Abstract
Biosynthetic gene clusters are common in microbial organisms, but rare in plants, raising questions regarding the evolutionary forces that drive their assembly in multicellular eukaryotes. Here, we characterize the biochemical function of a rice (Oryza sativa) cytochrome P450 monooxygenase, CYP76M7, which seems to act in the production of antifungal phytocassanes and defines a second diterpenoid biosynthetic gene cluster in rice. This cluster is uniquely multifunctional, containing enzymatic genes involved in the production of two distinct sets of phytoalexins, the antifungal phytocassanes and antibacterial oryzalides/oryzadiones, with the corresponding genes being subject to distinct transcriptional regulation. The lack of uniform coregulation of the genes within this multifunctional cluster suggests that this was not a primary driving force in its assembly. However, the cluster is dedicated to specialized metabolism, as all genes in the cluster are involved in phytoalexin metabolism. We hypothesize that this dedication to specialized metabolism led to the assembly of the corresponding biosynthetic gene cluster. Consistent with this hypothesis, molecular phylogenetic comparison demonstrates that the two rice diterpenoid biosynthetic gene clusters have undergone independent elaboration to their present-day forms, indicating continued evolutionary pressure for coclustering of enzymatic genes encoding components of related biosynthetic pathways.
INTRODUCTION
The production of small molecule antibiotics is a broadly conserved biological defense mechanism. If these bioactive natural products are separated from primary metabolism by multiple biosynthetic/enzymatic steps, the corresponding genes must be inherited together, along with the appropriate regulatory mechanisms (e.g., cotranscription), to provide positive selection pressure for their retention. Furthermore, if intermediate metabolites have deleterious effects, negative selection is exerted against inheritance of the corresponding subsets of these genes and/or inappropriate suppression of downstream enzymatic activity. This push-pull combination has led to the assembly of biosynthetic gene clusters for multistep specialized metabolism across a wide range of organisms (Fischbach et al., 2008). These include not only microbes, such as bacteria and fungi, in which horizontal gene transfer increases the selective pressure for clustering, but plants as well, where such clustering is much less common, with only a few such cases known (Frey et al., 1997; Qi et al., 2004; Field and Osbourn, 2008). The rarity of biosynthetic gene clusters in plants raises the question of how such clusters were assembled in these multicellular eukaryotic organisms.
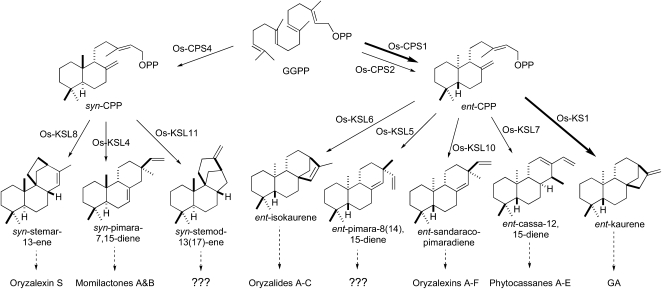
Rice (Oryza sativa) produces a complex mixture of antibiotic diterpenoid natural products in response to fungal infection (e.g., by the blast pathogen Magnaporthe grisea), as well as a separate set in response to bacterial infection (e.g., by the leaf blight pathogen Xanthomonas campestris), providing examples of both antifungal and antibacterial phytoalexins (Peters, 2006; Toyomasu, 2008). These phytoalexins all fall into the labdane-related diterpenoid subfamily, the founding members of which are the gibberellin (GA) phytohormones. Biosynthesis of these natural products is characteristically initiated by sequential cyclization reactions catalyzed by mechanistically distinct, yet phylogenetically related, diterpene synthases (Peters, 2006; Toyomasu, 2008). First, cyclization of the acyclic universal diterpenoid precursor [E,E,E]-geranylgeranyl diphosphate (GGPP) by class II diterpene cyclases, typically a labdadienyl/copalyl diphosphate (CPP) synthase (CPS), occurs. Second, this bicyclic intermediate is generally further cyclized by stereospecific class I diterpene synthases, sometimes termed kaurene synthase-like (KSL) because of their similarity to the corresponding enzyme in GA biosynthesis. Oxygen is then typically inserted into the resulting diterpene olefin by heme-thiolate cytochromes P450 (CYP) monooxygenases en route to the production of bioactive natural product(s). While the disparate CYP monooxygenases are all presumed to be, at least distantly, related to each other, they have been divided into numbered families and lettered subfamilies that are clearly homologous (≥40 and >55% amino acid sequence identity, respectively), with the number following the subfamily letter designating individual P450s (Werck-Reichhart and Feyereisen, 2000).
Given the importance of rice as a staple food crop and the knowledge of its genome sequence (Project, 2005), the metabolism of GA and other labdane-related diterpenoids has been extensively investigated in this model cereal plant (Peters, 2006). Although rice contains four CPS and 10 KSL, mutational analysis demonstrated that only one member of each of class of diterpene synthase (i.e., Os-CPS1 and Os-KS1) is involved in GA biosynthesis (Sakamoto et al., 2004). Biochemical characterization of the remaining CPS and KSL (Cho et al., 2004; Nemoto et al., 2004; Otomo et al., 2004a, 2004b; Prisic et al., 2004; Wilderman et al., 2004; Xu et al., 2004, 2007b; Kanno et al., 2006; Morrone et al., 2006) has assigned a unique metabolic function to each (Figure 1). By contrast, while P450 monooxygenases have been implicated in GA biosynthesis (Kato et al., 1995; Shimura et al., 2007), little is known about which of the >350 known rice CYPs are required and/or what their exact roles in diterpenoid phytoalexin biosynthesis are (Peters, 2006).
Figure 1.
Functional Map of Rice Diterpenoid Biosynthesis.
The cyclases and corresponding reactions are indicated, along with the downstream natural products, where known. Thicker arrows indicate enzymatic reactions specifically involved in GA metabolism; dashed arrows indicate multiple enzymatic reactions.
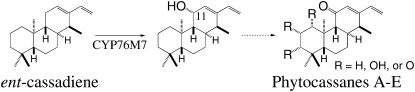
Intriguingly, the rice genome contains two gene clusters with genes encoding CPS, KSL, and CYP (Sakamoto et al., 2004). The smaller gene cluster on chromosome 4 is dedicated to the production of momilactones and contains the relevant, consecutively acting syn-CPP synthase (Os-CPS4) and syn-pimaradiene synthase (Os-KSL4) genes (Wilderman et al., 2004). In addition, this cluster contains a gene encoding a dehydrogenase that catalyzes the final step in the production of momilactone A (Os-MAS) and two closely related P450s (CYP99A2and3), one or both of which are required in an undefined role(s) in momilactone biosynthesis (Shimura et al., 2007). The larger cluster on chromosome 2 contains the gene encoding the phytoalexin-specific ent-CPP synthase Os-CPS2 (Prisic et al., 2004), three ent-CPP-specific KSLs (Os-KSL5-7), and several P450s from the CYP71 and 76 families. Although microarray transcriptional analysis has demonstrated that some of the coclustered CYP genes are coregulated with Os-CPS2 and Os-KSL7 (Okada et al., 2007), and members of the CYP71 and 76 families in other plant species function in terpenoid metabolism (Lupien et al., 1999; Collu et al., 2001; Ralston et al., 2001; Wang et al., 2001), a role for any of these monooxygenases in rice diterpenoid phytoalexin biosynthesis has remained conjectural. Here, we report that one of these coclustered and coregulated P450s, CYP76M7, is an ent-cassadiene-specific C11α-hydroxylase that seems to catalyze an early step in phytocassane biosynthesis and thus defines a second diterpenoid biosynthetic gene cluster, whose unique metabolic multifunctionality provides some insight into biosynthetic gene cluster assembly in plants.
