Abstract
Development of the flattened laminar structure in plant leaves requires highly regulated cell division and expansion patterns. Although tight regulation of these processes is essential during leaf development, leaf shape is highly diverse across the plant kingdom, implying that patterning of growth must be amenable to evolutionary change. Here, we describe the molecular identification of the classical tomato (Solanum lycopersicum) mutant lyrate, which is impaired in outgrowth of leaflet primodia and laminar tissues during compound leaf development. We found that the lyrate phenotype results from a loss-of-function mutation of the tomato JAGGED homolog, a well-described positive regulator of cell division in lateral organs. We demonstrate that LYRATE coordinates lateral outgrowth in the compound leaves of tomato by interacting with both the KNOX and auxin transcriptional networks and suggest that evolutionary changes in LYRATE expression may contribute to the fundamental difference between compound and simple leaves.
INTRODUCTION
Leaves are the principal organs produced during vegetative growth and provide a flattened surface allowing for efficient light capture during photosynthesis. Leaves begin at the flanks of the shoot apical meristem (SAM) as peg-like outgrowths that rapidly expand along the proximal/distal axis via cell division. As elongation proceeds, the archetypal dicot simple leaf begins lateral expansion of the blade from a continuous band of meristematic cells at the edge of the leaf primordium termed the marginal blastozone (Hagemann and Glesissberg, 1996). Such leaves are divided into two major developmental domains: the distal blade, where lateral growth is initiated, and the proximal petiole, which lacks blade outgrowth. Blade outgrowth continues until a wave of cell cycle arrest usually begins at the distal tip of the developing leaf and proceeds to the leaf base (Avery, 1933; Poethig, 1984; Nath et al., 2003). The remaining growth required to produce a mature leaf is accommodated by cell expansion. Cell division patterns during early leaf development may, in part, guide subsequent expansion, but it is well established that disruption in cell division patterns can lead to changes in expansion and roughly equivalent final organ shape (Tsukaya, 2006). It is therefore hypothesized that cell division-dependent and -independent pathways may dictate final organ shape. In simple leafed model species, such as Arabidopsis thaliana and snapdragon (Antirrhinum majus), these processes result in a flattened, continuous lamina subtended by a bladeless petiole.
Although coordination of cell division and expansion are critical factors directing continuous laminar development, simple leaves represent only a fraction of the total diversity in plant leaf morphology. Many plants produce leaves that are deeply lobed or are compound, consisting of multiple blade units termed leaflets. In compound leaves, the marginal blastozone initiates leaflet primordia that are discriminated from adjacent intercalary tissues by rapid cell division (Hagemann and Glesissberg, 1996). As leaf development progresses, laminar outgrowth is tightly regulated such that intercalary tissues and leaflet bases are bladeless, ultimately resulting in discrete leaflets (Kaplan, 1975). This pattern can be reiterated to produce secondary and tertiary leaflets. Thus, in comparison to simple leaves, compound leaves must undergo highly patterned growth during several stages of leaf development. Our understanding of the mechanism by which these subdomains within the compound leaf are specified remains in its infancy.
The plant hormone auxin, which is known to positively influence both cell division and expansion, has recently been shown to be important in patterning differential growth in several compound leafed species (DeMason and Chawla, 2004; Wang et al., 2005; Barkoulas et al., 2008; DeMason and Polowick, 2009; Koenig et al., 2009). In tomato (Solanum lycopersicum) and Cardamine hirsuta, auxin is transported into incipient leaflets where it activates response pathways leading to primordium outgrowth (Barkoulas et al., 2008; Koenig et al., 2009). Furthermore, this differential auxin distribution is critical to pattern blade outgrowth such that equalization of auxin distribution in the primordium results in ectopic blade outgrowth in intercalary tissues between leaflets (Koenig et al., 2009). This repression of auxin response in bladeless intercalary tissues of wild-type tomato compound leaves is accomplished by the auxin/indole-3-acetic acid (IAA) gene ENTIRE (Wang et al., 2005; Zhang et al., 2007). entire mutants initiate ectopic blade formation between leaflets leading to simple leaf development (Koenig et al., 2009). A similar phenotype is observed in the goblet mutant in tomato (Brand et al., 2007; Blein et al., 2008; Berger et al., 2009). GOB encodes an ortholog of the Arabidopsis CUC2 gene, which specifies organ boundaries in the SAM (Aida et al., 1997). In several compound leafed species, GOB specifies the boundaries between initiating leaflets (Blein et al., 2008; Berger et al., 2009). The relationship between auxin distribution, ENTIRE, and GOB remains unclear at this point, but auxin distribution and response is central to the patterning of growth in compound leaves.
To gain insight into how molecular regulation of cell division has been modified to generate differential growth in compound leaves, we cloned and analyzed the function of the JAGGED ortholog in tomato. In the simple leaved species Arabidopsis, JAGGED (JAG) and its paralog NUBBIN (NUB) are important positive regulators of cell division and outgrowth in lateral organs (Dinneny et al., 2004, 2006; Ohno et al., 2004). Both genes encode transcription factors consisting of a single C2H2-type zinc finger and a Pro-rich motif. JAG loss-of-function causes premature cessation of cell proliferation in lateral organs, ultimately resulting in irregular margins (Dinneny et al., 2004; Ohno et al., 2004). This phenotype is greatly enhanced in jag nub double mutants that display a large number of developmental defects, including deeply lobed leaves (Dinneny et al., 2006). Plants overexpressing JAG display ectopic blade outgrowth on leaf petioles and expansion of cryptic bracts, presumably by overcoming inhibition of cell division in these regions (Dinneny et al., 2004; Ohno et al., 2004). Thus, JAG and NUB are thought to pattern cell cycle activity in young leaf primordia to differentiate the bladed distal tissues and the bladeless proximal tissues. Based on this described role in Arabidopsis, we hypothesized that changes in JAGGED expression might contribute to differential growth in developing compound leaves.
RESULTS
Identification of the Tomato JAG Ortholog
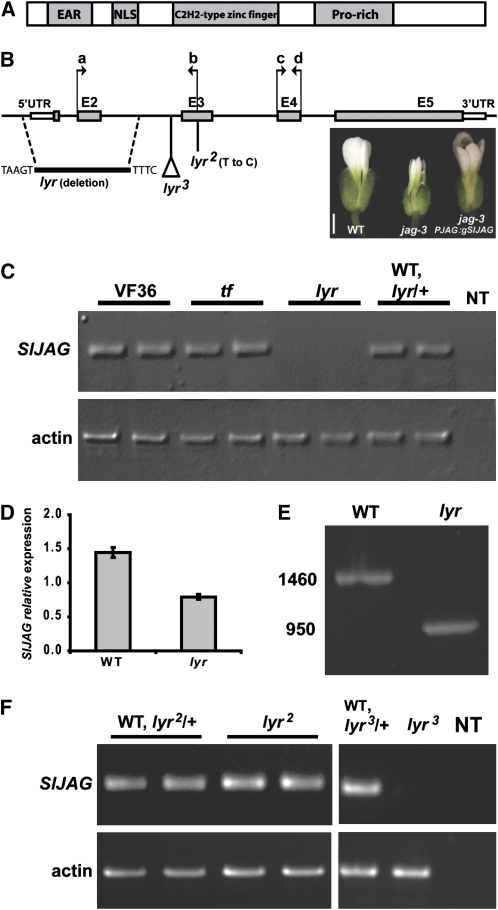
First, we identified the S. lycopersicum JAG homolog and showed it encodes a putative protein composed of 259 amino acids. Three conserved domains were readily identified in the predicted S. lycopersicum JAG sequence when compared with Arabidopsis JAG (Dinneny et al., 2004; Ohno et al., 2004): a putative nuclear localization signal sequence, a single C2H2-type zinc finger motif, and a Pro-rich motif (Figure 1A; see Supplemental Figure 1 online). Another motif, identified previously in Arabidopsis as an EAR motif (Ohno et al., 2004), was identical between Arabidopsis JAG and S. lycopersicum JAG (Figure 1A; see Supplemental Figure 1 online). Outside of these described domains, amino acid sequence conservation between S. lycopersicum JAG and Arabidopsis JAG was low, indicating possible functional dissimilarities (see Supplemental Figure 1 online). Nevertheless, the intron-exon structure of S. lycopersicum JAG (3134 bp) is completely conserved relative to Arabidopsis JAG (Figure 1B). DNA gel blot analysis demonstrated that S. lycopersicum JAG is a single-copy gene (see Supplemental Figure 2 online). In combination, these data demonstrate that S. lycopersicum JAG is a single-copy homolog of the Arabidopsis JAG gene.
