Abstract
Inefficient glycosylation caused by defective synthesis of lipid-linked oligosaccharide donor results in multi-systemic syndromes known as congenital disorders of glycosylation type I (CDG-I). Strong loss of function mutations are embryonic lethal, patients with partial losses of function are occasionally born but are very ill, presenting with defects in virtually every tissue. CDG-I clinical expression varies considerably and ranges from very mild to severe, and the underlying cause of the variable clinical features is not yet understood. We postulate that accompanying defects in an individual's genetic background enhance the severity of CDG-I clinical phenotypes. Since so many protein structures and functions are compromised in CDG-I illnesses, the gene products that are dependent on N-linked glycosylation which cause lethality or particular symptoms are difficult to resolve. The power of genetic silencing that is a characteristic of C. elegans has allowed us to systematically dissect the complex glycosylation phenotype observed in CDG-I patients into specific glycan-dependent gene products. To accomplish this, we inhibited glycosylation with a sub-phenotypic dose of tunicamycin, reduced single genes by RNA interference, and then sought loci where the combination caused a synthetic or dramatically enhanced phenotype. This screen has identified genes in C. elegans that require N-linked glycans to function properly as well as candidate gene homologues that may enhance the clinical severity of CDG-I disorders in humans.
Keywords: C. elegans, congenital disorders of glycosylation, lipid-linked oligosaccharide, RNAi, tunicamycin
Introduction
Protein N-glycosylation is an essential and pervasive event in eukaryotic development. Mild defects in this process lead to deficiencies in cellular growth and function and are associated with disease. Most notable are a group of inherited autosomal recessive human disorders called congenital disorders of glycosylation (CDG). CDGs are categorized as type I or type II depending on the location in the biochemical pathway where the defect occurs. Type I disorders are characterized as defects of enzymes in the lipid-linked oligosaccharide assembly pathway. Currently there are 12 known defects, or subtypes, in this category. Type II disorders are deficiencies of enzymes involved in the trimming and processing of the protein bound oligosaccharide. To date, there are eight defective enzymes within type II CDGs (Jaeken and Matthijs 2007; Zeevaert et al. 2008).
CDG is a rare disease with roughly 900 documented cases (Vodopiutz and Bodamer 2008). In addition to the infrequency of CDG, there is evidence suggesting a clear disequilibrium with the frequency of the disease in the population (Schollen et al. 2000). Two possibilities exist to explain the disequilibrium of CDG patients, either CDG illnesses are underdiagnosed or those afflicted die before birth (Matthijs et al. 2000; de Lonlay et al. 2001).
Individuals born with CDG express defective glycosylation enzymes that lead to absent and/or aberrant glycan structures (Mills et al. 2001). CDG patients suffer from psychomotor retardation, low muscle tone, incomplete brain development, visual problems, seizures, stroke-like episodes, coagulation disorders, endocrine abnormalities, and overall failure to thrive (Eklund and Freeze 2006). The biosynthetic pathway for glycoprotein formation is quite well understood and has provided a basis for establishing the etiology of CDG (Jaeken and Matthijs 2001). However, there are still cases of CDG with unknown etiology, where strong evidence points to CDG but the defective gene has yet to be identified (Prietsch et al. 2002).
Since all CDG-I etiologies alter the effectiveness and substrate availability of oligosaccharyltransferases, it might be expected that the disease would manifest consistently from patient to patient. This is not the case, as clinical CDG symptoms within types and subtypes vary considerably between individuals, making diagnosis difficult. Additionally, in any specific CDG illness, where the genetic mutation is the same, no consistent clinical manifestation exists and the severity of the illness ranges from very mild to severe (Aebi and Hennet 2001). To date clinical evidence fails to explain the exceeding inconsistency between the etiology and pathology in all CDG conditions.
We hypothesize that an individual's genetic background alters or aggravates a CDG glycosylation defect to give rise to the inconsistent clinical symptoms. Given the diversity of glycoproteins made by eukaryotes and the extent of N-glycosylation, additional biochemical defects directly or indirectly involved with a glycosylation defect could elicit variable phenotypes characteristic of CDG diseases. For example, the clinical severity of CDG-Ia, caused by phosphomannomutase deficiency, is enhanced by a defect in α1,3-glucosyltransferase, an enzyme also found in the lipid-linked oligosaccharide (LLO) pathway (Westphal et al. 2002). Defects in α-1,3-glucosyltransferase alone result in CDG-Ic illnesses. In addition to this example, previous reports allude to the possibility of genetic and/or environmental factors that contribute to the clinical spectrum observed in CDG-I patients (Freeze and Westphal 2001).
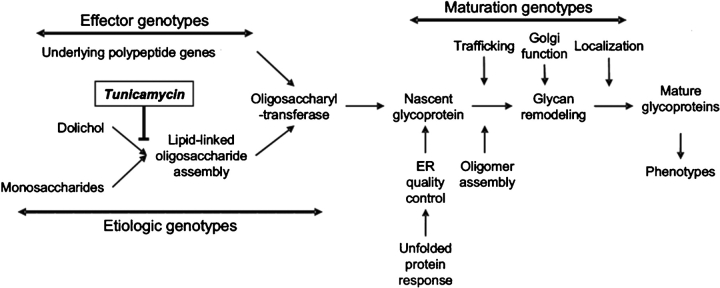
In broad terms, clinical heterogeneity among CDG patients could have four origins. An individual may have mutations in genes encoding (1) polypeptides that require N-glycans for function, (2) polypeptides that require N-glycans for structure, (3) genes responsible for glycoprotein maturation (i.e., unfolded protein response, secretion or Golgi function), (4) genes that are involved in the lipid-linked oligosaccharide assembly pathway. Any individual with CDG may have additional genetic defects in their genome that rely on N-linked glycosylation to function correctly. It is this interaction between the CDG glycosylation defect and a secondary genetic mutation or flaw that is the grounds for the variable pathology in CDG cases.
In this study, we explored genetic factors that may influence CDG pathogenesis. We postulated that for any cellular function that depends on N-glycosylation, a protein component must be involved (the underlying polypeptide of a glycoprotein, a lectin receptor, etc.). Due to the fact that so many structures and functions are compromised in CDG diseases, the gene products that contribute to lethality or particular symptoms are difficult to resolve. Here we have demonstrated a novel approach that combines RNA-mediated interference (RNAi) with an N-glycosylation inhibitor to systematically dissect this problem on a gene-by-gene basis in C. elegans. The annotated glycome of C. elegans, in addition to its well-characterized physiology, provides a fundamental starting point in studying complex developmental topics and offers a robust model system to examine CDG diseases (Schachter 2004; Berninsone 2006; Lehle et al. 2006; Paschinger et al. 2008).