RESULTS
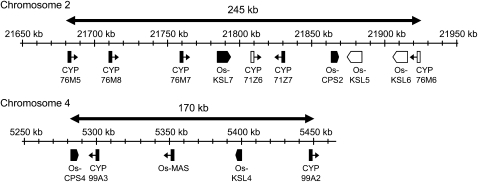
Extension of the Chromosome 4 Diterpenoid Biosynthetic Gene Cluster
It was originally suggested that the putative diterpenoid biosynthetic gene cluster on chromosome 2 contained four P450s (Sakamoto et al., 2004), which we found to correspond to CYP71Z6 and 7 and CYP76M6 and 7 by BLAST searches with the corresponding gene sequences against the cytochrome P450 database. Previous microarray/transcriptional analysis suggested that some of these CYP genes are also coregulated with Os-CPS2 and Os-KSL7, with their transcription being induced ∼4 h after elicitation of rice cell cultures with the fungal cell wall component chitin (Okada et al., 2007). A number of other CYP genes exhibit an analogous transcriptional induction pattern, and we found that two of these coregulated CYPs (CYP76M5 and 8) are immediately adjacent to the previously reported chromosome 2 diterpenoid biosynthetic gene cluster, meriting inclusion in this cluster. The resulting cluster then contains 10 genes and spans ∼245 kb on chromosome 2 and is comparable to the five-gene, ∼170-kb cluster on chromosome 4 (Figure 2). Notably, previous results have demonstrated that the transcription of some genes in the chromosome 2 cluster is not elicited by chitin (Okada et al., 2007). Strikingly, the coregulated genes are not grouped together.
Figure 2.
Schematic of Rice Diterpenoid Biosynthetic Gene Cluster.
Black boxes represent genes induced by chitin elicitation, while white boxes represent those not induced (Okada et al., 2007), with the arrowheads representing the direction of transcription. Note that no other genes appear to be present in these regions.
CYP76M7 Hydroxylates ent-Cassadiene
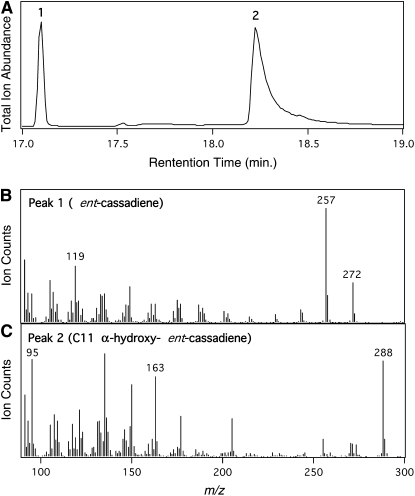
Given the coclustering and coregulation of four CYPs (CYP71Z7 and CYP76M5, 7, and 8) with the Os-CPS2 and Os-KSL7 known to act in phytocassane biosynthesis (Peters, 2006; Toyomasu, 2008), we hypothesized that these monooxygenases would similarly play a role in the production of this group of antifungal phytoalexins. Genes for all four of these CYPs were identified in the rice full-length cDNA sequencing project at KOME (Kikuchi et al., 2003) and obtained from this source. These were then expressed in the yeast (Saccharomyces cerevisiae) strain WAT11, in which the endogenous NADPH-cytochrome P450 reductase (CPR) had been replaced by a corresponding gene from Arabidopsis thaliana (At-CPR1; Urban et al., 1997). However, no P450 expression or activity was detected. To further assess the role of these CYPs, we attempted to express them in insect cells (Spodoptera frugiperda), which has proven to be a successful alternative method for the heterologous expression of other plant CYP (Jennewein et al., 2001). Using an insect cell (Sf21)-baculovirus (Autographa californica) expression system, we found that CYP76M7, although not the other three CYPs, reacts with ent-cassadiene (molecular mass = 272 D) fed to either microsomes or intact cells, with the resulting product appearing to be hydroxylated on the basis of its molecular mass (288 D). However, turnover was relatively limited, and it was not possible to obtain sufficient amounts of the hydroxylated product for structural analysis.
Identification of Hydroxylated Product as 11α-Hydroxy-ent-Cassadiene
To obtain larger amounts of the hydroxylated product, we employed the modular metabolic engineering system that we developed previously (Cyr et al., 2007) and attempted to coexpress CYP76M7 and a rice CPR (Os-CPR1) along with GGPP, ent-CPP, and ent-cassadiene synthases in Escherichia coli. However, while ent-cassadiene was produced in significant quantities, no hydroxylated derivatives were detected using the full-length CYP76M7. Based on our experience with recombinant expression of the kaurene oxidase CYP701A3 (D. Morrone and R.J. Peters, unpublished data), we modified the N terminus of CYP76M7 for functional bacterial expression. Specifically, we replaced the first 33 amino acids with a 10–amino acid Lys-rich sequence, based on a modification used for bacterial expression of the CYP2B subfamily of mammalian P450s (Scott et al., 2001). With this modified CYP76M7, we were able to detect significant hydroxylation (>50% conversion) of ent-cassadiene (Figure 3), resulting in the same product observed with the unmodified P450 expressed in insect cells. Accordingly, it was possible to produce and purify sufficient amounts of the hydroxylated ent-cassadiene for structural characterization by nuclear magnetic resonance (NMR). This analysis demonstrated that CYP76M7 performs C11α-hydroxylation of ent-cassadiene [forming 11-(S)-hydroxy-ent-cassa-12,15-diene]. In addition, coexpression of the modified CYP76M7 with other diterpene synthases demonstrated that this P450 is specific for ent-cassadiene, with only trace amounts of hydroxylation (<1% conversion) observed for ent-pimaradiene, ent-sandaracopimaradiene, ent-kaurene, ent-isokaurene, syn-pimaradiene, syn-stemarene, or syn-stemodene.
Figure 3.
Hydroxylation of ent-Cassadiene by CYP76M7.
(A) Gas chromatography (GC)-MS chromatogram of extract from E. coli engineered for production of ent-cassadiene and coexpressing CYP76M7 and rice CPR1 (1, ent-cassadiene; 2, 11α-hydroxy-ent-cassadiene).
(B) Mass spectra for peak 1 (ent-cassadiene).
(C) Mass spectra for peak 2 (11α-hydroxy-ent-cassadiene).
Enzymatic Characterization of Recombinant CYP76M7
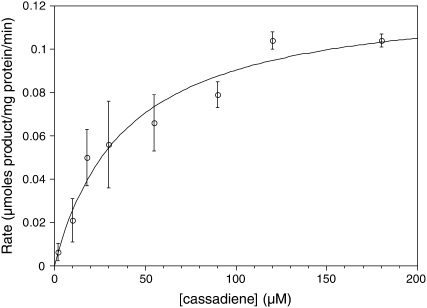
It was possible to measure the enzymatic activity of the modified recombinant CYP76M7 in vitro, which enabled more detailed enzymatic characterization. These in vitro assays required CPR supplementation, which was provided by the addition of exogenously produced At-CPR1. Intriguingly, in these assays, the production of an oxo-group (molecular mass = 286 D) was sometimes observed (see Supplemental Figure 1 online), in place of the expected C11α-hydroxy-ent-cassadiene. This oxo-containing compound also was produced when purified C11α-hydroxy-ent-cassadiene was fed to bacteria coexpressing CYP76M7 and Os-CPR1 (see Supplemental Figure 2 online), demonstrating that it represents further oxidation of the C11α-hydroxyl to a keto group. Uncoupling due to poor or limited association with recombinant CPR may result in the catalytic cycle being aborted after the initial oxidation. Kinetic constants were determined by simply measuring the rate of formation of C11α-hydroxy-ent-cassadiene in reactions where little of the C11-keto-ent-cassadiene product was made, whereupon CYP76M7 was found to exhibit a Km of 39 ± 10 μM and specific activity of 0.13 ± 0.02 μmoles product/mg protein/min (Figure 4). No activity was observed in vitro for the other diterpenes produced by rice (i.e., ent-pimaradiene, ent-sandaracopimaradiene, ent-kaurene, ent-isokaurene, syn-pimaradiene, syn-stemarene, or syn-stemodene).
