Abstract
Aluminum (Al) toxicity is the major limiting factor of crop production on acid soils, but some plant species have evolved ways of detoxifying Al. Here, we report a C2H2-type zinc finger transcription factor ART1 (for Al resistance transcription factor 1), which specifically regulates the expression of genes related to Al tolerance in rice (Oryza sativa). ART1 is constitutively expressed in the root, and the expression level is not affected by Al treatment. ART1 is localized in the nucleus of all root cells. A yeast one-hybrid assay showed that ART1 has a transcriptional activation potential and interacts with the promoter region of STAR1, an important factor in rice Al tolerance. Microarray analysis revealed 31 downstream transcripts regulated by ART1, including STAR1 and 2 and a couple of homologs of Al tolerance genes in other plants. Some of these genes were implicated in both internal and external detoxification of Al at different cellular levels. Our findings shed light on comprehensively understanding how plants detoxify aluminum to survive in an acidic environment.
INTRODUCTION
Ionic aluminum (mainly Al3+) inhibits root elongation rapidly at low concentrations (Kochian et al., 2004; Ma, 2007; Poschenrieder et al., 2008). Subsequent inhibition of water and nutrient uptake results in reduced crop production and increased susceptibility to environmental stresses on acid soils, where Al toxicity is the major limiting factor for crop production (von Uexkull and Mutert, 1995). Approximately 55, 39, and 37% of the soil in tropical America, tropical Africa, and tropical Asia, respectively, are acidic, the total area being 1.6 billion hectares (Sanchez and Salinas, 1981). Therefore, enhancing Al tolerance of crops has been considered a key to increasing crop productivity on acidic problem soils, which would subsequently help solve the problem of food shortage and biofuel production. Some plants have evolved strategies to detoxify Al. Elucidation of these strategies will help us generate crops with increased Al tolerance.
Some Al-tolerant plant species or cultivars are able to detoxify Al both internally and externally. Internal detoxification in Al-accumulating plants is achieved by sequestration of Al into the vacuoles and chelation with organic acids such as citrate and oxalate (Ma, 2007). The most well-documented mechanism for external detoxification is the secretion of organic acid anions, such as oxalate, citrate, and/or malate, from the roots in response to Al. These organic acid anions chelate toxic Al and thereby detoxify Al in the rhizosphere (Ryan et al., 2001; Kochian et al., 2004; Ma, 2007; Poschenrieder et al., 2008). Genes responsible for Al-induced secretion of malate (ALMT1) have been identified in wheat (Triticum aestivum), Arabidopsis thaliana, and rape (Brassica napus; Sasaki et al., 2004; Hoekenga et al., 2006; Ligaba et al., 2006). Recently, the genes, involved in Al-induced secretion of citrate have also been identified in barley (Hordeum vulgare), sorghum (Sorghum bicolor), and Arabidopsis (Furukawa et al., 2007; Magalhaes et al., 2007; Liu et al., 2009). All these genes encode a citrate efflux transporter that belongs to the multidrug and toxic compound extrusion (MATE) family.
Japonica cultivars of rice (Oryza sativa) show the most tolerance to Al among the small-grain cereal crops (Foy, 1988). Recently, two genes (STAR1 and 2) required for Al tolerance in rice have been cloned (Huang et al., 2009). STAR1 and 2 encode ATP binding and transmembrane domains of a novel ABC transporter, respectively. The complex between STAR1 and 2 transports UDP-glucose, which is used for modification of the cell wall although the exact mechanism remains unknown. Here, we report a gene (Al resistance transcription factor 1 [ART1]) that encodes a transcription factor that regulates 31 genes implicated in Al tolerance, including STAR1 and 2 in rice.
RESULTS
Isolation and Phenotypic Analysis of the art1 Mutant
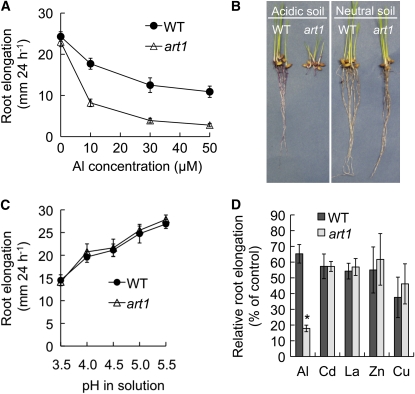
A mutant sensitive to Al rhizotoxicity (art1) was isolated by screening M3 lines derived from an Al-tolerant cultivar of rice (Koshihikari) irradiated with γ-rays according to the procedures described previously (Ma et al., 2005). There is no difference in the root and shoot morphology between the art1 and its wild type. In the absence of Al, the mutant showed root growth similar to that of the wild type (Figure 1A). However, in the presence of Al, the root elongation of art1 was inhibited significantly more than in the wild type. At 10, 30, and 50 μM Al, root elongation was inhibited by 64, 83, and 88%, respectively, in art1 and was inhibited by 27, 49, and 68%, respectively, in the wild type (Figure 1A). In neutral soil, both lines grew similarly (Figure 1B), while in acid soil, the root growth of art1 was completely inhibited. The wild type and art1 were equally sensitive to a low pH and to other metals, including Cd, La, Zn, and Cu (Figures 1C and 1D). In addition, when grown in a field at a pH of 6.5 (without Al toxicity stress, but with other natural biotic and abiotic stresses), the plant growth and grain yield did not differ significantly between the wild type and art1 (see Supplemental Figure 1 online). All these results indicate that art1 is a mutant specifically sensitive to Al.
Figure 1.
Phenotype of the art1 Mutant.
(A) Response to Al. Five-day-old seedlings of both the wild-type rice and art1 were exposed to a 0.5 mM CaCl2 solution containing 0, 10, 30, or 50 μM AlCl3, pH 4.5, for 24 h. Data are means ± sd (n = 10).
(B) Growth on acid soil. Germinated seeds were sowed on acidic soil, pH 4.5, or neutral soil, pH 6.5, and grown for 6 d.
(C) Response to different pHs. Seedlings were exposed to a buffered solution at different pHs for 24 h. Data are means ± sd (n = 10).
(D) Effect of toxic metals on root elongation. Five-day-old seedlings were exposed to a 0.5 mM CaCl2 solution, pH 4.5, containing 0, 30 μM Al, 20 μM Cd, 5 μM La, 100 μM Zn, or 0.5 μM Cu in their chloride form for 24 h. Root elongation was measured before and after the treatment and relative root elongation, (root elongation with metals)/(root elongation without metals) × 100. Data are means ± sd (n = 8 to 10). The asterisk shows a significant difference between the wild type and art1 (P < 0.05 by Student's t test).
