Abstract
Objectives
The aim of this study was to isolate and characterize methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-resistant coagulase-negative Staphylococcus spp. (MRCoNS) from marine water and intertidal beach sand from public beaches in Washington State, USA.
Methods
Fifty-one staphylococci from Washington State beaches were characterized using antimicrobial susceptibility testing, carriage of acquired tetracycline and/or macrolide resistance genes, staphylococcal cassette chromosome mec (SCCmec) typing, the BBL Crystal™ Gram-Positive ID System and/or 16S rRNA sequencing, coagulase test and multilocus sequence typing (MLST) for MRSA.
Results
Five multidrug-resistant MRSA SCCmec type I, of which three were MLST type ST45, one ST59 and one a new MLST type, ST1405, plus one susceptible non-typeable (NT) MRSA ST30 were characterized. Thirty-three MRCoNS isolates, representing 21 strains from 9 Staphylococcus spp., carried a range of SCCmec types [I (2), II (6), III (3), V (2), I/II (1) and NT (7)] and varied in their antibiotic susceptibility to other antibiotic classes and carriage of acquired tetracycline/macrolide resistance gene(s). MRSA and MRCoNS donors co-transferred tet(M) and erm(A) genes to an Enterococcus faecalis recipient at a frequency of 10−8.
Conclusions
This is the first report of MRSA and MRCoNS isolated from marine water and intertidal beach sand. The MLST types and antibiotic carriage of five MRSA isolates were similar to hospital MRSA isolates rather than US community-acquired MRSA isolates. Our results suggest that public marine beaches may be a reservoir for transmission of MRSA to beach visitors as well as an ecosystem for exchange of antibiotic resistance genes among staphylococci and related genera.
Keywords: MRSA, MRCoNS, recreational environment, SCCmec, MLST
Introduction
Staphylococcus aureus is a common cause of serious and life-threatening infections. The first methicillin-resistant S. aureus (MRSA) isolates were reported in England in 1961, 1 year after the introduction of methicillin.1 The prevalence of MRSA has increased rapidly over the last decade due in large part to the emergence of community-acquired MRSA (CA-MRSA) infections.2 Most (∼90%) CA-MRSA infections are skin and soft tissue infections; however, more serious and deadly infections do occur.2,3 Repeat MRSA infections are common, and a recent study found that 27% of the MRSA-positive hospital patients were still colonized with MRSA a year after they had been discharged from the hospital.4
S. aureus and MRSA are spread from fomite to person and from person to person5,6 yet few environmental reservoirs outside the healthcare setting, and closed communities such as schools, prisons and sports teams have been characterized.2 A 1987 report describes the isolation of Staphylococcus spp. from the Israeli coastal marine waters, while a second report described the isolation of S. aureus, Staphylococcus epidermidis and Staphylococcus hominis from North Eastern Atlantic ocean and estuarine waters.7,8 More recently both S. aureus and MRSA isolates have been shown to survive river, sea and swimming pool water under laboratory conditions.9 Elmir et al.10 found that 10 volunteers shed ∼6 × 106 S. aureus into the marine water column during a 15 min exposure, which agrees with earlier studies. Together these studies suggest that S. aureus and coagulase-negative Staphylococcus spp. (CoNS) can be isolated from marine environments and the possibility that MRSA and methicillin-resistant CoNS (MRCoNS) may also be present and could be a potential reservoir for transmission of MRSA to beach visitors.
Methicillin resistance in Staphylococcus spp. is due to the acquisition of an altered penicillin-binding protein PBP2a (PBP2′), encoded by the mecA gene.11 The mecA gene is carried on large staphylococcal cassette chromosome mec (SCCmec) elements, which are found in both MRSA and MRCoNS isolates.2 Eight different SCCmec types (I–VIII) have been recognized, which vary in size from 21 to 67 kb, and have different sets of ccr recombinase genes.6,12–14 The tetracycline resistance tet(K) gene, macrolide resistance erm(A) gene, mercury merA gene, transposons and/or plasmids are often integrated within the SCCmec elements.15 Some of the mecA elements contain DNA from other species of staphylococci and it has been hypothesized that these mecA elements are mobile and have been passed from MRCoNS to S. aureus, creating MRSA isolates.2 Multidrug-resistant MRSA are usually associated with hospital isolates that may also be resistant to aminoglycoside, sulfamethoxazole and trimethoprim.16,17
In this study, we determined the antimicrobial susceptibility and carriage of acquired tetracycline and macrolide resistance genes in isolates of MRSA, S. aureus, MRCoNS and CoNS from nine different public marine beaches in Washington State on the West Coast of North America. This study is the first to isolate and characterize MRSA from marine recreational waters and suggests that recreational marine beaches may be a reservoir for direct exposure of the public, with the potential for MRSA colonization and/or disease, and the data suggest that recreational beaches may provide an environment for such exchanges to occur.
