Abstract
Local efflux-dependent auxin gradients and maxima mediate organ and tissue development in plants. Auxin efflux is regulated by dynamic expression and subcellular localization of the PIN auxin-efflux proteins, which appears to be established not only through a self-organizing auxin-mediated polarization mechanism, but also through other means, such as cell fate determination and auxin-independent mechanisms. Here, we show that the Arabidopsis thaliana NO VEIN (NOV) gene, encoding a novel, plant-specific nuclear factor, is required for leaf vascular development, cellular patterning and stem cell maintenance in the root meristem, as well as for cotyledon outgrowth and separation. nov mutations affect many aspects of auxin-dependent development without directly affecting auxin perception. NOV is required for provascular PIN1 expression and region-specific expression of PIN7 in leaf primordia, cell type–specific expression of PIN3, PIN4, and PIN7 in the root, and PIN2 polarity in the root cortex. NOV is specifically expressed in developing embryos, leaf primordia, and shoot and root apical meristems. Our data suggest that NOV function underlies cell fate decisions associated with auxin gradients and maxima, thus establishing cell type–specific PIN expression and polarity. We propose that NOV mediates the acquisition of competence to undergo auxin-dependent coordinated cell specification and patterning, thereby eliciting context-dependent auxin-mediated developmental responses.
INTRODUCTION
In plants, the phytohormone auxin has been established as a key regulator of axial patterning processes. Local auxin gradients associated with auxin maxima mediate pattern formation in the root (Sabatini et al., 1999; Friml et al., 2002a; Blilou et al., 2005), lateral organ (Reinhardt et al., 2000, 2003; Benková et al., 2003), embryo (Liu et al., 1993; Steinmann et al., 1999; Friml et al., 2003), and vascular tissue (Sachs, 1991; Mattsson et al., 1999; Sieburth, 1999; Mattsson et al., 2003). By contrast, organ separation is associated with local reductions in auxin concentration (Heisler et al., 2005). Local auxin gradients direct formation of plant organs, regardless of their morphology or developmental origin (Benková et al., 2003). Cell type–specific expression and asymmetric subcellular localization of the PIN family of auxin-efflux proteins define auxin distribution, a central factor in the formation of auxin concentration gradients and maxima (Petrásek et al., 2006; Wisniewska et al., 2006). It has been shown that PIN expression and polarity are established not only through a self-organizing auxin-mediated feedback regulatory loop (Sieberer et al., 2000; Paciorek et al., 2005; Vieten et al., 2005; Sauer et al., 2006) but also through cell fate determination (Sauer et al., 2006; Xu et al., 2006), reversible phosphorylation of PIN proteins (Michniewicz et al., 2007), endocytic recycling (Geldner et al., 2001; Jaillais et al., 2007; Dhonukshe et al., 2008; Kleine-Vehn et al., 2008a), vesicle trafficking to vacuoles (Kleine-Vehn et al., 2008b; Shirakawa et al., 2009), protein turnover (Abas et al., 2006; Kleine-Vehn et al., 2008b; Spitzer et al., 2009), and auxin-independent mechanisms (Willemsen et al., 2003; Bennett et al., 2006). Physical stimuli also modulate PIN expression and polar localization, thus affecting plant architecture through triggering changes in efflux-dependent auxin distribution (Friml et al., 2002b; Ditengou et al., 2008; Laskowski et al., 2008; Laxmi et al., 2008). Feedback regulation between auxin signaling and transport constitutes a self-organizing auxin-mediated polarization and patterning mechanism. This mechanism links individual cell polarity with tissue and organ polarity (Sauer et al., 2006; Scarpella et al., 2006; Wenzel et al., 2007).
In roots, auxin transport and the auxin response are essential for correct expression of the cell fate regulators PLETHORA (PLT) and SCARECROW (SCR) (Friml et al., 2002a; Sabatini et al., 2003; Aida et al., 2004; Blilou et al., 2005), suggesting that auxin and auxin flow regulate cell fate. Reciprocally, the cell fate regulators appear to control the polarity of auxin flow in roots (Blilou et al., 2005; Xu et al., 2006). Therefore, in roots, cell fate determinants appear to be integrated into the positive feedback loop that controls the polarization of auxin flow and polarized organ development.
In shoots, auxin maxima mark sites of incipient primordia (Heisler et al., 2005) and leaf venation (Mattsson et al., 2003; Scarpella et al., 2006). Externally applied auxin initiates primordium outgrowth (Reinhardt et al., 2000) and vascular formation (Sachs, 1991; Sauer et al., 2006) de novo. The earliest sign of vascular differentiation in leaves is the expression of PIN1 in presumptive preprocambial cells (Scarpella et al., 2006; Bayer et al., 2009), which arises among seemingly equivalent subepidermal cells. Hereafter, we collectively refer to preprocambial and procambial cells as provascular cells. PIN1 directs auxin flow to converge in the marginal epidermis of developing leaf primordia. Concomitantly, this creates adjacent subepidermal auxin maxima, which give rise to the distal ends of presumptive provascular strands (Scarpella et al., 2006). Thus, it was proposed that the epidermal convergence points of auxin flow externally determine the sites of provascular formation and trigger auxin-mediated provascular patterning in the ground meristem. However, the molecular mechanism that establishes the provascular expression and polarity of PIN proteins remains largely unknown. Other important open questions are the mechanisms by which (1) auxin maxima are generated in internal tissues and (2) how auxin maxima induce cell specification and patterning.
Here, we describe the NO VEIN (NOV) gene of Arabidopsis thaliana. It encodes a novel plant-specific nuclear factor required for leaf vascular development, cellular patterning and stem cell maintenance in the root meristem, as well as for cotyledon outgrowth and separation. Embryonic apical-basal patterning, vascular continuity, and shoot-derived organ separation become defective when polar auxin transport is additionally impaired in nov-1, either chemically or genetically, further supporting the important role of NOV in auxin-mediated development. nov mutations affect many aspects of auxin-dependent development without directly affecting auxin perception. NOV is required for the cell type–specific expression and polarity of PIN proteins during leaf primordium and root development. NOV is specifically expressed in developing embryos, leaf primordia, lateral root primordia, and the meristematic region of shoots and roots. Our data suggest that NOV function underlies cell fate decisions associated with auxin gradients and maxima, thus establishing PIN expression and polarity in a cell type–specific manner. We propose that NOV is involved in endowing cells with the competence to undergo auxin-dependent coordinated cell specification and patterning, thereby eliciting context-dependent auxin-mediated developmental responses during formation of plant organs and tissues, regardless of their morphology and developmental origin.
RESULTS
NOV Is Required for Vascular Formation in Leaves
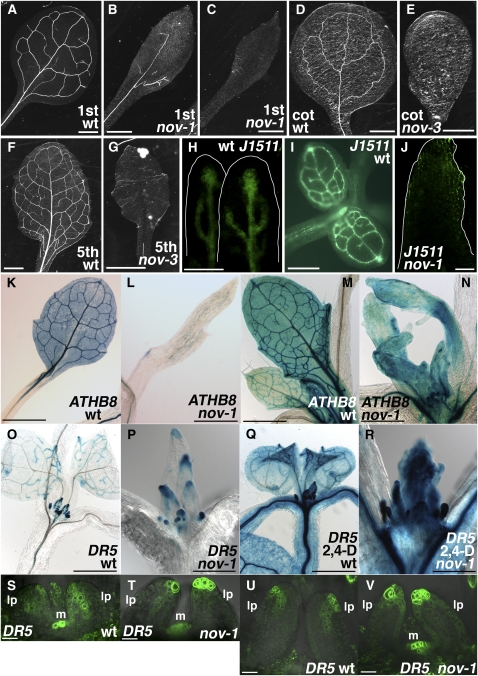
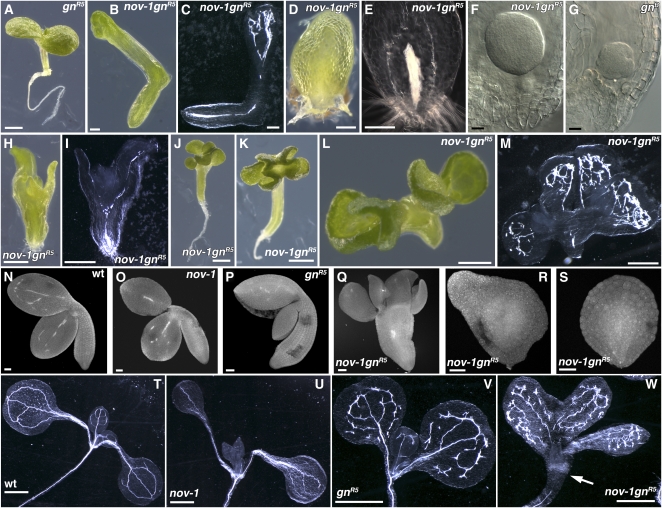
To understand the mechanisms of leaf provascular development, we isolated Arabidopsis mutants defective in leaf vein formation (see Methods). nov-1 was identified as a recessive mutant showing narrow rosette leaves with far fewer veins than those of the wild type (cf. Figures 1B and 1C with 1A). Occasionally, nov-1 plants never formed veins in their first two rosette leaves (Figure 1C). Four T-DNA insertion alleles with stronger phenotypes, nov-2, -3, -4, and -5, were also identified (Figure 8A). Hereafter, all four T-DNA alleles are collectively described as nov-2∼5. While nov-2∼5 usually exhibit an embryo-defective phenotype (Figure 3), nov-3 seedlings were infrequently detected among the progeny of heterozygous nov-3 plants. In nov-3 seedlings, no leaf vein was observed either in the first two rosette leaves, in the cotyledons (cf. Figure 1E with 1D), or in the adult-phase rosette leaves (cf. Figure 1G with 1F). These results suggest that NOV is required for vascular formation in cotyledons and leaves. In nov-1 leaf primordia, the expression of the pre-pro/procambium markers ARABIDOPSIS THALIANA HOMEOBOX 8 (ATHB8) and J1511-green fluorescent protein (GFP) was either downregulated or undetectable (cf. Figures 1J with 1H and 1I; cf. Figures 1L and 1N with 1K and 1M, respectively; cf. Supplemental Figure 1B with 1A online), suggesting that NOV is necessary for the formation of provascular cells in leaves.
Figure 1.
Leaf Phenotype in nov Mutants.
(A) to (G) Leaf venation pattern. Cleared samples of the first or second rosette leaves of 19-d-old wild type (C24 in [A]) and nov-1 ([B] and [C]). Cotyledons and fifth rosette leaves of 38-d-old wild type (Col in [D] and [F]) and nov-3 ([E] and [G]), respectively.
(H) to (N) Expression of provascular markers. Wild-type leaf primordia show J1511-GFP ([H] and [I]) and ATHB8pro:GUS ([K] and [M]) expression in provascular cells. nov-1 develops leaf primordia with no J1511-GFP (J) or ATHB8pro:GUS expression in provascular cells ([L] and [N]).
