Abstract
Relatively little is known about how metals such as iron are effluxed from cells, a necessary step for transport from the root to the shoot. Ferroportin (FPN) is the sole iron efflux transporter identified to date in animals, and there are two closely related orthologs in Arabidopsis thaliana, IRON REGULATED1 (IREG1/FPN1) and IREG2/FPN2. FPN1 localizes to the plasma membrane and is expressed in the stele, suggesting a role in vascular loading; FPN2 localizes to the vacuole and is expressed in the two outermost layers of the root in response to iron deficiency, suggesting a role in buffering metal influx. Consistent with these roles, fpn2 has a diminished iron deficiency response, whereas fpn1 fpn2 has an elevated iron deficiency response. Ferroportins also play a role in cobalt homeostasis; a survey of Arabidopsis accessions for ionomic phenotypes showed that truncation of FPN2 results in elevated shoot cobalt levels and leads to increased sensitivity to the metal. Conversely, loss of FPN1 abolishes shoot cobalt accumulation, even in the cobalt accumulating mutant frd3. Consequently, in the fpn1 fpn2 double mutant, cobalt cannot move to the shoot via FPN1 and is not sequestered in the root vacuoles via FPN2; instead, cobalt likely accumulates in the root cytoplasm causing fpn1 fpn2 to be even more sensitive to cobalt than fpn2 mutants.
INTRODUCTION
Iron is essential for plant growth, yet the redox properties that make iron biologically useful make free iron highly destructive. Consequently, iron uptake is highly regulated and iron metabolism highly compartmentalized. In Arabidopsis thaliana, soil Fe3+ is reduced by FERRIC REDUCTASE OXIDASE2 (FRO2) (Robinson et al., 1999) and then transported into the epidermal cells by the divalent metal transporter IRON REGULATED TRANSPORTER1 (IRT1; Vert et al., 2002) that also transports zinc, manganese, cadmium, cobalt (Korshunova et al., 1999), and nickel (Schaaf et al., 2006). Iron likely moves symplastically to the pericycle, where it then needs to be effluxed into the xylem to move to the shoot (Durrett et al., 2007). Despite progress in understanding iron uptake into the root in Arabidopsis, relatively little is known about the iron transporters required for movement from the root to the shoot.
In mammals, ferroportin is the sole iron efflux protein identified to date, functioning in both iron absorption in the intestine and iron recycling in macrophages (Muckenthaler et al., 2008). There are two closely related orthologs in Arabidopsis, IRON REGULATED1/Ferroportin 1 (IREG1/FPN1) and IREG2/FPN2. FPN2 was previously reported to be expressed in the roots of iron-deficient plants (Colangelo and Guerinot, 2004) and to localize to the vacuolar membrane (Schaaf et al., 2006). Although upregulated in response to iron deficiency, FPN2 was reported to function in nickel sequestration (Schaaf et al., 2006). As nickel is one of the divalent cations taken up by IRT1, this suggests that plants have strategies to deal with the influx of potentially toxic metals that enter the root during iron deficiency. A similar role has been described for MTP3, which is iron regulated, localizes to the vacuolar membrane, and is thought to sequester zinc in the vacuole during iron deficiency (Arrivault et al., 2006).
Cobalt is also transported by IRT1. It is not essential for plants and likely induces oxidative stress (Salnikow et al., 2000). Cobalt has been shown to disrupt iron homeostasis in a wide range of organisms, including Escherichia coli (Ranquet et al., 2007), Salmonella enterica (Thorgersen and Downs, 2007), yeast (Stadler and Schweyen, 2002), mice (Latunde-Dada et al., 2004), and mung beans (Liu et al., 2000). Cobalt likely competes with iron for access to transporters and has already been shown to bind the E. coli iron-sensing protein Fur (Adrait et al., 1999) and interfere with the assembly of iron-sulfur clusters (Ranquet et al., 2007) and heme (Watkins et al., 1980). As plants are sessile and possess a wide range of adaptations to soil stresses, it would seem likely that mechanisms exist to control cobalt levels and localization to minimize disruption of iron homeostasis.
