Abstract
Eukaryotic processing bodies (P-bodies) are implicated in mRNA storage and mRNA decapping. We previously found that a decapping complex comprising Decapping 1 (DCP1), DCP2, and Varicose in Arabidopsis thaliana is essential for postembryonic development, but the underlying mechanism is poorly understood. Here, we characterized Arabidopsis DCP5, a homolog of human RNA-associated protein 55, as an additional P-body constituent. DCP5 associates with DCP1 and DCP2 and is required for mRNA decapping in vivo. In spite of its association with DCP2, DCP5 has no effect on DCP2 decapping activity in vitro, suggesting that the effect on decapping in vivo is indirect. In knockdown mutant dcp5-1, not only is mRNA decapping compromised, but the size of P-bodies is also significantly decreased. These results indicate that DCP5 is required for P-body formation, which likely facilitates efficient decapping. During wild-type seed germination, mRNAs encoding seed storage proteins (SSPs) are translationally repressed and degraded. By contrast, in dcp5-1, SSP mRNAs are translated, leading to accumulation of their products in germinated seedlings. In vitro experiments using wheat germ extracts confirmed that DCP5 is a translational repressor. Our results showed that DCP5 is required for translational repression and P-body formation and plays an indirect role in mRNA decapping.
INTRODUCTION
Eukaryotic cells assemble messenger ribonucleoproteins (mRNPs) into cytoplasmic foci referred to as processing bodies (P-bodies; Sheth and Parker, 2003), whose functions include translational repression, mRNA storage, and mRNA decapping. Under stress conditions, yeast P-bodies contain mRNAs that are translationally repressed, suggesting the P-body assembly is associated with regulation of mRNA translatability. This translation arrest is reversible, as when favorable conditions return, certain mRNAs may reengage with ribosomes to resume translation (Brengues et al., 2005). The mechanisms that regulate decapping of selective stored mRNAs in P-bodies are still unknown.
In yeast, nontranslated mRNAs are sequestered in P-bodies, but the exact site of mRNA storage in mammalian cells is not clearly understood. Under adverse environmental conditions, mammalian cells accumulate stress granules, which are believed to be sites of mRNA triaged to storage, degradation, or translation reinitiation. Recently, P-bodies in yeast were found to be required for stress granule assembly, and stress granules are the only site of translation reinitiation (Buchan et al., 2008). Therefore, P-bodies may also function as mRNA storage sites in multicellular eukaryotes.
The decapping of mRNAs, which also occurs inside P-bodies, represents an irreversible step in mRNA degradation and therefore is highly regulated. We previously characterized an Arabidopsis thaliana decapping complex consisting of Decapping 1 (DCP1), DCP2, and Varicose (VCS), with only DCP2 possessing the pyrophosphatase activity specific for removing the m7GDP cap (Xu et al., 2006; Iwasaki et al., 2007). All three proteins colocalized in P-bodies, suggesting that plants may use a similar conserved mechanism to that found in yeast to regulate mRNA storage and degradation. However, since knockout mutants of DCP1, DCP2, and VCS display postembryonic lethality, and the abundance of a large number of transcripts is changed (upregulated and downregulated) in these mutants (Goeres et al., 2007), it is difficult to identify direct mRNA substrates for the decapping complex and to determine the biological significance of decapping in plants.
Yeast dcp1-1 mutants are temperature sensitive but viable; mutant cells are arrested in growth at 36°C (Beelman et al., 1996; Hatfield et al., 1996). By contrast, both Arabidopsis dcp1 and dcp2 mutants deficient in mRNA decapping are postembryonic lethal (Xu et al., 2006), indicating that functional P-bodies are required for plant postembryonic development. The mechanism of this lethality has not been explored. Although DCP1 and VCS have no known enzyme activities, they are still required for mRNA decapping in vivo. This observation suggests that the decapping process is highly regulated and P-body formation may be critical for decapping in vivo.
Assembly of yeast P-bodies is regulated by many proteins, including Like SM 4 (Lsm4p) and Lsm16/EDC3 (Decker et al., 2007). EDC3, which is an enhancer of decapping, belongs to the Lsm domain protein family (Albrecht and Lengauer, 2004; Kshirsagar and Parker, 2004). Notably, the Lsm domain protein family is conserved in eukaryotes and has been extensively investigated in many organisms. For example, Cytokinesis, apoptosis and RNA 1 (CAR-1) in Caenorhabditis elegans colocalized with conserved germline RNA helicase 1 (CGH-1), a DExD/H-box Helicase 1 (DHH1) homolog to RNA-containing P-granules, and is required for cytokinesis during early embryogenesis (Audhya et al., 2005). RNA-associated protein 55 (RAP55) was found to arrest mRNA translation in Xenopus laevis oocytes (Tanaka et al., 2006); the Drosophila melanogaster Trailer hitch (Tral) protein is required for proper dorsal-ventral patterning during oogenesis (Wilhelm et al., 2005), and RAP55/hlsm14 (human lsm14) is a P-body marker in human cells (Yang et al., 2006).
Various partners interacting with these Lsm proteins have been described, and information on their localizations and functions is available. Yeast Lsm13 protein Suppressor of clathrin deficiency (Scd6) was found to colocalize with Dcp2p under conditions of high cell density or nutrient starvation (Barbee et al., 2006). Genetic interaction between Scd6 and P-body components has been reported recently (Decourty et al., 2008). CAR-1/Tral/xRAP55 were found to colocalize and associate with CGH-1/Me31B/xP54, an RNA helicase that labels P-bodies (Audhya et al., 2005; Boag et al., 2005; Wilhelm et al., 2005; Tanaka et al., 2006). Structural analysis revealed that hEdc3 possesses an N-terminal Lsm domain for binding to DCP1, a FDF domain, and an YjeF-N domain mediating dimerization (Tritschler et al., 2007; Ling et al., 2008). Tral/CAR-1/xRAP55 carries an Lsm domain and a FDF domain flanked by a domain rich in charged residues, the RGG domain. The Lsm domain in Tral also mediates the interaction between Tral and DCP1. However, Tral was found not to associate with DCP2 in Drosophila, whereas Edc3 interacts with DCP2 though a linker region between the Lsm domain and the FDF motif (Tritschler et al., 2007, 2008). However, the exact function of these Lsm domain proteins in both P-body formation and mRNA decapping is largely unknown.
