Abstract
To sense ambient light conditions in order to optimize their growth and development, plants employ a battery of photoreceptors responsive to light quality and quantity. Essential for the sensing of red and far-red (FR) light is the phytochrome family of photoreceptors. Among them, phytochrome A is special because it mediates responses to different light conditions, including both very low fluences (very low fluence response [VLFR]) and high irradiances (high irradiance response [HIR]). In contrast with the FR-HIR signaling pathway, in which several intermediates of the signaling pathway have been identified, specific components of the VLFR pathway remain unknown. Here, we describe owl1 (for orientation under very low fluences of light), a mutant that is specific for the VLFR, suggesting that VLFR and HIR pathways are genetically distinct, although some common mechanisms can be observed. OWL1 codes for a ubiquitous J-domain protein essential for germination, cotyledon opening, hypocotyl elongation, and deviation of the direction of hypocotyl growth from the vertical under very low light conditions. Additionally, we observed a flowering phenotype suggesting a role for the VLFR during the whole life cycle of a plant. OWL1 interacts with the basic helix-loop-helix HFR1 (LONG HYPOCOTYL IN FAR-RED) transcription factor, previously characterized as a component of the FR-HIR pathway. Both proteins are involved in the agravitropic response under FR light. We propose a central function of OWL1 in the VLFR pathway, which is essential for plant survival under unfavorable light conditions.
INTRODUCTION
To optimize their growth and development, plants have evolved different photosensory systems to perceive light quality, quantity, duration, direction, and periodicity. Essential for sensing of red (R) and far-red (FR) light is the phytochrome (phy) family of photoreceptors (Chen et al., 2004; Bae and Choi, 2008). Phytochromes are encoded by a small gene family, and five phytochromes have been identified in the model plant Arabidopsis thaliana (Clack et al., 1994). Light stable phytochromes (phyB to phyE) mediate low fluence responses (LFRs) primarily to red light (Borthwick et al., 1952; Shinomura et al., 1996). The function of the light unstable phyA is more complex because it participates in at least three photosensory modes: the very low fluence response (VLFR) and the high irradiance response (HIR) to FR and, as recently shown, to R light as well (Casal et al., 2000, Franklin et al., 2007). The light treatments required to trigger the responses are distinct: the VLFR is activated by very low intensity light of any visible wavelength (fluences below 1 μmol m−2 of light) (Botto et al., 1996; Shinomura et al., 1996), whereas the FR-HIR is induced by higher fluences of continuous far-red light (Shinomura et al., 2000) and the R-HIR by strong red light (above 100 μmol m−2 s−1) (Franklin et al., 2007). Moreover, phyA in its VLFR mode antagonizes phyB operating in the LFR mode, whereas phyA in the FR-HIR mode enhances phyB action in the LFR (Cerdan et al., 1999; Hennig et al., 2001). Thus, all three signaling modes of phytochromes, VLFR, LFR, and HIR, are linked in a complex web of interacting signaling pathways.
Phytochromes exist as dimers with each subunit of ∼120 kD linked to a tetrapyrrol chromophore, responsible for light absorption (Chen et al., 2004; Bae and Choi, 2008). phyA, like all phytochromes, is synthesized in the cytoplasm, where it accumulates to high levels in darkness. Upon light activation, which leads to conformational changes of the chromophore transduced to the protein backbone (Pfr form), phyA moves from the cytoplasm to the nucleus where it localizes to nuclear foci (Kircher et al., 2002; Schwinte et al., 2008). The phyA photoreceptor itself is modulated by light on multiple levels. Its protein level is rapidly reduced in response to light via transcriptional and posttranslational mechanisms (Canton and Quail, 1999; Clough et al., 1999; Seo et al., 2004). Furthermore, the phosphorylation state of phyA and therefore its activity and stability is light dependent (Lapko et al., 1997; Kim et al., 2004; Rubio and Deng, 2005; Trupkin et al., 2007). In addition, there is evidence that the phosphorylation state of the phyA dimer could lead to a preferential induction either of the FR-HIR or the VLFR pathway (Ryu et al., 2005; Trupkin et al., 2007; Kneissl et al., 2008).
A number of signaling components downstream of phyA have been identified in Arabidopsis that are either specific for the FR-HIR (such as FAR-RED ELONGATED HYPOCOTYL [FHY3], PHYTOCHROME A SIGNAL TRANSDUCTION [PAT1], and Bell-like homeodomain; Wang and Deng, 2002; Bolle et al., 2004; Staneloni et al., 2009) or can mediate both the VLFR and FR-HIR (such as FHY1, LONG HYPOCOTYL IN FAR-RED [HFR1], SUPPRESSOR OF PHYA [SPA1], and PHYTOCHROME KINASE SUBSTRATE [PKS4]; Hoecker et al., 1999; Fairchild et al., 2000; Soh et al., 2000; Desnos et al., 2001; Zeidler et al., 2001; Schepens et al., 2008) (see Supplemental Figure 1 online). Pks1 and 2 mutants show enhanced phyA-mediated VLFR but are also involved in blue light–mediated phototropism (Lariguet et al., 2003, 2006). Although GIGANTEA (GI) was first characterized as playing a role in the circadian clock and deetiolation under red light conditions, recent work showed that this protein is also important for hypocotyl growth, cotyledon opening, and germination under VLF conditions (Oliverio et al., 2007).
Quantitative trait locus (QTL) mapping led to the identification of genetically unidentified loci that have been physiologically characterized as specific for the VLFR. Introgression of the Arabidopsis Columbia-0 (Col-0) ecotype with a reduced VLFR, to the ecotype Landsberg erecta (Ler), which shows a strong VLFR, led to the identification of vlf1 and 2 (Yanovsky et al., 1997). Additionally, QTLs specific for VLFR have been identified by recombinant inbred lines between the Ler and Cvi accessions (vlf3 to 7) (Botto et al., 2003) or by rescreening of dwarfish mutants (compacta3) (Quinn et al., 2002). None of them have so far been characterized on the molecular level, but the results suggest that FR-HIR and VLFR are at least partially genetically distinct.
Here, we describe the characterization of a positive regulator specific for the VLFR pathway, OWL1 (for orientation under very low fluences of light). Because this protein is essential for the sensing of very low fluences, the name alludes to the way owls can orientate themselves under minimal light conditions.
RESULTS
Molecular Characterization of owl1 Mutant Alleles
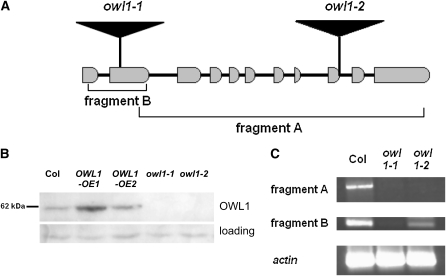
The owl1-1 mutant was identified using a screen designed to detect plants defective in phyA signaling (Bolle et al., 2000), and the insertion site of the T-DNA was identified by sequencing. OWL1 is a single-copy gene that consists of 11 exons and encodes a J-domain protein (Figure 1A). J-domains, first identified in Heat shock protein (Hsp)40/DNAJ proteins, are highly conserved and feature four α-helices and a HPD motif after the second helix (Kelley, 1998; Qiu et al., 2006). The J-domain is situated between residues 12 and 90 of the 538–amino acid OWL1 protein (see Supplemental Figure 2 online). Because OWL1 lacks the zinc finger and Cys repeats present in the C terminus of class I (Hsp40) and class II (Hsp40-like) J-domain proteins, it is relegated to class III. The gene encoding OWL1 has been previously isolated in a yeast screen for Arabidopsis cDNAs that render yeasts tolerant to thiol-oxidizing drugs (Kushnir et al., 1995). The human DNAJ C11 group, which so far has not been functionally characterized, shows the highest homology to OWL1 (20 to 30% amino acid identity; see Supplemental Figure 2 online). OWL1 is present in all higher plants analyzed so far, usually as a single-copy gene. It is also found in Ostreococcus lucimarinus, a green algae with a highly compact genome.
Figure 1.
Molecular Characterization of Two owl1 Alleles.
(A) Schematic representation of OWL1 with intron and exon structure and the position of the insertions in the mutant lines.
(B) Immunological detection of OWL1 in 3-week-old wild-type lines (Col), two overexpressing lines (OWL1-OE1 and OWL1-OE2), and two different owl1 alleles (owl1-1 and owl1-2) with a specific antibody against OWL1. A nonspecific band was used as loading control.
(C) RT-PCR with cDNA from the mutant lines (owl1-1 and owl1-2) and the wild type (Col) to test for the presence of the 5′-mRNA (fragment B) and the 3′-mRNA (fragment A) in the lines. Actin2 served as a control.