To determine how the uptake and homeostasis of cobalt and other metals are controlled, the Arabidopsis Ionomics project uses a high-throughput approach combining inductively coupled plasma–mass spectrometry (ICP-MS) and bioinformatic analysis to identify lines that have altered elemental profiles (Baxter et al., 2007). Approximately 10,000 Arabidopsis mutant lines and natural accessions have been screened for variation in elemental composition (data can be accessed at www.ionomicshub.org). Here, we report on several accessions of Arabidopsis that accumulate cobalt and identify FPN2 as the gene mutated in these accessions. We also present data supporting a role for FPN1 in cobalt transport to the shoot as well as a role for both FPN1 and FPN2 in iron efflux.
RESULTS
Increased Shoot Cobalt Is Seen in fpn2-1 and Natural Accessions with Truncated FPN2
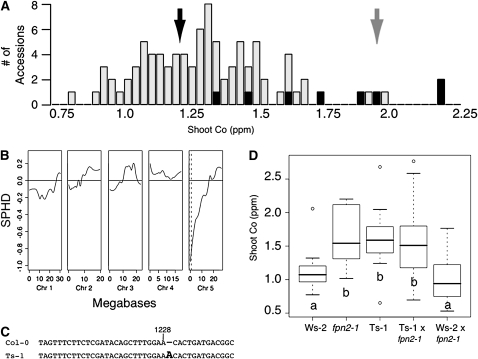
Shoot cobalt content was measured across 94 Arabidopsis accessions (Figure 1A). The Spanish accession Ts-1 was one of the top 10 accessions that showed elevated shoot cobalt relative to Columbia-0 (Col-0) (see Supplemental Table 1 online). Bulk segregant analysis mapped the high shoot cobalt locus in Ts-1 to the region of chromosome five containing the metal transporter gene FPN2 (Figure 1B). FPN2 was sequenced in Ts-1 and found to contain an adenine inserted after base pair 1228 of the genomic sequence (Figure 1C), upstream of five of the 10 predicted transmembrane domains (see Supplemental Figure 1C online; Aramemnon Plant Membrane Protein Database) and in the first third of the ferroportin domain shared with Hs FPN and At FPN1. This frame shift produces a stop codon 117 amino acids earlier than in Col-0 (see Supplemental Figure 1B online). Of the top seven cobalt accumulating accessions, five have the same adenine insertion after position 1228, resulting in truncation of FPN2 (see Supplemental Figure 1A online).
Figure 1.
Mapping the Shoot Cobalt Accumulation Locus to FPN2 in Ts-1.
(A) Shoot cobalt content across 94 accessions. Histogram of shoot cobalt content in 94 Arabidopsis accessions (Nordborg et al., 2005). Black bars indicate lines having the insertion that produces a frameshift in FPN2. The black arrow denotes Col-0 cobalt content, and the gray arrow indicates Ts-1. Shoot cobalt concentrations are normalized so that the average of the Col-0, Fab-2, Ts-1, and Cvi-0 means included in each growth tray are equivalent across all trays. Plants were grown in soil for 5 weeks. Data represent median values (n = 6) for each accession.
(B) Bulk segregant analysis of the high shoot cobalt content in an F2 population from a Col-0 × Ts-1 cross. Data are presented as a scaled pool hybridization difference (SPHD), representing the difference between the hybridization of the two pools at the single feature polymorphisms (SFPs), scaled so that a pure Col-0 pool would be at 1 and a pure Ts-1 pool would be at −1. The pools were prepared from F2 plants with a low cobalt content (n = 30) and F2 plants with a high cobalt content (n = 28). SFPs were scored after hybridization of genomic DNA prepared from these pools to Affymetrix ATH1 DNA microarrays. Dotted lines denote the region where the signal is larger than the 95% confidence threshold for unlinked loci.
(C) Alignment of FPN2 showing adenine inserted after position 1228 of the Ts-1 genomic sequence.
(D) Cobalt content of Ws-2, fpn2-1, Ts-1, and F1 plants from Ws-2 × fpn2-1 and Ts-1 × fpn2-1. Data are shown as a five-number summary (the minimum, 1st quartile, median, 3rd quartile, and maximum) for each line with outliers indicated by small circles and is summarized from an average of 10 replicate plants for each line. Lowercase letters denote groups that are not significantly different from each other at P < 0.01. Plants were grown in soil for 5 weeks.
