Abstract
Some inter- and intraspecific crosses may result in reduced viability or sterility in the offspring, often due to genetic incompatibilities resulting from interactions between two or more loci. Hybrid necrosis is a postzygotic genetic incompatibility that is phenotypically manifested as necrotic lesions on the plant. We observed hybrid necrosis in interspecific lettuce (Lactuca sativa and Lactuca saligna) hybrids that correlated with resistance to downy mildew. Segregation analysis revealed a specific allelic combination at two interacting loci to be responsible. The allelic interaction had two consequences: (1) a quantitative temperature-dependent autoimmunity reaction leading to necrotic lesions, lethality, and quantitative resistance to an otherwise virulent race of Bremia lactucae; and (2) a qualitative temperature-independent race-specific resistance to an avirulent race of B. lactucae. We demonstrated by transient expression and silencing experiments that one of the two interacting genes was Rin4. In Arabidopsis thaliana, RIN4 is known to interact with multiple R gene products, and their interactions result in hypersensitive resistance to Pseudomonas syringae. Site-directed mutation studies on the necrosis-eliciting allele of Rin4 in lettuce showed that three residues were critical for hybrid necrosis.
INTRODUCTION
During evolution, ancestral species can diverge into several derived species that become genetically isolated from each other due to pre- and postzygotic barriers, reducing the capacity for hybridization (Mallet, 2006; Rieseberg and Willis, 2007). In plants, one of the best described postzygotic barriers is hybrid necrosis (Bomblies and Weigel, 2007). This type of genetic incompatibility is manifested as necrotic lesions in seedlings or adult plants and is often associated with phenotypes such as wilting, chlorosis, stunted growth, and lethality. Hybrid necrosis has been reported in both interspecific and intraspecific plant crosses. As these phenotypes are not observed in the parental genotypes, hybrid necrosis must be a result of interactions between two or more genes (negative epistasis or Bateson-Dobzhansky-Muller incompatibilities) brought together from different species or genotypes (Bomblies and Weigel, 2007). However, the mechanisms underlying hybrid necrosis are still poorly understood. Studies on interspecific hybrids of tomatoes (Solanum lycopersicum) and intraspecific hybrids in Arabidopsis thaliana indicate that some forms of hybrid necrosis result from particular alleles encoding resistance (R) proteins inducing autoimmunity-like responses when combined with particular alleles of genes elsewhere in the genome (Krüger et al., 2002; Wulff et al., 2004; Bomblies et al., 2007; Alcázar et al., 2009).
We study disease resistance in interspecific hybrids between cultivated lettuce (Lactuca sativa) and a distantly related, wild species (Lactuca saligna). The latter is considered a nonhost species to Bremia lactucae, a causal agent of lettuce downy mildew (Bonnier et al., 1992). The sexually compatible gene pool of lettuce consists of Lactuca serriola, which is closely related and probably the progenitor of L. sativa, and more distantly related species, such as L. saligna and Lactuca virosa (De Vries, 1997; Koopman et al., 2001). These Lactuca species are autogamous and therefore predominantly homozygous. We generated two interspecific F2 populations (cross 1 and 2) between different accessions of L. saligna and cultivars of L. sativa (Jeuken et al., 2001). From cross 1, we developed a set of 29 backcross inbred lines (BILs), each with a single introgressed chromosomal segment from L. saligna in the L. sativa genetic backgrounds; this set of BILs represented 96% of the L. saligna genome (Jeuken et al., 2001, 2008; Jeuken and Lindhout, 2004; Zhang et al., 2009b).
In previous studies that were focused on disease resistance, we observed the possible functional correlation of several traits: lethality, temperature-dependent necrotic lesions on leaves, retarded growth, quantitative resistance, and complete resistance associated with hypersensitivity to downy mildew on the hybrid progeny and introgression lines; all of these traits mapped to one locus at the top of chromosome 9 at 9 centimorgans (cM) (hereafter referred to as the C9 locus; see Supplemental Figure 1 online) (Jeuken et al., 2001, 2008; Jeuken and Lindhout, 2002, 2004). Lethality was observed in seedlings that were homozygous for the L. saligna C9 introgression in the L. sativa genetic background (BIL9.1) (Jeuken and Lindhout, 2004). These seedlings were extremely necrotic and died within a week, suggesting a lethal homozygous allelic combination at two (or more) independent loci (Figure 1). Plants that were heterozygous for the C9 introgression (designated preBIL9.1; the “pre” suffix indicates that the introgression was heterozygous) were viable and fertile but developed necrotic lesions on their leaves and stems at a later stage and showed retarded growth (Jeuken and Lindhout, 2004). These phenotypes were more extreme in winter. Variable quantitative resistance to race Bl:14 of B. lactucae was observed in preBIL9.1 plants (Jeuken et al., 2008). Complete hypersensitive resistance to downy mildew (formerly designated R39) was observed in the interspecific F2 populations and in preBIL9.1 plants; this resistance was effective against race Bl:16 and not against race Bl:14 (Jeuken and Lindhout, 2002; Jeuken et al., 2008).
Figure 1.
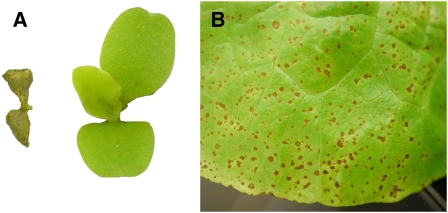
Hybrid Necrosis Phenotypes in Lettuce.
(A) Left: Completely necrotic seedling that is unable to survive; this backcross introgression line is homozygous for the L. saligna allele at the C9 locus and the L. sativa allele at the C6 locus (6sat9sal). Right: Normal wild-type seedling phenotype; genotype is homozygous L. sativa at both the C6 and C9 loci (6sat9sat). Both seedlings were grown at 15°C for 12 d.
(B) Detail of leaf with a high density of necrotic lesions in a 6-week-old plant of preBIL9.1, which was heterozygous at the C9 locus (6sat9het).
The availability of hybrid plant materials (F2 progeny and a set of BILs) allowed for the use of a forward genetic approach to dissect and validate the loci determining hybrid necrosis. In this article, we provide evidence for two interacting loci determining hybrid necrosis and the identification of Rin4 as one of the genes responsible. In addition, we demonstrate a relationship between hybrid necrosis and resistance to B. lactucae.
RESULTS
Two Epistatic Loci Are Involved in Hybrid Necrosis
To identify the epistatic loci determining hybrid necrosis, we first focused on the lethality phenotype. Genotyping had shown that the nonviable BIL9.1 plants were homozygous for the C9 introgression from L. saligna (Figure 1). Plants homozygous L. saligna for the C9 locus will be referred to as having the genotype 9sal, heterozygotes will be referred to as 9het, and homozygotes L. sativa for the C9 locus will be referred to as 9sat. In the original interspecific F2 populations (L. saligna × L. sativa crosses 1 and 2), we compared genotypic composition of all plants that were homozygous L. saligna for the C9 locus but were viable. We identified one (cross 1) and seven (cross 2) viable 9sal plants (see Supplemental Table 1A online, right column). None of these eight plants were homozygous for L. sativa alleles in a region from ∼30 to 35 cM on chromosome 6 (hereafter referred to as the C6 locus, and similar to C9, genotypes on chromosome 6 are referred to as 6sal, 6sat, and 6het; see Supplemental Table 1A online; equivalent to chromosome 8 of the consensus map for Lactuca spp; Truco et al., 2007). Thus, a homozygous combination of L. sativa alleles on the C6 locus and L. saligna alleles on the C9 locus (6sat9sal) was implicated as causing necrosis leading to lethality. Consistent with this, the C9 locus showed distorted segregation in cross 1 (deficiency of L. saligna alleles) and the C6 locus in cross 2 (deficiency of L. sativa alleles; see Supplemental Table 1A online).
