Abstract
The main trimeric light-harvesting complex of higher plants (LHCII) consists of three different Lhcb proteins (Lhcb1-3). We show that Arabidopsis thaliana T-DNA knockout plants lacking Lhcb3 (koLhcb3) compensate for the lack of Lhcb3 by producing increased amounts of Lhcb1 and Lhcb2. As in wild-type plants, LHCII-photosystem II (PSII) supercomplexes were present in Lhcb3 knockout plants (koLhcb3), and preservation of the LHCII trimers (M trimers) indicates that the Lhcb3 in M trimers has been replaced by Lhcb1 and/or Lhcb2. However, the rotational position of the M LHCII trimer was altered, suggesting that the Lhcb3 subunit affects the macrostructural arrangement of the LHCII antenna. The absence of Lhcb3 did not result in any significant alteration in PSII efficiency or qE type of nonphotochemical quenching, but the rate of transition from State 1 to State 2 was increased in koLhcb3, although the final extent of state transition was unchanged. The level of phosphorylation of LHCII was increased in the koLhcb3 plants compared with wild-type plants in both State 1 and State 2. The relative increase in phosphorylation upon transition from State 1 to State 2 was also significantly higher in koLhcb3. It is suggested that the main function of Lhcb3 is to modulate the rate of state transitions.
INTRODUCTION
To efficiently harvest solar energy, photosynthetic organisms use an array of light-harvesting antenna protein complexes that bind chlorophylls and carotenoids. In higher plants, these complexes are composed of a number of chlorophyll a/b binding proteins that are highly conserved through evolution; with only one exception, the same set of proteins is shared by all angiosperms and gymnosperms (Jansson, 1999; Klimmek et al., 2006). The Lhca proteins are associated with the light-harvesting complexes of photosystem I (PSI) and the Lhcb proteins are associated with those of photosystem II (PSII). The most abundant PSII-associated light-harvesting complex (LHCII) consists of homo- and heterotrimers of Lhcb1, Lhcb2, and Lhcb3 gene products. In vivo, the PSII core, composed of the D1, D2, CP47, and CP43 proteins, is a dimer (C2) that associates with two copies each of CP29 (Lhcb4), CP26 (Lhcb5), and a strongly bound LHCII trimer (S trimer) to form the C2S2 supercomplex (Boekema et al., 1995). Larger PSII supercomplexes that contain, in addition to the C2S2 unit, two copies each of CP24 (Lhcb6) and an LHCII trimer (M-trimer) bound with medium strength, represent the basic organization of PSII in Arabidopsis thaliana thylakoid membranes (Dekker and Boekema, 2005). Further LHCII trimers are loosely bound to the supercomplex (L-trimers), and some exist in LHCII-only regions (Dekker and Boekema, 2005). The C2S2M2 PSII supercomplexes are frequently found to associate in the lateral plane of the membrane into ordered semicrystalline domains.
The antenna complexes of PSII harvest light efficiently under light-limited conditions but dissipate excess energy under light-saturated conditions (Horton et al., 1996, 2008). Regulation of light harvesting occurs in three different ways. First, the amount of light-harvesting complexes is dependent upon both retrograde and anterograde communications between the nucleus and the chloroplasts (Fey et al., 2005) and the regulated turnover of the proteins (Yang et al., 1998; Zelisko et al., 2005). Second, two important regulatory mechanisms work on a rapid time scale. The ΔpH-induced and PsbS-mediated dissipation of excess light energy known as the qE type of nonphotochemical quenching (NPQ) of chlorophyll fluorescence operates on a timescale of seconds (Li et al., 2000; Niyogi et al., 2005). The molecular mechanism of qE has not been fully elucidated, although there is evidence for the involvement of conformational changes in LHCII trimers (Pascal et al., 2005; Ruban et al., 2007; Ilioaia et al., 2008) and/or the minor monomeric antenna complexes (Ahn et al., 2008).
A third regulatory mechanism operating under conditions of low light on a timescale of minutes is denoted by the term “state transitions” (Allen and Forsberg, 2001; Haldrup et al., 2001; Lunde et al., 2003). State transitions alleviate imbalances in the relative rates of excitation of the PSII and PSI and thereby optimize the rate of electron transport. The current mechanistic model for a State 1–State 2 transition is that an overreduced plastoquinone pool, a consequence of PSI being less excited than PSII, activates the Stn7 kinase that directly, or indirectly, phosphorylates LHCII (Bellafiore et al., 2005). Phosphorylation of the LHCII trimers alters their distribution in the thylakoid membrane (Kargul and Barber, 2008). Redistribution has been suggested to be caused by a modulation of the affinity of the LHCII trimers for PSI and PSII (by either molecular recognition or charge repulsion) or alternatively by modulation of the thylakoid macrostructure, resulting in an increased amount of grana margins (Danielsson et al., 2006).
The net result of the State 1–State 2 transition is a relative increase in the absorption cross section of PSI at the expense of PSII, thus correcting for the imbalance in the relative absorption cross section of each photosystem (Ruban and Johnson, 2009). Under conditions that favor PSI excitation, the process is reversed, increasing the relative absorption cross section of PSII at the expense of PSI (the State 2–State 1 transition). The reversal is caused by a higher LHCII phosphatase activity relative to the LHCII kinase activity (Allen, 2005). Mutants lacking Stn7 have no observable state transitions (Bellafiore et al., 2005), and a similar phenotype is also observed in antisense plants lacking Lhcb1 and Lhcb2 (Andersson et al., 2003), confirming that the ultimate target of the kinase cascade is the LHCII trimer (Allen, 2003).
Analysis of plants lacking the PsaH protein indicates that, in the absence of a docking site for the LHCII trimer on PSI, the association of phosphorylated LHCII with PSI is severely reduced; these plants do not show a reduction of the PSII cross section even though LHCII is phosphorylated (Lunde et al., 2000). Studies on detergent-solubilized thylakoid membranes in State 2 reveal an increase in the abundance of a specific PSI-LHCII particle and a concurrent reduction in the amount of C2S2M2 PSII supercomplexes in favor of C2S2M supercomplexes, implying the involvement of the M-trimer in state transitions (Kouril et al., 2005). In Chlamydomonas reinhardtii, state transitions provide more prominent changes in the relative antenna sizes of PSI and PSII, and the minor antenna complexes CP26 and CP29 also appear to be involved (Kargul et al., 2005; Takahashi et al., 2006; Kargul and Barber, 2008).
Unlike Lhcb1 and Lhcb2, Lhcb3 does not contain the N-terminal phosphorylation site involved in the state transitions. Lhcb3 is mainly present in a LHCII trimer with two copies of Lhcb1, but Lhcb1/Lhcb2/Lhcb3 trimers also exist, although in rather limited amounts. Again, unlike Lhcb1/Lhcb2, which accumulate strongly in low light and are depleted in high light, the level of Lhcb3 is affected less by light conditions (Bailey et al., 2001). It has been suggested that Lhcb3 is a subunit of the M-trimer of the PSII supercomplex (Boekema et al., 1999) on the basis that it is present in the C2S2M1-2 supercomplex but is absent in the C2S2 supercomplex (Hankamer et al., 1997). Additionally, Lhcb3 is frequently found in a CP29-CP24-LHCII trimer subcomplex (Peter and Thornber, 1991; Bassi and Dainese, 1992) and is absent from the peripheral loosely attached LHCII trimers (Peter and Thornber, 1991). The lower expression levels of Lhcb3 found in Arabidopsis plants lacking CP24 (koLhcb6) support this hypothesis (Kovacs et al., 2006). Indeed, the absence of CP24 is associated with a loss of the M-trimer from the PSII supercomplex in koLhcb6 plants. This disruption to the PSII macrostructure is accompanied by a reduced level of qE and an increased rate and extent of state transitions, again suggesting that the M-trimer is involved in these processes (Kovacs et al., 2006). This is an unexpected result given the absence of the phosphorylation site in Lhcb3 and the absence of Lhcb3 in PSI-LHCII preparations (Jansson et al., 1997).