To confirm that this FPN2 truncation was the cause of the elevated shoot cobalt phenotype, Ts-1 was crossed to the fpn2-1 loss-of-function mutant and its corresponding wild type, Wassilewskija-2 (Ws-2) (Ws-2 and Col have similar cobalt levels). When crossed to Ws-2, the cobalt accumulation phenotype is abolished; however, when Ts-1 is crossed to fpn2-1, the progeny still accumulate cobalt (Figure 1D). This indicates that the increased shoot cobalt phenotype seen in fpn2-1 can be rescued by a wild-type copy of Ws-2 FPN2; however, the truncated version of FPN2 found in Ts-1 cannot rescue fpn2-1. Thus, the disruption of FPN2 results in increased cobalt accumulation in the shoot.
Expression of FPN2 Is Iron Regulated, While FPN1 Expression Is Not
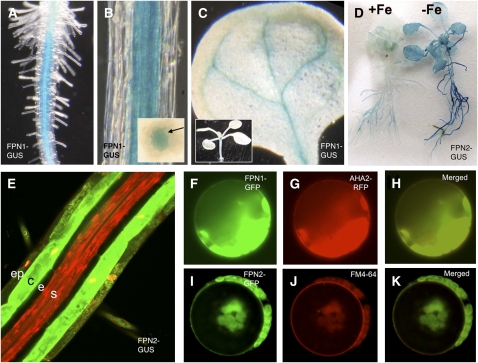
To understand how the loss of FPN2 could change cobalt homeostasis, we examined the tissue expression pattern of FPN2 and its paralog FPN1. Previous experiments have shown that FPN1 expression in the roots is low and not iron regulated (Colangelo and Guerinot, 2004; Dinneny et al., 2008); FPN2 transcript is present in iron-sufficient roots, and expression in roots is upregulated almost fourfold in response to iron deficiency (Colangelo and Guerinot, 2004; Schaaf et al., 2006; Dinneny et al., 2008). We fused the respective FPN promoters to the β-glucuronidase (GUS) reporter gene (Figure 2). The FPN1-GUS plants show staining in the vasculature of the root and shoot (Figures 2A to 2C). The FPN2-GUS plants show expression in the roots of iron-sufficient plants, with expression increasing under iron-deficient conditions (Figure 2D). Staining in the leaf veins also becomes evident in iron-deficient plants. When iron-deficient FPN2-GUS roots are stained with the fluorescent GUS substrate ImaGene Green, expression is limited to the outer two layers of the root (Figure 2E). Expression is strongest in the cortex but also present in the epidermis and root hairs and absent from the inner layers. This agrees well with the elevated level of FPN2 expression detected via transcriptomics in the cortex during iron starvation (Dinneny et al., 2008).
Figure 2.
Localization of FPN1 and FPN2.
Eleven-day-old FPN1-GUS plants show staining in the stele ([A] and [B]), root-shoot junction, and veins of the cotyledons (C). Inset in (B) shows staining in stele of root cross section. Inset in (C) shows staining in the seedling root-shoot junction. When 2-week-old FPN2-GUS plants are transferred from B5 to –Fe minimal medium for 3 d, GUS staining is very dark in the root and is also present in the shoot (D). Staining with propidium iodide (red) and the fluorescent GUS substrate, ImaGene Green, shows FPN2-GUS expression primarily in the cortex but also in the epidermis and root hairs (E). When FPN1 and FPN2 are fused to GFP and transiently expressed in protoplasts, FPN1-GFP localizes to the plasma membrane (F), as does the plasma membrane marker AHA2-RFP (G); FPN2-GFP localizes to the vacuole (I), as does the vacuole marker dye FM4-64 (J). Overlays of GFP and the markers are shown in (H) and (K).
FPN1 and FPN2 Are Localized to Different Membranes
FPN2 was previously found to localize to the vacuolar membrane (Schaaf et al., 2006). To determine the localization of FPN1, the gene was fused to green fluorescent protein (GFP) and transiently expressed in protoplasts. FPN1 localizes to the plasma membrane (Figures 2F to 2H), while FPN2 localizes to the vacuole as expected (Figures 2I to 2K). This suggests that while both ferroportins likely efflux metal from the cytoplasm, they play very different roles in metal homeostasis. FPN1 likely effluxes metals from the cytoplasm into the vasculature, allowing movement of metals from root to shoot; FPN2 effluxes into the vacuole, sequestering metals in the outer cell layers of the root, especially under iron deficiency.