We exploited C. elegans’ susceptibility to genetic silencing to conduct a genome-wide RNAi screen to identify genes that are dependent upon N-linked glycosylation (Fire et al. 1998). To accomplish this, we used a sub-phenotypic dose of the nucleoside antibiotic tunicamycin to mimic a very mild CDG-I-like condition. Tunicamycin inhibits the enzyme DPAGT1 in the first step of the lipid-linked oligosaccharide biosynthetic pathway (Barnes et al. 1984; Zhu et al. 1992). Hypomorphic mutations of the corresponding gene in humans are associated with CDG-Ij (Wu et al. 2003). Without the formation of dolichol pyrophosphate N-acetylglucosamine, no donor for oligosaccharyltransferase can be formed and protein N-glycosylation is absent.
In addition to tunicamycin treatment, we decreased the activity of an individual glycan-dependent protein via RNAi which rendered it hypersensitive to reduced glycosylation. Since this interaction is largely specific, glycan-dependent gene products can be identified. Under these conditions, gene-products that control glycosylation efficiency or glycoprotein maturation, or encode key glycoproteins that require glycosylation for their synthesis or activity, will manifest synthetic enhancer or suppressor phenotypes. The genome-wide RNAi screen with the ORFeome v1.1 library and its analysis in silico have revealed 512 tunicamycin-hypersensitive loci, some of which are characterized and cloned but not previously recognized to be N-glycosylation dependent.
Results
Effect of tunicamycin on C. elegans development
The phenotypic qualities of a CDG-I-like defect were characterized in C. elegans using 0, 3, and 5 μg/mL tunicamycin (Table I). Like CDG-I in humans, tunicamycin treatment in C. elegans resulted in pleiotropic traits. In the absence of tunicamycin, nearly all animals reached adulthood and exhibited no aberrant phenotype. At 3 μg/mL concentration, the consequences of tunicamycin treatment were more evident. Only ∼76% of the population was wild type and reached the adult stage. Nearly 10% of the larvae died. Interesting to note was the range of phenotypes present with a mild dose of drug, characteristic of CDG-I diseases. When the tunicamycin concentration was increased to 5 μg/mL almost all animals remained in the larval stage, of which ∼75% were lethal. Only 1% of animals scored grew to become wild-type adults.
Table I.
Tunicamycin treatment results in an increase in observable aberrant phenotypes. Tunicamycin concentration at 3 μg/mL results in a wide variety of phenotypes among N2 adults and larvae, emulating a CDG-I-like pleotropic effect. Concentrations at 5 μg/mL result predominantly in lethality.
| Tunicamycin (μg/mL) | ||||
|---|---|---|---|---|
| Stage | Phenotype | 0 | 3 | 5 |
| Adult | W.T. | 99.81% | 75.63% | 1.01% |
| Dpy | 0.00% | 3.39% | 1.72% | |
| Unc | 0.00% | 2.35% | 0.20% | |
| Sma | 0.00% | 2.04% | 0.00% | |
| Vab | 0.00% | 0.63% | 0.00% | |
| Egl | 0.00% | 0.57% | 0.00% | |
| Clr | 0.00% | 0.05% | 0.00% | |
| Dpy Unc | 0.00% | 0.05% | 0.00% | |
| Dpy Egl | 0.05% | 0.05% | 0.00% | |
| Larval | Gro | 0.14% | 1.51% | 14.34% |
| Dpy Gro | 0.00% | 1.10% | 7.95% | |
| Let | 0.00% | 9.71% | 74.77% | |
| n = | 2119 | 1916 | 1974 | |
Clr, clear patches; Dpy, dumpy; Egl, egg-laying defective; Gro, slow growth; Let, lethality; Sma, small; Unc, uncoordinated locomotion; Vab, variably abnormal morphology; W.T., wild type.
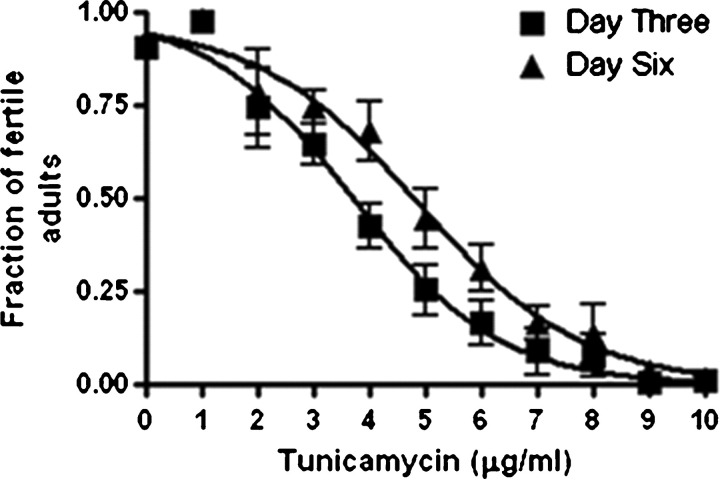
The success of the genome-wide RNAi screen relied on a low dose of the antibiotic tunicamycin to emulate a mild form of CDG-type Ij. In C. elegans high doses of tunicamycin caused adult and embryonic lethality, but partial inhibition by lower doses resulted in variable phenotypes. Tunicamycin concentration of 2 μg/mL or less did not significantly affect larval development, but higher concentrations caused survival rates to decrease (Figure 1). However, the presence of 2 μg/mL tunicamycin emulated a lesser “sub-phenotypic” form of CDG-Ij in C. elegans.
Fig. 1.
Tunicamycin concentrations greater than 2 μg/mL induce a dose-dependent lethality in N2 (Bristol) strains. Each line represents the same observable population, but at day 3 and day 6 respectively. Concentrations of 8 or greater result in nearly complete lethality of larvae. Results are based on triplicate experiments (n = 264) for each tunicmaycin treatment.