In prior work, we have successfully generated and analyzed antisense and/or T-DNA Arabidopsis knockout (KO) plants targeting almost all of the individual Lhcb and Lhca proteins (Zhang et al., 1997; Andersson et al., 2001, 2003; Ganeteg et al., 2001, 2004; Klimmek et al., 2005; Kovacs et al., 2006). Here, we use the same approach to investigate the function of Lhcb3. Analysis of T-DNA KO plants of Lhcb3 has been performed to determine the function of Lhcb3, testing its role in the PSII macrostructure and regulation of light harvesting.
RESULTS
Depletion of Lhcb3 Is Accompanied by Overexpression of Lhcb1 and Lhcb2
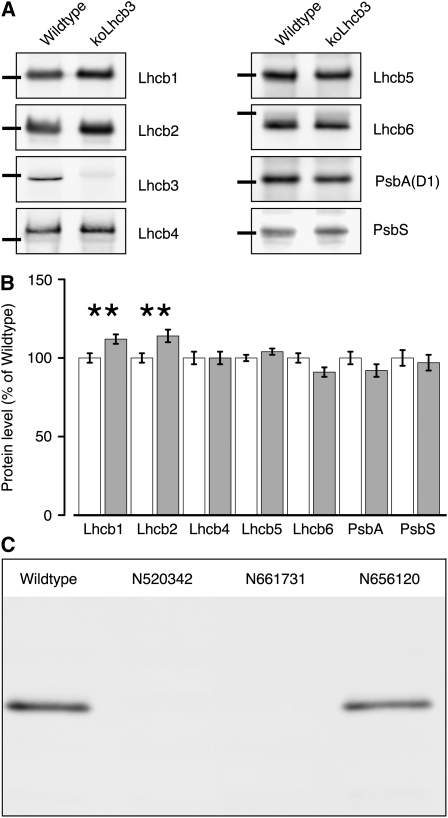
Arabidopsis plants lacking Lhcb3 were created by selfing of a SALK T-DNA KO line (N520342) and using PCR to screen for the absence of the wild-type allele of the gene coding for Lhcb3. The lack of a functional copy of the gene was confirmed by immunoblotting with an antibody specific for Lhcb3 The line, hereafter named koLhcb3, showed no visible phenotype and had a growth rate indistinguishable from the wild type. Immunoblotting with a set of monospecific antibodies against PSII antenna proteins was used to investigate if the absence of Lhcb3 induced other changes in the protein composition of PSII. As apparent from Figure 1A, the minor light-harvesting proteins (Lhcb4-6), the PSII core complex subunit (PsbA), and PsbS accumulated in the koLhcb3 plants at wild-type levels. The only observed difference was that the amounts of the major light-harvesting proteins Lhcb1 and Lhcb2 were slightly, but significantly, increased (Figure 1B). During the course of this work, two additional lines (N661731 and N656120) with a T-DNA insertion mapped to the Lhcb3 gene became available. One of these (N661731) lacked the Lhcb3 protein, whereas the other (N656120), which had the T-DNA insert in the promoter, contained wild-type levels of Lhcb3 (Figure 1C). The latter line behaved like the wild type in all observed aspects. The former line showed very similar phenotypic characteristics to the N520342. Below, we present data from the N520342 line, unless otherwise stated.
Figure 1.
Thylakoid Protein Composition of Wild-Type and koLhcb3 Plants.
(A) Immunoblotting was performed with monospecific antibodies against Lhcb1, Lhcb2, Lhcb3, Lhcb4 (CP29), Lhcb5 (CP26), Lhcb6 (CP24), PsbA, and PsbS. Lanes were loaded with 1.0 μg chlorophyll; black bars show the location of the 25-kD size marker.
(B) Quantification of immunoblot data. Error bars indicate se (n = 4), ** indicates statistically significant differences (P < 0.01) using the Student's t test.
(C) Lhcb3 protein levels in the wild type and three different putative koLhcb3 lines (N520342, N661731, and N656120).
PSII Supercomplex Structure Is Altered in Plants Lacking Lhcb3
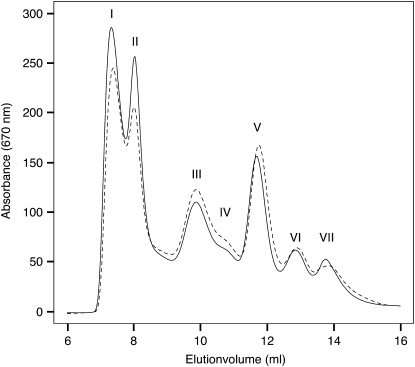
To characterize the effect of the absence of Lhcb3 on the organization and content of the pigment protein complexes, thylakoid membranes from koLhcb3 were solubilized in detergent and fractionated by fast protein liquid chromatography (FPLC) (Figure 2). There was a tendency for slightly enhanced solubilization in koLhcb3 membranes compared with the wild type, causing a relative decrease in the amount of PSII grana membrane fragments (Figure 2, peak I) and PSII supercomplexes (Figure 2, peak II) and a concurrent relative increase in the PSII core, LHCII trimer, and minor momomeric antenna peaks (Figure 2, peaks IV, V, and VI, respectively). The ratios of PSI to PSII and of LHCII trimers to PSII were unchanged in koLhcb3 compared with the wild type, indicating that PSII antenna size was unaltered, as was the stoichiometry of the two photosystems. In agreement with the similar composition of PSI, PSII, and LHCII trimers in koLhcb3 and wild-type plants, no significant differences in the pigment composition of their thylakoid membranes were detected (for example chlorophyll a/b ratio 2.50 ± 0.1 and 2.49 ± 0.1, respectively, and lutein 11.8 ± 0.1 and 11.7 ± 0.1 moles/100 moles chlorophyll, respectively).
Figure 2.
FPLC Elution Profile of Wild-Type (Solid Line) and koLhcb3 Line N520342 (Dashed Line) Stacked Thylakoid Membranes Solubilized in 0.85% α-DM.
The major peaks were identified by their absorption spectra (data not shown). (I) PSII membrane fragments (Tikkanen et al., 2006), (II) PSII supercomplexes, (III) PSI-LHCI, (IV) PSII core, (V) LHCII trimers, (VI) monomeric LHCII, and (VII) displaced pigments.
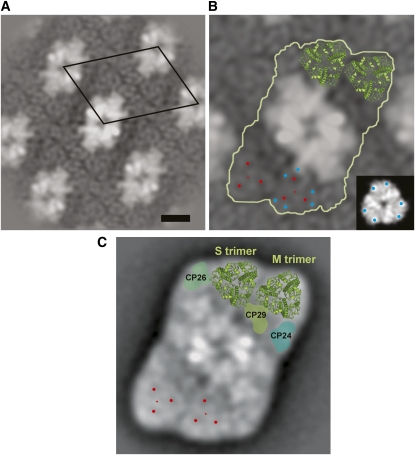
The macrostructure of PSII in koLhcb3 thylakoids was determined by electron microscopy and image analysis on the PSII grana membrane fragments derived from the FPLC fractionation (Figure 2, peak I). Crystalline areas were found in the membrane fragments from the koLhcb3 thylakoids, similar to those observed when wild-type plants are studied. However, the overall size of the crystals was somewhat larger in koLhcb3 than were those observed in the wild-type samples. Additionally, the crystals from koLhcb3 did not show mosaic packing consisting of several small crystals, as frequently observed in wild-type membranes (Figure 3A). About 2000 frames were selected from the micrographs and processed by single particle averaging. The aligned data set was statistically analyzed and classified, and the best resolved classes were combined. The final projection map is presented in Figure 3A. The repeating unit has dimensions of 26.4 × 18.8 nm with an angle of 67°, and the surface of the repeating unit is 457 nm2, similar to the surface of wild-type Arabidopsis crystals, which was previously found to be 482 nm2 (Ruban et al., 2003).
Figure 3.
Photosystem II Supercomplex Structure in Wild-Type and koLhcb3 Plants.
(A) Projection map of semicrystalline wild-type Arabidopsis PSII-LHCII membranes with the repeating unit (unit cell) indicated by the black diamond. Bar = 10 nm.
(B) koLhcb3 line N520342 crystal map showing the C2S2M2 supercomplex particle (white outline) and the position of the S- and M-trimers from the high-resolution x-ray map of LHCII is superimposed on the top trimers (green). The small red dots mark the position of the S- and M-trimers, and the larger red dots surrounding these show positions of high contrast. Blue dots indicate two recognizable densities at the periphery. Inset: A LHCII trimer with comparable densities in EM and x-ray maps indicated with blue dots.