Figure 1.
Molecular Characterization of S. lycopersicum JAG.
(A) Schematic of conserved domains of JAG. The four conserved regions are indicated in gray boxes (not to scale).
(B) JAG gene structure showing five exons (gray boxes) and untranslated regions (UTRs; white boxes). The deletion in the lyr allele is designated as a black line with the flanking nucleic acids. The estimated position of the putative insertion in the lyr3 allele is designated as well as the location of the point mutation in the lyr2 allele (not to scale). The arrows a and b indicate primer position for RT-PCR analysis described in (C), and the arrows c and d indicate primer position for quantitative RT-PCR analysis described in (D). Photo on the right shows flowers (from left to right) of wild-type, jag-3, and AtJAG:gSlJAG in jag-3, demonstrating rescue of the jag-3 flower phenotype by S. lycopersicum JAG. Bar = 1 mm.
(C) RT-PCR analysis of S. lycopersicum JAG in wild-type VF36, tf, lyr, and a mix of lyr/+ and wild-type plants. Each sample represents RNA that was isolated from two apices from two different plants. Two samples were tested for each line. Negative control was a PCR reaction without a template (NT), and actin mRNA was amplified as a positive control for RNA integrity.
(D) Quantitative RT-PCR analysis of the expression of JAG in wild-type and lyr tomato plants. Error bars indicate se over three technical replicates.
(E) PCR on genomic DNA of wild-type and lyr showing smaller JAG PCR product in lyr (numbers on left indicate band size in base pairs).
(F) RT-PCR analysis of Sl JAG in lyr2 and lyr3. Two samples were tested for lyr2 and one for lyr3. Negative control was a PCR reaction without a template (NT), and actin was amplified as a positive control.
[See online article for color version of this figure.]
The sequence conservation between the JAG and S. lycopersicum JAG proteins was low outside of the conserved motifs, making it crucial to determine the extent to which the two proteins are functionally similar. To resolve this question, we examined the ability of S. lycopersicum JAG to suppress the floral loss-of-function phenotype seen in jag-2 and jag-3 Arabidopsis plants (Ohno et al., 2004) by transforming both mutants with the S. lycopersicum JAG genomic fragment driven by the Arabidopsis JAG promoter (PAtJAG:gSlJAG). The floral phenotype of jag-2 was fully suppressed in 36 and partially suppressed in 16 out of 124 jag-2 transgenic plants (Figure 1B). Similar complementation was seen in the jag-3 background. In addition, we compared the effect of S. lycopersicum JAG overexpression in the Arabidopsis wild-type, jag-2, and jag-3 genetic backgrounds with the published effects of Arabidopsis JAG overexpression (Dinneny et al., 2004; Ohno et al., 2004) (see Supplemental Figure 3 online). The 35S:SlJAG transgenics displayed phenotypes highly reminiscent of JAG overexpressing Arabidopsis in all attributes except the elaboration of cryptic bracts. These results demonstrate that, despite substantial divergence in amino acid sequence, Arabidopsis JAG and S. lycopersicum JAG show a high degree of functional homology. This being said, S. lycopersicum JAG overexpression did not result in the expansion of cryptic bracts as seen in experiments using JAG, suggesting that some of this protein sequence divergence may have functional consequences.
lyrate Is an S. lycopersicum JAG Null Mutant
We next placed JAG on the tomato genetic map using a segregating S. lycopersicum × Solanum pennellii F2 population (Frary et al., 2004) and found that JAG maps to a 2-centimorgan region on the short arm of chromosome 5, between markers T1335 and CD64. Two recessive leaf shape mutants, lyrate (lyr) and trifoliate (tf), roughly map to this region of chromosome 5. We hypothesized that loss of JAG function might be responsible for one of these mutant phenotypes.
To determine if a mutation in S. lycopersicum JAG causes either lyr or tf phenotypes, we analyzed the level of JAG transcript in each mutant. RT-PCR analysis showed complete absence of JAG transcript in lyr relative to VF36 wild-type, tf, and pooled lyr/+ and +/+ segregants (Figure 1C). Confirmation of these results with quantitative RT-PCR analysis suggested that JAG expression in lyr is significantly reduced compared with the wild type (Figure 1D). To determine whether this change of expression was due to mutation of the JAG gene, we preformed DNA gel blot hybridization using a JAG probe on lyr and wild-type segregants. A restriction fragment length polymorphism at the 5′ end of JAG was detected in lyr when compared with wild-type hybridization (see Supplemental Figure 4 online). Sequence analysis of the JAG gene in lyr plants revealed a 509-bp deletion starting at position −265 and ending in intron 2 (Figures 1B and 1E). This deletion greatly reduces JAG transcription and likely interrupts translation of the JAG protein.
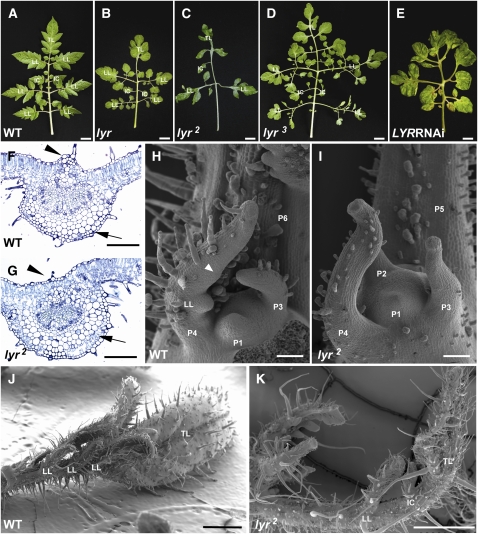
The lyr mutant (Figures 2A and 2B) was originally characterized by the fan-like appearance of its leaves. To verify that the mutation in JAG is the cause of the lyr phenotype, we examined JAG expression and sequence from two other lyr alleles, which show similar phenotypes. lyr2 (Figure 2C), a spontaneous allele, showed a normal level of JAG transcription (Figure 1F), but sequence analysis revealed a single nucleotide polymorphism (T to C) that results in substitution of the first highly conserved Cys in the C2H2-type zinc finger domain with an Arg (see Supplemental Figure 1 online, red arrowhead). lyr3 (n2731, Figure 2D) is derived from a fast neutron mutagenesis project, (Menda et al., 2004) (Y. Eshed, unpublished data) and showed a strong reduction in JAG transcript levels (Figure 1F). DNA gel blot analysis of lyr3 revealed a large insertion at the JAG locus (Figure 1B; see Supplemental Figure 4 online). Finally, to further confirm that reduction in JAG RNA results in a lyr-like phenotype, we generated RNA interference (RNAi) transgenics targeting the JAG locus. One of these transgenic lines exhibited phenotypes highly similar to that of lyr (1/16 transgenic plants; Figure 2E). We therefore named the tomato JAG homolog LYR.
Figure 2.
lyrate Leaf Phenotype.
(A) to (D) Mature fifth leaves of a wild-type plant (A) and the three lyr alleles ([B] to [D]).
