Abstract
Gaucher disease, the most common lysosomal storage disease, can be treated with enzyme replacement therapy (ERT), in which defective acid-β-glucosidase (GlcCerase) is supplemented by a recombinant, active enzyme. The X-ray structures of recombinant GlcCerase produced in Chinese hamster ovary cells (imiglucerase, Cerezyme®) and in transgenic carrot cells (prGCD) have been previously solved. We now describe the structure and characteristics of a novel form of GlcCerase under investigation for the treatment of Gaucher disease, Gene-ActivatedTM human GlcCerase (velaglucerase alfa). In contrast to imiglucerase and prGCD, velaglucerase alfa contains the native human enzyme sequence. All three GlcCerases consist of three domains, with the active site located in domain III. The distances between the carboxylic oxygens of the catalytic residues, E340 and E235, are consistent with distances proposed for acid–base hydrolysis. Kinetic parameters (Km and Vmax) of velaglucerase alfa and imiglucerase, as well as their specific activities, are similar. However, analysis of glycosylation patterns shows that velaglucerase alfa displays distinctly different structures from imiglucerase and prGCD. The predominant glycan on velaglucerase alfa is a high-mannose type, with nine mannose units, while imiglucerase contains a chitobiose tri-mannosyl core glycan with fucosylation. These differences in glycosylation affect cellular internalization; the rate of velaglucerase alfa internalization into human macrophages is at least 2-fold greater than that of imiglucerase.
Keywords: Gaucher disease, gene activation, glucocerebrosidase, glycans, mannose-6-phosphate receptor, site-specific glycosylation, X-ray structure
Introduction
Gaucher disease is caused by mutations in the gene encoding the lysosomal enzyme, acid-β-glucosidase (glucocerebrosidase, GlcCerase, E.C. 3.2.1.45) (Beutler and Grabowski 2001; Futerman and Zimran 2006). The most common treatment for Gaucher disease is enzyme replacement therapy (ERT), in which defective GlcCerase is supplemented with an active enzyme. ERT using imiglucerase, a recombinant analog of human GlcCerase expressed in Chinese hamster ovary (CHO) cells has been available for ∼15 years. After expression and purification, imiglucerase is modified by exo-glycosidase treatment (Friedman and Hayes 1996) to expose the core mannose residues that can be recognized by macrophages. Glycan remodeling greatly improves targeting to and internalization by macrophages, the main cell type affected in Gaucher disease (Futerman and Zimran 2006). An alternative means of producing GlcCerase (prGCD) in transgenic carrot root cells has been developed (Aviezer et al. 2009). The X-ray structures of imiglucerase and prGCD have been previously reported (Dvir et al. 2003; Shaaltiel et al. 2007).
In the current study, we have used gene activation in a well-characterized, continuous human cell line to produce gene-activated human acid-β-glucocerebrosidase (velaglucerase alfa). Gene activation refers to targeted recombination with a promoter that activates the endogenous GlcCerase gene in the selected human cell line. Velaglucerase alfa is secreted as a monomeric glycoprotein of approximately 63 kDa and is composed of 497 amino acids with a sequence identical to that of the natural human protein (Zimran et al. 2007). Glycosylation of velaglucerase alfa is altered by using kifunensine, a mannosidase I inhibitor, during cell culture, which results in the secretion of a protein containing predominantly high-mannose type glycans (Elbein et al. 1990).
Herein we describe the crystal structure of velaglucerase alfa, using a preparation that had been partially deglycosylated, and show that it is similar to that of imiglucerase (Dvir et al. 2003) and prGCD (Shaaltiel et al. 2007). Velaglucerase alfa differs from imiglucerase and prGCD as the latter two enzymes contain a mutation at residue 495 (an Arg to His substitution: R495H), and prGCD contains seven additional residues at the C terminus (DLLVDTM) and two additional residues at the N terminus (EF). Moreover, the kinetic parameters and specific activity of velaglucerase alfa are very similar to those of imiglucerase. We also compare the glycosylation patterns of velaglucerase alfa and imiglucerase by use of LC-MS and assess the impact of the different glycosylation patterns by analyzing internalization in human macrophages.
Results and discussion
X-ray structure
Diffraction-quality crystals of velaglucerase alfa were obtained after partial deglycosylation using N-glycosidase F, by a procedure similar to that previously described for imiglucerase (Dvir et al. 2003). Velaglucerase alfa crystallized in the same space group, C2221, as imiglucerase (Table I), and unit cell parameters were similar to the previously published GlcCerase structures (Dvir et al. 2003; Premkumar et al. 2005; Brumshtein et al. 2006). The asymmetric unit contained two copies of velaglucerase alfa, designated as molecules A and B. The root mean square deviation (RMSD) value between molecules A and B (<0.3 Å) shows that they are virtually identical. A comparison of the structures of imiglucerase, prGCD, and velaglucerase alfa demonstrates that these structures are very similar, with an RMSD of 0.35–0.46 Å (Table II).
Table I.
Data collection and refinement statistics
| Velaglucerase alfa | |
|---|---|
| Data collection | |
| Space group | C2221 |
| Cell dimensions | |
| a, b, c (Å) | 109.37, 285.55, 91.69 |
| abg (°) | 90.00, 90.00, 90.00 |
| Resolution (Å) | 19.9–2.7 (2.75–2.70)a |
| Rsym(%) | 15.7 (51.0) |
| I/s<I> | 14.4 (4.6) |
| Completeness (%) | 100 (100) |
| Redundancy | 7.5 (7.6) |
| Refinement | |
| Resolution (Å) | 19.9–2.7 |
| Number of reflections | 39,776 |
| Rwork/Rfree | 17.3/23.4 |
| Rms deviations | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.486 |
| Number of refined atoms | |
| Protein | 7871 |
| Carbohydrates | 70 |
| Ions | 90 |
| Solvent | 326 |
| Ramachandran outliers (%) | 0.4 |
aThe highest resolution shell is shown in parentheses.