(C) Projection map of the wild-type C2S2M2 supercomplex obtained from single particle averaging for comparison (Kouril et al., 2005). Positions within the S- and M-trimers compatible to the crystal two-dimensional map are indicated by red dots.
More recently, a single particle map of the wild type with higher resolution has been obtained, showing more details in the peripheral antenna moiety (Kouril et al., 2005). A comparison of the wild type and koLhcb3 projection maps shows that the repeating unit consists of a particle similar to the C2S2M2 supercomplex. Within the peripheral antenna, the S-trimer and M-trimer were resolved, as indicated by two sets of three stain-filled areas (marked in red, Figure 3B). The S- and M-trimers and the three minor antenna complexes are in the expected positions. However, a detailed comparison between the C2S2M2 complexes of koLhcb3 (Figure 3B) and the wild type (Figure 3C) showed that there are differences in the rotation of the M-trimer. The rotation can be determined by investigating (1) the outline of the trimer and (2) two densities at the periphery recognizable in both maps (blue dots in frame B). When the positions of the LHCII monomers (marked with red dots) were compared, the M-trimers of the koLhcb3 supercomplex were found to be rotated clockwise by ∼21°. By contrast, the S-trimers had the same location and rotation in both the wild type and the koLhcb3 mutant. We concluded that depletion of Lhcb3 led to a compensatory response in which Lhcb1 and Lhcb2 subunits replace the missing Lhcb3 subunit in the M trimer, causing the M trimer to rotate.
PSII Photosynthetic Functions and qE-type NPQ Are Unaffected in koLhcb3 Plants
The impact of the altered PSII macrostructure in koLhcb3 plants on the photosynthetic function was investigated. Table 1 shows that there were no differences in F0, Fm, Fv/Fm, qP, or ΦPSII, indicating that the function of PSII was not affected. PSII antenna connectivity (J parameter) was also unaffected. Similarly, analysis of the P700 oxidation rate indicated that there were no significant changes in the functional PSI antenna size in koLhcb3 plants compared with the wild type (Table 1). The formation and relaxation of the qE type of NPQ were investigated using two cycles of illumination. The lack of Lhcb3 did not significantly change either the kinetics or total amplitude of qE (see Supplemental Figure 1 online; Table 1).
Table 1.
Photosynthetic Parameters in Wild-Type and koLhcb3 Plants NPQ, Qp, and ΦPSII measured at 1000 μmol photons m−2 s−1 actinic light. No significant (P < 0.05) differences were detected. The standard deviation is indicated, n = 5.
| Wild Type | koLhcb3 | |
|---|---|---|
| F0 | 0.604 ± 0.015 | 0.595 ± 0.032 |
| Fm | 3.224 ± 0.219 | 3.255 ± 0.229 |
| Fv/Fm | 0.812 ± 0.010 | 0.817 ± 0.004 |
| NPQ | 2.033 ± 0.074 | 2.067 ± 0.320 |
| qP | 0.082 ± 0.006 | 0.080 ± 0.028 |
| ΦPSII | 0.047 ± 0.003 | 0.044 ± 0.014 |
| J (PSII) | 1.560 ± 0.100 | 1.480 ± 0.130 |
| P700 oxidation half-time (ms) |
2.510 ± 0.120 |
2.650 ± 0.130 |
The Rate of the State Transition Is Enhanced in koLhcb3 Plants
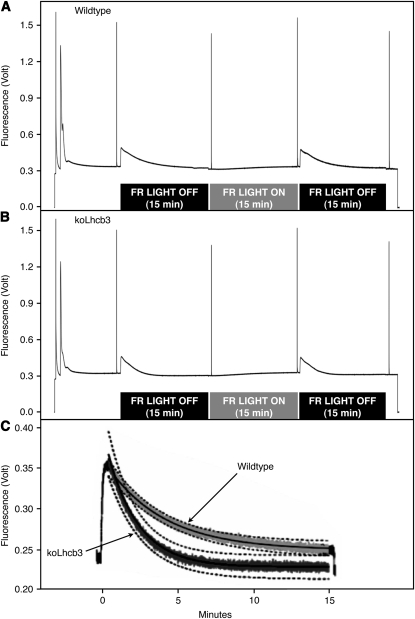
State transitions were measured in the koLhcb3 plants (Figure 4).When the fluorescence traces for the wild type (Figure 4A) and koLhcb3 line N520342 (Figure 4B) were compared (Figure 4C), it was observed that after Light 1 had been turned off, the decrease of fluorescence, indicating the transition to State 2, was considerably faster in the koLhcb3 plants. As apparent from Figure 4C, there was a discernible difference in fluorescence between the genotypes after 5 min of illumination: the koLhcb3 mutant displayed an almost complete State 1 to State 2 transition, whereas the corresponding value for the wild type was only ∼65%. Thus, although the final extent of the state transitions observed after a 15-min illumination was not significantly different between koLhcb3 and wild-type leaves, after only a 5-min light treatment, a significant difference between the genotypes was revealed (see Supplemental Table 1 online). Corresponding data for line N661731 are shown in Supplemental Figure 2 online. This difference was found when the state transitions were measured using either Fm or Fs (Table 2), although the latter (qS, a state transition parameter indicating the electron transport balance) gave a larger difference than the former (qT, reflecting photosystem II cross section changes). The average traces were fitted to a standard exponential decay function [f(x) = ((a-c)/(2^(x/b))+c)], where a is the initial level, b is the half-time of decay (t1/2), and c is the final level. The calculated t1/2 values for fluorescence decays associated with the State 1 to State 2 transitions were 149 s for wild-type leaves and 79 s for koLhcb3 leaves (Figure 4C).
Figure 4.
State Transitions in Wild-Type and koLhcb3 Plants.
(A) and (B) State transitions in the wild type (A) and koLhcb3 line N520342 (B) Arabidopsis plants. Average room temperature fluorescence traces. The black bar below the trace indicates far-red light OFF (State 2 inducing) treatment and the gray bar below the trace indicates far-red light ON treatment (State 1 inducing).
(C) State 1 to State 2 transition induced by PSII light treatment after 15 min PSI light (far red) illumination. Solid lines show exponential decay curve fits, and the dashed lines show se limits, n = 3. The top (light gray) trace shows the wild type, and the bottom (dark gray) trace shows koLhcb3.
Table 2.
State Transition Parameters in Wild-Type and koLhcb3 Plants Exposed to 5 min of State 2 Inducing Light Treatment
| Wild Type | koLhcb3 | P Value | |
|---|---|---|---|
| qS | 65% ± 2% | 91% ± 2% | 0.001*** |
| qT |
13% ± 0.07% |
16% ± 0.04% |
0.003*** |
The standard deviation is indicated, n = 3.