Figure 4.
Kinetic Plot for CYP76M7 Activity.
The measured initial rates from duplicate assays (data points are the average and errors bars represent the sd) and fit to the Michaelis-Menton equation (R2 = 0.96) are shown.
Physiological Relevance of CYP76M7 Activity
The physiological relevance of the C11α-hydroxy-ent-cassadiene and C11-keto-ent-cassadiene products of CYP76M7 was investigated by metabolite analysis of induced and noninduced rice leaves. Detection of C11α-hydroxy-ent-cassadiene and C11-keto-ent-cassadiene by liquid chromatography–tandem mass spectrometry (LC-MS/MS) was optimized using purified compound. Extracts of rice leaves induced with methyl jasmonate, which has been demonstrated to induce the production of antifungal phytoalexins, such as the phytocassanes (Peters, 2006), were found to contain C11α-hydroxy-ent-cassadiene (Table 1), although C11-keto-ent-cassadiene was not detected. By contrast, neither compound was found in noninduced leaf extracts. The relevance of these findings is not entirely clear, as C11α-hydroxy-ent-cassadiene might accumulate as a metabolic dead-end rather than as a biosynthetic intermediate, while C11-keto-ent-cassadiene would not accumulate if the subsequently acting enzyme were not rate limiting. Nevertheless, the identification of at least one of the CYP76M7 products demonstrates that the associated hydroxylation reaction reported here does occur in planta, with the observed accumulation of C11α-hydroxy-ent-cassadiene upon induction consistent with the transcriptional pattern previously reported for CYP76M7. Given the presence of a C11-keto group in all of the identified phytocassanes from rice, CYP76M7 presumably catalyzes an early step in phytocassane biosynthesis (Figure 5), and the timing of induction of both CYP76M7 transcription and activity matches that for the production of phytocassanes.
Table 1.
LC-MS/MS Analysis of C11α-Hydroxy-ent-Cassadiene in Induced Rice Leaf Extracts
| Sample | RT (min)a | MS (m/z, %) | MS/MS Fragments (m/z, %) |
|---|---|---|---|
| Standard | 19.0–19.4 | 289(16), 271(100) | 243(6), 229(9), 215(33), 201(45), 189(16), 175(50), 163(100), 149(65), 133(10), 121(49), 109(36), 95(15), 81(4) |
| Induced leaf extract |
19.0–19.1 |
289(10), 271(100) |
243(5), 229(26), 215(26), 201(49), 189(15), 175(51), 163(100), 149(71), 133(17), 121(43), 109(35), 95(33), 81(8) |
Retention time.
Figure 5.
Reaction Catalyzed by CYP76M7 and Its Putative Relationship to Phytocassane Biosynthesis.
The dashed arrow indicates the multiple biosynthetic steps leading to the mixture of phytocassanes A to E.
Molecular Phylogenetic Analysis of the Coclustered Enzymatic Gene Families
The results presented above extend the functionality of the diterpenoid phytoalexin biosynthetic gene cluster on chromosome 2 to include CYP (at least CYP76M7), along with the previously noted sequentially acting CPS and KSL diterpene synthases (Prisic et al., 2004; Wilderman et al., 2004), just as found in the cluster on chromosome 4 (Okada et al., 2007). Since the work presented above indicates that both of these diterpenoid biosynthetic gene clusters in rice contain sequentially acting CPS, KSL, and CYP (Figure 2), we performed molecular phylogenetic analysis of these shared enzymatic gene families to provide some insight into the assembly of these clusters.
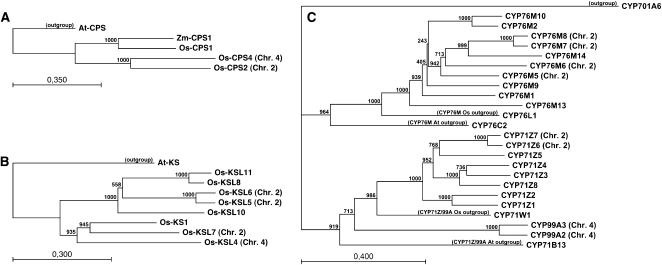
As we have previously noted (Prisic et al., 2004), the single class II diterpene cyclases found in each cluster, Os-CPS2 and Os-CPS4, are more closely related to each other than to the Os-CPS1 required for GA biynthesis. Indeed, Os-CPS1 is more closely related to a CPS found in Zea mays (Zm-CPS1) than to either Os-CPS2 or Os-CPS4. Thus, for molecular phylogenetic analysis, the CPS from the dicot Arabidopsis (At-CPS) was used as the outgroup sequence (Figure 6A; see Supplemental Figure 3 and Supplemental Data Set 1 online). This analysis demonstrates that there was a gene duplication following the separation of mono- and dicot plants that led to the Os-CPS2 and Os-CPS4 found in the clusters on chromosome 2 and 4, respectively.
Figure 6.
Molecular Phylogenetic Analysis of the Coclustered Rice Diterpenoid Biosynthetic Enzymatic Gene Families.
(A) Phylogenetic tree for cereal CPSs.
(B) Phylogenetic tree for Os-KSL.
(C) Phylogenetic tree for CYP71Z, CYP76M, and CYP99A subfamilies from rice.
The genes in the rice diterpenoid biosynthetic gene clusters are indicated by the corresponding chromosomal number (i.e., Chr. 2 or 4) in parentheses. The scale bars indicate the expected substitutions per site for the given branch length.
Molecular phylogenetic analysis of the class I diterpene synthase KSL family from rice (Os-KSL), using the ent-kaurene synthase (KS) from Arabidopsis (At-KS) as the outgroup sequence, indicates that there are two related groups of Os-KSL (Figure 6B). One of these groups is composed of Os-KS1, which is involved in GA biosynthesis, along with the ent-cassadiene producing Os-KSL7 found in the chromosome 2 cluster and the Os-KSL4 found in the chromosome 4 cluster. The other group includes the closely related and presumably recently duplicated Os-KSL5/Os-KSL6 pair of class I diterpene synthases, which are also found in the chromosome 2 cluster, indicating a separate evolutionary origin for these relative to the coclustered Os-KSL7.
A previous molecular phylogenetic analysis demonstrated that the CYP99 family falls within the CYP71 family and presumably forms a subfamily within the broader CYP71 family (Nelson et al., 2004). However, this does not necessarily mean that the CYP71Z subfamily members found in the chromosome 2 cluster and the CYP99A subfamily members found in the chromosome 4 cluster are closely related, as the CYP71/99 family is the largest CYP family in rice, being composed of 107 genes (although 31 are thought to be pseudogenes). Indeed, the majority of the CYP71Z subfamily members are scattered elsewhere in the genome. Furthermore, while the CYP71/99 and CYP76 families both fall within the CYP71 clan (Nelson et al., 2004), this relationship reflects an ancient division as both the CYP71/99 and CYP76 families are also found in Arabidopsis. Accordingly, our molecular phylogenetic analysis of the various CYP contained within the rice diterpenoid gene clusters includes not only the other subfamily group members scattered across the rice genome, but also the most closely related CYP from a different subfamily in both rice and Arabidopsis as outgroup sequences for each subfamily of interest and the rice kaurene oxidase (CYP701A6) as the outgroup sequence for the overall analysis (Figure 6C). This analysis further indicates some complexity in the CYP76M subfamily, as the subfamily members in the chromosome 2 cluster are closely related to the CYP76M14 that is found elsewhere in the rice genome. Examination of the CYP molecular phylogeny also indicates that several CYP genes within the clusters have undergone relatively recent tandem duplication (i.e., CYP99A2 and 3, CYP76M7 and 8, and CYP71Z6 and 7), much as has been hypothesized for Os-KSL5 and 6 (Xu et al., 2007a).