Table II.
RMS deviations of velaglucerase alfa compared to imiglucerase and prGCD. RMS deviations (Å) are shown for each of the two copies of the molecules in the asymmetric unit and were calculated using PyMol (www.pymol.org). The PDB codes for imiglucerase and pr-GlcCerase are 2J25 and 2V3F, respectively
| Imiglucerase-A | Imiglucerase–B | prGCD-A | prGCD-B | |
|---|---|---|---|---|
| Velaglucerase alfa-A | 0.39 | 0.35 | 0.36 | 0.40 |
| Velaglucerase alfa-B | 0.38 | 0.43 | 0.46 | 0.46 |
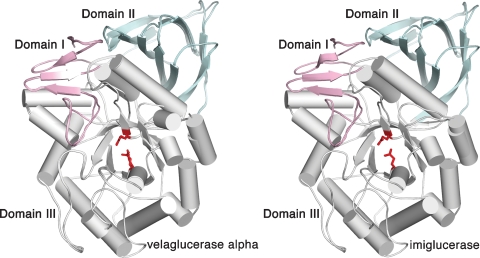
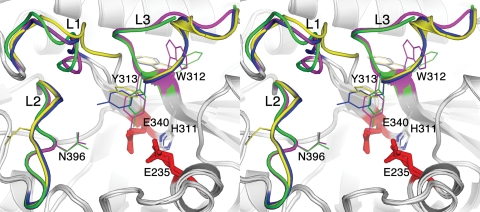
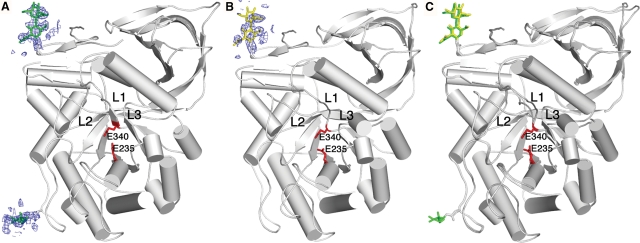
Velaglucerase alfa thus consists of three noncontiguous domains, with the catalytic site located in domain III (residues 76–381 and 416–430), which is a (β/α)8 (TIM) barrel (Figure 1). A more detailed analysis of the active site reveals that it is virtually identical to that of imiglucerase (Figure 2), with the distances between the carboxylic oxygens of the catalytic residues, E340 and E235 (5.2 Å in molecule A and 5.1 Å in molecule B), similar to those obtained previously (Brumshtein et al. 2006) and in agreement with the distances proposed for acid–base hydrolysis (Davies and Henrissat 1995). Moreover, the three loops (loop 1, residues 345–350; loop 2, residues 393–399; and loop 3, residues 312–319) observed in previous structures (reviewed in Kacher et al. (2008)) are also seen in velaglucerase alfa. Similarly to prGCD (Shaaltiel et al. 2007), loops 2 and 3 show differences in their backbone angles and side chain orientations in the two molecules of the asymmetric unit, whereas loop 1, since it makes crystal contacts, exhibits less pronounced conformational changes (Figure 2). In the case of loop 3, a helical conformation is seen in molecule B, whereas a coiled conformation is seen in molecule A (Figure 3), as previously reported for imiglucerase (Brumshtein et al. 2006). Although the crystal was cryo-protected with 25% ethylene glycol, we did not detect any ethylene glycol molecules in the electron density map.
Fig. 1.
Comparison of the crystal structures of velaglucerase alfa and imiglucerase. The three domains of the enzymes are colored pink (domain I, residues 1–29 and 383–414), blue (domain II, residues 30–75 and 431–497), and gray (domain III, residues 76–382 and 415–430).
Fig. 2.
Active site of velaglucerase alfa. Stereo representation of an overlay of the active sites of imiglucerase (blue and magenta) and velaglucerase alfa (yellow and green). Catalytic residues are shown as red sticks. Loops near the entrance to the active site are indicated (L1, loop 1; L2, loop 2; L3, loop 3).
Fig. 3.
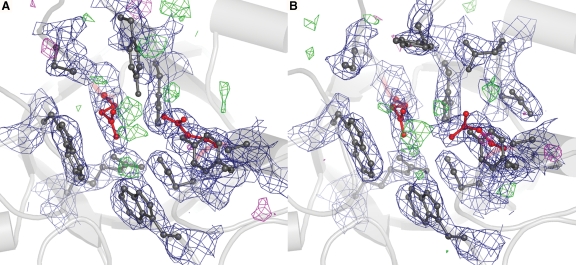
Electron density around the catalytic center. Catalytic residues E235 and E340 are shown as red balls and sticks and surrounding residues are in dark gray. Contours of the 2Fo–Fc map are shown as a blue mesh (at 1.2σ); contours of the Fo–Fc map are shown in green mesh (at 3σ) and in magenta (at −3σ). Several Fo–Fc peaks are visible in the active site, but they did not overlap with the 2Fo–Fc map, nor are they continuous; hence, at this resolution they appear to be noise. A and B show the catalytic centers of molecules A and B, respectively, in the asymmetric unit.
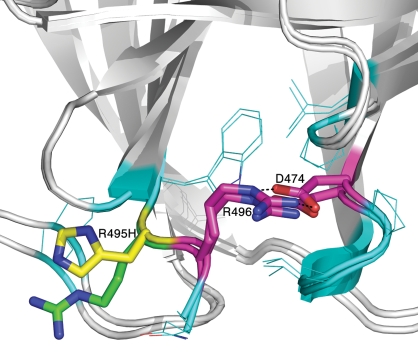
Imiglucerase and prGCD both contain an Arg to His mutation at residue 495, with H495 making an H-bond (2.6 Å) with the peptide carbonyl of F31. In contrast, velaglucerase alfa contains a sequence identical to that of the natural human enzyme, with an Arg at residue 495, which does not make a similar H-bond. No major structural differences were observed in velaglucerase alfa around residue R495, relative to imiglucerase or prGCD. Two mutations which cause Gaucher disease, R496 and D474 (Figure 4) (Kawame et al. 1992; Beutler et al. 1993; Choy et al. 1998), are in close proximity to R495 near the N-terminus of GlcCerase. D474 is at the end of a β-strand, and R496 is part of a coil with no clear secondary structure, and their side-chains form a salt bridge and hydrogen bonds with each other; mutations in either of these two residues would disrupt these interactions. By analyzing the geometry and the interactions between the side chains of these two residues, and the secondary structure of the region, we conclude that R496 or D474 may be involved in stabilizing the conformation of the N-terminus of the enzyme by their side chain interactions, with disruption of these bonds resulting in a flexible N-terminus and hence in a less stable structure. However, neither of these residues interacts with R495.
