Rap1-dependent activation of Rac1, and Rac1-mediated positive feedback activation of Rap1 and Rac1.
We explored the mechanism underlying the inactivation of Rap1 by knockdown of afadin and showed here that SPA-1 is involved in this process. Because afadin binds SPA-1, afadin might recruit SPA-1 to inactivate Rap1, since Su et al. (15) previously showed that AF6 (a human homologue of afadin) binds SPA-1 and that AF6/afadin enhances the SPA-1-induced decrease in the levels of GTP-bound Rap1. On the contrary, Zhang et al. (31) reported that overexpression of AF6/afadin increases the level of GTP-bound Rap1, whereas knockdown of AF6/afadin decreases it in the Jurkat T cell line. They concluded that AF6/afadin binds Rap1 and stabilizes Rap1 in the GTP-bound state. In agreement with the result of the study by Zhang et al. (31), we showed here that knockdown of afadin inhibits the activation of Rap1, indicating that the binding of afadin to SPA-1 and/or Rap1 may prevent SPA-1 from inactivating Rap1. We showed here that GFP-afadin was able to inhibit the Rap GAP activity of SPA-1, whereas GFP-afadin-ΔRA was unable to inhibit it. These results indicate that the interaction of afadin with Rap1 rather than with SPA-1 is probably responsible for the prevention of SPA-1 from inactivating Rap1. Because the interaction between afadin and SPA-1 was decreased in the presence of Rap1-CA, it is plausible that the binding of Rap1 to afadin may induce the dissociation of SPA-1 from the afadin-SPA-1 complex. Taken together, it is likely that SPA-1 prebound to afadin is dissociated upon binding of Rap1 to afadin and thereby induces the inactivation of Rap1 that is not bound to afadin.
We demonstrated that SPA-1 plays a pivotal role in the regulation of the Rap1 activity at the leading edges of moving NIH3T3 cells. In afadin-knockdown NIH3T3 cells, the Rap1-dependent formation of leading edge structures was suppressed by the action of SPA-1. Consistent with our results, Shimonaka et al. (32) demonstrated that SPA-1 effectively suppresses the chemokine-induced Rap1-dependent cell polarization in lymphocytes. We also showed that the phenotype of afadin-knockdown cells is restored by additional knockdown of SPA-1 and that the effect of knockdown of SPA-1 is fully restored by co-transfection of an siRNA-resistant mutant of SPA-1 but not SPA-1-ΔGAP with SPA-1 siRNA. Although the regulation of the Rap1 GAP activity of SPA-1 might vary depending on cell types and/or stimuli, and other Rap GAPs might play a role, our results suggest that SPA-1 regulates the Rap1 activity by competitively interacting with afadin.
Although either an expression of Rap1-CA or knockdown of SPA-1 in afadin-knockdown NIH3T3 cells restored the formation of leading edge structures, knockdown of SPA-1 but not an expression of Rap1-CA restored cell movement. The expression of Rap1-CA was not limited to the leading edges in GFP-Rap1-CA-transfected afadin-knockdown NIH3T3 cells. These results suggest that regional, rather than whole, inactivation of Rap1 by SPA-1 is responsible for the impaired formation of leading edge structures in afadin-knockdown NIH3T3 cells.
We demonstrated here that ARAP1 is expressed in NIH3T3 cells and is critical for the inactivation of RhoA at the leading edges of moving cells. ARAP, comprising ARAP1 to -3, consists of five pleckstrin homology domains, an RA domain, an Arf GAP domain, and a Rho GAP domain (16, 30). Arf GAP activity is increased by the binding of phosphatidylinositol 3,4,5-triphosphate to the pleckstrin homology domains in ARAP1. Unlike the Arf GAP activity, the Rho GAP activity is not affected by phosphatidylinositol 3,4,5-triphosphate (16). It has been reported that the Rho GAP activity of ARAP3 depends on the binding of Rap and that ARAP3 is an effector for Rap (29). Although the mechanism underlying the regulation of the Rho GAP activity of ARAP1 remained to be determined, we showed here that the Rap1-dependent Rho GAP activity is needed for the inactivation of RhoA, because knockdown of ARAP1 suppressed the Rap1-CA-induced inactivation of RhoA in afadin-knockdown NIH3T3 cells. Moreover, re-expression of HA-ARAP1r restored the phenotype of afadin- and ARAP1-double knockdown cells, whereas HA-ARAP1-ΔRBDr or HA-ARAP1-ΔRGDr failed to restore it. Taken together, these results indicate that Rap1 induces the inactivation of RhoA though ARAP1 in NIH3T3 cells in response to PDGF.