DISCUSSION
The results presented here demonstrate that CYP76M7 is a cytochrome P450 monooxygenase that catalyzes the insertion of oxygen at the C11α position in the labdane-related diterpene ent-cassadiene. The resulting C11α-hydroxy-ent-cassadiene can be found in planta upon methyl jasmonate induction, matching both the previously demonstrated mRNA transcript accumulation of CYP76M7 (Okada et al., 2007) and the inducible nature of phytocassane production (Koga et al., 1995), presumably reflecting a role for CYP76M7 in the biosynthesis of these phytoalexins. Given the typically conserved functionality among CYP subfamilies, these results further suggest that other CYP76M subfamily members are also involved in terpenoid biosynthesis, as has been found for the functionally characterized CYP71D subfamily members (Lupien et al., 1999; Ralston et al., 2001; Wang et al., 2001), which provides some general guidelines for further investigating the remaining nine CYP76M subfamily members in rice, as well as other CYP76M subfamily members identified in other plant species.
Definition of a role for CYP76M7 in phytoalexin diterpenoid biosynthesis also increases the number of CYP families acting in such specialized metabolism. In particular, the only previous angiosperm cytochrome P450 functionally identified as operating in specialized diterpenoid metabolism is the CYP71D16 involved in the production of cembratriendiol by tobacco (Nicotiana tabacum) trichomes (Wang et al., 2001). Interestingly, while both the CYP71 and 76 families fall within the large CYP71 clan, the cytochromes P450 from gymnosperms involved in specialized diterpenoid metabolism, including CYP720B1, which is involved in conifer resin acid biosynthesis (Ro et al., 2005) and the various CYP725 family members involved in taxoid biosynthesis (Jennewein et al., 2001; Schoendorf et al., 2001; Jennewein et al., 2003; Chau and Croteau, 2004; Chau et al., 2004) fall within the separate CYP85 clan. Nevertheless, the role for CYP76M7 in phytoalexin biosynthesis indicated here adds the CYP76 family to those involved in diterpenoid specialized metabolism. In addition, it seems likely that other CYP families from these broader clans will contain members and/or subfamilies dedicated to specialized diterpenoid metabolism.
Having provided strong evidence that CYP76M7 plays a role in phytocassane biosynthesis in rice by acting upon the ent-cassadiene product of the Os-CPS2 and Os-KSL7 diterpene synthases that are also encoded within the chromosome 2 cluster indicates that this cluster is functionally composed of consecutively acting CPS and KSL diterpene synthases and at least one CYP, much as previously shown for the diterpenoid biosynthetic gene cluster on chromosome 4 (Shimura et al., 2007). Accordingly, we performed molecular phylogenetic analysis to investigate the evolution of these two diterpenoid biosynthetic gene clusters from the rice genome.
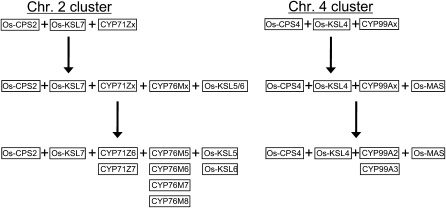
Intriguingly, the CPSs are the only genes in the rice diterpenoid biosynthetic gene clusters that are more closely related to those found in the other cluster than to paralogs found elsewhere in the genome (Figure 6). While some argument could be made that the evolutionary relationship between Os-KSL4 and Os-KSL7 reflects early incorporation of a common precursor in an ancestral cluster, both are more closely related to Os-KS1, which is involved in GA biosynthesis and is found elsewhere in the genome, than they are to each other. Furthermore, the closely related and presumably recently duplicated Os-KSL5 and 6 clearly have a separate evolutionary origin. Similarly, while the fact that CYP99 is actually a subfamily of CYP71 might be used to argue for early inclusion of a shared evolutionary precursor to the CYP71Z and CYP99A subfamily members in an ancestral cluster, this is undermined by the large numbers of other CYP71 family members (including the majority of CYP71Z subfamily members) found scattered elsewhere throughout the rice genome. In addition, the CYP76 family is sufficiently different to effectively rule out any recent shared origin for those family members found in the chromosome 2 cluster with either the coclustered CP71Z or the CYP99A found in the chromosome 4 cluster. Finally, the Os-MAS dehydrogenase found in the chromosome 4 cluster has no paralog in the chromosome 2 cluster. Thus, from our molecular phylogenetic analysis, it seems likely that these two diterpenoid biosynthetic gene clusters were independently assembled, although it is also possible that there was an ancestral cluster containing precursors to Os-CPS2 and 4, Os-KSL4 and 7, and CYP71Z and 99A. However, even if this latter case were true, further assembly would still have been required, with the chromosome 4 cluster picking up the MAS dehydrogenase, while the chromosome 2 cluster would have needed to add the KSL5 and 6 precursor, as well as CYP76M family members (Figure 7). In any case, such differential assembly implies that there was significant selective pressure driving this process.
Figure 7.
Hypothetical Assembly Process for the Observed Current Rice Diterpenoid Biosynthetic Gene Clusters Depicted in Figure 2.
Each box represents a gene in the cluster. In those cases where duplication has been postulated to occur after cluster assembly, a precursor gene is indicated (i.e., Os-KSL5/6 as the precursor to the current Os-KSL5 and 6, with “x” designating the precursor to the various CYP subfamily members found in the clusters).
Regardless of the precise assembly process, the chromosome 2 cluster defined here is a novel example of a multifunctional biosynthetic cluster. Os-KSL6 produces the ent-isokaurene precursor of the antibacterial oryzalides/oryzadiones, while Os-KSL7 and, as now indicated here, CYP76M7 are involved in biosynthesis of the antifungal phytocassanes (Figures 1 and 5). The previously reported difference in transcriptional regulation for these genes (Figure 2) also is consistent with their distinct roles in phytoalexin biosynthesis. This metabolic multifunctionality is unprecedented in the previously identified biosynthetic gene clusters from plants (Frey et al., 1997; Qi et al., 2004; Field and Osbourn, 2008) and helps define the underlying selective pressure leading to the at least partially independent differential assembly of the two rice diterpenoid biosynthetic gene clusters.
In particular, it is notable that Os-CPS2 is the only enzymatic gene from the chromosome 2 cluster that is clearly shared between phytocassane and oryzalide/oryzadione biosynthesis. While Os-CPS2 produces the enantiomeric form of CPP that could be an intermediate in GA phytohormone metabolism (Figure 1), knocking out Os-CPS1 is sufficient to essentially abolish GA production (Sakamoto et al., 2004). This rules out a role for Os-CSP2 in such primary metabolism, and its transcriptional regulatory pattern (Otomo et al., 2004a; Prisic et al., 2004) indicates that Os-CPS2 acts in more specialized (i.e., phytoalexin) metabolism instead. Accordingly, it appears that all the genes in both clusters are devoted to specialized metabolism, as none of the associated KSL enzymes make the ent-kaurene intermediate that is relevant to GA biosynthesis, and none of the characterized members of the CYP71/99 or CYP76 families have been found to be involved in primary metabolism. Thus, the selective force driving the differential assembly of the two rice diterpenoid biosynthetic gene clusters must be related to their role in more specialized metabolism. Indeed, in each of the identified cases, plant biosynthetic gene clusters are involved in specialized rather than primary metabolism.