(O) to (V) Expression of the auxin response marker DR5 in wild type (DR5pro:GUS in [O] and [Q]; DR5pro:GFP in [S] and [U]) and nov-1 (DR5pro:GUS in [P] and [R]; DR5pro:GFP in [T] and [V]). In the presence of the synthetic auxin, 2,4-D, DR5 is similarly expressed in wild type (Q) and nov-1 (R). Samples in (K) to (T) are taken from plants grown at 27°C, as nov-1 phenotype is strengthened at higher temperature. For comparison, ATHB8 expression at 27°C ([L] and [N]) and at 21 to 22°C (see Supplemental Figure 1 online) is shown. lp, leaf primordium; m, shoot apical meristem.
Bars = 1 mm in (A) to (G), (K), and (L), 50 μm in (H) and (J), 0.5 mm in (I), (M) to (O), and (Q), 0.25 mm in (P) and (R), and 20 μm in (S) to (V).
Figure 8.
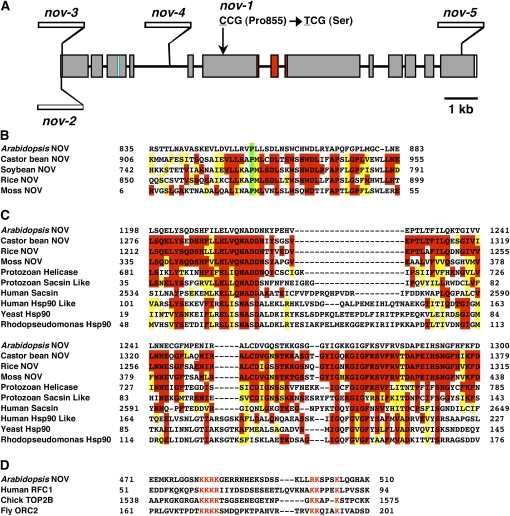
Schematic Structure of NOV Gene and Some Features of NOV Protein.
(A) NOV gene is composed of 13 exons and 12 introns. The missense mutation site in nov-1 is schematically indicated by an arrow. Sites of T-DNA insertions in nov-2, -3, -4, and -5 are shown. Regions coding the putative NLS and the putative GHKL-type ATPase domain are marked in blue and in red, respectively.
(B) Alignment of protein sequences around the 855th Pro residues (marked in green) of NOV homologs. The Pro residue is conserved among NOV homologs in Arabidopsis, castor bean, soybean, rice (O. sativa Japonica group), and moss.
(C) Comparison of the putative GHKL-type ATPase domain in plant NOV homologs, Cryptosporidium parvum superfamily I helicase (Protozoan Helicase), sacsin-like protein (Protozoan Sacsin Like), human sacsin (Human Sacsin), Hsp90-like TNF receptor-associated protein 1 (Human Hsp90 Like), yeast Hsp90 (Yeast Hsp90), and R. palustris Hsp90 (Rhodopseudomonas Hsp90). In (B) and (C), residues of amino acids identical and similar to those of Arabidopsis NOV are marked in red and in yellow, respectively.
(D) The putative NLS in NOV is compared with those that are experimentally confirmed to be a NLS in human replication factor C subunit 1 (RFC1), chicks DNA topoisomerase 2-β (TOP2B), and Drosophila origin recognition complex subunit 2 (ORC2). Amino acid residues marked in red are core sequences of K[RK]{3,5}x{11,18}[RK]Kx{2,3}K motif, where K[RK]{3,5} represents for K followed by 3 to 5 of R or K, x{11,18} for 11 to 18 of any amino acid residue, [RK]K for R or K followed by K and x{2,3}K for two to three of any amino acid residues followed by K.
Figure 3.
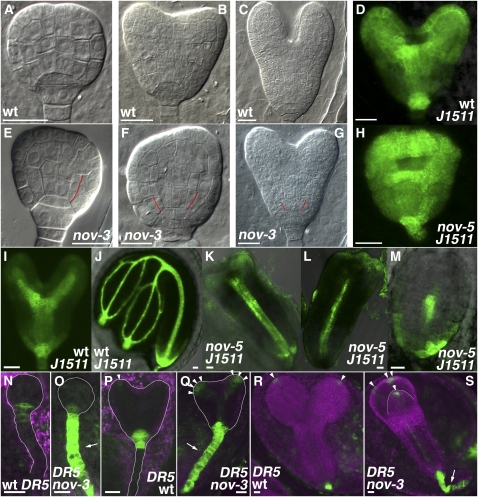
Embryonic Phenotype in nov Mutants.
(A) to (M) Embryos of the wild type ([A] to [D], [I], and [J]), nov-3 ([E] to [G]), and nov-5 ([H] and [K] to [M]). Wild-type embryos at the mid globular (A), triangular to early-heart (B), early-heart to mid-heart (D), mid-heart to late-heart (I), early-torpedo (C), and mature-embryo (J) stages are shown as controls. Embryos of nov-3 and nov-5 are defective in cotyledon growth ([F] to [H] and [K] to [M]) and separation ([G], [H], and [L]) and maintenance of stem cells for cortex/endodermal cells ([E] to [G]; cell boundaries presumably caused by precocious periclinal division of cortex/endodermal cells are indicated by red lines). Embryos in (D) and (H) to (M) harbor J1511-GFP. In nov-5, provascular expression of J1511-GFP in cotyledon primordia is repressed or missing ([K] to [M]).
(N) to (S) DR5pro:GFP expression in embryos of the wild type ([N], [P], and [R]) and nov-3 ([O], [Q], and [S]). In embryos of nov-3, ectopic expression of DR5 was detected in the suspensor (arrows in [O], [Q], and [S]) and tips of cotyledon primordia. DR5 expression in tips of cotyledon primordia is marked by arrowheads in (P) to (S).
Bars = 20 μm.
In wild-type plants, during leaf development, auxin/indole-3-acetic acid-auxin response factor-dependent transcription from the DR5 promoter is detected in provascular cells (Ulmasov et al., 1997; Mattsson et al., 2003; Figure 1O). In nov-1, DR5 expression in leaves was disrupted (cf. Figure 1P with 1O; cf. Figures 1T and 1V with 1S and 1U, respectively) and often confined to the epidermis of the leaf margin (Figures 1P and 1V). However, the ability to respond to exogenous auxin in nov-1 was comparable to that in the wild type (cf. Figure 1R with 1Q), indicating that NOV is not necessary for auxin/indole-3-acetic acid-auxin response factor–dependent gene expression. These data suggest that NOV is required for proper auxin distribution during leaf development.
NOV Is Required for Root Development
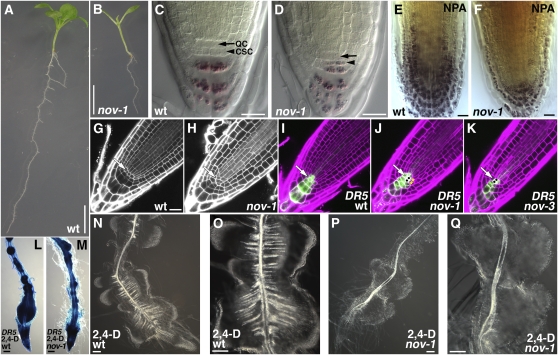
Roots of nov-1 were shorter than those of the wild type (cf. Figure 2B with 2A), implying that NOV also functions in root development. Compared with the wild type (Figures 2G and 2I), cellular organization of the root meristem was disarranged in nov-1 (Figures 2H and 2J) and nov-3 (Figure 2K). An auxin maximum, which establishes a distal organizer in the root (Sabatini et al., 1999), was detected at the quiescent center in nov-1 (Figure 2J) and nov-3 (Figure 2K) mutants, as in the wild type (Figures 2I). These results suggest that NOV is required for auxin-mediated cellular patterning in the root tip.
Figure 2.
Root Phenotype in nov Mutants.
(A) and (B) Seedlings of wild type (A) and nov-1 (B) grown for 12 d.
(C) and (D) Starch staining of root tips of the wild type (C) and nov-1 (D). Arrows and arrowheads indicate positions of the quiescent center (QC) and columella stem cells (CSC), respectively.
(E) and (F) Effect of NPA on root tips of the wild type (E) and nov-1 (F). Root tips were stained for starch.
(G) and (H) Propidium iodide staining of root tips of the wild type (G) and nov-1 (H).
(I) to (K) DR5pro:GFP expression in root tips of the wild type (I), nov-1 (J), and nov-3 (K). Cells resulting from an ectopic division in the quiescent center and cortex/endodermis stem cell are marked by black dots in (J) and (K) and by asterisks in (J), respectively. White arrows in (G) to (K) indicate positions of the quiescent center
(L) to (Q) Effect of the synthetic auxin 2,4-D on roots of the wild type ([L], [N], and [O]) and nov-1 ([M], [P], and [Q]). DR5pro:GUS expression in roots of the wild type (L) and nov-1 (M) treated with 2,4-D.
Bars = 5 mm in (A) and (B), 20 μm in (C) to (G) (equal scale in [G] to [K]), and 100 μm in (L) to (Q).
Mature columella root cap cells contain starch granules that are not seen in their stem cells (van den Berg et al., 1997; Figure 2C; see Supplemental Figure 2A online). In nov-1 (Figure 2D) and nov-3 (see Supplemental Figure 2B online), starch granules were also detected in columella stem cells, suggesting that NOV is required for columella stem cell maintenance. The maintenance of cortex/endodermis stem cells requires two successive cell divisions with the cortex/endodermis stem cell first undergoing an anticlinal cell division. The basal daughter cell is then maintained as a stem cell, and the apical cell divides periclinally, giving rise to cortex and endodermis cells. In nov-1 (Figure 2J, asterisks) and nov-3 (see Supplemental Figure 3B online), the cortex/endodermis stem cells underwent a periclinal division without a prior anticlinal division (cf. Figures 2G and 2I; see Supplemental Figure 3A online; see also Figures 3E to 3G). Therefore, NOV is required to maintain root stem cells for the cortex and endodermis. We observed ectopic divisions in the quiescent center in nov-1 and nov-3 (Figures 2J and 2K, black dots), presumably replacing the stem cells that failed to be maintained (see Supplemental Figure 5 online).
In wild-type plants, exogenous application of the synthetic auxin 2,4-D induced the formation of lateral root primordia, most of which were fused. These primordia had more xylem strands than the primary root (Figures 2N and 2O). Although 2,4-D also induced the formation of lateral root primordia in nov-1, xylem strand formation was either absent or greatly repressed in 2,4-D–induced lateral roots (Figures 2P and 2Q). There was no significant difference in auxin response (as detected by DR5 expression) between wild-type and nov-1 plants (Figures 2L and 2M). These results suggest that NOV is required for auxin-mediated vascular formation not only in leaves but also in roots.