Fig. 4.
Mutations at the C-terminus of GlcCerase. Imiglucerase and pr-GlcCerase contain a His at residue 495 (yellow), whereas velaglucerase alfa contains Arg (green). Mutations R496 and D474, which cause Gaucher disease, are shown in magenta. Residues within 4 Å distance of R495 and R496 are shown in cyan.
Kinetic analysis
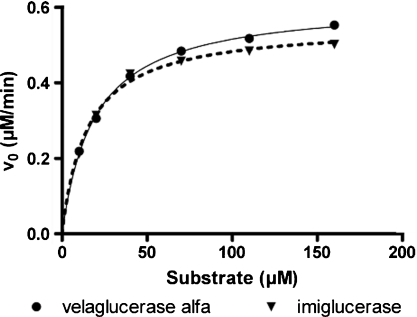
To further compare velaglucerase alfa and imiglucerase, and to determine if the mutation at residue 495 has any effect, kinetic parameters and specific activity were determined using a natural glucosylceramide (GlcCer) substrate, rather than a surrogate substrate typically used to assess enzyme activity. Velaglucerase alfa has a kcat of 2100 min−1, a Km of 19 μM, and a Vmax of 0.61 μM min−1. Imiglucerase has a kcat of 1900 min−1, a Km of 15 μM, and a Vmax of 0.56 μM min−1 (Figure 5). Similar Km values were reported in the literature; GlcCerase derived from brain tissue and fibroblasts both have a Km of 32 μM using GlcCer from Gaucher spleen (Vaccaro et al. 1982), while imiglucerase and prGCD have a Km of 15.2 and 20.7 μM, respectively, using a fluorescent GlcCer analog, C6-NBD-GlcCer (Shaaltiel et al. 2007). In addition, at a 210 μM GlcCer substrate concentration, velaglucerase alfa and imiglucerase have similar specific activities of 26 and 24 U/mg, respectively. These results demonstrate that human and CHO-cell derived GlcCerase, prepared by two different manufacturing processes, have similar enzymatic activities for the natural substrate.
Fig. 5.
Kinetic analysis of velaglucerase alfa and imiglucerase. Vmax and Km values were determined using a natural GlcCer substrate (n = 2).
Glycan composition
We next examined which sugars could be detected in the crystal structure of velaglucerase alfa. Even after partial deglycosylation using N-glycosidase F, two sugar residues were observed attached to residue N19 in both molecules A and B (Figure 6). One sugar was detected on N146 in molecule A whereas no sugars were detected on N146 in molecule B (Figure 6). As reported previously for imiglucerase, no sugars were detected attached to either N270 or N59 in velaglucerase alfa. It should be noted that sugars attached to N270 have not been seen in any of the crystal structures solved to date, and sugars have been seen only occasionally on N59 (Brumshtein et al. 2006). The inability to detect sugars on either N59 or N270 is most likely due to the high flexibility of the corresponding glycan chains since nano-liquid chromatography electrospray ionization tandem mass spectrometry (nano-LC-ESI-MS/MS) analysis of intact imiglucerase (Kacher et al. 2008), and of velaglucerase alfa (see below) showed that glycan chains were attached to both these residues.
Fig. 6.
Glycosylation sites seen in the crystal structure of velaglucerase alfa. 2Fo–Fc electron density maps are shown, which are contoured at 1.2σ in the vicinity of two of the putative glycosylation sites, N19 and N146 for molecule A, and N19 for molecule B. (A) Glycosylation sites detected in molecule A are shown in green. (B) Glycosylation site detected in molecule B is shown in yellow. (C) Superposition of the two individual molecules in the asymmetric unit reveals their similarity. In all three representations, catalytic residues E235 and E340 are shown as red sticks.
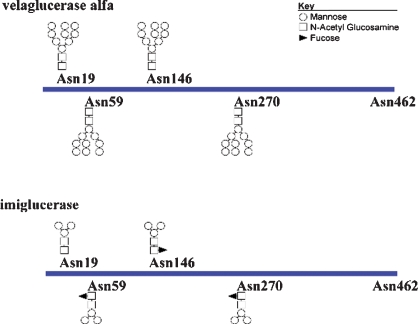
Velaglucerase alfa and imiglucerase bear distinctly different glycan chains due to the differences in their manufacture. In our comparative study of the carbohydrate content of unmodified velaglucerase alfa and imiglucerase by LC-ESI-MS, four of the five potential glycosylation sites, namely, N19, N59, N146, and N270, were observed to be fully occupied in both. As expected from the crystal structures, N462 is fully unoccupied in both, due to its buried location.
According to LC-ESI-MS analysis of glycopeptide maps, velaglucerase alfa contains primarily high-mannose type glycans, consisting of six to nine mannose units. Listed as the predominant structure in Table III, the most abundant ion present in the averaged spectra for each site corresponds to a glycan with nine mannose units. Glycan microheterogeneity was observed at each site and the less abundant structures are listed as other glycans. These other glycans consist of mannose residues with phosphorylation at the C-6 position to create a mannose-6-phosphate (M6P) residue. The lowest levels of M6P were at N19; N59 and N146 had similar but higher levels relative to N19, while N270 had the highest amount of M6P. Despite the site-specific variation in relative levels of M6P, nonphosphorylated glycans remained the predominant species for all four sites. Also observed on N59, N146 and N270 were mono-sialylated mono-antennary hybrid and complex-type structures with core fucosylation, which were quantified by glycan map analysis. These structures are consistent with a low percentage of glycosylation sites escaping kifunensine inhibition, resulting in glycan maturation and core fucosylation. In the case of hybrid-type glycans, only a single antenna maturated.
