Abstract
The antigen recognition site of antibodies consists of the heavy and light chain variable domains (VL and VH domains). VL domains catalyze peptide bond hydrolysis independent of VH domains (Mei, S., Mody, B., Eklund, S. H., and Paul, S. (1991) J. Biol. Chem. 266, 15571–15574). VH domains bind antigens noncovalently independent of VL domains (Ward, E. S., Güssow, D., Griffiths, A. D., Jones, P. T., and Winter, G. (1989) Nature 341, 544–546). We describe specific hydrolysis of fusion proteins of the hepatitis C virus E2 protein with glutathione S-transferase (GST-E2) or FLAG peptide (FLAG-E2) by antibodies containing the VH domain of an anti-E2 IgG paired with promiscuously catalytic VL domains. The hybrid IgG hydrolyzed the E2 fusion proteins more rapidly than the unpaired light chain. An active site-directed inhibitor of serine proteases inhibited the proteolytic activity of the hybrid IgG, indicating a serine protease mechanism. The hybrid IgG displayed noncovalent E2 binding in enzyme-linked immunosorbent assay tests. Immunoblotting studies suggested hydrolysis of FLAG-E2 at a bond within E2 located ∼11 kDa from the N terminus. GST-E2 was hydrolyzed by the hybrid IgG at bonds in the GST tag. The differing cleavage pattern of FLAG-E2 and GST-E2 can be explained by the split-site model of catalysis, in which conformational differences in the E2 fusion protein substrates position alternate peptide bonds in register with the antibody catalytic subsite despite a common noncovalent binding mechanism. These studies provide proof-of-principle that the catalytic activity of a light chain can be rendered antigen-specific by pairing with a noncovalently binding heavy chain subunit.
Antibodies (Abs)2 are composed of light and heavy chain subunits linked by intra- and inter-chain disulfide bonds. The noncovalent antigen binding site of Abs is formed mainly by amino acids located in the complementarity determining regions of the light and heavy chain variable domains (VL and VH domains). Physiological Ab-antigen binding reactions require both Ab subunits. The individual light and heavy chains can bind antigens independent of each other, but the binding affinity of the isolated subunits is often lower than the intact Abs from which they are derived (1–4). From crystallography analyses of Ab-antigen complexes, it appears that antigen contact areas with the VH domain are somewhat greater than the VL domain (5, 6). Recombinant IgG Abs composed of the heavy chain drawn from antigen-specific IgGs paired with irrelevant light chains retain antigen binding activity, albeit at reduced levels (1, 3).
Following the initial noncovalent antigen binding step, some Abs proceed to catalyze hydrolysis of peptide bonds (7–12). The chemical catalysis step entails nucleophilic attack on the electrophilic carbonyl of peptide bonds by serine protease-like sites present in Ab V domains followed by hydrolysis of the covalent reaction intermediate if a water molecule is available (13–15). Unlike reversible binding, the catalytic function offers a means to permanently inactivate the antigen by its hydrolysis into smaller fragments. Reversibly binding Abs bind the antigen stoichiometrically (e.g. 2 antigen molecules/IgG molecule). As catalysts are reusable, a single catalytic Ab molecule can hydrolyze multiple antigen molecules. This offers the possibility of increased antigen neutralizing potency. Therefore, there is considerable interest in developing catalytic Abs directed to individual polypeptide antigens. The serine protease-like activity is a heritable trait encoded by germline Ab V genes, and Abs in the preimmune repertoire can hydrolyze peptides with diverse sequence promiscuously (13, 14, 16, 17). However, the adaptive immune system has evolved to maximize noncovalent binding affinity of Abs over the course of B cell differentiation. Physiological immune mechanisms do not favor retention and improvement of the catalytic function. B cell clonal proliferation is driven by antigen binding to B cell receptors (Abs associated with signal transducing proteins). Antigen hydrolysis by catalytic B cell receptors is followed by release of the antigen fragments, resulting in reduced B cell receptor occupancy and loss of the proliferative stimulus for the cells. Therefore, unlike the noncovalent antigen binding activity, the catalytic function is poorly selectable. Indeed, other than Abs to autoantigen and B cell superantigen substrates, there are no examples of antigen-specific catalytic Abs generated by physiological adaptive mechanisms (18).
Much effort has been devoted to developing antigen-specific catalytic Abs by immune and protein engineering strategies. Based on the premise that binding to the transition state reduces the activation energy of the catalytic reaction, immunization with transition state analogs has been applied to raise Abs that catalyze ester bonds in small haptens (19). Attempts to improve the esterase activity by random mutagenesis followed by isolation of transition state analog-binding Abs have also been described (20). Developing antigen-specific proteolytic Abs, however, has been difficult because peptide bond hydrolysis is an energetically demanding reaction. Moreover, there is no viable engineering strategy available to render catalytic Abs specific for individual polypeptide antigens. We (8, 21, 22) and others (23, 24) have identified Ab light chains that hydrolyze peptide bonds promiscuously without participation from the heavy chain subunit. Disruption of the serine protease-like catalytic triad in an Ab light chain by site-directed mutagenesis was without effect on its ability to bind the polypeptide antigen by noncovalent means (13), and a discrete peptide epitope remote from the bond hydrolyzed by a proteolytic Ab preparation has been identified (25). This lead to a split-site model of proteolysis, in which distinct subsites present within the Ab combining site are responsible for initial noncovalent antigen binding and the ensuing peptide bond hydrolysis reaction (26). If this model is correct, it should be possible to develop hybrid proteolytic Abs specific for individual antigens by pairing light chains containing a promiscuous catalytic subsite with heavy chains that contribute the noncovalent subsite responsible for specific antigen binding. We describe proof-of-principle for this engineering approach using previously described catalytic light chains paired with the heavy chain of a monoclonal IgG that binds the hepatitis C virus (HCV) E2 coat protein. This protein is thought to be important in viral entry into hepatocytes and B cells by virtue of its ability to bind receptors expressed on the host cells (27, 28).
EXPERIMENTAL PROCEDURES
Recombinant Abs
VH and VL cDNA of the anti-E2-IgG1 CBH-7 were prepared by reverse transcriptase-PCR using as template total RNA from hybridoma cells (29). VL cDNAs of light chains HK13, HK14, and GG63 cloned in Escherichia coli were obtained similarly (respectively, GenBankTM accession numbers L43498, L43499, and AF352557) (21, 22). Anti-gp120 VL and VH domain cDNA was prepared from a single chain Fv clone GL2 isolated from a phage library (30). VL domains were cloned into the light chain expression vector on the 5′ side of the κ constant domain vector (pLC-huCκ vector; Ref. 31 and see supplemental Table S1 for PCR primers and method). VH domains were cloned into the heavy chain expression vector on the 5′ side of the γ1 constant domains (CH1, CH2, and CH3 domains; pHC-huCγ vector). To accommodate the EcoRI restriction site needed for cloning, the two N-terminal VH residues were changed from Gln-Val to Glu-Phe. Dideoxynucleotide sequencing of the VL and VH domains in the 5′ and 3′ directions yielded identical sequences corresponding to the parent VL and VH domains. Coexpression of the heavy and light chain vectors in NS0 cells afforded secretion of the desired Abs into the culture supernatant (31). Stable cell lines expressing full-length IgGs were obtained by electroporation of PvuI-linearized pLC-huCκ vectors containing various VL domains (20 μg) into 107 NS0 cells using a BTX ECM830 square wave electroporator in phosphate-buffered saline (10 mm sodium phosphate, 137 mm NaCl, 2.7 mm KCl, pH 7.4; Genetronics; one 15-ms pulse, 250 V). The cells were cultured in 96-well plates in Dulbecco's modified Eagle's medium supplemented with 4.5 g/liter of glucose, 200 mm l-glutamine, and 10% IgG-depleted fetal bovine serum (Complete Dulbecco's modified Eagle's medium; 5 × 104 cells/well). After 24 h, puromycin (3 μg/ml) was added. Positive transformants were identified by ELISA using immobilized polyclonal anti-human κ Ab for capture of secreted κ light chains (200 ng/well; Sigma) and peroxidase-conjugated goat anti-human κ chain Ab for detection of bound κ chains (1:1000; Sigma) (32). pHC-huCγ plasmids containing VH domains were electroporated into κ chain-expressing clones (>0.1 μg/ml) similarly, followed by selection with geneticin (1.5 mg/ml; Invitrogen). IgG expression was determined as above except that detection was with anti-human IgG1 Ab (Fc specific; 1:1000; Sigma). Culture supernatants were concentrated 10-fold (Centriprep YM10; Millipore) and purified by affinity chromatography on immobilized Protein G using a pH 2.7 buffer for elution (14). Electrophoresis was on SDS-polyacrylamide gels (4–20%; Bio-Rad), followed by electrotransfer to nitrocellulose membranes, staining with peroxidase-conjugated Abs to human IgG (Fc-specific; 1:1000; Sigma) or to κ chains (1:1000; Sigma), and detection with SuperSignal West Pico chemiluminescent substrate (Pierce) (22). Total proteins were stained with silver nitrate or Coomassie Blue. Band densities were computed using Quantity One image analysis software (33). Protein concentrations were determined by the Bradford method using the Coomassie Plus assay kit (Pierce).