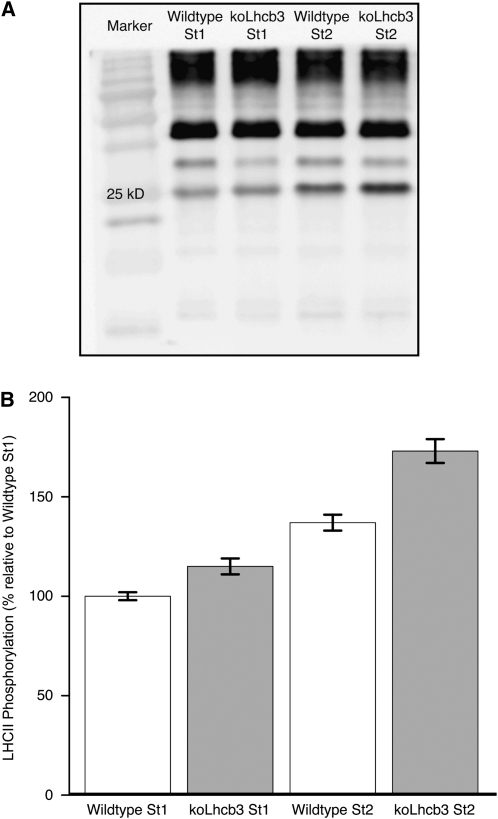
LHCII Trimers Are More Phosphorylated in Plants Lacking Lhcb3
The relative extent of LHCII phosphorylation was analyzed to test whether the increased rate of the State 1 to State 2 transition in koLhcb3 plants was accompanied by enhanced phosphorylation. The extent of phosphorylation was measured using the phosphothreonine antibody signal from the LHCII band on SDS-PAGE separated thylakoid samples (Figure 5A). Prior to thylakoid isolation, the leaf samples were exposed to 15 min of light that would induce either State 1 or State 2. Distinct differences were discovered. First, as expected, there were differences between the extent of LHCII phosphorylation in State 1 compared with State 2, in both wild-type and koLhcb3 plants. However, there was a significant difference between the relative phosphorylation of LHCII in State 1 and State 2, when the wild type and koLhcb3 (line N520342) were compared (Figure 5B): the wild type displayed a 37% increase in phosphorylation in State 2 relative to State 1, whereas the koLhcb3 plants showed a 48% increase. The koLhcb3 leaves showed 15 and 25% higher LHCII phosphorylation than the wild type in State 1 and State 2, respectively. Corresponding data for line N661731 are shown in Supplemental Figure 3 online. An analysis of variance test showed that not only were the differences between the genotypes and light treatments highly significant but also revealed a significant interaction between light treatment and genotype (see Supplemental Table 2 online). Thus, the increase in phosphorylation observed in the koLhcb3 mutant, going from State 1 to State 2, was significantly higher than the increase observed in the wild type.
Figure 5.
LHCII Phosphorylation in Wild-Type and koLhcb3 Plants.
(A) Phosphothreonine immunoblot detection of thylakoid protein phosphorylation. Lanes contain the wild type or the koLhcb3 line N520342 Arabidopsis plants in State 1 or State 2 standardized using 1.0 μg chlorophyll loaded per lane; “25 kD” shows the location of the 25 kD size marker.
(B) In vivo LHCII phosphorylation in the wild type (white bars) and koLhcb3 line N520342 (gray bars) Arabidopsis thylakoid isolated following State 1 and State 2 inducing light treatments. Results are normalized to wild-type state 1 phosphorylation, corrected relative to the CP47 phosphorylation signal. Error bars show se, n = 27.
Plants Lacking Lhcb3 Had a Seed Set Identical to That of Wild-Type Plants
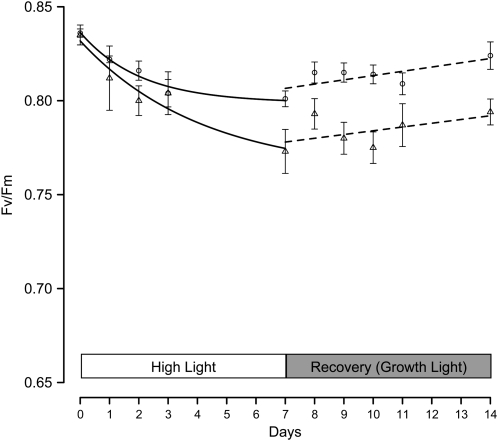
The fitness, growth, and vigor of koLhcb3 plants were investigated under natural conditions in an experimental garden. A randomized block design was used as described previously (Kulheim et al., 2002; Frenkel et al., 2008), and the growth rate and seed set were quantified and compared with wild-type plants. No altered growth phenotype was observed for the koLhcb3 plants under field conditions, and the seed set was identical to that found for the wild-type plants (see Supplemental Table 3 online). Finally, to see if koLhcb3 plants were impaired in their ability to acclimate to high light conditions, we followed changes in chlorophyll a/b ratios, Fv/Fm, qE, and state transitions during a shift from normal (150 μmol photons m−2 s−1) to high (550 μmol photons m−2 s−1) light. In koLhcb3, the increase in chlorophyll a/b ratio after 7 d in high light condition did not differ from the wild type (3.19 ± 0.02 to 3.55 ± 0.06 for the wild type and 3.15 ± 0.06 to 3.52 ± 0.03 for koLhcb3). The capacity for state transitions and qE did change after the shift, but there was no difference between the wild type and koLhcb3 in these parameters. However, the Fv/Fm ratio dropped significantly more in koLhcb3 than in wild type, and also after 7 d of recovery in normal light, koLhcb3 still had a lower Fv/Fm ratio, indicating that PSII in koLhcb3 experienced photoinhibition after transition to high light (Figure 6).
Figure 6.
Photoacclimation in Wild-Type and koLhcb3 Plants.
Fv/Fm ratios in the wild type (circles) and koLhcb3 line N520342 (triangles) of Arabidopsis plants after a shift from growth light (150 μmol photons m−2 s−1) to high light (550 μmol photons m−2 s−1) for 7 d and back to growth light to recovery. Error bars indicate se (n = 6); ratios in high light are fitted to a standard exponential decay function during recovery with a linear function.
DISCUSSION
We created plants lacking Lhcb3 to investigate the role of this protein in the organization and macrostructure of PSII and to test its involvement in the regulation of photosynthetic electron transport and light harvesting. Analysis of the levels of PSII antenna protein subunits in the mutant revealed a 15% increase in the levels of Lhcb1 and Lhcb2. Considering the amount of Lhcb3 present in wild-type plants (Dainese and Bassi, 1991; Peter and Thornber, 1991), the increase in Lhcb1 and 2 approximately corresponds to the amount needed to fully compensate for a lack of Lhcb3. The finding that the ratio of LHCII trimers to PSII was unchanged in the mutant supports the idea of a compensatory replacement of Lhcb3. The compensatory response is further evidence of the plasticity in the design of the PSII light-harvesting antenna, where the loss of one subunit causes a compensatory increase in another subunit. In the asLhcb2 antisense plants lacking Lhcb1 and 2, LHCII homotrimers of Lhcb5 and heterotrimers of Lhcb3 and Lhcb5 are formed in compensation, allowing the native PSII macrostructure to be maintained (Ruban et al., 2003, 2006). In the asLhcb2 plants, the PSII antenna size is smaller than it is in wild-type plants, indicating that Lhcb5 cannot completely substitute for Lhcb1 and 2. By contrast, in the absence of Lhcb3, the overall size of the antenna was unchanged.
Whereas the PSII macrostructure was not affected by the absence of Lhcb1 and Lhcb2 (Ruban et al., 2003), clear alterations were seen in the koLhcb3 plants. The M-trimer was rotated relative to its native position in the wild-type PSII macrostructure. The rotation suggests that the docking site of the M-trimer to the PSII supercomplex in the wild type may involve a specific surface interaction between Lhcb6 (CP24) and the Lhcb3-containing trimer. The loss of M-trimers in koLhcb6 plants (Kovacs et al., 2006) and the rotation of the M-trimer seen in koLhcb3 plants suggest that if either partner in this interaction is absent, the supercomplex structure is disturbed.
The alterations in the PSII macrostructure in koLhcb3 plants did not result in any detectable differences in photosynthetic fluorescence parameters or, indeed, in antenna connectivity. This is in contrast with the koLhcb6 mutant, in which the loss of the M-trimer both reduced connectivity and caused a rise in Fo. The maintenance of the M-trimer, albeit in a different orientation, clearly mitigates these effects in the koLhcb3 plants. No significant differences were detected either in the amplitude or kinetics of qE, again in contrast with the characteristics of the both the koLhcb6 and asLhcb2 plants. From the nearly unchanged reduction state of QA (qP), we conclude that photosynthetic electron transport also is not altered in the absence of Lhcb3. Consistent with these observations, no altered growth or fitness phenotype was observed as a result of the lack of Lhcb3. The evolutionary conservation of Lhcb3 indicates that there should be a fitness advantage associated with the presence of this protein. However, in the unshaded conditions in the experimental garden, the lack of Lhcb3 did not reveal any differences in the fitness of the mutant, although obviously this does not exclude the possibility that, under some other conditions, a growth or fitness phenotype would be revealed.