Table III.
Carbohydrate composition of velaglucerase alfa. Predominant structures are those observed to be most abundant at each N-linked glycosylation site. The other glycans consisted mostly of high-mannose type structures (some with M6P) and with the hybrid and complex types observed at low levels (∼2% of the total as determined by glycan map analysis)
| Glycosylation site | Predominant glycan | Other glycans |
|---|---|---|
| Asn19 | High mannose | High mannose |
| (Man)9(GlcNAc)2 | (Man)6-8(GlcNAc)2 | |
| Phosphorylated high mannose | ||
| (Phos)1(Man)8-9(GlcNAc)2 | ||
| GlcNAc-capped phosphate | ||
| (Phos)1(Man)8-9(GlcNAc)3 | ||
| Hybrid | ||
| (Hex)2(Man)3(GlcNAc)3(Fuc)1 | ||
| Asn59 | High mannose | High mannose |
| (Man)9(GlcNAc)2 | (Man)5-8(GlcNAc)2 | |
| Phosphorylated high mannose | ||
| (Phos)1(Man)7-9(GlcNAc)2 | ||
| GlcNAc-capped phosphate | ||
| (Phos)1(Man)8-9(GlcNAc)3 | ||
| Hybrid | ||
| (NeuAc)1(Gal)1(Man)5(GlcNAc)3(Fuc)1 | ||
| Complex | ||
| (NeuAc)0-2(Gal)2(Man)3(GlcNAc)4(Fuc)1 | ||
| (Gal)3(Man)3(GlcNAc)5(Fuc)1 | ||
| Asn146 | High mannose | High mannose |
| (Man)9(GlcNAc)2 | (Man)6-8(GlcNAc)2 | |
| Phosphorylated high mannose | ||
| (Phos)1(Man)7-9(GlcNAc)2 | ||
| GlcNAc-capped phosphate | ||
| (Phos)1(Man)9(GlcNAc)3 | ||
| Hybrid | ||
| (NeuAc)1(Gal)1(Man)5(GlcNAc)3(Fuc)1 | ||
| Asn270 | High mannose | High mannose |
| (Man)9(GlcNAc)2 | (Man)6-8(GlcNAc)2 | |
| Phosphorylated high mannose | ||
| (Phos)1(Man)6-9(GlcNAc)2 | ||
| GlcNAc-capped phosphate | ||
| (Phos)1(Man)9(GlcNAc)3 | ||
| Hybrid | ||
| (Gal)1(Man)7(GlcNAc)3(Fuc)1 | ||
| (NeuAc)1(Gal)1(Man)5(GlcNAc)3(Fuc)1 | ||
| Complex | ||
| (NeuAc)2(Gal)2(Man)3(GlcNAc)4(Fuc)1 | ||
| Asn462 | Not detected | Not detected |
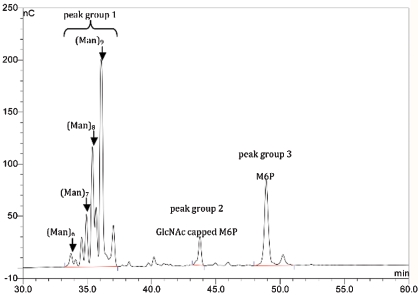
The results from site-specific glycan characterization were corroborated by glycan map analysis (Figure 8), which demonstrates high-mannose type glycans consisting of six to nine mannose units with a predominant nine-mannose structure. Estimates from glycan map analysis show that the mono-sialylated mono-antennary hybrid structures account for ∼2% of the total glycan pool. The map also demonstrates the presence of high-mannose glycans containing one GlcNAc-capped M6P, a result of incomplete glycan processing, as well as high-mannose glycans bearing a single M6P. Also consistent with these results were data obtained from monosaccharide compositional analysis that demonstrates approximately 0.8 mole of M6P per mole of velaglucerase alfa, and approximately 0.6 moles of M6P per mole of imiglucerase.
Fig. 8.
Glycan map analysis of velaglucerase alfa. Glycans released by N-glycosidase F were analyzed by anion-exchange chromatography with amperometric detection. The method resolves glycans based on negative charge where peak group 1 corresponds to high-mannose type neutral glycans that are resolved into multiple peaks according to the number of mannose units, peak group 2 corresponds to high-mannose type glycans with one M6P that retained its GlcNAc cap (one negative charge), and peak group 3 corresponds to high-mannose type glycans containing one fully processed M6P (two negative charges). In peak group 1, smaller peaks are resolved that correspond to positional isomers of the various oligomannose types observed.
Site-specific glycan analysis demonstrated that imiglucerase contains primarily complex-type glycans with core fucosylation that terminate with the chitobiose tri-mannosyl core (Table IV), with an exception at the N19 site, which was observed to be devoid of fucose. These structures are as expected for GlcCerase with exoglycosidase treatment to expose the core mannose residues. Imiglucerase also contains glycan microheterogeneity at each site of glycosylation, with lower levels of core structures terminating with N-acetylglucosamine (GlcNAc) that are likely a result of incomplete digestion with N-acetylglucosaminidase. At N146 and N270, high-mannose type glycans were observed containing five to six mannose units with one M6P.
Table IV.