Postembryonic development among C. elegans strains in the presence of tunicamycin
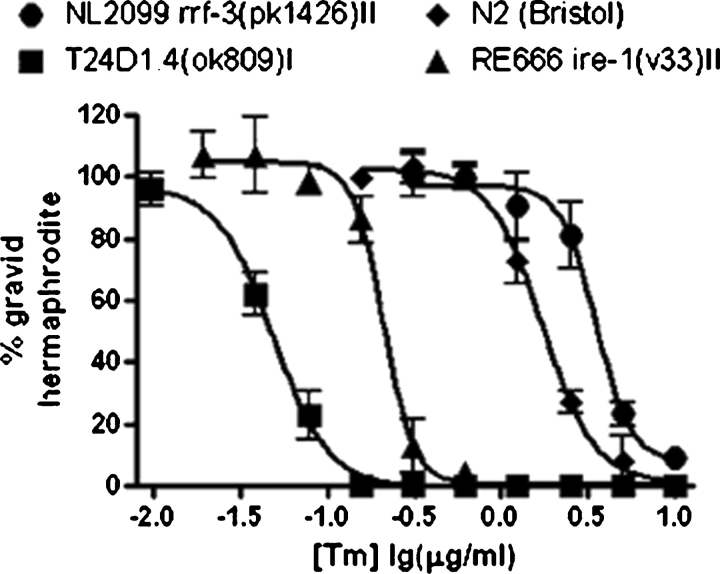
Tunicamycin dose-dependent lethality in C. elegans varied from one mutant strain to another (Figure 2). Four strains were grown in the presence of increasing concentrations of tunicamycin. The four test-strains included: (1) N2 (Bristol) wild-type strain, (2) VC569 tag-179 (ok809) which is a complete loss of function of glucosyltransferase activity required to add the terminal glucose to the LLO formed in the endoplasmic reticulum (ER), (3) RE666 ire-1 (v33) which is a loss of a serine-threonine kinase required for the unfolded protein response (UPR), (4) NL2099 rrf-3 (pk1426) which is a homozygous deletion of an RNA-directed RNA polymerase that inhibits somatic RNAi. In addition to N2 as a control, NL2099 was assayed for validation in the genome-wide screen because of its hypersensitivity to RNAi gene knockdown. The VC569 and RE666 strains were chosen because of their role in N-glycosylation. As expected, the C. elegans VC569 strain was exceedingly sensitive to tunicamycin than N2, possibly due to a decrease in substrate availability caused by blockage of DPAGT1 in the ER. Additionally, the strain RE666 was more sensitive to tunicamycin than N2 (Bristol) or NL2099 strains and reports show that elevated concentrations of tunicamycin can trigger the UPR in C. elegans (Shen et al. 2001; Calfon et al. 2002). Since the NL2099 strain was used in the genome-wide screen, it is necessary to note that the effect of 2 μg/mL tunicamycin between N2 and NL2099 was comparatively minor.
Fig. 2.
Tunicamycin affects postembryonic development among C. elegans strains. The percent of animals that reach fertile adulthood was measured as a function of tunicamycin concentration. Development of RE666 ire-1 (v33), which lacks a component of the unfolded protein response, is moderately affected when tunicamycin is present. Lethality is significantly increased in VC569 tag-179 (ok809), which encodes the enzyme responsible for the final step of LLO biosynthesis. Conversely, N2 (Bristol) and NL2099 rrf-3 (pk1426) strains are less affected by the presence of tunicamycin and are viable at 2 μg/mL.
Assessment of genes recognized to be tunicamycin hypersensitive
A series of RNAi trials were conducted to validate the legitimacy of the screen and the existence of tunicamycin-hypersensitive loci. A small number of genes that were suspected to be hypersensitive to tunicamycin were assayed on 2 μg/mL tunicamycin in an rrf-3 (pk1426) mutant background. These included six “etiologic” genes from the ER luminal phase of the LLO pathway (Table II) and six “maturation” genes in endoplasmic reticulum-Golgi functions (i.e., UPR or glycoprotein secretion) (Table III). Our results were also compared to previously described RNAi phenotypes with an rrf-3 (pk1426) mutant background of the same genes annotated in WormBase (http://www.wormbase.org, source for C. elegans RNAi and gene product descriptions). All “etiologic” results correlated with published RNAi phenotypes except in the case of sequence name C08H9.3 which encodes the α-1,3-glucosyltransferase responsible for the second glucose addition on the N2M9G1 LLO precursor. RNAi phenotypes in our hands produced a slow growth phenotype, whereas previous reports found larval arrest, embryonic lethality, and reduced brood size (Rual et al. 2004). Additionally, K09E4.2 generated a Gro RNAi phenotype in the absence of tunicamycin. Published reports found no observable phenotype via RNAi (Kamath et al. 2003; Rual et al. 2004). However, the slow growth phenotype was enhanced to embryonic lethal in the presence of 2 μg/mL tunicamycin.
Table II.
RNAi of loci in the LLO pathway presents no overt phenotype on NGM alone. In the presence of 2 μg/mL tunicamycin, RNAi silencing of etiologic genotypes enhances hypo-glycosylation that causes multiple phenotypes. Corresponding S. cerevisiae homologues and C. elegans RNAi phenotypes from WormBase were are also shown for comparison
| C. elegans | S. cerevisiae | ||||
|---|---|---|---|---|---|
| Sequence name | Homologue | Product | Wormbase | −Tn | +Tn |
| K09E4.2 | ALG3 | DppN2M6 | W.T. | Gro | Emb |
| C14A4.3 | ALG9 | DppN2M7/9 | W.T. | W.T. | Ste, Emb |
| ZC513.5 | ALG12 | DppN2M8 | W.T. | W.T. | Emb, Lvl, Lva |
| C08B11.8 | ALG6 | DppN2M9G1 | W.T. | W.T. | Gro |
| C08H9.3 | ALG8 | DppN2M9G2 | Emb, Lva | W.T. | Emb, Lvl, Lva |
| T24D1.4 | ALG10 | DppN2M9G3 | W.T. | W.T. | Brd, Gro |
| Empty vector | W.T. | W.T. |
Brd, small brood; Emb, embryonic lethality; Gro, slow growth; Lva, larval arrest; Lvl, larval lethal; Ste, sterile; W.T., wild type.
Table III.
RNAi silencing of genes involved in aspects of ER-Golgi networking exhibit no phenotype on NGM alone. However in the presence of 2 μg/mL tunicamycin, synthetic effects are evident in a range of phenotypes. Corresponding RNAi phenotypes from WormBase were also shown for comparison. Known cellular function is shown where available.
| Function | Gene | Locus | Wormbase | −Tn | +Tn |
|---|---|---|---|---|---|
| ERAD | K08E3.7 | pdr-1 | W.T. | W.T. | Ste,Emb,Gro |
| UPR | C41C4.4 | ire-1 | W.T. | W.T. | Emb,Lva |
| UPR | F46C3.1 | pek-1 | W.T. | W.T. | Gro |
| W02D7.7 | sel-9 | Gro,Unc,Pvl | W.T. | Ste,Gro | |
| Y60A3A.9 | Unc | W.T. | Let,Gro | ||
| F47G9.1 | Not tested | W.T. | W.T. | ||
| Empty vector | W.T. | W.T. |
Emb, embryonic lethality; Gro, slow growth; Let, lethal; Lva, larval arrest; Pvl, protruding vulva; Ste, sterile; Unc, uncoordinated locomotion; W.T., wild type.
To verify the presence of “maturation” tunicamycin-hypersensitive genes, trials with RNAi library plasmids representing genes in glycoprotein maturation (pdr-1, ire-1, pek-1, sel-9 and two p24 genes (sequence names Y60A3A.9 and F47G9.1)) and suspected of being tunicamycin hypersensitive were tested (Table III) (Link et al. 1992; Sundaram and Greenwald 1993; Belden and Barlowe 2001; Shen et al. 2001; Springer et al. 2005). The genes sel-9, Y60A3A.9, and F47G9.1 are members of the p24 protein family that are implicated in quality control trafficking between the endoplasmic reticulum and Golgi (Wen and Greenwald 1999). Results were consistent with those made by others with the exception of sel-9 which exhibited no phenotype in the absence of tunicamycin and had conflicting RNAi phenotype data (Simmer et al. 2003; Rual et al. 2004). By and large our RNAi assays were consistent with published results and also demonstrated a 10–30% replication variability of the genes tested which is common among laboratory-to-laboratory RNAi experiments in C. elegans (Simmer et al. 2003).