We show here that an Arabidopsis Lsm domain protein, called DCP5, indirectly regulates mRNA decapping through P-body formation. Interactions and self-dimerizations among DCP1, DCP2, and DCP5 lead to the formation of a complex with mRNA and final assembly into P-bodies. Knockdown mutants of DCP5 share developmental abnormalities with other decapping-deficient mutants in postembryonic development (Xu et al., 2006; Goeres et al., 2007; Iwasaki et al., 2007). We conclude that P-body formation mediated largely by DCP5 plays an important role in mRNA decapping during postembryonic development.
RESULTS
Arabidopsis DCP5 and DCP5-Like Belong to the LSM Domain Protein Family
We performed a BLAST search to identify proteins that may be associated with mRNA decapping in Arabidopsis. At least seven proteins share similar amino acid sequences with the Lsm14 domain protein RAP55 (Albrecht and Lengauer, 2004). Among them, DCP5, which belongs to the Lsm13 group, shares 48 and 47% similar amino acids with CAR-1 and human RAP55, respectively. Moreover, the overall organization of these proteins and their domain structures are also highly conserved. DCP5 possesses an N-terminal Lsm domain, a Ser/Thr-rich domain, and two RGG domains separated by an FDF domain in the C terminus. The Arabidopsis genome contains another predicted protein, which we named decapping 5-like (DCP5-L). DCP5-L, which shares 35% identical and 51% similar amino acids with DCP5, is the closest DCP5 homolog.
The DCP5-Deficient Mutant Shares Similar Phenotype with Other Decapping-Deficient Mutants
To characterize functions of DCP5 and DCP5-L, we obtained a T-DNA insertion line dcp5-1 with disruption in the DCP5 locus (Figure 1A). We also designed RNA interference (RNAi) constructs to specifically knockout each of these genes (see Methods). Among 25 transgenic lines of DCP5-L-RNAi, none was distinguishable from the wild type. However, ∼65% of the transgenic lines (n = 43) of DCP5-RNAi displayed a clear lethal phenotype, similar to the transgenic lines of DCP2-RNAi (56% of 68) (Figure 2A), dcp2-1, dcp1-1, and vcs-6 (Xu et al., 2006). We therefore decided to further investigate the function of DCP5.
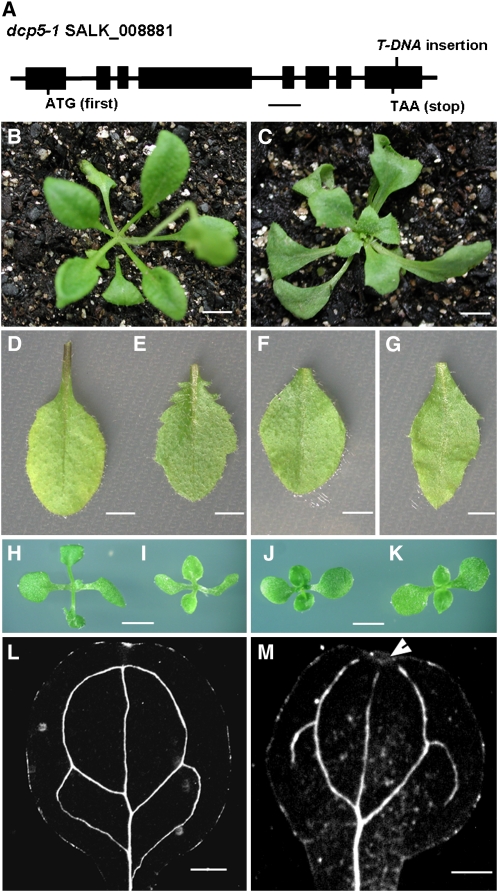
Figure 1.
Phenotypic Analysis of the dcp5-1 Mutant.
(A) Schematic presentation of T-DNA insertion confirmed in dcp5-1 (bar = 500 bp).
(B) Three-week-old wild-type plant.
(C) Three-week-old dcp5-1 plant (bar = 1 cm).
(D) Rosette leaf of the wild type.
(E) Rosette leaf of dcp5-1.
(F) Cauline leaf of the wild type.
(G) Cauline leaf of dcp5-1 (bar = 20 mm).
(H) to (K) Two-week-old dcp5-1 plant (I) was compared with wild-type (Columbia) plant (H), as well as a vcs-1 plant (K) and wild-type plant (J) both in the Landsberg erecta ecotype (bars = 0.5 cm).
(L) and (M) Light microscopy image of cotyledons from 6-d-old wild-type (L) and dcp5-1 (M) seedlings after clearing (bars = 1 mm) to show venation patterns. Arrowhead indicates discontinuity of the venation system.
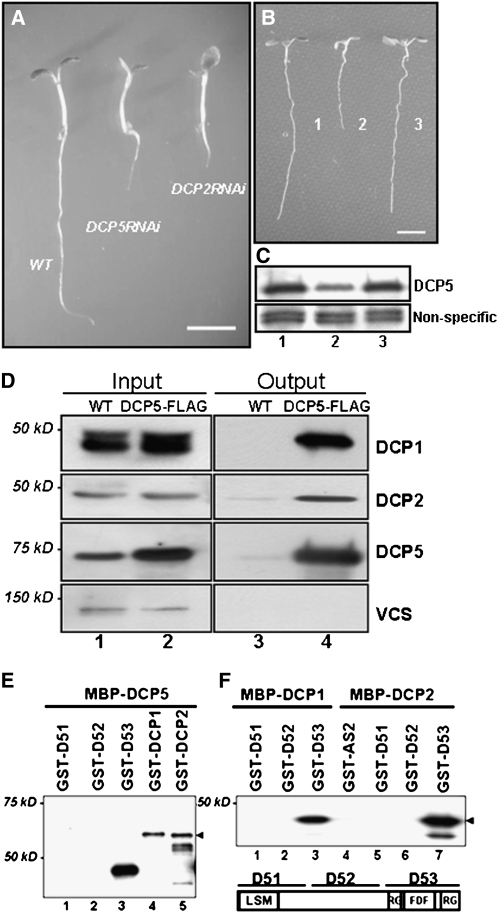
Figure 2.
Complementation of the dcp5-1 Mutant Phenotype and Interactions of DCP5 with Other Decapping Components.
(A) Six-day-old seedlings of the wild type, DCP5-RNAi, and DCP2-RNAi (bar = 0.5 cm).