Materials and methods
Sample collection
Marine water and intertidal sand samples were taken from 10 public marine beaches in Western Washington, USA from February to September 2008. Wet sand was collected below or at the tide line and 500 mL of marine water was collected during outgoing and low tide, and both were stored at 4°C and processed within 24 h of collection. Water (100 mL) was filtered thorough a GN-6 Metricel 47 mm sterile membrane filter (Pall Co., East Hills, NY, USA) and placed on a Bacto® Staphylococcus Medium 110 agar plate (Staph 110 agar; Difco Laboratories, Becton Dickinson & Co, Sparks, MD, USA) supplemented with 75 mg/L polymyxin B and 0.01% potassium tellurite (Sigma, St Louis, MO, USA). Duplicate filters were incubated at 36.5°C in 5% CO2 for a minimum of 48 h. Sand (5 g) was suspended in either brain heart infusion (BHI) or Bacto® m Staphylococcus Broth (Staph broth; Difco Laboratories) with either 5% NaCl or different concentrations of polymyxin plus 0.01% potassium tellurite. The combination that was superior in suppressing the growth of Gram-negative bacteria and enriching the growth of Gram-positive bacteria was Staph broth supplemented with 25 mg/L polymyxin B and 0.01% potassium tellurite (Sigma) incubated at 36.5°C in 5% CO2 until the broth was turbid (24–72 h), then serially diluted and plated onto Staph agar supplemented with 75 mg/L polymyxin B and 0.01% potassium tellurite with incubation at 36.5°C for 24–48 h. The black colonies were plated onto brucella agar supplemented with 5% sterile sheep blood incubated at 36.5°C in 5% CO2 to screen for the presence of β-haemolysin. Isolates were plated onto Staph 110 agar supplemented with 10 mg/L methicillin to determine methicillin susceptibility. Colonies that produced β-haemolysin were verified as S. aureus using the Remel Staphaurex® rapid latex test according to the manufacturer's instructions (Thermo Fisher Scientific Remel Products, Lenexa, KS, USA). The coagulase-negative isolates were speciated using the BD BBL Crystal™ Identification systems Rapid Gram-positive ID Kit (BD, Franklin Lakes, NJ, USA) and/or by 16S rRNA sequencing as previously described.18
Bacterial isolates
The study included 51 staphylococci isolated from both marine water and sand samples from 9 of 10 beach sites in Washington State (see Table 2).
Table 2.
Phenotypic and genotypic characteristics of MRSA, MSSA, MRCoNS and CoNS from Washington State beaches
| Isolate | Species | Location | Phenotypea | Sccmec type | PFGE | Genotypeb | MLSTc |
|---|---|---|---|---|---|---|---|
| MRSA n = 6 isolates | |||||||
| 10-55 | S. aureus | 1 | ERY, KAN, MET, SXT, TET | I | A | ccrB, erm(A), tet(K), tet(M) | ST1405 |
| 9-48 | S. aureus | 2 | ERY, MET | I | B | ccrB, erm(A) | ST45 |
| 10-62 | S. aureus | 3 | ERY, KAN, MET | I | B | ccrB, erm(A) | ST45 |
| 5-41 | S. aureus | 8 | ERY, KAN, MET, SXT, TET | I | B | ccrB, erm(A), tet(K) | ST45 |
| 10-579 | S. aureus | 3 | CHL, ERY, KAN, MET, SXT, TET | I | C | ccrB, erm(A), tet(M) | ST59 |
| OSS143-7 | S. aureus | 4 | MET | NT | D | none | ST30 |
| MSSA n = 4 strains | |||||||
| 10-51 | S. aureus | 1 | susceptible | ND | A | none | ST15 |
| 10-578 | S. aureus | 3 | susceptible | ND | C | none | ST59 |
| OSS143-6 | S. aureus | 4 | susceptible | ND | D | none | ST30 |
| 11-31, 11-33, 11-34 | S. aureus | 7 | susceptible | ND | D | none | ST59 |
| MRCoNS n = 21 strains | |||||||
| DP6-10 | S. capitis | 6 | CHL, ERY, KAN, MET, SXT, TET | NT | E | msr(A), tet(K) | |
| DP8-11, DP8-37 | S. capitis | 6 | ERY, MET, SXT | II | F | ccrB, erm(C), msr(A) | |
| 10-24 | S. capitis | 9 | ERY, KAN, MET, SXT, TET | I | ccrB, erm(A), tet(M) | ||