These tests established that RNAi, in combination with a low dose of tunicamycin, could generate synthetic phenotypes to identify genes that are tunicamycin hypersensitive. Despite the RNAi results of C08H9.3 (the glucosyltransferase that catalyzes the addition of the second glucose residue to the LLO), we were confident that the results validated the legitimacy of the screen (Table II). All of the above genes were included as positive controls in the genome-wide screen and at the culmination of the screen 8 of 12 control genes remained present in the final results.
Tunicamycin on RNAi effectiveness
As it is theoretically possible that tunicamycin causes a generalized increase in the effectiveness of the RNAi by the feeding method, we tested loci known to be refractory to RNAi (i.e., genes localized to neuronal cells) with and without 2 μg/mL tunicamycin and compared the RNAi phenotypes to the corresponding mutant phenotypes (Asikainen et al. 2005). It was expected that the six strains, which have a mutant “knock-out” phenotype, but were wild type in an RNAi screen would be unaltered by tunicamycin treatment. For example, sqv-1 which encodes an UDP-glucuronic acid decarboxylase had no observable RNAi phenotype (Hwang and Horvitz 2002; Kamath et al. 2003; Rual et al. 2004). However, the mutant phenotype was lethal. The RNAi phenotype in the presence of 2 μg/mL tunicamycin remained wild type. These results further supported the legitimacy of the screen by showing that tunicamycin does not increase the effectiveness of RNAi in C. elegans (Table IV).
Table IV.
Tunicamycin treatment does not alter RNAi effectiveness in C. elegans. Seven genes for which a phenotype is exhibited in the genetic mutants, but which have wild-type phenotypes in previously published RNAi screens were chosen to test RNAi in the presence of tunicamycin. All strains remained refractory to RNAi when tunicamycin was present, demonstrating that tunicamycin does not modify the effectiveness of RNAi
| Gene | Locus | Mutant | Wormbase | +Tn |
|---|---|---|---|---|
| F11C3.2 | unc-122 | UNC | W.T. | W.T. |
| W03A3.1 | ceh-10 | UNC | W.T. | W.T. |
| C52B9.9 | mec-18 | UNC | W.T. | W.T. |
| C17D12.2 | unc-75 | UNC | W.T. | W.T. |
| C52E12.3 | sqv-7 | LET | W.T. | W.T. |
| D2096.4 | sqv-1 | LET | W.T. | W.T. |
| ZC64.3 | ceh-18 | LET | W.T. | W.T. |
Genome-wide RNAi screen
The screen was divided into two phases: (1) a triage screen of the ORFeome v1.1 RNAi library for all genes that caused a visible synthetic phenotype in the presence of 2 μg/mL tunicamycin and (2) a follow-up screen where candidate genes were retested in the presence and absence of 2 μg/mL tunicamycin to separate tunicamycin-hypersensitive loci from single-gene phenotypes and false positives. During the triage screen, a criterion for selecting candidates for progression to the second phase was experimentally established (see Material and methods).
The completion of the phase I triage screen required 13,190 RNAi assays in the presence of 2 μg/mL tunicamycin. Of those genes tested in phase I, 4459 genes were candidates for the phase II follow-up screen based on the criteria mentioned above. Ultimately, 512 genes were found to be tunicamycin hypersensitive. These results were further analyzed to determine function location and the presence of Asn-X-Ser/Thr sequon for N-linked glycosylation (see Discussion and Supplementary Data). Additionally, qualified results were classified according to their eukaryotic orthologous group (KOG) assignments to determine cellular functions. In addition to KOG analysis, cellular location was determined with Proteome Analyst (http://path-a.cs.ualberta.ca, source for gene product cellular location), a web-based tool that can analyze the function and sub-cellular location of each sequence based on BLASTs for sequence alignment. Brief identifications and/or concise descriptions of each gene product were annotated manually with the aid of WormBase. We also examined our results with NetNglyc (http://www.cbs.dtu.dk/services/NetNGlyc, source for N-glycosylation site prediction), an online server that predicts N-glycosylation sites in proteins using artificial networks that examine the sequence context of Ans-Xaa-Ser/Thr sequons supplied in FASTA format (See supplementary data).
Discussion
Our hypothesis that mutations in “etiologic,” “maturation,” or “effector” loci interact genetically with defects in the LLO is the basis of the clinical diversity and heterogeneity in CDG-I diseases (Figure 5). By utilizing RNAi silencing in C. elegans, we identified tunicamycin-hypersensitive loci and dissected this phenomenon, effectively asking which genes enhance the severity of a CDG-I-like condition. The widespread understanding of C. elegans and its susceptibility to genetic silencing allowed us to systematically investigate CDG interactions on a gene-by-gene basis. Overall, each gene that was determined to be tunicamycin hypersensitive was tested at least three times: two assays with tunicamycin and one without. Therefore, the list of 512 genes was found to be decidedly tunicamycin hypersensitive.
Fig. 5.
Plausible genetic interactions that enhance defects in LLO biosynthesis that cause CDG-I diseases fall into three categories based on their role in N-glycosylation. RNAi knockdown of “effector,” “etiologic,” or “maturation” genes interact with a decrease in the LLO donor caused by tunicamycin treatment and result in visible phenotypes in C. elegans.
It is also important to consider that some tunicamycin-hypersensitive genes may be involved in drug metabolism and not directly interrelated with N-glycosylation. Effectively, knocking down these genes may increase the absorption or decrease detoxification of tunicamycin and would result in an increased drug effect. However, the quality of this experiment relied on precise drug treatment and only in the absence of drug can this possibility be excluded. A potential alternative to tunicamycin would utilize a double mutant containing both the RNAi sensitive rrf-3(pk1426) mutant background with an additional loss of function mutation in the LLO pathway, thereby effectively replacing drug treatment with a genetic condition. Additionally, it is possible that not all tunicamycin-hypersensitive genes would be identified from the ORFeome v1.1 library due to the inherent limitations of RNAi feeding methods (Kamath et al. 2001).
Currently, it is difficult to characterize each gene into its respective functional class (Figure 5), but some results can be drawn from previous works. This study cannot definitively assign each specific tunicamycin-hypersensitive gene to a particular function without confirmatory follow-up. However, this screen provides researchers with a list of genes that could be used to screen human CDG patients in the hope of identifying enhancer loci that contribute to the severity of the CDG-I diseases.