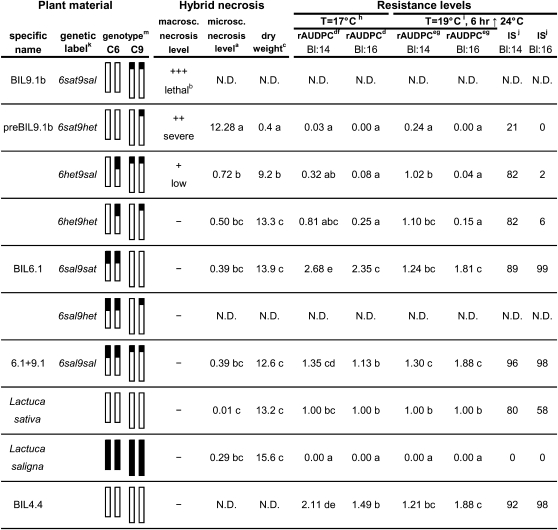
To confirm this interaction, we crossed two other introgressed lines derived from cross 1, preBIL9.1b (6sat9het) and BIL6.1 (6sal9sat), which differed for the putative interactive C6 and C9 loci and were homozygous L. sativa for the rest of the genome. Plant materials are diagrammed in Figure 2. BIL6.1 was homozygous for an L. saligna introgression from 0 to 40 cM on C6 (6sal9sat). PreBIL9.1b plants were 6sat9het and had identical phenotypes for hybrid necrosis and resistance to B. lactucae as preBIL9.1 plants but they had a smaller heterozygous C9 segment (0 to 11 cM instead of 0 to 48 cM). We identified an F1 plant that was heterozygous at the C6 and C9 loci. Its F2 progeny segregated for the two loci and produced the nine expected genotype classes, which were identified by DNA marker analysis (plant material column, Figure 2). Seeds were collected from eight different F2 genotypes. Plants of the ninth genotype class, 6sat9sal genotype (BIL9.1b), died after 1 week (Figure 2). Plants from six genotypic classes as well as the 6sat9sal plants (while they lived) were characterized for their phenotypes compared with the parental lines L. sativa cv Olof and L. saligna CGN05271. Of the six genotypes, four contained at least one interacting pair of C6 + C9 L. sativa and L. saligna alleles that was expected to lead to at least some degree of necrosis (Figure 2). The symptoms of hybrid necrosis (necrotic lesions and reduced growth) were extreme for the homozygous 6sat9sal genotype, severe for 6sat9het, low for 6het9sal, and lacking for 6het9het, 6sal9sat, and 6sal9sal genotypes (Figure 2). The most extreme and lethal hybrid necrosis phenotype was observed for the homozygous allelic combination with two L. sativa alleles at C6 and two L. saligna alleles at C9 (BIL9.1, 6sat9sal). The levels of necrosis in the two genotypes that had opposite heterozygous-homozygous allelic combinations, 6sat9het and 6het9sal, were not lethal but showed a large difference in the level of necrosis (∼20-fold, severe versus low, respectively; Figure 2). The genotype that was heterozygous at both loci, 6het9het, did not show symptoms of hybrid necrosis. These four levels of hybrid necrosis indicated a gene dosage effect of the alleles at the two interacting loci.
Figure 2.
Hybrid Necrosis and Resistance Levels.
Plant material is described in genotype labels and diagrams (columns 1 to 3). Hybrid necrosis symptoms are described as quantity of necrotic lesions at macroscopical and microscopical level and as retarded growth (columns 4 to 6). Resistance levels are described as relative area under disease progress curve (rAUDPC) and infection severity (IS) against B. lactucae races Bl:14 and Bl:16 at two temperatures (columns 7 to 12). Letters in common within a column indicate that the values are not significantly different (α = 0.05, Tukey honestly significant difference [HSD] procedure). N.D., not determined. a, Percentage of necrotic leaf area. Two leaf segments × three plants per genotype were examined. b, Seedling gets completely necrotic and shrivels after several days. c, Dry weight in grams from the shoots of 11-week-old plants grown in the greenhouse (n = 7). d, Relative AUDPC from YDT in climate chamber calculated from observations for infection severity at 8, 10, and 12 d after inoculation (DAI). L. sativa cv Olof was set at 1.00. e, Relative AUDPC from YDT in greenhouse calculated from observations for infection severity at 8, 9, 10, and 11 DAI. L. sativa cv Olof was set at 1.00. f, Similar results were observed in disease tests with race Bl:14 on cotyledons of these genotypes. g, Similar results were observed in four disease tests with races Bl:14 and Bl:16 on detached leaf discs from 9- and 12-week-old plants of these genotypes. h, In the climate chamber with day/night cycles of 19/12°C (block intervals) and an average temperature of 17°C. i, In the greenhouse with an average temperature of 19°C, minimal night temperature of 15°C, and the day temperature was 6 h above 24°C and 12 h above 22°C. j, Average infection severity level of downy mildew at 11 DAI on young plants (YDT) scored as percentage of leaf area showing sporangiophores. k, sat = homozygous L. sativa, sal = homozygous L. saligna, and het = heterozygous. m, graphical genotype of C6 and C9; white means L. sativa allele, and black means L. saligna allele. The C6 and C9 introgressions are 40 and 11 cM long, respectively.
A similar interaction was observed between C6 and C9 loci in the progeny of a BC4S1 line from cross 2; this line has the 6het9sal genotype and has been introgressed into the L. sativa background. This suggests the same epistatic interaction for hybrid necrosis as in progeny from cross 1, which were derived from a different accession of L. saligna.
Hybrid Necrosis Is Temperature Sensitive
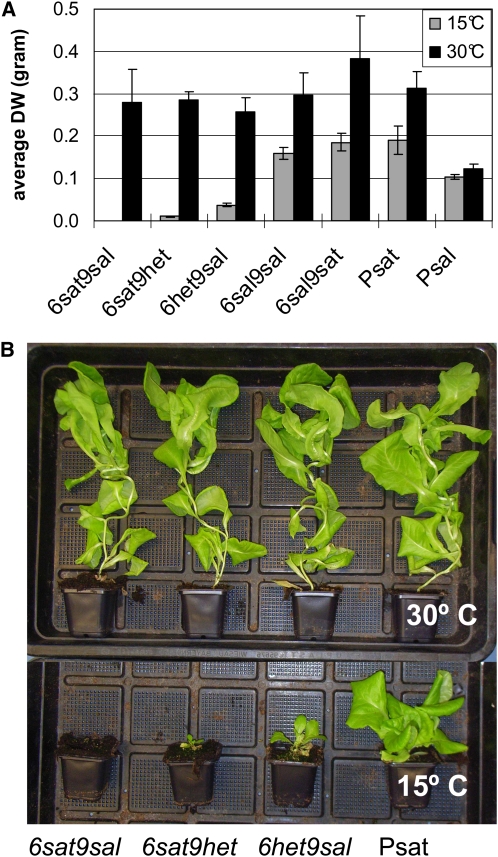
To check for the temperature sensitivity that had been observed earlier for resistance (Jeuken and Lindhout, 2004), we tested plants at 15 and 30°C. At 15°C, plants showed the degrees of hybrid necrosis as described above (Figure 2); however, at 30°C, all plants, even the lethal 6sat9sal genotype, grew normally, similar to L. sativa cv Olof (Figure 3). After 7 weeks, we transferred plants of four of the genotypes (6sat9sal, 6sat9het, 6sat9sat, and 6sal9sal) that had been grown at 30 to 15°C. The first symptoms of hybrid necrosis were brown necrotic lesions in the youngest leaf, particularly close to the major veins; these symptoms were seen 48 h after the transfer for 6sat9sal and at 80 h for 6sat9het (see Supplemental Figure 2 online). Ultimately, the 6sat9sal plants became completely necrotic and died after 8 d. Plants with the 6sat9het genotype died after 20 d (see Supplemental Figure 3 online).