An additional loss-of-function line, owl1-2, was identified by reverse genetics, and in both mutant alleles, OWL1 protein accumulation could not be detected (Figure 1B). Nevertheless, in owl1-2, the expression of the 5′-part of the mRNA could be observed by RT-PCR in agreement with the more 3′-terminal location of the T-DNA insertion (Figure 1C). To complement the physiological analysis, we generated cauliflower mosaic virus 35S promoter (35S CaMV) promoter-driven overexpression lines (OWL1-OE), which expressed the protein at approximately two- to fourfold higher levels (Figure 1B).
owl1 Mutant Alleles Are Impaired in VLFR but Not in HIR
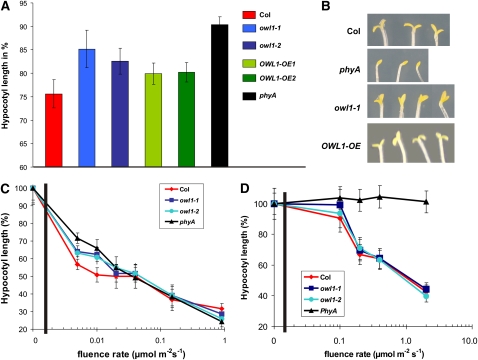
Because exposure of etiolated seedlings to light inhibits hypocotyl elongation and promotes cotyledon unfolding, the analysis of seedling development under different light conditions provides information on the functionality of the corresponding photoreceptor or its transduction pathway. To discriminate between the phyA-dependent VLFR and FR-HIR, we analyzed the phenotypes of the mutant lines under hourly pulses of FR (VLFR) and continuous FR (FR-HIR) light, respectively. Under pulses of FR light, hypocotyl elongation was partially inhibited in wild-type seedlings, whereas owl1 seedlings were significantly longer than the wild type (P < 0.005; Figure 2A). Statistical evaluation of the hypocotyl elongation of the overexpression lines showed that the OWL1-OE1 line, which overexpresses OWL1 to a higher level, was similar to the wild type, whereas the OWL1-OE2 line was more similar to the owl1 mutants, suggesting cosuppression effects. In a phyA mutant, hypocotyl elongation was not inhibited. Furthermore, cotyledons of phyA mutants did not expand under these conditions, and seedlings featured an apical hook (Figure 2B). owl1 alleles had an intermediate phenotype with partially unfolded cotyledons, while the overexpressing lines had completely opened cotyledons.
Figure 2.
OWL1 Is Specifically Involved in phyA-Dependent VLFR.
(A) Hypocotyl length under hourly pulses of FR light (0.5 μmol m−2 s−1, 5 min/h). Seedlings of the wild type (Col), two different owl1 alleles (owl1-1 and owl1-2), two overexpressing lines (OWL1-OE1 and OWL1-OE2), and phyA were grown for 4 d under these conditions. Hypocotyl elongation is displayed as a percentage of elongation relative to dark-grown seedlings of the same genotype.
(B) Cotyledon opening under hourly pulses of FR light (0.5 μmol m−2 s−1, 5 min/h).
(C) Fluence response curve under very low R light for 4 d. Hypocotyl length is displayed as percentage of elongation relative to dark-grown seedlings of the same genotype.
(D) Fluence response curve under continuous FR light. Seedlings were grown for 4 d under these conditions. Hypocotyl length is displayed as percentage of elongation relative to dark-grown seedlings of the same genotype. Error bars are sd.
As VLFRs are induced by not only FR light but also very low fluences of R light, we performed a fluence response assay measuring hypocotyl lengths to evaluate the effect of impaired VLFR in owl1. Hypocotyl elongation of wild-type seedlings was strongly inhibited up to fluence rates of ∼0.05 μmol m−2 s−1 when compared with the phyA mutant, which is devoid of VLFR. The two owl1 mutant alleles had significantly longer hypocotyls compared with Col-0, confirming the strongly reduced VLFR (P < 0.05; Figure 2C). Under higher fluence rates of R light, no statistically significant difference between wild type and owl1 could be observed, although owl1, OWL1 overexpressing lines and phyA were slightly shorter than Col-0 (Figure 2C; see Supplemental Figure 3 online).
On the other hand, under continuous FR light (HIR), no statistically significant difference between the wild type and owl1 was noted in hypocotyl elongation and cotyledon opening (Figure 2D). To exclude the possible involvement of OWL1 in signal pathways transduced by the blue light photoreceptors, hypocotyl elongation and cotyledon opening of seedlings grown under continuous blue (B) light was measured. None of these conditions led to statistically significant differences in growth from the wild type under the analyzed conditions, suggesting that the B light perception is not affected in owl1 (see Supplemental Figure 4A online). Furthermore, phototropic responses to lateral nonsaturating B light were not significantly altered in the owl1 mutant alleles, although a slight increase in curvature was observed (see Supplemental Figure 4B online). Additionally, no difference between dark-grown seedlings of the wild type and owl1 mutants could be detected. From these results, we concluded that in owl1 mutants, the regulation of hypocotyl elongation and cotyledon expansion is impaired in a VLFR-specific manner and that OWL1 is not important for the FR-HIR signal transduction.
Characterization of the VLFR in owl1
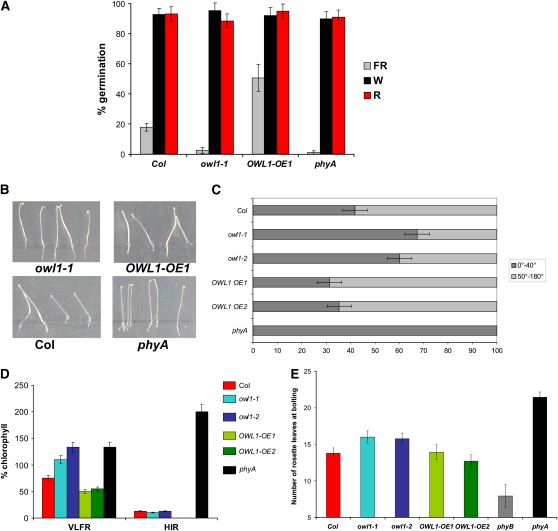
The VLFR has been implicated in several aspects of seedling development, including germination, agravitropic growth, hypocotyl elongation, and cotyledon opening. Germination efficiency was evaluated after the exposure to an FR light pulse (450 μmol m−2) applied ∼48 h after imbibitions, conditions that would allow for phyA accumulation in seed. Figure 3A shows that owl1 was not able to germinate under these conditions, similar to phyA but unlike the wild type. On the other hand, plants overexpressing OWL1 showed a 2.5-fold increase in the germination frequency compared with the wild type, suggesting an enhanced sensitivity toward very low fluences. LFRs do not lead to differences in the germination efficiency of owl1 and OWL1-OE tested after induction by saturating white (W) light (48,000 μmol m−2) or R light pulse (600 μmol m−2) applied 3 h after imbibition (Figure 3A), suggesting that only VLFR-mediated processes are impaired in the owl1 mutant.
Figure 3.
OWL1 Is Important for All Tested VLFRs.
(A) Germination efficiency after a FR light pulse (450 μmol m−2) applied 48 h after imbibition, after a R light pulse (600 μmol m−2) applied 3 h after imbibitions, or a saturating 6-h W light pulse.
(B) and (C) Orientation of growth on vertical plates under FR light (0.5 μmol m−2 s−1) for 3 d.
(C) Percentage of seedlings that grew between 0 and 40° from the perpendicular; n (Col) = 172, n (owl1-1) = 107, n (owl1-2) = 123, n (OWL1 OE1) = 65, n (OWL1 OE2) = 70, and n (phyA) = 66.
(D) Accumulation of chlorophyll in seedlings exposed to 3 d of FR light either as hourly pulses of 5 min (VLFR) or as continuous FR light (HIR) with 0.5 μmol m−2 s−1 and then transferred into W light (80 μmol m−2 s−1) for 3 d.
(E) Flowering time of mutants and overexpression lines under long-day conditions. The number of rosette leaves at bolting was counted. Error bars are sd.
Wild-type seedlings, grown under FR light conditions, exhibited agravitropic growth that deviated from the perpendicular axis, probably due to circumnutational movements during their growth (Figures 3B and 3C). Because phyA plants cannot sense FR light, their growth axis was not influenced by FR light and hence seedling growth was perpendicular, responding only to gravity. This behavior was analyzed on vertical agar plates, which allowed a momentary flash at the position of the hypocotyls after a certain time. The growth of OWL1-OE lines under these conditions was similar to the wild type (P > 0.4), whereas owl1 mutants exhibited a mostly perpendicular growth with 60 to 70% of the seedlings deviating only up to 40° from the perpendicular (P < 0.001; Figures 3B and 3C; see Supplemental Figure 5A online). This indicates that the gravitropic response can override the light response.
Under FR light, protochlorophyllide does not convert into chlorophyll because the light is not energetic enough to activate the protochlorophyllide oxidoreductase. Additionally, plants that are transferred into W light conditions after exposure to FR light (continuous light or pulses) are disturbed in their ability to green, in contrast with phyA mutants (van Tuinen et al., 1995; Barnes et al., 1996). This response is known as the far-red-killing effect. phyA mutants are able to retain their prolamellar body under FR light, thereby being able to produce chlorophyll upon induction with W light. Whereas wild-type plants fail to green in W light after continuous FR light, they can accumulate low amounts of chlorophyll after pulses of FR light. By contrast, owl1 was able to green efficiently after pulses of FR light (Figure 3D; see Supplemental Figure 5B online). On the other hand, OWL1-OE lines were even more sensitive to the FR light pulses than the wild type and accumulated less chlorophyll. After continuous FR light for 4 d (FR-HIR conditions), owl1 mutants did not accumulate chlorophyll, similar to the wild type, demonstrating that only after very low fluences was the ability to green retained in the owl1 mutants (Figure 3D). These data confirmed that OWL1 is specific for many or all developmental processes that are mediated by the VLFR.