Carbohydrate composition of imiglucerase. Predominant structures are those observed to be most abundant at each N-linked glycosylation site. The other glycans consisted mostly of core structures with additional GlcNAc and high-mannose structures with M6P
| Glycosylation | ||
|---|---|---|
| site | Predominant glycan | Other glycans |
| Asn19 | Complex | Complex |
| (Man)3(GlcNAc)2 | (Man)3(GlcNAc)3 | |
| Asn59 | Complex | Complex |
| (Man)3(GlcNAc)2(Fuc)1 | (Man)3(GlcNAc)3(Fuc)1 | |
| Asn146 | Complex | Complex |
| (Man)3(GlcNAc)2(Fuc)1 | (Man)3(GlcNAc)3-4(Fuc)1 | |
| Phosphorylated high mannose | ||
| (Phos)1(Man)5-6(GlcNAc)2 | ||
| GlcNAc-capped phosphate | ||
| (Phos)1(Man)5-6(GlcNAc)3 | ||
| Asn270 | Complex | Complex |
| (Man)3(GlcNAc)2(Fuc)1 | (Man)3(GlcNAc)3-4(Fuc)1 | |
| Phosphorylated high -mannose | ||
| (Phos)1(Man)5-6(GlcNAc)2 | ||
| GlcNAc-capped phosphate | ||
| (Phos)1(Man)5-6(GlcNAc)3 |
The glycan graphics shown in Figure 7 help to visualize the predominant structures for both forms of GlcCerase as described in Tables III and IV. These structures were consistent with glycan types and levels observed with glycan map analysis as well as with previous reports (Van Patten et al. 2007). In the current study, the glycans of prGCD were not characterized, but earlier studies demonstrated the presence of core α-(1,2)-xylose and core α-(1,3)-fucose (Shaaltiel et al. 2007), which are unique to plant-derived proteins and would not be expected to be present on either velaglucerase alfa or imiglucerase.
Fig. 7.
Glycan structures of velaglucerase alfa and imiglucerase. Predominant N-linked carbohydrate structures on velaglucerase alfa (top) and imiglucerase (bottom) are shown graphically at their relative positions along the protein backbone.
Internalization by macrophages
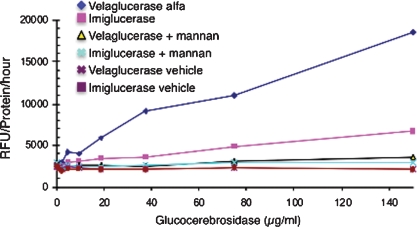
Internalization of proteins by endocytosis is highly dependent upon their carbohydrate composition and has been well characterized (Kornfeld 1986). A comparison of the internalization rate of velaglucerase alfa to that of imiglucerase in U937-derived macrophages demonstrated that velaglucerase alfa is internalized approximately 2.5-fold more efficiently than imiglucerase (Figure 9). Internalization of both enzymes could be inhibited by the addition of mannan to the culture medium, demonstrating that internalization was mediated via mannose receptors; moreover, U937 cells were shown by immunohistochemistry to express mannose receptors (CD206) (data not shown). It should be noted that during optimization of this assay, variations in results were obtained when different culture media were used. Therefore, additional research will be required to determine the exact nature of the uptake since different mannose receptors exist, which may be involved in this cellular internalization. In contrast, the addition of M6P to the culture medium had no effect, confirming that the M6P receptor is not involved in internalization (data not shown). Since velaglucerase alfa and imiglucerase display similar kinetic parameters, specific activities, and structural features, the different rates of internalization can be ascribed to differences in glycosylation patterns between velaglucerase alfa and imiglucerase, with the increased rate of internalization of velaglucerase alfa likely due to the expression of longer chain high-mannose type glycans compared to the core mannose structures found on imiglucerase.
Fig. 9.
Velaglucerase alfa and imiglucerase internalization into differentiated macrophages. The ordinate of the graph represents the fluorescence data normalized for the cellular protein concentration and incubation time (RFU/μg/h). The GlcCerase dose is shown on the abscissa.
Conclusions
In summary, the X-ray structure of velaglucerase alfa is very similar to those of recombinant GlcCerases produced in other expression systems, with the R495H mutations found in imiglucerase and prGCD having no effect on the secondary structure. The main difference between imiglucerase and velaglucerase alfa concerns their glycan structures, with the latter containing longer chain high-mannose type glycans compared to the core mannose structures found on imiglucerase. This difference in glycosylation appears to lead to the increased cellular uptake of velaglucerase alfa over imiglucerase. The role of protein glycosylation in cellular uptake is widely established in many cell types (Barton et al. 1991). However, while the function of the macrophage mannose receptor (MR; CD206) in internalization of mannosylated proteins is well characterized (East and Isacke 2002), a growing family of carbohydrate-binding receptors have been implicated in diverse macrophage functions including removal and disposal of endotoxin (Ono et al. 2006), utilization of secreted lysosomal enzymes (Abe et al. 2008), phagocytosis (Kang et al. 2005), and regulation of the innate immune response to microbial pathogen-associated structures (Garner et al. 1994). Thus, the differences in uptake observed between imiglucerase and velaglucerase-alfa can be attributed to differences in affinity for CD206, or alternatively could be due to differential uptake mediated by other macrophage mannose receptors such as Endo180. This observed increase in cellular uptake of velaglucerase-alfa over imiglucerase can be envisioned to lead to a more rapid time to improvement of clinical parameters and potentially increased therapeutic efficacy
Material and methods
Crystallization, structure determination, and refinement
Velaglucerase alfa was partially deglycosylated (Kacher et al. 2008) prior to crystallization, as previously described for imiglucerase (Dvir et al. 2003; Premkumar et al. 2005), using N-glycosidase F (88 h at 25°C), which removes carbohydrate chains from proteins and peptides by cleaving the amide bonds between Asn residues and N-acetylglucosamine (GlcNAc) (Han and Martinage 1992), but does not necessarily remove all carbohydrate chains from native proteins. Subsequent to N-glycosidase F-treatment, velaglucerase alfa was diluted in the crystallization buffer (10 mM citrate pH 5.5, 7% (v/v) ethanol, 0.02% (w/v) NaN3) and passed through a Centricon YM-30 centrifugal filter device with a molecular mass cut-off of ∼30 kDa, to give a final concentration of 4–5 mg/mL. Velaglucerase alfa crystals were obtained by micro-batch crystallization under oil (Chayen et al. 1990) using a Douglas Instruments Oryx6 robot. The crystallization solution had a 1:1 ratio of the concentrated enzyme solution and of 1 M (NH4)2SO4/0.1 M HEPES, pH 7.0, containing 0.5% (w/v) PEG8000. Crystallization was performed under Al's oil (D’Arcy et al. 1996) (1:1 ratio of paraffin and silicone liquid oils) for 5–14 days at 20°C. Data were collected on beam line ID14eh2 at the ESRF synchrotron (Grenoble, France). Crystals were cryo-protected with a 25% ethylene glycol solution, mounted, and flash cooled to 100 K. X-ray diffraction images were processed using HKL2000 and scaled with SCALEPACK (Otwinowski et al. 1997). The structure was solved using the molecular replacement method based on PDB 2J25 (Brumshtein et al. 2006) and refined with Refmac5 (Murshudov et al. 1997). During the course of refinement, the electron density map showed significant improvement, and putative sugars could be seen adjacent to N19 and N146 for molecule A, and adjacent to N19 for molecule B. Table I summarizes data collection and processing. Structures and structure factors were deposited in the PDB (code 2WKL).