(B) Ten-day-old seedlings of the wild type, dcp5-1, and the DCP5-FLAG complementation line (bar = 1.5 cm).
(C) Protein gel blot analysis of DCP5 protein levels in the wild type (lane 1), dcp5-1 (lane 2), and the DCP5-FLAG complementation line (lane 3) using anti-DCP5 antibody. Similar results were obtained with the other two biological replicates.
(D) DCP5 interacts with DCP1 and DCP2 in vivo. Coimmunopreciptitation was performed with extracts prepared from the wild type (lanes 1 and 3) and DCP5-FLAG line (lanes 2 and 4). Extracts were immunoprecitated with anti-FLAG antibody, and immunopreciptates were then analyzed using specific antibodies against DCP1, DCP2, DCP5, and VCS. Five percent of extract was loaded as input.
(E) and (F) In vitro interactions between DCP1, DCP2, DCP5, and various DCP5 domains. Arrowhead in (E) indicates full-length GST-DCP2. Arrowhead in (F) indicates full-length GST-D53. Purified proteins used in pull-down assays are indicated above each lane, and different portions of DCP5 used in the assays are illustrated in the right bottom panel. GST fusion proteins were detected by anti-GST monoclonal antibody in protein gel blots.
A DCP5 Knockdown Mutant, dcp5-1, Is Not Postembryonic Lethal
In contrast with dcp2-1, dcp1-1, and vcs-6, dcp5-1 did not display postembryonic lethality. Seed germination in dcp5-1 was severely perturbed, producing pale and weak cotyledons, which could be typically seen 6 d after germination. The seedling phenotype was similar to that of dcp2-1, dcp1-1, and vcs-6 (Xu et al., 2006). Seedlings of dcp2-1, dcp1-1, and vcs-6 ceased to develop further 6 d after germination. By contrast, dcp5-1 resumed growth soon after 6 d, producing flowers only slightly later than the wild type.
Vegetative growth of dcp5-1 was generally normal (Figure 1C), with visible morphological changes, including serration and pointed rosette and cauline leaves (Figures 1E and 1G). Leaf shape of dcp5-1 (Figure 1I) was similar to that of vcs-1 (Figure 1K), a weak allele of VCS, which survives under lower temperatures (Deyholos et al., 2003). Cotyledonary veins of dcp5-1 were disorganized, with disruptions at the distal end, in contrast with the closed loop seen in wild-type veins (Figures 1L and 1M). This abnormal venation phenotype was moderate compared with the stronger disturbances seen in cotyledons of dcp1-1, dcp2-1, and vcs-6 (Xu et al., 2006).
DNA sequencing analysis confirmed that the T-DNA was inserted in the 3′-untranslated region (UTR) of DCP5, 10 bp downstream of the stop codon. In dcp5-1, DCP5 transcripts were reduced by approximately fivefold as measured by quantitative RT-PCR (qRT-PCR; Table 1). This reduction presumably resulted from a destabilization of its transcripts carrying the altered 3′-UTR. DCP5 protein levels in dcp5-1 were decreased by approximately fivefold in comparison to wild-type levels (Figure 2C). A 5.7-kb genomic fragment corresponding to the DCP5 loci, with an insertion of a FLAG tag sequence directly 5′ upstream of the stop codon of DCP5, fully complemented dcp5-1 mutant phenotypes (Figures 2B and 2C). Hence, dcp5-1 is a DCP5 knockdown mutant.
Table 1.
qRT-PCR Analysis of Transcripts (Shown in Fold Change) in 6-d-Old Decapping Mutants
| AGI | Symbol | Wild Type | dcp5-1 | dcp1-1 | dcp2-1 | vcs-6 | Description |
|---|---|---|---|---|---|---|---|
| AT4G25140 | OLEO1 | 1 | 97.6 | 3.3 | 9 | 1.1 | Oleosin* |
| AT5G40420 | OLEO2 | 1 | 14.2 | 1.7 | 3.8 | 0.8 | Oleosin* |
| AT5G51210 | OLEO3 | 1 | 23.8 | 7.5 | 3.6 | 1.7 | Oleosin* |
| AT3G27660 | OLEO4 | 1 | 15.1 | 1.3 | 0.9 | 0.2 | Oleosin* |
| AT2G25890 | OLEO1L | 1 | 3.3 | 6.5 | 3.1 | 0.9 | Oleosin* |
| AT3G01570 | OLEO2L | 1 | 42.3 | 3.3 | 6.5 | 1.7 | Oleosin* |
| AT4G27140 | 1 | 43.5 | 6.3 | 10.9 | 0.8 | 2S seed storage protein 1* | |
| AT4G27150 | 1 | 251.2 | 26.4 | 37.6 | 3.3 | 2S seed storage protein 2* | |
| AT4G27160 | 1 | 389.2 | 41 | 71.6 | 6 | 2S seed storage protein 3* | |
| AT4G27170 | 1 | 305.2 | 21.4 | 55.1 | 2.2 | 2S seed storage protein 4* | |
| AT5G44120 | CRA1 | 1 | 259.5 | 53.8 | 120.5 | 10.5 | 12S seed storage protein* |
| AT1G03890 | CRA1L | 1 | 73.1 | 22.2 | 10.3 | 0.6 | 12S seed storage protein* |
| AT1G03880 | CRB | 1 | 149.1 | 12.7 | 11.3 | 0.4 | 12S seed storage protein* |
| AT4G28520 | CRC | 1 | 1377.1 | 304.9 | 311 | 17.2 | 12S seed storage protein * |
| AT1G32560 | 1 | 8.5 | 26.8 | 18.5 | 11.9 | Late embryogenesis abundant | |
| AT2G35300 | 1 | 9.8 | 40.7 | 24 | 11 | Late embryogenesis abundant | |
| AT1G48130 | 1 | 10.7 | 6.5 | 14.2 | 2.1 | Rehydrin | |
| AT2G41260 | 1 | 33.8 | 71.5 | 94.1 | 13.6 | Late embryogenesis abundant | |
| AT3G17520 | 1 | 6.7 | 117.5 | 32.2 | 53.2 | Late embryogenesis abundant | |
| AT3G15670 | 1 | 6.1 | 14.8 | 7.1 | 2.9 | Late embryogenesis abundant | |
| AT1G26110 |
DCP5 |
1 |
0.2 |
0.6 |
1 |
0.5 |
DCP5 (this work) |
The asterisks indicate SSPs. The means of three replicates of qRT-PCR are shown. Standard deviations are not shown. The expression level in the wild type (Col) was arbitrarily set to 1.0. Similar results were obtained when qRT-PCR was performed using a second set of samples.