(E) Mature leaf of LYR RNAi plant showing lyr phenotype. Bars = 2 cm.
(F) and (G) Cross sections of the midvein of the terminal leaflet of the wild type (F) and lyr2 (G). Arrowheads indicate position of the supportive tissue on the adaxial side of the midvein, and arrows indicate parenchyma cell layer position. Bars = 0.5 mm.
(H) to (K) Scanning electron microscopy analysis of wild-type ([H] and [J]) and lyr2 ([I] and [K]) apices. (H) and (I) show plastochron P1-P6, while (J) and (K) show the P6 leaf phenotype. Arrowhead in (H) indicates lobe formation. TL, terminal leaflet; LL, lateral leaflet; IC, intercalary. Bars = 100 μm in (H) and (I) and 1 mm in (J) and (K).
[See online article for color version of this figure.]
The lyr Phenotype
The naturally occurring mutant lyr was isolated from cultivated tomato in Central America. It is characterized by a broad reduction in outgrowth of lateral organs. The mature leaves of wild-type tomato plants are bipinnate, consisting of a serrated terminal leaflet, three or more lateral leaflet pairs, and a few intercalary leaflets (Figure 2A, Table 1). The base of each of the large primary leaflets will give rise to one or two secondary leaflets. By contrast, the adult lyr leaf consists of a deeply divided terminal leaflet, one or two lateral leaflets, and few intercalary leaflets (Figures 2B to 2D). All of these leaflet types exhibited reduced laminar outgrowth when compared with wild-type plants. Interestingly, secondary leaflet number on the first lateral leaflet pair was increased in all three lyr alleles (Figures 2B to 2D, Table 1). Increased dissection to produce tertiary leaflets was observed in lyr and lyr3 alleles but not in the lyr2 allele (Figures 2B to 2D, Table 1). lyr2 plants were in general more inhibited in blade outgrowth and leaflet outgrowth then the other alleles, suggesting possible differences in allelic strengths. Nevertheless, all three lyr alleles exhibited severely reduced outgrowth of laminar tissues in all leaflets (Figures 2B to 2D, Table 1).
Table 1.
Phenotypic Characterization of lyrate Mature Leaves
| Genotype | Leaf Length (cm) | Width of the First LL (cm) | Number of ILs between the TL and the First LL | TL Lobe Number | Number of LL | Number of SLs on the First LL | Number of TLs on the First LL |
|---|---|---|---|---|---|---|---|
| Wild type | 29.6 ± 0.55 | 15.9 ± 0.52 | 1.0 ± 0.31 | 0 | 6 ± 0 | 1.57 ± 0.81 | 0 |
| lyr | 24.9 ± 1.14* | 19.0 ± 0.82* | 2.5 ± 0.50* | 4.7 ± 0.76* | 3.33 ± 0.33* | 6.00 ± 0.58* | 0.83 ± 0.40* |
| Wild type | 29.7 ± 0.78 | 18.5 ± 0.97 | 3.7 ± 0.37 | 2.0 ± 0.37 | 6 ± 0 | 0.8 ± 0.29 | 0 |
| lyr2 | 22.5 ± 0.73* | 22.0 ± 1.07* | 3.6 ± 0.37 | 3.5 ± 0.27* | 2 ± 0* | 5.7 ± 0.54* | 0 |
| Wild type | 24.0 ± 0.71 | 18.5 ± 0.89 | 1.38 ± 0.64 | 1.5 ± 0.28 | 6 ± 0 | 1.75 ± 0.66 | 0 |
|
lyr3 |
22.9 ± 0.52* |
21.5 ± 1.09* |
4.30 ± 0.56* |
5.3 ± 0.47* |
3 ± 0.38* |
8.60 ± 0.37* |
4.7 ± 0.71* |
Measurements of leaf length were done from tip to petiole base. Measurements of leaf width were done from margin to margin of the first lateral leaflet pair. TL, terminal leaflet; LL, lateral leaflet; SL, secondary leaflet; TL, tertiary leaflet. Asterisk indicates that the difference between the wild type and lyr is statistically significant at P < 0.05.
In addition to the dramatic leaf phenotype, lyr floral development is abnormal. lyr flowers open prematurely and have reflexed petals (see Supplemental Figures 5A to 5C online). Wild-type tomato stamens are fused to form a cone surrounding the carpel, but lyr anthers are narrow and unfused curving inwardly toward the carpel (see Supplemental Figures 5D and 5E online). lyr carpels are composed of a thickened style and an enlarged ovary containing disorganized ovules (see Supplemental Figures 5F to 5H online). These floral phenotypes are reminiscent of those observed in jag nub mutants (Dinneny et al., 2006). Thus, the function of the LYR gene seems to be conserved with that of JAG and NUB as a broad regulator of aerial lateral organ development. Although the Arabidopsis jag, nub, or jag nub mutants are compromised in leaf development, lyr exhibits an even more dramatic leaf phenotype relative to wild-type tomato. This suggests that tomato compound leaf development is more sensitive to the disruption of LYR/JAG function than Arabidopsis simple leaf development.
Leaf Development in lyr
We examined early leaf development in lyr to determine the origin of its leaf shape alteration. lyr differed significantly from wild-type plants from the earliest stages of leaf development. Wild-type tomato leaves begin as peg-like structures that curve toward the SAM due to increased cell division on the abaxial leaf face relative to the adaxial face. Even in these young leaves (P2-P3, primordia numbered starting from the first visible primordia at the shoot apex) differentiation is apparent at the tip of the leaf. The clearest indicator of this is the initiation of trichome development at the tip of the leaf (Hagemann and Glesissberg, 1996; Chen et al., 1997). In equivalent lyr primordia, the angle of leaf curvature was reduced and trichome development was absent. In place of trichome differentiation at the leaf tip, lyr leaf primordia develop a globular ectopic outgrowth composed of large cells (Figures 2H and 2I). These results suggest a general disruption of cell division and differentiation patterns in lyr young leaf primordia.
In wild-type tomato, leaflet primordia begin to initiate in the late P3 or P4 stage, and subsequent leaflets develop basipetally (Coleman and Greyson, 1976; Dengler, 1984). Lobe formation in each leaflet occurs acropetally and is first evident in the terminal leaflet just before initiation of blade outgrowth in P4 stage primordia (Janssen et al., 1998) (Figure 2H, arrowhead). In contrast with wild-type plants, the first lateral leaflet primordium in lyr develops in P4-P5 instead of P3-P4 (Figures 2I and 2K). Initiation of the second set of lateral leaflets is also delayed, and additional primary leaflets rarely develop (Figure 2K). Lamina outgrowth in wild-type terminal leaflets is evident in P3, while it is restricted in lyr until P5 or later (Figures 2H to 2K). Petiole elongation occurs in P6-P7 in wild-type plants, while the petiole in lyr is already elongated by the P5 stage (Figures 2H to 2K). In combination, these results demonstrate that the lyr mutant is impaired in outgrowth of both blade and leaflets.
In Arabidopsis, JAG is proposed to regulate lateral organ morphology by activating and maintaining cell division (Dinneny et al., 2004; Ohno et al., 2004). The general reduction in lateral growth in lyr leaves might be explained by reduced cell cycle activity. To test this hypothesis, we examined the overall cell density in lyr and wild-type terminal leaflets (see Supplemental Figure 6 online). Despite a reduction in terminal leaflet area (see Supplemental Figures 6A, 6B, and 6E online), mesophyll cell density was lower in lyr (see Supplemental Figures 6C, 6D, and 6F online). In addition, mesophyll cell size was increased in lyr (see Supplemental Figures 6C and 6D online). These results combined demonstrate that the number of cell divisions that occur during blade formation in lyr is decreased.