Map-Based Cloning of ART1
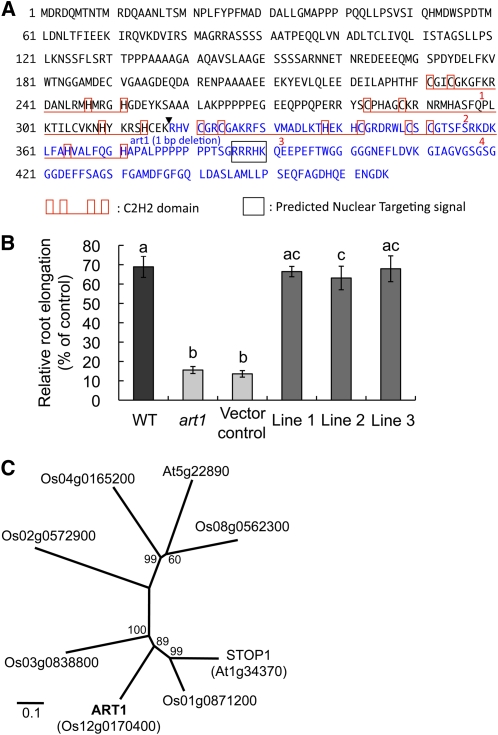
Genetic analysis with 164 F2 seedlings derived from a cross between the mutant and the wild type showed that plants tolerant and sensitive to Al segregated at a ratio of 3:1, indicating that a single recessive gene is responsible for the sensitivity of Al in the art1 mutant (see Supplemental Figure 2 online). To map the ART1 gene, we constructed an F2 population by crossing art1 with Kasalath, an indica cultivar. Bulked segregant analysis with 59 polymorphic InDel markers covering the whole rice genome showed that the C62896 marker on chromosome 12 was linked to the ART1 gene (see Supplemental Figure 3A online). Cosegregation analyses using 46 Al-sensitive F2 plants indicated that ART1 was located between MaOs1219 and MaOs1229 on the short arm of chromosome 12, with a distance of 1.1 and 4.5 centimorgans, respectively (see Supplemental Figure 3A online). By fine mapping with a large F2 population (986 plants), we located ART1 between MaOs1250 and MaOs1246, which covered two overlapping BAC clones (see Supplemental Figure 3B online). The candidate region between these two markers covers 38.5 kb and contains six candidate genes based on the prediction from the RAP-DB website. Sequencing the open reading frames (ORFs) of these genes in the mutant revealed that a putative gene Os12g0170400 has a 1-bp deletion at 953 bp from the ATG start codon in its coding sequence, suggesting that this gene is ART1 (see Supplemental Figure 3C online; Figure 2A).
Figure 2.
Sequence and Complementation Test of ART1.
(A) Amino acid sequence of ART1. Predicted C2H2 domain (red frame with underline), putative nuclear targeting signal (black frame), mutation site (reversed triangle), and defect region in art1 due to frame shift by 1-bp deletion (blue letters) are shown.
(B) Complementation test. Relative root elongation of the wild type, art1, transgenic rice with empty vector (vector control), and three independent transgenic lines with ART1 genomic region (lines 1 to 3) were measured. Data are means ± sd (n = 13 to 15 biological replicates). Different letters indicate significant differences at P < 0.05 by Tukey's test.
(C) Phylogenic tree of ART1-like C2H2 zinc finger proteins in rice (Os-) and Arabidopsis (At-). The sequence alignment used to generate the phylogeny is presented in Supplemental Data Set 1 online. Bootstrap values from 1000 trials are indicated. The 0.1 scale shows substitution distance.
[See online article for color version of this figure.]
Complementation Test
To confirm this mapping result, we performed a complementation test by introducing a 6.0-kb DNA fragment containing the candidate gene Os12g0170400 plus a 3.4-kb upstream region into art1 mutant by Agrobacterium tumefaciens–mediated transformation. The tolerance to Al in the three independent transgenic lines harboring the candidate gene recovered to the similar level of the wild type (Figure 2B), whereas the tolerance in the vector control line remains the same as the art1 mutant. These results indicate that the one-base deletion in Os12g0170400 is responsible for the Al hypersensitivity observed in the mutant art1. In addition, we checked transcript levels of these three independent complementation lines by quantitative RT-PCR. Although these levels include both endogenous ART1/art1 and transgenic ART1, one of the complementation lines, line 2, showed significantly higher expression of ART1, probably due to positional effect or copy number of the transgene (see Supplemental Figure 4 online). However, the Al tolerance of this line was not significantly different from other two complementation lines (Figure 2B). These results suggest that ART1 activity is probably also controlled by posttranslational activation as discussed later.
Phylogenic Analysis of ART1
ART1 is predicted to encode a putative C2H2 zinc finger protein with 465 amino acids (Figure 2A), which has four potential zinc finger domains. The 1-bp deletion in the mutant resulted in a frame shift and loss of the last two C2H2 domains. BLAST searches revealed that ART1 has five close homologs in rice (Figure 2C). There are two close homologs in Arabidopsis genome, and the closest one (At1g34370, STOP1) shared 41.2% identity with ART1. STOP1 has been identified as a putative transcriptional factor regulating both H+ and Al tolerance (Iuchi et al., 2007). However, ART1 is not the likely rice ortholog of STOP1, as the closest homolog of STOP1 in rice is Os01g0871200 (Figure 2C), not ART1. Also, STOP1 regulates H+ and Al tolerance in Arabidopsis, but ART1 specifically regulates Al-responsive genes in rice (Figure 1C). Indeed, the specific downstream genes regulated by ART1 and STOP1 are also different as discussed later.
Expression Patterns and Localization of ART1
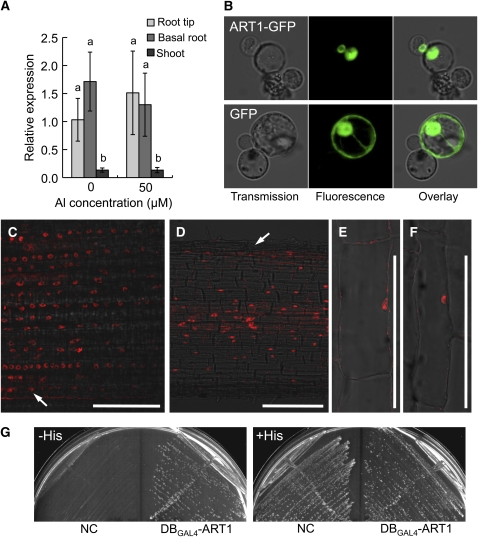
We investigated the expression pattern of ART1 with quantitative real-time RT-PCR. ART1 was mainly expressed in the roots (Figure 3A), and the expression level in the root tips (0 to 1 cm) was similar to that in the basal region roots (1 to 2 cm). Furthermore, the expression level of this gene was not significantly affected by Al (Figure 3A).
Figure 3.
Expression, Localization, and Transcriptional Activation Analysis of ART1.
(A) Relative mRNA expression of ART1 in different tissues. Five-day-old seedlings (cv Koshihikari) were exposed to a 0.5 mM CaCl2 solution containing 0 or 50 μM Al for 6 h, and then root tips (0 to 10 mm), basal roots (10 to 20 mm), and shoots were used for quantitative RT-PCR to determine relative expression of ART1. Histone H3 was used as an internal control. Expression relative to the root tip expression without Al treatment is shown. Data are means ± sd (n = 6; three biological replicates and two technical replicates). Different letters indicate significant differences at P < 0.05 by Tukey's test.
(B) Transient expression of the ART1-GFP fusion and GFP alone as control in rice callus protoplasts.
(C) and (D) Immunostaining of root longitudinal section with an anti-ART1 antibody at 1 mm (C) and 20 mm (D) from the tip treated with Al (50 μM, 6 h). Arrows indicate root epidermal cells with (C) or without (D) ART1 expression.
(E) and (F) Magnified images for single root cells at 20 mm from the tip treated with (F) or without (E) Al (50 μM, 6 h). Bars = 100 μm in (C) to (F).