Figure 3.
Temperature Sensitivity of Hybrid Necrosis.
(A) Plant growth as measured by average shoot dry weight (n = 5, 45 d old). Psat, L. sativa cv Olof (6sat9sat); Psal, L. saligna CGN05271 (6sal9sal); 95% confidence intervals are shown.
(B) Phenotypes of 7-week-old plants of indicated C6C9 genotypes, grown at 30 and 15°C.
Hybrid Necrosis and Resistance to B. lactucae Involve the Same Loci
The colocalization of race-specific resistance (formerly designated R39 and derived from L. saligna; Jeuken and Lindhout 2002) with the C9 hybrid necrosis locus and evidence from other studies of involvement of R genes in hybrid necrosis (Krüger et al., 2002; Bomblies et al., 2007; Alcázar et al., 2009) suggested a possible relationship between hybrid necrosis and this resistance. To investigate whether there is a relationship between resistance and hybrid necrosis, we inoculated the same range of C6 and C9 genotypes used to characterize hybrid necrosis with two races of B. lactucae, Bl:14 and Bl:16. These races differ in their virulence phenotypes and have been tested also on the two original interspecific F2 populations (see Introduction). Seedlings, young plants, and adult plants were tested. The results for young plants are shown in Figure 2. The results were very similar for all three plant stages.
In the disease tests with race Bl:14 at 17°C, the genotype 6sat9het showed low levels of infection severity, 6het9sal showed medium levels of infection severity, and 6het9het showed almost the same high level as the susceptible parent, L. sativa cv Olof (Figure 2). At a 2°C higher temperature, the severity of infection rose significantly for all genotypes, which resulted in only 6sat9het still exhibiting resistance, while 6het9sal and 6het9het were as susceptible as L. sativa cv Olof (Figure 2). Therefore, the resistance to Bl:14 was quantitative, showed different levels for different doses of alleles at the C6 and C9 loci, and depended upon temperature. Interestingly, the resistance levels to Bl:14 paralleled the different levels of hybrid necrosis (necrosis and reduced dry weight) observed in uninoculated plants (Figure 2). This is consistent with the digenic interaction triggering a resistance response resulting in necrotic leaf lesions and a decrease of the spread of pathogen.
In the disease tests with race Bl:16, only very low or very high levels of infection were observed. Surprisingly, the 6sal9sal genotype was susceptible to both races of B. lactucae, indicating that the L. saligna C9 allele does not lead to resistance per se. Complete resistance to Bl:16 was observed in the 6sat9het, 6het9sal, and 6het9het genotypes that did not parallel the levels of resistance to Bl:14 and hybrid necrosis and did not change with temperature. At least one L. sativa allele on C6 with at least one L. saligna allele on C9 was sufficient to give complete resistance to Bl:16. This race-specific resistance seemed not to be a consequence of the triggered autoimmune hypersensitive response (HR) but another outcome of the same allelic interaction. The 6het9het genotype was remarkable in that it was completely resistant to race Bl:16 but completely susceptible to race Bl:14 and did not show necrotic lesions.
In the F2 population derived from L. saligna × L. sativa cross 1, we detected only one peak logarithm of the odds (LOD) value for resistance to Bl:16 at the C9 locus and none at the C6 locus (Jeuken and Lindhout, 2002) (see Supplemental Figure 1 online). In hindsight, failure to detect a significant LOD peak on C6 in this F2 might have resulted from the distorted segregation of the C9 locus with a deficiency of L. saligna alleles (see Supplemental Table 1B online).
Rin4 Is the Candidate Gene on C9
The EST-derived marker most closely associated with the resistance to race Bl:16 in the interspecific F2 populations was LE0478 (see Supplemental Figure 1 online). Primers for this marker were based on a contig of lettuce ESTs (QG_CA_Contig7104; CGP1 database; The Compositae Genome Project; http://compgenomics.ucdavis.edu). This contig had the highest sequence similarity to RPM1 INTERACTING PROTEIN4 of Arabidopsis (RIN4; At3g25070) and was therefore designated Rin4. Extensive studies of RIN4 have provided support for the guard mechanism of action of R proteins that detect the activity of pathogen effectors indirectly by their effects on key proteins, such as RIN4 (Mackey et al., 2002, 2003). RIN4 is a 211–amino acid, acylated plasma membrane–associated protein that acts as a negative regulator of basal defense (Mackey et al., 2002; Kim et al., 2005b). It is a target of at least three effectors from Pseudomonas syringae (AvrRpm1, AvrB, and AvrRpt2) and is guarded by two independent R proteins (RPM1 and RPS2) (Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003; Kim et al., 2005a).
We determined the cDNA sequences of the Rin4 alleles from our parental lines of cross 1, L. sativa cv Olof and L. saligna CGN05271, and from the wild species L. virosa CGN05978. From each accession, we detected two versions of Rin4 transcripts with open reading frames of 735 and 732 bp (designated Transcript1 and Transcript2) due to a CAG indel at base pair position 705; the inferred amino acid sequences are consequently 244 and 243 amino acids long, respectively. Following sequencing of the genomic DNA of L. sativa cv Olof and L. saligna CGN05271, we detected only one version of Rin4, which included the CAG sequence. The CAG polymorphism between the transcripts occurred at an intron splice site; therefore, an alternative splicing event seems to be responsible for these two versions of this Rin4 transcript (see Supplemental Figure 4 online). We detected five synonymous single nucleotide polymorphisms, five nonsynonymous single nucleotide polymorphisms, and a 3-bp replacement (in addition to the indel) between the Rin4 cDNA sequences of L. sativa cv Olof and L. saligna CGN05271 resulting in inferred Rin4 amino acid sequences that differ for six amino acids (Figure 4A). At these six polymorphic sites, L. virosa CGN05978 encoded four amino acids that were identical to L. sativa and two that were identical to L. saligna (Figure 4A).
Figure 4.
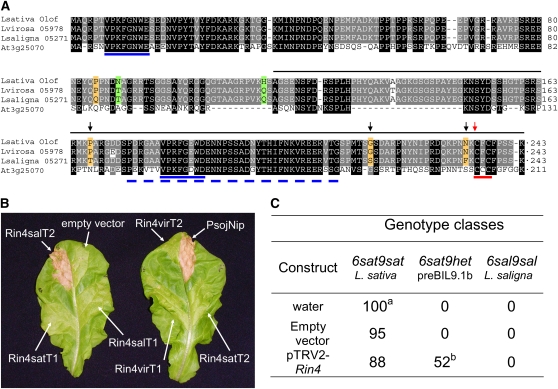
Rin4 Is the Candidate Gene on C9.
(A) Rin4 from L. sativa cv Olof, L. saligna CGN05271, L. virosa CGN05978, and RIN4 from Arabidopsis (At3g25070). The deduced amino acid sequences of lettuce Rin4 transcript2 were aligned using the ClustalW software. Transcript1 has one extra amino acid, a Gln, Q, between Lys-235 and Cys-236 (red arrow). Conserved residues between Arabidopsis and Lactuca species are shaded in black, and conserved residues within Lactuca species are shaded in gray. Orange-shaded amino acids highlight amino acid differences between L. saligna and L. sativa/L. virosa. Green-shaded amino acids highlight amino acid differences between L. saligna/L. virosa and L. sativa. Arrows point at critical residues for hybrid necrosis in Rin4salT2 based on site-directed mutagenesis studies. The solid blue lines represent AvrRpt2 cleavage sites in RIN4 of Arabidopsis, RCS1 and RCS2; the dashed blue line represents a RIN4 P142-G179 fragment that interacts with AvrB; the red line represents a RIN4 putative palmitoylation site at C203-C205 (Kim et al., 2005a). The black line marks the DNA sequence that was engineered into pTRV2 vector for VIGS experiments.