NOV Is Required for Embryonic Development
NOV is required for auxin-mediated development in the leaf (Figure 1) and root (Figure 2), suggesting that NOV has a common function in auxin-mediated developmental processes. Consistent with this idea, the stronger T-DNA insertion alleles, nov-2∼5 (described below; Figure 8A), exhibited an embryo-defective phenotype characterized by size reduction and frequent fusion of cotyledons (Figures 3F to 3H and 3K to 3M; cf. developing wild-type embryos in Figures 3A to 3D, 3I, 3J, 3N, 3P, and 3R) and abnormalities in the early steps of vascular development (see Supplemental Figure 4 online). A similar phenotype has been observed in mutants either defective in polar auxin transport (Steinmann et al., 1999; Friml et al., 2003; Vieten et al., 2005) or auxin signaling (Hardtke et al., 2004).
The expression pattern of the pre-pro/procambium marker J1511-GFP in nov-5 embryos further indicated fusion of cotyledons (cf. Figure 3H with 3D, early-heart to mid-heart stage embryos) and defective provascular development (cf. Figures 3K to 3M with 3I and 3J, mid-heart to late-heart stage and mature embryos). In nov-5 embryos in which the cotyledons did not grow, expression of J1511-GFP seen in the apical part of wild-type embryos (Figures 3D, 3I, and 3J) was missing or greatly repressed (Figure 3M), suggesting that NOV is required for J1511-GFP expression in the apical part of embryos, which may correlate with development of cotyledon primordia. On the other hand, nov-5 embryos showing some outgrowth of cotyledons exhibited J1511-GFP expression in the apical part, although provascular J1511-GFP expression tended to be missing or greatly repressed (Figures 3K and 3L). These results suggest that NOV is required for provascular formation in the apical part of embryos, which may promote development of cotyledon primordia. In wild-type embryos, from around the triangular stage, DR5 expression is found mainly in the tips of cotyledon primordia and the uppermost cell of the suspensor (Benková et al., 2003; Friml et al., 2003; Figures 3N, 3P, and 3R). In nov-3, however, the area of DR5 expression was expanded in the tips of cotyledon primordia and encompassed the entire suspensor (Figures 3O, 3Q, and 3S), suggesting that NOV is also required for proper auxin distribution in embryonic development.
In nov-2∼5 embryos, cortex/endodermis stem cells often underwent periclinal division without a prior anticlinal division and were lost (cf. Figures 3E to 3G with 3A to 3C; cf. Supplemental Figure 5C with 5A online). As was observed in roots, an ectopic division was observed in the quiescent center (cf. Supplemental Figures 5C and 5D with 5A and 5B online). Judging from the expression of SCR and SHORT ROOT (SHR) (see Supplemental Figure 6 online), radial patterning appears to be normal in nov-3 embryos.
Genetic Interaction of NOV and GNOM
GNOM is required for the proper subcellular localization of PIN proteins and therefore for polar auxin transport (Steinmann et al., 1999; Geldner et al., 2003; Kleine-Vehn et al., 2008a). To study the role of NOV in polar auxin transport-dependent organ development, the genetic interaction between NOV and GNOM was assessed. gnomR5 is a weak allele, invariably producing primary roots and displaying partial or complete fusion of cotyledons in about one-third of seedlings (Geldner et al., 2004; Figures 4A, 4P, and 4V). nov-1 gnomR5 occasionally lacked the root (Figures 4B to 4F, 4H, 4I, 4L, 4M, and 4R) and/or cotyledon (Figures 4D to 4F, 4R, and 4S) and resembled strong gnom alleles defective in apical-basal polarity (Mayer et al., 1993; Figure 4G). For comparison, plants of the wild type (Figures 4N and 4T) and nov-1 (Figures 4O and 4U) are shown. Also segregated among nov-1 gnomR5 plants were seedlings with four to six cotyledons that were, to a greater or lesser extent, fused to each other (Figures 4H to 4M and 4Q). This phenotype was not seen in nov-1, -2, -3, -4, or -5, and neither has it been reported in gnom alleles, suggesting that NOV and GNOM are required redundantly for the determination of cotyledon number. A similar phenotype was observed in the double mutant of nov-1 with another weak allele, gnomB/E (Geldner et al., 2004) (see Supplemental Figure 7 online). Defects in xylem strand continuity in gnomR5 (Geldner et al., 2004; Figure 4V) were enhanced in nov-1 gnomR5 (Figures 4C, 4I, 4M, and 4W). Here, in seedlings that produced roots, not only was xylem strand continuity repressed in the cotyledon, but also xylem differentiation was repressed at the cotyledon-hypocotyl junction (Figure 4W, arrow), where the protoxylem first differentiates (Pyo et al., 2004; cf. Figures 4T to 4V). All phenotypes described indicate that NOV is genetically redundant to GNOM in organ and tissue development associated with polar auxin transport.
Figure 4.
Embryonic and Seedling Phenotype in the nov-1 gnomR5 Double Mutant.
Embryonic and seedling phenotype in the wild type ([N] and [T]), nov-1 ([O] and [U]), weak allele gnomR5 ([A], [P], and [V]), strong allele gnomB (G), and nov-1 gnomR5 ([B] to [F], [H] to [M], [Q] to [S], and [W]). Samples were taken from seedlings ([A] to [E], [H] to [M], and [T] to [W]), developing ovules ([F] and [G]), and imbibed seeds ([N] to [S]). nov-1 gnomR5 is shown with either strong phenotype ([B] to [F], [H] to [M], and [Q] to [S]) or a relatively weak phenotype (W). Samples in (B), (D), (H), and (L) were cleared and shown in (C), (E), (I), and (M), respectively. nov-1 gnomR5 with strong phenotype failed to develop the primary root ([B] to [F], [H], [I], [L], [M], and [R]), cotyledons ([D] to [F], [R], and [S]), and, in some cases, developed multiple cotyledons either fused or not ([H] to [M] and [Q]). In nov-1 gnomR5 with a weak phenotype, development of lignified xylem strands is repressed at the cotyledon-hypocotyl junction (an arrow in [W]). gnR5, gnomR5; gnB, gnomB. Bars = 1 mm in (A), (J), (K), and (T) to (W), 0.2 mm in (B) to (E), 20 μm in (F) and (G), 0.5 mm in (H), (I), (L), and (M), and 50 μm in (N) to (S).
The Effect of Inhibition of Polar Auxin Transport on the nov-1 Mutant
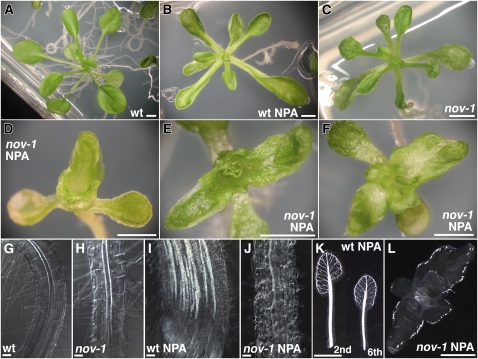
Further analysis of the roles of NOV in auxin-mediated development focused on the effect of 1-naphthylphthalamic acid (NPA), an inhibitor of polar auxin transport, in nov-1. In untreated plants, both leaf blades and petioles were significantly shorter in nov-1 when compared with the wild type (cf. Figure 5C with 5A; see Supplemental Table 1 online). The exogenous application of NPA exacerbated the difference in petiole length between the wild type and nov-1 (cf. Figures 5D to 5F with 5B). While leaf petioles of wild-type plants treated with NPA were shorter than those of untreated plants (Figures 5A and 5B; see Supplemental Table 1 online), in most nov-1 plants after NPA application (20/24 plants examined), the petiole was not recognizable (Figures 5D to 5F; see Supplemental Table 1 online). After NPA application, the fusion of rosette leaves was infrequently seen in wild-type plants (8/95 plants examined). In nov-1, however, fusion of rosette leaves was significantly induced (64/64 plants examined) (Figures 5D to 5F). In the wild type, NPA application greatly induced the formation of vascular strands in the leaf lamina and petioles, and pronounced unbroken vascular strands extended along the entire margin of the leaf lamina (Mattsson et al., 1999; Figure 5K). By contrast, in nov-1, increased formation of vascular strands did not occur, but rudimentary isolated vascular strands were discontinuously formed only in the marginal regions (Figure 5L).
Figure 5.
Phenotype of nov-1 Seedlings Treated with NPA.
(A) to (F) Aerial tissue phenotype. Wild type (A) and nov-1 (C) grown in the absence of NPA. The wild type (B) and nov-1 ([D] to [F]) grown in the presence of NPA.
(G) to (J) Root vascular tissue phenotype. In roots, increased formation of vascular strands seen in NPA-treated wild-type roots (I) is strongly repressed or abolished in nov-1 (J). Images in (G) to (J) are regions of cleared roots where the protoxylem cells have started to become lignified.
(K) and (L) Leaf vascular tissue phenotype. Shown in (K) are cleared samples of the second and sixth rosette leaves of the NPA-treated wild type. The NPA-treated nov-1 seedling in (F) was trimmed cotyledons, cleared and shown in (L). Seedlings were grown in the absence ([A], [C], [G], and [H]) or presence of NPA ([B], [D] to [F], and [I] to [L]) for 21 d. The wild type ([A], [B], [G], [I], and [K]) and nov-1 ([C] to [F], [H], [J], and [L]).
Bars = 2 mm in (A) to (F), (K), and (L) and 50 μm in (G) to (J).
In wild-type roots, NPA induces cell fate changes in epidermal and cortical cells, for example, inducing the development of starch-containing amyloplasts and a broadening of the columella domain (Sabatini et al., 1999; Blilou et al., 2005; Figure 2E). NPA treatment also increases vascular formation in the stele (Mattsson et al., 1999; Sabatini et al., 1999; cf. Figure 5I with 5G). In nov-1, cell fate changes in epidermal and cortical cells were much less pronounced, and broadening of the columella domain was repressed (Figure 2F). Furthermore, increased formation of vascular strands was strongly repressed or not seen (cf. Figure 5J with 5H for untreated nov-1 and with 5I for NPA-treated wild type). Therefore, the nov-1 mutation does not simply enhance the effect of NPA. When polar auxin transport is inhibited in nov-1 by NPA, certain aspects of the NPA-induced phenotype are enhanced, such as leaf organ fusion and the suppression of leaf petiole elongation. On the other hand, other aspects, such as NPA-induced vascular formation and root cell fate changes, are strongly repressed. These observations support the hypothesis that NOV is required for cell specification and patterning events associated with polar auxin transport.