(G) Transcriptional activation analysis in yeast. Yeast strain AH109 carrying fusion gene of GAL4 DNA binding domain and ART1 (DBGAL4-ART1) or DBGAL4 alone as control (NC) were cultured on SD medium with or without histidine (His) at 30°C for 3 d.
To examine the subcellular localization of ART1, we transiently introduced a gene (ART1-GFP) fused between ART1 and a gene encoding a green fluorescent protein (GFP) into protoplasts derived from rice callus. The ART1-GFP green fluorescence was observed only in the nuclei (Figure 3B), whereas GFP alone as a control was found in both the nuclei and cytoplasm. These results indicate that ART1 is localized to the nucleus.
We also performed immunostaining with the anti-ART1 antibody to investigate the cell specificity of localization of ART1. ART1 was localized to the nuclei of all cells in the root tip (Figure 3C) and all cells except the epidermal cells in the basal region (Figure 3D). Exposure to Al did not affect the subcellular localization of ART1 (Figures 3E and 3F).
Determination of Transcriptional Activation Potential
To examine whether ART1 has transcriptional activation potential, we conducted a modified yeast one-hybrid analysis using a chimeric protein with the DNA binding domain of the yeast GAL4 transcription factor and full-length ART1 (DBGAL4-ART1). If ART1 has transcriptional activation potential, DBGAL4-ART1 bound to the GAL1 upstream activating sequence will induce HIS3 reporter gene expression and accordingly complement the histidine requirement of the host yeast strain (AH109). The yeast carrying DBGAL4-ART1 grew well on the medium without histidine, but the control yeast carrying DBGAL4 alone (NC) did not (Figure 3G). These results indicate that ART1 has a transcriptional activation potential, at least in yeast.
Genes Regulated by ART1
To examine the genes regulated directly or indirectly by ART1, we compared genome-wide transcriptional profiles between treatments with Al and without Al in both the wild type and the mutant by microarray analysis with the rice 44 K oligo microarray. The roots were exposed for a short period (4 h) to a low Al concentration (10 μM), at which concentration the root elongation of the wild type was hardly inhibited so that genes affected by Al toxicity could be excluded. The microarray analysis identified differentially expressed genes between the wild type and mutant (false discovery rate [FDR] < 0.1). Then, genes upregulated by more than threefold in the wild type, but hardly changed in the mutant (less than twofold change) were picked up. As a result, a total of 31 genes were selected as candidates for genes regulated by ART1. Based on the annotation database, the candidate genes regulated by ART1 are classified into four groups: (1) cell wall maintenance and root elongation, (2) membrane protein, (3) metabolism, and (4) unknown (Table 1). These genes are probably involved in Al detoxification at different cellular levels in rice as discussed later.
Table 1.
Fold Changes in Gene Expression
| Fold Change by Al Treatment |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| RAP-DB | Arabidopsis Homolog | Description | Wild Type | sd | Mutant | sd | FDR | P Value | Harboring QTL |
| (1) Cell wall maintenance and root elongation | |||||||||
| Os01g0178300 | None | OsCDT3 | 4.42 | 0.45 | 1.10 | 0.06 | 0.02 | 0.000 | |
| Os01g0652100 | At1g29050 | PMR5-like DUF231 domain containing protein | 3.08 | 0.10 | 1.37 | 0.05 | 0.01 | 0.000 | |
| Os01g0860500 | At5g24090 | Chitinase | 8.69 | 0.54 | 1.15 | 0.08 | 0.02 | 0.000 | |
| Os03g0760800 | At2g39540 | Gibberellin-regulated Cys-rich protein family | 5.11 | 0.62 | 1.27 | 0.10 | 0.02 | 0.000 | |
| Os04g0583500 | EXP14/At3g03220 | Expansin-A10 | 3.38 | 0.95 | 1.11 | 0.13 | 0.06 | 0.004 | |
| Os09g0479900 | At5g59810 | Subtilisin-like Ser protease | 3.15 | 0.29 | 1.54 | 0.03 | 0.02 | 0.000 | |
| Os10g0524600 | SDD1/At1g04110 | Subtilisin-like Ser protease | 3.25 | 0.62 | 1.51 | 0.14 | 0.06 | 0.003 | |
| (2) Membrane protein | |||||||||
| Os01g0869200 | At3g19640 | Putative Mg2+ transporter | 4.19 | 0.22 | 1.26 | 0.05 | 0.01 | 0.000 | Ma et al. (2002) |
| Os02g0131800 | At1g80830 | Nramp | 5.43 | 0.25 | 1.96 | 0.10 | 0.02 | 0.000 | Ma et al. (2002) |
| Os02g0755900 | At1g22400 | UDP-glucuronosyl/UDP-glucosyltransferase | 3.43 | 0.98 | 1.15 | 0.10 | 0.07 | 0.004 | |
| Os03g0755100 | ALS1/At5g39040 | At ALS1 homolog | 3.06 | 0.09 | 1.21 | 0.05 | 0.01 | 0.000 | |
| Os05g0119000 | At2g37330 | STAR2 | 3.78 | 0.29 | 1.64 | 0.05 | 0.02 | 0.000 | |
| Os06g0695800 | At1g67940 | STAR1 | 3.60 | 0.24 | 1.07 | 0.06 | 0.02 | 0.000 | |
| Os09g0426800 | WAX2/At5g57800 | GLOSSY1-like | 4.48 | 0.92 | 1.60 | 0.11 | 0.04 | 0.001 | |
| Os10g0206800 | At1g51340 | MATE | 4.14 | 0.33 | 1.45 | 0.02 | 0.02 | 0.000 | |
| Os10g0578800 | At1g32080 | LrgB-like | 4.10 | 0.38 | 1.84 | 0.11 | 0.02 | 0.000 | |
| (3) Metabolism and detoxification | |||||||||
| Os01g0716500 | At5g10830 | SAM-dependent methyltransferase | 4.39 | 0.62 | 1.78 | 0.38 | 0.06 | 0.003 | |
| Os02g0186800 | At2g30750 | Cytochrome P450 family protein | 4.69 | 0.59 | 1.38 | 0.16 | 0.03 | 0.000 | |
| Os02g0770800 | NIA1/At1g77760 | Nitrate reductase | 9.39 | 2.63 | 1.96 | 0.44 | 0.02 | 0.000 | |
| Os12g0227400 | At3g03080 | Allyl alcohol dehydrogenase | 4.19 | 0.85 | 1.18 | 0.15 | 0.04 | 0.001 | |
| (4) Unknown | |||||||||
| Os01g0731600 | At1g78780 | Hypothetical protein | 11.62 | 1.19 | 1.19 | 0.21 | 0.02 | 0.000 | |
| Os01g0766300 | none | Hypothetical protein | 3.58 | 0.70 | 1.13 | 0.09 | 0.03 | 0.000 | |
| Os01g0919200 | none | Hypothetical protein | 3.10 | 0.88 | 1.29 | 0.19 | 0.08 | 0.008 | Wu et al. (2000) |
| Os03g0126900 | none | Hypothetical protein | 4.31 | 0.99 | 1.09 | 0.04 | 0.03 | 0.001 | |
| Os03g0304100 | At1g56320 | Hypothetical protein | 3.45 | 0.67 | 1.40 | 0.08 | 0.05 | 0.002 | |
| Os04g0419100 | none | Hypothetical protein | 3.36 | 0.35 | 1.34 | 0.10 | 0.02 | 0.000 | |
| Os04g0494900 | At5g11420 | Unknown function DUF642 family | 10.05 | 2.15 | 1.92 | 0.24 | 0.02 | 0.000 | |
| Os07g0493100 | none | Non-protein coding transcript | 10.12 | 1.70 | 1.16 | 0.18 | 0.02 | 0.000 | |
| Os07g0587300 | none | Hypothetical protein | 3.42 | 0.24 | 1.08 | 0.08 | 0.02 | 0.000 | Nguyen et al. (2003) |
| Os11g0488100 | none | Hypothetical protein | 3.14 | 0.63 | 1.06 | 0.04 | 0.04 | 0.001 | Nguyen et al. (2001) |
| Os11g0490100 |
At1g67330 |
Uncharacterized plant-specific DUF579 family |
5.42 |
0.87 |
1.35 |
0.25 |
0.03 |
0.001 |
Nguyen et al. (2001) |
Microarray analysis was performed with wild-type rice (wild type) and art1 mutant exposed to 10 μM Al for 4 h. Data are means ± sd from three independent biological replicates. Genes upregulated by Al more than threefold in the wild type but less than twofold change in the mutant (cutoff by FDR < 0.1 of Benjamini-Hochberg FDR method) were extracted.