We also examined the histology of lyr leaves to determine whether these defects extended to the organization of internal tissues. Paraffin sections through mature lyr blade tissue revealed relatively normal internal patterning, suggesting that lyr does not play a major role in patterning adaxial/abaxial polarity or differentiation (Figures 2F and 2G). Interestingly, histological analysis of the lyr terminal leaflet midrib revealed ectopic spongy mesophyll tissue replacing collenchyma cells just above the epidermis on the abaxial side of the leaf (Figures 2F and 2G, arrow). In addition, collenchyma cells are missing from the center of the adaxial side at the midrib region (Figures 2F and 2G, arrowhead). These results suggest that LYR is not a major factor patterning differentiation of internal tissues during leaf development but may play a minor role in specification of collenchyma identity in the midrib.
LYR Expression Pattern
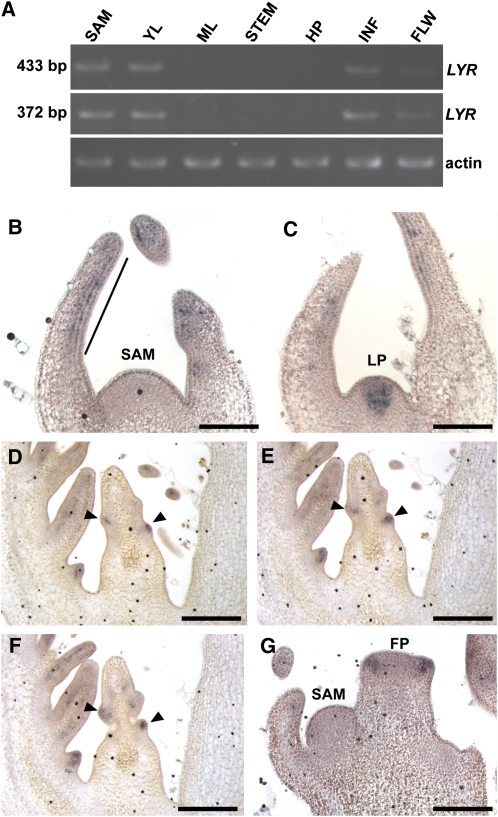
To further analyze the role of LYR in leaf development in tomato, we characterized the LYR expression pattern. RT-PCR analysis revealed LYR expression in shoots (including P0-P5), young leaves (4 to 7 mm in length), inflorescences, and flowers but not in the mature leaf, stem, or hypocotyl (Figure 3A). In vegetative apices, the LYR transcript localized to initiating leaf primordia and expanding leaves but was excluded from the SAM (Figures 3B to 3F; see Supplemental Figure 7 online). LYR expression was evident across the adaxial surface of the leaf primordia with more intense expression in distal tissues that give rise to the blade of the terminal leaflet. During these early stages of leaf development, the LYR expression domain resembles that of JAG in Arabidopsis young leaf primordia (Figures 3B and 3C; see Supplemental Figure 7 online) (Ohno et al., 2004). As the leaf transitions to leaflet initiation, LYR is strongly and specifically upregulated in leaflet primordia (Figures 3D to 3F). Expression was low or not detected in tissues adjacent to leaflets. This represents a significant divergence from reported JAG expression. In addition to expression in developing leaves, LYR was also detected in all floral organ primordia (Figure 3G). In summary, the LYR expression domain is consistent with a role in promoting active cell proliferation in developing lateral organs. Interestingly, LYR is strongly expressed in the actively proliferating cells that will give rise to leaflets and blade in tomato compound leaves. This result suggests that LYR expression might regulate the differential outgrowth necessary for dissected leaf development.
Figure 3.
LYR Expression Pattern in Wild-Type Tomato.
(A) RT-PCR analysis showing expression of LYR in several tissues collected from VF36 tomato plants. LYR was amplified by two different primer sets; length of each product is indicated on the left. Actin was amplified as a positive control for RNA integrity.
(B) to (G) In situ hybridization in VF36 meristems with LYR RNA probe.
(B) to (F) LYR expression pattern in developing leaves.
(B) LYR RNA is absent in the meristem and is localized to the adaxial domain of the distal part of leaf primordium (black line).
(C) LYR in leaf primordium (see Supplemental Figure 7 online for serial section of this meristem).
(D) to (F) Serial sections of young leaf showing LYR RNA localization in leaflet initiation sites (arrowheads).
(G) Floral primordium showing expression of LYR RNA in sepal primordia but no signal in SAM.
YL, young leaf; ML, mature leaf; HP, hypocotyl; INF, inflorescence; FLW, flowers; LP, leaf primordium; FP, flower primordium. Bars = 250 μm.
Ectopic Expression of LYR in Tomato Results in Ectopic Laminar Growth
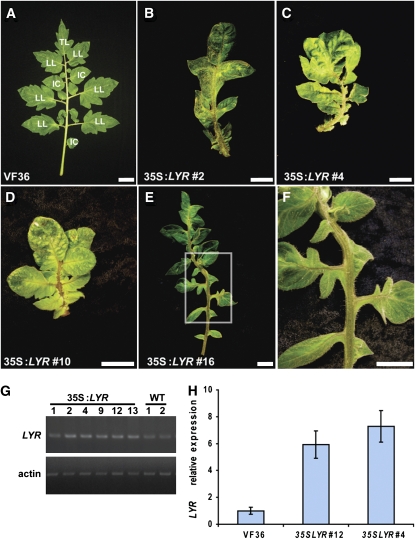
Our analysis suggested that differential expression of LYR in developing leaf primordia may be important to pattern outgrowth in dissected leaves. To determine whether this might be the case, we attempted to alter the LYR expression domain by generating transgenic plants that ubiquitously express LYR (35S:LYR). Analysis of 16 plants expressing 35S:LYR revealed eight plants with ectopic blade growth on petioles, rachis, and petiolules (Figures 4B to 4D). In some cases, ectopic outgrowth was along the entire rachis (Figure 4B), while in weaker events, the ectopic blade only partially covered the main rachis and petiolules (Figures 4C to 4F). Leaflet placement was irregular compared with that of the wild type, suggesting ectopic primordia initiation (Figure 4E). Secondary leaflet formation was not in evidence or was dramatically reduced (Figure 4). Occasionally, the leaves of these plants were bifurcated (see Supplemental Figure 8 online). Flowers of transgenic plants were larger than wild-type flowers (see Supplemental Figure 8 online). These plants rarely produced fruits, and when they did, the fruits were usually irregular in shape and mostly parthenocarpic. The increased expression of LYR, as detected by RT-PCR and quantitative RT-PCR analyses, could be correlated to these phenotypes (Figures 4G and 4H). In contrast with JAG overexpression in Arabidopsis, and in agreement with what was shown for 35S:LYR in Arabidopsis (see Supplemental Figure 3 online), bract formation was not evident in tomato plants expressing 35S:LYR. Together, these results suggest that LYR promotes blade outgrowth in tomato leaves. Furthermore, ubiquitous expression of LYR throughout the developing leaf primordia results in highly simplified leaf development. This result further suggests that punctuated LYR expression is important to specify differential outgrowth in dissected leaves.
Figure 4.
Ectopic Expression of LYR in Tomato.
(A) VF36 wild type fully developed leaf with the different leaflet type designation. TL, terminal leaflet; LL, lateral leaflet; IC, intercalary leaflet.
(B) to (F) 35S:LYR VF36 transgenic plants.
(B) to (D) Leaves with ectopic blade on rachis and petiolule of line numbers 2 (B), 4 (C), and 10 (D). Bars = 2 cm in (A) to (D).
(E) Ectopic blade outgrowth with an extra leaflet.
(F) Higher magnification of the frame in (E) showing ectopic blade outgrowth on rachis and petiolules.
(G) RT-PCR analysis of 35S:LYR transgenic plants showing upregulation of LYR in lines that show ectopic blade growth on leaves (numbers 2, 4, 9, 12, and 13) but not in a line that shows wild-type phenotype (number 1). Actin mRNA was amplified as a positive control for RNA integrity. Two technical replications were preformed for each sample.