Interactions between DCP1, DCP2, and DCP5
The phenotypic similarity of dcp5-1 and other decapping-deficient mutants prompted us to test for direct interactions between proteins encoded by these genes. Using a complementation line expressing DCP5-FLAG (hereafter referred to as the DCP5-FLAG line), we tested in vivo protein interactions by immunoprecipitation followed by protein gel blot analysis. Interactions could be observed between DCP5 and DCP1 and DCP2. Surprisingly, no interaction was detected between DCP5 and VCS (Figure 2D). DCP1 appeared as a doublet in SDS-PAGE gels. Interestingly, the upper band of DCP1 was more strongly associated with DCP5 than the lower band in the immunoprecipitates. This result suggested that DCP1 is associated with DCP5 in vivo, and a certain modified form of DCP1 may display a stronger affinity for DCP5. However, we could not detect VCS in the immunocomplexes, suggesting weak or indirect association between VCS and DCP5.
To test possible direct interactions between the three proteins in vitro, we purified recombination proteins MBP-DCP5, MBP-DCP2, MBP-DCP1, glutathione S-transferase (GST)-DCP2, and GST-DCP1 and three fragments of DCP5 (GST-DCP51, GST-DCP52, and GST-DCP53) from Escherichia coli extracts (see Methods). GST-DCP1 and GST-DCP2 could be pulled down by MBP-DCP5 with amylose resin (Figure 2E), confirming their direct interactions. Furthermore, using fragments containing various portions of DCP5, we found that DCP5 interacted with DCP1, DCP2, and itself mainly through the C-terminal region, which contains the RGG domains and FDF domain (Figures 2E and 2F). The self-association of DCP5 in vitro suggested that DCP5 may form a homodimer in vivo.
dcp5-1 Accumulates mRNAs with a Prolonged Half-Life
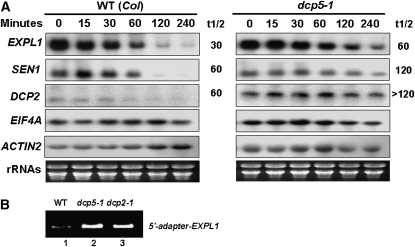
We previously reported that unstable mRNAs, like those of Expansin-Like1 (EXPL1) and Senescence-related 1 (SEN1), accumulate in the capped form in decapping mutants dcp1-1, dcp2-1, and vcs-6 (Xu et al., 2006). In dcp5-1, EXPL1 transcript was also stabilized, with an estimated half-life of 60 min compared with the half-life of 30 to 40 min found in control samples (Figure 3A). In contrast with a previously estimated half-life of EXPL1 transcripts of ∼100 min in dcp2-1, 120 min in dcp1-1, and 85 min in vcs-6, the prolongation of half-life of these transcripts in dcp5-1 was rather moderate. These results are consistent with the fact that dcp5-1 is a knockdown mutant with residual decapping activity, whereas decapping activity is completely abolished in knockout mutants dcp1-1, dcp2-1, and vcs-6. On the other hand, the steady state EXPL1 transcript level in dcp5-1 was only slightly higher than the control level, in contrast with the 10-fold higher level in dcp2-1, dcp1-1, and vcs-6 mutants. This could be the consequence of the moderate effect of DCP5 knockdown; alternatively, EXPL1 may not be the direct target of DCP5 at the developmental state tested. An increase in half-life similar to EXPL1 was observed with the other transcripts, SEN1 mRNA and DCP2 mRNA, confirming that the decapping activity in vivo was indeed attenuated in dcp5-1 (Figure 3A). Similar to our previous finding with dcp1-1, dcp2-1, and vcs-6, using modified rapid amplification of cDNA ends (RACE)-PCR, we confirmed that EXPL1 accumulated in the capped form in dcp5-1 (Figure 3B). Therefore, DCP5 is required for mRNA decapping in vivo.
Figure 3.
DCP5 Is Required for mRNA Decapping in Vivo.
(A) Transcript accumulation in dcp5-1. Individual transcript (labeled on the left) levels in control and dcp5-1 at different time points after cordycepin treatment, as shown by RNA gel blot using specific DNA probes. Estimated half-life (min) of specific mRNAs is shown to the right of each panel. Ethidium bromide–stained rRNA bands (bottom panel) are shown as loading controls.
(B) Modified RACE-PCR to detect capped EXPL1 transcripts in wild-type and different mutant plants.
Next, we tested if direct interaction among DCP5 and DCP2 would affect the decapping activity of DCP2 in vitro. However, DCP5 had no effect on DCP2 activity in decapping assays (see Supplemental Figure 1 online). This result indicates that interaction between DCP5 and DCP2 is functionally distinct from interactions between DCP1 and VCS and DCP2. DCP1 and VCS have been previously shown to modulate DCP2 activity directly in vitro (Xu et al., 2006).
DCP5 Is Required for P-Body Formation
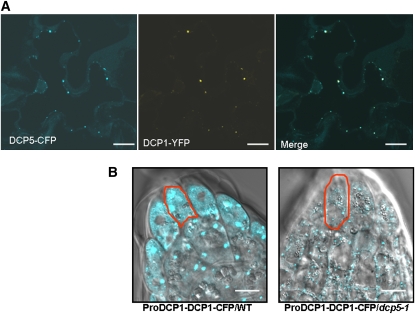
Since DCP5 does not affect DCP2 decapping activity in vitro, the accumulation of capped mRNAs in the dcp5-1 mutant may be an indirect effect of the decapping process in vivo. To explore this further, we decided to investigate intracellular localization of DCP5. We generated a construct expressing a DCP5-cyan fluorescent protein (CFP) fusion protein. In Nicotiana benthamiana leaf epidermal cells, DCP5-YFP colocalized with DCP1-yellow fluorescent protein (YFP) and DCP2-CFP (Figure 4A). These results, which confirmed that DCP5 localizes in P-bodies, are consistent with the observation that DCP5 directly interacts with DCP1 and DCP2 in vivo.
Figure 4.
DCP5 Is Required for P-Body Assembly.
(A) Subcellular localization of DCP5-CFP and DCP1-YFP in tobacco epidermal cells (bars = 10 μm); CFP fluorescence is pseudocolored in blue and YFP in yellow.