| CK962-8 | S. capitis | 9 | MET | NT | ccrB | ||
| 4-29 | S. epidermidis | 3 | ERY, MET, SXT | III | ccrB, erm(B), erm(C), msr(A) | ||
| 4-4, 4-5 | S. epidermidis | 1 | ERY, MET, SXT | V | G | ccrB, erm(C), msr(A) | |
| 5-12 | S. epidermidis | 8 | ERY, MET, SXT | III | ccrB, msr(A) | ||
| 5-13, 5-14 | S. epidermidis | 9 | ERY, MET, SXT | III | H | ccrB, erm(C), msr(A) | |
| 2-36 | S. epidermidis | 8 | ERY, MET, SXT | V | msr(A) | ||
| KSB125-5 | S. epidermidis | 5 | ERY, MET | NT | msr(A) | ||
| 8-50 | S. epidermidis | 9 | MET | NT | I | ccrB | |
| 11-25 | S. haemolyticus | 7 | ERY, MET | I/II | msr(A) | ||
| 28-10 | S. saccharolyticus | 6 | ERY, MET, TET | II | ccrB, erm(C), msr(A), tet(K), tet(M) | ||
| DP6-9 | S. saprophyticus | 6 | ERY, MET | NT | J | ccrB, erm(C), msr(A) | |
| DPW28-7 | S. saprophyticus | 6 | MET | NT | K | none | |
| DPW28-13d | S. saprophyticus | 6 | ERY, MET, SXT, TET | II | L | ccrB, erm(B), erm(C), msr(A), tet(K) | |
| DP8-1d | S. saprophyticus | 6 | ERY, MET, SXT | II | L | ccrB, erm(B), erm(C), msr(A) | |
| DP8-29, 8-59 | S. saprophyticus | 6 | ERY, MET, SXT | II | M | ccrB, erm(B), erm(C), msr(A) | |
| 16-1, 16-2, 16-3, 16-4, 3-3 | S. sciuri | 6 | MET | II | N | ccrB | |
| DPW28-1, 28-9, 28-12 | S. simulans | 6 | ERY, MET, SXT, TET | II | O | ccrB, erm(B), erm(C), msr(A), tet(K) | |
| 10-14, 10-56 | S. vitulinus | 8, 3 | ERY, KAN, MET, SXT, TET | I | ccrB, erm(A), tet(M) | ||
| CK9101-7 | S. xylosus | 9 | MET | NT | none | ||
| CoNS n = 4 strains | |||||||
| 8-13 | S. capitis | 8 | ERY, SXT | ND | erm(A) | ||
| KBS125-1, 125-7 | S. epidermidis | 5 | susceptible | ND | P | none | |
| KBS125-2e | S. epidermidis | 5 | ERY | ND | Q | erm(B), msr(A) | |
| KBS125-4e | S. epidermidis | 5 | ERY, SXT | ND | Q | msr(A) | |
| KBS125-3 | S. warneri | 5 | ERY | ND | erm(A) | ||
aCHL, chloramphenicol; ERY, erythromycin; KAN, kanamycin; MET, methicillin; SXT, trimethoprim/sulfamethoxazole; TET, tetracycline.
bAntibiotic resistance genes examined include: ccrB (site-specific recombinase); erm(A), erm(B), erm(C) and msr(A) (macrolide resistance); and tet(K) and tet(M) (tetracycline resistance).
c MLST type defined by the allelic profile of the seven S. aureus housekeeping genes arc, aro, glp, gmk, pta, tpi and ygi.
d,eCounted as one strain.
Antimicrobial susceptibility tests and detection of tetracycline and macrolide resistance genes
Antimicrobial susceptibilities to chloramphenicol, erythromycin, kanamycin, trimethoprim/sulfamethoxazole and tetracycline were determined by disc diffusion on blood Mueller–Hinton agar (Thermo Fisher Scientific Remel Products) according to CLSI (formerly NCCLS) guidelines.19 S. aureus ATCC 29213 and ATCC 25923 were used as susceptibility control strains. PCR assays were used to determine the presence of macrolide resistance genes, erm(A), erm(B), erm(C) and msr(A), and tetracycline resistance genes, tet(M) and tet(K), with verification by hybridization with an internal 32P-labelled probe as described previously.20 Cloned positive controls for the PCR assays were pEM9592 for erm(A), pJIR229 for erm(B), pBR328:33RV for erm(C), pJ13 for tet(M), pAT102 for tet(K) and non-cloned RN42201 for msr(A),20–22 and Enterococcus faecalis JH2-2 was used as a negative control as previously described.20
PCR assays for mecA and ccrB genes and Panton–Valentine leucocidin (PVL) genes, SCCmec typing and sequencing
Primers used in the study are listed in Table 1. Detection of the presence of the mecA gene was done by PCR assay with the following cycling parameters: initial denaturation at 96°C for 3 min; followed by 35 cycles of 30 s at 96°C, annealing for 1 min at 55°C and 2 min at 72°C; and an extension for 10 min at 72°C.
Table 1.