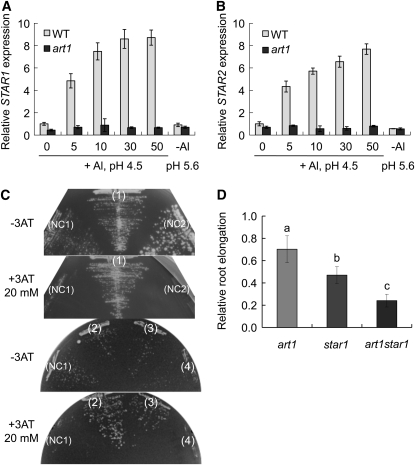
To confirm the microarray result, we examined the expression pattern of two genes, STAR1 and STAR2, belonging to group 2 (Table 1). Both genes have been demonstrated to be required for Al tolerance in rice (Huang et al., 2009). In the absence of Al, there was no difference between the wild type and the mutant in the expression level of STAR1 and 2 (Figures 4A and 4B). However, in the presence of Al, the expression levels of both STAR1 and 2 were upregulated in the wild type but not in the mutant (Figures 4A and 4B). Interestingly, we found that the expression of both STAR1 and 2 is not responsive to a low pH treatment as reported previously (Figures 5A and 5B) (Huang et al., 2009). Furthermore, we examined the direct interaction between the ART1 protein and the promoter region of STAR1 using a yeast one-hybrid assay. ART1 protein can interact with promoter regions of STAR1 (−939, −629, and −436 to −172 from the start codon), but we found no interaction in the region (−297 to −172) in the yeast assay (Figure 4C). These results suggest that a cis-acting element(s) recognized by the ART1 transcription factor is present in the region between −436 and −298 of the STAR1 promoter. The exact cis-element(s) in this region remains to be identified in the future.
Figure 4.
Regulation of STAR1 and STAR2 by ART1.
(A) and (B) Relative expression levels of STAR1 (A) and STAR2 (B) in root tip (0 to 10 mm) of the wild type and art1 mutant treated with 0 to 50 μM Al, pH 4.5, or without Al, pH 5.6, for 6 h, were determined by quantitative RT-PCR. Expression relative to the wild type expression without Al treatment, pH 4.5, is shown. Data are means ± sd (n = 3).
(C) Yeast one-hybrid assay. A pair of plasmids, (1 to 4) pHIS-ProSTAR1 [HIS reporter gene with STAR1 promoter, containing a region between −939 (1), −629 (2), −436 (3), or −297 (4) to −172 from STAR1 start codon] and pGAD-ART1 (fusion of GAL4 activation domain and ART1), (NC1) pHIS2.1 (without STAR1 promoter) and pGAD-ART1, (NC2) pHIS-ProSTAR1 and pGADT7 (GAL4 activation domain only) were introduced into yeast strain Y187 and cultured on SD medium containing 0 or 20 mM 3-amino-1,2,4-triazole, a competitor of HIS3 at 30°C for 3 d in the absence of His.
(D) Relative Al sensitivity of art1, star1, and the art1 star1 double mutant. Five-day-old seedlings were exposed to a 0.5 mM CaCl2 solution containing 0 or 5 μM Al, pH 4.5, for 24 h. Relative root elongation (root elongation with Al/root elongation without Al × 100) is shown. Data are means ± sd (n = 10). Different letters indicate significant differences at P < 0.05 by Student's t test.
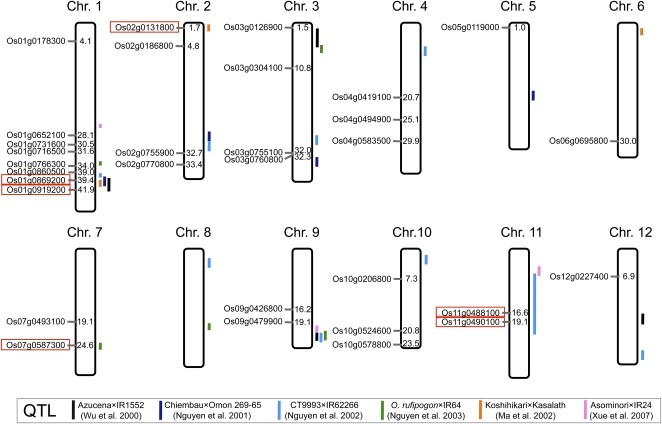
Figure 5.
Position of Candidate Genes Regulated by ART1 and QTLs for Al Tolerance.
Values indicate position of genes on each chromosome (Mb), and QTLs found by different studies are indicated by colored lines. Genes located at a similar position to the QTL are indicated with red boxes.
[See online article for color version of this figure.]
To further demonstrate that ART1 also regulates other genes involved in Al tolerance, we made a double mutant between star1 and art1. Al tolerance tests showed that the double mutant is more sensitive to Al than the single mutant star1 at 5 μM Al (Figure 4D). These results support that ART1 also regulates other Al tolerance genes, although STAR1 may play a major role in Al tolerance. The Al sensitivity of star1 is higher than art1 (Figure 4D). Interestingly, the distribution of Al in root cells observed by both morin and eriochrome cyanine R staining was different between star1 and art1 (see Supplemental Figure 5 online). Al reached all cortex cells in the star1 mutant root, but not in art1. These differences are due to the basic level expression of functional STAR1 and other downstream genes in the art1 mutant (Figure 4D). In other words, knockout of the ART1 transcription factor does not result in silence of the downstream genes (Figures 4A and 4B). This result also suggests that other downstream genes and their upregulation by ART1 in response to Al are required for Al tolerance in rice.