We showed here that the localization of Necl-5, integrin β3, and PDGFR at the leading edges was changed concomitantly with the alterations in cell morphology, which were associated with the activities of small G proteins, such as Rac1 and RhoA. However, the changes in these activities by transfection of various plasmids and siRNAs did not appear to be so significant despite the great alterations in cell morphology. This is not surprising and seems to be a limitation of the method to measure the activities of small G proteins in this study. We measured the activities of small G proteins by the pull-down assay, and this strategy was to evaluate these activities within whole cells but not limited to the leading edges. If the activities of small G proteins at the leading edges could be selectively measured, they might change more obviously. The development of a novel technique to selectively quantitate the activities of small G proteins at the leading edges is required.
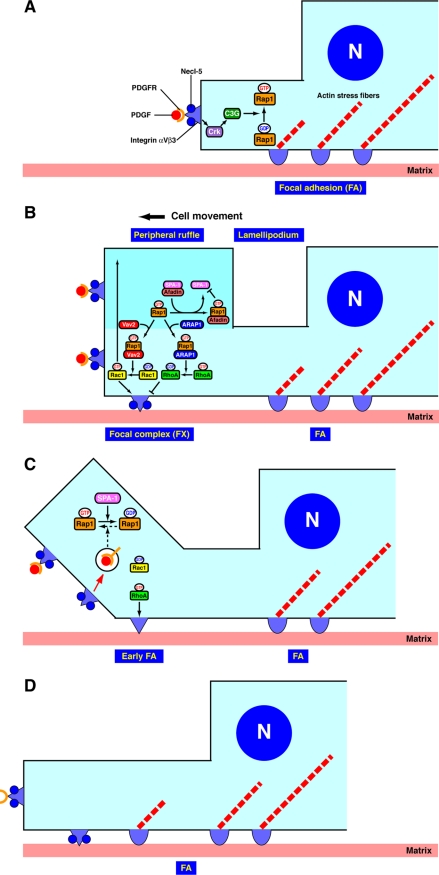
Cell motility is critically important during physiological events, such as embryogenesis and angiogenesis, as well as under pathological conditions, including metastasis of cancer cells. Our finding also provides insights into the molecular events that lead to embryonic lethality due to developmental defects in afadin-deficient mice (10). We propose a model for the regulation of the cyclical activation and inactivation of Rap1 and RhoA (Fig. 10). Rap1 is activated by virtue of binding of PDGF to its receptor (A). Activated Rap1 induces the activation of Rac1, whereas it binds ARAP1 and induces the inactivation of RhoA, resulting in the formation of lamellipodia, ruffles, and focal complexes at the leading edges (B). On the other hand, activated Rap1 binds afadin and induces the recruitment of afadin to the leading edges, which then results in the release of SPA-1 from afadin (B). SPA-1, unbound to afadin, then inactivates Rap1 at the leading edges, causing the inactivation of Rac1 and the activation of RhoA (C). The activation of RhoA then induces the activation of ROCK, thereby stimulating the transformation of focal complexes to focal adhesions (33). Thus, the cyclical activation and inactivation of these small G proteins facilitate the turnover of leading edge structures and the transformation of focal complexes to focal adhesions during cell movement (D). Afadin plays a pivotal role in this dynamic cyclical activation and inactivation of Rap1, Rac1, and RhoA by the coordinated regulation of SPA-1 and ARAP1.
FIGURE 10.
A schematic model for the regulation of the cyclical activation and inactivation of Rap1, Rac1, and RhoA. Details are described under “Discussion.”