(H) Quantitative RT-PCR analysis of LYR expression in wild-type and two 35S:LYR transgenic plants. GAPDH expression values were used to normalize the expression level of LYR. Error bars indicate se over three technical replicates.
[See online article for color version of this figure.]
LYR Negatively Regulates KNOX Gene Expression
The knotted-like homeobox genes have been shown to positively regulate compound leaf development by promoting a meristematic environment in developing leaves (Hareven et al., 1996; Chen et al., 1997; Parnis et al., 1997; Janssen et al., 1998), and overexpression of several KNOX genes in tomato results in a highly dissected leaf form. We investigated the possibility that the lyr phenotype might result from alteration in expression of members of the KNOX transcription factor gene family. Using quantitative RT-PCR, we examined the expression of two KNOX genes, LeT6 (the STM ortholog, also known as TKN2) and TKN1 (the bp ortholog) in wild-type and mutant apices (Hareven et al., 1996; Chen et al., 1997). Both LeT6 and TKN1 expression levels were upregulated in lyr when compared with the wild type, although only LeT6 upregulation was statistically significant (Figure 5A). These results suggest that LYR negatively regulates KNOX gene expression in developing tomato leaves. Thus, KNOX overexpression may partially explain the increase in leaf complexity seen in lyr.
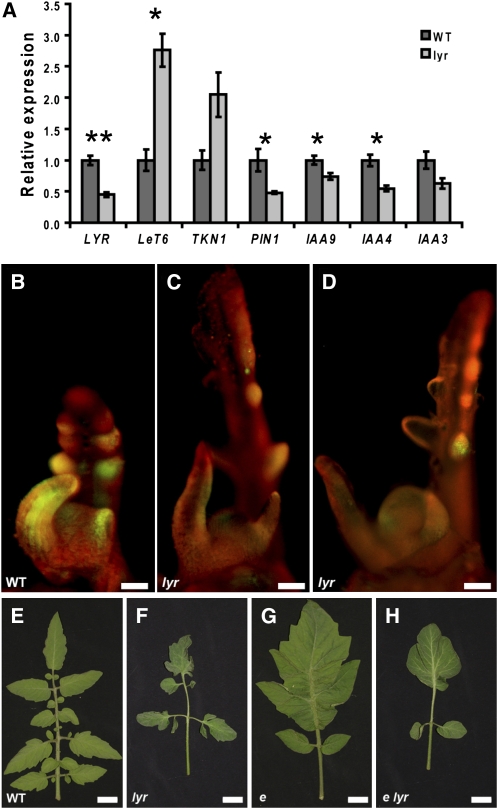
Figure 5.
LYR Affects KNOX and Auxin-Related Genes.
(A) Quantitative RT-PCR analyses showing relative expression of different genes in lyr versus wild-type apices. GAPDH expression values were used to normalize the expression level of the different genes. Error bars indicate se over three technical replicates.
(B) to (D) pPIN1:PIN1:GFP transgene expression in wild-type (B) and lyr ([C] and [D]) apices. Image shown in (D) was taken in high contrast to illustrate that GFP is upregulated in leaf and leaflet initiation sites.
(E) to (H) Leaf phenotypes observed in e × lyr F2 progeny.
Bars = 200 μm in (B) to (D) and 2 cm in (E) to (H). One asterisk indicates P < 0.05 and two asterisks indicate P < 0.01.
LYR Is Necessary for a Robust Auxin Response in Tomato Leaves
Auxin signaling has been shown to be important for leaflet initiation and for lamina outgrowth in tomato (DeMason and Chawla, 2004; Wang et al., 2005). Since lyr develops fewer leaflets and displays restricted lamina outgrowth, we examined whether the auxin response is altered in lyr. Using quantitative RT-PCR, we found that the expression of the auxin-responsive genes PIN1, IAA9, and IAA4 (Wang et al., 2005) was significantly downregulated in lyr when compared with wild-type plants (Figure 5A). IAA3 was similarly downregulated, albeit not to statistically significant levels (Figure 5A). These results suggest that LYR positively regulates the auxin response.
To further address this hypothesis, we examined the activity of the auxin-responsive transgene pPIN1:PIN1:GFP in the lyr mutant background. In agreement with previously published results, pPIN1:PIN1:GFP expression in a wild-type background is highest in initiating leaf and leaflet primordia (Figure 5B) (Koenig et al., 2009). Green fluorescent protein (GFP) was also upregulated in initiating leaves and leaflets of lyr but at much reduced levels (Figures 5C and 5D). This reduction in PIN1:GFP expression supports the quantitative RT-PCR results showing endogenous tomato PIN1 is downregulated in lyr (Figure 5A). In combination, these results suggest that LYR is necessary to condition high levels of auxin response in developing compound leaves.
We additionally examined the role of LYR in promoting the auxin response in leaflets by generating double mutants with the auxin response mutant entire (e) (Figures 5E to 5H). e mutants fail to inhibit the auxin response between developing leaflets, resulting in ectopic blade outgrowth along the rachis (Figure 5G) (Koenig et al., 2009). e lyr double mutants exhibited a somewhat intermediate phenotype relative to both single mutants (Figure 5H). The increased secondary dissection observed in lyr was eliminated in the e lyr doubles (Figure 5H), but ectopic blade outgrowth in e lyr was restricted distally when compared with e. This intermediate phenotype suggests that E and LYR regulate the auxin response independently.
DISCUSSION
We have described the identification of the classical tomato mutant lyr, which results from a mutation in the tomato ortholog of JAG. Like JAG, LYR acts to promote lateral outgrowth in aerial organs. LYR is expressed in a punctuated pattern during leaf development that coincides with the location of auxin response maxima and subsequent leaflet initiation. LYR promotes auxin response in the developing leaflet, first to drive primordia initiation, and later to promote blade outgrowth.
The Function of LYR in Patterning Dissected Leaves
During early development, the simple leaves of Arabidopsis are divided into two major domains that are delineated by JAG expression (Ohno et al., 2004). The distal region of the leaf primordia that will give rise to the blade is characterized by high expression of JAG, while the proximal bladeless petiole lacks JAG expression. In compound leaves, the patterning of outgrowth is significantly more complex, and our results reveal LYR as an important molecular component of this process.
A major difference between early compound and simple leaf development is the initiation of leaflet primordia. Simple leaves initiate outgrowth throughout the distal margin to promote development of a continuous blade. By contrast, initiation and delineation of leaflets in compound leaves requires unequal promotion of outgrowth along the margin. In accordance with this requirement, we find that LYR/JAG expression has diverged from the bipartite pattern that delineates blade and petiole in simple leaves to a punctuated pattern accommodating localized leaflet outgrowth. Additionally, the lyr mutant suppresses leaflet initiation along the primary axis of development. Loss of leaflet initiation is most evident in the lyr2 allele that results from a mutation of the LYR DNA binding domain. It is possible that the more severe phenotype of this allele results from a dominant-negative effect, while lyr1 and lyr3 are more representative of LYR loss of function. This hypothesis remains to be tested directly in future experiments. Regardless, all alleles reduced primary leaflet number to some extent, demonstrating that LYR is an important regulator of leaflet initiation.
The reduction in the number of leaflets in lyr plants seems to result from loss of one or two basal lateral pairs, suggesting that the distal pair may be less sensitive to LYR loss of function. This may result from the activity of a redundant factor in the distal leaf primordium, and in the absence of a tomato genome sequence, we cannot exclude the possibility of the existence of a NUB ortholog. It is worth noting that a common compound leaf morphology is trifoliate (as seen in several legumes), suggesting that production of a single pair of lateral leaflets may be fundamentally different from reiteration of this process to form additional lateral pairs. This hypothesis is supported by the tomato monogenic mutant tf, which produces trifoliate leaves that are otherwise similar to those of wild-type plants. Identification of the molecular nature of the tf mutant and examination of the genetic interaction between lyr and tf should add to our understanding of the process that regulates initiation of the first pair of leaflets.