(B) Transient expression with Rin4 alleles/transcripts infiltrated in L. sativa cv Olof 8 DAI. Five plants × two leaves per genotype class. Alleles: sat = L. sativa, sal = L. saligna, and vir = L. virosa. T1 and T2 are Rin4 transcript versions 1 and 2.
(C) Relative infection severity to B. lactucae Bl:16 of Rin4-silenced lettuce genotypes by VIGS. a The absolute infection severity of 6sat9sat, L. sativa cv Olof, was 82% at 10 DAI in the young plant disease test. b The infection severity was estimated for entire leaves, although we could not observe whether the entire leaf was silenced for Rin4. Therefore, the presented relative infection severity value may underestimate the effect of the silencing.
Only the Rin4salT2 Transcript Caused Necrosis
To examine whether one or both Rin4 proteins from L. saligna CGN05271 elicited the hybrid necrosis, different Rin4 transcripts were expressed in planta using Agrobacterium tumefaciens–mediated transient assays. The shorter Transcript2, but not Transcript1, of the L. saligna allele of Rin4 (Rin4salT2) caused a severe necrotic reaction in L. sativa plants harboring the L. sativa allele at the C6 locus (Figure 4B; see Supplemental Table 2A online). Neither of the Rin4 transcripts from L. sativa and L. virosa elicited necrosis.
We used site-directed mutagenesis of Rin4T2 to determine which of the six polymorphic amino acids between L. saligna and L. sativa were critical for hybrid necrosis. Replacement of six L. saligna residues by their corresponding L. sativa residues indicated that the last three polymorphic residues (positions 167, 217, and 234) are required for hybrid necrosis elicited by Rin4sal (Figure 4; see Supplemental Table 3 online). However, replacement of the last three polymorphic L. sativa residues in Rin4sat by their corresponding L. saligna residues indicated that the three residues alone or in combination were not sufficient to convert Rin4sat into a necrosis-inducing protein.
Silencing of Rin4 Impaired Resistance to Race Bl:16
To validate the involvement of Rin4 in the resistance in preBIL9.1b (6sat9het) to B. lactucae Bl:16, we reduced Rin4 transcript levels by virus-induced gene silencing (VIGS) and challenged these plants with race Bl:16. The silencing of Rin4 did not result in abnormal plant phenotypes in any of the tested genotypes. Silencing of Rin4 rendered preBIL9.1b (6sat9het) susceptible (Figure 4C). This result confirms that Rin4 is involved in this resistance. We conclude, therefore, that the L. saligna allele of Rin4 located at C9 is responsible for both the hybrid necrosis and the resistance to B. lactucae race Bl:16.
Rin4 Amino Acid Haplotypes in Lactuca spp
To learn more about the diversity of Rin4 amino acid haplotypes among species of Lactuca, we sequenced cDNAs of Rin4 from a diverse panel of 38 additional accessions comprising 12 Lactuca species (six to eight accessions for each of L. sativa, L. serriola, L. saligna, and L. virosa and one to two accessions for the other Lactuca species; see Supplemental Table 4 online). Sequence analysis of 41 accessions (38 + the previous three; Figure 4) showed that all species expressed both the T1 and T2 transcripts of Rin4; L. tatarica expressed two additional Rin4 transcripts (see Supplemental Figure 5 online). We focused on the T2 transcript because Rin4salT2 elicited hybrid necrosis. The accessions from L. sativa, L. serriola, L. aculeata, L. dregeana, and L. altaica were monomorphic for Rin4 at the inferred amino acid level (see Supplemental Figure 5 online), which was consistent with their close taxonomic affiliations. L. saligna showed intraspecific amino acid polymorphism; three Rin4T2 amino acid haplotypes were detected among eight accessions of L. saligna, with six or five polymorphic residues relative to L. sativa. The three haplotype groups reflected their origin (France-Italy, Russia-Georgia, and Israel). L. saligna CGN05271 (parent of cross 1) and L. saligna CGN11341 (parent of cross 2) both had the France-Italy haplotype that had six polymorphic residues relative to L. sativa. No intraspecific amino acid polymorphism was detected among the six accessions of L. virosa. L. virosa was the only species that had any (two) amino acids in common with L. saligna at the amino acid sites that were polymorphic between L. sativa and L. saligna (T89 and Q115, Figure 4; see Supplemental Figure 5 online).
No Necrosis Was Induced by Rin4salT2 in Species Other Than L. sativa
Agrobacterium-mediated transient expression of Rin4salT2 from accession CGN05271 in 36 accessions comprising nine species in the genus Lactuca and two species from genera related to Lactuca demonstrated that a severe necrotic reaction was observed in all 10 L. sativa cultivars but was not observed in any of the other eight Lactuca species nor in two species from genera related to Lactuca (see Supplemental Table 2B online). Therefore, the hybrid necrosis is the result of a specific interaction between the products of the Rin4sal and the L. sativa C6 alleles.
DISCUSSION
Hybrid necrosis and resistance to B. lactucae in an interspecific lettuce hybrid is due to the specific combination of an L. sativa allele on C6 and an L. saligna allele on C9 (hereafter referred to as the interacting allele pair). The C9 gene was shown to be Rin4, which interacts with an unknown gene on C6. In Arabidopsis, the phosphorylation or disappearance of RIN4 elicits a necrotic HR dependent on the R proteins RPM1 or RPS2 (Axtell and Staskawicz., 2003; Mackey et al., 2003). Therefore, the association of resistance with this digenic interaction suggests that the unknown C6 gene may also be an R gene. Even in the absence of infection, the interaction of C6sat with Rin4sal triggers a macroscopic necrotic phenotype. A similar resistance response has been observed in tomato; a specific interaction between alleles originating from different species at the loci encoding Rcr3 and the R protein Cf-2 results in weak necrosis (Krüger et al., 2002).
The Rin4sal and C6sat hybrid necrosis response is quantitative, temperature dependent, and causes increased resistance to the otherwise virulent Bl:14 race of B. lactucae in a gene dosage-dependent manner. Different combinations of interacting alleles elicited different levels of hybrid necrosis and resistance. The temperature dependence and association with increased pathogen resistance has been observed in several other instances of hybrid necrosis (Bomblies et al., 2007; Bomblies and Weigel, 2007; Alcázar et al., 2009). In addition, several resistances to pathogens are temperature sensitive (Wang et al., 2009).
The C6sat-Rin4sal pair elicits hybrid necrosis and confers complete HR-mediated resistance to infection with the incompatible Bl:16 race of B. lactucae. The double heterozygote 6het9het was sufficient to provoke complete resistance to Bl:16 (Figure 2); however, it was not sufficient to elicit hybrid necrosis. Resistance to Bl:16 was not dependent upon temperature and was only mildly dependent upon the level of hybrid necrosis and the doses of the interacting alleles. This resistance required the heterologous C6sat-Rin4sal pair of alleles; the homologous combinations of alleles C6sal-Rin4sal and C6sat-Rin4sat and the other heterologous combination, C6sal-Rin4sat, did not confer resistance. This is different from previously reported situations. In the case of interaction between alleles of Rcr3 and Cf-2 from Solanum pimpinellifolium and S. lycopersicum (formerly Lycopersicon esculentum), Cf-2pim-Rcr3esc caused an HR-mediated resistance to Avr2-containing races of Cladosporum fulvum and a weak autonecrosis; however, the allelic combination from S. pimpinellifolium, Cf-2pim-Rcr3pim, also conferred race-specific resistance to Avr2-containing races (Jones et al., 1993; Krüger et al., 2002; Rooney et al., 2005).