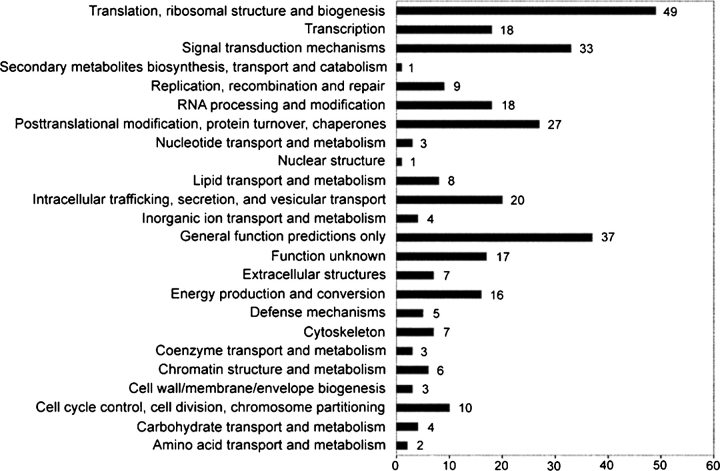
Results were evaluated by comparing our results to eukaryotic orthologous groups of proteins (KOGs) from sequenced genomes (Tatusov et al. 2003). This analysis classified our tunicamycin-hypersensitive genes into functional protein sets. Of the 512 tunicamycin-hypersensitive genes confirmed, 308 had KOG assignments and as a result 204 were omitted from analysis (Figure 6). The two largest known groups of KOG protein sets in our results were involved in translation, ribosomal structure and biogenesis (n = 49), and signal transduction mechanisms (n = 33). The significance of this result suggests that the combination of hypoglycosylation and decreased protein expression is the most detrimental interaction in CDG-I-like conditions in C. elegans. This result was not anticipated and does not principally fit the “etiologic,” “maturation,” or “effector” classification in Figure 5. Unsurprisingly, the subsequent two groups were posttranslational modification, protein turnover, chaperones (n = 27) and intracellular trafficking, secretion, and vesicular transport (n = 20). It is noteworthy that, 54 of the 308 KOG assignments fell under “general function predictions only” or “function unknown” classifications, which is a consequence of the uncharacterized status of many C. elegans genes. The KOG database serves principally to facilitate functional studies by utilizing 43 complete genomes. This analysis is not absolute, but we found that the KOG database was the most useful to designate tunicamycin-hypersensitive genes into functional classes over other available resources. Above all, this analysis does help to reasonably classify a substantial number of results into “effector,” “maturation,” or “etiologic” gene classes.
Fig. 6.
Analysis of eukaryotic orthologous groups (KOGs) classifies tunicamycin-hypersensitive genes into functional sets. Of 512 genes found to be hypersensitive to tunicamycin, 308 had KOG assignments.
Effector gene class
As mentioned above, we expected results to fall into three categories: “effector,” “etiologic,” and “maturation.” We postulated that “effector” gene products fall into two subclasses. “Formation” subclass members require N-glycosylation to fold, pass quality control, or to be trafficked within the cell. Those in the “function” subclass require N-glycosylation for their extracellular function; their catalytic, ligand–receptor, localization or stability properties are modulated by the presence and structure of the glycans.
There are several examples of “effector” class genes found in the results. The gene, eff-1 (epithelial fusion failure), is a known type I transmembrane glycoprotein required for epithelial cell fusion (Mohler et al. 2002). RNAi knockdown of eff-1 in the presence of 2 μg/mL tunicamycin caused larval arrest, sterility, uncoordinated movement, slow growth, and clear phenotypes. In the absence of tunicamycin, the eff-1 RNAi phenotype was wild type. A second “effector” type result is the mom-2 (more mesoderm) gene which is a member of the Wnt family of secreted signaling glycoproteins. In addition to being a glycoprotein, MOM-2 may also function as a ligand in Wnt signaling during embryonic development to specify the production of endoderm cells in the gut (Thorpe et al. 2000). Gene knockdown of mom-2 with tunicamycin generates embryonic lethality, small brood size, and sick phenotypes. In the absence of tunicamycin, the mom-2 RNAi phenotype was wild type.
Etiologic gene class
The comparatively low number of “etiologic” class genes (i.e., genes involved in the LLO pathway) in the results may be due to a general reduction in the number of possible candidate genes present in the C. elegans genome or their absence in the ORFeome v1.1 RNAi library. There are 16 genes in the C. elegans genome that correspond to enzymes in LLO biosynthesis (Berninsone 2006). Of those 16 genes, 11 are present in the RNAi library. Four genes of 11 were found to be tunicamycin hypersensitive and 2 genes produced robust phenotypes with and without drug. Only 4 of the 11 genes failed to qualify from the phase I triage screen, which was well within the false negative rate that is typical in genome-wide RNAi screens (Kamath et al. 2003; Simmer et al. 2003).
An example of a tunicamycin-hypersensitive positive result from the 16 C. elegans LLO genes mentioned above was sequence gene name T09A5.11, which encodes a 48 kD subunit of the oligosaccharyltransferase complex responsible for the en bloc transfer of the LLO oligosaccharide to nascent polypeptides in the endoplasmic reticulum. The RNAi phenotype of this gene knockdown was identical in the presence and absence of tunicamycin. However, when drug was present, the expressivity of the embryonic lethality phenotype increased from 25% to 75%.
Maturation gene class
“Maturation” gene classifications, defined by genes in ER-Golgi maturation and secretion, were highly abundant in the 512 gene results based on KOG analysis. The phenotype penetrance caused by 2 μg/mL tunicamycin on the RNAi control p24 genes (Y60A3A.9 and F47G9.1) demonstrated that maturation of nascent glycoproteins can influence the phenotype outcome of defects in N-glycosylation in C. elegans. Likewise, many lectins may act in intracellular systems necessary for the transport of immature glycoproteins through the ER-Golgi secretory pathway. Components of the Golgi responsible for the redistribution and proper localization of glycosylation enzymes were found to be tunicamycin hypersensitive in our screen. For example, RNAi of a subunit of the coatamer complex protein (COPI) which is involved in retrograde transport of Golgi enzymes was found to be hypersensitive to tunicamycin. RNAi of this gene in the presence of tunicamycin caused phenotypes in all four observable wells whereas in the absence of drug, only phenotypes in two wells were scored. Additionally, RNAi of the SNAP-25 (synaptosome-associated protein) component of the SNARE complex involved in Golgi trafficking was tunicamycin hypersensitive. The SNARE complex mediates fusion of Golgi transport vesicles and may interact with COPI vesicles directly during intercisternal Golgi transport (Ungar et al. 2006). RNAi of the SNAP-25 component with tunicamycin caused embryonic lethality and sterile progeny and produced no phenotypes in the absence of drug treatment.
Currently, the remarkable variability of CDG diseases is generally believed to be a consequence of insufficient glycosylation among many glycoproteins in different cells. In this study, we provided evidence that variable pathology may also be caused by the interaction between the primary CDG-I-like etiologic defect and modifier loci in the genetic background. We also demonstrated a novel approach that systematically dissects the complex glycosylation phenotype into specific glycan-dependent proteins and functions. As a result, we successfully delineate the conserved mechanisms that underlie the type of pathology observed in type I CDG and identified candidate gene homologous in C. elegans that may contribute to the severity of the disease.