Formation of Provascular Cells for the Midvein in Leaf Primordia
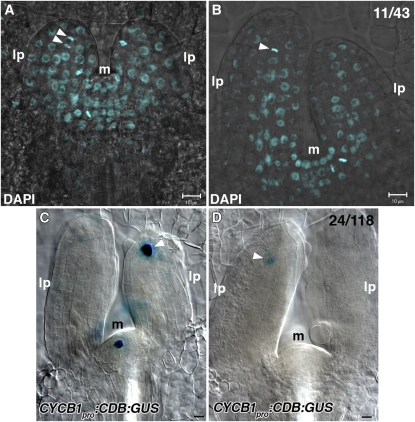
In wild-type plants, PIN1 expression in leaf provascular cells was first detected close to the center of young primordia, aligned toward the hypocotyl (Figures 6A and 6C). At the apical end of the provascular PIN1 expression domain, almost isodiametric cells exhibited nonpolar or less polar localization of PIN1 with more or less enhanced basal localization (Figure 6A, asterisks; see Supplemental Figure 8 online). A gradual shift from nonpolar to basal localization of PIN1 was observed from the apical to basal side of the midvein provascular strand, which accompanied gradual cell elongation and differentiation into procambial cells (Figures 6A and 6C). These observations raise the possibility that the apical isodiametric cells have progenitor cell–like properties and divide transversely to produce apical daughter cells to be self-maintained as vascular progenitor cells and basal daughter cells to mature into provascular cells. In this scenario, as leaf primordia grow and increase in cell number along the apical-basal axis, the putative vascular progenitor cells should divide transversely. Using 4',6-diamidino-2-phenylindole (DAPI), which allows detection of condensed nuclear DNA in the M phase, and cyclin B1 promoter:cyclin B1 destruction box:β-glucuronidase (CycB1pro:CDB:GUS), which marks the late G2 to M phase of cell cycle (Colón-Carmona et al., 1999), activity of cell division was examined in 3-d-old wild-type leaf primordia. Cell division activity was detected in the putative vascular progenitor cells for the midvein (Figure 7). DAPI staining allows us to predict the orientation of division plane as judged by direction of alignment of DAPI-stained condensed nuclear DNA. Ten out of the 11 division planes were transverse (Figures 7A and 7B). A transverse division plane accumulating PIN1 protein was also detected (Boutté et al., 2005; Figure 6A, arrow). These results suggest that cells at the apical end of midvein provascular cells have potential characters of midvein progenitor cells.
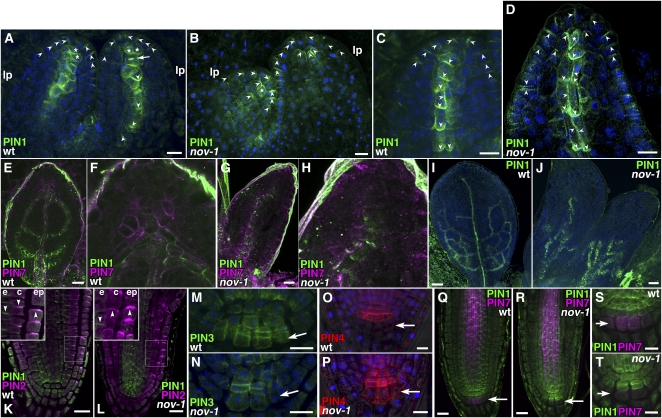
Figure 6.
PIN Expression and Localization in the Wild Type and nov-1.
(A) to (J) Immunostaining of PIN1 and PIN7 in leaf primordia. PIN1 (green in [A] to [J]), PIN7 (magenta in [E] to [H]) and DAPI (blue in [A] to [D], [I], and [J]). Images were obtained from the first two leaf primordia of the wild type ([A], [C], [E], [F], and [I]) and nov-1 ([B], [D], [G], [H], and [J]). Asterisks indicate cells with nonpolar PIN1. An arrow in (A) indicates PIN1 accumulation at a transverse cell division plane. lp, leaf primordium.
(K) and (L) Immunostaining of PIN1 and PIN2 in the root tip. Double staining of PIN1 (green) and PIN2 (magenta) in root tips of the wild type (K) and nov-1 (L). Insets show boxed areas enlarged. e, endodermis; c, cortex; epi, epidermis.
(M) and (N) Immunostaining of PIN3 in the root tip. PIN3 (green) and DAPI (blue) in the root stem cell niche of the wild type (M) and nov-1 (N).
(O) and (P) Immunostaining of PIN4 in the root tip. PIN4 (red) and DAPI (blue) in the root stem cell niche of the wild type (O) and nov-1 (P).
(Q) to (T) Immunostaining of PIN1 and PIN7 in the root tip. Double staining of PIN1 (green) and PIN7 (magenta) in root tips of the wild type ([Q] and [S]) and nov-1 ([R] and [T]). Arrowheads indicate the polarity of PIN proteins for clarity. Arrows in (M) to (T) indicate the tier three of columella root cap cells.
Bars = 10 μm in (A) to (D), (M) to (P), (S), and (T), 20 μm in (E), (G), (K), (L), (Q), and (R), and 50 μm in (I) and (J).
Figure 7.
DAPI Staining and CYCB1pro:CDB:GUS Expression in Wild-Type Leaf Primordia.
(A) and (B) Wild-type leaf primordia were stained with DAPI. Arrowheads indicate transversely aligned condensed nuclear DNA at the apical ends of provascular cells for the midvein.
(C) and (D) The wild type containing CYCB1pro:CDB:GUS was examined for GUS activity. Arrowheads indicate GUS expression at the apical ends of provascular cells for the midvein. All images were taken from the lateral view of leaf primordia. Shown in the top left corner of (B) and (D) are fractions of primordia with displayed features, respectively.
lp, leaf primordium; m, shoot apical meristem. Bars = 10 μm.
In epidermal cells of nov-1 leaf primordia, PIN1 is expressed and localized apically as in the wild type (Benková et al., 2003; Reinhardt et al., 2003; Figures 6B and 6D). Cells with opposite PIN1 polarity were also detected in the epidermis, suggesting that the convergence point of auxin flow is present in nov-1 leaf primordia. However, provascular PIN1 expression was impaired in nov-1, leading to the complete loss of provascular PIN1 expression (Figure 6B). Only in the tip region of leaf primordia was subepidermal PIN1 expression detected, in cells that elongated along the apical-basal axis with apical PIN1 polarity (Figure 6B). When PIN1 expression was detected in provascular cells (Figure 6D), cell elongation occurred much closer to the apical side compared with the wild type (Figure 6C), suggesting precocious vascular differentiation. The most apical provascular cells also elongated. Basal localization of provascular PIN1 was more or less repressed. Ectopic expression of PIN1 with apical polarity was found in the apical region of the ground tissue (Figures 6B and 6D).
As leaf primordia develop in the wild type, PIN1 is expressed in the looped secondary and tertiary provascular tissues, as well as to a lesser extent in the midvein provascular tissue (Scarpella et al., 2006; Figures 6E and 6I). At this stage, PIN7 was expressed in the region apically adjacent to the distal end of the midvein provascular strand and the region around the border of differentiating midvein procambial cells (Figures 6E and 6F), suggesting that PIN7 plays a role in auxin flow into provascular tissue. In nov-1 at the corresponding stage, we detected both leaf primordia without provascular PIN1 expression and leaf primordia with a rudimentary provascular PIN1 expression domain in the basal region of leaf primordia (Figures 6G and 6J). Leaf primordia without a PIN7 expression domain apically adjacent to the distal end of the midvein provascular strand were also seen in nov-1 (Figures 6G and 6H). Taken together, these results suggest that NOV is required from the initial stage of provascular formation and throughout the gradual differentiation of provascular cells. Furthermore, NOV is required for the development of surrounding ground tissues and, therefore, indirectly regulates auxin flow in developing leaf primordia.
Expression and Subcellular Localization of PIN Proteins Are Altered in nov-1 Root Tips
The requirement of NOV for the correct expression and polarity of PIN proteins was also tested in roots. In contrast with leaf primordia, in roots, no obvious difference in expression and polarity of PIN1 was detected between the wild type and nov-1 (Figures 6K, 6L, and 6Q to 6T). However, expression and polarity of other PIN members were affected in nov-1. In wild-type root tips, PIN2 is polarized apically in the epidermis and basally in the cortex (Friml et al., 2004; Figure 6K). In nov-1, PIN2 polarity in the cortex was not basal, but apical or nonpolar (Figure 6L). In the epidermis, PIN2 polarity was apical both in the wild type and nov-1. These indicate that NOV is essential for basal PIN2 polarity in the cortex but not for apical polarity in the epidermis. In the wild type, expression of PIN3 and PIN7 in columella cells is detected in tiers two and three of columella cells (Blilou et al., 2005; Paponov et al., 2005; Figures 6M, 6Q, and 6S). In nov-1, expression of PIN3 and PIN7 in these columella cells was decreased or almost absent (Figures 6N, 6R, and 6T). In the wild-type columella root cap, PIN4 expression is confined to tiers one and two of columella cells (Friml et al., 2002a; Blilou et al., 2005; Paponov et al., 2005; Figure 6O). In nov-1, PIN4 expression was expanded to the third tier of columella cells and possibly to the lateral root cap (Figure 6P). Differences in expression of PIN3, PIN4, and PIN7 in other tissues were not detected between the wild type and nov-1 (Figures 6Q to 6T for PIN7). Nor was any clear difference found in the polarity of PIN3, PIN4, and PIN7 in any tissues. These data indicate that NOV is required for proper cell-specific expression of PIN3, PIN4, and PIN7 in the columella root cap.
NOV Encodes a Novel, Plant-Specific Protein
NOV (At4g13750) was identified by map-based cloning. NOV comprises 13 exons and 12 introns (Figure 8A) and encodes a novel protein of a predicted 2729 amino acids. The missense mutation in nov-1 causes substitution of an amino acid from Pro to Ser, the 855th amino acid from the N terminus (Figures 8A and 8B). Four T-DNA insertion lines were identified: nov-2, nov-3, nov-4, and nov-5 (Figure 8A). All were allelic to nov-1. In Arabidopsis, another gene (At1g08300) designated NO VEIN-LIKE (NVL) was found to encode a protein of 746 amino acids, which is mainly composed of domains similar to the N-terminal (197 to 598) and C-terminal (2507 to 2729) parts of NOV with 72 and 69% sequence similarity (percentage in amino acid identity), respectively. NOV homologs were found in other plants, including rice (Oryza sativa), soybean (Glycine max), castor bean (Ricinus communis), and moss (Physcomitrella patens; Figures 8B and 8C) but not in animals, insects, yeasts, and bacteria, suggesting that NOV is specific to the plant kingdom. Sequence similarity of Arabidopsis NOV to other plant NOV homologs is 40% to rice NOV (2821 amino acids), 47% to soybean NOV (2711 amino acids), 48% to castor bean NOV (2833 amino acids), and 35% to moss NOV (1962 amino acids). The Physcomitrella NOV homolog does not contain a recognizable N-terminal domain. The aforementioned Pro residue, at which substitution to Ser occurs in nov-1, is conserved among the plant NOV homologs (Figure 8B). The deduced amino acid sequence of the NOV protein includes a putative GHKL-type-ATPase domain, which has been found in diverse protein families such as DNA gyrase, Hsp90, histidine kinase, and DNA mismatch repair enzyme MutL (Dutta and Inouye, 2000). Figure 8C shows a comparison among the putative GHKL-type-ATPase domains of NOV, plant NOV homologs, protozoan (Cryptosporidium parvum) superfamily I helicase and sacsin-like protein, human sacsin and Hsp90-like protein, yeast (Saccharomyces cerevisiae) Hsp90, and bacterial (Rhodopseudomonas palustris) Hsp90. It must be pointed out that no protein, other than plant NOV homologs and NVL, currently listed in publicly available databases has a significant similarity to NOV. A putative nuclear localization signal (NLS) found in NOV was similar to those that have been experimentally confirmed to be an NLS in human replication factor C subunit 1, chicks DNA topoisomerase 2-β, and fruitfly (Drosophila melanogaster) origin recognition complex subunit 2 (http://cubic.bioc.columbia.edu/services/predictNLS/; Figure 8D).