List of oligonucleotide primers used in this study
| Gene/SCC type/region | Primer name | Sequence (5′→3′) | Reference |
|---|---|---|---|
| mecA | MecAF | CAT TGA TCG CAA CGT TCA ATT T | this study |
| MecAR | CGG TTT TAA AGT GGA ACG AAG GT | this study | |
| MecAInt | TGG AAG TTA GAT TGG GAT CAT AGC GTC | this study | |
| SCCmec I | type I-F | GCT TTA AAG AGT GTC GTT ACA GG | 12 |
| type I-R | GTT CTC TCA TAG TAT GAC GTC C | 12 | |
| SCCmec II | type II-F | CGT TGA AGA TGA TGA AGC G | 12 |
| type II-R | CGA AAT CAA TGG TTA ATG GAC C | 12 | |
| SCCmec III | type III-F | CCA TAT TGT GTA CGA TGC G | 12 |
| type III-R | CCT TAG TTG TCG TAA CAG ATC G | 12 | |
| SCCmec IV | type IV-F | ACC AAC GTT TGT AGC GGG TT | this study |
| type IV-R | AAG CGT CCA CGT CAT CTT CA | this study | |
| SCCmec V | type V-F | GAA CAT TGT TAC TTA AAT GAG CG | 12 |
| type V-R | TGA AAG TTG TAC CCT TGA CAC C | 12 | |
| ccrB | αc | ATT GCC TTG ATA ATA GCC TTC T | 24 |
| βc | ATC TAT TTC AAA AAT GAA CCA | 24 | |
| PVL | luk-PV1 | ATC ATT AGG TAA AAT GTC TGG ACA TGA TCC A | 25 |
| luk-PV-2 | GCA TCA AST GTA TTG GAT AGC AAA AGC | 25 | |
| mecA-mecI (mA7-mI6) | mA7 | ATA TAC CAA ACC CGA CAA CTA CA | 26 |
| mI6 | CAT AAC TTC CCA TTC TGC AGA TG | 26 |
The ccr gene complex encodes site-specific recombinase products responsible for integration and excision of the SCCmec element.11 The ccrB gene was chosen as the target gene because it has highly conserved sequences compared with the ccrA and ccrC genes.23 All the mecA-positive isolates were tested for the ccrB gene, as described previously.24 The MRSA isolates were tested for the PVL gene using the PCR assay described previously.25
The SCCmec I–V typing was done using five pairs of multiplex primers and PCR conditions described previously.12 Some variants of SCCmec type II are negative in the SCCmec multiplex assay, thus all the isolates that were non-typeable (NT) by the multiplex PCR were tested with the PCR assay previously described.26 A clinical MRSA USA300 strain was used as a positive control and S. aureus ATCC 29213 was used as a negative control for all four PCR assays.
Sequencing of the mecA PCR products in the mecA–mecI region was done on all isolates that were NT by the multiplex SCCmec typing PCR to determine if they carried an SCCmec type II element.26 The PCR products were sequenced bi-directionally at the University of Washington, Genome Sciences High Throughput Sequencing Facility, Seattle, WA, USA. Homology searches were performed with BLAST (http://www.ncbi.nlm.nih.gov), multiple sequences were aligned with Clustal W (http://www.ebi.ac.uk/clustalw/index.html) and assembled using Vector NTI Advance 10™ (Invitrogen, Carlsbad, CA, USA).
PFGE
PFGE analyses were done on the six MRSA, six methicillin-susceptible S. aureus (MSSA) and 22 MRCoNS. The DNA blocks were prepared, digested separately with SmaI (Fermentas Inc., Glen Burnie, MD, USA) for 6 h at 28°C, and subjected to electrophoresis in a contour-clamped homogeneous electric field (CHEF DR II system; Bio-Rad Laboratories Inc., Hercules, CA, USA) for 21 h at 14°C with switch times of 5 s (initial) and 40 s (final) at 6 V/cm as described previously.27 The gels were stained with ethidium bromide, de-stained in distilled water and photographed under UV transillumination. The different PFGE patterns were labelled A–Q. Isolates were considered to have the same PFGE pattern if they had indistinguishable patterns or patterns that differed by ≤3 bands as previously described28 (see Table 2).
Multilocus sequence typing (MLST)
MLST was performed using previously published primer sequences and conditions for the PCR amplification of S. aureus housekeeping genes arc, aro, glp, gmk, pta, tpi and ygi.29 The PCR products were sequenced bi-directionally at the University of Washington, Genome Sciences High Throughput Sequencing Facility. Alleles were assigned by a comparison of their sequences with the corresponding loci in the S. aureus MLST database (www.mlst.net). Sequencing of the glp allele of MRSA isolate 10-55 was repeated three times bi-directionally to ensure reproducibility, and a new glp allele type 193 and ST1405 were assigned (www.mlst.net).
