Abstract
MicroRNAs (miRNAs) are a class of endogenous non-protein-coding RNAs that function as important regulatory molecules by negatively regulating gene and protein expression via the RNA interference (RNAi) machinery. MiRNAs have been implicated to control a variety of cellular, physiological, and developmental processes. Aberrant expressions of miRNAs are connected to human diseases such as cancer. Cancer stem cells are a small subpopulation of cells identified in a variety of tumors that are capable of self-renewal and differentiation. Dysregulation of stem cell self-renewal is a likely requirement for the initiation and formation of cancer. Furthermore, cancer stem cells are a very likely cause of resistance to current cancer treatments, as well as relapse in cancer patients. Understanding the biology and pathways involved with cancer stem cells offers great promise for developing better cancer therapies, and might one day even provide a cure for cancer. Emerging evidence demonstrates that miRNAs are involved in cancer stem cell dysregulation. Recent studies also suggest that miRNAs play a critical role in carcinogenesis and oncogenesis by regulating cell proliferation and apoptosis as oncogenes or tumor suppressors, respectively. Therefore, molecularly targeted miRNA therapy could be a powerful tool to correct the cancer stem cell dysregulation.
Key words: cancer stem cells, microRNAs, oncogenes, tumor suppressors
INTRODUCTION
In this review, we discuss the acknowledged functions and characteristics of microRNAs and cancer stem cells, focusing on the potential roles of the stem cell related microRNAs (miRNAs) in cancer stem cells regulation and the implications in developing novel and more effective molecular cancer therapies.
MicroRNA Biogenesis
MiRNAs and small interfering RNAs (siRNAs) are two key components of RNA interference within cells. siRNAs are derived by processing of long double-stranded RNAs and are often of exogenous origin and degrade mRNAs bearing fully complementary sequences (1). In contrast, miRNAs are endogenously encoded small noncoding RNAs, derived by processing of short RNA hairpins, which can inhibit the translation of mRNAs bearing partially complementary target sequences (1). miRNAs are endogenous and naturally generated in animal cells. For this reason, the use of miRNAs is more applicable in developing therapeutics that can regulate mRNA in animal cells.
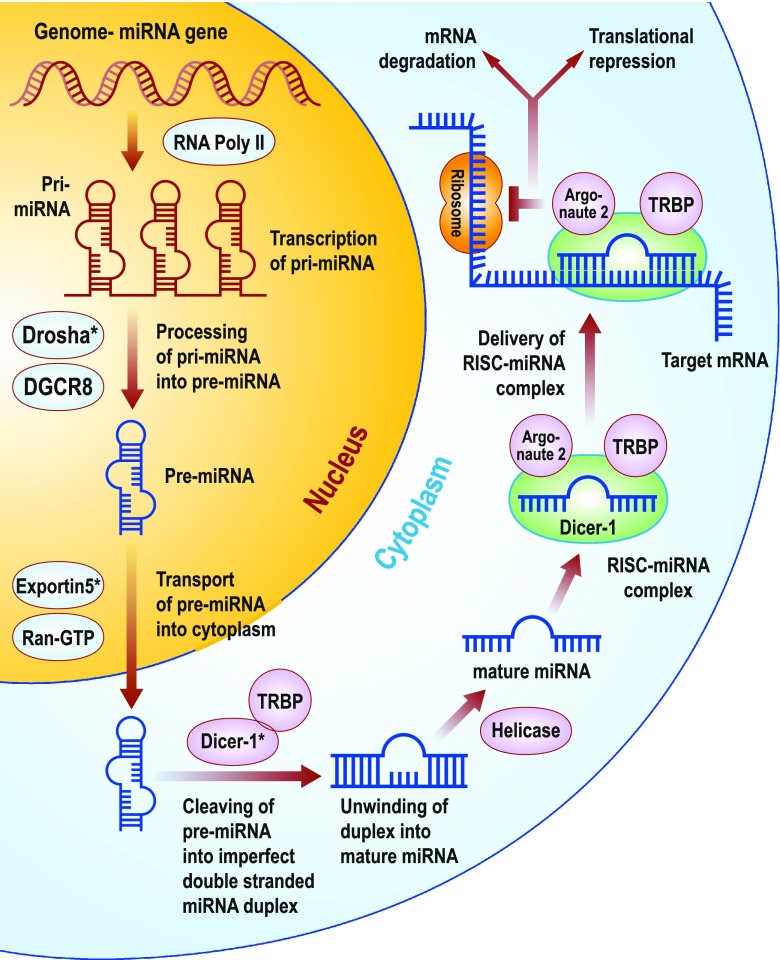
MiRNA biogenesis has been studied by many investigators. A schematic overview of miRNA biogenesis is given in Fig. 1. MiRNAs are transcribed by RNA polymerase II enzyme producing a long primary-miRNA (pri-miRNA) (2). These pri-miRNAs contain a cap structure at the 5′ end and are poly-adenylated at the 3′ end, suggesting that pri-miRNAs are structurally and functionally similar to mRNAs (3). Also, pri-miRNAs contain specific hairpin-shaped stem-loop structures of ~70 nucleotides that are recognized and cleaved by a ~650-kDa nuclear microprocessor complex consisting of the RNase III endonuclease Drosha and the essential DiGeorge syndrome critical region gene 8 (DGCR8) binding protein (4). The resulting ~70 nucleotide hairpin intermediate (pre-miRNA) is transported out of the nucleus and into the cytoplasm by Exportin-5 and its cofactor Ran-GTP (5). In the cytoplasm, the pre-miRNAs are further cleaved by a second RNase III endonulease Dicer-1 and its essential transactivating response RNA binding protein (TRBP) producing a short imperfect double-stranded miRNA duplex. The imperfect miRNA duplex is then unwound into a mature miRNA by helicase. Next, TRBP recruits the catalytic Argonaute 2 to the Dicer complex with the mature miRNA forming the RNA-induced silencing complex (RISC) (6, 7). RISC then regulates gene expression by mRNA degradation or translational repression (8–10). Therefore, miRNAs negatively regulate gene and protein expression via the RNA interference (RNAi) pathway. miRNA is different from siRNA in that miRNA represses mRNA with complementary sequence in the 3′-untranslated region (3′-UTR) (11), although miRNA may also target coding regions of mRNA, at least in animals (12).
Fig. 1.