DISCUSSION
Genes Regulated by ART1 Are Implicated in the Detoxification of Al at Different Cellular Levels
Our results clearly show that ART1 is a transcription factor localizing to the nucleus and having transcriptional activation potential (Figure 3). Furthermore, specific response of art1 mutant to Al, but not to low pH and other metals, suggests that genes regulated by ART1 are specifically implicated in Al tolerance (Figure 1). Although the exact mechanisms for Al-induced root inhibition have not been elucidated, it is clear that Al targets multiple cellular sites, including the cell walls, plasma membranes, and cellular processes, such as signal transduction pathways and homeostasis mechanisms (Barceló and Poschenrieder, 2002; Kochian et al., 2004; Ma, 2007; Poschenrieder et al., 2008). Therefore, plants have to detoxify Al at different cellular levels for survival. Among candidate genes regulated by ART1, most of them are implicated in the internal and external detoxification of Al (Table 1). Candidate genes in group 1 are probably involved in the cell wall maintenance and/or root elongation. For example, Os04g0583500 encodes an expansin, EXPA10. Expansins were identified as proteins that mediate extension of isolated cell walls (Lee and Kende, 2002), and the effect of Al on expansin has been reported (Gao et al., 2008). Os10g0524600 is the closest homolog of Arabidopsis subtilisin-like Ser protease, SDD1, which is exported to the apoplast and associates with the plasma membrane and then acts as a putative generator of extracellular signal (von Groll et al., 2002). Os01g0652100 encodes a close homolog of Arabidopsis PMR5. The pmr5 mutant exhibited pectin enrichment in the cell wall, although the function of PMR5 protein is still unknown (Vogel et al., 2004). Os01g0178300 encodes a small Cys-rich peptide, CDT3. A similar peptide, CDT1, was proposed to be localized in the cell wall, and overexpression of this peptide gave cadmium tolerance in transgenic Arabidopsis (Kuramata et al., 2009). Os01g0860500 is a close homolog of Arabidopsis putative chitinase At5g24090, which is a cell wall–associated protein in Arabidopsis suspension cells and probably involved in the mechanical properties of the cell wall (Kwon et al., 2005). Since >90% of Al is bound to the cell wall and affects cell wall extensibility (Horst et al., 1983; Ma et al., 2004), the upregulation of these genes by ART1 will likely contribute to Al tolerance, although further confirmation is required.
Group 2 includes nine membrane protein genes (Table 1). Among them, a bacterial-type ABC transporter composed of STAR1 and STAR2 has been demonstrated to play an essential role in detoxifying Al (Huang et al., 2009). Os03g0755100/ALS1 is a homolog of Arabidopsis ALS1, which has been reported as a putative half-type ABC transporter localized to the tonoplast and probably responsible for sequestration of Al into the vacuoles in Arabidopsis (Larsen et al., 2007). A putative citrate efflux transporter, Os10g020131800, which showed high similarity with an Al-activated citrate transporter (AACT1) in barley (Furukawa et al., 2007), may also play a role in conferring Al tolerance by secretion of citrate from the roots. Os01g0869200 and Os02g0131800 encode a putative Mg2+ transporter and an Nramp family protein, respectively. It is reported that Mg2+ transporters are one of the cellular targets of Al toxicity, and overexpression of the Arabidopsis transporter gene MGT1 in Nicotiana benthamiana confers Al tolerance on the plant (Deng et al., 2006). Nramp proteins exhibit functional divergence, and most Nramp proteins are able to transport multiple metal ions such as Fe, Mn, Zn, and Cd (Oomen et al., 2009). Therefore, Os01g0869200 and Os02g0131800 seem to be involved in maintenance of metal homeostasis under Al stress.
Genes in group 3 (Table 1) are predicted to be involved in metabolism and detoxification. However, the functions of most genes in this group and those in group 4 are unknown and remain to be examined.
Although the genes regulated by ART1 are upregulated by Al, they are not newly induced, but constitutively expressed at basal levels (Figures 4A and 4B). This is probably because, unlike temporary environmental stresses such as water and temperature stresses, Al toxicity stress is prevalent in acid soils. Therefore, it seems that plants have acquired basic strategies to constitutively detoxify Al at different cellular levels.
Different Pathways of Al Tolerance between Rice and Arabidopsis
Rice and Arabidopsis differ greatly in the tolerance to Al toxicity (Ma et al., 2002; Iuchi et al., 2007). Recently, STOP1, a C2H2 zinc finger-type putative transcriptional factor, was reported as a responsible gene for both proton and Al hypersensitivity in Arabidopsis (Iuchi et al., 2007). ART1 is not the closest homolog of STOP1 in the rice genome (Figure 2C), and ART1 and STOP1 only share 41.2% identity. Comparison between ART1 and STOP1 shows that they differ in the response to stresses and downstream genes. The stop1 mutant showed increased sensitivity to both Al and low pH (Iuchi et al., 2007), whereas art1 only showed increased sensitivity to Al, but not to low pH (Figure 1C). The fact that STAR1 and STAR2 do not respond to low pH (Figures 4A and 4B) (Huang et al., 2009) also supports this conclusion. Most importantly, the downstream genes regulated by STOP1 and ART1 are different except two genes (STAR2/ALS3 and MATE) (Sawaki et al., 2009; Table 1). STOP1 and ART1 each regulate an Nramp gene (Table 1; Sawaki et al., 2009), but the two Nramp genes belong to different branches of the Nramp family. These differences may contribute to different mechanisms for Al tolerance. The major Al tolerance mechanism in Arabidopsis is the secretion of malate from the roots, whereas rice does not secrete malate in response to Al (Ma et al., 2002). Furthermore, Al tolerance in rice is much higher than that in Arabidopsis; one possible mechanism for higher tolerance is that more tolerance genes are required in rice. In addition, rice is very tolerant to low pH independent of ART1 (Figure 1C), whereas Arabidopsis is very sensitive to low pH (Iuchi et al., 2007). Another possibility for the difference is that the experimental conditions are different between different studies. To avoid genes induced by Al toxicity, we exposed the rice roots to a low Al concentration (10 μM) for a short period (4 h). However, Sawaki et al. (2009) exposed Arabidopsis to 10 μM Al at pH 5.0 for 24 h. Under such conditions, the root growth of Arabidopsis is severely inhibited even in the wild type (Iuchi et al., 2007). Therefore, most of genes extracted might not be directly regulated by STOP1 but might be induced indirectly by Al toxicity. It would be interesting to extract genes regulated by STOP1 under an experimental condition with low Al concentration and short exposure and compare these genes with those regulated by ART1.
ART1-Regulated Genes May Be Involved in Genotypic Variation of Al Tolerance
There is a genotypic variation in Al tolerance in rice. Usually, japonica cultivars show higher Al tolerance than indica cultivars, which are cultivated on acidic soil areas. More than 10 quantitative trait loci (QTL) for Al tolerance have been detected (Wu et al., 2000; Nguyen et al., 2001, 2002, 2003; Ma et al., 2002; Xue et al., 2007), but the genes for these QTLs have not been cloned so far. Among the genes regulated by ART1 (Table 1), several genes, Os01g0869200, Os01g0919200, Os02g0131800, Os07g0587300, Os11g0488100, and Os11g0490100, are located at the similar positions of Al tolerance QTLs (Figure 5). Therefore, these genes may be involved in genotypic differences in Al tolerance in rice, although further confirmation is required in the future.