Conjugation transfer studies
Mating experiments were performed as described previously.20 MRSA isolate 10-55 and MRCoNS isolates Staphylococcus vitulinus 10-14 and Staphylococcus capitis 10-24 were used as donors with recipient E. faecalis JH2-2 that had previously been selected for chromosomal resistance to streptomycin (500 mg/L), rifampicin (25 mg/L), nalidixic acid (25 mg/L) and fusidic acid (25 mg/L).20 E. faecalis transconjugants were selected on BHI agar (Difco Laboratories) supplemented with 10 mg/L tetracycline and 25 mg/L fusidic acid, or 10 mg/L erythromycin and 25 mg/L fusidic acid. Transconjugants were verified as E. faecalis and transferred resistance genes confirmed by PCR.
Results
Antimicrobial susceptibility and characterization of MRSA and S. aureus
Six MRSA isolates were isolated from five different beach sites. Five of these isolates were resistant to erythromycin, carried an erm(A) gene, were SCCmec type I, carried a ccrB gene and were isolated from four different beaches in urban environments. Three of these isolates, 9-48, 10-62 and 5-41, had in addition the same PFGE pattern, B, and the same MLST type, ST45, though they differed in phenotypic and genotypic characteristics, with isolate 5-41 being tetracycline resistant and carrying the tet(K) gene (Table 2). The remaining two SCCmec type I isolates, 10-55 and 10-579, differed in their phenotypic and genotypic characteristics and had different PFGE and MLST types, though both were tetracycline resistant and carried the tet(M) gene. Isolate 10-55 also carried a tet(K) gene and had a novel MLST type ST1405 while 10-579 was MLST type ST59. Isolate OSS143-7 was phenotypically susceptible to all other classes of antibiotics tested, and was SCCmec NT, had distinct PFGE patterns and did not carry the ccrB gene. None of the MRSA isolates carried the PVL genes.
Six MSSA representing four strains were isolated from four beach sites. All MSSA were susceptible to all five other antibiotics tested and PCR negative for antibiotic resistance genes (Table 2). The MSSA 10-578 and MRSA 10-579 isolated from the same beach and MSSA OSS143-6 and MRSA OSS143-7 isolated from the same beach had the same PFGE and MLST pattern, suggesting that each pair of isolates from the same beach were probably genetically related. In contrast, MSSA 10-51 and MRSA 10-55, both isolated from the same beach, had indistinguishable PFGE patterns but differed in their MLST type, ST15 versus new ST1405. Similarly, the MSSA 11-31 group of isolates were all ST59 yet had the same PFGE pattern D as OSS143-6 and OSS143-7, which were both ST30 (Table 2).
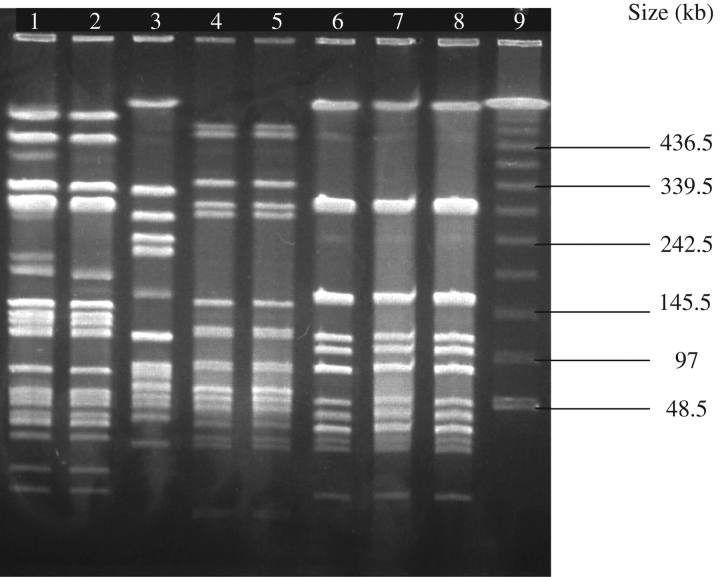
See Figure 1 for PFGE with SmaI enzyme of MRSA and MSSA.
Figure 1.
PFGE with the SmaI enzyme of MRSA and MSSA. PFGE pattern A: lane 1, MRSA 10-55; and lane 2, MSSA 10-51. PFGE type B: lane 3, MRSA 9-48. PFGE type C: lane 4, MRSA 10-579; and lane 5, MSSA 10-578. PFGE type D: lane 6, MRSA OSS143-7; lane 7, MSSA OSS143-6; and lane 8, MSSA 11-31. Lane 9, MW standards.