Acknowledgments
We thank Prof. N. Minato and Dr. K. Miura for the generous gifts of plasmids and antibodies.
This work was supported by grants-in-aid for Scientific Research and for Cancer Research and, in part, by the Targeted Proteins Research Program of the Ministry of Education, Culture, Sports, Science, and Technology, Japan (2006–2008) and a research grant from the Japan Foundation for Applied Enzymology.
- GAP
- GTPase-activating protein
- PDGF
- platelet-derived growth factor
- PDGFR
- PDGF receptor
- RA
- Ras association
- CA
- constitutively active
- DMEM
- Dulbecco's modified Eagle's medium
- Ab
- antibody
- mAb
- monoclonal antibody
- pAb
- polyclonal antibody
- BSA
- bovine serum albumin
- PBS
- phosphate-buffered saline
- RT
- reverse transcription
- GFP
- green fluorescent protein
- EGFP
- enhanced GFP
- aa
- amino acid(s)
- siRNA
- small interfering RNA
- KD
- knockdown
- HA
- hemagglutinin.
REFERENCES
- 1.Ridley A. J., Hall A. (1992) Cell 70, 389–399 [DOI] [PubMed] [Google Scholar]
- 2.Nobes C. D., Hall A. (1995) Cell 81, 53–62 [DOI] [PubMed] [Google Scholar]
- 3.Ballestrem C., Hinz B., Imhof B. A., Wehrle-Haller B. (2001) J. Cell Biol. 155, 1319–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rottner K., Hall A., Small J. V. (1999) Curr. Biol. 9, 640–648 [DOI] [PubMed] [Google Scholar]
- 5.Minami Y., Ikeda W., Kajita M., Fujito T., Amano H., Tamaru Y., Kuramitsu K., Sakamoto Y., Monden M., Takai Y. (2007) J. Biol. Chem. 282, 18481–18496 [DOI] [PubMed] [Google Scholar]
- 6.Amano H., Ikeda W., Kawano S., Kajita M., Tamaru Y., Inoue N., Minami Y., Yamada A., Takai Y. (2008) Genes Cells 13, 269–284 [DOI] [PubMed] [Google Scholar]
- 7.Takahashi M., Rikitake Y., Nagamatsu Y., Hara T., Ikeda W., Hirata K., Takai Y. (2008) Genes Cells 13, 549–569 [DOI] [PubMed] [Google Scholar]
- 8.Mandai K., Nakanishi H., Satoh A., Obaishi H., Wada M., Nishioka H., Itoh M., Mizoguchi A., Aoki T., Fujimoto T., Matsuda Y., Tsukita S., Takai Y. (1997) J. Cell Biol. 139, 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takai Y., Nakanishi H. (2003) J. Cell Sci. 116, 17–27 [DOI] [PubMed] [Google Scholar]
- 10.Ikeda W., Nakanishi H., Miyoshi J., Mandai K., Ishizaki H., Tanaka M., Togawa A., Takahashi K., Nishioka H., Yoshida H., Mizoguchi A., Nishikawa S., Takai Y. (1999) J. Cell Biol. 146, 1117–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhadanov A. B., Provance D. W., Jr., Speer C. A., Coffin J. D., Goss D., Blixt J. A., Reichert C. M., Mercer J. A. (1999) Curr. Biol. 9, 880–888 [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto T., Harada N., Kano K., Taya S., Canaani E., Matsuura Y., Mizoguchi A., Ide C., Kaibuchi K. (1997) J. Cell Biol. 139, 785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linnemann T., Geyer M., Jaitner B. K., Block C., Kalbitzer H. R., Wittinghofer A., Herrmann C. (1999) J. Biol. Chem. 274, 13556–13562 [DOI] [PubMed] [Google Scholar]
- 14.Boettner B., Govek E. E., Cross J., Van Aelst L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9064–9069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su L., Hattori M., Moriyama M., Murata N., Harazaki M., Kaibuchi K., Minato N. (2003) J. Biol. Chem. 278, 15232–15238 [DOI] [PubMed] [Google Scholar]