As presented by Field and Osbourn (2008), the assembly of such specialized biosynthetic gene clusters in plants (as well as other organisms) could be driven by a variety of factors. These include the need for coregulation and inheritance of the full set of enzymatic genes necessary for biosynthesis of a bioactive natural product, as well as avoidance of toxic intermediate accumulation (i.e., from inheritance of only the corresponding subset of enzymatic genes). Notably, the lack of any recognizable regulatory pattern within the chromosome 2 cluster reported here suggests that coregulation was not a primary factor driving cluster assembly in at least this particular case, as does the coclustering of two functionally distinct biosynthetic pathways (i.e., enzymatic genes involved in either antifungal phytocassane or antibacterial oryzalide/oryzadione biosynthesis), which are differentially regulated (i.e., induced in response to either fungal or bacterial infections). Accordingly, we hypothesize that biosynthetic gene cluster assembly in plants is driven by the need for inheritance of complete biosynthetic pathways. In particular, the inheritance of complete biosynthetic pathways is important because the selective pressure for conservation of any enzymatic gene solely involved in specialized metabolism requires coinheritance of all the enzymatic genes necessary for production of the relevant bioactive natural product without the accumulation of toxic intermediates.
METHODS
General Procedures
Unless otherwise noted, chemicals were purchased from Fisher Scientific, and molecular biology reagents from Invitrogen. Gene mapping was based on the annotated rice genome sequence at GenBank. CYP nomenclature was determined via BLAST searches at the Cytochrome P450 homepage maintained by David Nelson (http://drnelson.utmem.edu/CytochromeP450.html). GC was performed with a Varian 3900 GC with Saturn 2100 ion trap mass spectrometer in electron ionization mode (70 eV). Samples (1 μL) were injected in splitless mode at 50°C and, after holding for 3 min at 50°C, the oven temperature was raised at a rate of 14°C/min to 300°C, where it was held for an additional 3 min. MS data from 90 to 600 m/z were collected starting 12 min after injection until the end of the run. LC was performed with Agilent 1100 series HPLC instruments, with LC-MS/MS analysis performed by a coupled Agilent MSD ion trap mass spectrometer in positive ion mode on an instrument located in the W.M. Keck Metabolomics Research Laboratory located at Iowa State University.
Recombinant Constructs
Genes for the four CYPs that are coclustered and coregulated with Os-CPS2 and Os-KSL7 (CYP71Z7 and CYP76M5, 7, and 8) were obtained from the KOME rice cDNA databank (Kikuchi et al., 2003). These were transferred into the Gateway vector system via PCR amplification, with incorporation of a consensus Kozak sequence and 6xHis tag at the 5′ end and directional topoisomerase-mediated insertion into pENTR/SD/D-TOPO and verified by complete sequencing. The resulting genes were then transferred via directional recombination to the yeast expression vector pYES-DEST52, the insect cell expression vector pDEST8, and the T7-promoter expression vector pDEST14. CYP76M7 was modified for functional bacterial expression in a two-stage PCR process, first removing 33 codons from the 5′ end of the open reading frame and then adding 10 new codons (encoding the amino acid sequence MAKKTSSKGK). The resulting construct was inserted into pENTR/SD/D-TOPO and verified by complete sequencing. For bacterial coexpression of the requisite redox partner, we cloned a rice CPR (Os-CPR1), also obtained from KOME, into the second multiple cloning site of the pCDFDuet dual expression vector using the NdeI and KpnI restriction sites (Novagen). For insertion of the various CYPs, a DEST cassette was then inserted into the first multiple cloning site using the NcoI and NotI restriction sites to enable transfer via directional recombination with Gateway pENTR-derived constructs.
Recombinant Expression in Yeast
Yeast CYP expression was performed following the manufacturer's directions for pYES-DEST52. Briefly, the pYES-DEST52 CYP constructs were transformed into the WAT11 strain of yeast (Urban et al., 1997), with selection on SC-Ura media. For expression, overnight cultures were inoculated (1:20) into SC-Ura with 2% (w/v) galactose, and cells were harvested at various time points from 0 to 24 h after inoculation. To examine CYP expression, cell extracts were run on 12% SDS polyacrylamide gels, and the separated proteins transferred to Hybond-P polyvinylidene difluoride membrane (Amersham Biosciences) for protein gel blot analysis using standard procedures (Harlow and Lane, 1988). Dilutions of 1:1500 and 1:2000 were used for the primary rabbit anti-His antiserum and secondary goat anti-rabbit Ig-peroxidase conjugate (Sigma-Aldrich), respectively. Bound antibodies were visualized with the ECL Plus Western Blotting Detection System (Amersham Biosciences). In none of the examined cases was CYP expression detected.
Recombinant Expression in Insect Cells
Sf21 cells (Vaughn et al., 1977) were maintained in TC-100 insect cell medium (Sigma-Aldrich) supplemented with FBS (Gibco-BRL) to a final concentration of 10% and antibiotics (1 unit penicillin/mL and 1 mg streptomycin/mL; Sigma-Aldrich). The cell cultures were maintained at 28°C as monolayers in screw-capped plastic flasks (Falcon).
Recombinant baculoviruses were constructed with the Bac-to-Bac baculovirus expression system (Invitrogen). Briefly, the CYPs were transferred from the pDEST8 vectors by recombination with the bMON14272 baculovirus shuttle vector in Escherichia coli following the manufacturer's instructions, but using 100 μg/mL kanamycin, 14 μg/mL gentamicin, and 20 μg/mL tetracycline for selection, and the resulting recombinant bacmid DNA was then isolated and transfected into Sf21 cells. The resulting recombinant viruses were amplified and their titer determined in Sf21 cells using standard procedures (O'Reilly et al., 1992).
Expression of CYP proteins by the recombinant baculoviruses was performed in Sf21 insect cells (Vaughn et al., 1977; Wickham et al., 1992). The cells were seeded into 150-cm2 cell culture flasks and infected with recombinant baculoviruses at a multiplicity of infection of 10 plaque-forming units/cell. Cells were harvested 72 h after infection, with CYP expression readily detected by protein gel blot analyses performed as described above.
Microsomes from the harvested cells were prepared by differential centrifugation. Briefly, cells were pelleted by centrifugation at 3000g at 4°C for 10 min. The pellets were resuspended in half a cell culture volume of 100 mM sodium phosphate buffer, pH 7.8, and repelleted at 3000g for 10 min, washed in ice-cold cell lysate buffer (100 mM sodium phosphate, pH 7.8, 1.1 mM EDTA, 0.1 mM DTT, 0.5 mM PMSF, 1/1000 v/v Sigma-Aldrich protease inhibitor cocktail, and 20% glycerol), repelleted at 3000g for 10 min, and resuspended in 1/50 cell culture volume of cold cell lysate buffer. The cells were lysed by sonification twice for 30 s on ice and by vortexing for 15 s. The lysate was centrifuged at 10,000g for 20 min at 4°C in a microcentrifuge, and the supernatant was further centrifuged in a Ti-70 rotor at 120,000g for 1 h in a Beckman-Coulter Optima ultracentrifuge to pellet the microsomes. The microsomal pellet was resuspended in 500 μL cold cell lysate buffer and used immediately or flash-frozen in liquid N2 and stored at −80°C for up to 1 month.
In vitro assays were performed in a 500-μL assay mixture containing the isolated microsomes (500 μg), 0.4 mM NADPH, 300 μM substrate, 2 μM reductase, 1 mM DTT, 5 μM FMN, 5 μM FAD, as well as NADPH regenerating system comprised of 2 mM glucose-6-phosphate and 0.5 units of glucose-6-phosphate dehydrogenase. The reaction mixture was incubated at 28°C for 6 h. The resultant product was extracted thrice with an equal volume of ethyl acetate, dried under a gentle stream of N2 gas, and dissolved in hexane for GC-MS analysis.
Whole-cell assays were performed by feeding 10 μM ent-cassadiene to CYP-expressing sf21 cells 2 d after infection. The cells were grown for two more days, the media and cells were sonicated, and diterpenoids were extracted for GC-MS analysis as above.