Antimicrobial susceptibility and characterization of MRCoNS and CoNS isolates
Thirty-three MRCoNS belonging to nine different Staphylococcus spp. were isolated from seven beach sites (Table 2). All isolates were phenotypically and genotypically characterized and SCCmec typed. A number of the isolates from the same site were identified as the same species and had the same phenotype and genotype, as well as the same SCCmec type, suggesting that they could represent the same strain. To verify whether this hypothesis was correct, 22 of the MRCoNS including 3 S. capitis, 5 S. epidermidis, 6 Staphylococcus saprophyticus, 5 Staphylococcus sciuri and 3 Staphylococcus simulans were PFGE typed. Eleven distinct PFGE patterns (types E–O) were identified from these 22 MRCoNS isolates. The isolates with indistinguishable PFGE patterns were grouped together. However, not all six S. saprophyticus isolates from beach site 6 represented a single strain since four different PFGE patterns, J, K, L and M, were identified, suggesting that the six isolates represented four strains. DPW28-13 and DP8-1 had the same PFGE pattern, L, but DPW28-13 was tetracycline resistant and carried the tet(K) gene, while DP8-1 was tetracycline susceptible; therefore, the two were considered variants of the same strain. Thus the number of different MRCoNS strains isolated was reduced to 21.
Six MRCoNS strains carried SCCmec type II, two carried type I, three carried type III, two carried type V, one isolate carried two mecA elements (type I and type II) and seven strains were NT. All the strains carried the ccrB gene except one type V strain (2-36), one type I/II strain (11-25) and four NT strains (Table 2).
Of the 21 MRCoNS strains, S. capitis CK962-8, S. epidermidis 8-50, S. saprophyticus DPW28-7, S. sciuri 16-1 group and Staphylococcus xylosus CK9101-7 were susceptible to all the other antibiotic classes tested. The CK962-8, 8-50, DPW28-7 and CK9101-7 strains were NT, while the S. sciuri 16-1 group were type II. In contrast, the 16 (76%) multidrug-resistant MRCoNS, from 8 of 9 species, were erythromycin resistant, with 5 positive for msr(A), 2 positive for erm(A), 5 positive for both erm(C) and msr(A) and 4 positive for erm(B), erm(C) and msr(A). Twelve strains were trimethoprim/sulfamethoxazole resistant, though the specific genes carried were not determined. Six strains, S. capitis DP6-10 and 10-24, Staphylococcus saccharolyticus 28-10, S. saprophyticus DPW28-13, S. simulans DPW28-1 group and the S. vitulinus 10-14 group, carried tet(K) (three strains), tet(M) (two strains) or both tet(K) and tet(M) (one strain). Three strains, S. capitis DP6-10 and 10-24 and S. vitulinus 10-14 group, were kanamycin resistant. The S. capitis DP6-10 strain was resistant to chloramphenicol but which gene was carried was not determined.
Five (83%) of the six methicillin-susceptible CoNS isolates were isolated from a single beach and four of the isolates were S. epidermidis. S. epidermidis KBS125-1 and KBS125-7 were susceptible to all antibiotics tested, had the indistinguishable PFGE pattern P and represented a single strain (Table 2). The other two, S. epidermidis KBS125-2 and KBS125-4, are listed separately because KBS125-4 was erythromycin and trimethoprim/sulfamethoxazole resistant and positive for the msr(A) gene, while KBS125-2 was erythromycin resistant and positive for both the msr(A) and erm(B) genes. However, the two isolates had indistinguishable PFGE pattern Q and most probably represent a single strain. The erythromycin- and trimethoprim/sulfamethoxazole-resistant S. capitis 8-13 was isolated at a different beach site and carried the erm(A) gene, while the erythromycin-resistant Staphylococcus warneri KBS125-3 carried the erm(A) gene (Table 2).
Conjugal gene transfer
MRSA isolate 10-55 [erm(A), tet(K), tet(M)], S. capitis 10-24 [erm(A), tet(M)] and S. vitulinus 10-14 [erm(A), tet(M)] were used as donors with E. faecalis JH2-2 as the recipient. All three donors were able to co-transfer their erm(A) and tet(M) genes at transfer rates ranging between 2.7 × 10–8 and 5.8 × 10–8 per recipient. Donor 10-55 carried both the tet(K) and tet(M) genes but only the tet(M) gene transferred to the E. faecalis.
Discussion
This is the first report of identification and characterization of MRSA isolated from marine waters and intertidal beach sand in the literature and the first report of MLST type ST1405. We have also verified that the staphylococci in the beach environment carry tetracycline and macrolide resistance genes on mobile elements, and gene exchange of mobile antibiotic resistance genes is possible. The source of the MRSA in the study is not clear, though the five multidrug-resistant MRSA SCCmec type I carried the erm(A) gene and were PVL negative, and four of the five MRSA strains carried previously described MLST types, all characteristics commonly associated with hospital MRSA, though the beach sites were not near hospitals.30 We cannot rule out the possibility that these multidrug-resistant MRSA SCCmec type I strains came from swimmers; however, the normal surface water temperature at these beaches ranges from 15 to 18°C while the deeper water temperature ranges from 8 to 12°C, limiting the likelihood of swimming or complete immersion by beach visitors in these waters. Three of the multidrug-resistant MRSA isolates had common PFGE and MLST types, but were isolated from three beach sites located within 16 km of each other. Although these three strains differ in their antibiotic resistance patterns and the number of antibiotic resistance genes carried, we cannot rule out the possibility that these three came from a common source. The multidrug-resistant MRSA strains 10-55 and 10-579 were unique, while the MRSA strain OSS143-7 was susceptible to other antibiotic classes, isolated from rural sites with limited populations and healthcare institutions. OSS143-7 was MLST type ST30, which is one of the most successful widespread genetic lineages found among MSSA described.31 This strain did not have traits commonly associated with either hospital MRSA or USA300, the most common strain of CA- MRSA in the USA.