- 16.Miura K., Jacques K. M., Stauffer S., Kubosaki A., Zhu K., Hirsch D. S., Resau J., Zheng Y., Randazzo P. A. (2002) Mol. Cell 9, 109–119 [DOI] [PubMed] [Google Scholar]
- 17.Fukuyama T., Ogita H., Kawakatsu T., Fukuhara T., Yamada T., Sato T., Shimizu K., Nakamura T., Matsuda M., Takai Y. (2005) J. Biol. Chem. 280, 815–825 [DOI] [PubMed] [Google Scholar]
- 18.Yamada T., Sakisaka T., Hisata S., Baba T., Takai Y. (2005) J. Biol. Chem. 280, 33026–33034 [DOI] [PubMed] [Google Scholar]
- 19.Ikeda W., Nakanishi H., Tanaka Y., Tachibana K., Takai Y. (2001) Oncogene 20, 3457–3463 [DOI] [PubMed] [Google Scholar]
- 20.Nakahara H., Otani T., Sasaki T., Miura Y., Takai Y., Kogo M. (2003) Genes Cells 8, 1019–1027 [DOI] [PubMed] [Google Scholar]
- 21.Nakata S., Fujita N., Kitagawa Y., Okamoto R., Ogita H., Takai Y. (2007) J. Biol. Chem. 282, 37815–37825 [DOI] [PubMed] [Google Scholar]
- 22.Ikeda W., Kakunaga S., Itoh S., Shingai T., Takekuni K., Satoh K., Inoue Y., Hamaguchi A., Morimoto K., Takeuchi M., Imai T., Takai Y. (2003) J. Biol. Chem. 278, 28167–28172 [DOI] [PubMed] [Google Scholar]
- 23.Yatohgo T., Izumi M., Kashiwagi H., Hayashi M. (1988) Cell Struct. Funct. 13, 281–292 [DOI] [PubMed] [Google Scholar]
- 24.Fujito T., Ikeda W., Kakunaga S., Minami Y., Kajita M., Sakamoto Y., Monden M., Takai Y. (2005) J. Cell Biol. 171, 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukuhara T., Shimizu K., Kawakatsu T., Fukuyama T., Minami Y., Honda T., Hoshino T., Yamada T., Ogita H., Okada M., Takai Y. (2004) J. Cell Biol. 166, 393–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikeda W., Kakunaga S., Takekuni K., Shingai T., Satoh K., Morimoto K., Takeuchi M., Imai T., Takai Y. (2004) J. Biol. Chem. 279, 18015–18025 [DOI] [PubMed] [Google Scholar]
- 27.van der Flier A., Sonnenberg A. (2001) Biochim. Biophys. Acta 1538, 99–117 [DOI] [PubMed] [Google Scholar]
- 28.Sander E. E., ten Klooster J. P., van Delft S., van der Kammen R. A., Collard J. G. (1999) J. Cell Biol. 147, 1009–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krugmann S., Williams R., Stephens L., Hawkins P. T. (2004) Curr. Biol. 14, 1380–1384 [DOI] [PubMed] [Google Scholar]
- 30.Krugmann S., Anderson K. E., Ridley S. H., Risso N., McGregor A., Coadwell J., Davidson K., Eguinoa A., Ellson C. D., Lipp P., Manifava M., Ktistakis N., Painter G., Thuring J. W., Cooper M. A., Lim Z. Y., Holmes A. B., Dove S. K., Michell R. H., Grewal A., Nazarian A., Erdjument-Bromage H., Tempst P., Stephens L. R., Hawkins P. T. (2002) Mol. Cell 9, 95–108 [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z., Rehmann H., Price L. S., Riedl J., Bos J. L. (2005) J. Biol. Chem. 280, 33200–33205 [DOI] [PubMed] [Google Scholar]
- 32.Shimonaka M., Katagiri K., Nakayama T., Fujita N., Tsuruo T., Yoshie O., Kinashi T. (2003) J. Cell Biol. 161, 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagamatsu Y., Rikitake Y., Takahashi M., Deki Y., Ikeda W., Hirata K., Takai Y. (2008) J. Biol. Chem. 283, 14532–14541 [DOI] [PubMed] [Google Scholar]