An overview of miRNA biogenesis. MiRNAs are a class of endogenous non-protein-coding RNAs that negatively regulate gene and protein expression via the RNAi pathway. Figure modified from Chen, C.Z., et al. N Engl J Med, 2005; 353(17):1768–71
MicroRNA and Regulation of Stem Cells
MiRNAs have been proposed to be important factors in stem cell function. One reason for this is that expression levels of certain miRNAs in stem cells are different from other normal tissues (13). This implies that miRNAs may have a unique role in stem cell regulation. In order to confirm that miRNAs do indeed regulate stem cell function, many investigators have used Dicer-1 (dcr-1) mutants. Dicer-1 plays a specialized role in the biogenesis of miRNAs and therefore can offer great insight into the role of miRNAs in stem cells. Loss of dcr-1 function in mouse models resulted in animal death early in development and depletion of stem cells in mouse embryos, suggesting that the disrupted miRNA pathway plays a large role in maintaining the stem cell population (14). Mutated dcr-1 gene in embryonic stem cells in mice leads to a reduced expression of miRNAs and displayed severe defects in embryonic stem cell differentiation in vivo and in vitro. Re-expression of Dicer-1 in the knockout cells rescued the phenotypes (15). Another study observed a reduction in cyst production in Drosophila germline stem cell mutants for dcr-1 and a delay in the transition from G1 to S phase, which is dependent on the cyclin-dependent kinase inhibitor Dacapo (16). This finding that miRNAs are required for stem cell division suggests that miRNAs are needed to make stem cells insensitive to environmental signals in order to overcome the normal G1/S checkpoint (16). The use of dcr-1 mutants has resulted in a great multitude of data and findings that implicate the important role that miRNAs play in stem cell function.
Cancer Stem Cell Hypothesis
Stem cells are defined by their ability to undergo self-renewal, as well as multi-lineage differentiation (17). Adult stem cells are found in numerous tissues of the body and play a role in tissue development, replacement, and repair (18). In this review, cancer cells are defined as cells that are part of a malignancy. Recently, there has been a dramatic increase in research geared towards a small subpopulation of cells identified in cancers that have stem cell properties. The cancer stem cell hypothesis proposes that cancers are derived from a small fraction of cancer cells that constitute a reservoir of self-sustaining cells with the exclusive ability to self-renew and initiate/maintain the tumor (19). Thus, according to the cancer stem cell hypothesis, these cancer stem cells are tumor-initiating cells that proliferate through their unique self-renewal ability. Cancer stem cells were first identified in leukemia (20, 21). Recently, many investigators have identified cancer stem cells in solid tumors including breast, brain, pancreas, colon, and head and neck cancers (22–35). These new discoveries provide further support for the cancer stem cell hypothesis. In some types of human cancers, such as melanoma, such tumorigenic cells may not be rare (36).
The cancer stem cell hypothesis established a basis for future studies, as well as presented a better understanding of the biology and intricacies of cancer and tumor formation. To be maximally effective, cancer therapy must also be directed against both the resting cancer stem cells and the proliferating cancer cells (37). This may be possible if specific stem cell signals are inhibited using molecular therapy, while at the same time attacking proliferating cells by conventional therapies (38, 39).
Self-Renewal
Using different systems, many investigators have demonstrated that only a small minority of cells in human cancers are capable of self-renewal. Self-renewal is distinguished from other proliferating processes in that at least one of the progeny is identical to the initial stem cell (22). Specifically, asymmetric stem cell self-renewal produces two different progenies. The first progeny is identical to the original stem cell—thus, maintaining stem cell number—and the other progeny produced is a committed progenitor cell, which undergoes cellular differentiation (22). Both the self-renewal and differentiation of normal stem cells are regulated by the stem cell microenvironment, which has been termed the stem cell niche (39, 40). In order to study the self-renewal potential, a mammosphere assay has been developed that plates cells in a serum-free medium with growth factor supplementation on a non-adherent substrata followed by quantification of sphere formation (41). Using this method, one study found that secondary mammospheres from the human breast cancer Lin−CD29HCD24H cell subgroup as determined from seven independent tumors were larger in size and number compared with all other subpopulations, suggesting the ability for tumor-initiating cells to undergo self-renewal (42). Therefore, a certain subpopulation of cancer cells is able to self-renew and initiate tumor formation, thus coining the term “cancer stem cells.” The central feature of cancer stem cells is this relatively unlimited asymmetric self-renewal (22). Self-renewal of cancer stem cells could be a likely cause of resistance of current cancer treatment, as well as relapse in cancer patients. One recent study provided the first clinical evidence for the implication of a “glioma stem cell” or “self-renewal” phenotype in treatment resistance of glioblastoma (43). It is believed that genetic alterations cause dysregulation in cancer stem cells, resulting in unlimited self-renewal capabilities. Abnormal stem cell self-renewal is a likely requirement for the initiation, formation and resistance of cancer.
Signaling Pathways of the “Stem Cell Genes”
There is growing evidence that illustrates that many pathways classically connected with cancer may also regulate normal stem cell development (44). The pivotal signaling pathways of the “stem cell genes” Notch, Hedgehog, Wnt/β-catenin, HMGA2, Bcl-2, and Bmi-1 are involved in the regulation of self-renewal, differentiation, and survival of cancer stem cells (44–46). These key signaling pathways, which may be dysregulated in cancer stem cells, offer great promise for future cancer therapies and treatments.
Notch
The Notch signaling pathway is a short-range communication transducer that is involved in regulating many cellular processes during development and renewal of adult tissues. Notch signaling has been highlighted as a pathway that aids in development of the breast and is frequently dysregulated in invasive breast cancer (18). It was also demonstrated that Notch signaling can act on mammary stem cells to promote self-renewal and on early progenitor cells to promote their proliferation (28). These effects were also shown to be completely inhibited by either a Notch 4 antibody or a gamma secretase inhibitor that blocks Notch processing (28). These findings suggest that atypical Notch signaling could lead to dysregulation of the self-renewal properties of cancer stem cells, thus resulting in carcinogenesis and oncogenesis (47).
Hedgehog
The importance of Hedgehog signaling in carcinogenesis revolves around Hedgehog’s effect on cancer stem cell self-renewal. Hedgehog (specifically Sonic Hedgehog) signaling has been implicated in the regulation of self-renewal characteristics by the finding that populations enriched for human hematopoietic stem cells exhibit increased self-renewal in response to Sonic Hedgehog stimulation in vitro, albeit in combination with other growth factors (44, 48). In humans, several distinctive cancers, including basal-cell carcinoma, result from mutations that aberrantly activate Hedgehog signal transduction (49). It has been shown that Drosophila ovarian stem cells cannot proliferate as stem cells in the absence of Hedgehog signaling, whereas excessive Hedgehog signaling produces supernumerary stem cells, implying that Hedgehog is a stem-cell factor (49). This suggests that human cancers due to excessive Hedgehog signaling might result from dysregulated self-renewal properties of cancer stem cells.