Flowering Time Is Affected by OWL1
One of the few phyA-dependent phenotypes described in adult plants is that phyA mutants flower later than the wild type (Johnson et al., 1994). In owl1 mutants, flowering time under long-day conditions was also moderately, but statistically significantly, delayed as determined by the number of leaves at bolting (Col 13.8 ± 0.7; owl1-1 16.0 ± 0.9, owl1-2 15.8 ± 0.7; P < 0.01; Figure 3E) and the number of days until bolting, whereas OWL1-OE lines were similar to the wild type. So far, flowering time has not been attributed to any specific fluence rates, but the data suggest that VLFR is at least partially involved in regulating the transition from the vegetative state to the reproductive state.
OWL1 Is a Ubiquitously Expressed Protein
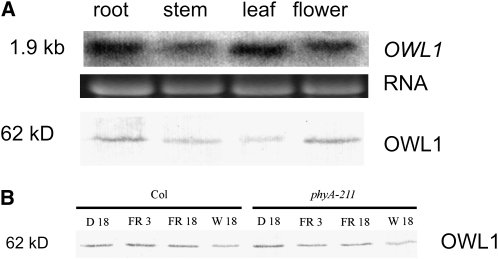
In order to elucidate the site of action of OWL1, OWL1 transcript and its protein accumulation were investigated by RNA and protein blot analyses. Figure 4A shows that OWL1 mRNA and protein were present in root, stem, leaf, and flower tissues and that the protein accumulation level did not vary strongly between tissues. The ubiquitous presence of OWL1 mRNA was confirmed by microarray data (AtGenExpress). No difference in protein levels was detected during development from seedlings up to 6-week-old plants, suggesting a role for OWL1 during the whole lifespan of a plant (see Supplemental Figure 6 online). Because the phenotype of owl1 prevailed in the seedling stage, we also tested OWL1 accumulation in dark-grown seedlings and etiolated seedlings illuminated with 3 or 18 h of FR light and 18 h W light, but no differences were seen (Figure 4B). Similar results with phyA mutant seedlings indicated that OWL1 accumulation was also not dependent on the presence of phyA.
Figure 4.
Expression and Protein Accumulation of OWL1.
(A) RNA gel blot analysis of OWL1 accumulation (top, with RNA shown as a loading control) and immunological detection of OWL1 (bottom) in different tissues. Total protein was quantified by amido black assay, and equal amounts of protein were loaded.
(B) OWL1 accumulation is not changed in phyA mutants. Protein was extracted from dark-grown (D 18) seedlings and etiolated seedlings illuminated with 3 or 18 h of FR light (FR 3 or FR 18) and or with 18 h W light (W 18) and immunologically detected. Total protein was quantified by amido black assay, and equal amounts of protein were loaded.
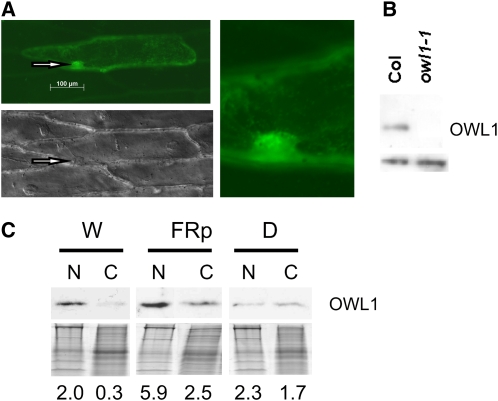
Fluorescence microscopy was employed to determine the intracellular localization of OWL1-GFP (green fluorescent protein) fusion proteins. Figure 5A shows onion epidermis cells bombarded with an OWL1-GFP construct driven by the 35S CaMV promoter. An accumulation of OWL1-GFP in the nucleus and cytoplasm could be observed with a punctuated pattern of green fluorescence. This expression pattern was independent of the N- or C-terminal position of the fusion protein (see Supplemental Figure 7 online). Subcellular fractionation confirmed the localization of OWL1 within the nucleus (Figure 5B). To evaluate the distribution of OWL1 between the nucleus and the cytoplasm, we fractionated plant extracts into a nuclear and a soluble (presumably cytosolic) fraction. Seedlings grown in darkness showed an even distribution of OWL1 between the nucleus and the soluble fraction, whereas in light-grown seedlings, OWL1 accumulated in the nucleus, although the protein was still detectable in the soluble fraction (Figure 5C). Also, pulses of FR light were sufficient to cause protein accumulation in the nucleus.
Figure 5.
Subcellular Localization of OWL1.
(A) Onion epidermis cells bombarded with a OWL1-GFP fusion driven by the 35S cauliflower mosaic virus promoter analyzed by fluorescence microscopy. To visualize the localization of the nucleus (arrows) differential interference contrast microscopy was performed (bottom panel). Detail of GFP signal in the area of the nucleus is enlarged.
(B) Immunodetection of OWL1 in nuclear preparations of W light–grown wild type and owl1-1 Arabidopsis confirmed the nuclear localization of OWL1. Bottom, cross-reacting band indicates equal loading.
(C) The subcellular localization of OWL1 was followed by immunodetection in nuclear (N) or the cytoplasmic (C) fraction. Arabidopsis seedlings were grown under W light (80 μmol m−2 s−1), FR light pulses (FRp; 0.5 μmol m−2 s−1, 5 min/h), or in darkness for 4 d. Coomassie blue–stained gel is shown below the blot as loading control. Quantified intensity of immunoblot corrected with the loaded protein amount is given.
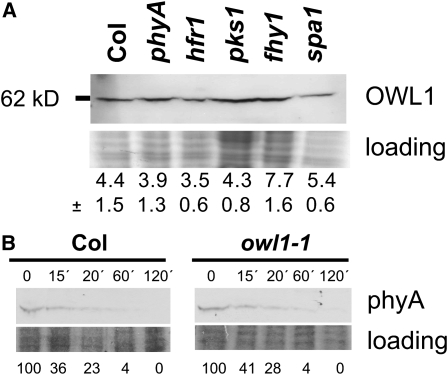
Changes in the abundance of OWL1 could be responsible for a modified VLFR in phyA or in phyA signaling mutants. Especially the SPA1 protein together with CONSTITUTIVE PHOTOMORPHOGENIC (COP1) has been shown to be involved in the protein stability of several phyA signaling intermediates (Hoecker and Quail, 2001). However, our analyses of different mutants, such as hfr1, pks1, fhy1, and spa1, that show changed VLFR, did not uncover any drastic changes in OWL1 levels, albeit in fhy1, OWL1 levels appeared slightly increased. Nor could a change of the protein level in the phyA mutant be detected. This was verified for seedlings grown under pulses of FR light (Figure 6A) and for 3-week-old plants grown in W light (see Supplemental Figure 8 online).
Figure 6.
Biochemical Role of OWL1.
(A) OWL1 accumulation is not affected in different phyA signaling mutants. Seedlings were grown under VLFR conditions for 3 d (FRp; 0.5 μmol m−2 s−1, 5 min/h) and OWL1 detected immunologically. Coomassie blue–stained gel is shown as loading control. Quantified intensity of protein gel blot corrected with the loaded protein amount is given as a mean from three independent blots and sd is added.
(B) The stability of phyA under R light (10 μmol m−2 s−1) is not changed in the owl1 mutant compared with Col. PhyA was detected immunologically between 0 and 120 min of treatment. Coomassie blue–stained gel is shown as loading control. Percentage of accumulation relative to time 0 is given.
It is well established that phyA levels rapidly decline upon induction with red light (Hennig et al., 1999). One reason for the observed insensitivity toward VLF in the owl1 mutants could be a faster degradation or a lower steady state level of the phyA photoreceptor. Protein levels of phyA were therefore analyzed in etiolated seedlings treated with different times of R light (fluence rate of 10 μmol m−2 s−1). In wild-type seedlings, phyA started to degrade after 30 min and no protein was detectable after 2 h. Figure 6B shows that phyA stability was not affected by the presence or absence of OWL1, suggesting that OWL1 is a true signaling component that could either be part of the signaling cascade proper or be involved in feedback regulation.
OWL1 Interacts with HFR1
To place OWL1 in the signal transduction chain, we performed a yeast two-hybrid assay using OWL1 as a bait against a cDNA library. One of the interacting partners that was confirmed by retesting in the yeast system was HFR1 (Figure 7A). HFR1 is a putative transcription factor with a basic helix-loop-helix domain acting as a positive regulator downstream of both phyA and the cryptochrome (cry; Fairchild et al., 2000; Duek and Fankhauser, 2003). Although the hfr1 mutant is deficient in several FR-HIR responses, its agravitropic behavior under FR light has also been described, indicating some role in the VLFR (Fairchild et al., 2000). OWL1 fused to a DNA binding domain (BD) interacts with HFR1 fused to the activation domain (AD) as determined in the yeast two-hybrid assay under rising concentrations of 3-amino-1,2,4-triazole (3AT) (Figure 7A). OWL1-BD was not able to induced yeast growth on selection media by itself via intrinsic transactivation domains, and HFR1-AD could not interact with the BD. The reverse experiment was not performed due to an intrinsic ability of HFR1 to transactivate.