(B) P-bodies as visualized by DCP1-CFP expressed from a native promoter in root tip cells of wild-type (left) and dcp5-1 (right) Arabidopsis seedlings (bars = 20 μm). A typical cell from each line was outlined in red.
A construct harboring DCP1-CFP driven by a native DCP1 promoter (ProDCP1-DCP1-CFP) was introduced into the dcp5-1 mutant and wild-type plants, and roots of 3-d-old transgenic seedling were examined. We found that the size of the P-body marked by DCP1-CFP was significantly decreased in the dcp5-1 mutant compared with the wild type (Figure 4B). As dcp5-1 is a knockdown mutant, we also tested plants cotransformed with a DCP5-RNAi or a DCP2-RNAi construct, along with DCP1-CFP. In roots of cotransformants of DCP5-RNAi and DCP1-CFP, which displayed a severe lethal phenotype, the punctuate pattern of DCP1-CFP was lost and DCP1-CFP signal was mostly dispersed in the cytosol. However, in DCP2-RNAi and DCP1-CFP cotransformants, DCP1-CFP continued to be localized to discrete cytoplasmic foci (see Supplemental Figure 2 online). From these results we concluded that DCP5 was required for P-body formation.
dcp5-1 Seedlings Accumulate SSP mRNAs during Germination
To determine which specific mRNAs are processed by the DCP5/2/1 complex in vivo, we performed a preliminary genomic transcript profiling of dcp5-1 and wild-type seedlings by microarray analysis. Six-day-old dcp5-1 seedlings were used because of their distinctive phenotype compared with wild-type seedlings. About 3% of all detectable transcripts accumulated at more than fivefold higher levels in dcp5-1 compared with the wild type, and 0.02% of all transcripts were reduced by more than fivefold in dcp5-1 compared with the wild type, and this list included DCP5 transcripts. Interestingly, the most highly accumulated transcripts (∼30%) were mRNAs encoding seed storage proteins (SSPs), which are expressed during seed maturation and stored in dry seeds but are degraded during seed germination.
We then focused on these SSP mRNAs and used qRT-PCR to compare their transcript levels in wild-type, dcp5-1, dcp1-1, dcp2-1, and vcs-6 plants (Table 1). Nineteen out of 20 mRNAs tested were increased by more than fivefold in dcp5-1 compared with the wild type, with the OLEO1L (OLEO, Oleosin) mRNA increased by 3.3-fold. Similar to dcp5-1, most of the 20 mRNAs also accumulated by more than fivefold in dcp1-1 and dcp2-1 compared with the wild type. This is consistent with the notion that mRNAs are regulated by the decapping complex comprising DCP5/2/1. We also observed differential accumulation of mRNA levels among these mutants. In particular, dcp5-1 accumulated transcripts encoding many SSP (as marked with an asterisk in Table 1) at a level much higher than in dcp2-1 and dcp1-1. Because dcp5-1 is a knockdown mutant, this result indicates that OLEO mRNAs and the other SSP mRNAs are more sensitive to the DCP5 deficiency than to the loss of DCP1 and DCP2. Hence, DCP5 operates upstream of DCP2 and DCP1 to regulate these mRNAs. The vcs-6 mutant accumulated many LEA (later embryogenesis abundant) mRNAs but not SSP mRNAs in similar levels as in dcp2-1 and dcp1-1, suggesting that decapping of these LEA mRNAs also required VCS.
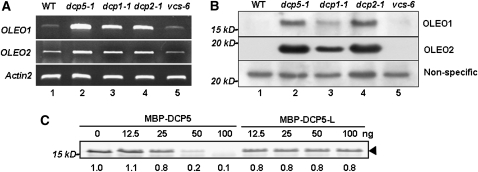
DCP5 Arrests OLEO1 mRNA Translation in Vivo and in Vitro
The overaccumulation of OLEO1/2 in dcp1-1, dcp2-1, and dcp5-1 was reconfirmed by RT-PCR (Figure 5A). To test if the accumulated transcripts are capable of returning to polysomes for translation, we determined protein levels of OLEO1/2. Using specific antibodies (Shimada et al., 2008), we found that OLEO1 and OLEO2 accumulated to high levels in dcp1-1, dcp2-1, and dcp5-1, compared with in the wild type and vcs-6 (Figure 5B). The overaccumulation of OLEO proteins largely correlated with the accumulation of their cognate mRNAs in these decapping-deficient mutants. This result indicates that OLEO mRNAs, if not subject to decapping, are capable of returning to polysomes for translation. This result also suggests that OLEO mRNAs are both translationally repressed and decapped in P-bodies containing at least DCP5/2/1. The vcs-6 mutant expressed about the same level of OLEO mRNA as the wild type, and OLEO protein levels in vcs-6 were comparable to those in the wild type. These observations rule out the possibility that VCS is involved in the translational repression of OLEO1 and OLEO2 mRNAs.
Figure 5.
DCP5 Is Required for Translation Repression of OLEO1/2 mRNAs.
(A) Transcript levels of OLEO1 and OLEO2 in 6-d-old plants were determined by RT-PCR (qRT-PCR results are shown in Table 1). Actin2 levels were used as loading controls.
(B) OLEO1 and OLEO2 levels in 6-d-old plants were measured by protein gel blots using specific antibody. A nonspecific band was used as a loading control.
(C) Effects of DCP5 and DCP5-L on OLEO1 mRNA translation in vitro. MBP-DCP5 and MBP-DCP5-L were added to a wheat germ extract translation system containing the same amount of OLEO1 mRNA. Amounts of proteins are indicated above each lane. Arrowhead indicates the OLEO1 protein.
To test if DCP5 can directly arrest translation of OLEO mRNAs, we generated capped and polyA-containing OLEO1 mRNAs through an in vitro transcription system. Using a wheat germ translation system, we found that, with the same amount of mRNA input, MBP-DCP5 protein isolated from E. coli extracts was capable of arresting the translation of OLEO1 mRNA (Figure 5C). MBP-DCP5-L, which was purified by the same procedure from E. coli extracts, however, had no significant effect on the translation (Figure 5C). The latter result excluded the possibility that the translation arrest was due to MBP or contaminating E. coli proteins and provided evidence that DCP5 functions in vitro to suppress translation. In a separate experiment, we monitored levels of labeled OLEO1 mRNA in the reaction mixtures (see Methods). We found that the amounts of OLEO1 mRNA were not affected by the addition of MBP-DCP5 or MBP-DCP5-L (see Supplemental Figure 3 online), indicating that these proteins did not affect mRNA stability under the experimental conditions.