ART1 Specifically Regulates Al-Responsive Genes in Higher Plants
Since these genes are implicated in the detoxification of Al at different cellular levels, further elucidation of these gene functions will help to understand comprehensively plant Al tolerance mechanisms, whose molecular bases are still poorly understood. Moreover, ART1 also provides an invaluable clue to identify plant sensing and signaling pathway for Al toxicity in the future. ART1 is constitutively expressed in the roots (Figure 3A), and nuclear localization of ART1 protein is not affected by Al treatment (Figures 3E and 3F), while genes regulated by ART1 are upregulated by Al. These observations suggest that posttranslational regulation of ART1, such as protein phosphorylation or interaction with some other factors, is required to activate ART1. This is supported by the finding that there was no correlation between the expression of ART1 and Al tolerance in transgenic complementation lines (Figure 2B; see Supplemental Figure 4 online).
METHODS
Isolation of the art1 Mutant
M3 seeds of rice (Oryza sativa cv Koshihikari) irradiated with γ-rays were used for isolation of Al-sensitive mutants in 2002, following the procedures described previously (Ma et al., 2005). After three rounds of screening based on root elongation inhibition, we obtained a mutant sensitive to Al, which we named art1 based on the features of the responsible gene described in the text.
Physiological Analysis of the Mutant
Seeds of the mutant (art1) and its wild-type rice were soaked in deionized water overnight at 30°C in the dark and then transferred to nets that were floated on a 0.5 mM CaCl2 solution in a 1.5-liter plastic container. After growth at 25°C for 4 or 5 d, the seedlings were used for subsequent experiments. To compare Al sensitivity, seedlings were exposed to a 0.5 mM CaCl2, pH 4.5, containing various AlCl3 concentrations for 24 h, and then relative root elongation (RRE) was used to evaluate Al sensitivity of each line. RRE was calculated as follows: (root elongation with Al treatment)/(root elongation without Al) ×100.
To further evaluate the sensitivity to Al, we grew both wild-type and mutant plants in acid soil (Andosol) at pH 4.5 and an alluvial soil at pH 6.5. After 6 d, the root length was measured.
The effect of pH on root elongation was investigated in a 0.5 mM CaCl2 solution buffered with 10 mM Homo-PIPES at pH ranging from 3.5 to 5.5. Root length of 10 seedlings each from the wild type and art1 was measured after 24 h. The root elongation was also compared by exposing seedlings to a 0.5 mM CaCl2 solution, pH 4.5, containing 0, 30 μM AlCl3, 20 μM CdCl2, 100 μM ZnCl2, 0.5 μM CuCl2, or 5 μM LaCl3 for 24 h.
Both wild-type rice (cv Koshihikari) and art1 mutants were cultivated in a field (soil pH 6.5) at an experimental farm of Okayama University in 2006 as described previously (Tamai and Ma, 2008). Three replicates of a plot (1 × 0.4 m) were made for each line. Plant growth and yield were investigated at harvest.
Genetic Analysis
Seeds of F1 plants were derived from a cross between the mutant and wild-type rice plants. F2 seedlings (164) were exposed to a 0.5 mM CaCl2 solution for 24 h and then transferred to a 0.5 mM CaCl2 solution containing 20 μM AlCl3 for another 24 h. RRE (see above) was used to evaluate Al sensitivity of each seedling.
Map-Based Cloning of ART1
For mapping the responsible gene, we constructed an F2 population derived from a cross between art1 mutant and Kasalath. First, we performed a bulked segregant analysis to determine the molecular markers linked to ART1 (Michelmore et al., 1991). Rough mapping of this gene was then done using 46 F2 Al-sensitive plants. For fine mapping of this gene, we used 986 F2 plants from the art1/Kasalath population. Polymorphic InDel markers were developed based on the rice InDel database at http://shenghuan.shnu.edu.cn/ricemarker (Shen et al., 2004) and the comparison of the genomic sequence of Nipponbare with that of 93-11 by BLASTn searches in Genbank (see Supplemental Table 1 online). Twenty-five crossover plants between InDel markers MaOs1219 and MaOs1237 from the art1/Kasalath population were further evaluated for their Al sensitivity in the F3 generation. ART1 was finally defined to 38.5-kb region, and the six candidate genes were sequenced by BigDye Terminators V3.1 cycle sequencing kit and ABI PRISM 310 genetic analyzer. The putative C2H2 zinc finger was independently sequenced twice in the mutant.
Phylogenetic Analysis
Alignment was performed with ClustalW using default setting (http://clustalw.ddbj.nig.ac.jp/), and the phylogenetic tree was constructed using the neighbor-joining algorithm with MEGA version 4 (Tamura et al., 2007) with 1000 bootstrap trials.
RNA Isolation and Quantitative Real-Time RT-PCR
Seedlings of the wild type and art1 were exposed to a 0.5 mM CaCl2 solution, pH 4.5, containing 0 to 50 μM Al for 6 h, and the roots tips (0 to 10 mm), basal roots (10 to 20 mm), and the shoots were harvested. Total RNA was extracted using the RNeasy mini kit (Qiagen). One microgram of total RNA was used for first-strand cDNA synthesis using a SuperScript kit (Invitrogen) following the manufacturer's instructions with an oligo(dT)12-18 primer. One microliter of 10-fold dilution cDNA from each sample was used for the quantitative analysis of gene expression with SYBR Premix Ex Taq (Takara). Data were collected in accordance with using the 7500 Real Time PCR System (Applied Biosystems). HistoneH3 was used as an internal control. Primer sequences used are as follows: STAR1, 5′-TCGCATTGGCTCGCACCCT-3′ (forward) and 5′-TCGTCTTCTTCAGCCGCACGAT-3′ (reverse); STAR2, 5′-ACCTCTTCATGGTCACCGTCG-3′ (forward) and 5′-CCTCAGCTTCTTCATCGTCACC-3′ (reverse); ART1, 5′- CAGTGCTTCTCGTGGGTCTT-3′ (forward) and 5′- CCTGTGCGTGAAGAACCACT-3′ (reverse); HistoneH3, 5′- AGTTTGGTCGCTCTCGATTTCG-3′ (forward) and 5′- TCAACAAGTTGACCACGTCAC-3′ (reverse).
Generation of Transgenic Rice
For the complementation test of ART1, we obtained a BAC clone (BAC AL731761) containing ART1 by screening of BAC library of Koshihikari. After partial enzymatic cutting with Sau3AI, we selected a 6.1-kb clone containing ART1 and its promoter (from 3.4 kb upstream of start codon). This clone was inserted into pPZP2H-lac vector (Fuse et al., 2001) and then transformed into Agrobacterium tumefaciens (strain EHA101). Calluses derived from the rice mutant art1 were transformed by Agrobacterium-mediated transformation (Hiei et al., 1994). We obtained five independent transgenic lines. T2 seeds from three independent lines were further used to test the Al tolerance with a vector control line. The root elongation was measured for 24 h in a 0.5 mM CaCl2 solution without Al and then to the same solution containing 20 μM Al, pH 4.5, for a further 24 h. Relative root elongation was calculated as (root elongation with Al/root elongation without Al) × 100.
Construction of Fluorescent Gene Fusion and Transient Expression
For construction of a translational ART1-GFP fusion, the ORF of ART1 except the stop codon was amplified by PCR from rice (cv Koshihikari) root cDNA. Primer pairs used for amplification and introduction of restriction sites were 5′-AGATCACTCGAGATTATTCAGAAGCTTGCA-3′ and 5′-TAATCATGACTGATCCCTTGTCACCATTCTCCTCCTG-3′. The ORF was inserted between cauliflower mosaic virus 35S promoter and GFP-NOS terminator in pBluescript vector. Plasmid DNA was transiently introduced into rice callus protoplasts using the polyethylene glycol method according to Kamiya et al. (2006). After overnight incubation, GFP fluorescence was observed with a fluorescence microscope (Axio Imager with Apotome; Carl Zeiss).