Two of the four MSSA strains shared PFGE and MLST types in common with the MRSA isolates from the same beaches. In contrast, 10-51 and 10-55 shared PFGE patterns, as did the 11-31 group with OSS143-6 and OSS143-7, but differed in their MLST types, ST15 versus ST1405, and ST59 versus ST30, respectively. Disparity between PFGE and MLST typing has previously been reported for both MRSA and MSSA isolates.32
We characterized 21 different strains of MRCoNS isolates, of which 14 carried one of the known SCCmec types. Sixteen of the MRCoNS were multidrug resistant, and it is likely that many of these antibiotic resistance genes are associated with mobile elements as illustrated by the ability of the S. capitis 10-24 and S. vitulinus 10-14 strains to transfer their erm(A) and tet(M) genes to a susceptible recipient. We have found that the tet(M) gene readily goes into recipients that are from genera different to the donor,33 thus demonstrating that the erm(A) and tet(M) genes are associated with mobile elements, suggesting that these elements may also be transferred into other staphylococci. Therefore we have hypothesized that the environmental staphylococci carrying antibiotic resistance genes could also act as donors in the environment and transfer antibiotic resistance genes to other environmental staphylococci.
The level of risk to the public of acquiring MRSA when visiting public beaches where MRSA is isolated is unknown. However, a recent study where >27 000 people were interviewed34 found an increased incidence of subsequent illness following digging or being buried in the sand. Our results indicate that public marine beaches may be a potential reservoir for transmission of MRSA to beach visitors, especially those with skin lesions. The results presented in this study are unlikely to be unique to Washington State or North America and further studies are needed to determine the distribution and level of MRSA in public marine and fresh water beaches.
Funding
This study was supported in part by the National Institute of Environmental Health Sciences grant 5R25ES016150-02.
Transparency declarations
None to declare.
Acknowledgements
This study was presented in part at the Forty-ninth Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, 2009 (C2-146).
References
- 1.Jevons MP. “Celbenin”-resistant staphylococci. BMJ. 1961;1:124–5. [Google Scholar]
- 2.Eady EA, Cove JH. Staphylococcal resistance revisited: community-acquired methicillin resistant Staphylococcus aureus—an emerging problem for the management of skin and soft tissue infections. Curr Opin Infect Dis. 2003;16:103–24. doi: 10.1097/00001432-200304000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Miller LG, Diep BA. Colonization, fomites, and virulence: rethinking the pathogenesis of community-associated methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46:452–60. doi: 10.1086/526773. [DOI] [PubMed] [Google Scholar]
- 4.Datta R, Huang SS. Risk of infection and death due to methicillin-resistant Staphylococcus aureus in long-term carriers. Clin Infect Dis. 2008;47:176–81. doi: 10.1086/589241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khivsara A, Sushma TV, Dhanashree B. Typing of Staphylococcus aureus from mobile phones and clinical samples. Curr Sci. 2006;90:910–12. [Google Scholar]
- 6.Zhang K, McClure JA, Elsayed S, et al. Staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53:531–40. doi: 10.1128/AAC.01118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshpe-Purer Y, Golderman S. Occurrence of Staphylococcus aureus and Pseudomonas aeruginosa in Israeli costal water. Appl Environ Microbiol. 1987;53:1138–41. doi: 10.1128/aem.53.5.1138-1141.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunn BA, Singleton FL, Peele ER, et al. A note on the isolation and enumeration of Gram-positive cocci from marine and estuarine waters. J Appl Bacteriol. 1982;53:127–9. doi: 10.1111/j.1365-2672.1982.tb04742.x. [DOI] [PubMed] [Google Scholar]
- 9.Tolba O, Loughrey A, Goldsmith CE, et al. Survival of epidemic strains of healthcare (HA-MRSA) and community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) in river-, sea-, and swimming pool water. Int J Hyg Environ Health. 2008;211:398–402. doi: 10.1016/j.ijheh.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Elmir SM, Wright ME, Abdelzaher A, et al. Quantitative evaluation of bacteria released by bathers in a marine water. Water Res. 2007;41:3–10. doi: 10.1016/j.watres.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito T, Katayama Y, Asada K, et al. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1323–36. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang K, McClure JA, Elsayed S, et al. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–33. doi: 10.1128/JCM.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berglund C, Ito T, Ikeda M, et al. Novel type of staphylococcal cassette chromosome mec in a methicillin-resistant Staphylococcus aureus strain isolated in Sweden. Antimicrob Agents Chemother. 