Wnt/β-catenin
Another pathway that regulates both self-renewal and oncogenesis in different tissues is the Wnt/β-catenin signaling pathway (44). Activation of the Wnt receptor causes an accumulation of β-catenin and other Wnt gene family proteins in the cytoplasm, which eventually translocates into the nucleus. The nuclear translocation of β-catenin drives the expression of genes associated with self-renewal. Over-expression of activated β-catenin expands the pool of stem cells (50). Dysregulation in the Wnt/β-catenin signaling pathway contributes to the onset of cancer. Gain or loss-of-function mutations of several members of this pathway have been found in many types of human tumors (51, 52). Another study showed that, in chronic myelogenous leukemia, β-catenin accumulates in the nuclei of granulocyte–macrophage progenitors, seemingly enhancing the self-renewal activity and leukemic potential of these cells (53). Thus, dysregulation of this pathway within cancer stem cells may be associated with the acquisition of self-renewal properties.
HMGA2
HMGA2 has also been implicated in survival and self-renewal of cancer stem cells. HMGA2 is thought to play a role in modulating macromolecule complexes that are involved in many biological processes, including binding directly to the DNA and aiding in the regulation of many genes (54). The expression of HMGA proteins during embryogenesis suggests that they have important functions in development (54). Moreover, the HMGA2 gene is suggested to control growth, proliferation, and differentiation (54). HMGA2 has also been implicated in cancer. HMGA2 overexpression has been found in lung and pancreatic carcinomas (55, 56). HMGA2 protein overexpression is usually met with the presence of metastasis and reduced survival of the cancer patient (54). Thus, HMGA2’s role in embryogenesis and aggressive cancers suggests that human cancers due to excessive HMGA2 signaling might result from dysregulated cell survival and self-renewal properties of cancer stem cells.
Bmi-1
The significance of Bmi-1 signaling in carcinogenesis revolves around Bmi-1’s effect on cancer stem cell self-renewal. Bmi-1 was shown to be expressed in neural stem cells and proliferating progenitor cells, but not in differentiated cells (45). Loss of Bmi-1 resulted in a drastic decrease in neural stem cell proliferation and self-renewal (45). This suggests that Bmi-1 is necessary for stem cell self-renewal. Bmi-1 has also been implicated in cancer. Bmi-1 was identified to promote the generation of lymphomas (57, 58). This demonstrates that Bmi-1 plays a role in cancer development. Bmi-1 seems to be important in both stem cells and cancer. Bmi-1 was found to be activated in human breast “cancer stem cells” characterized as CD44+CD24−/lowLin− (59). Furthermore, Bmi-1 was found to mediate the mammosphere-initiating cell number and mammosphere size, supporting a role in the regulation of self-renewal of normal and tumorigenic human mammary stem cells (59). Therefore, dysregulation of this Bmi-1 pathway within cancer stem cells may be associated with the acquisition of self-renewal properties.
Bcl-2
Bcl-2 has been researched by many investigators because of its role within cancer cells as a proto-oncogene. Bcl-2 is over-expressed in many cancers, leading to a prevention of apoptosis. It has been shown that this obstacle to apoptosis due to over-expression of Bcl-2 results in an increased number of stem cells in vivo (60). This suggests that apoptosis plays a role in regulating the microenvironments of stem cells (61). Therefore, the Bcl-2 signaling pathway is very important to the survival of stem cells, especially cancer stem cells, because of the overexpression of Bcl-2 in cancers (62).
Link Between miRNA and Cancer Stem Cells
Aberrant expressions of miRNAs are connected to human diseases, such as cancer. Tumors analyzed by miRNA profiling have shown significantly different miRNA profiles (for mature and/or precursor miRNAs) compared with normal cells from the same tissue (63). It has also been shown by convincing evidence that miRNAs are important factors in stem cell biology. Thirty-six unique miRNAs (from 32 stem-loops) have been identified by cDNA cloning to be specifically expressed in human embryonic stem cells relative to their differentiated embryoid bodies (13). The obvious parallel that can be drawn is that undifferentiated stem cells display miRNA expression profiles reminiscent of cancer cells (19). There is also overwhelming evidence that a distinct subpopulation of cancer cells have the ability of self-renewal, and thus act as cancer stem cells within tumors (64). Knowing that aberrant gene function and expression are key characteristics in cancer, it is thought that acquired epigenetic abnormalities participate in genetic alterations, causing dysregulation in cancer stem cells (51). This dysregulation allows them to escape the restrictions of the stem cell niche, resulting in unlimited self-renewal ability and potential. It is believed that microenvironmental factors or signals account for the epigenetic abnormalities in cancer stem cells. These signals interfere with gene expressions, resulting in the silencing of some genes. Therefore, there must be some underlying sub-cellular process that accounts for this dysregulation in cancer stem cells.
One pathway of investigative relevance is the RNAi pathway. The RNAi pathway is important because it silences gene expression at transcription or translation. MiRNAs have been implicated in the RNAi pathway by negatively regulating gene and protein expression at the post-transcriptional level. Altered expression of specific miRNA genes contributes to the initiation and progression of cancer (11). Disruption of miRNA expression levels in tumor cells may result from distorted epigenetic regulation of miRNA expression, abnormalities in miRNA processing genes or proteins, and the location of miRNAs at cancer-associated genomic regions (63). Clearly, miRNAs play a critical role in carcinogenesis and oncogenesis. Emerging evidence suggests that certain abnormal miRNA expression levels cause cancer stem cell dysregulation, resulting in unlimited self-renewal and cancer progression. Therefore, miRNA expression is a vital key to cancer stem cell dysregulation.
In addition, a number of miRNAs have been identified within cancers to function as either oncogenes or tumor suppressors (65). These miRNAs offer great promise for cancer therapy because they might have the potential to regulate aberrant miRNA expression. Therefore, miRNA therapy could be a powerful tool to address cancer stem cell dysregulation and its resulting self-renewal and cancer progression in patients.
Examples of Potential Links Between miRNA and Cancer Stem Cells
Oncogenes
Many events can trigger cancer formation within the body. Over the past few years, research in the area of cancer has shown that there are aberrant levels of certain miRNAs in cancer stem cells, resulting in dysregulation of these cancer stem cells. This cancer stem cell dysregulation may explain how carcinogenesis and oncogenesis progress. MiRNAs either act as oncogenes or tumor suppressors in cancer cells. Oncogenic miRNAs are often called oncomiRs and are up-regulated in the cancer cells.