Recombinant E2 Fusion Proteins
A stable cell line coexpressing E1 and FLAG-E2 (residues 406–746 of E2 HCV H77 polyprotein with an N-terminal FLAG peptide tag) was prepared by transfection with the plasmid pCDNA3/E1FLAGE2. This plasmid was constructed from vectors kindly provided by Dr. L. Cocquerel (by transferring the 450-bp BamHI/AscI fragment from pTM1/E1FLAGE2 vector containing the C terminus of E1, FLAG tag, and N terminus of E2 to pcDNA3/E1E2 vector containing the e1 and e2 genes (34). After PvuI linearization, pcDNA3/E1FLAGE2 was electroporated into Chinese hamster ovary cells and transformants were selected with geneticin as above in Complete Dulbecco's modified Eagle's medium supplemented with non-essential amino acids. Lysates containing FLAG-E2 were prepared by treating the cells with 50 mm Tris, 150 mm NaCl, pH 7.4 (Tris-buffered saline), containing 1% Triton X-100, 10 mm CaCl2, 20 mm iodoacetamide, and a protease inhibitor mixture (35). The lysate (40 ml from ∼2 × 108 cells) was fractionated using anti-FLAG M1 Ab conjugated to agarose (0.5 ml of gel; >0.6 mg of FLAG peptide binding capacity/ml of gel; Sigma). Unbound proteins were removed by washing with Tris-buffered saline, 1 m CaCl2 containing 0.1 mm CHAPS. FLAG-E2 was eluted using 100 μg/ml of FLAG peptide (Sigma). This procedure enriched the FLAG-E2 only partially (estimated FLAG-E2 purity, ∼1%, determined by silver staining of SDS-electrophoresis gels). Repeat anti-FLAG or Galanthus nivalis lectin conjugated to agarose chromatography did not yield pure FLAG-E2, presumably because of its hydrophobic character and tendency to associate with other proteins. GST-E2, corresponding to full-length E2 (residues 384–746 of HCV polyprotein) with an N-terminal glutathione S-transferase tag, produced in a baculovirus insect cell expression system was purchased from Immunodiagnostics Inc.
Hydrolysis Assays
The enriched FLAG-E2 preparation (70 ng of FLAG-E2/ml; 1 nm) was incubated with electrophoretically homogenous IgG in 20 μl of 100 mm glycine-HCl, 50 mm Tris, 0.1 mm CHAPS at 37 °C. Following SDS electrophoresis under reducing conditions, the gels were electroblotted and FLAG-E2 was stained with: (a) goat anti-E2 polyclonal IgG (1:2000; BioDesign) followed by peroxidase-conjugated rabbit anti-goat IgG (1:1000; Pierce), (b) mouse anti-E2 monoclonal IgG (1:2000; Sigma) followed by peroxidase-conjugated goat anti-mouse IgG (1:2000; Sigma), or (c) peroxidase-conjugated mouse anti-FLAG monoclonal IgG (1:2000; Sigma). The intensity of the intact FLAG-E2 band (68 kDa) was quantified in arbitrary volume units (AVU, pixel intensity × band area) (36). Hydrolysis was computed as ((AVUDiluent − AVUIgG) × 100/(AVUDiluent)). Hydrolysis of GST-E2 (10 μg/ml; 130 nm) was determined similarly except that reactions were conducted in buffer containing skim milk (150 μg/ml) and human IgG denatured by boiling for 10 min (150 μg/ml) to preclude adsorptive protein loss on the reaction vessel surface. The intact 72-kDa GST-E2 band was stained with peroxidase-conjugated anti-GST Ab (1:1000; Sigma). For N-terminal sequencing of product bands, GST-E2 incubated with IgG without protein additives was subjected to SDS electrophoresis and the gels electroblotted onto a polyvinylidene fluoride membrane were stained with Coomassie Blue. Protein bands not present in control reactions containing GST-E2 alone or hybrid IgG alone were excised and their N termini sequenced by Edman's degradation using a CLC Capillary 492 Protein Sequencing System (Applied Biosystems) with a phenylthiohydantoin-derivative mixture as standard (2 pmol each; sensitivity of detection of individual amino acids, 0.04–0.10 pmol). To determine substrate specificity, IgG was incubated with GST devoid of the E2 polypeptide (Sigma), biotinylated bovine serum albumin, or biotinylated C2 domain of Factor VIII (biotinylated-C2) prepared as described (33). GST hydrolysis was quantified using anti-GST Ab. Hydrolysis of biotinylated proteins was determined with peroxidase-conjugated streptavidin. Syntheses of the serine protease inhibitors diphenyl N-[2-(biotinamido)ethylamido-3,30-dithiodipropionyl]amino(4-amidinophenyl)methanephosphonate (E-hapten 1), N-[2-(biotinamido)ethylamido-3,30-dithiodipropionyl]amino(4-amidinophenyl)methanephosphonate (NE-hapten), and diphenyl N-(benzyloxycarbonyl)amino(4-amidinophenyl)methanephosphonate (E-hapten 2) have been described (37). Hydrolysis of the amide bond linking 7-amino-4-methylcoumarin (AMC) to the C-terminal amino acid in peptide-AMC substrates was measured in 50 mm Tris-HCl, 0.1 m glycine, 0.1 mm CHAPS, pH 7.7, at 37 °C in 96-well plates by fluorimetry (λex 360 nm, λem 470 nm) (17). Authentic aminomethylcoumarin was used to construct a standard curve. Kinetic parameters were obtained by fitting the rate data obtained at increasing concentrations of peptide-AMC substrates to the Michaelis-Menten-Henri equation: V = kcat·[Ab] · [S]/(Km + [S]).
RESULTS
Hybrid Abs
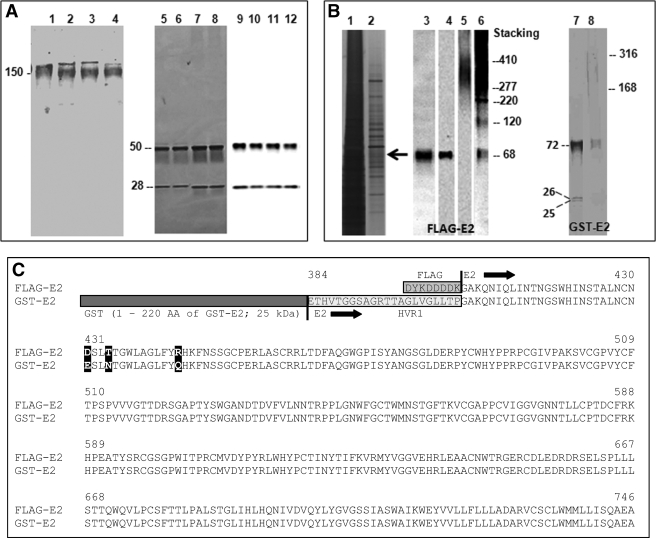
Human IgG molecules containing catalytic VL domain paired with the VH domain of an anti-HCV E2 Ab were purified from supernatants of NS0 cells. The VL domains were from catalytic light chain clones GG63, HK13, and HK14 isolated from phage display libraries. Light chain GG63 hydrolyzes various tripeptide model substrates (22). Light chains HK14 and HK13 hydrolyze vasoactive intestinal polypeptide (21) and amyloid-β peptide (38), substrates unrelated in sequence to the E2 fusion proteins. This catalyst engineering strategy is predicated on the hypothesis that the specificity of hybrid IgGs can be derived from the noncovalent antigen binding activity contributed by the VH domain. The VH domain is from the monoclonal IgG1 CBH-7, an Ab that recognizes HCV E2 with high affinity (Kd ∼ 10 nm) and neutralizes infection by HCV pseudovirions expressing E2 (39, 40). The precise epitope bound noncovalently by the wild type anti-E2 IgG CBH-7 (VH domain donor) is not known, but studies with E2 deletion mutants suggest that the epitope is a conformational determinant outside the hypervariable 1 region composed of amino acids distant from each other in the linear protein sequence (39, 41). Recombinant IgG containing the natural VL-VH pair of anti-E2 Ab (29) and the irrelevant anti-gp120 IgG GL2 were prepared as controls. The recombinant IgGs are intended to recapitulate the structure of natural IgGs containing 2 heavy chains disulfide bonded to 2 light chains. Like natural IgGs purified from blood, each of the hybrid and wild type IgGs purified by chromatography displayed a molecular mass of ∼150 kDa on non-reducing SDS-electrophoresis gels (Fig. 1A, lanes 1–4). Under reducing conditions, silver staining and immunoblotting of the gels revealed the anticipated 50-kDa heavy chain and 28-kDa light chain bands (Fig. 1A, lanes 5–12). The electrophoresis results indicated that the hybrid IgGs have been assembled as full-length, tetrameric IgG molecules. Expression levels for wild type anti-E2 IgG, hybrid GG63 IgG, hybrid HK13 IgG, and hybrid HK14 IgG were, respectively, 0.9, 2, 0.4, and 1.3 mg/liter.