Dynamic Changes of DCP Protein Levels during Seed Development
The accumulation of SSP mRNAs in decapping-deficient mutants prompted us to test if DCPs are dynamically linked to seed-related developmental phases. Protein gel blot analysis showed that DCP5 and DCP1 gradually accumulated during seed maturation and reached their peak levels in dry seeds (Figure 6). Upon germination, however, levels of DCP5 gradually decreased along with a reduction in DCP1 (Figure 6). DCP2 was not detected during seed maturation processes and in dry seeds; however, it was immediately induced after seed germination. These dynamic changes of DCPs suggest that different constituent proteins are assembled into P-bodies during plant development.
Figure 6.
Dynamic Changes in DCP1, DCP2, and DCP5 Protein Levels during Seed Maturation and Seed Germination.
Seeds from siliques of 6-week-old plants were labeled in order, with the bottom (i.e., oldest) silique being labeled first (left panel). Each lane represents proteins from two fresh seeds (left panel) or two dried seeds (middle panel). Protein levels in germinating seeds are shown in order of days after imbibition (right panel).
DISCUSSION
DCP5 Is a Novel Component of the Arabidopsis Decapping Complex
Previously, we reported that DCP1, DCP2, and VCS form a decapping complex required for postembryonic development (Xu et al., 2006). Here, we identified DCP5 as an additional component of the Arabidopsis decapping complex. First, DCP5 knockdown mutants or DCP5-RNAi lines deficient in DCP5 proteins share similar phenotypes with those of the other decapping-deficient mutants. Second, DCP5 associates with DCP1 and DCP2 in vivo and in vitro. Third, mRNA decapping is compromised in the dcp5-1 mutant. Interestingly, there is no direct association between VCS and DCP5, suggesting the possible differential utilization of P-body constituents to generate different decapping complexes.
The Lsm13 protein DCP5 belongs to the conserved Lsm13-15 protein family in eukaryotes. DCP5 homologs, including SCD6, RAP55, and CAR1, have not yet been shown to function in decapping, although most of them are localized to P-bodies. It would be interesting to test if these proteins also act as a component of the decapping complex. In yeast, an Lsm16 family protein Edc3 targets yeast Rps28b and YRA1 mRNAs for deadenylation-independent decapping (Badis et al., 2004; Dong et al., 2007). However, the Arabidopsis genome does not appear to contain any close homolog of Edc3 because the distinctive YjeF_N domain of Edc3 is only found in several putative pyridoxamine 5′-phosphate oxidases (Albrecht and Lengauer, 2004). The function of Edc3 appears to have been supplanted by DCP5 in plants.
Deletion analysis of Edc3 demonstrated that the Lsm16 domain and a fragment encompassing the low-complexity linker region interacts with DCP1 and DCP2, respectively (Tritschler et al., 2007). Tral interacts with DCP1 and localizes to mRNA P-bodies via the Lsm15 domain, but it does not associate with DCP2 (Tritschler et al., 2008). By contrast, we found that DCP5 interacts with DCP1 and DCP2 through its C terminus, which contains the FDF and RGG domains. These differences may be attributed to different domain organizations between Edc3 and DCP5 and different Lsm domains between Tral and DCP5. Although there is no report on the in vitro effect of Edc3 on decapping, we found that DCP5 has no direct effect on DCP2 decapping activity in vitro. The perturbed mRNA decapping in vivo in dcp5-1 is more likely due to a defect in translation repression and P-body formation.
DCP5 Is Required for Both Translation Repression and P-Body Formation
Translation and decapping of mRNA are competitive processes. Before mRNAs are made available for decapping, certain translation factors have to be released from mRNAs through translation repression (Beelman and Parker, 1994; Schwartz and Parker, 1999, 2000). Several decapping activators have been found to promote translation repression. For example, Dhh1 triggers translational silencing and promotes the assembly of individual mRNPs into P-bodies through an interaction with Edc3 (Decker et al., 2007). Similar to RAP55-mediated translation arrest of mRNAs in X. laevis oocytes (Tanaka et al., 2006), we found that DCP5 is capable of inhibiting OLEO1 translation both in vivo and in vitro. To our knowledge, no other plant proteins have been reported to act as a translation repressor. Our in vitro results provide direct evidence that a plant protein can act as a translation repressor. The function of DCP5 at this level suggests that P-body formation is a consequence of the active repression of mRNA translation.
Not much is known regarding the detailed mechanism of P-body formation in eukaryotes. Edc3 and Lsm4 are known to regulate P-body formation in yeast, and the Yjef_N dimerization domain of Edc3 and the Q/N-rich domain of Lsm4 are critical for this function (Decker et al., 2007; Reijns et al., 2008). However, these two domains are absent from their plant counterparts. DCP5 does not possess an Yjef_N domain, and the Q/N-rich domain is substituted with a RGG domain in Arabidopsis Lsm4. It is plausible that the mechanism underlying P-body formation in plants is different from that in yeast and different protein domains are used for mRNP assembly. Notably in mammalian cells, small interfering RNA-mediated silencing of RAP55 also led to a depletion of P-bodies (Yang et al., 2006). Under what evolutionary pressure plants and mammals select against the Yjef_N domain in P-body formation is still a question to be addressed.
DCP5 Is Essential for P-Body Function and Postembryonic Development
A recent kinetic model proposed by Franks and Lykke-Andersen (2008) suggests that at least three steps are involved in P-body function. First, mRNAs are translationally arrested and sequestered from the polysomes. Second, sequestered mRNAs initiate the formation of mRNPs with P-body protein constituents and subsequently mRNPs are assembled into P-bodies. Third, mRNAs are decapped and then degraded. Based on this model, we propose the following sequence of events in Arabidopsis during postembryonic development. DCP5 may arrest mRNA translation first and then recruit DCP1 and DCP2 to assemble the mRNPs into P-bodies. P-body formation mediated by DCP5 is required for efficient decapping in vivo, although DCP5 has no direct effect on DCP2 decapping activity in vitro.