Cobalt Accumulation Changes in Ferroportin Mutants
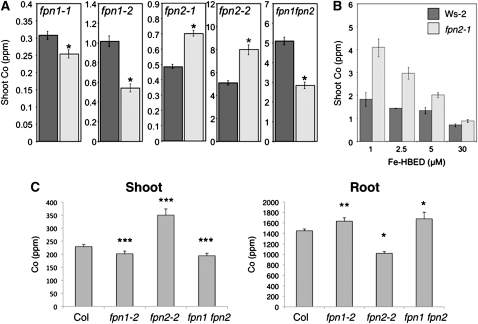
To confirm that plants carrying the fpn2-2 allele in the Col-0 background also accumulate cobalt, and to determine how the loss of FPN1, as well as the loss of both FPN1 and FPN2, would affect metal accumulation, ICP-MS was used to analyze the shoots of soil-grown plants (Figure 3A). The shoots of fpn2-1 and fpn2-2 have elevated cobalt, while fpn1-1, fpn1-2, and the fpn1 fpn2 double mutant have decreased shoot cobalt compared with the wild type. Despite the role FPN2 plays in nickel tolerance (Schaaf et al., 2006), no change in shoot nickel accumulation was observed in ferroportin mutants grown under our soil conditions (0.02 μmol Ni g−1 dry weight soil). Because FPN2 localizes to the vacuoles in the root, it likely sequesters cobalt in the root; its loss allows cobalt to move to the shoot, as seen in fpn2-1, fpn2-2, Ts-1, and other Arabidopsis accessions that accumulate cobalt. FPN1 localizes to the plasma membrane of stele cells, suggesting that it loads cobalt into the vasculature; its loss reduces the amount of cobalt moving to the shoot. In the double mutant, cobalt is not sequestered in the vacuoles of the root via FPN2 but is unable to enter the vasculature via FPN1.
Figure 3.
Ferroportin Mutants Have Altered Shoot Cobalt Accumulation.
(A) Shoot concentrations of cobalt in wild-type (dark gray) and mutant (light gray) soil-grown plants. Data represent mean ± se. For fpn1-1, wild type n = 11 and mutant n = 12; for fpn1-2, wild type n = 12 and mutant n = 7; for fpn2-1, wild type n = 30 and mutant n = 29; for fpn2-2, wild type n = 20 and mutant n = 18; and for fpn1 fpn2, wild type n = 20 and mutant n = 18. All plants were grown, sampled, and analyzed as individuals. Asterisks represent samples that are significantly different from the paired wild type (P < 0.01).
(B) Shoot concentrations of cobalt in wild-type (dark gray) and fpn2-1 (light gray) soil-grown plants after supplying plants with various concentrations of Fe provided as Fe-HBED. Data represent mean ± se, wild type n = 3 to 10 and fpn2-1 n = 11 to 12. Plants grown, sampled, and analyzed as individuals.
(C) Col-0 plants carrying FPN alleles were grown on B5 for 2 weeks and then transferred to a hydroponic solution of quarter-strength Hoagland, supplemented with 2.5 μM CoCl2. After 3 weeks, the plants were harvested and separated into shoot and roots and analyzed using ICP-MS. Values represent mean ± se. Significant differences from the wild type are represented by * P < 0.05, ** P < 0.01, and *** P < 0.0001; n = 21 plants, except for fpn1 fpn2, n = 17.
To determine how the changes in shoot cobalt related to metal levels in the roots, the mutant lines were grown hydroponically in medium supplemented with cobalt (to ensure a robust ICP-MS signal). The plants were harvested and the roots and shoots analyzed for cobalt accumulation and distribution using ICP-MS (Figure 3C). The loss of FPN2 produces a decrease in the concentration of cobalt in the root but an increase in the shoot. This supports the hypothesis that FPN2 sequesters cobalt in the root, and its loss allows cobalt to escape the root and accumulate in the shoot. The fpn1 and fpn1 fpn2 mutants have the reverse phenotype: both show increased cobalt concentrations in the root and decreased cobalt concentrations in the shoot. This further suggests that FPN1 loads cobalt into the root vasculature for transport to the shoot. We also measured the levels of Fe and Ni (see Supplemental Table 2 online). The only significant difference in Fe levels was for the fpn2 mutant that accumulated more Fe than the wild type in its roots. There were no significant differences between the wild type and the fpn mutants for Ni in either roots or shoots.