Figure 7.
OWL1 Interacts with HFR1.
(A) OWL1-HFR1 interaction in yeast two-hybrid assays on plates without His and with 10 and 15 mM 3AT. As a control, yeast growth on media without Leu and Trp is shown (0).
(B) In vitro pull-down assays showing the interaction between OWL1 and HFR1. Recombinant HIS-HFR1 bound to Ni-NTA agarose was used in pull-down assays with overexpressed OWL1-GST or PAT1-GST (as control). Furthermore, noninduced (n.i.) cultures of OWL1-GST were used as controls. In each pair, the first lane shows the input proteins and the second the eluate. Arrows marks the OWL1-GST or PAT1-GST band, respectively. Samples were separated by SDS-PAGE gel, and the blots were probed with anti-OWL or anti-GST antibodies. Experiments were repeated three times with similar results.
To provide evidence for a direct interaction between HFR1 and OWL1, we performed in vitro pull-down assays using overexpressed HIS and glutathione S-transferase (GST) fusion proteins. Figure 7B shows that HIS-HFR1 bound to Ni-NTA agarose interacted with GST-OWL1. As a negative control, GST-PAT1, a FR-HIR phyA signaling component (Bolle et al., 2000), was used, which has been shown not to interact with HFR1 (Jang et al., 2007). Additionally, GST alone was also not able to interact with HFR1. These results indicated that OWL1 and HFR1 can indeed directly interact.
DISCUSSION
The physiological characterization of owl1 mutant alleles and overexpression lines clearly showed that OWL1 plays an important role in the VLFR branch, but not the FR-HIR branch, of the phyA-specific signaling pathway. Our data suggest that FR-HIR and VLFR are indeed genetically separable pathways, as also indicated by the identification of several QTLs in the Arabidopsis ecotype Ler responsible for the VLFR (vlf1-7; Yanovsky et al., 1997; Botto et al., 2003). In addition to these VLFR-specific components, several other signaling intermediates have been demonstrated to be important for both FR-HIR and VLFR. These include FHY1 and FHY1-LIKE, two proteins necessary for the import of phyA into the nucleus upon light activation (see Supplemental Figure 1 online; Zeidler et al., 2004; Hiltbrunner et al., 2005, 2006; Zhou et al., 2005; Rosler et al., 2007; Genoud et al., 2008) and some members of the PKS1 and SPA1 protein family. The cytoplasmic PKS1 and 2 proteins, which can interact with phyA, show enhanced phyA-mediated VLFR and are also involved in blue light–dependent phototropism (Fankhauser et al., 1999; Lariguet et al., 2003). SPA1, which together with COP1 is needed for the degradation of several light signaling factors, is also important for VLFR as the hypersensitive phenotype of spa1 can also be observed under very low fluences (Hoecker et al., 1999; Fittinghoff et al., 2006). Interestingly though, the OWL1 protein level is not regulated by SPA1 (Figure 6A). GI, a small nuclear protein whose biochemical function is still not understood, was first characterized for its role in promoting flowering under long-day conditions and circadian clock. However, loss-of-function mutants also show an increased hypocotyl elongation under R and B light (Fowler et al., 1999; Park et al., 1999; Huq et al., 2000; Martin-Tryon et al., 2007). Recently, GI has been implicated in many VLFR, but not in HIR under continuous FR light (Oliverio et al., 2007). The involvement of brassinosteroids as positive regulators in the phytochrome signaling cascade was established by the analysis of deetiolated2 and the identification of the brassinosteroid biosynthesis mutant diminuto/dwarf1 as an enhanced very-low-fluence response1 mutant (Luccioni et al., 2002). Additional to the hypersensitive phenotype under very low fluence conditions, the mutant displayed a reduced LFR and HIR, suggesting a crosstalk between the VLFR and LFR/HIR.
owl1 is indeed a bona fide VLFR mutant because we could not detect any blue or red light–dependent responses except under very low fluence conditions. Whereas the germination of owl1 mutants was strongly inhibited by FR light similar to phyA, other owl1 phenotypes such as inhibition of hypocotyl elongation and deviation of the hypocotyl growth from the vertical were intermediate between phyA and the wild type. This suggests that additional components are necessary for signal transduction under these conditions or functional contributions from other pathways are necessary for these responses. It would be intriguing to link this observation with the fact that the hormones most important for germination responses are gibberellins and abscisic acid, whereas for cell elongation responses such as hypocotyl elongation and agravitropic movements, the responsible hormone is mainly auxin (Collett et al., 2000; Finkelstein et al., 2008). Additionally, gibberellins and ethylene have been shown to affect hypocotyl elongation independently of auxin (Collett et al., 2000, Saibo et al., 2003; Vandenbussche et al., 2007). Therefore, the germination response and the growth responses could target very different endpoints and overlap with distinct hormonal pathways. The interaction with the different hormones could differentiate the effectiveness of the OWL1-dependent responses.
Compared with the wild type, the OWL1 overexpression lines are more sensitive to FR light pulses with respect to germination and chlorophyll accumulation, but no significant difference was detected between the wild type and overexpression lines for agravitropic responses and flowering time. When seeds were germinated under FR light pulses, hypocotyls of the overexpressing lines were even slightly longer than wild-type hypocotyls, resulting in a hyposensitive phenotype. This differential pattern could be attributed to the fact that all these physiological responses are induced by different endpoints in the signaling cascade. High amounts of OWL1, which is expressed from a 35S CaMV promoter, could lead to inhibitory effects. It is possible that this could be the case for inhibition of hypocotyl elongation, but not for the other responses described. It is likely that OWL1 might interact with different factors to modulate different VLFRs.
OWL1 is unlikely to be one of the loci identified by QTL mapping in the Ler ecotype (Yanovsky et al., 1997). Like VLF1, OWL1 is located on chromosome 2 but maps to a different location. A divergence of Col-0 backgrounds seems to exist because the Col background used by Yanovsky et al. (1997) to identify VLFR loci between Col and Ler retains negligible VLFR, whereas we clearly observed VLFR in the Col-0 strain used for the generation of the transgenic T-DNA lines used in this study. These differences could also result from variations in growth conditions. Nevertheless, it is possible that the intermediate phenotype observed for hypocotyl elongation could also be due to the fact that Col-0 has reduced sensitivity toward very low fluences with respect to hypocotyl elongation compared with germination.
The ubiquitous presence of OWL1 in the plant and the VLFR-specific owl1 phenotypes observed mainly at the seedling stage offer two possible explanations for OWL1 function. (1) OWL1 could have additional functions, which have not yet been detected. On the other hand, no obvious disturbance of development can be observed under greenhouse conditions. Furthermore, because OWL1 was initially isolated in a yeast screen for plant genes in the oxidative stress response (Kushnir et al., 1995), we tested the mutants and the overexpression lines on media with different stress elicitors to induce oxidative (0.5 and 1 μM methyl viologen), salt (100 mM NaCl), osmotic (250 mM mannitol), and temperature stress (14 and 42°C). No divergence from the wild-type phenotype could be observed in any instance. (2) The VLFR could play a more significant role in plant development than previously assumed. This is supported by a role for OWL1 in flowering time (which is delayed in owl1 mutants), a process previously not attributed to VLFR. The observed light-induced relocation of OWL1 from the cytoplasm to the nucleus could be one way to regulate the amount of active protein available for a distinct process; therefore, we favor this hypothesis. The increased sensitivity to light mediated by components of the VLFR, such as OWL1, can only be observed experimentally under very low fluences, which does not exclude a modulating role of this protein at every stage of a plant's life. These variations might not be visible under standard greenhouse conditions. Nevertheless, mechanisms that allow plants to adjust their sensitivity to light signals are of utter importance under the ever-changing light conditions in the natural environment. Being able to sense even low amounts of light provides a kind of safety net for plant development that allows, for example, germination under unfavorable conditions or induces deviation of the direction of hypocotyl growth from the vertical, probably also performing circumnutational movements for seedlings to locate better light conditions.
Because OWL1 does not affect the stability of phyA, yet specifically impairs phyA-dependent VLFR, we assume that OWL1 acts downstream of phyA after the divergence into the FR-HIR and VLFR pathways. DNAJ/Hsp40 proteins have been characterized as molecular chaperones and are found ubiquitously in all organisms (Kelley, 1998; Miernyk, 2001; Qiu et al., 2006).The J-domain is crucial for the interaction with Hsp70, regulating the activity of Hsp70 proteins by stimulating their ATPase activity. The type III J-domain proteins, to which OWL1 belongs, represent a functionally distinct group from the DNAJ proteins and are very heterogenous. They have been suggested not to act as chaperones and seem not to bind to non-native polypeptides, although they are still able to recruit Hsp70 proteins, as the binding motifs are highly conserved (Walsh et al., 2004; Hennessy et al., 2005).