We have also shown that during postembryonic development, these three steps are dynamically linked and are largely reversible before the committed step of mRNA decapping. DCP5 and DCP1 accumulate during seed maturation and in drying seeds, but they decrease over time upon seed germination. However, there is no proportionate or synchronous increase of DCP2 along with DCP5 and DCP1. This uncoupling in DCP2 accumulation from that of DCP5/1 may facilitate the storage function of the P-body in dry seeds, where mRNAs are translationally repressed and stored in a quiescent state. DCP2 seems to be recruited for decapping and degradation of the stored mRNAs upon seed germination. These results support the notion that dynamic P-body formation modulates mRNA storage and mRNA decapping. Signaling pathways involved in translational arrest, mRNA decapping, and assembly/disassembly of P-bodies are interesting challenges for the future.
During seed germination of decapping-deficient mutants, mRNAs encoding seed storage proteins are not decapped. These mRNAs reengage with polysomes for translation, which explains why mutant seedlings still retain a seed protein repertoire 6 d after germination. The occupancy of polysomes with seed-stored mRNAs (i.e., SSP mRNAs) preempts translation of newly synthesized mRNAs, whose products are required for growth. This competition leads to the arrest of postembryonic development and consequential lethality. With respect to mRNA storage, it is conceivable that SSP mRNAs are likely equivalent to maternal mRNAs in metazoan oocytes, which are translationally suppressed and stabilized by CGH-1 (Rajyaguru and Parker, 2009). Eukaryotic P-bodies in general may play an important role in cells that are maintained in a quiescent state but can be subsequently reactivated for growth and development.
METHODS
Plant Materials and Constructs
dcp5-1 (SALK_008881) was obtained from the ABRC. Plants were grown on half-strength Murashige and Skoog (MS) medium for 18 d before being transferred to a greenhouse under similar conditions (22°C, 16/8 h photoperiod cycle). Agrobacterium tumefaciens–mediated genetic transformation of Arabidopsis thaliana was by the floral dip method (Clough and Bent, 1998).
DCP5-RNAi and DCP2-RNAi vectors were generated through a LR recombination system (Invitrogen) using binary vector pH7WGIGW2(II) (Karimi et al., 2002) with the entry clones. The entry clones were inserted with a 449-bp fragment (510th to 959th nucleotides from the first ATG) and a 550-bp fragment (14th to 564th nucleotides from the first ATG) using Topo cloning technology (Invitrogen).
Complementation of dcp5-1 was accomplished using the binary vector pBA002a, which contained a 5669-bp genomic fragment, including 2028 bp upstream of the first ATG (promoter region) and 677 bp downstream of the stop codon (3′-UTR, etc.). A DNA fragment encoding the FLAG tag (DYKDDDDK) was inserted in frame 5′ to the stop codon to express the fusion protein DCP5-FLAG.
Expression of Recombinant Proteins and Generation of Antibodies
pMBP-DCP5 was generated by the LR recombination system. GST fusion proteins were expressed using pGEX-4T1 (GE Bioscience) carrying different cDNA fragments encoding truncated DCP5 derivatives, D51 (1 to 200 amino acids), D52 (201 to 439 amino acids), and D53 (440 to 612 amino acids). All insertions and changes in plasmids were confirmed by sequencing. His-tag fusion proteins were expressed using pET28b(+) (Novagen). Anti-DCP5, anti-DCP1, and anti-DCP2 were generated in rabbits immunized with purified native proteins D53-His, DCP1-His, and DCP2-His, respectively (Cocalico Biologicals).
Coimmunoprecipitation and Pull-Down Assays
Coimmunoprecipitation experiments were performed as described (Xu et al., 2006). For immunoprecipitation of FLAG-tagged proteins, an EZview Red ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich) was used.
For in vitro pull-down assays, purified recombinant proteins (2 μg each) were preabsorbed with amylase resin for 0.5 h at room temperature. Clarified extracts were then incubated with 2 μg of each of the target proteins at room temperature for 2 h in 1 mL of binding buffer (50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 0.2% glycerol, 1% Triton X-100, and 0.5 mM β-mercaptoethanol). Amylose resin beads were added and the incubation continued under the same conditions. The beads were then washed six times with the binding buffer. Pulled-down proteins were resolved by 10% SDS-PAGE and analyzed by protein gel blots using the appropriate antibody.
Microscopy Analysis
Transient expression of fusion proteins in Nicotiana benthamiana leaves was assayed 2 d after agroinfiltration. Roots of 3-d-old transgenic Arabidopsis seedlings were incubated in 5% glycerol and imaged for subcellular localization of fusion proteins. Confocal laser scanning microscopy images were collected in the Rockefeller University Bioimaging Center using an Axiovert 200 microscope (Zeiss) with the LSM510 META laser scanning module and processed by the LSM Image Browser.
Microarray Analysis, qRT-PCR, and RACE-PCR
Microarray analysis was performed at the Rockefeller University Genomics Resources Center using GeneChip Arabidopsis ATH1 Genome Array. Total RNAs were extracted from 6-d-old wild-type and dcp5-1 using the RNeasy plant mini kit (Qiagen). The total RNA was used for qRT-PCR for quantifying candidate mRNAs. RT was performed using oligo(dT) primer and 5 μg of each total RNA after DNase treatment. cDNA was then mixed with SYBR green master mix (Applied Biosystems), following the manufacturer's instructions. qRT-PCR was performed in the Rockefeller University Genomics Resource Center using the ABI 7900HT fast real-time PCR system (Applied Biosystems). The comparative CT method was used to quantify the relative amounts of target gene transcripts. All qRT-PCR reactions were performed with an annealing temperature of 55°C and a total of 40 cycles of amplification. Primer sequences are presented in Supplemental Table 1 online. RACE-PCR was performed essentially as described (Xu et al., 2006).
Cordycepin Treatments and RNA Gel Blot Analysis
Six-day-old seedlings were incubated in Murashige and Skoog medium with cordycepin (3′-deoxyadenosine; Sigma-Aldrich) or cycloheximide (0.5 mM). Total RNA was extracted from samples harvested at various time points using TRIZOL reagent (Invitrogen). For RNA gel blot analysis, 15 μg of total RNA was fractionated on a 1.2% (w/v) agarose gel and then transferred to a Hybond-XL membrane. DNA probes were labeled with [α-32P]dCTP by a random prime labeling system (GE Healthcare).