2008;52:3512–6. doi: 10.1128/AAC.00087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliveira DC, Milheirico C, de Lencastre H. Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob Agents Chemother. 2006;50:3457–9. doi: 10.1128/AAC.00629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grundmann H, Aires-de-Sousa M, Boyce J, et al. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;368:874–85. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 16.Donnio PY, Preney L, Gautier-Lerestif AL, et al. Changes in staphylococcal cassette chromosome type and antibiotic resistance profile in methicillin-resistant Staphylococcus aureus isolates from a French hospital over an 11 year period. J Antimicrob Chemother. 2004;53:808–13. doi: 10.1093/jac/dkh185. [DOI] [PubMed] [Google Scholar]
- 17.Novotna G, Adamkova V, Janata J, et al. Prevalence of resistance mechanisms against macrolides and lincosamides in methicillin-resistant coagulase-negative staphylococci in the Czech Republic and occurrence of an undefined mechanism of resistance to lincosamides. Antimicrob Agents Chemother. 2005;49:3586–9. doi: 10.1128/AAC.49.8.3586-3589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bosshard PP, Abels S, Zbinden R, et al. Ribosomal DNA sequencing for identification of aerobic Gram-positive rods in the clinical laboratory (an 18-month evaluation) J Clin Microbiol. 2003;41:4134–40. doi: 10.1128/JCM.41.9.4134-4140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Committee for Clinical and Laboratory Standards. Performance Standards for Antimicrobial Disk Susceptibility Tests-Eighth Edition: Approved Standard M2-A8. Wayne, PA, USA: NCCLS; 2003. [Google Scholar]
- 20.Luna VA, Heiken M, Judge K, et al. Distribution of mef(A) in gram-positive bacteria from healthy Portuguese children. Antimicrob Agents Chemother. 2002;46:2513–7. doi: 10.1128/AAC.46.8.2513-2517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luna VA, Roberts MC. The presence of the tetO gene in a variety of tetracycline-resistant Streptococcus pneumoniae serotypes from Washington State. J Antimicrob Chemother. 1998;42:613–9. doi: 10.1093/jac/42.5.613. [DOI] [PubMed] [Google Scholar]
- 22.Pang Y, Bosch T, Roberts MC. Single polymerase chain reaction for the detection of tetracycline-resistant determinants Tet K and Tet L. Mol Cell Probes. 1994;8:417–22. doi: 10.1006/mcpr.1994.1059. [DOI] [PubMed] [Google Scholar]
- 23.Yang JA, Park DW, Sohn JW, et al. Novel PCR-restriction fragment length polymorphism analysis for rapid typing of staphylococcal cassette chromosome mec elements. J Clin Microbiol. 2006;44:236–8. doi: 10.1128/JCM.44.1.236-238.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibrahem S, Salmenlinna S, Lyytikäinen O, et al. Molecular characterization of methicillin-resistant Staphylococcus epidermidis strains from bacteraemic patients. Clin Microbiol Infect. 2008;14:1020–7. doi: 10.1111/j.1469-0691.2008.02080.x. [DOI] [PubMed] [Google Scholar]
- 25.Lina G, Piémont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–32. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 26.Kondo Y, Ito T, Ma XX, et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51:264–74. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDougal LK, Steward CD, Killgore GE, et al. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–20. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts MC, Soge OO, Giardino MA, et al. Vancomycin-resistant Enterococcus spp. in marine environments from the West Coast of the USA. J App Microbiol. 2009;107:300–7. doi: 10.1111/j.1365-2672.2009.04207.x. [DOI] [PubMed] [Google Scholar]
- 29.Enright MC, Day NP, Davies CE, et al. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–15. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson DA, Enright MC. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47:3926–34. doi: 10.1128/AAC.47.12.3926-3934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Lencastre H, Oliveira DC, Tomasz A. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr Opin Microbiolog. 2007;10:428–35. doi: 10.1016/j.mib.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faria NA, Carrico JA, Oliveria DC, et al. Analysis of typing methods for epidemiological surveillance of both methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Clin Microbiol. 2008;46:136–44. doi: 10.1128/JCM.01684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agerso Y, Jensen LB, Givskov M, Roberts MC. The identification of a tetracycline resistance gene tet(M), on a Tn916-like transposon, in the Bacillus cereus group. FEMS Microbiol Lett. 2002;214:251–6. doi: 10.1111/j.1574-6968.2002.tb11355.x. [DOI] [PubMed] [Google Scholar]
- 34.Heaney CD, Sams E, Wing S, et al. Contact with beach sand among beachgoers and risk of illness. Am J Epidemiol. 2009;170:164–72. doi: 10.1093/aje/kwp152. [DOI] [PubMed] [Google Scholar]