The koLhcb3 plants did possess one altered phenotype: a faster state transition. This phenotype is similar to that found in the koLhcb6 plants, and, as discussed above, both mutants possess alterations in PSII macrostructure affecting the LHCII M-trimer, which is implicated as the mobile complex involved in state transitions (Kouril et al., 2005). In the case of the koLhcb6 mutant, the faster state transition is attributed to the displacement of the M-trimer from the PSII macrostructure, enhancing its ability to migrate to PSI in State 2 (Kovacs et al., 2006). It is possible that, in the koLhcb3 mutant, the rotation of the M-trimer and its changed polypeptide composition similarly increases its ability to migrate to PSI. Indeed, an enhanced degree of detergent-induced solubilization of koLhcb3 membranes compared with the wild type suggests a somewhat looser association of the PSII macrostructure.
Alternatively, the biochemical analysis showed that the in vivo phosphorylation state of the LHCII trimers differed significantly between the wild type and koLhcb3 in State 1 and even more so in State 2. It is possible that an enhanced level of phosphorylation of LHCII trimers, caused by the increase in content of Lhcb1 and Lhcb2, increases the repulsive forces between phosphorylated LHCII and PSII, which is suggested to be the driving force for the state transition (for example, see Allen, 2003). Thus, the M-trimer would have three potential phosphorylation sites in the koLhcb3 plants but only two such sites in the wild type. This could explain how the higher level of phosphorylation in koLhcb3 plants causes an increased rate, but not extent, of state transitions. What remains to be addressed in future studies is the apparent contradiction between the involvement of the Lhcb3-containing M-trimer in state transitions and absence of evidence that Lhcb3-containing trimers attach to PSI.
Our data suggest that Lhcb3 is involved in reducing the rate of state transitions. By contrast, the first literature on the Lhcb2 protein referred to this protein as the “rapidly phosphorylated 25 kD protein,” which was the first to be involved in the lateral LHCII movements after phosphorylation in vitro. Although that work was performed on spinach (Spinacia oleracea) rather than Arabidopsis, it is an intriguing possibility that the main, or perhaps only, functional difference between the Lhcb1, Lhcb2, and Lhcb3 proteins is that, due to differences in phosphorylation kinetics, they control state transitions (i.e., Lhcb2 increases and Lhcb3 decreases their rate). Hence, the relative subunit composition of LHCII would determine the availability of phosphorylation sites, modulating the state transition rate.
If the main function of Lhcb3 is to limit the rate of state transitions, it is not surprising that no fitness reduction was recorded for the koLhcb3 plants under field conditions. Even stn7 plants that lack the LHCII kinase and therefore do not perform state transitions at all under similar conditions show only a limited fitness reduction (Frenkel et al., 2008). Clearly, the experimental garden experiment does not pose a sufficient challenge to Arabidopsis plants with respect to state transitions. However, it is unclear under what conditions slower state transitions would be beneficial. Perhaps Lhcb3 containing trimers in the M position are associated more tightly to PSII, conferring in some way an evolutionary advantage, the effect on state transitions being secondary and with no physiological importance.
Conversely, it has been speculated that there is less need for fast state transitions in land plants compared with aquatic ones (Finazzi, 2005); therefore, it may be significant that Lhcb3 appeared in evolution at about the time when plants inhabited the terrestrial environment (Alboresi et al., 2008). Although the evolution of a protein that slows down a regulatory process seems counterintuitive, the possibility that there may be an advantage associated with “dampened” state transitions in land plants cannot be ruled out. One possibility might relate to the role of phosphorylation and redox control in the long-term adjustment in the content of LHCII, the process of photoacclimation (Dietzel et al., 2008). There is evidence that prolonged exposure to Light 2 results in a reduction in level of LHCII trimers, as a result of increased turnover (Yang et al., 1998). Although the time threshold for this response has not be determined, it is logical to suggest that such effects should not be initiated by exposure to only a few minutes of change; rather, a sustained change in light conditions should be needed. Assuming the rate-limiting step in LHCII turnover is the availability of LHCII dissociated from PSII, slowing state transitions might prevent excessive LHCII turnover.
Alternatively, and oppositely, since the plastoquinone redox state is such an important signal for changing gene expression in photosynthetic acclimation (Allen, 2005; Dietzel et al., 2008), perhaps there is some advantage for light-induced changes in redox state not to be corrected too quickly by the state transitions. The finding that the Fv/Fm ratio dropped more in koLhcb3 after a shift to high light points to a role of Lhcb3 in photoacclimation, although further investigations are needed to understand the mechanistic details behind it.
METHODS
Plant Material
Arabidopsis thaliana seeds (N520342) with a T-DNA insertion in the Lhcb3 gene (AT5G54270) were obtained from the ABRC. A homozygous KO mutant line was obtained by selfing the plants and screening the progeny with PCR according to the manufacturer's protocol (Finnzymes). For amplification of the wild-type allele of Lhcb3, PCR primers with the sequences 5′-CCAGAAGATGAATTTAGGAGACTGAAGC-3′ (LP) and 5′-TTATCCGCAGACCCTGAAGCC-3′ (RP) were used, giving rise to a 900-bp fragment. For identification of the knockout allele, the primers 5′-CCAGAAGATGAATTTAGGAGACTGAAGC-3′ (LP) and 5′-TGGTTCACGTAGTGGGCCATCG-3′ (LBa1) were used, yielding a 700-bp fragment. Plant lines showing only the T-DNA knockout fragment were selfed, and the segregation of the progeny was characterized with PCR, as above. F2 generation plant lines that did not segregate were characterized by immunoblotting using an Lhcb3 antibody (Andersson et al., 2001) to detect if the T-DNA knockout mutation removed the Lhcb3 protein. Multiple lines were produced and all showed the absence of the Lhcb3 protein. One line was selected for further analysis. Two additional homozygous Lhcb3 T-DNA KO lines N661731 and N656120 were obtained from the ABRC. To obtain material for biochemical and fluorometric analysis, plants were grown for 6 to 7 weeks with an 8-h photoperiod at a light intensity of 150 μmol photons m−2 s−1 and a day/night temperature of 23/18°C. For the high-light experiments, fully grown plants were irradiated with 550 μmol photons m−2 s−1.
Pigment and Immunoblot Analysis of Thylakoid Membrane Proteins
Chlorophylls were extracted from leaf discs with 80% (v/v) acetone and assayed spectrophotometrically using extinction coefficients according to Porra et al. (1989). Immunoblot analysis of thylakoid membrane proteins was performed as described by Ganeteg et al. (2001), with slight modifications according to antibody manufacturer's instructions. Five- to six-week-old leaves were homogenized and filtered using a nylon mesh with a mesh size of 20 μm (Millipore). The filtered homogenate was pelleted and resuspended in hypotonic buffer to break the chloroplasts. The thylakoid membranes were pelleted and then resuspended in storage buffer containing 20% glycerol. All buffers contained 10 mM NaF to inhibit phosphatase activity and maintain the in vivo phosphorylation state (Bellafiore et al., 2005). All steps of the preparation were performed on ice or in a cold room (4°C) under a green safe light.