NOV Is a Nuclear Protein and Specifically Expressed in Developing Organs
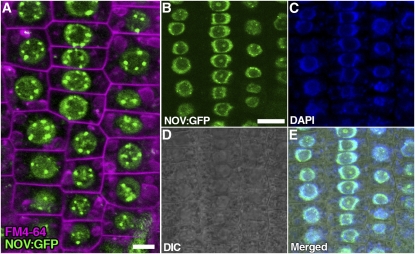
We created transgenic plants in which NOV:GFP, NOV:GUS, and GFP:NOV translational fusion genes are expressed under control of the NOV promoter. NOVpro:NOV:GFP and NOVpro:GFP:NOV were able to complement nov-1 and nov-3, indicating that NOV:GFP and GFP:NOV are functional. Both NOV:GFP (Figures 9, 10B, 10C, and 10I to 10N) and GFP:NOV (see Supplemental Figure 9 online) were specifically localized in the nucleus, with GFP fluorescent signals of higher intensity detected as speckles within the nucleus and the nucleolus, suggesting that NOV is a nuclear protein and may be localized to a specific subnuclear structure.
Figure 9.
Nuclear Localization of NOV:GFP.
(A) to (E) Subcellular localization of NOV:GFP was examined in root epidermal cells of seedlings containing NOVpro:NOV:GFP (NOV:GFP). Shown in (A) is a confocal image of root epidermal cells for NOV:GFP (green) and FM4-64 staining (magenta, plasma and endocytic membranes). NOV:GFP (green in [B]), DAPI (blue in [C]), differential interference contrast (DIC) optic image (D), and merged image (E) are also shown. NOV:GFP is specifically localized in the nucleus. Bars = 5 μm in (A) and 10 μm in (B) (equal scale in [B] to [E]).
Figure 10.
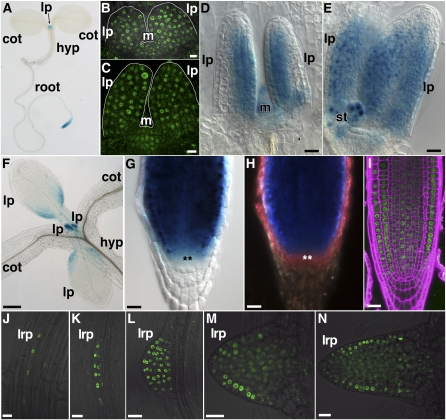
Expression of NOV in Seedlings, Leaf Primordia, and Roots.
(A) to (F) Expression of NOVpro:NOV:GUS ([A] and [D] to [F]) and NOVpro:NOV:GFP ([B] and [C]) in seedlings. A whole seedling (A) and leaf primordia ([B] to [F]). Images in (B) to (D) were taken from the lateral view of leaf primordia. Leaf primordia shown in (A) to (E) and in (F) are of the first and second rosette leaves and of the first to fourth rosette leaves, respectively.
(G) to (N) Expression of NOVpro:NOV:GUS ([G] and [H]) and NOVpro:NOV:GFP ([I] to [N]) in roots. Primary root tips ([G] to [I]) and lateral root primordia ([J] to [N]). (H) is a dark-field image of the sample in (G). In (G) and (H), cells of the quiescent centers are marked by asterisks. The root tip in (I) was stained with propidium iodide (magenta) for detecting cell boundary. Samples were taken from seedlings grown for 3 d (B), 3.5 d (C), 4 d (A), 5 d ([D], [E], and [I]), 7 d ([F] to [H] and [L] to [N]), and 8 d ([J] and [K]) after germination. cot, cotyledon; hyp, hypocotyl; lp, leaf primordium; lrp, lateral root primordium; m, shoot apical meristem; st, stipule.
Bars = 10 μm in (B), (C), and (J) to (N), 20 μm in (D), (E), and (G) to (I), and 0.2 mm in (F).
To clarify further NOV function, the NOV expression pattern was analyzed. In shoots, NOV was expressed in developing leaf primordia and weakly in the shoot apical meristem (Figures 10A to 10F). This expression pattern is supported by a recent comprehensive study of gene expression in the Arabidopsis shoot apical meristem stem cell niche (Yadav et al., 2009). NOV was initially expressed throughout young leaf primordia (Figures 10B and 10C). As leaf primordia develop, NOV expression became restricted to the adaxial side of primordia and then toward the basal side (Figures 10D to 10F). We detected NOV expression in developing embryos and postembryonic NOV expression in provascular cells of cotyledons at the early stage of vascular differentiation (see Supplemental Figure 10 online). In roots, NOV was expressed in the root apical meristem with weak expression in the quiescent center and columella initials but not in mature root tissues (Figures 10A and 10G to 10I). NOV was also expressed during primordial development of lateral roots (Figures 10J to 10N). Consistent with the nov phenotype, NOV is expressed in developing organs and tissues especially at their early developing and differentiating stages.
DISCUSSION
We have shown here a function, mediated by the NOV gene, regulating PIN expression and polarity in a cell type–specific manner. nov mutations affect many aspects of auxin-dependent development without directly affecting auxin perception. Our data suggest that NOV functions in potentiating cells for competence to undergo auxin-dependent coordinated cell specification and patterning, thereby establishing expression and polarity of PIN proteins.
The Role of NOV in Auxin-Mediated Vascular Formation
Epidermal auxin convergence at the tip of an incipient primordium precedes primordium outgrowth (Reinhardt et al., 2000; Heisler et al., 2005). After this convergence, auxin is thought to be channeled from the marginal epidermis into internal tissues in a process that triggers PIN1 expression and polarization in the ground tissue and leads to the formation of provascular cells. This process has also been proposed to instruct the gradual selection of narrow strands of PIN1-expressing provascular cells in developing leaves (Scarpella et al., 2006). Our data indicate that midvein provascular cells become progressively more differentiated toward the basal side of leaf primordia. Relatively isodiametric cells with nonpolar PIN1 at the apical end of the midvein provascular strand divide transversely, stacking daughter cells along the apical-basal axis. A new basal daughter cell therefore abuts both a new apical daughter cell and an older basal daughter cell, which has already initiated gradual basal polarization and differentiation. Thus, in addition to the auxin gradient, the automatic placement of new basal daughter cells into an existing cell polarity gradient may contribute to its position-dependent polarity. It may also serve to initiate developing vascular cells' gradual differentiation, connecting production of preprocambial cells with their polarity acquisition and continuity. These data suggest that the cells with nonpolarly localized PIN1 in the tip of leaf primordium are progenitor cells for the midvein. This environment with an auxin maximum resembles that of the root stem cell niche, although the auxin flow is topologically opposite.
It has been recently reported that, before leaf primordia bulge out from the surface of the meristem, subepidermal PIN1 is transiently polarized toward the PIN1 convergence point in the epidermis in a process that could underlie the formation of the auxin maximum at the tip of the incipient primordium (Bayer et al., 2009). Subsequently, in bulging leaf primordia, midvein provascular cells with basal PIN1 polarity emerge. In nov-1 leaf primordia, subepidermal PIN1 expression and polarity are impaired to such an extent that provascular PIN1 expression is completely missing, and though PIN1 in the apical subepidermal region is apically polarized, epidermal PIN1 expression and polarity remain normal. Consistent with these data, in nov-1 leaf primordia, DR5 expression tends to be confined to the marginal epidermis, especially at the tip. Furthermore, the first two rosette leaves in nov-1 were occasionally observed entirely without veins. These leaves developed from primordia without provascular tissues (as judged by expression of J1511-GFP and ATHB8). It is worth noting here that the apical polarity of subepidermal PIN1 in nov-1 primordia is topologically similar to the aforementioned apical polarity of subepidermal PIN1 seen in wild-type incipient primordia before they express PIN1 in provascular cells. Our observations suggest that NOV is required for provascular PIN1 expression and polarization in the ground meristem after an auxin convergence point is formed in the epidermis at the tip of an incipient primordium. Furthermore, subsequent termination of leaf provascular formation and repressed basal polarization of midvein provascular PIN1 also appear to occur in nov-1. In nov-1 and nov-3 leaves, ground tissue cells, where the vascular path might be predicted in the wild type, either do not include elongated vascular cells or include elongated cells with or without differentiated xylem cells (see Supplemental Figure 11 online). These data suggest that NOV is required for provascular formation throughout leaf primordial development for the proper differentiation of provascular cells and their development into xylem cells. This hypothesis is further supported by the fact that NOV is postembryonically expressed in provascular cells of cotyledons at early stages of vascular differentiation.
In nov-1, the 2,4-D–induced lateral root failed to develop vascular cells despite the presence of a functional auxin response. When grown in the presence of NPA, an inhibitor of polar auxin transport, the increased formation of vascular tissue seen in leaves and roots of wild-type seedlings is greatly repressed in nov-1, although organ fusion is greatly enhanced. NOV is also required for early provascular development during embryogenesis. These results suggest that NOV facilitates cells' acquisition and maintenance of the competence to differentiate into vascular cells in response to auxin. We therefore suggest that in leaf primordia, NOV establishes provascular expression and polarity of PIN1.
Vascular continuity is achieved through reiterative coordinated cell polarization for provascular development. Vascular continuity defects seen in gnomR5 are enhanced in nov-1 gnomR5 seedlings, further indicating that NOV is important for vascular development. GNOM is directly involved in regulated intracellular vesicle trafficking of PIN1 (Geldner et al., 2003), a function that is considered to be instrumental for the coordinated polarization of PIN1 (Steinmann et al., 1999; Geldner et al., 2004; Kleine-Vehn et al., 2008a). Therefore, it is likely that in nov-1 gnomR5, auxin-mediated vascular development is synergistically impaired both by a decreased competence of cells for vascular differentiation due to the nov-1 mutation and by defective coordinated polarization of PIN1 due to the gnomR5 mutation.
The Role of NOV in Auxin-Mediated Organ Formation during Embryonic and Leaf Development
It has been shown that efflux-dependent auxin gradients are required for apical-basal patterning during embryonic development and that both the pin7 mutation and NPA treatment disrupts the apical-basal pattern of auxin distribution in the embryo, giving an ectopic auxin response in the suspensor (Friml et al., 2003). nov-3 embryos, whose phenotype is similar to those of mutants defective in efflux-dependent polar auxin transport (Steinmann et al., 1999; Friml et al., 2003; Vieten et al., 2005), exhibit an ectopic auxin response in the tips of cotyledon primordia and in the suspensor. These suggest that efflux-dependent auxin gradients are impaired in embryos of strong nov alleles, nov-2∼5. Thus, NOV may be required for establishment of PIN expression and polarity during embryonic development.