FIGURE 1.
Hybrid Abs and HCV E2 fusion protein substrates. A, expression and purification of full-length hybrid IgGs. Coomassie Blue-stained nonreducing SDS-electrophoresis gels of wild type IgG CBH-7, hybrid IgG HK13, hybrid IgG HK14, and hybrid IgG GG63 (respectively, lanes 1–4); silver-stained reducing SDS gels of these IgGs (respectively, lanes 5–8) and immunoblots of reducing SDS gels of these IgGs stained with a mixture of anti-heavy chain and anti-light chain Abs (respectively, lanes 9–12). IgGs were purified from culture supernatants by affinity chromatography on Protein G-agarose. The 150-kDa band corresponds to intact tetrameric IgGs. The 50- and 28-kDa bands are the heavy and light chains, respectively. B, E2 fusion protein substrates. Shown are reducing SDS gels of the crude lysate from stably transfected Chinese hamster ovary cells coexpressing FLAG-E2 and E1 (lane 1) and the lysate after fractionation on immobilized anti-FLAG Ab stained with silver (lane 2), an anti-E2 Ab (lane 3), or anti-FLAG Ab under reducing (lanes 4) or non-reducing conditions (lane 5 and its overexposed version, lane 6). Lanes 7 and 8 are, respectively, reducing and non-reducing SDS gels of GST-E2 stained with Coomassie Blue. C, FLAG-E2 and GST-E2 primary structure. The E2 fusion proteins have identical sequences except: (a) GST (amino acid residues 1–220 of GST-E2) and FLAG tags (residues 1–8 of FLAG-E2); (b) the hypervariable 1 region is deleted in FLAG-E2; and (c) mismatches at positions 431, 434, and 444 (bold, numbering corresponding to HCV H77 polyprotein).
E2 Substrate Properties
E2 is thought to be expressed on the HCV surface in association with E1 as a 66–70-kDa glycosylated transmembrane protein (42). Noncovalent E2-E1 complexes as well as disulfide-linked oligomers have been identified on the pseudovirion particles, the closest approximation of infectious HCV available for biochemical studies (43, 44). Soluble recombinant E2 versions linked to various polypeptide tags have been proposed as models for native E2 (35, 45). We studied two E2 fusion proteins as substrates for hybrid Abs. E2 bearing the N-terminal FLAG peptide tag (FLAG-E2) coexpressed with E1 in a mammalian cell line was evident as a band with a nominal 68-kDa mass stainable in reducing SDS gels stained with anti-E2 and anti-FLAG Abs (Fig. 1B, respectively, lanes 3 and 4). In nonreducing SDS gels, FLAG-E2 migrated primarily as complexes of 277–410 kDa mass along with a small amount of the monomeric protein (Fig. 1B, lanes 5 and 6). The alternate substrate was E2 bearing a GST tag at the N terminus obtained from a baculovirus expression system (GST-E2). Unlike FLAG-E2, most of the GST-E2 migrated as a 72-kDa monomer under nonreducing conditions, and only faint staining of aggregates was observed (168–316-kDa smear; Fig. 1B, lane 8). Two minor bands at 26 and 25 kDa were also visible, presumably degradation products formed during purification or sample handling (Fig. 1B, lane 7). All 3 bands were stained with anti-GST Abs.
FLAG-E2 and GST-E2 are derived from the same HCV genotype (genotype 1a). However, they contain several structural differences that may alter their antigenic reactivity with antibodies. The differences are (Fig. 1C): (a) the 21-residue hypervariable 1 region present in GST-E2 is deleted in FLAG-E2; (b) residues 431, 434, and 444 of the two substrates are non-identical; (c) GST-E2 contains the large 26-kDa GST tag, whereas FLAG-E2 contains the small FLAG tag (8 amino acids, ∼1 kDa); and (d) glycosylation of the two substrates may be different. GST-E2 is expressed in insect cells, and FLAG-E2 in mammalian cells known to support superior glycosylation (46, 47). From the primary structure, the mass values for the non-glycosylated E2 polypeptide and GST are 39 and 26 kDa, respectively. The observed GST-E2 mass (72 kDa) suggests limited glycosylation corresponding to ∼7 kDa (observed mass-predicted polypeptide mass). For FLAG-E2, the predicted polypeptide mass and observed mass values are 39 and 68 kDa, respectively, suggesting a higher glycosylation level (∼29 kDa/molecule). Proteins expressed in insect cells (used to obtain GST-E2) are deficient in complex-type glycan structures with terminal sialic acid residues (47, 48), consistent with poor GST-E2 glycosylation. Yet another factor is coexpression of FLAG-E2 with the E1 protein. E2-E1 association can influence the conformation of E2 antigenic epitopes and may allow improved mimicry of native E2 (42, 49).
Hybrid Ab Hydrolytic Activity
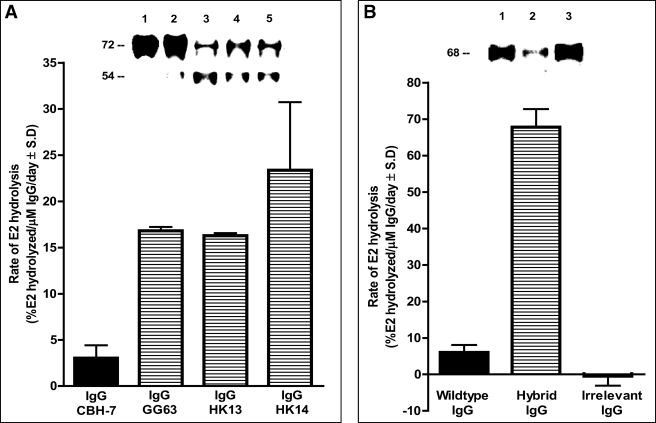
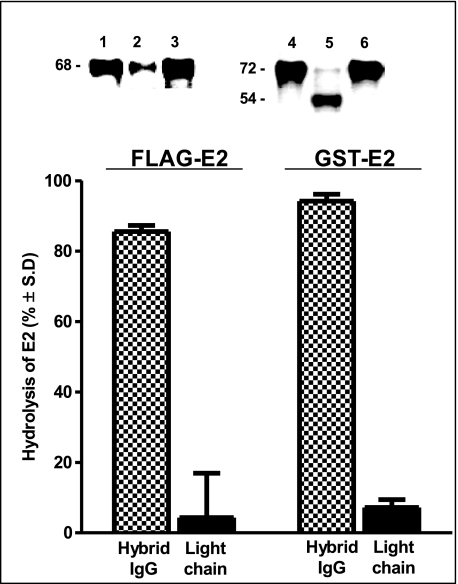
Treatment of GST-E2 with electrophoretically homogenous hybrid IgG preparations containing the VH domain from anti-E2 Ab clone CBH-7 resulted in slow cleavage of the intact 72-kDa protein band and appearance of a smaller 54-kDa product band stainable with anti-GST Ab (Fig. 2A, lanes 3–5; IgG clones GG63, HK13, and HK14). There was little or no cleavage of intact GST-E2 following treatment with wild type anti-E2 IgG CBH-7 purified from the culture supernatants by an identical experimental protocol (<3% decrease of GST-E2 cleavage; background rate + 2 S.D. observed for GST-E2 treated with diluent, 4.5%; n = 6 determinations). Further studies were done using hybrid IgG HK14. Three independent preparations of the hybrid IgG displayed reproducible hydrolytic activity. The gels do not allow determination of the full product profile or quantitative comparisons, as the fragmentation reaction may result in altered or even complete loss of reactivity with the anti-GST Ab used for detection. Treatment with the hybrid IgG resulted in reproducible depletion of the intact 68-kDa FLAG-E2 band stainable with a polyclonal anti-E2 Ab preparation in reducing SDS gels (see Fig. 2B, lane 2; see under “E2 Cleavage Regions” for evidence that the depletion is due to a cleavage reaction). No FLAG-E2 depletion was evident in reaction mixtures containing an irrelevant IgG purified from the culture supernatants by the same protocol as hybrid IgG HK14 (Fig. 2B, lane 3). FLAG-E2 depletion in the presence of wild type anti-E2 IgG CBH-7 was within or close to the error limit in several assays (Fig. 2B, background rate + 2 S.D. for FLAG-E2 treated with diluent, 8%; n = 6 determinations).
FIGURE 2.