In Vitro Translation of OLEO1 mRNA
An 893-bp Oleo1 cDNA with the T7 promoter sequence attached at its 5′ end was amplified by PCR using cDNAs derived from dcp5-1 seedlings. Primer sequences are presented in Supplemental Table 1 online. OLEO1 mRNA was synthesized by in vitro transcription using the mMESSAGE mMACHINE Kit (Ambion) and subsequently polyadenylated using the Poly(A) Tailing Kit (Ambion). Then, 0.5 pmol OLEO1 mRNA was added to each 25-μL wheat germ extract (Promega) with different concentrations of DCP proteins for in vitro translation. The FluoroTect GreenLys labeling system (Promega) and Typhoon 9400 (GE healthcare) were used for the detection of translation products and image acquisition, following the manufacturers' instructions. To test if OLEO1 mRNA levels are affected by the addition of different proteins, the OLEO1 mRNAs were trace-labeled by in vitro transcription using [α-32P]UTP. After incubation with reaction mixture, mRNAs were recovered, subjected to agarose gel electrophoresis, and visualized by autoradiography.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers At1g26110 (DCP5) and At5g45330 (DCP5-L).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Effect of DCP5 on DCP2 Decapping Activity in Vitro.
Supplemental Figure 2. P-Bodies in DCP5-RNAi and DCP2-RNAi Lines.
Supplemental Figure 3. Effect of MBP-DCP5 and MBP-DCP5-L on OLEO1 mRNA Levels in Wheat Germ Extract.
Supplemental Table 1. List of Primers and Their Uses.
Supplementary Material
Acknowledgments
We thank Ikuko Hara-Nishimura for providing the antisera to OLEO1 and OLEO2. We thank Qi-wen Niu, Jun-yi Yang, and other lab members for helpful discussions. This work was supported by National Institutes of Health Grant GM44640 to N.-H.C.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Nam-Hai Chua(chua@mail.rockefeller.edu).
Online version contains Web-only data.
References
- Albrecht, M., and Lengauer, T. (2004). Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett. 569 18–26. [DOI] [PubMed] [Google Scholar]
- Audhya, A., Hyndman, F., McLeod, I.X., Maddox, A.S., Yates III, J.R., Desai, A., and Oegema, K. (2005). A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J. Cell Biol. 171 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis, G., Saveanu, C., Fromont-Racine, M., and Jacquier, A. (2004). Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell 15 5–15. [DOI] [PubMed] [Google Scholar]
- Barbee, S.A., et al. (2006). Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman, C.A., and Parker, R. (1994). Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 269 9687–9692. [PubMed] [Google Scholar]
- Beelman, C.A., Stevens, A., Caponigro, G., LaGrandeur, T.E., Hatfield, L., Fortner, D.M., and Parker, R. (1996). An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382 642–646. [DOI] [PubMed] [Google Scholar]
- Boag, P.R., Nakamura, A., and Blackwell, T.K. (2005). A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans. Development 132 4975–4986. [DOI] [PubMed] [Google Scholar]
- Brengues, M., Teixeira, D., and Parker, R. (2005). Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310 486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan, J.R., Muhlrad, D., and Parker, R. (2008). P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Decker, C.J., Teixeira, D., and Parker, R. (2007). Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decourty, L., Saveanu, C., Zemam, K., Hantraye, F., Frachon, E., Rousselle, J.-C., Fromont-Racine, M., and Jacquier, A. (2008). Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc. Natl. Acad. Sci. USA 105 5821–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyholos, M.K., Cavaness, G.F., Hall, B., King, E., Punwani, J., Van Norman, J., and Sieburth, L.E. (2003). VARICOSE, a WD-domain protein, is required for leaf blade development. Development 130 6577–6588. [DOI] [PubMed] [Google Scholar]
- Dong, S., Li, C., Zenklusen, D., Singer, R.H., Jacobson, A., and He, F. (2007). YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol. Cell 25 559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks, T.M., and Lykke-Andersen, J. (2008). The control of mRNA decapping and P-body formation. Mol. Cell 32 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeres, D.C., Van Norman, J.M., Zhang, W., Fauver, N.A., Spencer, M.L., and Sieburth, L.E. (2007). Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell 19 1549–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield, L., Beelman, C.A., Stevens, A., and Parker, R. (1996). Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 16 5830–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki, S., Takeda, A., Motose, H., and Watanabe, Y. (2007). Characterization of Arabidopsis decapping proteins AtDCP1 and AtDCP2, which are essential for post-embryonic development. FEBS Lett. 581 2455–2459. [DOI] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kshirsagar, M., and Parker, R. (2004). Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics 166 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, S.H.M., Decker, C.J., Walsh, M.A., She, M., Parker, R., and Song, H. (2008). Crystal structure of human Edc3 and its functional implications. Mol. Cell. Biol. 28 5965–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajyaguru, P., and Parker, R. (2009). CGH-1 and the control of maternal mRNAs. Trends Cell Biol. 19 24–28. [DOI] [PubMed] [Google Scholar]
- Reijns, M.A., Alexander, R.D., Spiller, M.P., and Beggs, J.D. (2008). A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 121 2463–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D.C., and Parker, R. (1999). Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 5247–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D.C., and Parker, R. (2000). mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 20 7933–7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, U., and Parker, R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, L.T., Shimada, T., Takahashi, H., Fukao, Y., and Hara-Nishimura, I. (2008). A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J. 55 798–809. [DOI] [PubMed] [Google Scholar]
- Tanaka, K.J., Ogawa, K., Takagi, M., Imamoto, N., Matsumoto, K., and Tsujimoto, M. (2006). RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J. Biol. Chem. 281 40096–40106. [DOI] [PubMed] [Google Scholar]
- Tritschler, F., Eulalio, A., Helms, S., Schmidt, S., Coles, M., Weichenrieder, O., Izaurralde, E., and Truffault, V. (2008). Similar modes of interaction enable trailer hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol. Cell. Biol. 28 6695–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler, F., Eulalio, A., Truffault, V., Hartmann, M.D., Helms, S., Schmidt, S., Coles, M., Izaurralde, E., and Weichenrieder, O. (2007). A divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting. Mol. Cell. Biol. 27 8600–8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm, J.E., Buszczak, M., and Sayles, S. (2005). Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev. Cell 9 675–685. [DOI] [PubMed] [Google Scholar]
- Xu, J., Yang, J.Y., Niu, Q.W., and Chua, N.H. (2006). Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18 3386–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W.H., Yu, J.H., Gulick, T., Block, K.D., and Bloch, D.B. (2006). RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA 12 547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.