It has been shown that miRNA miR-21 is over-expressed in breast tumor tissues and functions as an oncogene by modulating tumorigenesis through the regulation of Bcl-2 and Programmed Cell Death 4 (PDCD4) (66–69). Thus, miR-21 over-expression leads to up-regulation of Bcl-2, which results in increased tumor growth and decreased apoptosis (69).
The miR-17-92 cluster, which is comprised of seven miRNAs, is markedly over-expressed in lung cancers and could play a role as an oncogene (70). Enforced expression of the mir-17-92 cluster acted with c-Myc expression to accelerate tumor development in a mouse B-cell lymphoma model (71). Introduction of miR-17-92 into hematopoietic stem cells was shown to significantly accelerate the formation of lymphoid malignancies (70). Other evidence has proposed the potential targets of miR-17-92 include E2F1 (which promotes cell proliferation) and the tumor-suppressor genes PTEN (which promotes apoptosis) and RB2 (72). Interestingly, a functional relationship between miR-17-92 and the Sonic Hedgehog signaling pathway was studied. In engineered medulloblastomas, miR-17-92-induced tumors were shown to have activated the Sonic Hedgehog signaling pathway (73). This is thought to result in increased self-renewal. Taken together, these studies implicate the miR-17-92 cluster as a potential human oncogene that plays a role in cancer stem cells.
miR-135 has an oncogenic role within cancer as well. miR-135a and miR-135b were found to be greatly up-regulated in colorectal adenomas and carcinomas (74). APC, a gene found to lead to truncated proteins that have lost their β-catenin binding sites, was down-regulated in cancers with increased expression of miR-135a&b (74). If APC is not expressed to the proper level, an accumulation of β-catenin would occur, leading to the activation of self-renewal genes. Thus, these oncogenes miR-135a&b play a vital role in controlling Wnt signaling pathway. Consequently, miR-135a and miR-135b may play vital roles in cancer stem cells themselves. Over-expression of these oncomiRs leads to further cancer progression.
Tumor Suppressors
In oncogenesis, some miRNAs’ expression is decreased in cancerous cells (8). These miRNAs are called tumor suppressor miRNAs. In this review, tumor suppressor miRNAs are termed TSmiRs. TSmiRs are supposed to prevent tumor development; however, their expression in cancer is down-regulated, resulting in increased progression of the disease.
One example of tumor suppressor miRNAs is let-7. Let-7 expression levels are reduced in various lung cancer cell lines and pulmonary tumors, relative to normal lung samples (75, 76). Let-7 expression is lower in lung tumors than in normal lung tissue, while RAS protein is significantly higher in lung tumors, suggesting that let-7 negatively regulates RAS protein (77). RAS appears to be important for self-renewal since silencing RAS reduces mammosphere formation, clonal expansion, and tumorigenicity (78). Chromosomal translocations previously associated with human tumors disrupt repression of HMGA2 by let-7 miRNA, suggesting that let-7 also negatively regulates HMGA2 (79). The let-7 family is not expressed in breast tumor-initiating cells (78). By expressing let-7 in breast tumor-initiating cells, it was found that let-7 regulates the key features of breast cancer stem cells—self renewal in vitro, multipotent differentiation, and the ability to form tumors (78). Thus, let-7 is a tumor suppressor that negatively regulates RAS protein and HMGA2 and plays an important role in the self-renewal potential of cancer stem cells.
miR-15a and miR-16-1 are both tumor suppressors. In the majority of leukemic cells, both miR-15a and miR-16-1 were expressed at low levels and Bcl-2 was over-expressed (80). It was also shown that down-regulation of Bcl-2 by miR-15a and miR-16-1 triggers apoptosis and that the levels of expression of these two miRNAs are important (80). Also in prostate cancer cells, down regulation of these two miRNAs resulted in an up regulation of WNT3A, a Wnt gene family protein (81). WNT3A help promote cancer cell proliferation and invasiveness of their respective tumors (81). These TSmiRs are critical to suppressing tumor growth and progression. Thus, miR-15a and miR-16-1 play extremely vital roles in both the Bcl-2 and Wnt signaling pathways, which are essential for self-renewal potential in cancer stem cells.
MiRNA-128 is also a tumor suppressor involved in cancer stem cells. In high-grade gliomas, miR-128 levels were significantly reduced, suggesting tumor suppressor properties (82). Glioma cell proliferation and growth were inhibited by miR-128 introduction (82). Later, the mechanism behind miR-128’s tumor suppressor characteristics was found. Expression of miR-128 caused a down regulation of Bmi-1 signaling pathway levels (82). Thus, miR-128 specifically blocked glioma self-renewal via Bmi-1 down-regulation. miR-128’s regulation of the Bmi-1 signaling pathway demonstrates the importance of it in the self-renewal capacity in cancer stem cells.
MiRNA-199b-5p is a tumor suppressor of great intrigue. Expression of miR-199b-5p was shown to be lost in metastatic cancer patients (83). In medulloblastoma cells, miR-199b-5p was found to down-regulate the expression of HES1, a transcription factor of the Notch signaling pathway (83). Thus, miR-199b-5p should lead to a decrease of the self-renewal properties of cancer stem cells. Introduction of an over-expression of miR-199b-5p did indeed block Notch signaling, as well as decrease the medulloblastoma stem-cell-like (CD133+) subpopulation of cells (83). Therefore, the tumor suppressor miR-199b-5p is extremely important to the cancer stem cell self-regulation potential via the Notch signaling pathway.
Furthermore, miR-125b, miR-326, and miR-324-5p are all tumor suppressors. Using miRNA profile screening of human medulloblastoma cells, these miRNAs were found to be down-regulated where the Hedgehog signaling pathway was elevated (84). miR-125b, miR-326, and miR-324-5p were all shown to suppress Smo, an activator component of the Hedgehog pathway, and only miR-324-5p was also found to suppress Gli-1, another activator component of the Hedgehog signaling pathway (84). All three of these miRNAs were found to inhibit cancer cell growth (84). This is consistent with the fact that the Hedgehog signaling pathway is involved in the self-renewal potential of cancer stem cells.