In plants, very few type III J-domain proteins have been biologically characterized. Two Arabidopsis proteins, ALTERED RESPONSE TO GRAVITY1 (ARG1) and its paralog ARG1-LIKE2 (ARL2), are required for root and hypocotyl gravitropism, influencing the distribution of auxin upon stimulation (Sedbrook et al., 1999; Boonsirichai et al., 2003; Guan et al., 2003). At least ARL2 localizes to the plasma membrane. By contrast, we could not detect any difference in the gravitropic behavior of owl1 mutant alleles, such as bending of the stem and root tip curvature in response to gravity and no exclusive localization to the membranes of OWL1, suggesting that OWL1 has a different biochemical function from ARG1 and ARL2.
Functions have been established for several type III J-domain proteins from species other than plants, but no consensus picture has yet emerged. Some type III J-domain proteins are involved in the recruitment of an Hsp70 protein to discrete sublocalizations within a compartment. Examples of this are the J-domain containing auxilins that are associated with clathrin-coated vesicles in the cytoplasm and that allow docking for the disassembly of clathrin coats (Lemmon, 2001). The localization of OWL1-GFP in distinct speckles could suggest a role in similar processes. In other cases, the Hsp70 and type III J-domain protein are recruited independently to the site of action, where the J-domain protein stimulates ATP hydrolysis by the partner Hsp70. This can be observed for the import machinery of mitochondria and chloroplasts (Westermann and Neupert, 1997; Becker et al., 2004; Kozany et al., 2004; Qbadou et al., 2007). The divergence of the C terminus among the type III J-domain proteins and their uniqueness suggests the possibility that they bind very different proteins. This way, varying proteins can be brought into proximity of each other, with the J-domain protein acting as a scaffold assembling a multisubunit protein complex. The Simian Virus 40 large T antigen contains a functional J-domain, which can bind an Hsp70 protein. An additional factor, such as the Retinoblastoma protein (Rb), can be bound to the C-terminal part of the J-domain protein. The Hsp70 protein in turn recruits a factor (e.g., E2F1) that is a substrate for Rb (Kim et al., 2001). Thus, E2F1 and Rb are brought together in close proximity and can interact.
OWL1 could therefore be important for the presence or activity of factors that are crucial for VLFR. One example is the confirmed interaction between OWL1 and HFR1, a putative transcription factor. HFR1 (long hypocotyl in far-red 1) is a positively acting component of phyA-dependent FR-HIR and the cry1 signaling pathway, as the mutant exhibits a reduction in seedling responsiveness specifically to continuous FR and blue light (Duek and Fankhauser, 2003; Fairchild et al., 2000). This basic helix-loop-helix protein with an atypical basic region binds also to another basic helix-loop-helix protein, PHYTOCHROME INTERACTING FACTOR3, and the complex can bind the Pfr form of phyA and phyB (Duek et al., 2004). HFR1 also interacts with the myb transcription factor LONG AFTER FAR-RED LIGHT1, which is involved in FR-HIR (Jang et al., 2007). Furthermore, HFR1 interacts with COP1 and SPA1, leading to its ubiquitin-dependent degradation in darkness, thereby abolishing photomorphogenesis (Duek et al., 2004; Jang et al., 2005; Yang et al., 2005a, 2005b). HFR1 phosphorylation is promoted by light, and the protein is stabilized under these conditions. The tight regulation of protein levels of transcription factors has emerged as a key event to regulate and modulate signal transduction pathways and to desensitize a pathway so that it is responsive to new signals. As both OWL1 and HFR1 seem to have a crucial role in the integration of VLFR and light signals to modulate hypocotyl growth and growth direction, OWL1 could be needed to recruit HFR1 for this specific subset of responses. Other factors might be recruited by OWL1 for different VLFRs, such as germination or flowering time. We note that several transcription factors, such as HFR1 and ELONGATED HYPOCOTYL5, are capable of transducing signals derived from high irradiance and very low fluences of light, in addition to mediating other signal transduction pathways, such as the response to B light or hormones. It is possible that OWL1 is important to impose specific VLF-dependent activation on these more general downstream effectors, thereby achieving specificity and modulation.
METHODS
Plant Materials
Wild-type and mutant plants used in this study were all in the Col-0 ecotypes. A phyA-null mutant in ecotype Col (phyA-211; Reed et al., 1994) was used as a control. owl1-1 was isolated from the Koncz T-DNA collection in a screen for phyA-defective mutants (Koncz et al., 1989; Bolle et al., 2000) and owl1-2 from the GABI_KAT collection (091G03; Rosso et al., 2003). The gene disrupted in owl1-1 was identified by plasmid rescue (Koncz et al., 1994). Mutants were backcrossed and selfed to generate homozygous lines. Homozygous mutants were confirmed by genotyping using PCR amplification with gene- and insertion-specific primers (see Supplemental Table 1 online). spa1-3 was kindly provided by U. Hoecker, University of Cologne (Hoecker et al., 1999), and pks1, phyA-211, phot1, and nph3 by the Nottingham Arabidopsis Stock Centre (NASC; Fankhauser et al., 1999; Liscum and Briggs, 1995). The mutants hfr1-201 and fhy1-3 have been described by Soh et al. (2000) and Zeidler et al. (2001), respectively.
Growth conditions, light sources, and phenotypical characterization were performed as described previously (Kneissl et al., 2008). All physiological experiments were repeated at least three times.
Statistical Analysis
Where appropriate, the physiological experiments were evaluated with t tests between the wild-type and the mutant line. P values are given. Furthermore, multiple comparisons among means have been tested using Tukey and Scheffe tests. To test for statistical significance of distributions, χ2 tests were applied.
Sequence Analysis
Proteins homologous to Arabidopsis thaliana OWL1 were identified by Protein BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequence alignments were performed with MegAlign (Lasergene7; DNAStar) with default settings.
Constructs
For overexpression, the full-length open reading frame of OWL1 was amplified by PCR from a cDNA library with the primers 5′-CAGATCTGATGATGGGCCAAGAAGCAGCTCCG-3′ adding a BglII site and 5′-AAAACTAGTCACTGGCCTTCTTGTGGTATACT-3′ adding a SpeI site. The fragment was cloned into pGEM-T easy (Promega), checked for mutations by DNA sequencing, and digested with the appropriate restriction endonucleases for cloning into a pVIP vector with the additional restriction sites AscI and PacI in the multiple cloning site and the CaMV 35S promoter (van der Krol and Chua, 1991). The insertion was rechecked by sequencing. This binary construct was introduced into the pGV3101 strain of Agrobacterium tumefaciens and transformed into Col-0 using the floral dip transformation method to generate overexpression lines (Clough and Bent, 1998). Transformants were selected on kanamycin-containing medium and tested for overexpression on the protein level.
For GFP fusion, OWL1 cDNA was amplified with the primers 5′-CACCATGATGGGCCAAGAAGCAGC-3′ and 5′-TCACTGGCCTTCTTGTGGTATACT-3′ (for N-terminal fusion of GFP) or 5′-CTGGCCTTCTTGTGGTATACTC-3′(for C-terminal fusion without a TGA codon) from a cDNA library, cloned into the pENTR/D-TOPO vector (Invitrogen), and inserted into the pK2FWG2 or pK7GWF2 GFP fusion vector (Karimi et al., 2002) with LR clonase (Invitrogen).
RNA Extractions and RT-PCR
For gene expression analyses, total RNA was extracted from 2-week-old seedlings using the Qiagen RNeasy Mini columns (Qiagen). RNA (1 μg) was used for first-strand cDNA synthesis using the Omniscript System (Qiagen) with a poly(T)18 primer, and 1.0 μL of this reaction was used as template for PCR amplification. Primer sets were used for amplification of the fragment A (5′-CACCATGATGGGCCAAGAAGCAGC-3′ and 5′-AGTGGGTTGAAATGAGCCATCTTC-3′) and fragment B (5′-CACCATGGAAGGATTGAATTCAGGA-3′ and 5′-CTGGCCTTCTTGTGGTATACT-3′) of OWL1 and actin2 (5′-CAGCACAATACCGGTTGTACGAC-3′ and 5′-CTCTTTCTTTCCAAGCTCATAAAAAATG-3′).
For RNA gel blots, total RNA was quantified photometrically, and 20 μg of RNA was loaded on a MOPS formaldehyde gel, size-fractionated, and subsequently transferred to a nylon membrane. After hybridization in 50% formamide with random prime-labeled fragments, membranes were washed with 0.1× saline sodium citrate and 0.1% SDS at 45°C. The result was documented with a PhosphorImager (Molecular Dynamics). As a probe for OWL1, the full-length cDNA fragment from the pGEM-T easy vector was excised and radioactively labeled.
Protein Extraction and Immunoblotting
To generate a specific antibody against OWL1, the full-length OWL1 cDNA was amplified by PCR from a cDNA library with the following primers: 5′-GCATGCATGATGGGCCAAGAAGCAGCTCCG-3′ adding an SphI site and 5′-GGTACCTCACTGGCCTTCTTGTGGTATAC-3′ adding a Kpn I site and cloned into pGEM-T easy. The fragment was checked for mutations by DNA sequencing. The vector was digested with the appropriate restriction endonucleases, and the fragment was cloned in frame behind a histidine hexapeptide in the pQE30 vector (Qiagen) using the SphI and KpnI sites. Escherichia coli was transformed with this construct. Positive clones were induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) and harvested 2 h after induction. A French press extract was divided after centrifugation into French press supernatant, the 8 M urea soluble fraction, and the 8 M urea insoluble fraction. The largest part of His-tagged protein was in the fraction of proteins soluble in 8 M urea. This fraction was loaded on a Ni2+ agarose column. By washing the column with 8 M urea of stepwise descending pH, the His-tagged protein was purified and finally eluted from the column at pH 4.4.