Thylakoid proteins were prepared for immunoblot analysis by addition of Laemmli denaturation buffer and incubation at 90°C for 10 min (Laemmli, 1970). The samples were normalized using chlorophyll content and then diluted to 20 μL volume before loading. One microgram of chlorophyll was loaded per lane, and the proteins were separated in a 16% denaturing SDS-PAGE gel (non-urea buffers) using the Bio-Rad Mini Protean III system. The proteins were blotted on nitrocellulose membranes (Bio-Rad; 0.40 μm), using a Bio-Rad wet blotting system with methanol-containing buffers, according to manufacturer's instructions. The nitrocellulose membranes were blocked using 5% BSA in PBS-T buffer with 0.1% Tween 20 for 1 h (Sigma-Aldrich Sweden AB) and incubated using rabbit primary antibodies against photosynthetic proteins (Jansson et al., 1996; Andersson et al., 2001; Ganeteg et al., 2001) (Agrisera) at a 1:3000 dilution for 1 h in PBS-T buffer with 0.1% Tween 20 and 2% BSA. In the case of PsbS, a chicken primary antibody was used at a 1:5000 dilution. The membranes were washed three times for 5 min in PBS-T buffer 0.05% Tween 20 and incubated in anti-rabbit donkey antibody horseradish peroxidase (HRP) conjugate (GE Healthcare Bio-Sciences) for 1 h at a 1:10,000 dilution in PBS-T buffer with 0.1% Tween 20 and 2% BSA. The PsbS chicken antibody was detected using an anti-chicken donkey antibody HRP conjugate (Pierce). For immunodetection of Lhcb3, a new antibody was developed. A peptide was synthesized with the sequence RINGLDGVGEGND, derived from amino acids 132 to 144 of the mature Arabidopsis Lhcb3 protein, at the end of the second helix and the loop connecting helix 2 and 3. It was conjugated to BSA and injected into rabbits. This antibody was detected with the same specificity as one used previously (Andersson et al., 2001), but the signal strength was considerably higher. Immunoblotted membranes were incubated for 2 min in ECL plus HRP substrate (GE Healthcare Bio-Sciences), and chemiluminescence was then detected using a LAS-3000 cooled CCD camera. Optimal exposure ranged from 5 to 10 min, and image quantification for each antibody used identical exposure time. Images were recorded using the ImageReader software with 1 min incremental recording and standard CCD sensitivity (Fujifilm Medical Systems). Image processing and quantification were performed in the Multi Gauge application (Fujifilm Medical Systems). Profile lane quantification was used with automatic background subtraction and band detection. Standard parameters for peak detection according to the manufacturer's instructions were used. The linearity of the antibody signals for Lhcb1 and Lhcb2 in the concentration range used is shown in Supplemental Figures 4 and 5 online, respectively. The samples for Lhcb and PsbS detection consisted of 15 biological replicates per genotype and, for LHCII phosphorylation detection, four biological replicates for each state and genotype. Statistical analyses using the Student's t test and data normalcy tests were performed using the R application package (Ihaka and Gentleman, 1996).
Analysis of Pigment-Protein Complexes
For FPLC analysis, stacked thylakoid membranes were diluted to a final chlorophyll concentration of 1.0 mg/mL and solubilized by the addition of n-dodecyl-α-d-maltoside to a final concentration of 0.85%, as described by Ruban et al. (2006). The samples were vortexed thoroughly for 1 min, left to stand on ice for 10 min, and then centrifuged for 1 min at 16,000g. The supernatant was filtered through a 0.45-μm nylon filter and subjected to gel filtration chromatography using a Superdex 200 HR 10/30 gel filtration column in a GE Healthcare AKTA purifier system. The flow rate was 0.4 mL min−1 with a running buffer containing (20 mM Bis-Tris, 0.03% n-dodecyl-α-d-maltoside, pH 6.5), and 0.5-mL fractions were collected.
Electron Microscopy
Samples were negatively stained with 2% uranyl acetate on glow-discharged carbon-coated copper grids. Electron microscopy was performed with a Philips CM120 electron microscope equipped with LaB6 filament operating at 120 kV. Images were recorded with a Gatan 4000 SP 4K slow-scan CCD camera at 80,000× magnification at a pixel size (after binning the images) of 0.375 nm at the specimen level, using GRACE software for semiautomated specimen selection and data acquisition (Oostergetel et al., 1998). Crystalline domains were analyzed by single particle analysis using GRIP software (http://www.rug.nl/gbb/research/researchgroups/electronmicroscopy/manuals/index). They were divided into frames of 72 × 72 nm, aligned by with multireference and nonreference alignment procedures, treated with multivariate statistical analysis, and classified as previously described (Boekema et al., 1999).
Room Temperature Fluorescence Measurements
Chlorophyll fluorescence was measured with a Dual PAM 100 chlorophyll fluorescence photosynthesis analyzer (Heinz Walz). The plants were adapted in the dark for 30 min prior to measurement. Actinic illumination was provided by arrays of 635-nm LEDs illuminating both the adaxial and abaxial surfaces of the leaf. F0 (the fluorescence level with PSII reaction centers open) was measured in the presence of a 10 μmol photons m−2 s−1 measuring beam. The maximum fluorescence levels in the dark-adapted state (Fm), during the course of actinic illumination (Fm’) and in the subsequent dark relaxation periods (Fm’’) were determined using a 0.8-s saturating light pulse (4000 μmol photons m−2 s−1). The fluorescence parameters Fv/Fm, qP, and ΦPSII were calculated using the DualPAM software. The reversible component of NPQ (relaxing 5 min after actinic illumination) was assigned to energy-dependent NPQ (qE) and calculated as (Fm/Fm’) – (Fm/Fm’’).
Estimation of the Functional Antenna Sizes of PSII and PSI
The kinetics of chlorophyll fluorescence induction and P700 oxidation were recorded simultaneously using a Dual PAM100 chlorophyll fluorescence photosynthesis analyzer (Heinz Walz) with the instrument operated in the fast kinetics mode. Detached, dark-adapted leaves were vacuum infiltrated with 100 μM DCMU and 50 μM methyl viologen before measurement. Actinic illumination (1000 μmol photons m−2 s−1) was provided by two arrays of 635-nm LEDs illuminating both the adaxial and abaxial surfaces of the leaf, applied 1 ms after turning on the measuring beam. The P700 oxidation traces were fitted with first-order kinetics, and the rate of oxidation was determined. Under these experimental conditions, the rate of P700 oxidation is proportional with the antenna size (Ghirardi and Melis, 1984). For fluorescence induction, the data obtained during the 20-ms induction curves (2000 points) were numerically fitted by the function F(t, I, σPSII, J) based on a sigmoidal fluorescence induction model (Koblizek et al., 2001), where J is the PSII connectivity parameter that determines the shape of the induction curve.
Measurement of the State Transitions
State transition measurement and calculations were performed according to Haldrup et al. (2001). Preferential PSII excitation was provided by illumination with red light with an intensity of 8 μmol photons m−2 s−1 provided by a KL1500 lamp equipped with a 650-nm interference filter (Light 2, red light). Excitation of PSI was achieved using far-red light with an intensity of 4 μmol photons m−2 s−1 provided by a KL1500 lamp equipped with 720-nm interference filter (Light 1, far-red light, ø = 20 nm). PSII fluorescence was recorded using the PAM-100/PDA-100 setup described previously (Kovacs et al., 2006). Light intensities were measured using a LI-COR LI-250 light meter equipped with a Quantum sensor (LI-COR). Attached leaves were dark adapted for 30 min and first preilluminated with both Light 1 and Light 2 for 5 min. The leaves were then treated with Light 2 only for 5 or 15 min to induce a State 1 to State 2 transition. This treatment was followed by a 15-min illumination with both Light 1 and Light 2 to recover State 1. After each light treatment, an 800-ms saturating light pulse (4000 μmol photons m−2 s−1) was applied to determine the maximum fluorescence level (Fm). The extent of the state transition was calculated, using both the steady state fluorescence (Fs) and Fm, according to Lunde et al. (2000). The state transition parameter indicating the electron transport balance (qS) was calculated as [(Fs1' − Fs1) − (Fs2' − Fs2)]/(Fs1' − Fs1), where Fs1 and Fs2 were the steady state fluorescence levels in the presence of Light 1 in State 1 and State 2, respectively, while Fs1' and Fs2' designate fluorescence in the absence of Light 1 in State 1 and State 2, respectively. The state transition parameter reflecting the PSII cross section changes (qT) was calculated as (Fm1 − Fm2)/Fm1, where Fm1 and Fm2 designate the maximal fluorescence yield in State 1 and State 2, respectively.
LHCII Phosphorylation Analysis
Two separate filtered light sources were used to induce either State 1 or State 2: Light 1 was 4 μmol photons m−2 s−1 filtered through a 720-nm interference filter (20-nm wide), and Light 2 was 8 μmol photons m−2 s−1 filtered through a 650-nm interference filter (20-nm wide). Light intensity was measured at the leaf surface, and the cone of light evenly illuminated the entire leaf rosette. State 2 was induced by treating the dark-adapted plants with both Light 1 and Light 2 for 10 min, followed by 15 min of Light 2. State 1 was induced by treating the dark-adapted plants with both Light 1 and Light 2 for 10 min, followed by 15 min of Light 2, and finally with 15 min of treatment with both Light 1 and Light 2. At the end of each treatment, the leaves of the plant were homogenized and thylakoids were isolated immediately, as described above, using buffers containing 10 mM NaF to preserve the in vivo phosphorylation state of the proteins. The phosphorylation level of the thylakoid proteins was quantified using a P-thr rabbit antibody (Cell Signaling Technology). Detection was done using anti-rabbit donkey antibody HRP conjugate and ECL Plus HRP substrate as described above (GE Healthcare Bio-Sciences). The relative LHCII trimer phosphorylation was calculated after normalization to the intensity of the CP47 phosphorylation signal (CP47 phosphorylation is unaltered by state transition and lack of Lhcb3).