Expression of J1511-GFP was impaired in nov-1 leaf primordia and nov-5 embryos. In the J1511 line, an enhancer-trap T-DNA (Laplaze et al., 2005) was found to be inserted in an intergenic region between At1g19840 (11.4 kb away) and At1g19850 (2.1 kb away). At1g19840 encodes an auxin-responsive family protein and At1g19850 encodes MONOPTEROS (MP). In fact, the spatial distribution of J1511-GFP expression is similar to that of MP mRNA (Hardtke and Berleth, 1998; Hardtke et al., 2004; Wenzel et al., 2007). During embryogenesis, J1511-GFP is almost uniformly expressed from the mid to late globular stages (see Supplemental Figure 12A online). The expression starts to become restricted to the central region from around the heart stage (Figures 3D and 3I; see Supplemental Figure 12B online) and is then confined to the provascular tissue and root cap at later stages of development (Figure 3J; see Supplemental Figures 12C and 12D online). These data suggest that the expression of J1511-GFP may reflect that of MP, which is known to partially regulate the provascular expression of PIN1 in the leaf primordium (Wenzel et al., 2007). It was previously reported that a strong loss-of-function mp mutant, mpG12, exhibits no cotyledon growth and has discontinuous vascular strands and that MP antisense lines have fewer higher order veins in rosette leaves (Hardtke et al., 2004). nov-5 embryos devoid of cotyledon do not express J1511-GFP in the apical part of embryos. nov-5 embryos with some outgrowth of cotyledons exhibit J1511-GFP expression in the apical part, although provascular J1511-GFP expression tends to be missing or strongly repressed. nov-1 leaf primordia also exhibit loss of provascular J1511-GFP expression. Thus, if expression of J1511-GFP reflects that of MP, NOV function may partly underlie the establishment of MP expression, thereby supporting provascular development in cotyledons and leaf primordia.
In nov-1 gnomR5 embryos and seedlings, apical-basal patterning defects, including complete loss of the cotyledon and/or root, are seen. The same phenotype classes are also seen in strong gnom alleles (Mayer et al., 1993). GNOM directly regulates the polar localization of PIN1 and therefore controls auxin distribution. This report establishes that NOV plays an important role in directing PIN expression and polarity through cell fate decisions. We hypothesize that, although each weak mutation causes relatively mild effects on embryonic development, when combined in nov-1 gnomR5 embryos, auxin distribution is synergistically impaired to the extent that apical-basal patterning is severely disrupted in a similar manner to that seen in strong gnom alleles, where coordinated PIN1 polarity for embryo axis formation is severely disrupted (Steinmann et al., 1999).
In addition to cotyledon growth, separation of cotyledon primordia are repressed in nov-2∼5 embryos. When grown in the presence of the polar auxin transport inhibitor NPA, even in the weak nov-1 allele, leaf organ separation is repressed. It has been suggested that auxin distribution in the apical part of the embryo is important for cotyledon separation (Furutani et al., 2004). 2,4-D treatment causes broader distribution of auxin in the developing embryo (Friml et al., 2003) and induces cotyledon fusion (Furutani et al., 2004). Separation of shoot-derived lateral organs is associated with a local reduction of auxin, which may be induced by divergence of auxin flow (Heisler et al., 2005). In nov-1 leaf primordia and nov-3 cotyledon primordia, as provascular development is impaired, one might expect that auxin is not efficiently drained through the primordium. Thus, as ectopic epidermal DR5 expression is detected in nov-1 leaf and nov-3 cotyledon primordia, a broader auxin distribution would be expected to form in the apical regions of leaf and cotyledon primordia of nov mutants. This may enhance the inhibitory effect of NPA on the separation of lateral organ primordia. Therefore, we think that NOV function contributes indirectly to lateral organ separation. A similar scenario may be applied to the multiple-cotyledon phenotype of nov-1 gnomR5 and nov-1 gnomB/E, which is neither seen in nov nor in gnom strong alleles. Broadly increased cellular auxin concentration in the apical part of embryos and enhanced PIN polarity defects in nov-1 gnomR5 and nov-1 gnomB/E may increase the probability of multiple auxin maxima forming as cotyledon primordia start to develop, leading to the outgrowth of four to six cotyledons. On the other hand, a contrasting scenario may be applied to the leaf petiole phenotype of nov-1 treated with NPA. Auxin positively regulates petiole elongation (Pierik et al., 2009). As auxin flow from the apical to basal region of the leaf primordia is synergistically decreased both by impaired provascular development and by chemical inhibition of auxin transport, auxin concentration would be expected to decrease in the basal region of leaf primordia, potentially abrogating auxin-mediated petiole elongation and petiole formation. In summary, whether they are primary consequences of nov mutations or their secondary effects, the defects associated with nov mutations described here highlight the importance of NOV in many aspects of auxin-mediated development.
The Role of NOV in Auxin-Mediated Cell Specification and Patterning in Roots
We have shown that NOV is required for auxin-mediated cell specification and patterning not only in the leaf, but also in the root. It has been shown that SCR (Sabatini et al., 2003), PLT (Aida et al., 2004), and WUSCHEL-RELATED HOMEOBOX5 (Sarkar et al., 2007) control stem cell maintenance in roots and that regulation of PLT expression by the GCN5 histone-acetyltransferase complex is essential for root stem cell maintenance (Kornet and Scheres, 2009). PLT genes are considered to be cell fate regulators in auxin-mediated cell specification and patterning, redundantly controlling expression of PIN genes in specific domains (Aida et al., 2004; Blilou et al., 2005; Galinha et al., 2007). In the plt1 plt2 double mutant, PIN4 mRNA, which is normally expressed in the root tip, is absent. Also, the expression of PIN3 and PIN7 mRNA is normal in columella cells, but markedly reduced in the provascular cells of roots (Blilou et al., 2005). A contrasting situation is seen in nov-1. Here, expression of PIN4 in columella cells is expanded and encompasses one more outer tier of columella cells, whereas expression of PIN3 and PIN7 is absent or markedly reduced in columella cells, while PIN7 is expressed normally in provascular cells. The strong reduction of PIN3 and PIN7 expression in columella cells of nov-1 might not simply be due to hastened differentiation of columella cells because differentiation of columella stem cells also occurs in plt1 plt2 (Aida et al., 2004). An expansion of the PIN4 expression domain may be due to the compensatory mechanism of PIN redundancy (Vieten et al., 2005). It should be noted that, in nov-1, in contrast with the internal tissue in leaf primordia, there is no obvious defect in PIN1 expression and polarity in roots. In addition, PIN7 expression in columella cells and the region immediately distal to the leaf midvein provascular strand is strongly repressed in nov-1, while the PIN7 expression persists along the differentiating procambium in the leaf and root. These data suggest regional specificity for the requirement of NOV control over the expression of PIN1 and PIN7, as well as for PIN1 polarity.
A basal-to-apical shift in polarity of PIN2 is observed in the root cortex of nov-1. A similar polarity defect of PIN2 in the root cortex is caused also by overexpression of PINOID (PID) protein kinase (Friml et al., 2004) and by loss-of-function mutations in genes encoding protein phosphatase 2A (PP2A) (Michniewicz et al., 2007). However, PID overexpression and PP2A mutations result in basal-to-apical shifts not only for PIN2 in the cortex but also for PIN1 and PIN4 in their respective expression domains. PID and PP2A both partially colocalize with PINs and antagonistically regulate apical-basal localization of PINs (Michniewicz et al., 2007). On the other hand, in nov-1, the basal-to-apical shift was detected only for PIN2 in the cortex. NOV, as a nuclear protein, does not colocalize with PINs and is therefore unlikely to directly influence their polarity. It is of course possible that NOV controls the expression of factors that do control PIN polarity, possibly including PID and PP2A.
PLT, SCR, and SHR genes appear to regulate indirectly expression of PIN genes through determination or stabilization of cell fate in the root meristem (Sabatini et al., 2003; Aida et al., 2004; Blilou et al., 2005; Xu et al., 2006). Our data suggest that NOV also indirectly regulates expression and polarity of PIN proteins through mechanisms that include the determination and/or stabilization of cell fate in the root meristem.
NOV Mediates Cell Acquisition of the Competence to Undergo Auxin-Mediated Cell Specification and Patterning in the Embryo, Shoot, and Root
Local, efflux-dependent auxin gradients have emerged as a unifying mechanism underlying plant organ formation (Benková et al., 2003). The auxin-efflux pattern is regulated by dynamic expression and asymmetric subcellular localization of PIN auxin-efflux proteins during plant organogenesis (Benková et al., 2003; Friml et al., 2003; Reinhardt et al., 2003; Blilou et al., 2005; Scarpella et al., 2006; Bayer et al., 2009). Thus, the question of how the expression and subcellular localization of PIN proteins are controlled goes to the heart of plant development. We have shown here that (1) NOV is required for PIN expression and polarity in a cell type–specific manner, for provascular PIN1 expression and region-specific expression of PIN7 in leaf primordia, for cell type–specific expression of PIN3, PIN4, and PIN7 in the root stem cell niche, and for PIN2 polarity in the root cortex. (2) NOV helps cells to acquire and maintain their ability to differentiate into vascular cells in response to auxin. (3) NOV is required for normal cellular organization and stem cell maintenance in the root stem cell niche. (4) NOV has an important role in auxin-mediated embryonic development. (5) NOV function may partly underlie establishing MP expression for growth of cotyledon primordia and provascular development in cotyledon and leaf primordia. (6) NOV encodes a plant-specific nuclear factor specifically expressed in developing organs and tissues. From the data presented in this report, we propose that NOV is a competence factor for auxin-dependent coordinated cell specification and patterning during plant organ formation. That is, NOV is involved in mediating cell acquisition of competence to undergo auxin-dependent polar development, thereby establishing PIN expression and polarity, developmentally regulated auxin distribution, and possibly MP expression, all of which are intertwined to form a positive feedback loop for auxin-mediated polar development. What is the cellular and molecular basis for such acquisition of competence? This question remains unanswered, but knowledge of the biochemical function of NOV and the relationship of NOV with other known cell fate regulators may lead us to an answer. This knowledge will be uncovered by the identification of factors interacting physically with the NOV protein and genetically with the NOV gene. Since NOV is a nuclear protein, NOV may act primarily in transcriptional or posttranscriptional regulation of nuclear gene expression. Thus, assessment of transcriptional outputs in nov mutants would also be beneficial to understanding the primary function of NOV. Furthermore, future studies on NOV may shed new light on the fundamental mechanisms by which auxin regulates the formation of plant organs and tissues, regardless of their fate and origin.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotypes C24, Columbia (Col), and Landsberg erecta were used as wild-type controls according to the genetic backgrounds of mutants and plant lines. The following mutants were used: nov-1 (C24), nov-2, -3, -4, and -5 (Col), gnomR5 (Landsberg erecta), and gnomB/E (Col) (Geldner et al., 2004). nov-2 (emb2597), nov-3 (salk_096246), nov-4 (salk_049619), and nov-5 (salk_028690) were obtained from the ABRC. Marker lines used were J1511 (C24) (from ABRC), DR5pro:GUS (Col) (Ulmasov et al., 1997), DR5pro:GFP (Col) (Ottenschläger et al., 2003), ATHB8pro:GUS (Col) (Baima et al., 1995), CYCB1pro:CDB:GUS (Colón-Carmona et al., 1999), SHRpro:GFP (Col) (Helariutta et al., 2000), and SCRpro:GFP (Col) (Wysocka-Diller et al., 2000).