Enzyme kinetics and specific activity
The novel enzymatic activity assay described below measures the ability of GlcCerase to release glucose from GlcCer obtained from Gaucher spleen (Matreya LLC, PA, Cat. no. 1057). Velaglucerase alfa (drug substance lot EP06-003) and imiglucerase (commercial product lot C7036C01) were assayed. The released glucose was quantified by anion-exchange chromatography equipped with a pulsed amperometric detector. The appropriate amount of GlcCer in chloroform/methanol (2:1, v/v) was dried by a SpeedVac in the presence of 0.2 M taurocholic acid in methanol and 20% (v/v) oleic acid in chloroform/methanol (2:1). The dried pellet was reconstituted in the 0.1 M citrate/0.2 M phosphate buffer (pH 5.0) and diluted to the desired concentrations. Enzyme samples were diluted to a concentration of 0.2 ng/μL with the dilution buffer (50 mM sodium citrate, pH 6.0 with 0.75 mg/mL BSA) and 2 ng of enzyme was incubated for 30 min at 37°C with serial dilutions of GlcCer in a 110 μL reaction volume. The reaction was stopped by heat denaturing samples at 100°C for 5 min. Sample manipulations were internally controlled by adding 100 μL of a galactosamine (GalN) solution to the reaction mixture. Dionex OnGuard II RP cartridges were used to remove the detergent and lipid. The analysis was carried out on a Dionex high-performance anion-exchange chromatography device, coupled with a pulsed amperometric detection apparatus (HPAE-PAD), using a CarboPac PA-10 analytical column equipped with a CarboPac PA-10 guard column. An isocratic flow of 6 mM NaOH at 0.25 mL/min for 25 min was used to separate monosaccharides (Glc and GalN). The amount of glucose (Glc) was calculated from linear regression analysis of GalN and Glc standards in the range of 10–480 pmol per injection. The assay was carried out in a range of substrate concentrations of 5–150 μM, and obeyed Michaelis–Menten kinetics, thus permitting assignment of Km and Vmax values.
Site-specific characterization of glycans
Velaglucerase alfa (drug substance lot EP06-003, Shire Human Genetic Therapies, Hampshire, UK) and imiglucerase (commercial product lot HA163BL) were prepared for enzymatic digestion by reductive denaturation with DTT, followed by and cysteine alkylation with iodoacetic acid. Alkylated samples were digested first with the endoproteinase Lys-C (Roche Diagnostics GmbH, Mannheim, Germany) (1:42 enzyme to substrate ratio, w/w, for 6 h at 37°C), followed by digestion with endoproteinase Glu-C (1:25 enzyme to substrate ratio, w/w, for 16 h at room temperature). Digested samples were analyzed by peptide mass mapping using reversed phase chromatography with in-line UV (214 nm) and electrospray ionization with mass spectrometric detection (LC-ESI-MS). By comparing the peptide maps before and after glycan release using N-glycosidase F (New England Biolabs, Ipswich, MA), the five potential glycosylation sites were identified. The glycan mass was calculated by subtracting the expected peptide mass from the observed glycopeptide masses. Using software to match the observed glycan masses with potential monosaccharide compositions, glycan compositions for each site were determined. To verify monosaccharide compositions, treatments (according to manufacturer's recommendations) with neuraminidase (Roche Diagnostics GmbH), alkaline phosphatase (Roche Diagnostics GmbH, Mannheim, Germany), and α-mannosidase (Glyko, Inc., Hayward, CA) were used to verify the presence of sialic acid, phosphate, and alpha-linked mannose, respectively. MS/MS fragmentation analysis was used to verify glycan phosphorylation.
Glycan map analysis
The procedure involves heat denaturation of the protein at 100°C for 3–4 min in the presence of 0.5% SDS, followed by enzymatic release of glycans with N-glycosidase F (Prozyme, San Leandro, CA). Velaglucerase alfa (drug substance lot EP06-001, Shire Human Genetic Therapies) was incubated with N-glycosidase F (30 mU/3 μL) for 4–6 h at 37°C with 0.9% NP40, followed by a second addition of N-glycosidase F, and an additional 17–19 h incubation at 37°C. Analysis of the released glycans was performed by HPAE-PAD, using a CarboPac PA-1 analytical column equipped with a CarboPac PA-1 guard column (Dionex, Sunnyvale, CA). Glycans were applied to the column in 12 mM sodium acetate/100 mM NaOH, followed by elution with a 12–300 mM sodium acetate gradient (6.4 mM/min) in 100 mM NaOH in 45 min. Using a flow rate of 1 mL/min and the column at ambient room temperature, glycans elute in the order of increasing negative charge.