MiRNA-34 is a tumor suppressor of great interest. This TSmiR is down-regulated in several types of cancer (85). We found that, in p53-deficient human gastric and pancreatic cancer cells, restoration of functional miR-34 inhibits cell growth and induces G1 block and apoptosis, indicating that miR34 may restore p53 function (62, 86). miR-34 restoration inhibited tumorsphere growth in vitro and tumor initiation in vivo, which is reported to be correlated to the self-renewal of cancer stem cells (62, 86). The mechanism of miR-34-mediated suppression of self-renewal appears to be related to the direct modulation of downstream targets—Bcl-2, Notch, and HMGA2—indicating that miR-34 may be involved in gastric cancer cells’ self-renewal/differentiation decision-making (62, 86). Thus, miR-34 is a significant tumor suppressor of cancer stem cells by regulating both apoptosis and self-renewal properties. Decreased expression of these TSmiRs leads to further cancer progression.
Examples Support the Role of “Stem Cells miRNAs” in Oncogenesis and the Implications in Molecular Cancer Therapy
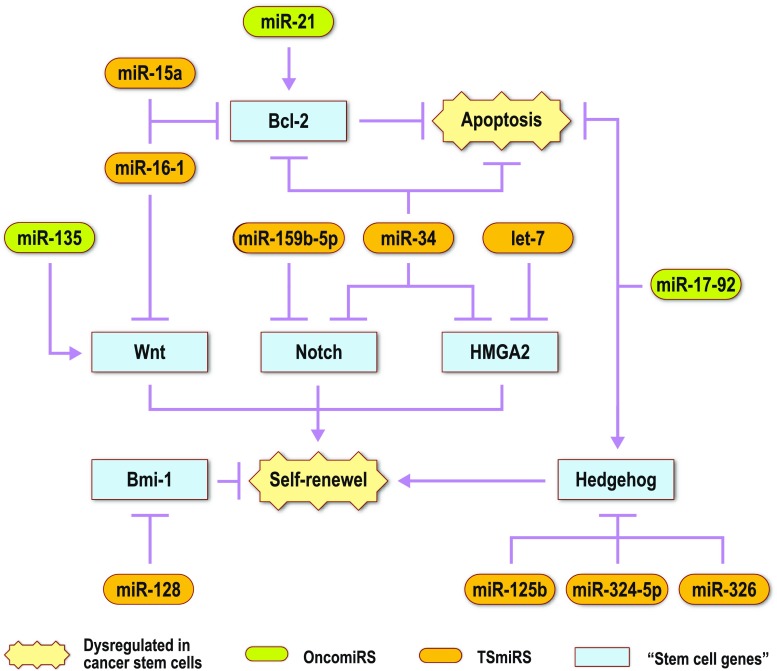
It is clear that these “stem cell miRNAs” play a vital purpose in the regulation of the discussed “stem cell genes” and their subsequent signaling pathways in cancer. Figure 2 provides a schematic view of these “stem cell miRNAs” and their interactions with “stem cell genes” in cancer stem cells.
Fig. 2.
Potential “stem cell miRNAs” that modulate “stem cell genes” related to cancer stem cells. Certain miRNAs have been shown to be aberrantly expressed in cancer. OncomiRs, which initiate cancer development, are over-expressed. TSmiRs, which prevent tumor development, are decreased. These miRNAs regulate genes that are implicated in stem cells. The aberrant expression of these potential “stem cell miRNAs” in cancer suggests that dysregulation of “stem cell genes” leads to increased levels of self-renewal and decreased levels of apoptosis within cancer stem cells. This results in further cancer progression
These “stem cell miRNAs” support the potential link between miRNAs and cancer stem cells. Figure 3 outlines this potential link between miRNAs and cancer stem cells. All of these examples suggest that miRNAs have a pivotal function in carcinogenesis and oncogenesis by regulating self-renewal and apoptosis via cancer stem cell signaling pathways as oncogenes or tumor suppressors, respectively. These oncogenic and tumor suppressor miRNAs lead to a better understanding of cancer stem cell biology, and thus, a greater knowledge of how cancer starts and progresses into malignant tumor formation. Dysregulation of cancer stem cells allows them to escape the restrictions of the stem cell niche, resulting in unlimited self-renewal ability and potential, which results in further cancer development and resistance to current treatments. It has been demonstrated that aberrant expression of certain miRNAs are not only connected to cancers in general, but these aberrant miRNA expressions are involved in cancer stem cell dysregulation. Functional studies of specific miRNAs within the cancer stem cells of various cancers are crucial for the elucidation of the mechanisms behind oncogenesis in various cancers (87). Some miRNAs are up-regulated in cancer stem cells and act as oncogenes. These oncogenes should be targeted with treatments that knockdown their expression. Other miRNAs suppress cell proliferation by nature, acting as tumor suppressors, but are down-regulated in cancer stem cells, resulting in cancer progression. These tumor suppressors should be targeted with therapies that restore their tumor suppressor capabilities within the cancer stem cells. Therefore, addressing these abnormal miRNA expression levels with molecular miRNA therapy could be a powerful tool to tackle cancer stem cell dysregulation and, hence, oncogenesis.
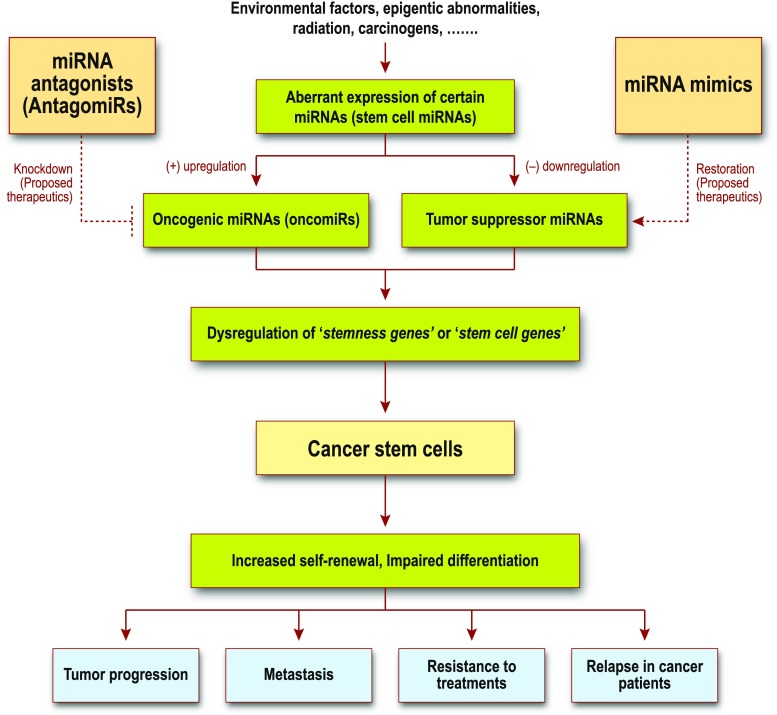
Fig. 3.