Hydrolysis of E2 fusion proteins by hybrid IgG Abs. A, GST-E2 hydrolysis. Inset, reducing SDS gels stained with anti-GST Ab showing GST-E2 (130 nm) incubated for 5 days with diluent (lane 1), wild type IgG CBH-7 (lane 2), hybrid IgG GG63 (lane 3), hybrid IgG HK13 (lane 4), or hybrid IgG HK14 (lane 5; all IgG samples tested at 1 μm). The bar plot shows GST-E2 hydrolysis rates computed by densitometry of the intact GST-E2 band. A reaction product at 54 kDa is evident. B, FLAG-E2 hydrolysis. Inset, reducing SDS gels stained with polyclonal anti-E2 Abs showing FLAG-E2 (1 nm) incubated for 2 days with wild type IgG CBH-7 (lane 1), hybrid IgG HK14 (lane 2), or irrelevant anti-gp120 IgG GL2 (lane 3; IgG samples tested at 0.5 μm). Bars show hydrolysis of FLAG-E2 determined as cleavage of the intact 68-kDa protein band as in panel A. Data in panels A and B are mean ± S.D. of three independent reactions.
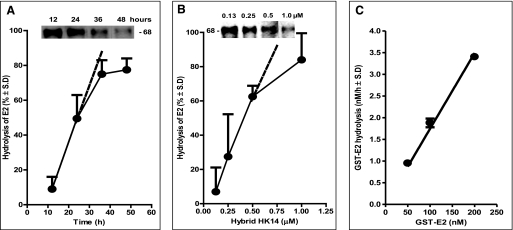
Hydrolysis of GST-E2 (not shown) and FLAG-E2 increased progressively with increasing duration of incubation (Fig. 3A) and increasing concentrations (Fig. 3B) of the hybrid IgG. FLAG-E2 was hydrolyzed about 3-fold more rapidly than GST-E2 by hybrid IgG HK14 (respectively, 65 ± 8 and 23.4 ± 7.4% hydrolysis/μm IgG/24 h; mean ± S.D. of three independent experiments). If E2 expressed on the surface of HCV is hydrolyzed at the rate observed for FLAG-E2, 4.3 and 65% of the viral protein will be degraded, respectively, by 150 μg/ml of hybrid IgG (1 μm IgG) in 1 and 24 h (computed as % hydrolysis = 100(1 − e(−kobs×t)), where kobs and t represent the first-order rate constant and time, respectively; kobs = ln(100/(100 − % cleavage))/t = ln(100/(100-65))/24 = 0.0437 h−1). The rate increased linearly at 50–200 nm GST-E2 concentrations (Fig. 3C). The concentration of FLAG-E2 in the hydrolysis assays was ∼1 nm (computed as: total protein content of the FLAG-E2 preparation × (intensity of the 68-kDa FLAG-E2 band/sum of intensity of all silver-stained bands observed in Fig. 1B, lane 2)). As the E2 fusion protein concentrations were insufficient to saturate the hybrid IgG, the reaction rate was determined by the strength of noncovalent substrate binding along with the catalytic rate constant.
FIGURE 3.
Characteristics of E2 fusion protein hydrolysis by hybrid HK14. A, time dependence of FLAG-E2 hydrolysis. Hydrolysis of FLAG-E2 (1 nm) by the hybrid IgG (0.5 μm) was measured as described in the legend to Fig. 2B. Mean ± S.D. of two reactions is shown. Inset, reducing SDS gels stained with polyclonal anti-E2 Abs showing cleavage of the 68-kDa FLAG-E2 band after incubation with hybrid IgG for 12, 24, 36, and 48 h. B, hybrid IgG concentration dependence. FLAG-E2 (1 nm) hydrolysis determined after incubation for 3 days with increasing hybrid IgG concentrations. Mean ± S.D. of two reactions is shown. Inset, reducing SDS gels stained with polyclonal anti-E2 Abs showing cleavage of the 68-kDa FLAG-E2 band after incubation with 0.13, 0.25, 0.5, and 1.0 μm hybrid IgG HK14. C, substrate concentration dependence. Increasing GST-E2 concentrations were incubated for 48 h with the hybrid IgG (0.5 μm). Hydrolysis was determined from cleavage of the intact 72-kDa protein band stainable with polyclonal anti-E2 Abs. Mean ± S.D. of two reactions is shown. The linear increase in rate confirms that substrate is present at concentrations that do not saturate the IgG, permitting measurement of the effect of noncovalent substrate recognition on the reaction rate.
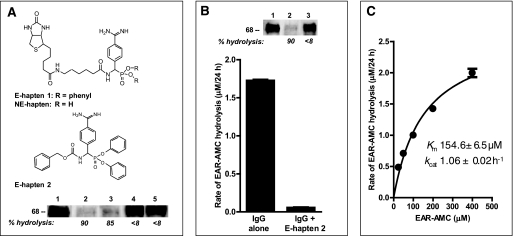
The light chains employed to construct the hybrid IgG hydrolyze peptide bonds by a serine protease-like mechanism (13, 22). Hydrolysis of FLAG-E2 by the hybrid IgG was inhibited completely by an active site-directed phosphonate inhibitor of serine proteases (>99 and 97% inhibition by E-hapten 1 and E-hapten 2, respectively, Fig. 4A). This suggests acquisition by the hybrid IgG of the serine protease-like property contributed by the light chain. Small model peptide substrates conjugated to a fluorophore have been employed to study Ab proteolytic activities free of the influence of noncovalent epitope recognition (17). The proteolytic activity of the hybrid IgG was detectable at an excess concentration of the tripeptide peptide-AMC substrate, Glu-Ala-Arg-AMC. Hydrolysis of Glu-Ala-Arg-AMC was inhibited by E-hapten 2, confirming the serine protease-like mechanism (Fig. 4B). Degradation of FLAG-E2 was inhibited by an excess concentration of the alternate substrate Glu-Ala-Arg-AMC, indicating that a common catalytic site is responsible for hydrolysis of both substrates (Fig. 4B, inset). From rate data at increasing Glu-Ala-Arg-AMC concentrations, the apparent catalytic rate constants (kcat) and Km for the reaction were, respectively, 1.06 ± 0.02 h−1 and 154 μm (Fig. 4C). Light chains generally hydrolyze peptide bonds on the C-terminal side of basic residues. Studies on a panel of 10 peptide-AMC substrates indicated preferential hydrolysis of the Arg/Lys-AMC bond by the hybrid IgG (Table 1). No hydrolysis of substrates containing acidic and neutral residues linked to AMC was evident. Hydrolysis of Gly-Gly-Arg-AMC and Gly-Gly-Leu-AMC was compared to confirm the requirement for a basic residue at the cleavage site. Gly-Gly-Arg-AMC was hydrolyzed, but Gly-Gly-Leu-AMC was not. These substrates are identical except for Arg-AMC/Leu-AMC linkage, eliminating the possibility of confounding remote residue effects.
FIGURE 4.
Serine protease inhibitor effect and kinetics of Glu-Ala-Arg-AMC hydrolysis. A, phosphonate diester hapten inhibition of FLAG-E2 hydrolysis. Reducing SDS gels showing FLAG-E2 (1 nm) incubated with diluent (lane 1) or hybrid IgG HK14 (0.5 μm) in the presence of 2% dimethyl sulfoxide (lane 2), the non-electrophilic compound NE-hapten (lane 3), E-hapten 1 (lane 4), or E-hapten 2 (lane 5). Inhibitor concentrations: E-hapten 1 and NE-hapten, 500 μm; E-hapten 2, 200 μm. Reaction time was 48 h. Staining was with polyclonal anti-E2 Abs. Numbers below the lanes indicate % hydrolysis compared with diluent control (lane 1). Top, structures of NE-hapten, E-hapten 1, and E-hapten 2. B, phosphonate diester inhibition of Glu-Ala-Arg-AMC hydrolysis. Shown are the hydrolysis rates of the substrate (400 μm) incubated with hybrid IgG (0.1 μm) for 24 h in the presence or absence of E-hapten 2 (200 μm). Mean ± S.D. of three reactions is shown. Inset, inhibition of FLAG-E2 hydrolysis by the alternate substrate Glu-Ala-Arg-AMC. Shown are reducing SDS gels showing FLAG-E2 (1 nm) incubated with the hybrid IgG (0.5 μm) in the absence (lane 2) or presence (lane 3) of Glu-Ala-Arg-MCA (200 μm) for 48 h. Lane 1, diluent control without IgG. Staining with anti-E2 Ab. Numbers below the lanes indicate % hydrolysis compared with diluent control (lane 1). Mean ± S.D. of two reactions is shown. C, kinetic parameters for hybrid IgG catalyzed Glu-Ala-Arg-AMC hydrolysis. IgG was 0.1 μm. Initial velocities (V) fitted to the Michaelis-Menten equation by nonlinear regression (r2 = 0.98). Data are means of three reactions computed as slopes of plots of rate versus time at each substrate concentration.
TABLE 1.