Material and methods
General materials, methods and strains
The C. elegans strains NL2099 rrf-3 (pk1426), VC569 tag-179 (ok809), RE666 ire-1 (v33), and N2 (Bristol) were obtained from the Caenorhabditis Genetics Center, University of Minnesota, USA. The VC569 strain, which originally contained a genetic balancer, was outcrossed with N2 (Bristol) to generate a viable strain that was ok809 homozygous. The balancer was removed in the fourth generation and was confirmed by PCR. General methods used for cultivating, handling, and genetic manipulation of C. elegans are as described (Brenner 1974). Genome-wide RNAi screening protocols were performed by feeding in NL2099 rrf-3 (pk1426), with minor adaptations from previous works (Simmer et al. 2003). The ORFeome v1.1 RNAi library was supplied from the Vidal lab at the Dana-Farber Cancer Institute (Rual et al. 2004). Tunicamycin (Calbiochem, Darmstadt, Germany) was dissolved in DMSO to 50 mg/mL stock concentration. To obtain synchronous populations of larval stage 1 hatchlings, eggs were acquired by digesting populations containing gravid hermaphrodites with alkaline hypochlorite and washing the egg pellet three times by centrifugation with the M9 buffer (Hope 1999). Eggs hatched overnight in the M9 buffer or were transferred to 35 mm petri dishes. C. elegans strains were cultivated at 20°C unless otherwise indicated. Observations were made using a Leica MS 5 stereomicroscope at 10×–40× magnification (Leica Microsystems, Wetzlar, Germany).
Tunicamycin phenotype assays
Larval Lethality Assay (Figure 1)
Individual N2 hatchlings were placed in each well of a 24-well cluster plate containing NGM supplemented with 0–10 μg/mL tunicamycin (n = 264). On day three and day six, plates were scored for death (no pumping, twitching, or movement when prodded with the platinum wire) of the founder hermaphrodite or the appearance of hatched progeny. Y-Axis values are shown as a fraction of fertile adult N2 animals. The N2 larval lethality experiment was repeated twice.
Larval Phenotype Assay (Table I)
Following egg preparation, 15 μL of hatchlings (∼200 L1 Larvae) in the buffer solution were pipetted onto seeded 60 mm NGM plates containing 0–5 μg/mL tunicamycin. Animals were counted and phenotypes were scored after 3 days.
Tunicamycin Dose Response (Figure 2)
Separate populations of NL2099 rrf-3 (pk1426), RE666 ire-1 (v33), VC569 tag-179 (ok809), and N2 (Bristol) were synchronized by the alkaline hypochlorite egg preparation method. Five microliters of hatchings (∼150 L1 Larvae) in the M9 buffer were placed on 35 mm Petri dishes containing 0–10 μg/mL tunicamycin. Animal survival was scored based on the ability of larvae to reach gravid hermaphrodites after 3 days.
RNAi
The ORFeome v1.1 library of recombinant E. coli strains, consisting of 11,942 constructs, was arrayed in microtiter plates stored at −80°C. Bacteria were cultured in 2×YT containing 50 μg/mL ampicillin and 12.5 μg/mL tetracycline for 72 h at 22°C to ensure the presence of the plasmid (ampicillin selection) and the DE3 lysogen carrying the IPTG inducible T7 RNA polymerase (tetracycline selection). From these, 50 μL liquid cultures were grown overnight in 2×YT supplemented with 50 μg/mL ampicillin at 37°C with shaking. Fifteen microliters aliquots of the overnight culture were used to grow bacterial lawns on 24-well clusters of NGM supplemented with 50 μg/mL ampicillin and 1 mM IPTG and 2 μg/mL tunicamycin. These cultures were grown at room temperature for 48 h to create a bacterial food source expressing double-stranded RNA.
The genome-wide RNAi screen consists of two phases. In phase I, all RNAi constructs were assayed on 2 μg/mL tunicamycin. The second phase consisted of phase I candidates retested with and without 2 μg/mL tunicamycin. However, before the culmination of the first phase, criteria for selecting candidates for the second phase were established (see Candidate selection for Phase II).
In both phases of the screen, 24 well clusters were used as the platform for C. elegans analysis. Each column, consisting of four wells, was used to assess one RNAi construct. In this manner, each gene tested provides four observable wells. Initially, 7–10 NL2099 rrf-3 (pk1426) animals were placed in the top row of each plate to generate progeny that have only existed in the presence of each RNAi construct and 2 μg/mL tunicamycin. After 2 days of “priming,” a single gravid hermaphrodite from the top row was transferred to each of the other lawns and allowed us to elaborate F1 progeny. During the phase I screen, genes where at least two of the three replicates displayed concordant phenotypes in any single F1 progeny were regarded as candidates. Any phenotypes observed in the priming well were also scored, and data collected were weighed with lesser significance when selecting phase II candidates. Phenotypes in the three remaining wells, including Po and F1 progeny, were given a value of 10. Phenotypes in the “priming” well was marked a value of 5.
Candidate selection for phase II
The screen requires strict criteria for candidate selection for the second phase to limit the accumulation of false positives. A set of 100 genes that gave rise to phenotypes in 0, 1, 2, 3 and 4 observatory wells out of 4 in the triage screen were tested in the presence and absence of 2 μg/mL tunicamycin (n = 500 total genes tested). These sets were selected at random in the early stages of the phase I screen when at least 100 genes were available for each set.
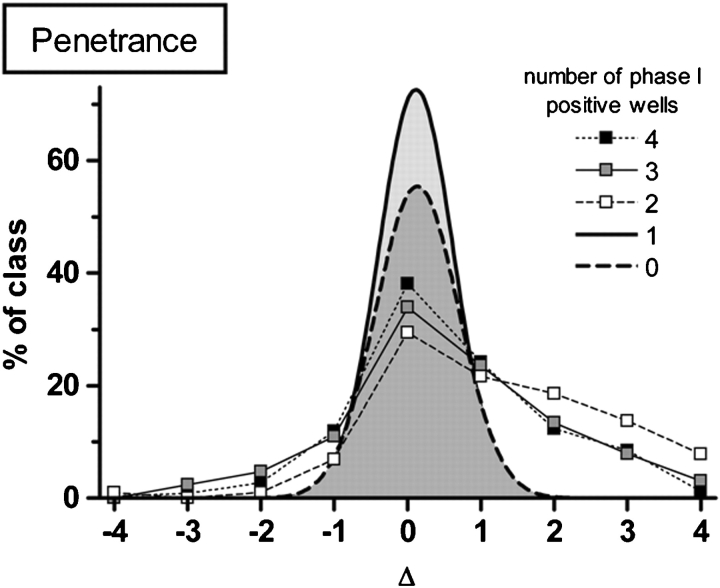
The penetrance was assessed by subtracting the number of wells showing any phenotype(s) with tunicamycin from the number in its absence (Δ). The larger the value of Δ, the more dependent the phenotype penetrance is on tunicamycin; i.e., a locus that induced phenotypes in all four wells containing tunicamycin but none in its absence would have Δ = 4. The delta values were plotted against the percent total in each of the five sets (100 genes per set) tested (Figure 3).