Ferroportin Mutants Are More Sensitive to Cobalt and Nickel
To determine if the observed changes in cobalt distribution alter metal tolerance, the mutant lines were grown on solid media supplemented with cobalt and nickel (Figure 4). Previously, fpn2-2 was found to be more sensitive to nickel than wild-type Col-0 (Schaaf et al., 2006). On B5 medium alone, the mutant lines have no visible phenotype. However, when grown on B5 supplemented with 20 μM CoCl2 or 50 μM NiCl2, fpn2-2 is more sensitive than the wild type. Similarly, fpn2-1 is also more sensitive to cobalt and nickel than Ws (Figure 4B), as are Ts-1 and Se-0 (Figure 4A). The fpn1-2 mutant is not visibly affected by cobalt or nickel, yet when crossed with fpn2-2, the double mutant is more sensitive than fpn2-2 and dramatically more sensitive than the wild type. This demonstrates that FPN1 contributes to cobalt and nickel sensitivity, but this can only be seen in the absence of FPN2.
Figure 4.
The Loss of FPN2 Increases Sensitivity to Cobalt and Nickel.
Plants were germinated and grown on B5 solid media, amended with either cobalt or nickel, for 3 weeks under 16-h-light/8-h-dark cycle at 21°C.
(A) Col-0 alleles and the accessions Ts-1 and Se-0.
(B) Ws-2 alleles.
Combined with the shoot ICP-MS data, this indicates that without FPN2, cobalt is not sequestered in the root vacuoles, allowing its movement to the shoot, resulting in sensitivity to cobalt. In the double mutant, the loss of FPN1 prevents cobalt export to the shoot, causing its concentration to rise in the root. Because the double mutant also lacks a functioning FPN2, the increased root cobalt cannot be detoxified by compartmentalization into the vacuole. This results in the extreme sensitivity to cobalt observed in the double mutant fpn1-2 fpn2-2.
FPN1 Is Required for Cobalt Movement from Root to Shoot
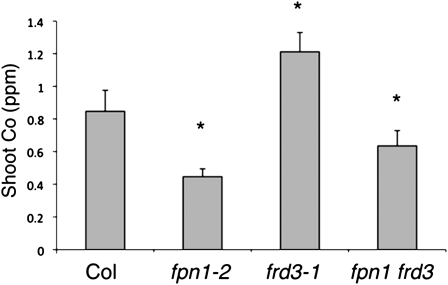
Because the fpn1 and fpn1 fpn2 mutants have decreased cobalt accumulation in the shoot, we further investigated whether FPN1 loaded cobalt into the vasculature. To do this, we crossed fpn1-2 with the frd3-1 mutant. The frd3 mutant expresses IRT1 constitutively and thus accumulates significantly more cobalt in the shoot (Lahner et al., 2003). If FPN1 is required for cobalt movement to the shoot, the loss of FPN1 in frd3-1 would abolish the shoot cobalt accumulation phenotype. When shoots of soil-grown plants are analyzed with ICP-MS (Figure 5), fpn1-2 shows significantly less shoot cobalt than the wild type, while frd3-1 accumulates much more than the wild type. In the fpn1 frd3 double mutant, shoot cobalt is much lower than the wild type, as is seen in fpn1-2. This confirms that FPN1 is essential for cobalt movement from root to shoot and indicates that FPN1 plays a significant role in cobalt loading into the vasculature.
Figure 5.
FPN1 Is Required for Cobalt Movement to the Shoot.
Shoots of soil-grown plants analyzed by ICP-MS. Mean ± se is shown. Significant differences from the wild type are indicated by * P < 0.0001. For Col-0, n = 12 plants; for fpn1-2, n = 36 plants; for frd3-1, n = 24 plants; and for fpn1 frd3, n = 36 plants.