Hybrid IgG cleavage site preference
Reaction conditions were IgG, 100 nm; peptide-AMC substrates, 200 μm, except GGR-AMC and GGL-AMC, 12.5 μm. Values are means of 3 replicates ± S.D. determined as slopes of rate versus reaction time plots.
| Substrate | μm AMC released/24 h |
|---|---|
| EAR-AMC | 1.68 ± 0.13 |
| PFR-AMC | 1.07 ± 0.31 |
| IEGR-AMC | 0.25 ± 0.06 |
| EKK-AMC | 3.45 ± 0.09 |
| VLK-AMC | 4.06 ± 0.80 |
| AE-AMC | NDa |
| AAA-AMC | ND |
| AAPF-AMC | ND |
| IIW-AMC | ND |
| GGR-AMC | 0.56 ± 0.02 |
| GGL-AMC | ND |
a ND, not detected (<0.1 μm AMC/24 h).
Specificity Derived from Noncovalent Binding
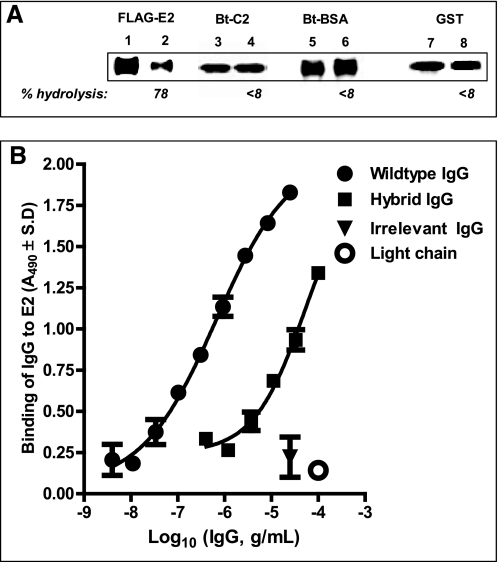
No cleavage of intact GST-E2 or FLAG-E2 was observed following treatment with the catalytic HK14 light chain alone or the wild type anti-E2 IgG CBH-7 under conditions affording robust hydrolysis by hybrid IgG HK14 (Fig. 5). The hybrid IgG did not hydrolyze irrelevant proteins (biotinylated C2 domain of Factor FVIII and bovine serum albumin; Fig. 6A, lanes 4 and 6, respectively). Importantly, GST devoid of the E2 protein was not hydrolyzed by hybrid IgG under conditions permitting GST-E2 hydrolysis, indicating that recognition of the E2 component of GST-E2 is essential for the hydrolytic reaction (Fig. 6A, lane 8). The wild type anti-E2 IgG was selected as the VH domain donor because of its ability to bind E2 noncovalently. In ELISA tests, hybrid IgG HK14 displayed readily detectable FLAG-E2 binding, although at reduced levels compared with the wild type anti-E2 IgG (by 90-fold; computed from IgG concentrations displaying a A490 value 1.0; Fig. 6B). Light chain HK14 did not bind FLAG-E2 (Fig. 6B). Therefore, the anti-E2 VH domain alone is sufficient to impart noncovalent E2 binding activity to the hybrid IgG despite pairing with a catalytic VL domain devoid of this activity. Taken together, these observations suggest that specific FLAG-E2 and GST-E2 hydrolysis by the hybrid IgG entails noncovalent recognition of the E2 component of the fusion proteins by contacts occurring at the VH domain followed by peptide bond hydrolysis by a catalytic site located in the VL domain. Reduced E2 binding by the hybrid IgG cannot be explained by diminished E2 availability due to its hydrolysis, as the rate of hydrolysis is slow (predicted FLAG-E2 hydrolysis at 100 μg of IgG/ml over the duration of the ELISA, 5.7%; assuming the same rate observed for soluble protein hydrolysis). Previous studies have also reported reduced antigen binding by heavy chains paired with irrelevant light chains compared with the wild type Ab (2, 3). This suggests that E2 recognition by the wild type anti-E2 IgG is dependent at least in part on noncovalent antigen interactions occurring at its VL domain. Unless E2 is present at an excess concentration (≫Kd), its reduced noncovalent recognition will also reduce the rate of catalysis.
FIGURE 5.
Hydrolytic specificity of hybrid IgG: failure of the light chain alone to hydrolyze E2 proteins. Hydrolysis of FLAG-E2 (left) or GST-E2 (right) by hybrid IgG HK14 (1 μm, stippled bars) or light chain HK14 alone (2 μm, solid bars) was measured as described in the legend to Fig. 2 (24 and 96 h incubation, respectively, for FLAG-E2 and GST-E2). Data are mean ± S.D. of two reactions. Insets, reducing SDS gels showing FLAG-E2 incubated with wild type IgG CBH-7 (lane 1), hybrid IgG HK14 (lane 2), or light chain HK14 (lane 3) and GST-E2 incubated with wild type IgG CBH-7 (lane 4), hybrid IgG HK14 (lane 5), or light chain HK14 (lane 6).
FIGURE 6.
Hydrolytic specificity of hybrid IgG: failure to hydrolyze irrelevant proteins and noncovalent E2 binding activity. A, substrate specificity. Reducing SDS gels showing reaction mixtures of hybrid IgG HK14 (0.5 μm) or diluent and FLAG-E2 (respectively, lanes 1 and 2), biotinylated Factor VIII C2 domain (respectively, lanes 3 and 4), biotinylated bovine serum albumin (respectively, lanes 5 and 6), or GST devoid of the E2 polypeptide (respectively, lanes 7 and 8). FLAG-E2 was 1 nm. Other substrates were 10 μm; reaction time was 48 h. FLAG-E2 staining was with anti-FLAG Ab, biotinylated protein staining with peroxidase-conjugated streptavidin, and GST staining with anti-GST Ab. Numbers below the lanes indicate % hydrolysis compared with diluent control. B, FLAG-E2 binding. Binding of hybrid IgG HK14, wild type IgG CBH-7, irrelevant anti-gp120 IgG GL2, and light chain HK14 to FLAG-E2 (4 ng/well) was measured by ELISA. Shown are A490 values versus Ab concentration curves fitted to A490 = bottom + (top − bottom)/(1 + 10((log EC50 − X) × Hill slope)) (r2 > 0.97). Mean ± S.D. of two wells each.
E2 Cleavage Regions
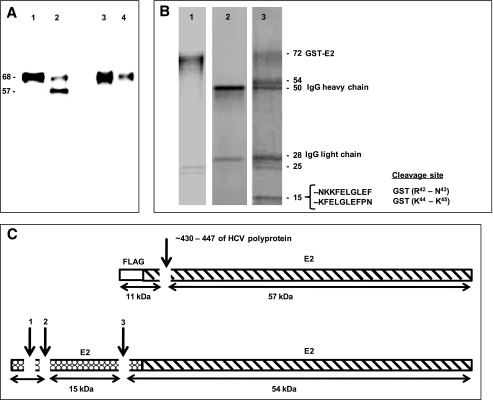
Pure FLAG-E2 was not available to us, precluding analysis of its fragmentation profile by total protein staining or product sequencing methods. However, a 57-kDa E2 product band was identified by staining the reaction mixture of FLAG-E2 and hybrid IgG with a commercially available monoclonal anti-E2 Ab (Fig. 7A, lane 2). The FLAG epitope is 8 amino acids in length. The predicted mass ranges of products released from the N- and C-terminal sides of hydrolysis at any peptide bond in the FLAG peptide are 0.1–1.0 and 67.0–67.9 kDa respectively. The observed product mass (57 kDa) indicates that the hybrid IgG must hydrolyze a peptide bond in the E2 polypeptide (Fig. 7C). This product was not stained with anti-FLAG Ab (Fig. 7A, lane 4). As the product does not contain the FLAG epitope, it must correspond to the fragment on the C-terminal side of the hydrolytic site. The deduced region of hydrolysis is ∼11 kDa from the N terminus of the fusion protein (∼10 kDa from E2 N terminus, corresponding to residue 406 of the HCV polyprotein in Fig. 7C). The FLAG epitope-containing product on the N-terminal side of the cleavage site (predicted mass ∼11 kDa) was not detected by immunoblotting. A possible explanation is its inefficient electrotransfer. Small polypeptides often remain undetected due to their poor transfer in immunoblotting procedures (50).
FIGURE 7.
E2 fusion protein cleavage regions. A, reducing SDS gel showing reaction mixtures of FLAG-E2 (1 nm) incubated with wild type IgG CBH-7 or hybrid IgG HK14 (0.5 μm) for 48 h and stained with anti-E2 monoclonal Ab (respectively, lanes 1 and 2) or anti-FLAG Ab (respectively, lanes 3 and 4). Note the 57-kDa anti-E2 stainable product band generated by the hybrid IgG (lane 2) that was not stained by anti-FLAG Ab (lane 4). B, reducing SDS gel lanes of GST-E2 alone (lane 1), the hybrid IgG alone (lane 2), and the reaction mixture of GST-E2 and hybrid IgG HK14 (lane 3) stained with Coomassie Blue. GST-E2 was 3 μm and hybrid IgG was 1 μm with incubation for 5 days. The N-terminal 10 residues of the 15-kDa band determined by Edman's degradation and the deduced scissile bonds are indicated. Yields of the phenylthiohydantoin-derivitized amino acids in individual sequencing cycles were 0.53–1.61 pmol. C, schematic representation of FLAG-E2 (top) and GST-E2 (bottom) showing the cleavage sites deduced from the mass and immunoreactivity of the FLAG-E2 57-kDa product, and from amino acid sequencing and mass of the GST-E2 15-kDa product (see “Results”).