Fig. 3.
Delta values were calculated by subtracting the number of wells showing any phenotype with tunicamycin from the number in its absence. Larger delta values correspond to a greater dependence of the phenotype penetrance on tunicamycin. Phenotypes in ≥2 of 4 possible observable wells behave nonrandomly and are candidates for further testing with and without drug in phase II. Delta values greater than 2 exhibit non-Gaussian distribution and indicate tunicamycin-hypersensitive loci. Sets of 100 genes that caused observable phenotypes in 0, 1, 2, 3 and 4 observatory wells in the initial stages of the phase I screen were used for this assay (n = 500 genes tested).
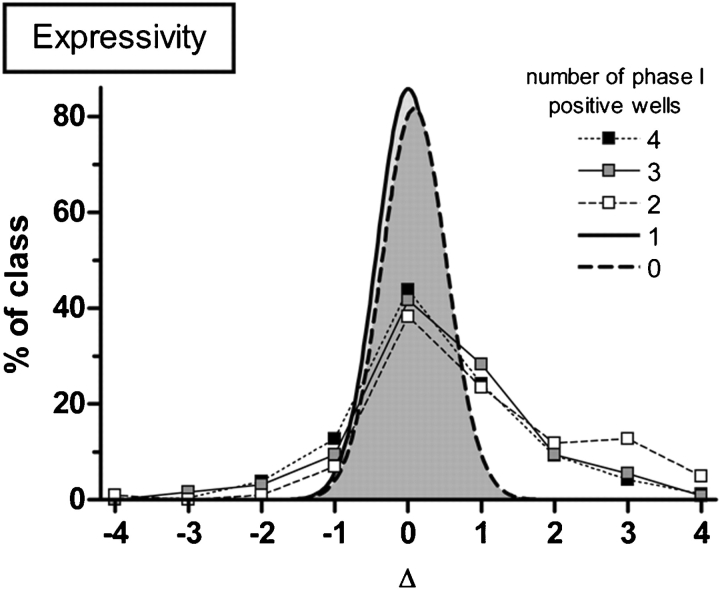
The effect of expressivity was assessed similarly except that Δ is calculated using the number of wells showing non-viable phenotypes under each condition. In this case, the larger the value of Δ, the more severe the phenotype becomes when tunicamycin is present; i.e., a locus that induced a dumpy_ phenotype in all four wells in the absence of tunicamycin but lethal_ in all wells when drug was present would have Δ = 4. Under this analysis, false positives and loci causing single-gene phenotypes score Δ = 0, but those genes that are tunicamycin-hypersensitive score Δ > 0 (Figure 4).
Fig. 4.
Delta values were calculated using the number of wells showing nonviable phenotypes under each condition. Larger delta values correspond to an increase in phenotype severity when tunicamycin is present. Phenotypes in ≥2 of 4 possible observable wells behave nonrandomly and are candidates for further testing with and without drug in phase II. Delta values greater than 2 demonstrate non-Gaussian distribution and indicate tunicamycin-hypersensitive loci. Sets of 100 genes that caused observable phenotypes in 0, 1, 2, 3 and 4 observatory wells in the initial stages of the phase I screen were used for this assay (n = 500 genes tested).
Loci causing phenotypes in ≤1 well during the triage screen behave randomly during retesting and the group results fitted to a Gaussian curve centered on 0 as expected. However, loci that caused phenotypes in ≥2 wells during the triage screen behaved non-randomly in the second round with a substantial fraction scoring Δ ≥ 2 which was two standard deviations from the mean and represented tunicamycin-hypersensitive loci.
Genome-wide RNAi screen
Preparation of media and bacterial culture were followed as stated previously. Twenty-four well cluster plates were used for both the triage and follow-up screens (phases I and II). A single 24-well cluster plate yielded results for six genes. Every column corresponded to a specific RNAi gene (four wells total). Larval stage 3 NL2099 rrf-3(pk1426) animals (7–10) were plated by hand to each well in the top row of each cluster. After 48 h at 15°C to “prime” these parents, single gravid hermaphrodites were transferred to each of the three replicate wells in the column and allowed us to segregate progeny at 22°C for 72 h. All wells were scored for any visible phenotypes in Po and F1 generations and noted using a controlled vocabulary. Additionally, data analysis similar to that completed for candidate selection for phase II determined the extent of penetrance and expressivity based on the recorded phenotypes (supplementary Table).
Supplementary Data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
NIH grant RR016459.
Supplementary Material
Acknowledgments
We thank Theresa Stiernagle at the Caenorhabditis Genetics Center for the C. elegans strains used in this study. We also thank Marc Vidal at the Dana-Farber Cancer Institute for the ORFeome v1.1 RNAi library.
Conflict of interest statement
None declared.