T-DNA insertion alleles, nov-2, -3, -4, and -5, were characterized at both ends of each insertion and confirmed to be integrated into the transcriptional unit of NOV gene (Figure 8A). As the first nucleotide of the initiator ATG is numbered as +1, insertion positions of nov-2, -3, -4, and -5 T-DNA are as follows: nov-2 T-DNA, between −41 (5′ untranslated region) and +12 with 51-bp deletion in the first exon; nov-3 T-DNA, between +12 and +33 with 20-bp deletion in the first exon; nov-4 T-DNA, between +3204 and +3211 with 6-bp deletion in the fourth intron; and nov-5 T-DNA, between +11987 and +12006 with 18-bp deletion in the thirteenth exon. nov-1∼5 mutants were backcrossed at least twice, respectively.
For growing plants on plates, seeds were sterilized and plated on agar-solidified basal medium containing 0.5× Murashige and Skoog salts, pH 5.7, 1% sucrose, and 0.7 or 1.5% agar. Before germination, seeds were placed at 4°C in the dark for 3 to 5 d. Plants were grown on horizontally placed 0.7% agar plates or vertically on the surface of 1.5% agar plates. Unless written otherwise, growth temperature is 21 to 22°C. For immunohistochemical detection of PIN proteins, seedlings grown at 27°C were used.
Mutagenesis and Screening
The enhancer-trap line J1511, in which reporter GFP is expressed in the provascular cells, was mutagenized with 0.1% (v/v) ethyl methanesulfonate (Sigma-Aldrich). Leaf vein patterns were observed in living M2 seedlings by detecting GFP expression in the provascular cells under a fluorescent dissection microscope.
Phenotypic Analyses
For observation of venation patterns, dissected leaves or seedlings were fixed in ethanol/acetic acid (6:1). Fixed samples were washed several times in 70% ethanol and then whole mounted in chloral hydrate solution (8 g of chloral hydrate, 1 mL of glycerol, and 2 mL of water). Developing embryos were observed as described previously (Tsugeki et al., 1996). For visualization of starch granules in root tips, roots excised from plants vertically grown for 8 d were incubated in Lugol solution (Sigma-Aldrich) and mounted in the chloral hydrate solution.
GUS Staining
Samples were first placed in ice-cold acetone for 15 min and then in GUS staining solution containing 0.5 mg/mL X-Gluc, 0.1 M sodium phosphate buffer, pH 7.0, 10 mM EDTA, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 0.1% Triton X-100. Samples in the GUS staining solution were placed under vacuum and incubated either at room temperature (16 to 18 h for CYCB1pro:CDB:GUS, DR5pro:GUS in leaves, and ATHB8pro:GUS) or at 37°C (2 h for DR5pro:GUS in roots and 18 to 24 h for NOVpro:NOV:GUS). Stained samples were processed as described above.
Immunolocalization of PIN Proteins
Samples were fixed and processed as described previously (Gälweiler et al., 1998). Polyclonal antibodies raised against PIN1, PIN2, PIN3, PIN4, and a monoclonal antibody raised against PIN7 were used as primary antibodies. As secondary antibodies, Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 555 anti-mouse IgG were used, respectively. Processed samples were mounted in SlowFade Gold antifade reagent with or without DAPI (Invitrogen) and subjected for confocal microscopy.
Microscopy
Images of seedlings were captured on a Leica M420 Macroscope equipped with Nikon COOLPIX 990 or Leica DFC280 digital camera system. GFP fluorescence in living seedlings was detected on the fluorescence dissection microscope MZFLIII (Leica) equipped with a digital camera system (C4742-95; Hamamatsu). Observation of mature embryos by aniline blue staining was performed as previously described (Bougourd et al., 2000). For DAPI staining, fixed samples were stained in 0.2 mg/L DAPI for 15 min, washed three times, and mounted on a glass slide. Optical sections were obtained using a Zeiss Axioplan 2 microscope equipped LSM 5 PASCAL (Carl Zeiss). The 405-nm excitation line of a 25-mW blue diode laser was used, and signals were collected through a band-pass filter of 420 to 480 nm. For confocal microscopy on leaf primordia, fluorescent signals were checked throughout the tested primordia.
Positional Cloning, Plasmid Construction, and Plant Transformation
The F2 population of the F1 hybrid between nov-1 (C24) and Col was used for mapping. By fine mapping, NOV was mapped between At4g13670 and At4g13800. An ∼17-kb genomic fragment containing the At4g13750 gene with 3.3-kb upstream and 1.4-kb downstream regions (from 7,971,914 to 7,989,006 bp on chromosome 4) was cloned into pGW-NB1 (Nakagawa et al., 2007). The resulting construct was able to complement nov-1, nov-3, or nov-5. G3GFP (Kawakami and Watanabe, 1997) or GUS gene was inserted into the aforementioned 17-kb genomic fragment in such a way that translational fusion genes NOV:GFP, NOV:GUS, and GFP:NOV would be expressed under the NOV promoter. To determine the exon-intron structure of At4g13750, RT-PCR analysis was performed based on the gene annotation data.
Hormone and Chemical Treatments
Seedlings were incubated either in water containing 10 μM 2,4-D or on agar-solidified medium containing 1 μM 2,4-D. For detecting DR5pro:GUS expression in leaves and roots, seedlings were first grown in the absence of 2,4-D and then incubated with 10 μM 2,4-D for 1 d and with 1 μM 2,4-D for 3 d, respectively. For analysis of 2,4-D–induced lateral roots, seedlings were grown in the presence of 1 μM 2,4-D for 11 d. For NPA treatment, seedlings were grown on agar-solidified medium containing 10 μM NPA for 8 d (starch staining of root tips) and for 21 d (phenotypic analysis in Figure 5).
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource (http://www.Arabidopsis.org/) or GenBank/EMBL databases under the following accession numbers: NOV, At4g13750; NVL, At1g08300; castor bean (Ricinus communis) NOV, EEF44191.1; soybean (Glycine max) NOV, AAQ62582.1; rice (Oryza sativa Japonica group) NOV, EEE60127.1; moss (Physcomitrella patens) NOV, XP_001777863.1; protozoan (Cryptosporidium parvum) superfamily I helicase, XP_626083; protozoan Sacsin-like protein, XP_627068; human Sacsin, NP_055178 (Engert et al., 2000); human Hsp90-like protein (TNF receptor-associated protein 1), NP_057376; human replication factor C subunit 1 (RFC1), P35251; yeast (Saccharomyces cerevisiae) Hsp90, P02829; Rhodopseudomonas palustris Hsp90, ZP_02301386; chicks DNA topoisomerase 2-β (TOP2B), O42131; Drosophila origin recognition complex subunit 2 (ORC2), Q24168.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ATHB8pro:GUS Expression in nov-1.
Supplemental Figure 2. Maintenance of Columella Root-Cap Stem Cells Is Defective in nov-3.
Supplemental Figure 3. Maintenance of Cortex/Endodermis Stem Cells Is Defective in nov-3 Root Tips.
Supplemental Figure 4. Abnormality in Formation of Vascular Stem Cells in nov Mutants during Embryogenesis.
Supplemental Figure 5. Ectopic Cell Division in the Quiescent Center of nov-2 during Embryogenesis.
Supplemental Figure 6. Expression of SCRpro:GFP and SHRpro:GFP in Embryos with Strong nov Alleles.
Supplemental Figure 7. Seedling Phenotype in nov-1 gnomB/E Double Mutant.
Supplemental Figure 8. Subepidermal Cells at the Apical End of the Midvein Provascular Strand Exhibit Nonpolar PIN1 Localization.
Supplemental Figure 9. Nuclear Localization of GFP:NOV.
Supplemental Figure 10. NOVpro:NOV:GUS Expression in Embryos and in Cotyledons of Developing Seedlings.
Supplemental Figure 11. Vascular and Ground-Tissue Phenotypes in nov-1 and nov-3 Leaves.
Supplemental Figure 12. J1511-GFP Expression during Embryogenesis.
Supplemental Table 1. Leaf and Petiole Lengths of the First Two Rosette Leaves of Seedlings Grown in the Absence or Presence of NPA.
Supplementary Material
Acknowledgments
We thank Philip Benfey for SCRpro:GFP and SHRpro:GFP, Peter Doerner for CYCB1pro:CDB:GUS, Tom J. Guilfoyle for DR5pro:GUS, Gerd Jürgens for gnomR5 and gnomB/E, Giorgio Morelli for ATHB8pro:GUS, Tsuyoshi Nakagawa for pGWB-NB1, Yuichiro Watanabe for G3GFP, the ABRC for J1511 and nov-2, -3, -4, and -5, Toshiharu Shikanai for critically reading the manuscript, Sumie Ishiguro, Noritaka Matsumoto, and Nana Tanaka for stimulating discussions, and Shiho Terada and Irina Kneuper for technical assistance. This work was supported in part by a Grant-in-Aid for Scientific Research (13740455 and 21657014 to R.T.) from the Japan Society for the Promotion of Science, by a Grant-in-Aid for Creative Scientific Research (19GS0315 to K.O. and R.T.) and a Grant-in-Aid for Scientific Research on Priority Areas (19060004 to K.O.) from the Ministry of Education, Culture, Sports, Science, and Technology, by the Core Research for Evolutional Science and Technology program of the Japan Science and Technology Agency to K.O., and by the Excellence Initiative of the German Federal and State Governments (EXC 294), SFB 592, and the Landestiftung to K.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ryuji Tsugeki (rtsugeki@ok-lab.bot.kyoto-u.ac.jp).
Online version contains Web-only data.