Molecular Considerations
Only six residues in Rin4 were polymorphic between L. saligna and L. sativa that might render the innocuous Rin4sat isoform into an autoimmunity-inducing, Bl:16-incompatible isoform in the C6sat-Rin4sal combination. Mutant studies with reciprocal replacement of these six residues in one Rin4 parental protein by their corresponding residues in the other Rin4 parental isoform showed that three residues were critical but not sufficient individually or in combination for hybrid necrosis (see Supplemental Table 3 online). It remains to be determined which of the additional polymorphic residues are required for hybrid necrosis and which of the polymorphic residues are critical to convert the Bl:16-susceptible C6sat-Rin4sat combination into a resistant C6sat-Rin4sal combination. Interestingly, none of these polymorphic residues are at positions previously characterized as being important for the function of RIN4 in Arabidopsis (Kim et al., 2005a). However, the last critical polymorphic residue in Rin4sal at position 234 is two residues away from the putative palmitoylation site at C203 to C205 in Arabidopsis RIN4 and therefore may influence membrane localization. Also, the polymorphisms at positions 167 and 217 in lettuce Rin4 flank by seven amino acids the region in Arabidopsis RIN4 that was identified as interacting with AvrB using yeast two-hybrid analysis (Figure 4A; Kim et al., 2005a).
Transcript1 of Rin4 is one amino acid longer than transcript 2 and is present in all the Lactuca species tested. It therefore seems to be the product of an evolutionarily conserved alternative splicing event. The functional significance of the two protein products remains unknown. One of several possibilities is that one isoform might fulfill the role in resistance of a decoy target in effector perception as hypothesized in the decoy model (Van der Hoorn and Kamoun, 2008).
Genetic Aspects of Hybrid Necrosis Levels
Four specific C6sat-Rin4sal allelic combinations caused different levels of hybrid necrosis. These four levels seemed to depend on the dose of the necrosis-inducing heterologous C6sat-Rin4sal pair of alleles. Decreasing numbers of combinations of C6sat and Rin4sal alleles, four (6sat9sal), three (6sat9het and 6het9sal), and two (6het9het), showed decreasing levels of hybrid necrosis from lethal to severe to low to nil, respectively (Figure 2). The lack of hybrid necrosis in the 6het9het genotype that contains one pair of interacting alleles requires further explanation. Assuming a guard-guardee relationship between a putative C6 R protein and the Rin4 protein, four possible guard-guardee associations could occur in the 6het9het genotype: the non-necrosis–inducing C6sal-Rin4sal, C6sal-Rin4sat, and C6sat-Rin4sat and the necrosis-inducing C6sat-Rin4sal. If pairs of the guard-guardee isoforms from the same species (C6sat-Rin4sat and C6sal-Rin4sal) preferentially interact compared with heterologous pairs of isoforms, the 6het9het would tend not to develop hybrid necrosis (see Supplemental Figure 6 online). In the other three genotypes with a pair of heterologous necrosis-inducing C6sat-Rin4sal alleles (6het9sal, 6sat9het, and 6sat9sal), one or both of the non-necrosis–inducing isoforms, C6sal and Rin4sat, are absent. As a consequence, C6sat or Rin4sal have no alternative but to form a guard-guardee association with a heterologous protein, leading to hybrid necrosis. The lower the doses of non-necrosis–inducing alleles the more heterologous guardee-guard associations are formed, leading to higher levels of hybrid necrosis (see Supplemental Figure 6 online).
In conclusion, we showed specific isoforms of Rin4 induced hybrid necrosis and resistance in combination with a heterologous gene product from a marginally sexually compatible species. Together with previous studies of other species, our findings raise several interesting questions. Is the C6sat allele a functional R gene encoding a nucleotide binding–leucine-rich repeat protein? Has it evolved to protect against B. lactucae or other pathogen species? Why is it widespread in L. sativa but lacking in other species?
METHODS
Lettuce Material
Cross 1 comprised Lactuca saligna CGN05271 × Lactuca sativa cv Olof. Cross 2 comprised L. saligna CGN11341 × L. sativa cv Norden. From both crosses one F1 plant was selfed and backcrossed to its recurrent L. sativa parent. F2 populations consisted of 126 and 54 plants from crosses 1 and 2, respectively. The F2 genetic linkage map and the infection severity levels in adult F2 plants to downy mildew were described previously (Jeuken et al., 2001; Jeuken and Lindhout, 2002). Both BC1 populations were further backcrossed with the respective cultivated L. sativa parent until the BC4 generation. For cross 2, we selfed the BC4 and genotyped the BC4S1. For cross 1, we developed a set of BILs from the BC4S1-2 and BC5S1-2 using marker-assisted selection. This resulted in a set of 29 BILs with a total of 96% of the L. saligna genome introgressed into L. sativa (Jeuken and Lindhout, 2004; Jeuken et al., 2008). BILs were genotyped with >700 DNA markers (amplification fragment length polymorphism [AFLP], ESTs, and simple sequence repeats (SSRs)). Most of these BILs contained one homozygous introgression fragment of L. saligna with an average genetic length of 33 cM (∼20 to 40% of a chromosome) in an L. sativa cv Olof background. For some lines it was not possible to obtain the introgression in a homozygous state and the best alternative, a line with the introgression in heterozygous state, was used. We designated such lines preBILs. An overview of the L. sativa × L. saligna hybrid material is shown in Supplemental Table 5 online.
In this study, we focused on preBIL9.1b and BIL6.1. PreBIL9.1b showed hybrid necrosis and contained one heterozygous introgression, from 0 to 11 cM on chromosome 9. It was derived from preBIL9.1 (introgression from 0 to 48 cM). BIL6.1 harbored one homozygous introgression from 0 to 40 cM on chromosome 6.
Pathogen Material
The virulence phenotypes of Bremia lactucae races Bl:14 and Bl:16 have been previously described (Jeuken and Lindhout, 2002) and in the evaluation report by the International Bremia Evaluation Board (http://www.plantum.nl/ibeb.html). Pathogen maintenance, inoculum preparation, and the method of inoculation were performed as described previously (Jeuken and Lindhout, 2002).
Linkage Analyses, Genotyping, and Quantitative Trait Loci Mapping
Analyses of additional markers were performed on the F2 population of cross 1 to saturate and improve the genetic linkage map of the L. saligna × L. sativa cross (Jeuken et al., 2001), especially near the interacting loci. Markers consisted of AFLP markers from two primer combinations, E48M59 (primer+CAC and Primer+CTA) and E33M59 (Primer+AAG and Primer+CTA) and SSR markers and candidate gene markers that were developed from lettuce EST sequences as part of the Compositae Genome Project (see Supplemental Table 6 online; http://compgenomics.ucdavis.edu/; McHale et al., 2009). Polymorphisms between L. saligna and L. sativa in EST and SSR markers were visualized by the size differences of their PCR products on agarose gels (directly or after enzymatic digestion) as previously described (Jeuken et al., 2008) or by high-resolution melting curve differences visualized on a LightScanner System (Idaho Technology).
Linkage analyses were performed using JoinMap 4.0 software (Van Ooijen, 2006) on the F2 population of cross 1 with the following mapping conditions. For grouping, regression mapping was used with weak linkages recombination and LOD thresholds of 0.45 and 0.05. Markers were assigned to nine linkage groups at a LOD threshold of 8. Calculations of the linkage maps were done using all pairwise recombination estimates smaller than 0.40, LOD scores higher than 1, a jump threshold of 5, and Haldane's mapping function. As the integration of former linkage maps for F2 populations from cross 1 and 2 showed high colinearity with respect to marker order and distance (Jeuken et al., 2001), we consider the linkage map for cross 2 to be identical. To fine-map the resistance to B. lactucae Bl:16 to the new F2 linkage map, we performed quantitative trait loci mapping procedures like simple interval mapping and approximate multiple quantitative trait loci mapping using MapQTL 5.0 (Van Ooijen, 2004).