Cellular internalization
Human U937 cells were cultured in growth media containing RPMI 1640 with 2 mM l-glutamine, 10 mM HEPES, 1 mM sodium pyruvate, 4.5 g/L glucose, 1.5 g/L sodium bicarbonate, and 10% FBS. Treatment with phorbol myristate acetate (PMA) for 3 days was used to induce differentiation into macrophages (Amento et al. 1984). The U937-derived macrophages were seeded into 96-well microtiter plates at 50,000 cells per well in growth medium, and allowed to adhere to the plates for 48 h. Seeded macrophages were incubated for 3 h with equimolar preparations of velaglucerase alfa (drug substance lot FEC06-003, Shire Human Genetic Therapies) or imiglucerase (Cerezyme®; commercial product lot C7036C01, Genzyme, Cambridge, MA) at pH 7.5, in growth medium containing RPMI 1640 devoid of phosphate, 0.1% BSA, 10 mM HEPES, pH 7.5, 2 mM l-glutamine, 1 mM DTT, and 10 mM CaCl2. In all assays, the cells were treated with GlcCerase for a 3-h duration which was previously determined to be in the linear range of internalization. For dose response curves utilized to demonstrate mannose-receptor specificity, 10 mg/mL mannan was used to antagonize the receptor. After a series of wash steps (wash buffer: 0.05 M Tris, 0.138 M NaCl, 0.0027 M KCl, with 0.05% Tween 20, 0.5% BSA, pH 8.0), the cells were lysed (lysis buffer: 10 mM Tris pH 8.0, 0.5% NP40, 0.2% deoxycholate, Complete Mini Protease Inhibitor Cocktail Tablets in EASYpacks and PhosSTOP Phosphatase Inhibitor Cocktail Tablets in EASYpacks, Roche Applied Science), and the internalized GlcCerase was quantified by an assay employing the synthetic substrate, 4-methylumbelliferyl-β-d-glucopyranoside (4-MU-glc), which releases a fluorescent product upon cleavage. The protein content in the well was determined (BCA method according to the manufacturer's protocol) and was used to normalize the assay signal to total protein from each sample. The assay signal for the GlcCerase samples was tested in vitro to determine the extent of activity or signal disparity between the two drugs, and there was no difference in activity (data not shown). For these assays, 2-fold serial dilutions of velaglucerase alfa and imiglucerase (starting at 30 nM enzyme) were made in the assay lysis buffer and tested using the 4-MU-glc enzymatic activity assay. Plates were read with a Perkin Elmer Envision multi-label plate reader.
Funding
Shire Human Genetic Therapies, Inc.
Acknowledgments
J.L. Sussman is the Morton and Gladys Pickman Professor of Structural Biology, and A.H. Futerman is the Joseph Meyerhoff Professor of Biochemistry at the Weizmann Institute of Science. The contribution of Meng Wu, for technical assistance is gratefully acknowledged. We are grateful to Dr. Hilary Voet (Faculty of Agriculture, The Hebrew University, Rehovot) for invaluable discussions concerning the statistical analysis of the choice of space groups.
Conflict of interest statement
None declared.
Abbreviations
- CHO
Chinese hamster ovary
- ERT
enzyme replacement therapy
- GA-GCB
velaglucerase alfa
- GlcCer
glucosylceramide
- GlcCerase
acid-β-glucosidase
- M6P
mannose-6-phosphate
- prGCD
GlcCerase expressed in transgenic carrot cells
- RMSD
root mean square deviation
References
- Abe A, Kelly R, Kollmeyer J, Hiraoka M, Lu Y, Shayman JA. The secretion and uptake of lysosomal phospholipase A2 by alveolar macrophages. J Immunol. 2008;181:7873–7881. doi: 10.4049/jimmunol.181.11.7873. [DOI] [PubMed] [Google Scholar]
- Amento EP, Bhalla AK, Kurnick JT, Kradin RL, Clemens TL, Holick SA, Holick MF, Krane SM. 1 alpha,25-dihydroxyvitamin D3 induces maturation of the human monocyte cell line U937, and, in association with a factor from human T lymphocytes, augments production of the monokine, mononuclear cell factor. J Clin Invest. 1984;73:731–739. doi: 10.1172/JCI111266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviezer D, Brill-Almon E, Shaaltiel Y, Hashmueli S, Bartfeld D, Mizrachi S, Liberman Y, Freeman A, Zimran A, Galun E. A plant-derived recombinant human glucocerebrosidase enzyme—A preclinical and phase I investigation. PLoS ONE. 2009;4:e4792. doi: 10.1371/journal.pone.0004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NW, Brady RO, Dambrosia JM, Di Bisceglie AM, Doppelt SH, Hill SC, Mankin HJ, Murray GJ, Parker RI, Argoff CE, et al. Replacement therapy for inherited enzyme deficiency—Macrophage-targeted glucocerebrosidase for Gaucher's disease. N Engl J Med. 1991;324:1464–1470. doi: 10.1056/NEJM199105233242104. [DOI] [PubMed] [Google Scholar]
- Beutler E, Gelbart T, West C. Identification of six new Gaucher disease mutations. Genomics. 1993;15:203–205. doi: 10.1006/geno.1993.1035. [DOI] [PubMed] [Google Scholar]
- Beutler E, Grabowski GA. Gaucher disease. In: Scriver CR, Sly WS, et al., editors. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill Inc.; 2001. pp. 3635–3668. [Google Scholar]
- Brumshtein B, Wormald MR, Silman I, Futerman AH, Sussman JL. Structural comparison of differently glycosylated forms of acid-beta-glucosidase, the defective enzyme in Gaucher disease. Acta Crystallogr D Biol Crystallogr. 2006;62:1458–1465. doi: 10.1107/S0907444906038303. [DOI] [PubMed] [Google Scholar]
- Chayen NE, Shaw Stewart PD, Maeder DL, Blow DM. An automated system for micro-batch protein crystallization and screening. J Appl Crystallogr. 1990;23:297–302. [Google Scholar]
- Choy FY, Humphries ML, Ben-Yoseph Y. Gaucher type 2 disease: Identification of a novel transversion mutation in a French-Irish patient. Am J Med Genet. 1998;78:92–93. [PubMed] [Google Scholar]
- D’Arcy A, Elmore C, Stihle M, Johnston JE. A novel approach to crystallising proteins under oil. J Crystal Growth. 1996;168:175. [Google Scholar]
- Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- Dvir H, Harel M, McCarthy AA, Toker L, Silman I, Futerman AH, Sussman JL. X-ray structure of human acid-beta-glucosidase, the defective enzyme in Gaucher disease. EMBO Rep. 2003;4:704–709. doi: 10.1038/sj.embor.embor873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East L, Isacke CM. The mannose receptor family. Biochim Biophys Acta. 2002;1572:364–386. doi: 10.1016/s0304-4165(02)00319-7. [DOI] [PubMed] [Google Scholar]
- Elbein AD, Tropea JE, Mitchell M, Kaushal GP. Kifunensine, a potent inhibitor of the glycoprotein processing mannosidase I. J Biol Chem. 1990;265:15599–15605. [PubMed] [Google Scholar]
- Friedman B, Hayes M. Enhanced in vivo uptake of glucocerebrosidase. 1996. US Patent 5549892.