Fitness Analysis
The reproductive fitness of the plants was determined by estimating seed production as described by Kulheim et al. (2002) and Frenkel et al. (2008) The plants were grown in an experimental garden in randomized block design as described previously (Frenkel et al., 2008). At maturity, seed production was estimated by counting the number of siliques on all plants and estimating the number of seeds per siliques by counting five siliques per plant. The average number of seeds per plant was then estimated (Frenkel et al., 2008).
Accession Numbers
The Lhcb3 gene has Arabidopsis Genome Initiative locus identifier AT5G54270. The KO lines mentioned in the article can be obtained from the ABRC under the stock numbers N520342, N661731, and N656120.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. NPQ Induction and Dark Relaxation in Wild-Type and koLhcb3 Arabidopsis Plants.
Supplemental Figure 2. State 1 to State 2 Transition Induced in Wild-Type and koLhcb3 Lines N520342 and N661731 by PSII Light Treatment after 15 min PSI Light (Far-Red) Illumination.
Supplemental Figure 3. In Vivo LHCII Phosphorylation in Wild-Type and koLhcb3 Lines N520342 and N661731 Following State 1 and State 2 Inducing Light Treatments
Supplemental Figure 4. Lhcb1 Antibody Linear Range.
Supplemental Figure 5. Lhcb2 Antibody Linear Range.
Supplemental Table 1. ANOVA Analysis of the Significance of the Interaction between Genotype (Wild Type or koLhcb3) and Treatment (State 1 or State 2 Light) with Regard to PSII Antennae Size.
Supplemental Table 2. ANOVA Analysis of the Significance of the Interaction between Genotype (Wild Type or koLhcb3) and Treatment (State 1 or State 2 Light) with Regard to LHCII Phosphorylation.
Supplemental Table 3. Seed Set of Wild-Type and koLhcb3 Arabidopsis Plants in the Field.
Supplementary Material
Acknowledgments
This work was supported by EC framework 6 Marie Curie Research Training Network “Interdisciplinary Network for Training and Research on Photosystem 2 (INTRO2)” Project MRTNCT-2003-505069 and by grants from the Swedish Research Council and the Swedish Research Council for Environment, Agricultural Sciences, and Spatial Planning, the Biotechnology and Biological Sciences Research Council of the United Kingdom (BB/E009743/1), and the University of London Central Research Fund (CRFT1G3R).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Stefan Jansson (stefan.jansson@plantphys.umu.se).
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Ahn, T.K., Avenson, T.J., Ballottari, M., Cheng, Y.-C., Niyogi, K.K., Bassi, R., and Fleming, G.R. (2008). Architecture of a charge-transfer state regulating light harvesting in a plant antenna protein. Science 320 794–797. [DOI] [PubMed] [Google Scholar]
- Alboresi, A., Caffarri, S., Nogue, F., Bassi, R., and Morosinotto, T. (2008). In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: Identification of subunits which evolved upon land adaptation. PLoS One 3 e2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, J.F. (2003). State transitions - A question of balance. Science 299 1530–1532. [DOI] [PubMed] [Google Scholar]
- Allen, J.F. (2005). Photosynthesis: The processing of redox signals in chloroplasts. Curr. Biol. 15 929–932. [DOI] [PubMed] [Google Scholar]
- Allen, J.F., and Forsberg, J. (2001). Molecular recognition in thylakoid structure and function. Trends Plant Sci. 6 317–326. [DOI] [PubMed] [Google Scholar]
- Andersson, J., Walters, R.G., Horton, P., and Jansson, S. (2001). Antisense inhibition of the photosynthetic antenna proteins CP29 and CP26: Implications for the mechanism of protective energy dissipation. Plant Cell 13 1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, J., Wentworth, M., Walters, R.G., Howard, C.A., Ruban, A.V., Horton, P., and Jansson, S. (2003). Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II - effects on photosynthesis, grana stacking and fitness. Plant J. 35 350–361. [DOI] [PubMed] [Google Scholar]
- Bailey, S., Walters, R.G., Jansson, S., and Horton, P. (2001). Acclimation of Arabidopsis thaliana to the light environment: The existence of separate low light and high light responses. Planta 213 794–801. [DOI] [PubMed] [Google Scholar]
- Bassi, R., and Dainese, P. (1992). A supramolecular light-harvesting complex from chloroplast photosystem-II membranes. Eur. J. Biochem. 204 317–326. [DOI] [PubMed] [Google Scholar]
- Bellafiore, S., Barneche, F., Peltier, G., and Rochaix, J.D. (2005). State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433 892–895. [DOI] [PubMed] [Google Scholar]
- Boekema, E.J., Hankamer, B., Bald, D., Kruip, J., Nield, J., Boonstra, A.F., Barber, J., and Rogner, M. (1995). Supramolecular structure of the photosystem II complex from green plants and cyanobacteria. Proc. Natl. Acad. Sci. USA 92 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekema, E.J., van Roon, H., Calkoen, F., Bassi, R., and Dekker, J.P. (1999). Multiple types of association of photosystem II and its light-harvesting antenna in partially solubilized photosystem II membranes. Biochemistry 38 2233–2239. [DOI] [PubMed] [Google Scholar]
- Dainese, P., and Bassi, R. (1991). Subunit stoichiometry of the chloroplast photosystem II antenna system and aggregation state of the component chlorophyll a/b binding proteins. J. Biol. Chem. 266 8136–8142. [PubMed] [Google Scholar]
- Danielsson, R., Suorsa, M., Paakkarinen, V., Albertsson, P.-A., Styring, S., Aro, E.-M., and Mamedov, F. (2006). Dimeric and monomeric organization of photosystem II: Distribution of five distinct complexes in the different domains of the thylakoid membrane. J. Biol. Chem. 281 14241–14249. [DOI] [PubMed] [Google Scholar]
- Dekker, J.P., and Boekema, E.J. (2005). Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706 12–39. [DOI] [PubMed] [Google Scholar]
- Dietzel, L., Bräutigam, K., and Pfannschmidt, T. (2008). Photosynthetic acclimation: State transitions and adjustment of photosystem stoichiometry – functional relationships between short-term and long-term light quality acclimation in plants. FEBS J. 275 1080–1088. [DOI] [PubMed] [Google Scholar]
- Fey, V., Wagner, R., Bräutigam, K., and Pfannschmidt, T. (2005). Photosynthetic redox control of nuclear gene expression. J. Exp. Bot. 56 1491–1498. [DOI] [PubMed] [Google Scholar]
- Finazzi, G. (2005). The central role of the green alga Chlamydomonas reinhardtii in revealing the mechanism of state transitions. J. Exp. Bot. 56 383–388. [DOI] [PubMed] [Google Scholar]
- Frenkel, M., Johansson Jankanpaa, H., Moen, J., and Jansson, S. (2008). An illustrated gardener's guide to transgenic Arabidopsis field experiments. New Phytol. 180 545–555. [DOI] [PubMed] [Google Scholar]
- Ganeteg, U., Klimmek, F., and Jansson, S. (2004). Lhca5 – An LHC-type protein associated with photosystem I. Plant Mol. Biol. 54 641–651. [DOI] [PubMed] [Google Scholar]
- Ganeteg, U., Strand, A., Gustafsson, P., and Jansson, S. (2001). The properties of the chlorophyll a/b-binding proteins Lhca2 and Lhca3 studied in vivo using antisense inhibition. Plant Physiol. 127 150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardi, M.L., and Melis, A. (1984). Photosystem electron-transport capacity and light-harvesting antenna size in maize chloroplasts. Plant Physiol. 74 993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldrup, A., Jensen, P.E., Lunde, C., and Scheller, H.V. (2001). Balance of power: A view of the mechanism of photosynthetic state transitions. Trends Plant Sci. 6 301–305. [DOI] [PubMed] [Google Scholar]
- Hankamer, B., Nield, J., Zheleva, D., Boekema, E., Jansson, S., and Barber, J. (1997). Isolation and biochemical characterisation of monomeric and dimeric photosystem II complexes from spinach and their relevance to the organisation of photosystem II in vivo. Eur. J. Biochem. 243 422–429. [DOI] [PubMed] [Google Scholar]