The C6 and C9 plant material was genotyped with a minimum of eight DNA markers (combinations of EST, SSR, and AFLP markers) per introgression segment at an early plant stage to select the desired genotypes.
Phenotyping Hybrid Necrosis
Plants were phenotyped by observing cotyledons or leaves for macroscopically visible necrotic lesions. Plants were categorized based on abundance and size of necrotic lesions compared with other plants under the same conditions.
We quantified the level of necrotic leaf area through microscopy evaluation. Three plants per genotype, randomly placed, were grown in a greenhouse at 15°С for 5 weeks. The 4th true leaf was then sampled from each by cutting two leaf segments near the mid-leaf, 1 × 2 cm in size. Leaf segments were discolored for 3 d in acetic acid/ethanol solution (v:v = 1:3), cleared and stored in saturated chloral hydrate solution (5g/2 mL), and finally mounted in 70% glycerol. Slides were observed under the light microscope, and the necrotic areas were recognized by cytoplasm granulation and darkening with a yellow or brown color. Each necrotic area was visualized through a digital camera and measured using AxioVison LE 4.6 (Carl Zeiss) in μm2. The percentage of necrotic area per leaf segment was calculated. For multiple comparisons of the percentage of necrotic area per genotype, we used two-way analysis of variance (ANOVA) and the Tukey HSD test (α = 0.05).
To quantify the retardation of growth, the shoot dry weights of 11-week-old plants that had been grown in a randomized block design in a greenhouse were measured in grams. Seven plants per genotype were examined. Aboveground parts were harvested and dried for 16 h at 105°C.
For multiple comparisons of the shoot dry weights between genotypes, we used one-way ANOVA and the Tukey HSD test (α = 0.05).
Disease Tests
The levels of infection severity in response to B. lactucae races Bl:14 and Bl:16 were determined using seedling disease tests (SDTs) (14 seedlings per genotype), young plant disease tests (YDTs) (eight plants per genotype), and adult plant disease tests in the greenhouse (ADTG) (seven plants × eight leaf discs per genotype). For each genotype, these tests were performed on plants at three different developmental stages as described previously (Jeuken and Lindhout, 2002; Zhang et al., 2009a).
For all three disease tests (SDT, YDT, and ADTG) infection severity levels were scored daily between 8 and 11 DAI as the percentage of sporulating area per cotyledon/representative leaf/leaf disc. For each test, the area under disease progress curve (AUDPC) was calculated. The relative AUDPC was calculated relative to the susceptible control parent, L. sativa cv Olof, for which the AUDPC was set at 1.00. For multiple comparisons of the AUDPC data between genotypes, we used one-way ANOVA and Tukey HSD tests (α = 0.05).
For YDTs, attached leaves were tested from young, 3- to 4-week-old plants. One test with race Bl:14 and one test with Bl:16 were performed in a climate chamber. In parallel, one test with Bl:14 and one test with Bl:16 were performed in a greenhouse compartment. The same inoculum was given for the tests with Bl:14 and for the tests with Bl:16. The temperature was controlled during growth and after inoculation at both locations but was inevitably more variable in the greenhouse due to the natural day/night cycle (17 h day/7 h night in June during the tests) and natural light conditions. In the greenhouse, the temperature varied gradually between a low point of 15°C during the night to a peak of 29°C at noon; the average was 18.6°C. In the climate chamber, the temperature shifted directly from 19.6°C during the artificial day of 16 h to 12.3°C in the artificial night, with an average temperature of 17.2°C. Representative leaves of young plants were scored (as described above) at 8, 9, 10, and 11 DAI.
Rin4 Sequences
The 3′ end of a lettuce Rin4 was identified from an EST sequence (QG_CA_Contig7104; CGP1 database; The Compositae Genome Project; http://compgenomics.ucdavis.edu) based on sequence similarity to RIN4 from Arabidopsis thaliana. We used the Qiagen RNeasy plant mini kit with a DNase column treatment per the manufacturer's directions for RNA isolation and iScript enzyme (Bio-Rad). The SMART RACE cDNA Amplification kit (Clontech) was used according to the manufacturer's instructions to identify the 5′ cDNA sequence from L. sativa cv Salinas. Oligonucleotide primers in the 5′ and 3′ untranslated regions (Rin4_UTR) as well as in the internal DNA sequence (Rin4_INT1, Rin4 INT2, and Rin4_INT3) were designed to the RIN4 cDNA sequence (see Supplemental Table 7 online). Genomic DNA from three lettuce genotypes, L. sativa cv Salinas, L. serriola accession UC96US23, and L. saligna CGN5322, were amplified with these oligonucleotides using Amplitaq Gold Polymerase with a 57°C annealing temperature (Applied Biosystems). Products were sequenced. Additional cDNA sequences of ESTs from L. virosa and L. saligna that showed sequence similarity to these Rin4 cDNA sequences were selected by BLAST analysis from the Compositae Genome Project Database (http://compgenomics.ucdavis.edu/). Based on all of these sequences, two oligonucleotide primers at the start and the end of the cDNA sequence of the RIN4 gene (Rin4_TOT) were designed (see Supplemental Table 7 online). In subsequent PCR experiments, the cDNA and genomic sequences spanning the open reading frames of Rin4 were obtained for L. sativa cv Olof, L. saligna CGN05271, and L. virosa CGN05978. In the same way, Rin4 homologous cDNA sequences from 38 additional accessions comprising 12 Lactuca species were obtained (see Supplemental Table 4 online). Four to six clones were sequenced per accession using Sanger sequencing. The inferred amino acid sequences of RIN4 transcripts were aligned using the ClustalW software from DNASTAR Lasergene8.
Phylogenetic Analysis
A midpoint rooted neighbor-joining phylogenetic tree was generated using MEGA version 4 and bootstrap value 1000 (Tamura et al., 2007).
Agrobacterium tumefaciens–Mediated Transient Assays
Transient assays were executed to overexpress different wild-type and mutant Rin4 transcripts. Total RNA isolation and cDNA syntheses were performed as described above. Full-length cDNAs encoding the Rin4 alleles (L. sativa = sat, L. saligna = sal, and L. virosa = vir) and transcript versions (T1 and T2) were amplified from lettuce cDNA prepared from leaves of L. sativa cv Olof, L. saligna CGN05271, and L. virosa CGN05978. The rin4 mutants were created using the Quick-Change site-directed mutagenesis kit (Stratagene), according to the manufacturer's instructions. PCR products, prepared using the proofreading enzyme Phusion DNA polymerase (Finnzymes), were cloned into pENTR/D-TOPO entry vector (Invitrogen) and then recombined into the GATEWAY T-DNA binary vector pK7WG2 (Karimi et al., 2002) using LR clonase (Invitrogen). The resulting binary vectors, pK7WG2 with target genes under the control of a 35S promoter, were electroporated into A. tumefaciens strain C58C1 (pGV2260).The A. tumefaciens strain C58C1 (pGV2260) and the strain containing the PsojNIP gene cloned in the binary vector pB7WG2 were provided by G. Van den Ackerveken (Utrecht University). PsojNIP is a necrosis-inducing protein from Phytophthora sojae (Qutob et al., 2002). Luria-Bertani medium (10 g/L bacteriological peptone, 10g/L NaCl, and 5 g/L yeast extract) was used for liquid and solid (15 g/L agar) bacterial cultures. Spectinomycin (50 mg/L) was used to maintain pB7WG2 in A. tumefaciens.