The role of LYR in tomato compound leaf development bears a striking resemblance to that seen for auxin (Koenig et al., 2009). Both LYR and auxin accumulate in initiating leaflets and are absent in the bladeless tissues separating leaflets. Furthermore, exogenous application of auxin to leaf primordia results in ectopic blade production on the rachis, reminiscent of the LYR overexpression phenotype. This phenotype is also mimicked by ectopic activation of auxin response in the simple leafed tomato mutant e. Our results show that these similarities are not a coincidence; LYR promotes a strong auxin response, suggesting that the lyr phenotype might result from a reduction in the magnitude of transcriptional response to auxin. The intermediate phenotype of e lyr suggests that the two genes regulate auxin response independently. An additional patterning factor, GOB, has also been shown to act in at least partial independence of E. Thus, it is difficult at this time to establish a linear relationship between the initial auxin signal and the downstream patterning elements required for leaf dissection in tomato. Although auxin distribution can influence E activity directly via degradation (Koenig et al., 2009), the relationship between GOB and LYR expression and auxin distribution remains unclear. Future experiments testing the phenotypic outcome of auxin treatment in lyr and the effect of auxin treatment on LYR and GOB transcript levels will help resolve this issue.
Function of LYR in Leaflet Indeterminacy
KNOX genes are key regulators in maintaining indeterminacy in the SAM and in the developing compound leaf (Bharathan et al., 2002). Increases in both leaflet number and the order of leaf complexity are seen in tomato mutants or transgenics that overexpress KNOX genes (Hareven et al., 1996; Chen et al., 1997). The lyr loss-of-function mutants also showed an increase in leaflet number, and expression analysis showed a modest increase in KNOX transcript levels (LeT6 and TKN1) in lyr (Figure 5). It is possible that lyr produces additional leaflets that are due to this increase in KNOX expression. Furthermore, both KNOX overexpression and lyr loss of function result in delayed trichome differentiation during leaf development. In the absence of loss-of-function mutations in tomato KNOX genes, it is difficult to definitively establish their contribution to the lyr phenotype. However, it is clear that even moderate KNOX overexpression in tomato transgenics and naturally occurring mutants results in highly ramified leaves along all leaf axes (Janssen et al., 1998). It is therefore likely that upregulation of KNOX gene expression in lyr contributes in part to the production of additional leaflets.
Secondary Morphogenesis and the Relationship between Simple and Compound Leaves
The homology between compound and simple leaves has long been a subject of botanical debate. The resemblance of the leaflets of compound leaves to an entire simple leaf has led some classical botanists to conclude that compound leaves possess a shoot-like identity (Arber, 1950). An alternative hypothesis suggests that compound leaves are simply the result of dissection of the simple leaf blade into smaller units (Kaplan, 1975). Although such mutually exclusive arguments are unlikely to accurately represent biological reality, they serve as useful starting points to interpret comparative developmental analyses.
Compound leaf morphogenesis is characterized by two basic developmental processes; the elaboration of leaflet primordia during early leaf development and restriction of blade outgrowth during late leaf development that maintains the independence of these leaflets. The KNOX genes are known to be important for the former process, and KNOX gene expression in young leaf primordia is closely correlated with initiation of leaflet primordia across most of flowering plants (however, see Champagne et al. [2007] for an exception). Since KNOX genes are important regulators of meristem identity, it is generally thought that this result provides evidence for homology between shoots and compound leaf primordia. Although the expression of KNOX genes in leaves and the subsequent formation of leaflet primordia may be prerequisites for compound leaf development in most species, it is clear that they are not sufficient. Many species show both of these characteristics, but still produce simple leaves by initiating blade outgrowth throughout the leaf margin, resulting in a continuous lamina (Bharathan et al., 2002). This process, by which blade outgrowth contributes to leaf shape, is termed secondary morphogenesis.
The mere existence of leaf simplification during secondary morphogenesis suggests that compound leaves are generated in part by dissecting the simple leaf blade, but several other lines of evidence support this idea. Specification of a complex pattern of growth such as that found during dissected leaf development requires the asymmetric distribution of molecular or physiological signals. Auxin appears to be one such signal, as its distribution during compound leaf development is in part responsible for delineating blade outgrowth and leaflet initiation (Barkoulas et al., 2008; Koenig et al., 2009). Our results demonstrate a close association between the function of LYR and auxin during compound leaf development. Much like the proposed function of auxin, LYR promotes outgrowth of both leaflets and blade, and its ubiquitous expression results in the formation of ectopic blade on petiolules and rachis. When considering the homology of compound and simple leaves, it is important to note that auxin peaks also form in the margins of simple leaves (guiding vascular and serration development) (Hay et al., 2006; Scarpella et al., 2006), while no such peaks have been reported for JAG or NUB expression. Thus, changes of JAG/LYR expression pattern represent a clear difference between simple and compound leaf development. It is therefore possible that alteration in auxin distribution, response pathways, or LYR expression pattern might be responsible for the natural occurrence of leaf simplification through secondary morphogenesis. Interestingly, both currently known promotive factors (auxin and LYR) in this network regulate both leaflet initiation and blade outgrowth. This suggests that all outgrowths from the marginal blastozone have some homology at the genetic and molecular levels and calls into question the idea that leaflet outgrowth is inherently different from blade outgrowth. It may be that the difference between these two fates is determined purely by the localization and/or intensity of the promotive signal during initiation. Previous experiments with auxin have suggested that this is the case (Koenig et al., 2009), and the fact that LYR overexpression can result in ectopic blade outgrowth and additional leaflet initiation also supports this hypothesis. Combined, these results reveal an intricate network of molecular and physiological regulators of growth that determine leaf shape in tomato, and it will be interesting to determine how these factors may influence the patterning of the myriad of leaf morphologies yet to be explored.
METHODS
Plant Material and Growth Conditions
Solanum lycopersicum cv VF36 was used for all experiments described in this article. The tomato mutants lyr (LA0763), lyr2 (LA2923), and tf (LA0579) were kindly provided by Tim Wills and Roger Chetelat from the C.M. Rick Tomato Genetics Resource Center (http://tgrc.ucdavis.edu/). The tomato mutant lyr3 was kindly provided by Yuval Eshed (Weizmann Institute of Science, Israel) and the Genes that Make Tomatoes mutant database (http://zamir.sgn.cornell.edu/mutants/).
The tomato transgenic line expressing pPIN1:PIN1:GFP was kindly provided by Cris Kuhlemeier (University of Bern). All tomato plants were grown in a growth chamber at 22°C with 75% relative humidity and a daylength of 16 h.
Arabidopsis thaliana plants were grown in a growth chamber at 20°C and a daylength of 18 h. All plants were in the Landsberg erecta background. Seeds of jag-2 and jag-3 (Ohno et al., 2004) were kindly provided by Carolyn Ohno and Elliot Meyerowitz (California Institute of Technology).
Cloning S. lycopersicum JAG
The Arabidopsis JAG sequence was used to search the SOL Genomics Network (http://www.sgn.cornell.edu) identifying a single EST sequence from tomato (EST242380) with high homology to JAG. Total RNA was extracted from tomato SAM using the plant RNeasy mini kit (Qiagen) and subjected to poly(A) selection using the PolyATtract kit (Promega). Primers SlJAG5′RACE (5′-CTCTTGACGGTGGCGGTTCATGTGTCCCCC-3′) and SlJAG3′RACE (5′-GGGGGACACATGAACCGCCACCGTCAAGAG-3′) were used to clone the full-length S. lycopersicum JAG cDNA using the BD SMART rapid amplification of cDNA ends kit (RACE; Clontech). The assembled cDNA sequence was used to design primers for genomic DNA amplification from genomic DNA of the tomato cultivar VF36. Nucleotide and amino acids sequences were aligned to the Arabidopsis JAG sequences using the Vector NTI Advance 9 Suite (Invitrogen).