Forty micrograms of purified protein per injection used for immunization of mice (Cocalico Biologicals). The first boost was given 21 d after initial inoculation, the second boost 49 d after initial inoculation, and exsanguination took place 59 d after initial inoculation.
Protein extraction, SDS-PAGE, and immunodetection were performed as described previously (Kneissl et al., 2008). The specific antibody was diluted 1:2000, and a goat anti-mouse antibody conjugated to horseradish peroxidase (Invitrogen), diluted 1:10,000, was used as a secondary antibody. For quantification, the program BioDocAnalyze (Biometra) was applied. PhyA degradation assay was performed according to Büche et al. (2000). For this assay, seedlings were pregrown in D then incubated in R light. Protein was extracted at the indicated time points and separated on SDS-PAGE, and phyA was detected immunologically.
Amido black assays were used to quantify protein amounts in extracts before loading (Popov et al., 1975).
Subcellular Localization
OWL1-GFP was introduced into onion epidermis cells via particle bombardment as described by Torres-Galea et al. (2006). Samples were incubated over night in the dark and analyzed with an Axioskop microscope (Carl Zeiss). GFP fluorescence was detected with the filter set 38HE.
Isolation of Nuclei
Seedling tissue (500 mg) was homogenized in 1 mL extraction buffer (2.5% ficoll 400 [w/v], 5% dextran T40 [w/v], 400 mM sucrose, 25 mM Tris/HCl [pH 7.4], and 10 mM MgCl2). Extract was filtered through Miracloth and supplied with Triton X-100 to a final concentration of 0.5% (v/v). After a 15-min incubation on ice, extract was sedimented (1500 rfc, 5 min, 4°C). The supernatant was used as the cytosolic fraction. The sediment was washed with extraction buffer containing 0.1% Triton X-100 (v/v) and resuspended in 1 mL of extraction buffer. Differential centrifugation steps removed cellular debris (100 rfc, 15 min, 4°C) and pelleted the nuclei (1800 rfc, 5 min, 4°C).
Yeast Two-Hybrid Assays
For the yeast two-hybrid screen, the ProQuest reverse two-hybrid system (Invitrogen) was used according to manufacturer's instructions. The full-length open reading frame for OWL1 in the pENTR/D-TOPO vector (see GFP fusion) was inserted in frame with the GAL4-BD into the pDEST32 vector (Invitrogen) using the respective attR sites. Plasmids containing these constructs were transformed into competent MaV203 yeast cells as described by Gietz and Woods (2002). Yeast cells were plated on SC-Leu-His + 10 mM 3AT to test for autoactivation activity associated with OWL1, which was minimal.
Yeast two-hybrid screen was performed using OWL1 in pDEST32 and an Arabidopsis cDNA library (SuperScript Arabidopsis cDNA library; Invitrogen) inserted into the pDEST22 vector (Invitrogen), both transformed into competent MaV203 yeast cells. Putative interactors were isolated on plates containing SC-Leu-Trp-His + 10 mM 3AT. Fifty-six independent transformants were selected for putative positive interaction. Plasmids were extracted from positive yeast clones using the QIAprep Spin Miniprep Kit (Qiagen). Lysis of the yeast cells was achieved by vortexing with the P1 buffer and added glass beads. The eluted vector DNA was transformed into E. coli, reisolated, and sequenced.
The full-length open reading frame of HFR1 was amplified with the primers 5′-CACCATGTCGAATAATCAAGCTTTCATGG-3′ and 5′-TCATAGTCTTCTCATCGCATGGG-3′ and inserted into the pENTR/D-TOPO vector (Invitrogen). The fragment was transferred in frame with GAL4-AD into the pDEST22 vector (Invitrogen) using LR clonase (Invitrogen), and yeast was transformed with the bait and the prey. Because HFR1 is autoactivating, the reverse experiment was not performed. Strength of interaction was tested on plates containing SC-Leu-Trp-His+10 mM/15 mM/20 mM 3AT.
Pull-Down Assays
Full-length OWL1 cDNA was amplified as for the GFP fusion but inserted into the pENTR/SD-TOPO vector (Invitrogen), and the insertion was then transferred, using the LR clonase, into pGEX-4T-1 (GE Healthcare) with an additional GATEWAY cassette to obtain the GST fusion. PAT1 cDNA was amplified with the primers 5′-CACCATGTACAAGCAGCCTAGAC-3′ and 5′-TCATTTCCAAGCACACGAGGC-3′, and the fragment was cloned accordingly into pGEX-4T-1.The vectors were transformed into the E. coli strain BL21(DE3)pLysS (Stratagene), and liquid cultures of single colonies were grown at 37°C to a A600 = 0.8. Expression of the GST fusion proteins was then induced with 0.2 mM IPTG, and cells were incubated for 3 h at 37°C. The full-length HFR1 cDNA was amplified as described above and inserted into the pENTR/SD-TOPO vector (Invitrogen), and the insertion was then transferred, utilizing the LR clonase, into pGEX-4T-1 (GE Healthcare). Using the primers 5′-CACCACCACAAGGAGCCCTTCACCATGTCG-3′ and 5′-GTGGTGGTGCATGAATACTGTTTCCTGTGT-3′, the vector was amplified using a proofreading polymerase (Finnzymes), thereby excising the sequence coding for the GST-tag and substituting it with an N-terminal 6xHIS-tag. The vector was transformed into the E. coli strain BL21(DE3)pRIPL codon plus (Stratagene), and liquid cultures of single colonies were grown at 37°C to A600 = 0.8. Expression of HIS-HFR1 was then induced with 1 mM IPTG, and cells were incubated for 3 h at 37°C.
The cultures were harvested and cells lysed under denaturing conditions by resuspending in buffer B (50 mM NaH2PO4, 10 mM Tris·HCl, and 8 M urea, pH 8.0). Cells were stirred for 30 min at room temperature and centrifuged at 10,000g for 30 min at room temperature to pellet the cellular debris. The proteins in the supernatant were dialyzed overnight against 50 mM NaH2PO4, 50 mM NaCl, 10 mM imidazole, 1 mM PMSF, and 1 mM ascorbate.
For pull-down assay, HIS-HFR1 was bound to Ni-NTA agarose (Qiagen) and washed according to the manufacturer's instructions. One microgram of OWL1-GST or PAT1-GST was applied to 1 μg bound HIS-HFR1 and incubated for 1 h at 4°C. Samples were washed using 50 mM NaH2PO4, 50 mM NaCl, 25 mM imidazole, 1 mM PMSF, and 1 mM ascorbate and eluted according to the manufacturer's instructions.
Samples were separated using SDS-PAGE, and immunodetection was performed with anti-OWL1 (dilution 1:2000), anti-Penta-HIS (Qiagen; dilution 1:10,000), and anti-GST antibody (Sigma-Aldrich; 1:10,000) as primary antibodies. As secondary antibodies, a goat anti-mouse antibody conjugated to horseradish peroxidase (Invitrogen), diluted 1:10,000, was applied. Blocking buffer was substituted with 3% nonfat milk powder, only to detect the HIS-tag 1% casein was used. The signal was detected by chemiluminescence, and for quantification the program BioDocAnalyze (Biometra) was used.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: OWL1 (At2g35720), HFR1 (AT1G02340), PAT1 (AT5G48150), OsOWL1 (Os10g0507800), OlOWL1 (XP_001419313), HsDNAJ c11C(NP_060668), DmDNAJ c11 (NP_610945), and CeDNAJ 9 (NP_494872).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Depiction of the VLFR and HIR Signaling Pathways.
Supplemental Figure 2. Alignment of the OWL1 Protein with Its Orthologs.
Supplemental Figure 3. Hypocotyl Elongation under LFR and FR-HIR.
Supplemental Figure 4. OWL1 Plays No Role in Blue Light Signaling.
Supplemental Figure 5. OWL1 Is Important for Agravitropic Growth and the Far-Red Light Killing Response.
Supplemental Figure 6. Developmental Time Course of OWL1 Protein Levels.
Supplemental Figure 7. Subcellular Localization of OWL1.
Supplemental Figure 8. OWL1 Protein Levels Are Not Affected in Different phyA Signaling Mutants.
Supplemental Table 1. Summary of Oligonucleotide Primers Used for Genotyping.
Supplementary Material
Acknowledgments
We thank C. Koncz for the T-DNA lines, GABI-KAT for owl1-2, U. Hoecker for spa1 seeds, NASC for pks1, phyA-211, phot1, and nph3, and E. Schäfer for the phyA antibody. We thank D. Leister for financial support and critical reading of the manuscript, R. Foster for editing the manuscript, and Martina Reymers and Ingrid Duschanek for technical assistance. C.B. was supported by grants from the Deutsche Forschungsgemeinschaft, and V.W. was supported by the Friedrich-Ebert-Stiftung. Work done at The Rockefeller University was supported by National Institutes of Health Grant GM44640 to N.-H.C.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Cordelia Bolle (c.bolle@bio.lmu.de).