Recombinant Expression in E. coli
For bacterial expression, we combined coexpression of Os-CPR1 and CYP76M7 via the pCDFDuet vector construct described above with our previously described modular diterpene metabolic engineering system (Cyr et al., 2007). Specifically, we coexpressed these genes with genes encoding GGPP and ent-CPP synthases on the cocompatible pGGeC vector and the ent-cassadiene synthase (Os-KSL7) gene as a glutathione S-transferase fusion expressed from the also cocompatible pDEST15. While expression of the full-length CYP76M7 in this context did not lead to production of any hydroxylated ent-cassadiene, expression with the modified CYP76M7 led to conversion of >50% of the endogenously produced ent-cassadiene to the same hydroxylated derivative observed with insect cell–expressed CYP76M7 fed ent-cassadiene. Substituting other diterpene synthases for Os-KSL7 and/or the ent-CPP synthase (as appropriate; see Cyr et al., 2007) in this bacterial system did not result in significant conversion of the corresponding various other diterpenes, which were all produced in good yield, to any recognizable oxidized derivatives.
Diterpenoid Production
To produce sufficient amounts of the novel enzymatic product for NMR analysis, we increased the isoprenoid precursor supply by incorporating the bottom half of the mevalonate-dependent isoprenoid pathway from yeast in the engineered bacteria, using the previously described pMBI vector and supplementation with 20 mM mevalonolactone (Martin et al., 2003), much as previously described (Morrone et al., 2008). Diterpenoids were extracted from a 3-liter culture (media and cells) solution consisting of an equal volume of ethyl acetate and hexanes (50:50). These organic extracts were pooled and dried by rotary evaporation. The residue was dissolved in 5 mL 45% methanol/45% acetonitrile/10% deionized water and the diterpenoids purified by HPLC with a Supelcosil LC-18 column (4.6 × 250 mm, 5 μm) and a 0.5 mL/min flow rate. After binding, the column was washed with 20% acetonitrile/water (0 to 5 min) and eluted with 20 to 100% acetonitrile (5 to 15 min), followed by a 100% acetonitrile wash (15 to 30 min). The fraction containing the novel diterpenoid (retention time: 21.5 to 22 min) was dried under a gentle stream of N2 gas and then dissolved in 0.5 mL deuterated chloroform (CDCl3; Sigma-Aldrich). This evaporation resuspension process repeated two more times to completely remove the protonated acetonitrile solvent, resulting in a final estimated ∼2.5 mg of the novel diterpenoid.
Chemical Structure Analysis
NMR spectra for both the ent-cassadiene precursor and hydroxylated product were recorded at 25°C in chloroform-d1. NMR spectra were collected using a Bruker Avance 700 spectrometer equipped with a 5-mm HCN cryoprobe. Structural analysis was undertaken using 1D 1H, 1D 13C, DQF-COSY, HSQC, HMBC, ROESY (tmix = 400 ms), and HMQC-COSY spectra acquired using standard experimental protocols. Chemical shifts were referenced to trimethyl-silane. Chemical shifts for the ent-cassadiene substrate were assigned and found to be in agreement with published values (Yajima et al., 2004). Proton (700.13 MHz) chemical shifts and assignments for 11-(S)-hydroxy-ent-cassa-12,15-diene were as follows: δ 2.053 ppm (1H, d, 13.0 Hz, H1a), 1.086 (1H, t, 13.6, H1b), 1.574 (1H, d, m, H2a), 1.434 (1H, d, 13.2, H2b), 1.396 (1H, d, 13.4, H3a), 1.141 (1H, t, 13.9,H3b), 0.782 (1H, d, 12.5, H5), 1.541 (1H, d, 13.1, H6a), 1.336 (1H, m, H6b), 1.700 (1H, m, H7a), 1.433 (1H, dd, 4.0, 13.0, H7b), 1.919 (1H, m, H8), 1.169 (1H, d, 14.9, H9), 4.335 (1H, 7, 4.6, H11), 5.648 (1H, s, H12), 2.441 (1H, m, H14), 6.203 (1H, dd, 19.8, 11.2 H15), 5.209 (1H, d, 19.7, H16a), 5.054 (1H, d, 10.7, H16b), 0.893 (3H, d, 7, H17), 0.870 (3H, s, H18 or H19), 0.854 (3H, s, H18 or H19), and 1.118 (3H, s, H20). Carbon (174.05 MHz) chemical shifts and assignments for 11-(S)-hydroxy-ent-cassa-12,15-diene were as follows: δ (ppm) 40.33 (C1), 18.84 (C2), 42.08 (C3), 33.34 (C4), 55.77 (C5), 21.63 (C6), 29.87 (C7), 30.45 (C8), 49.48 (C9), 37.86 (C10), 66.13 (C11), 129.51 (C12), 143.84 (C13), 32.41 (C14), 138.62 (C15), 113.36 (C16), 14.98 (C17), 22.67 (C18 or C19), 34.30 (C18 or C19), and 18.56 (C20). The hydroxyl group was not directly observed in the 1H NMR spectra. It was assigned to the C11 position on the basis of the proton multiplicity of C11 (i.e., one), the characteristic 13C chemical shift of C11, and comparison with the corresponding NMR spectra for the ent-cassadiene precursor. The stereochemical configuration at C11 was determined as (S) (or α) on the basis of nuclear Overhauser effect (NOE) cross-peaks observed in a ROESY spectrum, using the known configurations of C9 and C20. The observed NOE correlations to H11 (and their relative intensities) are H1a (s), H1b (w), H9 (s), H12 (m), and H20 (w). NOE connectivity to H8 was not observed, although that spectral region was free from overlap. The mass spectral data for 11-(S)-hydroxy-ent-cassa-12,15-diene is as follows, EIMS m/z (%): 288(93), 255(17), 205(37), 177(39), 163(77), 150(70), 135(100), 121(45), 117(31), 107(40), 95(94), and 91(60) (see Figure 3C).
Kinetic Analysis
Truncated and modified CYP76M7 was expressed in E. coli C41 using the pCDFDuet vector and Os-CPR1 described above. The E. coli C41 cells used to produce CYP76M7 for in vitro kinetic analysis were also transformed with pGro7, an arabinose-inducible, groES-groEL protein chaperone expressing plasmid (TaKaRa) to improve the yield of active P450. Expression cultures were grown in TB and induced with 1 mM isopropyl β-d-1-thiogalactopyranoside and 2 mg/mL arabinose upon reaching an OD A600 of 0.8. Cultures were induced for 40 h at 28°C and complemented with 1 mM thiamine, 2 mg/L riboflavin, and 75 mg/L δ-amino levulinic acid at the time of induction. Upon completion of induction, cultures were centrifuged and the cell pellet resuspended in 5% culture volume cold membrane prep buffer (MPB), consisting of 0.1 M Tris-HCl, pH 7.2, 20% glycerol, 0.5 mM EDTA, 1 mM DTT, and 1× protease cocktail inhibitor. Lysozyme was added to 0.2 mg/mL and stirred for 10 min at 4°C. Spheroblasts were pelleted at 5000g for 10 min and subsequently resuspended in 10% culture volume of cold MPB. Very brief sonication was used to open spheroblasts. Concentrated spheroblasts were used immediately. Kinetic assays were performed in vitro by determining the initial velocity for various substrate concentrations following spheroblast preparation. However, the amount of cytochrome P450 present could not be quantified due to the lack of the typical peak at 450 nm in CO binding difference spectra. To ensure a constant supply of reducing equivalents, the NADPH regeneration system described above was used. Additionally, At-CPR1 was added to enhance reducing potential. Concentrated spheroblasts were diluted into 5-mL aliquots of MPB; 1-mL aliquots were removed at the various time points and the reactions halted by the addition of 1 mL 1 M HCl, immediately followed by vortexing, with a 4 mL ethyl acetate overlay added later with additional vortexing to assist extraction. The organic extract was removed and the reaction further extracted three times with 5 mL of hexanes. An internal standard of ent-copalol was used to standardize extraction efficiency, and authentic C11α-hydroxy-ent-cassadiene was used to generate a standard curve for quantification. Product identification was confirmed by GC-MS, and quantification was performed by GC-flame ionization detection. The reaction appeared to be in the linear range for the first 15 min. Following precipitation with 1% (v/v) trichloroacetic acid and washing of spheroblasts, protein quantification was performed using the Bio-Rad Protein Assay, which is based on the Bradford method, and 8 ± 4 mg protein was found to be present.