Immunohistological Staining
Antibodies against ART1 were obtained by immunizing rabbits with the synthetic peptide C-EQFAGDHQEENGDK (positions 452 to 465 of ART1). The roots of 1-week-old seedlings (cv Koshihikari) treated with or without Al (50 μM, 6 h) were used for immunostaining as described previously (Yamaji and Ma, 2007).
Yeast One-Hybrid Assay
The yeast one-hybrid assay was performed using MATCHMAKER GAL4 Two-Hybrid System 3 (Clontech) and MATCHMAKER One-Hybrid Library Construction and Screening Kit (Clontech) to examine the transcriptional activation potential of ART1. The ORF of ART1 was amplified by PCR from rice (cv Koshihikari) root cDNA. Primer pairs used for amplification and introduction of restriction sites were 5′-AGATCACtCGAGATTATTCAGAAGCTTGCA-3′ and 5′-CCGCTCGAGTCACTTGTCACCATTCTCC-3′. The ORF was cloned in frame after the DNA binding domain of yeast GAL4 transcription factor (without activation domain) in pGBKT7 vector (pGBK-ART1). These plasmids, pGBK-ART1 and control pGBKT7, were introduced into yeast strain AH109 that carried the GAL4-responsive GAL1 promoter and HIS3 reporter gene and cultured on SD medium with or without histidine (His) at 30°C for 3 d according to the manufacturer's manual.
To investigate the interaction between ART1 protein and STAR1 promoter, we amplified the promoter sequence of STAR1 (−939, −629, −436, or −297 to −172 bp from the start codon) by PCR from rice (cv Koshihikari) genomic DNA. Primer pairs used for amplification and introduction of restriction sites were 5′-TTCTAGATAGCATCTGGATAATGATAATC-3′ (from -939), 5′-AGAATTCAATTGGAGTTCTCTTTGCGG-3′ (from −629), 5′-AGAATTCCGTCGTACCGGTGATAAC-3′ (from −436), 5′-AGAATTCCCACACAGATCCACGGCA-3′ (from −297) and 5′-TTCTAGATCGCCGTGGTCGGTTTGGA-3′. Two tandem copies of the amplified promoter region was cloned upstream of the HIS3 reporter gene in pHIS2.1 vector (pHIS-ProSTAR1). The ORF of ART1 described above was cloned in frame after transcriptional activation domain of yeast GAL4 transcription factor (without DNA binding domain) in pGADT7 (pGAD-ART1). A pair of these plasmids, pHIS-ProSTAR1 and pGAD-ART1, or control pHIS2.1 and pGADT7 were introduced into yeast strain Y187 and cultured on SD medium without His containing 0 to 20 mM 3-amino-1,2,4-triazole (a competitive inhibitor of HIS3) at 30°C for 3 d according to the manufacturer's manual.
Microarray Analysis
One-week-old seedlings of the wild type and art1 were treated with 10 μM Al, pH 4.5, or without Al, pH 4.5, in 0.5 mM CaCl2 solution for 4 h. RNA samples were prepared from root segments between 0 and 15 mm from the apex using the RNeasy Plant Mini Kit (Qiagen). Microarray analysis was performed according to Agilent Oligo DNA Microarray Hybridization protocols using the Rice Oligo DNA Microarray 44K RAP-DB (G2519F#15241; Agilent Technologies) with three biological replicates and color swap for each replicate. The hybridized slides were scanned using a DNA microarray scanner (Agilent Technologies). Signal intensities were extracted by Feature Extraction software (Agilent Technologies). For statistic analysis, we excluded genes with signal intensities below 100 in all the experiments after correction of the dye effect by averaging of the two color swaps. Significance test was performed by unpaired t test using GeneSpringGX10 (Agilent Technologies). The Benjamini-Hochberg FDR method was used to obtain P values corrected for multiple testing. Fold change and sd of each probe by Al treatment were calculated using the average of three biological replicates. Genes upregulated by Al more than threefold in the wild type but less than twofold change in the mutant (cutoff by FDR < 0.1) were extracted. The function of these genes was categorized based on the annotation database.
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession number AB379846 (ART1).
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Air-Dried Weight of Shoot and Grain Yield of Both Wild Type Rice and art1 Grown in a Field.
Supplemental Figure 2. Frequency Distributions of Al Sensitivities in an F2 Population from a Backcross between the Mutant (art1) and Wild Type.
Supplemental Figure 3. Map-Based Cloning of ART1.
Supplemental Figure 4. Expression Levels of ART1/art1 in Complementation Lines and the Control Plants.
Supplemental Figure 5. Root Al Staining in Wild Type, star1, and art1.
Supplemental Table 1. Primers for InDel Markers Used in Mapping ART1.
Supplemental Data Set 1. Alignment Used to Generate the Phylogeny Presented in Figure 2C.
Supplementary Material
Acknowledgments
We thank Kazuko Ono for technical assistance in generating transgenic rice and Ritsuko Motoyama for help in microarray analysis. This research was supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation IPG-0006 and QT3006 to J.F.M.) and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (21248009 to J.F.M. and 21780057 to N.Y.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jian Feng Ma (maj@rib.okayama-u.ac.jp).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Barceló, J., and Poschenrieder, C. (2002). Fast root growth responses, root exudates, and internal detoxification as clues to the mechanisms of aluminium toxicity and resistance: A review. Environ. Exp. Bot. 48 75–92. [Google Scholar]
- Deng, W., Luo, K., Li, D., Zheng, X., Wei, X., Smith, W., Thammina, C., Lu, L., Li, Y., and Pei, Y. (2006). Overexpression of an Arabidopsis magnesium transport gene, AtMGT1, in Nicotiana benthamiana confers Al tolerance. J. Exp. Bot. 57 4235–4243. [DOI] [PubMed] [Google Scholar]
- Foy, C.D. (1988). Plant adaptation to acid, aluminum-toxic soils. Commun. Soil Sci. Plant Anal. 19 959–987. [Google Scholar]
- Furukawa, J., Yamaji, N., Wang, H., Mitani, N., Murata, Y., Sato, K., Katsuhara, M., Takeda, K., and Ma, J.F. (2007). An aluminum-activated citrate transporter in barley. Plant Cell Physiol. 48 1081–1091. [DOI] [PubMed] [Google Scholar]
- Fuse, T., Sasaki, T., and Yano, M. (2001). Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol. 18 219–222. [Google Scholar]
- Gao, Q., Zhao, M., Li, F., Guo, Q., Xing, S., and Wang, W. (2008). Expansins and coleoptile elongation in wheat. Protoplasma 233 73–81. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Hoekenga, O.A., et al. (2006). AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 103 9738–9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst, W.J., Wagner, A., and Marschner, H. (1983). Effect of aluminum on root growth, cell-division rate and mineral element contents in roots of Vigna unguiculata genotypes. Z. Pflanzenphysiol. 109 95–103. [Google Scholar]