Online version contains Web-only data.
References
- Bae, G., and Choi, G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59 281–311. [DOI] [PubMed] [Google Scholar]
- Barnes, S.A., Nishizawa, N.K., Quaggio, R.B., Whitelam, G.C., and Chua, N.H. (1996). Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell 8 601–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, T., Hritz, J., Vogel, M., Caliebe, A., Bukau, B., Soll, J., and Schleiff, E. (2004). Toc12, a novel subunit of the intermembrane space preprotein translocon of chloroplasts. Mol. Biol. Cell 15 5130–5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle, C., Koncz, C., and Chua, N.H. (2000). PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 14 1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Boonsirichai, K., Sedbrook, J.C., Chen, R., Gilroy, S., and Masson, P.H. (2003). ALTERED RESPONSE TO GRAVITY is a peripheral membrane protein that modulates gravity-induced cytoplasmic alkalinization and lateral auxin transport in plant statocytes. Plant Cell 15 2612–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borthwick, H.A., Hendricks, S.B., Parker, M.W., Toole, E.H., and Toole, V.K. (1952). A reversible photoreaction controlling seed germination. Proc. Natl. Acad. Sci. USA 38 662–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto, J.F., Alonso-Blanco, C., Garzaron, I., Sanchez, R.A., and Casal, J.J. (2003). The Cape Verde Islands allele of cryptochrome 2 enhances cotyledon unfolding in the absence of blue light in Arabidopsis. Plant Physiol. 133 1547–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto, J.F., Sanchez, R.A., Whitelam, G.C., and Casal, J.J. (1996). Phytochrome A mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis. Plant Physiol. 110 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büche, C., Poppe, C., Schafer, E., and Kretsch, T. (2000). eid1: A new Arabidopsis mutant hypersensitive in phytochrome A-dependent high-irradiance responses. Plant Cell 12 547–558. [PMC free article] [PubMed] [Google Scholar]
- Canton, F.R., and Quail, P.H. (1999). Both phyA and phyB mediate light-imposed repression of PHYA gene expression in Arabidopsis. Plant Physiol. 121 1207–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal, J.J., Yanovsky, M.J., and Luppi, J.P. (2000). Two photobiological pathways of phytochrome A activity, only one of which shows dominant negative suppression by phytochrome B. Photochem. Photobiol. 71 481–486. [DOI] [PubMed] [Google Scholar]
- Cerdan, P.D., Yanovsky, M.J., Reymundo, F.C., Nagatani, A., Staneloni, R.J., Whitelam, G.C., and Casal, J.J. (1999). Regulation of phytochrome B signaling by phytochrome A and FHY1 in Arabidopsis thaliana. Plant J. 18 499–507. [DOI] [PubMed] [Google Scholar]
- Chen, M., Chory, J., and Fankhauser, C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38 87–117. [DOI] [PubMed] [Google Scholar]
- Clack, T., Mathews, S., and Sharrock, R.A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25 413–427. [DOI] [PubMed] [Google Scholar]
- Clough, R.C., Jordan-Beebe, E.T., Lohman, K.N., Marita, J.M., Walker, J.M., Gatz, C., and Vierstra, R.D. (1999). Sequences within both the N- and C-terminal domains of phytochrome A are required for PFR ubiquitination and degradation. Plant J. 17 155–167. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Collett, C.E., Harberd, N.P., and Leyser, O. (2000). Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiol. 124 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnos, T., Puente, P., Whitelam, G.C., and Harberd, N.P. (2001). FHY1: A phytochrome A-specific signal transducer. Genes Dev. 15 2980–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek, P.D., Elmer, M.V., van Oosten, V.R., and Fankhauser, C. (2004). The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol. 14 2296–2301. [DOI] [PubMed] [Google Scholar]
- Duek, P.D., and Fankhauser, C. (2003). HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J. 34 827–836. [DOI] [PubMed] [Google Scholar]
- Fairchild, C.D., Schumaker, M.A., and Quail, P.H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14 2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284 1539–1541. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R., Reeves, W., Ariizumi, T., and Steber, C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59 387–415. [DOI] [PubMed] [Google Scholar]
- Fittinghoff, K., Laubinger, S., Nixdorf, M., Fackendahl, P., Baumgardt, R.L., Batschauer, A., and Hoecker, U. (2006). Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J. 47 577–590. [DOI] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin, K.A., Allen, T., and Whitelam, G.C. (2007). Phytochrome A is an irradiance-dependent red light sensor. Plant J. 50 108–117. [DOI] [PubMed] [Google Scholar]
- Genoud, T., Schweizer, F., Tscheuschler, A., Debrieux, D., Casal, J.J., Schafer, E., Hiltbrunner, A., and Fankhauser, C. (2008). FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 4 e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and Woods, R.A. (2002). Screening for protein-protein interactions in the yeast two-hybrid system. Methods Mol. Biol. 185 471–486. [DOI] [PubMed] [Google Scholar]
- Guan, C., Rosen, E.S., Boonsirichai, K., Poff, K.L., and Masson, P.H. (2003). The ARG1-LIKE2 gene of Arabidopsis functions in a gravity signal transduction pathway that is genetically distinct from the PGM pathway. Plant Physiol. 133 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy, F., Nicoll, W.S., Zimmermann, R., Cheetham, M.E., and Blatch, G.L. (2005). Not all J domains are created equal: Implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 14 1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig, L., Buche, C., Eichenberg, K., and Schafer, E. (1999). Dynamic properties of endogenous phytochrome A in Arabidopsis seedlings. Plant Physiol. 121 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig, L., Poppe, C., Sweere, U., Martin, A., and Schafer, E. (2001). Negative interference of endogenous phytochrome B with phytochrome A function in Arabidopsis. Plant Physiol. 125 1036–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltbrunner, A., Tscheuschler, A., Viczian, A., Kunkel, T., Kircher, S., and Schafer, E. (2006). FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 47 1023–1034. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner, A., Viczian, A., Bury, E., Tscheuschler, A., Kircher, S., Toth, R., Honsberger, A., Nagy, F., Fankhauser, C., and Schafer, E. (2005). Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 15 2125–2130. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., and Quail, P.H. (2001). The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J. Biol. Chem. 276 38173–38178. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Tepperman, J.M., and Quail, P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284 496–499. [DOI] [PubMed] [Google Scholar]
- Huq, E., Tepperman, J.M., and Quail, P.H. (2000). GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 97 9789–9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, I.C., Yang, J.Y., Seo, H.S., and Chua, N.H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, I.C., Yang, S.W., Yang, J.Y., and Chua, N.H. (2007). Independent and interdependent functions of LAF1 and HFR1 in phytochrome A signaling. Genes Dev. 21 2100–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E., Bradley, M., Harberd, N.P., and Whitelam, G.C. (1994). Photoresponses of light-grown phyA mutants of Arabidopsis (phytochrome A is required for the perception of daylength extensions). Plant Physiol. 105 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kelley, W.L. (1998). The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 23 222–227. [DOI] [PubMed] [Google Scholar]
- Kim, H.Y., Ahn, B.Y., and Cho, Y. (2001). Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 20 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.I., Shen, Y., Han, Y.J., Park, J.E., Kirchenbauer, D., Soh, M.S., Nagy, F., Schafer, E., and Song, P.S. (2004). Phytochrome phosphorylation modulates light signaling by influencing the protein-protein interaction. Plant Cell 16 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Gil, P., Kozma-Bognar, L., Fejes, E., Speth, V., Husselstein-Muller, T., Bauer, D., Adam, E., Schafer, E., and Nagy, F. (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneissl, J., Shinomura, T., Furuya, M., and Bolle, C. (2008). A rice phytochrome A in Arabidopsis: The role of the N-terminus under red and far-red light. Mol. Plant 1 84–102. [DOI] [PubMed] [Google Scholar]
- Koncz, C., Martini, N., Mayerhofer, R., Koncz-Kalman, Z., Korber, H., Redei, G.P., and Schell, J. (1989). High-frequency T-DNA-mediated gene tagging in plants. Proc. Natl. Acad. Sci. USA 86 8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., Martini, N., Szabados, L., Hrouda, M., Bachmair, A., and Schell, J. (1994). Specialized vectors for gene tagging and expression studies. In Plant Molecular Biology Manual, Vol. B2, S.B. Gelvin and R.A. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic Press), pp. 1–22.