Rin4 alleles were tested in two independent experiments performed with 10 replications each on L. sativa cv Olof, L. sativa cv Norden, L. saligna CGN05271, L. saligna CGN11341, BIL6.1, and BIL6.1 +9.1. A third experiment was performed on 36 accessions comprising nine Lactuca species and two other species (Cichorium intybus and Mycelis muralis) and with four replications of each transient test. The rin4 mutants were tested in at least two independent experiments on at least five plants each from L. sativa cv Olof and BIL6.1. Plants were grown in a greenhouse at 21°С in the daytime and 19°С at night until they attained the sixth to seventh leaf stage. The 5th and 6th true leaves were then infiltrated; the culture preparations and leaf infiltrations were performed as described by Wroblewski et al. (2005).
VIGS
In initial experiments to develop a VIGS system for lettuce, we examined the ability of the tobacco rattle virus (TRV)-based VIGS vector from D. Kumar (Yale University) to suppress the expression of the endogenous phytoene desaturase gene of lettuce (PDS) in L. sativa and L. saligna, following the protocol used for tomato (Solanum lycopersicum; Liu et al., 2002a, 2002b). Lettuce is a host for tobacco rattle virus (Mojtahedi et al., 2003). Two independent sets of infiltrations were performed, with 20 L. sativa and 20 L. saligna plants infiltrated with a mixture of Agrobacterium culture containing the pTRV2-PDS and pTRV1. The same numbers of plants were also infiltrated with the pTRV1 and empty pTRV2 (empty vector control) and mock-infiltrated with water. Twenty-three days after agroinfiltration, the expected photobleaching phenotype, caused by inhibition of carotenoid synthesis, was observed on the 4th to 14th leaves in L. sativa and on the 3rd to 10th leaves in L. saligna on at least 75% of the plants (see Supplemental Figure 7 online). The effect of PDS suppression was visible uniformly throughout each entire leaf (see Supplemental Figure 7 online), especially for leaves 5 to 8 of L. sativa and 3 to 5 of L. saligna. We validated the silencing by measuring the transcript levels for PDS using quantitative RT-PCR. Primers that anneal to the PDS gene outside the region targeted for silencing were used (primer pair Ls-PDS-RT3; see Supplemental Table 7 online). The experiment was conducted using an iCycler MyiQ detection system (Bio-Rad), using the iQ SYBR Green Super mix (Bio-Rad). Assays were done in duplicate. Relative quantification of the PDS transcript level was normalized to results obtained using the lettuce ubiquitin transcript as a control by applying the 2−ΔCt formula (Livak and Schmittgen, 2001). Three plants representing each group and RNA target were analyzed. The Tukey HSD test with α = 0.05 was applied for pairwise multiple comparisons between the groups.
After confirming the effectiveness of the VIGS approach (see Supplemental Figures 7 and 8 online), a new TRV construct, pTRV2-Rin4, was made with a 285-bp fragment of lettuce Rin4 (Figure 4). In several independent experiments, plants were agroinfiltrated with pTRV2-Rin4, and 30 d after agroinfiltration, the plants were challenged with B. lactucae race Bl:16 as in standard disease tests of young plants (see description of disease tests above). Infection severities were measured and analyzed as described above. Similar trends were observed in two experiments. Detailed results of one experiment are presented (see Results).
VIGS Plasmid Constructions
pTRV1 and pTRV2 VIGS vectors have been described previously (Liu et al., 2002a). pTRV2-PDS was created as follows. A 315-bp fragment of a cDNA fragment corresponding to bases 1334 to 1648 of the lettuce PDS gene (contig CLS_S3_Contig8919; CGP1 database; The Compositae Genome Project, http://compgenomics.ucdavis.edu) was PCR amplified from L. sativa cv Olof cDNA using Taq DNA polymerase and the primer pair Ls-PDS1 (see Supplemental Table 7 online). The resulting PCR product was cloned into the pGEM-T Easy vector as described by the manufacturer (Promega) and later ligated into EcoRI-cut pTRV2.
pTRV2-Rin4 was created as follows. A 285-bp fragment corresponding to bases 451 to 735 of lettuce Rin4satT1 cDNA (Figure 4) was PCR amplified from lettuce cDNA using Taq DNA polymerase and the primer pair Rin4_TOT (see Supplemental Table 7 online). The resulting PCR product was digested with EcoRI, and the 285-bp fragment was ligated into EcoRI-cut pTRV2 using standard methods.
Agroinfiltrations
For Agrobacterium-mediated virus infection, cultures of A. tumefaciens GV3101 containing pTRV1, empty pTRV2 vector control, and each of the constructs derived from pTRV2 were grown, harvested, and subsequently infiltrated as described (Van der Hoorn et al., 2000; Bai et al., 2008). The infiltration was performed on the abaxial side of both cotyledons of each lettuce seedling at 9 d after sowing using a needleless syringe. The infiltrated plants were grown under normal greenhouse conditions (20°C daytime and 18°C nighttime) and were checked for virus symptoms at regular intervals. The only symptom of TRV infection observed was restricted plant growth compared with mock-infiltrated plants.
Temperature Sensitivity Tests
Eight plants per genotype class were grown in a randomized design at 15 and 30°C in identical climate chambers with identical conditions except for the temperature. Plants were observed weekly for necrotic lesions or any other aberrant phenotypes. After 45 d, the shoot dry weight of five plants per genotype was measured as described above. Forty-nine days after sowing, plants of four genotypes, grown at 30°C, were transferred to room temperature for 21 h and then to 15°C. The plants were monitored every 12 h for the next 14 d for necrotic lesions or any other aberrant phenotypes.
Accession Numbers
Sequence data from Rin4T2 from 41 Lactuca accessions from this article can be found in the GenBank/EMBL data libraries under accession numbers GQ497773 to GQ497814.
Author Contributions
M.J.W.J. designed and performed research, analyzed data, and wrote the article. K.P., L.K.M., and E.D.B. performed research. N.W.Z. designed and performed research and analyzed data. P.L. and R.G.F.V. contributed to biological interpretation, and R.E.N. and R.W.M. contributed to biological interpretation and writing.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Map Positions of Hybrid Necrosis Loci.
Supplemental Figure 2. Hybrid Necrosis Symptoms in Youngest Leaf after Temperature Shift.
Supplemental Figure 3. Hybrid Necrosis Symptoms in Whole Plants after Temperature Shift.
Supplemental Figure 4. Proposed Alternative Splicing of Rin4 Transcripts.
Supplemental Figure 5. Rin4 Amino Acid Haplotypes in Lactuca spp.
Supplemental Figure 6. Proposed Genetic and Molecular Model for Hybrid Necrosis Levels in Lettuce.
Supplemental Figure 7. Silencing of PDS in Lettuce by VIGS.
Supplemental Figure 8. Real-Time PCR Expression Data of PDS in Lettuce Leaves.
Supplemental Table 1. F2 Segregation Ratios and Infection Severities.
Supplemental Table 2. Transient Expression of Rin4 Alleles and Transcripts.
Supplemental Table 3. Transient Expression of rin4 Mutants.
Supplemental Table 4. Lactuca Diversity Panel.
Supplemental Table 5. L. sativa-L. saligna Hybrid Material.
Supplemental Table 6. DNA Markers at C6 and C9 Loci.
Supplemental Table 7. Primer Pairs for Rin4 Sequencing, Cloning, and RT-PCR.
Supplemental Data Set 1. Text File Corresponding to the Alignment in Supplemental Figure 5.
Supplementary Material
Acknowledgments
This research is supported by the Dutch Technology Foundation Stichting voor de Technische Wetenschappen, Applied Science Division of Nederlandse Organisatie voor Wetenschappelijk Onderzoek, and the Technology Program of the Ministry of Economic Affairs. The EST data were obtained from the Compositae Genome Project website at compgenomics.ucdavis.edu, which is supported by the USDA Integrated Food and Farming Systems and the National Science Foundation Plant Genome programs. The SSRs were developed and provided by Syngenta Company. The Centre for Genetic Resources, the Netherlands (http://www.cgn.wur.nl), provided seeds of wild Lactuca species.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Marieke J.W. Jeuken (marieke.jeuken@wur.nl).