- Horton, P., Johnson, M.P., Perez-Bueno, M.L., Kiss, A.Z., and Ruban, A.V. (2008). Photosynthetic acclimation: Does the dynamic structure and macro-organisation of photosystem II in higher plant grana membranes regulate light harvesting states? FEBS J. 275 1069–1079. [DOI] [PubMed] [Google Scholar]
- Horton, P., Ruban, A.V., and Walters, R.G. (1996). Regulation of light harvesting in green plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 655–684. [DOI] [PubMed] [Google Scholar]
- Ihaka, R., and Gentleman, R. (1996). A language for data analysis and graphics. J. Comput. Graph. Statist. 5 299–314. [Google Scholar]
- Ilioaia, C., Johnson, M.P., Horton, P., and Ruban, A.V. (2008). Induction of efficient energy dissipation in the isolated light-harvesting complex of photosystem II in the absence of protein aggregation. J. Biol. Chem. 283 29505–29512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson, S. (1999). A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 4 236–240. [DOI] [PubMed] [Google Scholar]
- Jansson, S., Andersen, B., and Scheller, H.V. (1996). Nearest-neighbor analysis of higher-plant photosystem I holocomplex. Plant Physiol. 112 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson, S., Stefansson, H., Nystrom, U., Gustafsson, P., and Albertsson, P.-A. (1997). Antenna protein composition of PS I and PS II in thylakoid sub-domains. Biochimi. Biophys. Acta 1320 297–309. [Google Scholar]
- Kargul, J., and Barber, J.T. (2008). Photosynthetic acclimation: Structural reorganisation of light harvesting antenna – role of redox-dependent phosphorylation of major and minor chlorophyll a/b binding proteins. FEBS J. 275 1056–1068. [DOI] [PubMed] [Google Scholar]
- Kargul, J., Turkina, M.V., Nield, J., Benson, S., Vener, A.V., and Barber, J. (2005). Light-harvesting complex II protein CP29 binds to photosystem I of Chlamydomonas reinhardtii under State 2 conditions. FEBS J. 272 4797–4806. [DOI] [PubMed] [Google Scholar]
- Klimmek, F., Ganeteg, U., Ihalainen, J.A., van Roon, H., Jensen, P.E., Scheller, H.V., Dekker, J.P., and Jansson, S. (2005). Structure of the higher plant light harvesting complex I: in vivo characterization and structural interdependence of the Lhca proteins. Biochemistry 44 3065–3073. [DOI] [PubMed] [Google Scholar]
- Klimmek, F., Sjodin, A., Noutsos, C., Leister, D., and Jansson, S. (2006). Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol. 140 793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblizek, M., Kaftan, D., and Nedbal, L. (2001). On the relationship between the non-photochemical quenching of the chlorophyll fluorescence and the photosystem II light harvesting efficiency. A repetitive flash fluorescence induction study. Photosynth. Res. 68 141–152. [DOI] [PubMed] [Google Scholar]
- Kouril, R., Zygadlo, A., Arteni, A.A., de Wit, C.D., Dekker, J.P., Jensen, P.E., Scheller, H.V., and Boekema, E.J. (2005). Structural characterization of a complex of photosystem I and light-harvesting complex II of Arabidopsis thaliana. Biochemistry 44 10935–10940. [DOI] [PubMed] [Google Scholar]
- Kovacs, L., Damkjaer, J., Kereiche, S., Ilioaia, C., Ruban, A.V., Boekema, E.J., Jansson, S., and Horton, P. (2006). Lack of the light-harvesting complex CP24 affects the structure and function of the grana membranes of higher plant chloroplasts. Plant Cell 18 3106–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulheim, C., Agren, J., and Jansson, S. (2002). Rapid regulation of light harvesting and plant fitness in the field. Science 297 91–93. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Li, X.P., Bjorkman, O., Shih, C., Grossman, A.R., Rosenquist, M., Jansson, S., and Niyogi, K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403 391–395. [DOI] [PubMed] [Google Scholar]
- Lunde, C., Jensen, P.E., Haldrup, A., Knoetzel, J., and Scheller, H.V. (2000). The PSI-H subunit of photosystem I is essential for state transitions in plant photosynthesis. Nature 408 613–615. [DOI] [PubMed] [Google Scholar]
- Lunde, C., Jensen, P.E., Rosgaard, L., Haldrup, A., Gilpin, M.J., and Scheller, H.V. (2003). Plants impaired in state transitions can to a large degree compensate for their defect. Plant Cell Physiol. 44 44–54. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K., Li, X.P., Rosenberg, V., and Jung, H.S. (2005). Is PsbS the site of non-photochemical quenching in photosynthesis? J. Exp. Bot. 56 375–382. [DOI] [PubMed] [Google Scholar]
- Oostergetel, G.T., Keegstra, W., and Brisson, A. (1998). Automation of specimen selection and data acquisition for protein electron crystallography. Ultramicroscopy 74 47–59. [Google Scholar]
- Pascal, A.A., Liu, Z., Broess, K., van Oort, B., van Amerongen, H., Wang, C., Horton, P., Robert, B., Chang, W., and Ruban, A. (2005). Molecular basis of photoprotection and control of photosynthetic light-harvesting. Nature 436 134–137. [DOI] [PubMed] [Google Scholar]
- Peter, G.F., and Thornber, J.P. (1991). Biochemical composition and organization of higher plant photosystem II light-harvesting pigment-proteins. J. Biol. Chem. 266 16745–16754. [PubMed] [Google Scholar]
- Porra, R.J., Thompson, W.A., and Kriedmann, P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochem. Biophys. Acta 975 384–394. [Google Scholar]
- Ruban, A.V., Berera, R., Ilioaia, C., van Stokkum, I.H., Kennis, J.T., Pascal, A.A., van Amerongen, H., Robert, B., Horton, P., and van Grondelle, R. (2007). Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 450 575–578. [DOI] [PubMed] [Google Scholar]
- Ruban, A.V., and Johnson, M.P. (2009). Dynamics of higher plant photosystem cross-section associated with state transitions. Photosynth. Res. 99 173–183. [DOI] [PubMed] [Google Scholar]
- Ruban, A.V., Solovieva, S., Lee, P.J., Ilioaia, C., Wentworth, M., Ganeteg, U., Klimmek, F., Chow, W.S., Anderson, J.M., Jansson, S., and Horton, P. (2006). Plasticity in the composition of the light harvesting antenna of higher plants preserves structural integrity and biological function. J. Biol. Chem. 281 14981–14990. [DOI] [PubMed] [Google Scholar]
- Ruban, A.V., Wentworth, M., Yakushevska, A.E., Andersson, J., Lee, P.J., Keegstra, W., Dekker, J.P., Boekema, E.J., Jansson, S., and Horton, P. (2003). Plants lacking the main light-harvesting complex retain photosystem II macro-organization. Nature 421 648–652. [DOI] [PubMed] [Google Scholar]
- Takahashi, H., Iwai, M., Takahashi, Y., and Minagawa, J. (2006). Identification of the mobile light-harvesting complex II polypeptides for state transitions in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 103 477–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen, M., et al. (2006). State transitions revisited-a buffering system for dynamic low light acclimation of Arabidopsis. Plant Mol. Biol. 62 779–793. [DOI] [PubMed] [Google Scholar]
- Yang, D.H., Webster, J., Adam, Z., Lindahl, M., and Andersson, B. (1998). Induction of acclimative proteolysis of the light-harvesting chlorophyll a/b protein of photosystem II in response to elevated light intensities. Plant Physiol. 118 827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelisko, A., Garcia-Lorenzo, M., Jackowski, G., Jansson, S., and Funk, C. (2005). AtFtsH6 is involved in the degradation of the light-harvesting complex II during high-light acclimation and senescence. Proc. Natl. Acad. Sci. USA 102 13699–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Goodman, H.M., and Jansson, S. (1997). Antisense inhibition of the photosystem I antenna protein Lhca4 in Arabidopsis thaliana. Plant Physiol. 115 1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.