Coomassie Blue staining of the alternate substrate GST-E2 treated with hybrid IgG revealed product bands at 54 and 15 kDa that were absent in GST-E2 alone or IgG alone (Fig. 7B). Also visible were the IgG heavy and light chain bands (respectively, 50- and 28-kDa bands) and the presumptive GST-E2 fragments present in the starting GST-E2 preparation employed as substrate (25- and 26-kDa bands). N-terminal sequencing identified two peptide sequences present in the 15-kDa band, corresponding to residues Asn-Lys-Lys-Phe-Glu-Leu-Gly-Leu-Glu-Phe and Lys-Phe-Glu-Leu-Gly-Leu-Glu-Phe located in the GST tag (Fig. 7B). This indicated hydrolysis of the Arg42–Asn43 and Lys44–Lys45 bonds in the GST component of GST-E2 (arrows 1 and 2, Fig. 7C). As GST devoid of the E2 polypeptide was not hydrolyzed by the hybrid IgG (Fig. 6A, lane 8), it may be concluded that noncovalent E2 recognition by IgG facilitates hydrolysis at the GST tag (see “Discussion” for a proposed reaction model). The small products on the N-terminal side of Arg42–Asn43 and Lys44–Lys45 cleavage sites were not visible (predicted mass, 5.2 and 5.4 kDa; residues 1–42 and 1–44 of the fusion protein), presumably because they migrated with the gel dye front. The 15-kDa product is too small to qualify as the entire polypeptide fragment on the C-terminal side of the Arg42–Asn43 and Lys44–Lys45 cleavage sites (predicted mass from primary structure ∼52 kDa), suggesting that another site within the GST tag is hydrolyzed by the IgG (∼6 kDa upstream of the GST C terminus; arrow 3, Fig. 7C). The predicted mass of this fragment is close to the observed 54-kDa product band. N-terminal sequencing of the 54-kDa product did not yield an identifiable amino acid sequence, suggesting a blocked N terminus, a common problem in protein sequencing studies. Further chemical analyses was not undertaken, as information about the identity of the 54-kDa product is not needed to validate the hypothesis underlying the present study, i.e. the hybrid IgG acquires antigen-specific catalytic activity driven by noncovalent binding interactions contributed by the VH domain.
DISCUSSION
These findings indicate specific hydrolysis of the two E2 fusion proteins containing FLAG and GST tags by hybrid IgGs composed of promiscuous catalytic VL domains paired with the VH domain of a noncatalytic E2-binding IgG. The hybrid IgG did not hydrolyze proteins unrelated to E2, indicating specific E2 recognition. Under conditions affording hydrolysis by the hybrid IgG, the light chain used to construct the hybrid IgG failed to hydrolyze FLAG-E2. Similarly, the hybrid IgG displayed E2 binding in ELISA tests and the light chain alone did not. Previous reports have indicated that the heavy chain alone can bind the antigen (4, 51). Taken together, these findings suggest accelerated E2 hydrolysis by the hybrid IgG facilitated by noncovalent E2 binding.
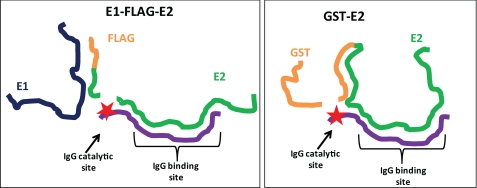
The V domain hybridization principle derives from the split-site model of Ab catalysis (13, 26). The model states that distinct Ab subsites are responsible for weak noncovalent binding and the subsequent nucleophilic hydrolytic step (Fig. 8). The hybrid IgG expressed hydrolytic properties similar to its component light chain. Like free light chains (8), the hybrid IgG hydrolyzes Arg/Lys-X bonds preferentially. An active site-directed inhibitor of serine proteases inhibited the hydrolysis of FLAG-E2 and a model peptide substrate by the hybrid IgG. This is consistent with previous reports that Ab VL domains contain serine protease-like catalytic sites (13, 22). The sites are composed of triads and diads of amino acids in which intramolecular hydrogen bonding imparts nucleophilic reactivity to Ser/Thr residues. Noncovalent binding, on the other hand, results from cumulative weak contacts between the antigen at a comparatively large number of V domain amino acids (15–22 residues) (6). The antigen-specific hydrolytic activity reported here indicates the feasibility of effective V domain pairing permitting initial noncovalent binding at VH domain residues coordinated with subsequent peptide bond hydrolysis at a site belonging to the VL domain.
FIGURE 8.
Split-site model for shared noncovalent E2 binding specificity and divergent hydrolytic specificity of hybrid IgG. The noncovalent antigen binding pocket of the hybrid IgG is occupied by the E2 epitope. The hydrolytic subsite of the IgG (★) is located distant from the binding site. If the conformations of the substrates (FLAG-E2, GST-E2) are divergent, different peptide bonds can be placed in register with the hydrolytic subsite despite noncovalent IgG binding to the same epitope in the substrates.
E2 is expressed on the HCV surface as a heterodimer in association with E1. E2 binding by a host cell receptor is thought to initiate infection. Host proteins advanced as potential E2 receptors include CD81 (41, 52), scavenger receptor class B type I (53), and the low-density lipoprotein receptor (54). E1 associated to E2 is thought to be essential for the next step in infection, virus-host cell membrane fusion (55). Certain structural details of the HCV coat have been elucidated by electron microscopy (56). However, the three-dimensional structures of native E2 and E1 have not been defined. Various E2 fusion proteins have been cloned as models of the native E2 protein. The conformation of the E2 polypeptide is sensitive to various microenvironmental factors, exemplified by changes in its reaction with various Abs (35, 57). Of the two E2 fusion protein tested in the present study for FLAG-E2 and GST-E2, the former likely approximates more closely the native protein structure. Unlike GST-E2, FLAG-E2 is co-expressed with E1, and the small FLAG tag is less likely to introduce conformational disturbances in the E2 polypeptide compared with the large GST tag. Moreover, FLAG-E2 is expressed in mammalian cells, which may produce a glycosylation pattern closer to the native protein. E2 contained in the FLAG-E2 was hydrolyzed by the hybrid IgG ∼11 kDa downstream from its N terminus. The precise identity of the scissile bond is not available. However, if the glycosylation sites of FLAG-E2 are identical to those determined previously for an E2 preparation obtained from a similar mammalian expression system (58), the E2 region containing the scissile bond can be narrowed to the HCV polyprotein region between glycosylated residues Asn430 and Asn447 (deduced assuming an average mass of 2.7 kDa/glycan chain (58); this region is located 10.8–12.8 kDa downstream from the FLAG-E2 protein N terminus, approximately corresponding to the deduced cleavage region). Further studies are necessary to determine whether the hybrid IgG recognizes viral E2 and influences viral infectivity. Studies by other groups have suggested that the CD81 binding site of E2 is a conformational determinant with contributions from E2 residues 474–492, 522–551, and 612–619 (52, 59, 60). Importantly, the hydrolytic reaction may influence the functions of E2 peptide regions remote from the cleavage site, as backbone breakage often produces fragments that differ in overall conformation compared with the corresponding intact protein regions.
Like other irrelevant polypeptides, GST devoid of the E2 polypeptide was not hydrolyzed by the hybrid IgG. This indicates that GST-E2 hydrolysis is a specific reaction. Interestingly, however, GST-E2 was hydrolyzed at peptide bonds located in the GST tag. In the split-site model, noncovalent binding is the basis of substrate specificity, and the catalytic subsite hydrolyzes the substrate at peptide bonds distant from the noncovalently bound region of the substrate (Fig. 8). The hydrolytic activity of hybrid IgG itself is a promiscuous function, evident from the hydrolysis of small peptide substrates tested at excess concentrations (400 μm). Provided contact is established in the transition state of the reaction, the Ab catalytic site is free to hydrolyze alternate peptide bonds in the substrate. This reaction model has been verified experimentally by epitope mapping, mutagenesis, and alternate scissile bond studies for autoantibody catalyzed hydrolysis of the neuropeptide VIP (13, 25). Unlike GST-E2, FLAG-E2 was unambiguously hydrolyzed at a site within the E2 polypeptide. Evidently, noncovalent E2 binding by the hybrid IgG positions different peptide regions of FLAG-E2 and GST-E2 in register with the catalytic subsite in the hydrolytic step of the reaction. Contacts between the Ab hydrolytic site and the substrate backbone are determined by the three-dimensional folding pattern of the substrate and the spatial distance between the noncovalent and hydrolytic sites (Fig. 8). The differing hydrolysis pattern of FLAG-E2 and GST-E2 may derive from conformational differences between the two proteins. FLAG-E2 and GST-E2 exist in differing aggregation states, and they also present additional structural variations (Fig. 1, B and C).