Online version contains Web-only data.
References
- Alcázar, R., García, A.V., Parker, J.E., and Reymond, M. (2009). Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc. Natl. Acad. Sci. USA 106 334–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell, M.J., and Staskawicz, B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112 369–377. [DOI] [PubMed] [Google Scholar]
- Bai, Y., Pavan, S., Zheng, Z., Zappel, N.F., Reinstadler, A., Lotti, C., De Giovanni, C., Ricciardi, L., Lindhout, P., Visser, R., Theres, K., and Panstruga, R. (2008). Naturally occurring broad-spectrum powdery mildew resistance in a central american tomato accession is caused by loss of Mlo function. Mol. Plant Microbe Interact. 21 30–39. [DOI] [PubMed] [Google Scholar]
- Bomblies, K., Lempe, J., Epple, P., Warthmann, N., Lanz, C., Dangl, J.L., and Weigel, D. (2007). Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5 e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies, K., and Weigel, D. (2007). Hybrid necrosis: Autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 8 382–393. [DOI] [PubMed] [Google Scholar]
- Bonnier, F.J.M., Reinink, K., and Groenwold, R. (1992). New sources of major gene resistance in Lactuca to Bremia lactucae. Euphytica 61 203–211. [Google Scholar]
- De Vries, I.M. (1997). Origin and domestication of Lactuca sativa L. Genet. Resour. Crop Evol. 44 165–174. [Google Scholar]
- Jeuken, M., and Lindhout, P. (2002). Lactuca saligna, a non-host for lettuce downy mildew (Bremia lactucae), harbors a new race-specific Dm gene and three QTLs for resistance. Theor. Appl. Genet. 105 384–391. [DOI] [PubMed] [Google Scholar]
- Jeuken, M., van Wijk, R., Peleman, J., and Lindhout, P. (2001). An integrated interspecific AFLP map of lettuce (Lactuca) based on two L. sativa x L. saligna F2 populations. Theor. Appl. Genet. 103 638–647. [Google Scholar]
- Jeuken, M.J.W., and Lindhout, P. (2004). The development of lettuce backcross inbred lines (BILs) for exploitation of the Lactuca saligna (wild lettuce) germplasm. Theor. Appl. Genet. 109 394–401. [DOI] [PubMed] [Google Scholar]
- Jeuken, M.J.W., Pelgrom, K., Stam, P., and Lindhout, P. (2008). Efficient QTL detection for nonhost resistance in wild lettuce: backcross inbred lines versus F2 population. Theor. Appl. Genet. 116 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.A., Dickinson, M.J., Balint-Kurti, P.J., Dixon, M.S., and Jones, J.D.G. (1993). Two complex resistance loci revealed in tomato by classical and RFLP mapping of the Cf-2, Cf-4, Cf-5, and Cf-9 genes for resistance to Cladosporium fulvum. Mol Plant Microbe Interact. 6 348–357. [Google Scholar]
- Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAY(TM) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kim, H.-S., Desveaux, D., Singer, A.U., Patel, P., Sondek, J., and Dangl, J.L. (2005. a). The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc. Natl. Acad. Sci. USA 102 6496–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.G., da Cunha, L., McFall, A.J., Belkhadir, Y., DebRoy, S., Dangl, J.L., and Mackey, D. (2005. b). Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121 749–759. [DOI] [PubMed] [Google Scholar]
- Koopman, W.J.M., Zevenbergen, M.J., and Van den Berg, R.G. (2001). Species relationships in Lactuca SL (Lactuceae, Asteraceae) inferred from AFLP fingerprints. Am. J. Bot. 88 1881–1887. [PubMed] [Google Scholar]
- Krüger, J., Thomas, C.M., Golstein, C., Dixon, M.S., Smoker, M., Tang, S., Mulder, L., and Jones, J.D.G. (2002). A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296 744–747. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., and Dinesh-Kumar, S.P. (2002. a). Virus-induced gene silencing in tomato. Plant J. 31 777–786. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S.P. (2002. b). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30 415–429. [DOI] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108 743–754. [DOI] [PubMed] [Google Scholar]
- Mallet, J. (2006). What does Drosophila genetics tell us about speciation? Trends Ecol. Evol. 21 386–393. [DOI] [PubMed] [Google Scholar]
- McHale, L.K., Truco, M.J., Kozik, A., Wroblewski, T., Ochoa, O.E., Lahre, K.A., Knapp, S.J., and Michelmore, R.W. (2009). The genomic architecture of disease resistance in lettuce. Theor. Appl. Genet. 118 565–580. [DOI] [PubMed] [Google Scholar]
- Mojtahedi, H., Boydston, R.A., Thomas, P.E., Crosslin, J.M., Santo, G.S., Riga, E., and Anderson, T.L. (2003). Weed hosts of Paratrichodorus allius and tobacco rattle virus in the Pacific Northwest. Am. J. Potato Res. 80 379–385. [Google Scholar]
- Qutob, D., Kamoun, S., and Gijzen, M. (2002). Expression of a Phytophthora sojae necrosis-inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32 361–373. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L.H., and Willis, J.H. (2007). Plant speciation. Science 317 910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney, H.C.E., van't Klooster, J.W., van der Hoorn, R.A.L., Joosten, M.H.A.J., Jones, J.D.G., and de Wit, P.J.G.M. (2005). Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308 1783–1786. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Truco, M.J., Antonise, R., Lavelle, D., Ochoa, O., Kozik, A., Witsenboer, H., Fort, S.B., Jeuken, M.J.W., Kesseli, R.V., Lindhout, P., Michelmore, R.W., and Peleman, J. (2007). A high-density, integrated genetic linkage map of lettuce (Lactuca spp.). Theor. Appl. Genet. 115 735–746. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn, R.A.L., and Kamoun, S. (2008). From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 20 2009–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoorn, R.A.L., Laurent, F., Roth, R., and De Wit, P.J.G.M. (2000). Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant Microbe Interact. 13 439–446. [DOI] [PubMed] [Google Scholar]
- Van Ooijen, J.W. (2004). MapQTL 5: Software for the Mapping of Quantitative Trait Loci in Experimental Populations. (Wageningen, The Netherlands: Kyazma).
- Van Ooijen, J.W. (2006). JoinMap 4: Software for the Calculation of Genetic Linkage Maps in Experimental Populations. (Wageningen, The Netherlands: Kyazma).
- Wang, Y., Bao, Z., Zhu, Y., and Hua, J. (2009). Analysis of temperature modulation of plant defense against biotrophic microbes. Mol. Plant Microbe Interact. 22 498–506. [DOI] [PubMed] [Google Scholar]
- Wroblewski, T., Tomczak, A., and Michelmore, R. (2005). Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 3 259–273. [DOI] [PubMed] [Google Scholar]
- Wulff, B.B.H., Kruijt, M., Collins, P.L., Thomas, C.M., Ludwig, A.A., De Wit, P.J.G.M., and Jones, J.D.G. (2004). Gene shuffling-generated and natural variants of the tomato resistance gene Cf-9 exhibit different auto-necrosis-inducing activities in Nicotiana species. Plant J. 40 942–956. [DOI] [PubMed] [Google Scholar]
- Zhang, N.W., Lindhout, P., Niks, R.E., and Jeuken, M.J.W. (2009. a). Genetic dissection of Lactuca saligna nonhost resistance to downy mildew at various lettuce developmental stages. Plant Pathol. 58 923–932. [Google Scholar]
- Zhang, N.W., Pelgrom, K., Niks, R.E., Visser, R.G.F., and Jeuken, M.J.W. (2009. b). Three combined quantitative trait loci from nonhost Lactuca saligna are sufficient to provide complete resistance of lettuce against Bremia lactucae. Mol. Plant Microbe Interact. 22 1160–1168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.