Link between miRNA and cancer stem cells. Aberrant expressions of miRNAs, either as oncogenic or tumor suppressor miRNAs, can lead to dysregulation of stem cell genes, causing increased self-renewal potential and impaired differentiation in cancer stem cells. This dysregulation subsequently results in carcinogenesis and oncogenesis. It is proposed that miRNA antagonists can knockdown the effects of oncogenic miRNAs and miRNA mimics can restore the capabilities of tumor suppressor miRNAs. Therefore, miRNA could be a vital tool in addressing cancer stem cell dysregulation. MiRNA-based molecular therapy could hold great therapeutic potential against cancer progression, resistance, and relapse
MicroRNA Therapeutics
MiRNAs are very promising as therapeutic targets for anti-cancer treatments because their aberrant expressions are linked to cancer stem cell dysregulation and, thus, oncogenesis. MiRNA-based molecular cancer therapy should eliminate the self-renewal capabilities of the cancer stem cells and greatly reduce the resistance of current cancer treatment, as well as relapse in cancer patients.
Development of miRNA/RNAi-based therapeutics requires several critical experimental steps, which include: (1) miRNA profiling of cancer versus healthy tissue, and especially cancer stem cells versus the differentiated cells, (2) functional analysis of dysregulated miRNAs, and (3) in vivo studies with use of different RNAi-based therapeutic methods address aberrant miRNA expressions (19).
For oncogenic miRNAs, which promote cancer when over-expressed, an antagomiR should be used to block the effects of the oncomiR (88). The antagomiR knocks down the oncogenic properties of the miRNA, resulting in cancer suppression and decreased progression. For example, to knock down the expression of the oncogene miR-21, an anti-miR-21 oligonucleotide was transfected into breast cancer MCF-7 cells (69). It was demonstrated that the anit-miR-21 suppressed both cell growth in vitro and tumor growth in a xenograft mouse model by increasing apoptosis and decreasing cell proliferation (69). Therefore, antagomiRs are promising as therapeutic targets for oncogenic miRNA-based cancer stem cell dysregulation.
For tumor suppressor miRNAs, which promote cancer when under-expressed, miRNA mimics or lentiviruses should be used to restore the tumor suppressors’ natural potential, resulting in decreased cancer development. For example, to re-introduce miR-34 and its tumor suppressor capabilities, we transfected miR-34 mimics into cancer cells, and the mimic was shown to block the cell cycle in the G1 phase, significantly increase activation of caspase-3, and knock down its downfield targets of bcl-2, Notch, and HMGA2 (86). The miRNA mimic, thus, restored miR-34 with its tumor suppressor potential; however, the transfection of the miR-34 mimics can only last a couple of days and the long-term biological effects were not observed very effectively. To overcome this dilemma, the cancer cells were infected with a lentivirus that expressed miR-34a. This generated stable cells expressing miR-34a. The lentiviral miR-34a was found to be able to inhibit cancer cell growth and tumorsphere formation (86). The lentiviral system restored the tumor suppressor effect of miR-34 in pancreatic cancer stem cells as well (62). Therefore, miRNA mimics and lentiviral miRNAs show great potential in restoring tumor suppressor miRNAs to correct the dysregulation of “stem cell genes” in cancer stem cells.
The Challenge of miRNA—Therapeutics: Delivery, Delivery, Delivery
However, from a clinical/translational research point of view, for the miRNA-based therapeutics to be effective, the efficient and functional delivery of miRNA mimics and/or antagonists to tumor remains a great challenge.
Current approaches to deliver gene- and RNAi-based therapeutics employ either viral or non-viral vector systems (89). Viral vector-directed methods show high gene transfer efficiency but are deficient in several areas. The limitations of a viral approach are related to their lack of tumor targeting and to residual viral elements that can be immunogenic, cytopathic, or recombinogenic (89). Non-viral gene transfer vectors could circumvent some of the problems associated with viral vectors. Progress has been made toward developing non-viral, pharmaceutical formulations of gene therapeutics for in vivo human therapy, particularly cationic liposome-mediated gene transfer systems (89, 90). Cationic liposomes are composed of positively charged lipid bilayers and can be complexed to negatively charged, naked DNA by simple mixing of lipids and DNA such that the resulting complex (lipoplex) has a net positive charge (90). The lipoplex is easily bound and taken up by cells with relatively high transfection efficiency. Features of cationic liposomes that make them versatile and attractive for DNA delivery include: simplicity of preparation; the ability to complex large amounts of DNA; versatility in use with any type and size of DNA or RNA; the ability to transfect many different types of cells, including non-dividing cells; and lack of immunogenicity or biohazardous activity (89, 90). There are multiple clinical trials now underway using cationic liposomes for gene delivery, and liposomes for delivery of chemotherapeutics such as doxorubicin are already on the market for breast cancer chemotherapy.
One disadvantage of cationic liposomes is that they lack tumor specificity and have relatively low transfection efficiencies as compared to viral vectors. However, this can be dramatically increased when the lipoplexes bear a ligand recognized by a cell surface receptor (89, 90). Receptor-mediated endocytosis represents a highly efficient internalization pathway in eukaryotic cells. The presence of a ligand on a lipoplex facilitates the entry of DNA into cells through initial binding of ligand by its receptor on the cell surface followed by internalization of the bound lipoplex (89). Once internalized, sufficient DNA escapes the endocytic pathway to be expressed in the cell nucleus. A variety of ligands have been examined for their lipoplex-targeting ability (89, 90).
Recently, we developed tumor-specific, ligand-targeting, self-assembled, nanoparticle–DNA lipoplex systems designed for systemic gene therapy of cancer (US Patent No. 6,749,863, European Patent No. EP 1,154,756) (91, 92). These nanovector systems employ transferrin (Tf) or scFv against transferrin receptor (TfR), which is over-expressed in the majority of human cancers, as tumor-targeting ligand (91, 92). When using Tf as a targeting ligand, we obtained the self-assembled nanovectors at the sizes of 50–90 nm, with highly compact structure and favorite surface charge (91). These nanovectors have novel nanostructure that resembles a virus particle with a dense core enveloped by a membrane coated with Tf molecules spiking on the surface (91). This nanovector system shows promising efficiency and specificity in targeted delivery of various genes and anti-sense oligonucleotides to cancer in vivo but not normal tissues (93, 94). Systemic p53 gene therapy using these nanovector systems demonstrated long-term therapeutic efficacy in animal models of human cancers (90–92, 95). Tf- and TfR-scFv-targeted nanovectors were recently approved by the FDA for clinical testing, and the first Phase-I clinical trial for non-viral systemic p53 gene therapy is ongoing (www.ClinicalTrials.gov). The success of these nanovectors for systemic p53 gene therapy, and more recently Her-2 siRNA therapy (96–98), provide a promising, tumor-targeted delivery system for novel RNAi-based therapies, such as miRNA-therapeutics discussed above.