Although driven by noncovalent E2 binding, hydrolysis at the GST tag is an unintended reaction that highlights the pitfalls of employing structurally divergent substrates (FLAG-E2 and GST-E2) in efforts to identify useful catalysts. Controlling the site of substrate hydrolysis by split-site hybrid IgGs is difficult. However, this criticism is valid only at the initial proteolytic activity analysis stage. Proteins mostly adopt stable conformations. Once a proteolytic IgG that hydrolyzes the native substrate at a functionally significant bond is identified, the hydrolytic specificity will be preserved.
Pegylated interferon and ribavirin are the mainstays of current therapy for chronic HCV infection (61). These drugs control infection only in ∼50% of patients and they can exert toxic side effects. Additional studies are needed to determine whether catalytic hybrid IgGs can be developed for immunotherapy of HCV infection. If FLAG-E2 and native E2 are recognized similarly by the hybrid IgG HK14, certain predictions concerning inactivation of the viral protein are possible. HCV is present at 4 × 104 to 7 × 107 copies/ml in the blood of infected patients (62), corresponding to the E2 concentration of 1.2 × 10−17 to 2.2 × 10−14 m if each virus particle is assumed to express 180 E2 copies (56). At viral E2 concentrations smaller than the catalytic IgG concentration, the rate equation for hydrolysis of FLAG-E2 applies (see “Results”). From this equation, 50 and 90% viral E2 in the blood will be hydrolyzed, respectively, in 16 and 53 h by 0.5 μm hybrid IgG (75 μg/ml; IgG concentrations in this range are readily attained in currently used passive immunotherapy protocols, e.g. natalizumab, bevacizumab, and cetuximab (63)). The rate is slower than required for rapid interruption of infection mediated by high affinity HCV interactions with host receptors (by about 1–2 log orders). However, IgG circulates in peripheral blood with half-life on the order of 14–21 days (64), and substantial proportions of E2 expressed on the viral surface can be hydrolyzed over the life of IgG in circulation. More advanced engineering strategies that preserve noncovalent antigen binding activity and improve the rate of catalysis may eventually enable the development of hybrid IgGs that interrupt the infection more rapidly. In the present study, the hybrid IgG displayed 90-fold reduced noncovalent E2 binding compared with the wild type anti-E2 IgG. Pairing of different VH domains from antigen-specific Abs with non-physiological VL domains results in variable loss of the antigen binding activity (2, 3). Identifying VH domains that bind E2 with unimpaired affinity despite pairing with a catalytic VL domain may yield catalytic IgGs with improved noncovalent E2 binding activity. We recently identified unpaired VL domains with exceptional catalytic activity from a phage-displayed Ab library by an electrophilic selection procedure (65). Using a highly catalytic VL domain to construct the hybrid IgG may improve the rate of catalysis.
The generation of antigen-specific proteolytic Abs by natural immunological mechanisms is a rare event. The studies reported here provide the first proof-of-principle that the catalytic activity of VL domains can be rendered antigen specific by pairing with an antigen binding VH domain. This engineering principle is not limited to E2 Abs. If the hybrid IgGs express sufficient levels of specific proteolytic activity, they can be conceived as viable candidates for immunotherapeutic applications.
Supplementary Material
Acknowledgments
We thank Dr. Hiroaki Taguchi and Dr. Gary McLean for helpful suggestions, and Dr. Runsheng Wang for technical assistance. Certain plasmids used for preparing FLAG-E2 were kindly provided by Dr. Laurence Cocquerel.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL079381.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- Ab
- antibody
- AMC
- 7-amino-4-methylcoumarin
- AVU
- arbitrary volume units
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- GST
- glutathione S-transferase
- HCV
- hepatitis C virus
- V domain
- variable domain
- gp
- glycoprotein
- ELISA
- enzyme-linked immunosorbent assay.
REFERENCES
- 1.Sun M., Li L., Gao Q. S., Paul S. (1994) J. Biol. Chem. 269, 734–738 [PubMed] [Google Scholar]
- 2.Edelman G. M., Olins D. E., Gally J. A., Zinder N. D. (1963) Proc. Natl. Acad. Sci. U.S.A. 50, 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roholt O., Onoue K., Pressman D. (1964) Proc. Natl. Acad. Sci. U.S.A. 51, 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward E. S., Güssow D., Griffiths A. D., Jones P. T., Winter G. (1989) Nature 341, 544–546 [DOI] [PubMed] [Google Scholar]
- 5.Wilson I. A., Stanfield R. L., Rini J. M., Arevalo J. H., Schulze-Gahmen U., Fremont D. H., Stura E. A. (1991) CIBA Found. Symp. 159, 13–28; 28–39 [DOI] [PubMed] [Google Scholar]
- 6.Davies D. R., Padlan E. A., Sheriff S. (1990) Annu. Rev. Biochem. 59, 439–473 [DOI] [PubMed] [Google Scholar]
- 7.Paul S., Volle D. J., Beach C. M., Johnson D. R., Powell M. J., Massey R. J. (1989) Science 244, 1158–1162 [DOI] [PubMed] [Google Scholar]
- 8.Paul S., Li L., Kalaga R., Wilkins-Stevens P., Stevens F. J., Solomon A. (1995) J. Biol. Chem. 270, 15257–15261 [DOI] [PubMed] [Google Scholar]
- 9.Polosukhina D. I., Kanyshkova T. G., Doronin B. M., Tyshkevich O. B., Buneva V. N., Boiko A. N., Gusev E. I., Favorova O. O., Nevinsky G. A. (2004) J. Cell. Mol. Med. 8, 359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lacroix-Desmazes S., Moreau A., Sooryanarayana , Bonnemain C., Stieltjes N., Pashov A., Sultan Y., Hoebeke J., Kazatchkine M. D., Kaveri S. V. (1999) Nat. Med. 5, 1044–1047 [DOI] [PubMed] [Google Scholar]
- 11.Hifumi E., Morihara F., Hatiuchi K., Okuda T., Nishizono A., Uda T. (2008) J. Biol. Chem. 283, 899–907 [DOI] [PubMed] [Google Scholar]
- 12.Ponomarenko N. A., Durova O. M., Vorobiev I. I., Belogurov A. A., Jr., Kurkova I. N., Petrenko A. G., Telegin G. B., Suchkov S. V., Kiselev S. L., Lagarkova M. A., Govorun V. M., Serebryakova M. V., Avalle B., Tornatore P., Karavanov A., Morse H. C., 3rd, Thomas D., Friboulet A., Gabibov A. G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 281–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao Q. S., Sun M., Rees A. R., Paul S. (1995) J. Mol. Biol. 253, 658–664 [DOI] [PubMed] [Google Scholar]
- 14.Planque S., Bangale Y., Song X. T., Karle S., Taguchi H., Poindexter B., Bick R., Edmundson A., Nishiyama Y., Paul S. (2004) J. Biol. Chem. 279, 14024–14032 [DOI] [PubMed] [Google Scholar]
- 15.Ramsland P. A., Terzyan S. S., Cloud G., Bourne C. R., Farrugia W., Tribbick G., Geysen H. M., Moomaw C. R., Slaughter C. A., Edmundson A. B. (2006) Biochem. J. 395, 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalaga R., Li L., O'Dell J. R., Paul S. (1995) J. Immunol. 155, 2695–2702 [PubMed] [Google Scholar]
- 17.Mitsuda Y., Planque S., Hara M., Kyle R., Taguchi H., Nishiyama Y., Paul S. (2007) Mol. Biotechnol. 36, 113–122 [DOI] [PubMed] [Google Scholar]
- 18.Paul S., Nishiyama Y., Planque S., Taguchi H. (2006) Immunol. Lett. 103, 8–16 [DOI] [PubMed] [Google Scholar]
- 19.Schultz P. G., Yin J., Lerner R. A. (2002) Angew. Chem. Int. Ed. Engl. 41, 4427–4437 [DOI] [PubMed] [Google Scholar]