DNA Gel Blot Analysis
Genomic DNA was extracted from mature leaf tissue using the CTAB extraction protocol with minor modifications. DNA gel blot analysis was performed as described by Chen et al. (1997). A LYR genomic fragment of 1.9 kb that resulted from digestion by both SacI and PacI restriction enzymes was used as a probe.
Construct Preparation
PAtJAG:gSlJAG
The JAG promoter was amplified from Arabidopsis (Landsberg erecta) using primers T26Bf_b 5′-GGAATAGAGCTGATGTAGTAGCCGTG-3′ and AtJAG_4 5′-GTTAAAGAAGAGAGGTTCGAAAGTTTTCTATCAG-3′ and cloned into the BJ36 vector using the EcoRI site. The genomic fragment of LYR was amplified from the VF36 tomato cultivar by primers HindIIILYRATE, 5′-CCCAAGCTTATCACTCAAAGTCTCAAACTCAAACTC-3′, and LYRATEBamHI, 5′-GCGGATCCGGTAACTCTTCATATTAAGTAAT-3′, and cloned into PCR2.1 using the TopoTA Kit (Invitrogen). The gLYRATE was then subcloned into BJ36;PAtJAG vector using HindIII and BamHI sites. The PAtJAG:gLYRATE cassette was subcloned into pMLBart using NotI sites and transformed into Agrobacterium tumefaciens ASE.
LYR RNAi
A 200-bp cDNA fragment of LYR was amplified from a cDNA library, which was made from SAM, by PCR using primers LYRATE_11a, 5′-CTCCACATCACCTTAGTTGTCC-3′, and LYRATE_11b, 5′-TACGCGGTGGAGAGGC-3′. The resulting cDNA was cloned into the pCR8/GW/TOPO entry vector (Gateway; Invitrogen) following the manufacturer's protocol. The LR reaction was performed between the entry vector and the destination vector pTKO2 as described in the Gateway cloning technology instruction manual. The pTKO2;LYRATE plasmid was introduced into A. tumefaciens ASE.
35S:LYR
The cDNA fragment of LYR was amplified from a cDNA library, which was made from SAM, by PCR using primers LYRATE_F, 5′-ATCACTCAAAGTCTCAAACTCAAACTC-3′, and LYRATE_R, 5′-TTGGAAAGGTAACTCTTCATATTAAG-3′. The resulting cDNA was cloned into the pCR8/GW/TOPO entry vector following the manufacturer's protocol. The attL × aatR reaction was performed between the entry vector and the destination vector pK2GW7 as described in the Gateway cloning technology instruction manual. The pK2GW7;LYR plasmid was introduced into A. tumefaciens ASE.
Transgenic Plants
35S:LYR and PAtJAG:gLYRATE constructs were transformed into jag-2 and jag-3 (Landsberg erecta) plants by the floral dipping method (Clough and Bent, 1998). 35S:LYR and LYR RNAi were transformed into tomato (S. lycopersicum cv VF36) at the Ralph M. Parsons Foundation Plant Transformation Facility (University of California, Davis).
Mapping S. lycopersicum JAG
JAG was mapped using a cleaved amplified polymorphic sequence marker in intron 2, using an F2 mapping population of tomato (S. lycopersicon × Solanum pennellii) that was provided by S. Tanksley (Cornell University, Ithaca, NY).
RT-PCR Analysis
Total RNA (1 μg) was extracted from tomato tissue using the Plant RNeasy mini kit (Qiagen) followed by RQ1 DNase (Promega) treatment and reverse transcription using random primer hexamers of the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen). In order to obtain results within the linear range, PCR conditions were 94°C for 5 min followed by 30 cycles of 94°C for 20 s, 53°C for 20 s, and 72°C for 45 s using primers LYRATE_RTPCR_F (5′-GAGTCCAGAAAGAAATCCACTTGA-3′) and LYRATE_RTPCR_R (5′-CCCCCAAGTGCTTGAGATT-3′) for LYR; tomato actin primers (5′-CCTCTTAACCCGAAGGCTAA-3′ and 5′-GAAGGTTGGAAAAGGACTTC-3′) were used as internal control for all RT-PCR reactions.
Quantitative RT-PCR Analysis
Quantitative real-time PCR analysis was performed using IQ SYBR Green Supermix (Bio-Rad) with a Bio-Rad icycler iQ. cDNA was made as described above and was diluted 20 times before being subjected to quantitative RT-PCR analysis. Two independent biological experiments were used for each sample, while quantifications were performed in triplicates. Specific gene expression was normalized to the internal control gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and a negative control without cDNA was also performed for each primer set. The gene expression value of the wild type was used as a control and set at 1.0. For primer sequences, see Supplemental Table 1 online.
Scanning Electron Microscopy
Tomato apices were fixed as described by Bharathan et al. (2002). Electronic images were obtained with a Hitachi S-3500 N scanning electron microscope (Hitachi Science Systems).
Histology
Tissues were fixed in FAA (formaldehyde 1.85%, glacial acetic acid 5%, and ethanol 63%), dehydrated through an ethanol series (70, 80, 90, and 100%, 30 min each), embedded in paraffin, sectioned using a Microtome HM 340 E (MICROM International), and stained with 0.1% toluidine blue O. The sectioned material was observed using a NIKON Eclipse E600 microscope, and digital images were taken using a SPOT RT camera (Diagnostic Instruments).
In Situ Hybridization Analysis
In situ analysis was performed as previously described (Long et al., 1996). A 560-bp fragment that was amplified from LYR cDNA by 5′-ACATGAACCGCCAC-3′ and 5′-AGTCCATGCCCTATTG-3′ primers was introduced into PCR2.1 and used as a DNA template to make RNA antisense probe by T7 RNA polymerase.
Mesophyll Cell Density Measurement
Leaf 1 was collected from wild-type and lyr plants with three expanded leaves. Photographs of all leaves were taken using an Olympus E600 camera. Measurement of leaf area was performed using the Image J software package. Terminal leaflet blade tissue was cleared by boiling in 80% ethanol until most chlorophyll was removed. The tissue was then transferred to 5% sodium hydroxide for 12 h, followed by chloral hydrate treatment overnight. The tissue was mounted and examined under a Nikon Eclipse SP-500UZ microscope. Each tissue sample was randomly photographed three times for technical replication from which a mean cell density was calculated for each biological replicate. Biological variation was then used to compare the genotypes.
Double Mutant Construction
The e homozygous plants were used as males and crossed to lyr heterozygotes to generate F1 plants. Plants were genotyped for lyr to identify heterozgotes, which were then selfed to generate segregating progeny. The novel e lyr double phenotype could be identified in these F2 plants but was confirmed by PCR for lyr and direct sequencing for e.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EU490614 for LYR and AAR30036 for Arabidopsis JAG.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Protein Alignment of JAG and Sl JAG.
Supplemental Figure 2. DNA Gel Blot Analysis of Sl JAG in Tomato.
Supplemental Figure 3. Function of Sl JAG in Arabidopsis Transgenic Plants.
Supplemental Figure 4. DNA Gel Blot Analysis Revealing Altered Sl JAG gene in lyr and lyr3 Tomato Mutants.
Supplemental Figure 5. lyr Flower Phenotype.
Supplemental Figure 6. lyr Leaf Mesophyll Cell Density.
Supplemental Figure 7. LYR Expression Pattern in an Apex of Wild-Type Tomato.
Supplemental Figure 8. Additional 35S:LYR Phenotypes.