CONCLUSIONS
Abnormal miRNA expressions are connected to cancer stem cell dysregulation. This dysregulation leads to the initiation, development, and progression of cancer. Consequently, molecular miRNA therapy is very important to addressing oncogenesis linked with cancer stem cell dysregulation. For this reason, future research should be aimed at validating the link between miRNAs and cancer stem cells, investigating miRNAs’ role in cancer stem cells’ self-renewal pathways, as well as studying therapeutic potential of miRNAs against cancer progression, resistance, and relapse.
Acknowledgements
We wish to thank Mr. Steven Kronenberg for graphical support and expertise in producing the figures. This review was supported in part by NIH grants CA121830, CA128220, and CA134655 (to L. X.). J. D. is a University of Michigan Undergraduate Research Opportunity Program (UROP) student.
References
- 1.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci U S A. 2003;100:9779–84. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–66. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, et al. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–7. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–4. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chekanova JA, Belostotsky DA. MicroRNAs and messenger RNA turnover. Methods Mol Biol. 2006;342:73–85. doi: 10.1385/1-59745-123-1:73. [DOI] [PubMed] [Google Scholar]
- 11.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 12.Rigoutsos I. New tricks for animal microRNAS: targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res. 2009;69:3245–8. doi: 10.1158/0008-5472.CAN-09-0352. [DOI] [PubMed] [Google Scholar]
- 13.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–98. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 15.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–8. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 17.Dontu G, Al-Hajj M, Abdallah WM, Clarke MF, Wicha MS. Stem cells in normal breast development and breast cancer. Cell Prolif. 2003;36(Suppl 1):59–72. doi: 10.1046/j.1365-2184.36.s.1.6.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farnie G, Clarke RB. Mammary stem cells and breast cancer—role of Notch signalling. Stem Cell Rev. 2007;3:169–75. doi: 10.1007/s12015-007-0023-5. [DOI] [PubMed] [Google Scholar]
- 19.Papagiannakopoulos T, Kosik KS. MicroRNAs: regulators of oncogenesis and stemness. BMC Med. 2008;6:15. doi: 10.1186/1741-7015-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dick JE. Normal and leukemic human stem cells assayed in SCID mice. Semin Immunol. 1996;8:197–206. doi: 10.1006/smim.1996.0025. [DOI] [PubMed] [Google Scholar]
- 21.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167–74. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 22.Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274–82. doi: 10.1038/sj.onc.1207947. [DOI] [PubMed] [Google Scholar]
- 23.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–51. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 24.Lawson DA, Witte ON. Stem cells in prostate cancer initiation and progression. J Clin Invest. 2007;117:2044–50. doi: 10.1172/JCI32810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang DG, Patrawala L, Calhoun T, Bhatia B, Choy G, Schneider-Broussard R, et al. Prostate cancer stem/progenitor cells: identification, characterization, and implications. Mol Carcinog. 2007;46:1–14. doi: 10.1002/mc.20255. [DOI] [PubMed] [Google Scholar]
- 26.Patrawala L, Calhoun-Davis T, Schneider-Broussard R, Tang DG. Hierarchical organization of prostate cancer cells in xenograft tumors: the CD44 + alpha2beta1+ cell population is enriched in tumor-initiating cells. Cancer Res. 2007;67:6796–805. doi: 10.1158/0008-5472.CAN-07-0490. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Chen X, Calhoun-Davis T, Claypool K, Tang DG. PC3 human prostate carcinoma cell holoclones contain self-renewing tumor-initiating cells. Cancer Res. 2008;68:1820–5. doi: 10.1158/0008-5472.CAN-07-5878. [DOI] [PubMed] [Google Scholar]
- 28.Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004;6:R605–15. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104:973–8. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 32.Szotek PP, Pieretti-Vanmarcke R, Masiakos PT, Dinulescu DM, Connolly D, Foster R, et al. Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian Inhibiting Substance responsiveness. Proc Natl Acad Sci U S A. 2006;103:11154–9. doi: 10.1073/pnas.0603672103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 34.Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6:425–36. doi: 10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- 35.Bussolati B, Grange C, Sapino A, Camussi G. Endothelial Cell Differentiation of Human Breast Tumor Stem/Progenitor Cells. J Cell Mol Med. 2009;13:309–19. doi: 10.1111/j.1582-4934.2008.00338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–8. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rich JN. Cancer stem cells in radiation resistance. Cancer Res. 2007;67:8980–4. doi: 10.1158/0008-5472.CAN-07-0895. [DOI] [PubMed] [Google Scholar]
- 38.Al-Hajj M. Cancer stem cells and oncology therapeutics. Curr Opin Oncol. 2007;19:61–4. doi: 10.1097/CCO.0b013e328011a8d6. [DOI] [PubMed] [Google Scholar]
- 39.Wicha MS. Cancer stem cells and metastasis: lethal seeds. Clin Cancer Res. 2006;12:5606–7. doi: 10.1158/1078-0432.CCR-06-1537. [DOI] [PubMed] [Google Scholar]
- 40.Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang M, Behbod F, Atkinson RL, Landis MD, Kittrell F, Edwards D, et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 2008;68:4674–82. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–24. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 44.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 45.Zencak D, Lingbeek M, Kostic C, Tekaya M, Tanger E, Hornfeld D, et al. Bmi1 loss produces an increase in astroglial cells and a decrease in neural stem cell population and proliferation. J Neurosci. 2005;25:5774–83. doi: 10.1523/JNEUROSCI.3452-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 47.Hambardzumyan D, Becher OJ, Holland EC. Cancer stem cells and survival pathways. Cell Cycle. 2008;7:1371–8. doi: 10.4161/cc.7.10.5954. [DOI] [PubMed] [Google Scholar]
- 48.Bhardwaj G, Murdoch B, Wu D, Baker DP, Williams KP, Chadwick K, et al. Sonic hedgehog induces the proliferation of primitive human hematopoietic cells via BMP regulation. Nat Immunol. 2001;2:172–80. doi: 10.1038/84282. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Kalderon D. Hedgehog acts as a somatic stem cell factor in the Drosophila ovary. Nature. 2001;410:599–604. doi: 10.1038/35069099. [DOI] [PubMed] [Google Scholar]
- 50.Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–14. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 51.Zhao RC, Zhu YS, Shi Y. New hope for cancer treatment: Exploring the distinction between normal adult stem cells and cancer stem cells. Pharmacol Ther. 2008;119:74–82. doi: 10.1016/j.pharmthera.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Luu HH, Zhang R, Haydon RC, Rayburn E, Kang Q, Si W, et al. Wnt/beta-catenin signaling pathway as a novel cancer drug target. Curr Cancer Drug Targets. 2004;4:653–71. doi: 10.2174/1568009043332709. [DOI] [PubMed] [Google Scholar]