The Loss of FPN1 Leads to Chlorosis
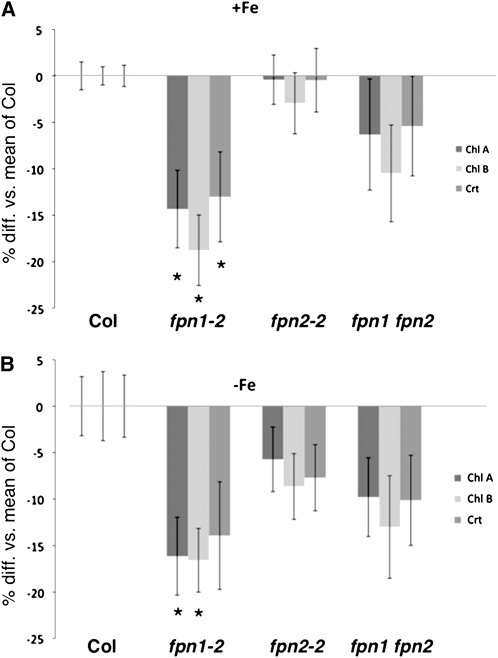
Because both ferroportins were found to affect cobalt movement from root to shoot, we examined whether the ferroportins also played a role in long-distance iron transport. To determine if iron was reaching the shoot efficiently, we assayed shoot chlorophyll concentrations; if a mutant had impaired iron movement, then it would likely become chlorotic and have lower chlorophyll levels. Plants were grown on B5 plates for 2 weeks and then transferred to minimal media that was either iron sufficient or deficient. Under both conditions, fpn1 is more chlorotic than the wild type: chlorophyll and carotenoid levels were decreased by 13 to 18% (Figure 6). This indicates that FPN1 plays a role in iron movement to the shoot; based on its localization, it most likely loads iron into the vasculature. The fpn2 mutant is not chlorotic under iron sufficiency, but its chlorophyll and carotenoid levels are slightly lower than the wild type on iron-deficient medium. The fpn1 fpn2 double mutant shows a smaller decrease in chlorophyll and carotenoid levels than fpn1-2 during both iron sufficiency and iron deficiency. This suggests the fpn1 chlorosis phenotype is ameliorated by the loss of FPN2, perhaps because iron is no longer being sequestered in the vacuoles of the outer root layers.
Figure 6.
Ferroportin Mutants Are Chlorotic.
The chlorophyll (Chl) and carotenoid (Crt) concentrations in ferroportin mutants were measured. Plants were grown on B5 solid medium for 2 weeks and then transferred to +Fe (A) or −Fe (B) media. Bars represent the average percentage difference among individual plants versus the mean of Col (±se). Significant differences from the wild type are indicated by * P < 0.05. For +Fe, n = 15 plants; for –Fe, n = 18 plants.
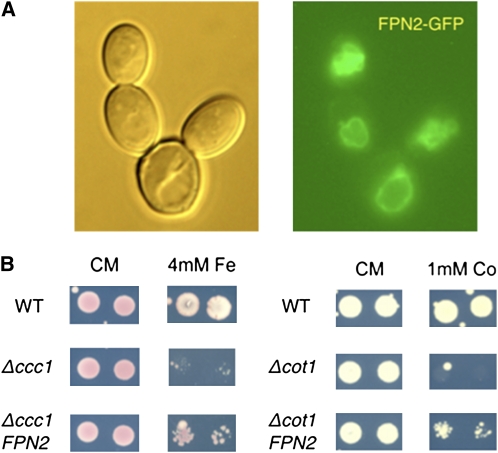
FPN2 Can Transport Iron and Cobalt When Expressed in Yeast
While FPN is the sole iron efflux protein identified to date in humans, the first ferroportin phenotypes we found in Arabidopsis were nickel and cobalt related. To determine whether the Arabidopsis ferroportins could also transport iron, FPN1 and FPN2 were expressed in yeast. FPN1 did not localize properly to the plasma membrane in yeast, and we could detect no transport activity. FPN2-GFP localizes to the yeast vacuole membrane, as in Arabidopsis (Figure 7A). To determine if FPN2 transports iron into the vacuole, it was expressed in Δccc1 yeast. CCC1 is an iron transporter expressed on the yeast vacuolar membrane that sequesters excess cytoplasmic iron; the Δccc1 mutant is thus iron sensitive and dies when grown on high iron concentrations (Li et al., 2001). When FPN2 is expressed in Δccc1, it rescues the mutant at otherwise toxic iron levels, presumably by pumping iron into the vacuole (Figure 7B). This indicates that FPN2, like Hs FPN, is an iron effluxer. To confirm that FPN2 also transports cobalt, it was expressed in Δcot1 yeast (Figure 7B). COT1 is a cobalt and zinc transporter that localizes to the yeast vacuolar membrane, sequestering those metals in the vacuole; the Δcot1 mutant is sensitive to cobalt (Conklin et al., 1992). Expression of FPN2 improves cobalt tolerance of Δcot1 mutant yeast, indicating that FPN2 can transport cobalt into the yeast vacuole and likely does the same in Arabidopsis.