- Futerman AH, Zimran A. Gaucher Disease. Boca Raton (FL): Taylor and Francis Group; 2006. [Google Scholar]
- Garner RE, Rubanowice K, Sawyer RT, Hudson JA. Secretion of TNF-alpha by alveolar macrophages in response to Candida albicans mannan. J Leukoc Biol. 1994;55:161–168. doi: 10.1002/jlb.55.2.161. [DOI] [PubMed] [Google Scholar]
- Han KK, Martinage A. Post-translational chemical modification(s) of proteins. Int J Biochem. 1992;24:19–28. doi: 10.1016/0020-711x(92)90225-p. [DOI] [PubMed] [Google Scholar]
- Kacher Y, Brumshtein B, Boldin-Adamsky S, Toker L, Shainskaya A, Silman I, Sussman JL, Futerman AH. Acid-β-glucosidase: Insights from structural analysis and relevance to Gaucher disease therapy. Biol Chem. 2008;389:1361–1369. doi: 10.1515/BC.2008.163. [DOI] [PubMed] [Google Scholar]
- Kang PB, Azad AK, Torrelles JB, Kaufman TM, Beharka A, Tibesar E, DesJardin LE, Schlesinger LS. The human macrophage mannose receptor directs Mycobacterium tuberculosis lipoarabinomannan-mediated phagosome biogenesis. J Exp Med. 2005;202:987–999. doi: 10.1084/jem.20051239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawame H, Hasegawa Y, Eto Y, Maekawa K. Rapid identification of mutations in the glucocerebrosidase gene of Gaucher disease patients by analysis of single-strand conformation polymorphisms. Hum Genet. 1992;90:294–296. doi: 10.1007/BF00220082. [DOI] [PubMed] [Google Scholar]
- Kornfeld S. Trafficking of lysosomal enzymes in normal and disease states. J Clin Invest. 1986;77:1–6. doi: 10.1172/JCI112262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Ono K, Nishitani C, Mitsuzawa H, Shimizu T, Sano H, Suzuki H, Kodama T, Fujii N, Fukase K, Hirata K, et al. Mannose-binding lectin augments the uptake of lipid A, Staphylococcus aureus, and Escherichia coli by Kupffer cells through increased cell surface expression of scavenger receptor A. J Immunol. 2006;177:5517–5523. doi: 10.4049/jimmunol.177.8.5517. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W, Carter CW., Jr . Methods in Enzymology. New York: Academic Press; 1997. Processing of X-ray diffraction data collected in oscillation mode; p. 307. [DOI] [PubMed] [Google Scholar]
- Premkumar L, Sawkar AR, Boldin-Adamsky S, Toker L, Silman I, Kelly JW, Futerman AH, Sussman JL. X-ray structure of human acid-beta-glucosidase covalently bound to conduritol-B-epoxide. Implications for Gaucher disease. J Biol Chem. 2005;280:23815–23819. doi: 10.1074/jbc.M502799200. [DOI] [PubMed] [Google Scholar]
- Shaaltiel Y, Bartfeld D, Hashmueli S, Baum G, Brill-Almon E, Galili G, Dym O, Boldin-Adamsky SA, Silman I, Sussman JL, et al. Production of glucocerebrosidase with terminal mannose glycans for enzyme replacement therapy of Gaucher's disease using a plant-cell system. Plant Biotech J. 2007;5:579–590. doi: 10.1111/j.1467-7652.2007.00263.x. [DOI] [PubMed] [Google Scholar]
- Vaccaro A, Kobayashi T, Suzuki K. Comparison of a synthetic and natural glucosylceramidase as substrate for glucosylceramidase assay. Clinica Chimica Acta. 1982;118:1–7. doi: 10.1016/0009-8981(82)90221-2. [DOI] [PubMed] [Google Scholar]
- Van Patten SM, Hughes H, Huff MR, Piepenhagen PA, Waire J, Qiu H, Ganesa C, Reczek D, Ward PV, Kutzko JP, et al. Effect of mannose chain length on targeting of glucocerebrosidase for enzyme replacement therapy of Gaucher disease. Glycobiology. 2007;17:467–478. doi: 10.1093/glycob/cwm008. [DOI] [PubMed] [Google Scholar]
- Zimran A, Loveday K, Fratazzi C, Elstein D. A pharmacokinetic analysis of a novel enzyme replacement therapy with gene-activated human glucocerebrosidase (GA-GCB) in patients with type 1 Gaucher disease. Blood Cells Mol Dis. 2007;39:115–118. doi: 10.1016/j.bcmd.2007.02.008. [DOI] [PubMed] [Google Scholar]