Abbreviations
- CDG
congenital disorders of glycosylation
- DPAGT1
UDP-GlcNAc dolichol-phosphate N-acetylglucosamine 1-phosphate transferase
- Dpp
dolichol pyrophosphate
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- G
glucose
- LLO
lipid-linked oligosaccharide
- M
mannose
- N
N-acetylglucosamine
- RNAi
RNA-mediated interference
- Tn
tunicamycin
- UPR
unfolded protein response
References
- Aebi M, Hennet T. Congenital disorders of glycosylation: Genetic model systems lead the way. Trends Cell Biol. 2001;11:136–141. doi: 10.1016/s0962-8924(01)01925-0. [DOI] [PubMed] [Google Scholar]
- Asikainen S, Vartiainen S, Lakso M, Nass R, Wong G. Selective sensitivity of Caenorhabditis elegans neurons to RNA interference. Neuroreport. 2005;16:1995–1999. doi: 10.1097/00001756-200512190-00005. [DOI] [PubMed] [Google Scholar]
- Barnes G, Hansen WJ, Holcomb CL, Rine J. Asparagine-linked glycosylation in Saccharomyces cerevisiae: Genetic analysis of an early step. Mol Cell Biol. 1984;4:2381–2388. doi: 10.1128/mcb.4.11.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Barlowe C. Deletion of yeast p24 genes activates the unfolded protein response. Mol Biol Cell. 2001;12:957–969. doi: 10.1091/mbc.12.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninsone PM. 2006. Carbohydrates and glycosylation (December 18, 2006) In: The C. elegans Research Community, editor. WormBook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- de Lonlay P, Seta N, et al. A broad spectrum of clinical presentations in congenital disorders of glycosylation I: A series of 26 cases. J Med Genet. 2001;38:14–19. doi: 10.1136/jmg.38.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund EA, Freeze HH. The congenital disorders of glycosylation: A multifaceted group of syndromes. NeuroRx. 2006;3:254–263. doi: 10.1016/j.nurx.2006.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Freeze HH, Westphal V. Balancing N-linked glycosylation to avoid disease. Biochimie. 2001;83:791–799. doi: 10.1016/s0300-9084(01)01292-5. [DOI] [PubMed] [Google Scholar]
- Hope IA. C. elegans: A Practical Approach. Oxford, New York: Oxford University Press; 1999. [Google Scholar]
- Hwang HY, Horvitz HR. The SQV-1 UDP-glucuronic acid decarboxylase and the SQV-7 nucleotide-sugar transporter may act in the Golgi apparatus to affect Caenorhabditis elegans vulval morphogenesis and embryonic development. Proc Natl Acad Sci USA. 2002;99:14218–14223. doi: 10.1073/pnas.172522199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeken J, Matthijs G. Congenital disorders of glycosylation. Annu Rev Genomics Hum Genet. 2001;2:129–151. doi: 10.1146/annurev.genom.2.1.129. [DOI] [PubMed] [Google Scholar]
- Jaeken J, Matthijs G. Congenital disorders of glycosylation: A rapidly expanding disease family. Annu Rev Genomics Hum Genet. 2007;8:261–278. doi: 10.1146/annurev.genom.8.080706.092327. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehle L, Strahl S, et al. Protein glycosylation, conserved from yeast to man: A model organism helps elucidate congenital human diseases. Angew Chem Int Ed Engl. 2006;45:6802–6818. doi: 10.1002/anie.200601645. [DOI] [PubMed] [Google Scholar]
- Link CD, Silverman MA, Breen M, Watt KE, Dames SA. Characterization of Caenorhabditis elegans lectin-binding mutants. Genetics. 1992;131:867–881. doi: 10.1093/genetics/131.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijs G, Schollen E, Bjursell C, Erlandson A, Freeze H, Imtiaz F, Kjaergaard S, Martinsson T, Schwartz M, Seta N, et al. Mutations in PMM2 that cause congenital disorders of glycosylation, type Ia (CDG-Ia) Hum Mutat. 2000;16:386–394. doi: 10.1002/1098-1004(200011)16:5<386::AID-HUMU2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Mills P, Mills K, Clayton P, Johnson A, Whitehouse D, Winchester B. Congenital disorders of glycosylation type I leads to altered processing of N-linked glycans, as well as underglycosylation. Biochem J. 2001;359:249–254. doi: 10.1042/0264-6021:3590249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler WA, Shemer G, del Campo JJ, Valansi C, Opoku-Serebuoh E, Scranton V, Assaf N, White JG, Podbilewicz B. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev Cell. 2002;2:355–362. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- Paschinger K, Gutternigg M, Rendic D, Wilson IB. The N-glycosylation pattern of Caenorhabditis elegans. Carbohydr Res. 2008;343:2041–2049. doi: 10.1016/j.carres.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Prietsch V, Peters V, Hackler R, Jakobi R, Assmann B, Fang J, Korner C, Helwig-Rolig A, Schaefer JR, Hoffmann GF. A new case of CDG-x with stereotyped dystonic hand movements and optic atrophy. J Inherit Metab Dis. 2002;25:126–130. doi: 10.1023/a:1015628810892. [DOI] [PubMed] [Google Scholar]
- Rual JF, Ceron J, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter H. Protein glycosylation lessons from Caenorhabditis elegans. Curr Opin Struct Biol. 2004;14:607–616. doi: 10.1016/j.sbi.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Schollen E, Kjaergaard S, Legius E, Schwartz M, Matthijs G. Lack of Hardy–Weinberg equilibrium for the most prevalent PMM2 mutation in CDG-Ia (congenital disorders of glycosylation type Ia) Eur J Hum Genet. 2000;8:367–371. doi: 10.1038/sj.ejhg.5200470. [DOI] [PubMed] [Google Scholar]
- Shen X, Ellis RE, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- Simmer F, Moorman C, et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer W, Hoppe T, Schmidt E, Baumeister R. A Caenorhabditis elegans Parkin mutant with altered solubility couples alpha-synuclein aggregation to proteotoxic stress. Hum Mol Genet. 2005;14:3407–3423. doi: 10.1093/hmg/ddi371. [DOI] [PubMed] [Google Scholar]
- Sundaram M, Greenwald I. Suppressors of a lin-12 hypomorph define genes that interact with both lin-12 and glp-1 in Caenorhabditis elegans. Genetics. 1993;135:765–783. doi: 10.1093/genetics/135.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe CJ, Schlesinger A, Bowerman B. Wnt signalling in Caenorhabditis elegans: Regulating repressors and polarizing the cytoskeleton. Trends Cell Biol. 2000;10:10–17. doi: 10.1016/s0962-8924(99)01672-4. [DOI] [PubMed] [Google Scholar]
- Ungar D, Oka T, Krieger M, Hughson FM. Retrograde transport on the COG railway. Trends Cell Biol. 2006;16:113–120. doi: 10.1016/j.tcb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Vodopiutz J, Bodamer OA. Congenital disorders of glycosylation—A challenging group of IEMs. J Inherit Metab Dis. 2008;31:267–269. doi: 10.1007/s10545-008-0849-2. [DOI] [PubMed] [Google Scholar]
- Wen C, Greenwald I. p24 proteins and quality control of LIN-12 and GLP-1 trafficking in Caenorhabditis elegans. J Cell Biol. 1999;145:1165–1175. doi: 10.1083/jcb.145.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal V, Kjaergaard S, Schollen E, Martens K, Grunewald S, Schwartz M, Matthijs G, Freeze HH. A frequent mild mutation in ALG6 may exacerbate the clinical severity of patients with congenital disorder of glycosylation Ia (CDG-Ia) caused by phosphomannomutase deficiency. Hum Mol Genet. 2002;11:599–604. doi: 10.1093/hmg/11.5.599. [DOI] [PubMed] [Google Scholar]
- Wu X, Rush JS, Karaoglu D, Krasnewich D, Lubinsky MS, Waechter CJ, Gilmore R, Freeze HH. Deficiency of UDP-GlcNAc:dolichol phosphate N-acetylglucosamine-1 phosphate transferase (DPAGT1) causes a novel congenital disorder of glycosylation type Ij. Hum Mutat. 2003;22:144–150. doi: 10.1002/humu.10239. [DOI] [PubMed] [Google Scholar]
- Zeevaert R, Foulquier F, Jaeken J, Matthijs G. Deficiencies in subunits of the Conserved Oligomeric Golgi (COG) complex define a novel group of congenital disorders of glycosylation. Mol Genet Metab. 2008;93:15–21. doi: 10.1016/j.ymgme.2007.08.118. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zeng Y, Lehrman MA. Evidence that the hamster tunicamycin resistance gene encodes UDP-GlcNAc:dolichol phosphate N-acetylglucosamine-1-phosphate transferase. J Biol Chem. 1992;267:8895–8902. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.