References
- Abas, L., Benjamins, R., Malenica, N., Paciorek, T., Wisniewska, J., Moulinier-Anzola, J.C., Sieberer, T., Friml, J., and Luschnig, C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8 249–256. [DOI] [PubMed] [Google Scholar]
- Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., Nussaume, L., Noh, Y.-S., Amasino, R., and Scheres, B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119 109–120. [DOI] [PubMed] [Google Scholar]
- Baima, S., Nobili, F., Sessa, G., Lucchetti, S., Ruberti, I., and Morelli, G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121 4171–4182. [DOI] [PubMed] [Google Scholar]
- Bayer, E.M., Smith, R.S., Mandel, T., Nakayama, N., Sauer, M., Prusinkiewicz, P., and Kuhlemeier, C. (2009). Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev. 23 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, T., Sieberer, T., Willett, B., Booker, J., Luschnig, C., and Leyser, O. (2006). The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 16 553–563. [DOI] [PubMed] [Google Scholar]
- Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, E., Jürgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. [DOI] [PubMed] [Google Scholar]
- Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., Heidstra, R., Aida, M., Palme, K., and Scheres, B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433 39–44. [DOI] [PubMed] [Google Scholar]
- Bougourd, S., Marrison, J., and Haseloff, J. (2000). An aniline blue staining procedure for confocal microscopy and 3D imaging of normal and perturbed cellular phenotypes in mature Arabidopsis embryos. Plant J. 24 543–550. [DOI] [PubMed] [Google Scholar]
- Boutté, Y., Crosnier, M.-T., Carraro, N., Traas, J., and Satiat-Jeunemaitre, B. (2005). The plasma membrane recycling pathway and cell polarity in plants: Studies on PIN proteins. J. Cell Sci. 119 1255–1265. [DOI] [PubMed] [Google Scholar]
- Colón-Carmona, A., You, R., Hamimovitch-Gal, T., and Doerner, P. (1999). Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20 503–508. [DOI] [PubMed] [Google Scholar]
- Ditengou, F.A., Teale, W.D., Kochersperger, P., Flittner, K.A., Kneuper, I., van der Graaff, E., Nziengui, H., Pinosa, F., Li, X., Nitschke, R., Laux, T., and Palme, K. (2008). Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105 18818–18823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe, P., et al. (2008). Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature 456 962–966. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dutta, R., and Inouye, M. (2000). GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 25 24–28. [DOI] [PubMed] [Google Scholar]
- Engert, J.C., et al. (2000). ARSACS, a spastic ataxia common in northeastern Québec, is caused by mutations in a new gene encoding an 11.5-kb ORF. Nat. Genet. 24 120–125. [DOI] [PubMed] [Google Scholar]
- Friml, J., Benková, E., Blilou, I., Wisniewska, J., Hamann, T., Ljung, K., Woody, S., Sandberg, G., Scheres, B., Jürgens, G., and Palme, K. (2002. a). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108 661–673. [DOI] [PubMed] [Google Scholar]
- Friml, J., Wisniewska, J., Benková, E., Mendgen, K., and Palme, K. (2002. b). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415 806–809. [DOI] [PubMed] [Google Scholar]
- Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., Offringa, R., and Jürgens, G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 147–153. [DOI] [PubMed] [Google Scholar]
- Friml, J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 862–865. [DOI] [PubMed] [Google Scholar]
- Furutani, M., Vernoux, T., Traas, J., Kato, T., Tasaka, M., and Aida, M. (2004). PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131 5021–5030. [DOI] [PubMed] [Google Scholar]
- Galinha, C., Hofhuis, H., Luijten, M., Willemsen, V., Blilou, I., Heidstra, R., and Scheres, B. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449 1053–1057. [DOI] [PubMed] [Google Scholar]
- Gälweiler, L., Guan, C., Müller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282 2226–2230. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A., and Jürgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112 219–230. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Friml, J., Stierhof, Y.-D., Jürgens, G., and Palme, K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413 425–428. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Richter, S., Vieten, A., Marquardt, S., Torres-Ruiz, R.A., Mayer, U., and Jürgens, G. (2004). Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131 389–400. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., Ckurshumova, W., Vidaurre, D.P., Singh, S.A., Stamatiou, G., Tiwari, S.B., Hagen, G., Tom, J., Guilfoyle, T.J., and Berleth, T. (2004). Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131 1089–1100. [DOI] [PubMed] [Google Scholar]
- Heisler, M.G., Ohno, C., Das, P., Sieber, P., Reddy, G.V., Long, J.A., and Meyerowitz, E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15 1899–1911. [DOI] [PubMed] [Google Scholar]
- Helariutta, Y., Fukaki, H., Wysocka-Diller, J., Nakajima, K., Jung, J., Sena, G., Hauser, M.-T., and Benfey, P.N. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101 555–567. [DOI] [PubMed] [Google Scholar]
- Jaillais, Y., Santambrogio, M., Rozier, F., Fobis-Loisy, I., Miege, C., and Gaude, T. (2007). The retromer protein VPS29 links cell polarity and organ initiation in plants. Cell 130 1057–1070. [DOI] [PubMed] [Google Scholar]
- Kawakami, S., and Watanabe, Y. (1997). Use of green fluorescent protein as a molecular tag of protein movement in vivo. Plant Biotechnol. 14 127–130. [Google Scholar]
- Kleine-Vehn, J., Dhonukshe, P., Sauer, M., Brewer, P.B., Wisniewska, J., Paciorek, T., Benková, E., and Friml, L. (2008. a). ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr. Biol. 18 526–531. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn, J., Leitner, J., Zwiewka, M., Sauer, M., Abas, L., Luschnig, C., and Friml, J. (2008. b). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105 17812–17817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornet, N., and Scheres, B. (2009). Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 21 1070–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze, L., Parizot, B., Baker, A., Ricaud, L., Martiniere, A., Auguy, F., Franche, C., Nussaume, L., Bogusz, D., and Haseloff, J. (2005). GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J. Exp. Bot. 56 2433–2442. [DOI] [PubMed] [Google Scholar]
- Laskowski, M., Grieneisen, V.A., Hofhuis, H., ten Hove, C.A., Hogeweg, P., Marée, A.F.M., and Scheres, B. (2008). Root system architecture from coupling cell shape to auxin transport. PLoS Biol. 6 2721–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxmi, A., Pan, J., Morsy, M., and Chen, R. (2008). Light plays an essential role in intracellular distribution of auxin efflux carrier PIN2 in Arabidopsis thaliana. PLoS One 3 e1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C., Xu, Z., and Chua, N.H. (1993). Auxin polar transport is essential for the establishment of bilateral symmetry during early plant embryogenesis. Plant Cell 5 621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, J., Ckurshumova, W., and Berleth, T. (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson, J., Sung, Z.R., and Berleth, T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126 2979–2991. [DOI] [PubMed] [Google Scholar]
- Mayer, U., Büttner, G., and Jürgens, G. (1993). Apical-basal pattern formation in the Arabidopsis embryo: studies on the role of the gnom gene. Development 117 149–162. [Google Scholar]
- Michniewicz, M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130 1044–1056. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T., et al. (2007). Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71 2095–2100. [DOI] [PubMed] [Google Scholar]
- Ottenschläger, I., Wolff, P., Wolverton, C., Bhalerao, R.P., Sandberg, G., Ishikawa, H., Evans, M., and Palme, K. (2003). Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 100 2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek, T., Zazímalová, E., Ruthardt, N., Petrásek, J., Stierhof, Y.-D., Kleine-Vehn, J., Morris, D.A., Emans, N., Jürgens, G., Geldner, N., and Friml, J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435 1251–1256. [DOI] [PubMed] [Google Scholar]
- Paponov, I.A., Teale, W.D., Trebar, M., Blilou, I., and Palme, K. (2005). The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci. 10 170–177. [DOI] [PubMed] [Google Scholar]
- Petrásek, J., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312 914–918. [DOI] [PubMed] [Google Scholar]
- Pierik, R., Djakovic-Petrovic, T., Keuskamp, D.H., de Wit, M., and Voesenek, L.A.C.J. (2009). Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and DELLA proteins in Arabidopsis. Plant Physiol. 149 1701–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo, H., Demura, T., and Fukuda, H. (2004). Spatial and temporal tracing of vessel differentiation in young Arabidopsis seedlings by the expression of an immature tracheary element-specific promoter. Plant Cell Physiol. 45 1529–1536. [DOI] [PubMed] [Google Scholar]
- Reinhardt, D., Mandel, T., and Kuhlemeier, C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Pesce, E.-R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260. [DOI] [PubMed] [Google Scholar]
- Sabatini, S., Beis, D., Wolkenfelt, H., Murfett, J., Guilfoyle, T., Malamy, J., Benfey, P., Leyser, O., Bechtold, N., Weisbeek, P., and Scheres, B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99 463–472. [DOI] [PubMed] [Google Scholar]
- Sabatini, S., Heidstra, R., Wildwater, M., and Scheres, B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, T. (1991). Cell polarity and tissue patterning in plants. Dev. Suppl. 1 83–93. [Google Scholar]
- Sarkar, A.K., Luijten, M., Miyashima, S., Lenhard, M., Hashimoto, T., Nakajima, K., Scheres, B., Heidstra, R., and Laux, T. (2007). Conserved factors regulates signaling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446 811–814. [DOI] [PubMed] [Google Scholar]
- Sauer, M., Balla, J., Luschnig, C., Wisniewska, J., Reinöhl, V., Friml, J., and Benková, E. (2006). Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 20 2902–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella, E., Marcos, D., Friml, J., and Berleth, T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa, M., Ueda, H., Shimada, T., Nishiyama, C., and Hara-Nishimura, I. (2009). Vacuolar SNAREs function in the formation of the leaf vascular network by regulating auxin distribution. Plant Cell Physiol. 50 1319–1328. [DOI] [PubMed] [Google Scholar]
- Sieberer, Y., Seifert, G.J., Hauser, M.-T., Grisafi, P., Fink, G.R., and Luschnig, C. (2000). Post-transcriptional control of the Arabidopsis auxin efflux carrier EIR1 requires AXR1. Curr. Biol. 10 1595–1598. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E. (1999). Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiol. 121 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer, C., Reyes, F.C., Buono, R., Sliwinski, M.K., Haas, T.J., and Otegui, M.S. (2009). The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell 21 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Gälweiler, L., Palme, K., and Jürgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286 316–318. [DOI] [PubMed] [Google Scholar]
- Tsugeki, R., Kochieva, E.Z., and Fedoroff, N.V. (1996). A transposon insertion in the Arabidopsis SSR16 gene causes an embryo-defective lethal mutation. Plant J. 10 479–489. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg, C., Willemsen, V., Hendriks, G., Weisbeek, P., and Scheres, B. (1997). Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390 287–289. [DOI] [PubMed] [Google Scholar]
- Vieten, A., Vanneste, S., Wisniewska, J., Benková, E., Benjamins, R., Beeckman, T., Luschnig, C., and Friml, J. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132 4521–4531. [DOI] [PubMed] [Google Scholar]
- Wenzel, C.L., Schuetz, M., Yu, Q., and Mattsson, J. (2007). Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in Arabidopsis thaliana. Plant J. 49 387–398. [DOI] [PubMed] [Google Scholar]
- Willemsen, V., Friml, J., Grebe, M., van den Toorn, A., Palme, K., and Scheres, B. (2003). Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15 612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska, J., Xu, J., Seifertová, D., Brewer, P.B., Ruzicka, K., Blilou, I., Rouquié, D., Benková, E., Scheres, B., and Friml, J. (2006). Polar PIN localization directs auxin flow in plants. Science 312 883. [DOI] [PubMed] [Google Scholar]
- Wysocka-Diller, J.W., Helariutta, Y., Fukaki, H., Malamy, J.E., and Benfey, P.N. (2000). Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127 595–603. [DOI] [PubMed] [Google Scholar]
- Xu, J., Hofhuis, H., Heidstra, R., Sauer, M., Friml, J., and Scheres, B. (2006). A molecular framework for plant regeneration. Science 311 385–388. [DOI] [PubMed] [Google Scholar]
- Yadav, R.K., Girke, T., Pasala, S., Xie, M., and Reddy, G.V. (2009). Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl. Acad. Sci. USA 106 4941–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.