Supplemental Table 1. Primers Used in This Study.
Supplemental References.
Supplementary Material
Acknowledgments
We thank Carolyn K. Ohno and Elliot M. Meyerowitz for providing Arabidopsis jag-2 and jag-3 seeds. We thank Yuval Eshed for lyr3 (n2731) seeds and information (http://zamir.sgn.cornell.edu/mutants/). We thank Cris Kuhlemeier for providing the pPIN1:PIN1:GFP tomato line. We also thank Tim Wills and Roger Chetelat from the Tomato Genetics Resource Center (University of California, Davis) for providing lyr (LA0763), lyr2 (LA2923), and tf2 (LA0579) seeds. We thank Steve Tanksley for providing us with the 80 F2 individuals from the cross L. esculentum LA925 × S. pennellii LA716. We thank Sakuntala Karunairetnam (HortResearch, New Zealand) for the gift of the pTKO2 plasmid. R.D.-S. was supported by Vaadia-BARD Postdoctoral Fellowship Award FI-343-2003 from BARD, the U.S.–Israel Binational Agricultural Research and Development. This work was funded by the National Science Foundation Developmental Mechanism Awards 0344743 and 0641696 (to N.R.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Neelima R. Sinha (nrsinha@ucdavis.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber, A. (1950). The Natural Philosophy of Plant Form. (Cambridge, UK: Cambridge University Press).
- Avery, G.S., Jr. (1933). Structure and development of the tobacco leaf. Am. J. Bot. 20 565–592. [Google Scholar]
- Barkoulas, M., Hay, A., Kougioumoutzi, E., and Tsiantis, M. (2008). A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 40 1136–1141. [DOI] [PubMed] [Google Scholar]
- Berger, Y., Harpaz-Saad, S., Brand, A., Melnik, H., Sirding, N., Alvarez, J.P., Zinder, M., Samach, A., Eshed, Y., and Ori, N. (2009). The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136 823–832. [DOI] [PubMed] [Google Scholar]
- Bharathan, G., Goliber, T.E., Moore, C., Kessler, S., Pham, T., and Sinha, N.R. (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296 1858–1860. [DOI] [PubMed] [Google Scholar]
- Blein, T., Pulido, A., Vialette-Guiraud, A., Nikovics, K., Morin, H., Hay, A., Johansen, I.E., Tsiantis, M., and Laufs, P. (2008). A conserved molecular framework for compound leaf development. Science 322 1835–1839. [DOI] [PubMed] [Google Scholar]
- Brand, A., Shirding, N., Shleizer, S., and Ori, N. (2007). Meristem maintenance and compound-leaf patterning utilize common genetic mechanisms in tomato. Planta 226 941–951. [DOI] [PubMed] [Google Scholar]
- Champagne, C.E., Goliber, T.E., Wojciechowski, M.F., Mei, R.W., Townsley, B.T., Wang, K., Paz, M.M., Geeta, R., and Sinha, N.R. (2007). Compound leaf development and evolution in the legumes. Plant Cell 19 3369–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.-J., Janssen, B.-J., Williams, A., and Sinha, N. (1997). A gene fusion at a homeobox locus: Alternations in leaf shape and implications for morphological evolution. Plant Cell 9 1289–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Coleman, W.K., and Greyson, R.I. (1976). The growth and development of the leaf in tomato (Lycopersicon esculentum). II. Leaf ontogeny. Can. J. Bot. 54 2704–2717. [Google Scholar]
- DeMason, D.A., and Chawla, R. (2004). Roles for auxin during morphogenesis of the compound leaves of pea (Pisum sativum). Planta 218 435–448. [DOI] [PubMed] [Google Scholar]
- DeMason, D.A., and Polowick, P.L. (2009). Patterns of Dr5:GUS expression in organs of pea (Pisum sativum). Int. J. Plant Sci. 170 1–11. [Google Scholar]
- Dengler, N.G. (1984). Comparison of leaf development in normal (+/+), entire (e/e), and Lanceolate (La/+) plants of tomato, Lycopersicon esculentum ‘Ailsa Craig’. Bot. Gaz. 145 66–77. [Google Scholar]
- Dinneny, J.R., Weigel, D., and Yanofsky, M.F. (2006). NUBBIN and JAGGED define stamen and carpel shape in Arabidopsis. Development 133 1645–1655. [DOI] [PubMed] [Google Scholar]
- Dinneny, J.R., Yadegari, R., Fischer, R.L., Yanofsky, M.F., and Weigel, D. (2004). The role of JAGGED in shaping lateral organs. Development 131 1101–1110. [DOI] [PubMed] [Google Scholar]
- Frary, A., Fritz, L.A., and Tanksley, S.D. (2004). A comparative study of the genetic bases of natural variation in tomato leaf, sepal, and petal morphology. Theor. Appl. Genet. 109 523–533. [DOI] [PubMed] [Google Scholar]
- Hagemann, W., and Glesissberg, S. (1996). Organogenetic capacity of leaves: The significance of marginal blastozones in angiosperms. Plant Syst. Evol. 199 121–152. [Google Scholar]
- Hareven, D., Gutfinger, T., Parnis, A., Eshed, Y., and Lifschitz, E. (1996). The making of a compound leaf: Genetic manipulation of leaf architecture in tomato. Cell 84 735–744. [DOI] [PubMed] [Google Scholar]
- Hay, A., Barkoulas, M., and Tsiantis, M. (2006). ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 133 3955–3961. [DOI] [PubMed] [Google Scholar]
- Janssen, B.-J., Lund, L., and Sinha, N. (1998). Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 117 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, D.R. (1975). Comparative developmental evaluation of the morphology of Unifacial leaves in the monocotyledons. Bot. Jahrb. Syst. 95 1–105. [Google Scholar]
- Koenig, D., Bayer, E., Kang, J., Kuhlemeier, C., and Sinha, N. (2009). Auxin patterns Solanum lycopersicum leaf morphogenesis. Development 136 2997–3006. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Menda, N., Semel, Y., Peled, D., Eshed, Y., and Zamir, D. (2004). In silico screening of a saturated mutation library of tomato. Plant J. 38 861–872. [DOI] [PubMed] [Google Scholar]
- Nath, U., Crawford, B.C., Carpenter, R., and Coen, E. (2003). Genetic control of surface curvature. Science 299 1404–1407. [DOI] [PubMed] [Google Scholar]
- Ohno, C.K., Reddy, G.V., Heisler, M.G., and Meyerowitz, E.M. (2004). The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131 1111–1122. [DOI] [PubMed] [Google Scholar]
- Parnis, A., Cohen, O., Gutfinger, T., Hareven, D., Zamir, D., and Lifschitz, E. (1997). The dominant developmental mutants of tomato, Mouse-ear and Curl, are associated with distinct modes of abnormal transcriptional regulation of a Knotted gene. Plant Cell 9 2143–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig, R.S. (1984). Cellular parameters of leaf morphogenesis in maize and tobacco. In Contemporary Problems in Plant Anatomy, R.A. White and W.S. Dickison, eds (Orlando, FL: Academic Press), pp. 235–259.
- Scarpella, E., Marcos, D., Friml, J., and Berleth, T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya, H. (2006). Mechanism of leaf-shape determination. Annu. Rev. Plant Biol. 57 477–496. [DOI] [PubMed] [Google Scholar]
- Wang, H., Jones, B., Li, Z., Frasse, P., Delalande, C., Regad, F., Chaabouni, S., Latche, A., Pech, J.C., and Bouzayen, M. (2005). The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17 2676–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Chen, R., Xiao, J., Qian, C., Wang, T., Li, H., Ouyang, B., and Ye, Z. (2007). A single-base deletion mutation in SlIAA9 gene causes tomato (Solanum lycopersicum) entire mutant. J. Plant Res. 120 671–678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.