- Kozany, C., Mokranjac, D., Sichting, M., Neupert, W., and Hell, K. (2004). The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat. Struct. Mol. Biol. 11 234–241. [DOI] [PubMed] [Google Scholar]
- Kushnir, S., Babiychuk, E., Kampfenkel, K., Belles-Boix, E., Van Montagu, M., and Inze, D. (1995). Characterization of Arabidopsis thaliana cDNAs that render yeasts tolerant toward the thiol-oxidizing drug diamide. Proc. Natl. Acad. Sci. USA 92 10580–10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapko, V.N., Jiang, X.Y., Smith, D.L., and Song, P.S. (1997). Posttranslational modification of oat phytochrome A: Phosphorylation of a specific serine in a multiple serine cluster. Biochemistry 36 10595–10599. [DOI] [PubMed] [Google Scholar]
- Lariguet, P., Boccalandro, H.E., Alonso, J.M., Ecker, J.R., Chory, J., Casal, J.J., and Fankhauser, C. (2003). A growth regulatory loop that provides homeostasis to phytochrome a signaling. Plant Cell 15 2966–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariguet, P., Schepens, I., Hodgson, D., Pedmale, U.V., Trevisan, M., Kami, C., de Carbonnel, M., Alonso, J.M., Ecker, J.R., Liscum, E., and Fankhauser, C. (2006). PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proc. Natl. Acad. Sci. USA 103 10134–10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon, S.K. (2001). Clathrin uncoating: Auxilin comes to life. Curr. Biol. 11 R49–R52. [DOI] [PubMed] [Google Scholar]
- Liscum, E., and Briggs, W.R. (1995). Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luccioni, L.G., Oliverio, K.A., Yanovsky, M.J., Boccalandro, H.E., and Casal, J.J. (2002). Brassinosteroid mutants uncover fine tuning of phytochrome signaling. Plant Physiol. 128 173–181. [PMC free article] [PubMed] [Google Scholar]
- Martin-Tryon, E.L., Kreps, J.A., and Harmer, S.L. (2007). GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 143 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk, J.A. (2001). The J-domain proteins of Arabidopsis thaliana: An unexpectedly large and diverse family of chaperones. Cell Stress Chaperones 6 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliverio, K.A., Crepy, M., Martin-Tryon, E.L., Milich, R., Harmer, S.L., Putterill, J., Yanovsky, M.J., and Casal, J.J. (2007). GIGANTEA regulates phytochrome A-mediated photomorphogenesis independently of its role in the circadian clock. Plant Physiol. 144 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, D.H., Somers, D.E., Kim, Y.S., Choy, Y.H., Lim, H.K., Soh, M.S., Kim, H.J., Kay, S.A., and Nam, H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285 1579–1582. [DOI] [PubMed] [Google Scholar]
- Popov, N., Schmitt, M., Schulzeck, S., and Matthies, H. (1975). Eine störungsfreie Mikromethode zur Bestimmung des Proteingehalts in Gewebshomogenaten. Acta Biol. Med. Germ. 34 1441–1446. [PubMed] [Google Scholar]
- Qbadou, S., Becker, T., Bionda, T., Reger, K., Ruprecht, M., Soll, J., and Schleiff, E. (2007). Toc64-a preprotein-receptor at the outer membrane with bipartide function. J. Mol. Biol. 367 1330–1346. [DOI] [PubMed] [Google Scholar]
- Qiu, X.B., Shao, Y.M., Miao, S., and Wang, L. (2006). The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63 2560–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn, M.H., Oliverio, K., Yanovsky, M.J., and Casal, J.J. (2002). CP3 is involved in negative regulation of phytochrome A signalling in Arabidopsis. Planta 215 557–564. [DOI] [PubMed] [Google Scholar]
- Reed, J.W., Nagatani, A., Elich, T.D., Fagan, M., and Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosler, J., Klein, I., and Zeidler, M. (2007). Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc. Natl. Acad. Sci. USA 104 10737–10742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Rubio, V., and Deng, X.W. (2005). Phy tunes: Phosphorylation status and phytochrome-mediated signaling. Cell 120 290–292. [DOI] [PubMed] [Google Scholar]
- Ryu, J.S., et al. (2005). Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell 120 395–406. [DOI] [PubMed] [Google Scholar]
- Saibo, N.J., Vriezen, W.H., Beemster, G.T., and Van Der Straeten, D. (2003). Growth and stomata development of Arabidopsis hypocotyls are controlled by gibberellins and modulated by ethylene and auxins. Plant J. 33 989–1000. [DOI] [PubMed] [Google Scholar]
- Schepens, I., Boccalandro, H.E., Kami, C., Casal, J.J., and Fankhauser, C. (2008). PHYTOCHROME KINASE SUBSTRATE4 modulates phytochrome-mediated control of hypocotyl growth orientation. Plant Physiol. 147 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinte, P., Foerstendorf, H., Hussain, Z., Gartner, W., Mroginski, M.A., Hildebrandt, P., and Siebert, F. (2008). FTIR study of the photoinduced processes of plant phytochrome phyA using isotope-labeled bilins and density functional theory calculations. Biophys. J. 95 1256–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook, J.C., Chen, R., and Masson, P.H. (1999). ARG1 (altered response to gravity) encodes a DnaJ-like protein that potentially interacts with the cytoskeleton. Proc. Natl. Acad. Sci. USA 96 1140–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H.S., Watanabe, E., Tokutomi, S., Nagatani, A., and Chua, N.H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura, T., Nagatani, A., Hanzawa, H., Kubota, M., Watanabe, M., and Furuya, M. (1996). Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93 8129–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura, T., Uchida, K., and Furuya, M. (2000). Elementary processes of photoperception by phytochrome A for high-irradiance response of hypocotyl elongation in Arabidopsis. Plant Physiol. 122 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staneloni, R.J., Rodriguez-Batiller, M.J., Legisa, D., Scarpin, M.R., Agalou, A., Cerdán, P.D., Meijer, A.H., Ouwerkerk, P.B., and Casal, J.J. (2009). Bell-like homeodomain selectively regulates the high-irradiance response of phytochrome A. Proc. Natl. Acad. Sci. USA 106 13624–13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh, M.S., Kim, Y.M., Han, S.J., and Song, P.S. (2000). REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell 12 2061–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Galea, P., Huang, L.F., Chua, N.H., and Bolle, C. (2006). The GRAS protein SCL13 is a positive regulator of phytochrome-dependent red light signaling, but can also modulate phytochrome A responses. Mol. Genet. Genomics 276 13–30. [DOI] [PubMed] [Google Scholar]
- Trupkin, S.A., Debrieux, D., Hiltbrunner, A., Fankhauser, C., and Casal, J.J. (2007). The serine-rich N-terminal region of Arabidopsis phytochrome A is required for protein stability. Plant Mol. Biol. 63 669–678. [DOI] [PubMed] [Google Scholar]
- Vandenbussche, F., Vancompernolle, B., Rieu, I., Ahmad, M., Phillips, A., Moritz, T., Hedden, P., and Van Der Straeten, D. (2007). Ethylene-induced Arabidopsis hypocotyl elongation is dependent on but not mediated by gibberellins. J. Exp. Bot. 58 4269–4281. [DOI] [PubMed] [Google Scholar]
- van der Krol, A.R., and Chua, N.H. (1991). The basic domain of plant B-ZIP proteins facilitates import of a reporter protein into plant nuclei. Plant Cell 3 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tuinen, A., Kerckhoffs, L.H., Nagatani, A., Kendrick, R.E., and Koornneef, M. (1995). Far-red light-insensitive, phytochrome A-deficient mutants of tomato. Mol. Gen. Genet. 246 133–141. [DOI] [PubMed] [Google Scholar]
- Walsh, P., Bursac, D., Law, Y.C., Cyr, D., and Lithgow, T. (2004). The J-protein family: Modulating protein assembly, disassembly and translocation. EMBO Rep. 5 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., and Deng, X.W. (2002). Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J. 21 1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann, B., and Neupert, W. (1997). Mdj2p, a novel DnaJ homolog in the mitochondrial inner membrane of the yeast Saccharomyces cerevisiae. J. Mol. Biol. 272 477–483. [DOI] [PubMed] [Google Scholar]
- Yang, J., Lin, R., Hoecker, U., Liu, B., Xu, L., and Wang, H. (2005a). Repression of light signaling by Arabidopsis SPA1 involves post-translational regulation of HFR1 protein accumulation. Plant J. 43 131–141. [DOI] [PubMed] [Google Scholar]
- Yang, J., Lin, R., Sullivan, J., Hoecker, U., Liu, B., Xu, L., Deng, X.W., and Wang, H. (2005b). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17 804–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., Casal, J.J., and Luppi, J.P. (1997). The VLF loci, polymorphic between ecotypes Landsberg erecta and Columbia, dissect two branches of phytochrome A signal transduction that correspond to very-low-fluence and high-irradiance responses. Plant J. 12 659–667. [DOI] [PubMed] [Google Scholar]
- Zeidler, M., Bolle, C., and Chua, N.H. (2001). The phytochrome A specific signaling component PAT3 is a positive regulator of Arabidopsis photomorphogenesis. Plant Cell Physiol. 42 1193–1200. [DOI] [PubMed] [Google Scholar]
- Zeidler, M., Zhou, Q., Sarda, X., Yau, C.P., and Chua, N.H. (2004). The nuclear localization signal and the C-terminal region of FHY1 are required for transmission of phytochrome A signals. Plant J. 40 355–365. [DOI] [PubMed] [Google Scholar]
- Zhou, Q., Hare, P.D., Yang, S.W., Zeidler, M., Huang, L.F., and Chua, N.H. (2005). FHL is required for full phytochrome A signaling and shares overlapping functions with FHY1. Plant J. 43 356–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.