Metabolite Analysis
Rice plants (Orzya sativa ssp Nipponbare) were cultivated to the 6th leaf stage in growth chambers under cycles of 12 h light at 28°C and 12 h dark at 24°C cycles. For induction, plants were treated with 1 mL 0.2% methyl jasmonate per plant and incubated for 72 h, while control plants were only treated with the 0.1% Tween 20 carrier solution. The leaves were then clipped off (2 g), frozen, and ground to powder in liquid nitrogen. This plant material was then extracted with 50 mL ethyl acetate by stirring overnight at room temperature. The mixture was clarified by centrifugation, and the ethyl acetate extract dried under nitrogen gas and redissolved in 0.1 mL 50% methanol/water, from which 10 μL was injected for LC-MS/MS analysis. The LC-MS/MS analysis was performed using an Agilent ZORBAX Eclipse XDB-C8 column (4.6 × 150 mm, 5 μm) and a 0.5 mL/min flow rate and the same elution program described above for purification. Purified compounds were easily detected and used to optimize selective MS/MS ion monitoring. Specifically, for C11α-hydroxy-ent-cassadiene, the base peak (m/z = 271 [MH-H2O]+) from the molecular ion (m/z = 289 [MH]+) was selected for further fragmentation, with the resulting mass spectra compared with that found in the induced rice leaf extract at the same retention time.
Phylogenetic Analysis
Molecular phylogenetic analysis was performed with the CLC Sequence Viewer (Version 6.2) software package (CLC Bio). The alignments were performed using the very accurate alignment option, with a gap open cost of 10, gap extension cost of 1, and the end gap cost set to cheap. The phylogenetic trees were built using the neighbor-joining algorithm with bootstrap analysis of 1000 replicates, leading to the values shown for each node in Figure 6. For clarity, in Figure 6C, the root was set to the node above the CYP701A6 outgroup.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: At-CPS, U11034; Zm-CPS1, L37750; Os-CPS1, AK100333; Os-CPS2, AY602991; Os-CPS4, AY530101; At-KS, AF034774; Os-KS1, AY347876; Os-KSL4, AY616862; Os-KSL5, DQ823352; Os-KSL6, DQ823353; Os-KSL7, DQ823354; Os-KSL8, AB118056; Os-KSL10, DQ823355; Os-KSL11, DQ100373; CYP701A6, AK066285; CYP76M1, NM_001068532; CYP76M2, AK102040; CYP76M5, AK059010; CYP76M6, AK101003; CYP76M7, AK105913; CYP76M8, AK069701; CYP76M9, AK106229; CYP76M10, AF488521; CYP76M13, AK288730; CYP76M14, AK069494; CYP76L1, NM_001069848; CYP76C2, NM_130119; CYP71Z1, NM_001059031; CYP71Z2, AK06571; CYP71Z3, NM_001071222; CYP71Z4, AK287479; CYP71Z5, NM_001053551; CYP71Z6, AK107418; CYP71Z7, AK070167; CYP71Z8, AK067591; CYP71Z9, NM_001071223; CYP71W1, NM_001057124; CYP71B13, NM_122423; CYP99A2, AK071546; CYP99A3, AK071864; At-CPR1, X66016; Os-CPR1, AK243608.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. GC-MS Data for in Vitro Production of C11-Keto-ent-Cassadiene.
Supplemental Figure 2. GC-MS Data for Conversion of C11α-Hydroxy-ent-Cassadiene to C11-Keto-ent-Cassadiene.
Supplemental Figure 3. Alignments Used to Generate the Phylogenetic Trees in Figure 6.
Supplemental Data Set 1. Protein Sequences Used to Generate the Alignments in Supplemental Figure 1.
Supplementary Material
Acknowledgments
This work was supported by grants from the USDA-CSREES-NRI (2008-35318-05027) and the National Science Foundation (MCB-0416948) to R.J.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Reuben J. Peters (rjpeters@iastate.edu).
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Chau, M., and Croteau, R.B. (2004). Molecular cloning and characterization of a cytochrome P450 taxoid 2alpha-hydroxylase involved in Taxol biosynthesis. Arch. Biochem. Biophys. 427 48–57. [DOI] [PubMed] [Google Scholar]
- Chau, M., Jennewein, S., Walker, K., and Croteau, R.B. (2004). Taxol biosynthesis: Molecular cloning and characterization of a cytochrome P450 taxoid 7 beta-hydroxylase. Chem. Biol. 11 663–672. [DOI] [PubMed] [Google Scholar]
- Cho, E.-M., et al. (2004). Molecular cloning and characterization of a cDNA encoding ent-cassa-12,15-diene synthase, a putative diterpenoid phytoalexin biosynthetic enzyme, from suspension-cultured rice cells treated with a chitin elicitor. Plant J. 37 1–8. [DOI] [PubMed] [Google Scholar]
- Collu, G., Unver, N., Peltenburg-Looman, A.M., van der Heijden, R., Verpoorte, R., and Memelink, J. (2001). Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. FEBS Lett. 508 215–220. [DOI] [PubMed] [Google Scholar]
- Cyr, A., Wilderman, P.R., Determan, M., and Peters, R.J. (2007). A modular approach for facile biosynthesis of labdane-related diterpenes. J. Am. Chem. Soc. 129 6684–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field, B., and Osbourn, A.E. (2008). Metabolic diversification - Independent assembly of operon-like gene clusters in different plants. Science 320 543–547. [DOI] [PubMed] [Google Scholar]
- Fischbach, M.A., Walsh, C.T., and Clardy, J. (2008). The evolution of gene collectives: How natural selection drives chemical innovation. Proc. Natl. Acad. Sci. USA 105 4601–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, M., Chomet, P., Glawischnig, E., S tettner, C., Grun, S., Winklmair, A., Eisenreich, W., Bacher, A., Meeley, R.B., Briggs, S.P., Simcox, K., and Gierl, A. (1997). Analysis of a chemical plant defense mechanism in grasses. Science 277 696–699. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Jennewein, S., Rithner, C.D., Williams, R.M., and Croteau, R.B. (2001). Taxol biosynthesis: Taxane 13α-hydroxylase is a cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. USA 98 13595–13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennewein, S., Rithner, C.D., Williams, R.M., and Croteau, R.B. (2003). Taxoid metabolism: Taxoid 14beta-hydroxylase is a cytochrome P450-dependent monooxygenase. Arch. Biochem. Biophys. 413 262–270. [DOI] [PubMed] [Google Scholar]
- Kanno, Y., Otomo, K., Kenmoku, H., Mitsuhashi, W., Yamane, H., Oikawa, H., Toshima, H., Matsuoka, M., Sassa, T., and Toyomasu, T. (2006). Characterization of a rice gene family encoding type-A diterpene cyclases. Biosci. Biotechnol. Biochem. 70 1702–1710. [DOI] [PubMed] [Google Scholar]
- Kato, H., Kodama, O., and Akatsuka, T. (1995). Characterization of an inducible P450 hydroxylase involved in the rice diterpene phytoalexin biosynthetic pathway. Arch. Biochem. Biophys. 316 707–712. [DOI] [PubMed] [Google Scholar]
- Kikuchi, S., et al. (2003). Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301 376–379. [DOI] [PubMed] [Google Scholar]
- Koga, J., Shimura, M., Oshima, K., Ogawa, N., Yamauchi, T., and Ogasawara, N. (1995). Phytocassanes A, B, C, and D, novel diterpene phytoalexins from rice, Oryza sativa L. Tetrahedron 51 7907–7918. [Google Scholar]
- Lupien, S., Karp, F., Wildung, M., and Croteau, R. (1999). Regiospecific cytochrome P450 limonene hydroxylases from mint (Mentha) species. cDNA isolation, characterization, and functional expression of (-)-4S-limonene-3-hydroxylase and (-)-4S-limonene-6-hydroxylase. Arch. Biochem. Biophys. 368 181–192. [DOI] [PubMed] [Google Scholar]
- Martin, V.J.J., Pitera, D.J., Withers, S.T., Newman, J.D., and Keasling, J.D. (2003). Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21 796–802. [DOI] [PubMed] [Google Scholar]
- Morrone, D., Xu, M., Fulton, D.B., Determan, M.K., and Peters, R.J. (2008). Increasing complexity of a diterpene synthase reaction with a single residue switch. J. Am. Chem. Soc. 130 5400–5401. [DOI] [PubMed] [Google Scholar]
- Morrone, D., Jin, Y., Xu, M., Choi, S.-Y., Coates, R.M., and Peters, R.J. (2006). An unexpected diterpene cyclase from rice: Functional identification of a stemodene synthase. Arch. Biochem. Biophys. 448 133–140. [DOI] [PubMed] [Google Scholar]
- Nelson, D.R., Schuler, M.A., Paquette, S.M., Werck-Reichhart, D., and Bak, S. (2004). Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol. 135 756–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto, T., et al. (2004). Stemar-13-ene synthase, a diterpene cyclase involved in the biosynthesis of the phytoalexin oryzalexin S in rice. FEBS Lett. 571 182–186. [DOI] [PubMed] [Google Scholar]
- Okada, A., Shimizu, T., Okada, K., Kuzuyama, T., Koga, J., Shibuya, N., Nojiri, H., and Yamane, H. (2007). Elicitor induced activation of the methylerythritol phosphate pathway towards phytoalexin biosynthesis in rice. Plant Mol. Biol. 65 177–187. [DOI] [PubMed] [Google Scholar]
- O'Reilly, D.R., Miller, L.K., and Luckow, V.A. (1992). Baculovirus Expression Vectors: A Laboratory Manual. (New York: Freeman).