- 53.Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–67. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 54.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 55.Abe N, Watanabe T, Suzuki Y, Matsumoto N, Masaki T, Mori T, et al. An increased high-mobility group A2 expression level is associated with malignant phenotype in pancreatic exocrine tissue. Br J Cancer. 2003;89:2104–9. doi: 10.1038/sj.bjc.6601391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer B, Loeschke S, Schultze A, Weigel T, Sandkamp M, Goldmann T, et al. HMGA2 overexpression in non-small cell lung cancer. Mol Carcinog. 2007;46:503–11. doi: 10.1002/mc.20235. [DOI] [PubMed] [Google Scholar]
- 57.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–56. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 58.Haupt Y, Alexander WS, Barri G, Klinken SP, Adams JM. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell. 1991;65:753–63. doi: 10.1016/0092-8674(91)90383-A. [DOI] [PubMed] [Google Scholar]
- 59.Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–71. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Domen J, Gandy KL, Weissman IL. Systemic overexpression of BCL-2 in the hematopoietic system protects transgenic mice from the consequences of lethal irradiation. Blood. 1998;91:2272–82. [PubMed] [Google Scholar]
- 61.Domen J, Cheshier SH, Weissman IL. The role of apoptosis in the regulation of hematopoietic stem cells: Overexpression of Bcl-2 increases both their number and repopulation potential. J Exp Med. 2000;191:253–64. doi: 10.1084/jem.191.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS ONE. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 64.Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U, et al. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A. 2004;101:14228–33. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wiemer EA. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43:1529–44. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 66.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed Cell Death 4 (PDCD4) is an important functional target of the MicroRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–33. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 67.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 68.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 70.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 71.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17–92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–53. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, et al. The miR-17 92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci U S A. 2009;106:2812–7. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, et al. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 75.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 76.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–64. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 77.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 78.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 79.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–9. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, et al. The miR-15a-miR-16–1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–7. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 82.Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–30. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 83.Garzia L, Andolfo I, Cusanelli E, Marino N, Petrosino G, De Martino D, et al. MicroRNA-199b–5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS ONE. 2009;4:e4998. doi: 10.1371/journal.pone.0004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferretti E, De Smaele E, Miele E, Laneve P, Po A, Pelloni M, et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. Embo J. 2008;27:2616–27. doi: 10.1038/emboj.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.He X, He L, Hannon GJ. The guardian’s little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 2007;67:11099–101. doi: 10.1158/0008-5472.CAN-07-2672. [DOI] [PubMed] [Google Scholar]
- 86.Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, et al. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hatfield S, Ruohola-Baker H. microRNA and stem cell function. Cell Tissue Res. 2008;331:57–66. doi: 10.1007/s00441-007-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 89.Pirollo KF, Xu L, Chang EH. Non-viral gene delivery for p53. Curr Opin Mol Ther. 2000;2:168–75. [PubMed] [Google Scholar]
- 90.Xu L, Pirollo KF, Chang EH. Tumor-targeted p53-gene therapy enhances the efficacy of conventional chemo/radiotherapy. J Control Release. 2001;74:115–28. doi: 10.1016/S0168-3659(01)00324-8. [DOI] [PubMed] [Google Scholar]
- 91.Xu L, Frederik P, Pirollo KF, Tang WH, Rait A, Xiang LM, et al. Self-assembly of a virus-mimicking nanostructure system for efficient tumor-targeted gene delivery. Hum Gene Ther. 2002;13:469–81. doi: 10.1089/10430340252792594. [DOI] [PubMed] [Google Scholar]
- 92.Xu L, Huang CC, Huang W, Tang WH, Rait A, Yin YZ, et al. Systemic tumor-targeted gene delivery by anti-transferrin receptor scFv-immunoliposomes. Molecular Cancer Therapeutics. 2002;1:337–46. [PubMed] [Google Scholar]
- 93.Xu L, Pirollo KF, Tang WH, Rait A, Chang EH. Transferrin-liposome-mediated systemic p53 gene therapy in combination with radiation results in regression of human head and neck cancer xenografts. Hum Gene Ther. 1999;10:2941–52. doi: 10.1089/10430349950016357. [DOI] [PubMed] [Google Scholar]
- 94.Xu L, Pirollo KF, Chang EH. Transferrin-liposome-mediated p53 sensitization of squamous cell carcinoma of the head and neck to radiation in vitro. Hum Gene Ther. 1997;8:467–75. doi: 10.1089/hum.1997.8.4-467. [DOI] [PubMed] [Google Scholar]
- 95.Xu L, Tang WH, Huang CC, Alexander W, Xiang LM, Pirollo KF, et al. Systemic p53 gene therapy of cancer with immunolipoplexes targeted by anti-transferrin receptor scFv. Mol Med. 2001;7:723–34. [PMC free article] [PubMed] [Google Scholar]
- 96.Pirollo KF, Rait A, Zhou Q, Hwang SH, Dagata JA, Zon G, et al. Materializing the potential of small interfering RNA via a tumor-targeting nanodelivery system. Cancer Res. 2007;67:2938–43. doi: 10.1158/0008-5472.CAN-06-4535. [DOI] [PubMed] [Google Scholar]
- 97.Pirollo KF, Zon G, Rait A, Zhou Q, Yu W, Hogrefe R, et al. Tumor-targeting nanoimmunoliposome complex for short interfering RNA delivery. Hum Gene Ther. 2006;17:117–24. doi: 10.1089/hum.2006.17.117. [DOI] [PubMed] [Google Scholar]
- 98.Hogrefe RI, Lebedev AV, Zon G, Pirollo KF, Rait A, Zhou Q, et al. Chemically modified short interfering hybrids (siHYBRIDS): nanoimmunoliposome delivery in vitro and in vivo for RNAi of HER-2. Nucleosides Nucleotides Nucleic Acids. 2006;25:889–907. doi: 10.1080/15257770600793885. [DOI] [PubMed] [Google Scholar]