- Huang, C.F., Yamaji, N., Mitani, N., Yano, M., Nagamura, Y., and Ma, J.F. (2009). A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi, S., Koyama, H., Iuchi, A., Kobayashi, Y., Kitabayashi, S., Kobayashi, Y., Ikka, T., Hirayama, T., Shinozaki, K., and Kobayashi, M. (2007). Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. USA 104 9900–9905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, T., Akahori, T., Ashikari, M., and Maeshima, M. (2006). Expression of the vacuolar Ca2+/H+ exchanger, OsCAX1a, in rice: Cell and age specificity of expression, and enhancement by Ca2+. Plant Cell Physiol. 47 96–106. [DOI] [PubMed] [Google Scholar]
- Kochian, L.V., Hoekenga, O.A., and Pineros, M.A. (2004). How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 55 459–493. [DOI] [PubMed] [Google Scholar]
- Kuramata, M., Masuya, S., Takahashi, Y., Kitagawa, E., Inoue, C., Ishikawa, S., Youssefian, S., and Kusano, T. (2009). Novel cysteine-rich peptides from Digitaria ciliaris and Oryza sativa enhance tolerance to cadmium by limiting its cellular accumulation. Plant Cell Physiol. 50 106–117. [DOI] [PubMed] [Google Scholar]
- Kwon, H.-K., Yokoyama, R., and Nishitani, K. (2005). A proteomic approach to apoplastic proteins involved in cell wall regeneration in protoplasts of Arabidopsis suspension-cultured cells. Plant Cell Physiol. 46 843–857. [DOI] [PubMed] [Google Scholar]
- Larsen, P.B., Cancel, J., Rounds, M., and Ochoa, V. (2007). Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 225 1447–1458. [DOI] [PubMed] [Google Scholar]
- Lee, Y., and Kende, H. (2002). Expression of α-expansin and expansin-like genes in deepwater rice. Plant Physiol. 130 1396–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligaba, A., Katsuhara, M., Ryan, P.R., Shibasaka, M., and Matsumoto, H. (2006). The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiol. 142 1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Magalhaes, J.V., Shaff, J., and Kochian, L.V. (2009). Aluminum-activated citrate and Malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 57 389–399. [DOI] [PubMed] [Google Scholar]
- Ma, J.F. (2007). Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int. Rev. Cytol. 264 225–252. [DOI] [PubMed] [Google Scholar]
- Ma, J.F., Nagao, S., Huang, C.F., and Nishimura, M. (2005). Isolation and characterization of a rice mutant hypersensitive to Al. Plant Cell Physiol. 46 1054–1061. [DOI] [PubMed] [Google Scholar]
- Ma, J.F., Shen, R., Nagao, S., and Tanimoto, E. (2004). Aluminum targets elongating cells by reducing cell wall extensibility in wheat roots. Plant Cell Physiol. 45 583–589. [DOI] [PubMed] [Google Scholar]
- Ma, J.F., Shen, R., Zhao, Z., Wissuwa, M., Takeuchi, Y., Ebitani, T., and Yano, M. (2002). Response of rice to Al stress and identification of quantitative trait loci for Al tolerance. Plant Cell Physiol. 43 652–659. [DOI] [PubMed] [Google Scholar]
- Magalhaes, J.V., et al. (2007). A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genet. 39 1156–1161. [DOI] [PubMed] [Google Scholar]
- Michelmore, R.W., Paran, I., and Kesseli, R.V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, B.D., Brar, D.S., Bui, B.C., Nguyen, T.V., Pham, L.N., and Nguyen, H.T. (2003). Identification and mapping of the QTL for aluminum tolerance introgressed from the new source, Oryza rufipogon Griff., into indica rice (Oryza sativa L.). Theor. Appl. Genet. 106 583–593. [DOI] [PubMed] [Google Scholar]
- Nguyen, V.T., Burow, M.D., Nguyen, H.T., Le, B.T., Le, T.D., and Paterson, A.H. (2001). Molecular mapping of genes conferring aluminum tolerance in rice (Oryza sativa L.). Theor. Appl. Genet. 102 1002–1010. [Google Scholar]
- Nguyen, V.T., Nguyen, B.D., Sarkarung, S., Martinez, C., Paterson, A.H., and Nguyen, H.T. (2002). Mapping of genes controlling aluminum tolerance in rice: Comparison of different genetic backgrounds. Mol. Genet. Genomics 267 772–780. [DOI] [PubMed] [Google Scholar]
- Oomen, R.J.F.J., Wu, J., Lelievre, F., Blanchet, S., Richaud, P., Barbier-Brygoo, H., Aarts, M.G.M., and Thomine, S. (2009). Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator Thlaspi caerulescens. New Phytol. 181 637–650. [DOI] [PubMed] [Google Scholar]
- Poschenrieder, C., Gunsé, B., Corrales, I., and Barceló, J. (2008). A glance into aluminum toxicity and resistance in plants. Sci. Total Environ. 400 356–368. [DOI] [PubMed] [Google Scholar]
- Ryan, P.R., Delhaize, E., and Jones, D.L. (2001). Function and mechanism of organic anion exudation from plant roots. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 527–560. [DOI] [PubMed] [Google Scholar]
- Sanchez, P.A., and Salinas, J.G. (1981). Low-input technology for managing oxisols and ultisols in tropical America. Adv. Agron. 34 279–406. [Google Scholar]
- Sasaki, T., Yamamoto, Y., Ezaki, B., Katsuhara, M., Ahn, S.J., Ryan, P.R., Delhaize, E., and Matsumoto, H. (2004). A wheat gene encoding an aluminum-activated malate transporter. Plant J. 37 645–653. [DOI] [PubMed] [Google Scholar]
- Sawaki, Y., Iuchi, S., Kobayashi, Y., Kobayashi, Y., Ikka, T., Sakurai, N., Fujita, M., Shinozaki, K., Shibata, D., Kobayashi, M., and Koyama, H. (2009). STOP1 regulates multiple genes which protect Arabidopsis from proton and aluminum toxicities. Plant Physiol. 150 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y.J., et al. (2004). Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol. 135 1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai, K., and Ma, J.F. (2008). Reexamination of silicon effects on rice growth and production under field conditions using a low silicon mutant. Plant Soil 307 21–27. [Google Scholar]
- Tamura, K., Dudleym, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Vogel, J.P., Raab, T.K., Somerville, C.R., and Somerville, S.C. (2004). Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 40 968–978. [DOI] [PubMed] [Google Scholar]
- von Groll, U., Berger, D., and Altmann, T. (2002). The subtilisin-like serine protease SDD1 mediates cell-to-cell signaling during Arabidopsis stomatal developent. Plant Cell 14 1527–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Uexkull, H.R., and Mutert, E. (1995). Global extent, development and economic impact of acid soils. Plant Soil 171 1–15. [Google Scholar]
- Wu, P., Liao, C.Y., Hu, B., Yi, K.K., Jin, W.Z., Ni, J.J., and He, C. (2000). QTLs and epistasis for aluminum tolerance in rice (Oryza sativa L.) at different seedling stages. Theor. Appl. Genet. 100 1295–1303. [Google Scholar]
- Xue, Y., Jiang, L., Su, N., Wang, J.K., Deng, P., Ma, J.F., Zhai, H.Q., and Wan, J.M. (2007). The genetic basic and fine-mapping of a stable quantitative-trait loci for aluminium tolerance in rice. Planta 227 255–262. [DOI] [PubMed] [Google Scholar]
- Yamaji, N., and Ma, J.F. (2007). Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol. 143 1306–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.