- Otomo, K., Kanno, Y., Motegi, A., Kenmoku, H., Yamane, H., Mitsuhashi, W., Oikawa, H., Toshima, H., Itoh, H., Matsuoka, M., Sassa, T., and Toyomasu, T. (2004. b). Diterpene cyclases responsible for the biosynthesis of phytoalexins, momilactones A, B, and oryzalexins A-F in rice. Biosci. Biotechnol. Biochem. 68 2001–2006. [DOI] [PubMed] [Google Scholar]
- Otomo, K., Kenmoku, H., Oikawa, H., Konig, W.A., Toshima, H., Mitsuhashi, W., Yamane, H., Sassa, T., and Toyomasu, T. (2004. a). Biological functions of ent- and syn-copalyl diphosphate synthases in rice: key enzymes for the branch point of gibberellin and phytoalexin biosynthesis. Plant J. 39 886–893. [DOI] [PubMed] [Google Scholar]
- Peters, R.J. (2006). Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry 67 2307–2317. [DOI] [PubMed] [Google Scholar]
- Prisic, S., Xu, M., Wilderman, P.R., and Peters, R.J. (2004). Rice contains disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol. 136 4228–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Project, I.R.G.S. (2005). The map-based sequence of the rice genome. Nature 436 793–800. [DOI] [PubMed] [Google Scholar]
- Qi, X., Bakht, S., Leggett, M., Maxwell, C., Melton, R., and Osbourn, A. (2004). A gene cluster for secondary metabolism in oat: Implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. USA 101 8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston, L., Kwon, S.T., Schoenbeck, M., Ralston, J., Schenk, D.J., Coates, R.M., and Chappell, J. (2001). Cloning, heterologous expression, and functional characterization of 5-epi-aristolochene-1,3-dihydroxylase from tobacco (Nicotiana tabacum). Arch. Biochem. Biophys. 393 222–235. [DOI] [PubMed] [Google Scholar]
- Ro, D.-K., Arimura, G.-I., Lau, S.Y.W., Piers, E., and Bohlmann, J. (2005). Loblolly pine abietadieneol/abietadienal oxidase PtAO (CYP720B1) is a multifunctional, multisubstrate cytochrome P450 monooxygenase. Proc. Natl. Acad. Sci. USA 102 8060–8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, T., et al. (2004). An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134 1642–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoendorf, A., Rithner, C.D., Williams, R.M., and Croteau, R.B. (2001). Molecular cloning of a cytochrome P450 taxane 10 beta-hydroxylase cDNA from Taxus and functional expression in yeast. Proc. Natl. Acad. Sci. USA 98 1501–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, E.E., Spatzenegger, M., and Halpert, J.R. (2001). A truncation of 2B subfamily cytochromes P450 yields increased expression levels, increased solubility, and decreased aggregation while retaining function. Arch. Biochem. Biophys. 395 57–68. [DOI] [PubMed] [Google Scholar]
- Shimura, K., et al. (2007). Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282 34013–34018. [DOI] [PubMed] [Google Scholar]
- Toyomasu, T. (2008). Recent advances regarding diterpene cyclase genes in higher plants and fungi. Biosci. Biotechnol. Biochem. 72 1168–1175. [DOI] [PubMed] [Google Scholar]
- Urban, P., Mignotte, C., Kazmaier, M., Delorme, F., and Pompon, D. (1997). Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J. Biol. Chem. 272 19176–19186. [DOI] [PubMed] [Google Scholar]
- Vaughn, J.L., Goodwin, R.H., Tompkins, G.J., and McCawley, P. (1977). The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera: Noctuidae). In Vitro 13 213–217. [DOI] [PubMed] [Google Scholar]
- Wang, E., Wang, R., DeParasis, J., Loughrin, J.H., Gan, S., and Wagner, G.J. (2001). Suppression of a P450 hydroxylase gene in plant trichome glands enhances natural-product-based aphid resistance. Nat. Biotechnol. 19 371–374. [DOI] [PubMed] [Google Scholar]
- Werck-Reichhart, D., and Feyereisen, R. (2000). Cytochromes P450: A success story. Genome Biol. 1: reviews3003.3001–3003.3009. [DOI] [PMC free article] [PubMed]
- Wickham, T.J., Davis, T., Granados, R.R., Shuler, M.L., and Wood, H.A. (1992). Screening of insect cell lines for the production of recombinant proteins and infectious virus in the baculovirus system. Biotechnol. Prog. 8 391–396. [DOI] [PubMed] [Google Scholar]
- Wilderman, P.R., Xu, M., Jin, Y., Coates, R.M., and Peters, R.J. (2004). Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 135 2098–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, M., Hillwig, M.L., Prisic, S., Coates, R.M., and Peters, R.J. (2004). Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J. 39 309–318. [DOI] [PubMed] [Google Scholar]
- Xu, M., Wilderman, P.R., Morrone, D., Xu, J., Roy, A., Margis-Pinheiro, M., Upadhyaya, N., Coates, R.M., and Peters, R.J. (2007. b). Functional characterization of the rice kaurene synthase-like gene family. Phytochemistry 68 312–326. [DOI] [PubMed] [Google Scholar]
- Xu, M., Wilderman, P.R., and Peters, R.J. (2007. a). Following evolution's lead to a single residue switch for diterpene synthase product outcome. Proc. Natl. Acad. Sci. USA 104 7397–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima, A., Mori, K., and Yabuta, G. (2004). Total synthesis of ent-cassa-12,15-diene, a putative precursor of rice phytoalexins, phytocassanes A-E. Tetrahedron Lett. 45 167–169. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.