- 20.Takahashi N., Kakinuma H., Liu L., Nishi Y., Fujii I. (2001) Nat. Biotechnol. 19, 563–567 [DOI] [PubMed] [Google Scholar]
- 21.Tyutyulkova S., Gao Q. S., Thompson A., Rennard S., Paul S. (1996) Biochim. Biophys. Acta 1316, 217–223 [DOI] [PubMed] [Google Scholar]
- 22.Paul S., Tramontano A., Gololobov G., Zhou Y. X., Taguchi H., Karle S., Nishiyama Y., Planque S., George S. (2001) J. Biol. Chem. 276, 28314–28320 [DOI] [PubMed] [Google Scholar]
- 23.Hifumi E., Hatiuchi K., Okuda T., Nishizono A., Okamura Y., Uda T. (2005) FEBS J. 272, 4497–4505 [DOI] [PubMed] [Google Scholar]
- 24.Matsuura K., Yamamoto K., Sinohara H. (1994) Biochem. Biophys. Res. Commun. 204, 57–62 [DOI] [PubMed] [Google Scholar]
- 25.Paul S., Volle D. J., Powell M. J., Massey R. J. (1990) J. Biol. Chem. 265, 11910–11913 [PubMed] [Google Scholar]
- 26.Paul S. (1996) Mol. Biotechnol. 5, 197–207 [DOI] [PubMed] [Google Scholar]
- 27.Helle F., Dubuisson J. (2008) Cell Mol. Life Sci. 65, 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poumbourios P., Drummer H. E. (2007) Antivir. Chem. Chemother. 18, 169–189 [DOI] [PubMed] [Google Scholar]
- 29.Chan C. H., Hadlock K. G., Foung S. K., Levy S. (2001) Blood 97, 1023–1026 [DOI] [PubMed] [Google Scholar]
- 30.Karle S., Planque S., Nishiyama Y., Taguchi H., Zhou Y. X., Salas M., Lake D., Thiagarajan P., Arnett F., Hanson C. V., Paul S. (2004) Aids 18, 329–331 [DOI] [PubMed] [Google Scholar]
- 31.McLean G. R., Nakouzi A., Casadevall A., Green N. S. (2000) Mol. Immunol. 37, 837–845 [DOI] [PubMed] [Google Scholar]
- 32.Karle S., Nishiyama Y., Taguchi H., Zhou Y. X., Luo J., Planque S., Hanson C., Paul S. (2003) Vaccine 21, 1213–1218 [DOI] [PubMed] [Google Scholar]
- 33.Paul S., Karle S., Planque S., Taguchi H., Salas M., Nishiyama Y., Handy B., Hunter R., Edmundson A., Hanson C. (2004) J. Biol. Chem. 279, 39611–39619 [DOI] [PubMed] [Google Scholar]
- 34.Fournillier Jacob A., Cahour A., Escriou N., Girad M., Wychowski C. (1996) J. Gen. Virol. 77, 1055–1064 [DOI] [PubMed] [Google Scholar]
- 35.Cocquerel L., Kuo C. C., Dubuisson J., Levy S. (2003) J. Virol. 77, 10677–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Planque S., Mitsuda Y., Taguchi H., Salas M., Morris M. K., Nishiyama Y., Kyle R., Okhuysen P., Escobar M., Hunter R., Sheppard H. W., Hanson C., Paul S. (2007) AIDS Res. Hum. Retroviruses 23, 1541–1554 [DOI] [PubMed] [Google Scholar]
- 37.Nishiyama Y., Taguchi H., Luo J. Q., Zhou Y. X., Burr G., Karle S., Paul S. (2002) Arch. Biochem. Biophys. 402, 281–288 [DOI] [PubMed] [Google Scholar]
- 38.Rangan S. K., Liu R., Brune D., Planque S., Paul S., Sierks M. R. (2003) Biochemistry 42, 14328–14334 [DOI] [PubMed] [Google Scholar]
- 39.Keck Z. Y., Op De Beeck A., Hadlock K. G., Xia J., Li T. K., Dubuisson J., Foung S. K. (2004) J. Virol. 78, 9224–9232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Op De Beeck A., Voisset C., Bartosch B., Ciczora Y., Cocquerel L., Keck Z., Foung S., Cosset F. L., Dubuisson J. (2004) J. Virol. 78, 2994–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hadlock K. G., Lanford R. E., Perkins S., Rowe J., Yang Q., Levy S., Pileri P., Abrignani S., Foung S. K. (2000) J. Virol. 74, 10407–10416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Op De Beeck A., Cocquerel L., Dubuisson J. (2001) J. Gen. Virol. 82, 2589–2595 [DOI] [PubMed] [Google Scholar]
- 43.Dubuisson J., Rice C. M. (1996) J. Virol. 70, 778–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flint M., Logvinoff C., Rice C. M., McKeating J. A. (2004) J. Virol. 78, 6875–6882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee K. J., Suh Y. A., Cho Y. G., Cho Y. S., Ha G. W., Chung K. H., Hwang J. H., Yun Y. D., Lee D. S., Kim C. M., Sung Y. C. (1997) J. Biol. Chem. 272, 30040–30046 [DOI] [PubMed] [Google Scholar]
- 46.Rindisbacher L., Cottet S., Wittek R., Kraehenbuhl J. P., Corthésy B. (1995) J. Biol. Chem. 270, 14220–14228 [DOI] [PubMed] [Google Scholar]
- 47.Tomiya N., Narang S., Lee Y. C., Betenbaugh M. J. (2004) Glycoconj. J. 21, 343–360 [DOI] [PubMed] [Google Scholar]
- 48.Kost T. A., Condreay J. P., Jarvis D. L. (2005) Nat. Biotechnol. 23, 567–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cocquerel L., Meunier J. C., Op de Beeck A., Bonte D., Wychowski C., Dubuisson J. (2001) J. Gen. Virol. 82, 1629–1635 [DOI] [PubMed] [Google Scholar]
- 50.Kurien B. T., Scofield R. H. (2003) J. Immunol. Methods 274, 1–15 [DOI] [PubMed] [Google Scholar]
- 51.Davies J., Riechmann L. (1995) Biotechnology 13, 475–479 [DOI] [PubMed] [Google Scholar]
- 52.Roccasecca R., Ansuini H., Vitelli A., Meola A., Scarselli E., Acali S., Pezzanera M., Ercole B. B., McKeating J., Yagnik A., Lahm A., Tramontano A., Cortese R., Nicosia A. (2003) J. Virol. 77, 1856–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jia Z. S., Du D. W., Lei Y. F., Wei X., Yin W., Ma L., Lian J. Q., Wang P. Z., Li D., Zhou Y. X. (2008) J. Int. Med. Res. 36, 1319–1325 [DOI] [PubMed] [Google Scholar]
- 54.Molina S., Castet V., Fournier-Wirth C., Pichard-Garcia L., Avner R., Harats D., Roitelman J., Barbaras R., Graber P., Ghersa P., Smolarsky M., Funaro A., Malavasi F., Larrey D., Coste J., Fabre J. M., Sa-Cunha A., Maurel P. (2007) J. Hepatol. 46, 411–419 [DOI] [PubMed] [Google Scholar]
- 55.Russell R. S., Kawaguchi K., Meunier J. C., Takikawa S., Faulk K., Bukh J., Purcell R. H., Emerson S. U. (March17, 2009) J. Viral. Hepat. 10.1111/j.1365-2893.2009.01111.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu X., Qiao M., Atanasov I., Hu Z., Kato T., Liang T. J., Zhou Z. H. (2007) Virology 367, 126–134 [DOI] [PubMed] [Google Scholar]
- 57.Owsianka A., Clayton R. F., Loomis-Price L. D., McKeating J. A., Patel A. H. (2001) J. Gen. Virol. 82, 1877–1883 [DOI] [PubMed] [Google Scholar]
- 58.Iacob R. E., Perdivara I., Przybylski M., Tomer K. B. (2008) J. Am. Soc. Mass. Spectrom. 19, 428–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yagnik A. T., Lahm A., Meola A., Roccasecca R. M., Ercole B. B., Nicosia A., Tramontano A. (2000) Proteins 40, 355–366 [DOI] [PubMed] [Google Scholar]
- 60.Owsianka A. M., Timms J. M., Tarr A. W., Brown R. J., Hickling T. P., Szwejk A., Bienkowska-Szewczyk K., Thomson B. J., Patel A. H., Ball J. K. (2006) J. Virol. 80, 8695–8704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayashi N., Takehara T. (2006) J. Gastroenterol. 41, 17–27 [DOI] [PubMed] [Google Scholar]
- 62.Glynn S. A., Wright D. J., Kleinman S. H., Hirschkorn D., Tu Y., Heldebrant C., Smith R., Giachetti C., Gallarda J., Busch M. P. (2005) Transfusion 45, 994–1002 [DOI] [PubMed] [Google Scholar]
- 63.Fracasso P. M., Burris H., 3rd, Arquette M. A., Govindan R., Gao F., Wright L. P., Goodner S. A., Greco F. A., Jones S. F., Willcut N., Chodkiewicz C., Pathak A., Springett G. M., Simon G. R., Sullivan D. M., Marcelpoil R., Mayfield S. D., Mauro D., Garrett C. R. (2007) Clin. Cancer Res. 13, 986–993 [DOI] [PubMed] [Google Scholar]
- 64.Lobo E. D., Hansen R. J., Balthasar J. P. (2004) J. Pharmacol. Sci. 93, 2645–2668 [DOI] [PubMed] [Google Scholar]
- 65.Taguchi H., Planque S., Sapparapu G., Boivin S., Hara M., Nishiyama Y., Paul S. (2008) J. Biol. Chem. 283, 36724–36733 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.