Figure 7.
FPN2 Sequesters Iron and Cobalt in the Vacuole.
FPN2 expressed in yeast. FPN2-GFP localizes to vacuole membrane (A), and FPN2 expression rescues the iron-sensitive phenotype of the yeast Δccc1 mutant and the cobalt-sensitive phenotype of the Δcot1 mutant (B), presumably by loading iron and cobalt into the vacuole, respectively.
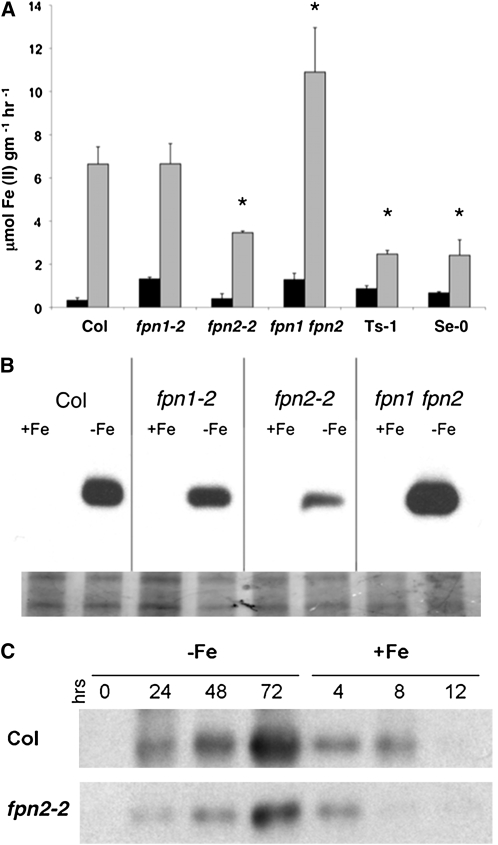
Ferroportins Affect the Iron Deficiency Response
Because we found that FPN2 transports iron and that its expression is iron regulated, we examined how the loss of ferroportins affects the iron deficiency response. During iron deficiency in Arabidopsis, FRO2 activity is highly induced in the root epidermis to generate ferrous iron for uptake via IRT1 (Figure 9). The fpn1-2 mutant shows no change in ferric chelate reductase activity compared with the wild type (Figure 8A). The loss of FPN2, however, results in a significant decrease in ferric chelate reductase activity: fpn2-2 reduces about half as much ferric iron as Col (Figure 8A), while fpn2-1 reduces about half as much as Ws-2 (see Supplemental Figure 2 online). Two Spanish accessions with the frameshift mutation in FPN2, Ts-1 and Se-0, also show significantly less ferric chelate reductase activity than Col-0 and Ws-2 in response to iron deficiency. Overexpression of FPN2 using the 35S promoter produces the opposite phenotype: ferric chelate reductase activity under both iron-sufficient and iron-deficient conditions is >40% greater than in the wild type. This is similar to results seen with yeast where overexpression of FPN2 results in cytoplasmic iron depletion as evidenced by induction of the iron-responsive FET3-lacZ reporter (see Supplemental Figure 3 online).
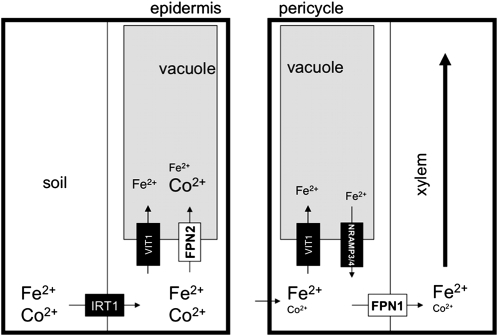
Figure 9.
Model of Ferroportin Function during Iron Deficiency.
IRT1 is expressed in response to iron deficiency, transporting iron and cobalt, among other metals, into the root epidermis. At the same time, FPN2 is expressed in the root epidermis and cortex and transports iron and cobalt into the vacuole. This buffers the influx of highly reactive iron in the outermost root layers and sequesters the nonessential element cobalt in the root. In the stele, NRAMP3 and NRAMP4 are also expressed and pump iron out of the vacuole (previously loaded by VIT1). This is loaded into the vasculature by FPN1, along with any cobalt that was not sequestered by FPN2. For simplicity, we have only shown Fe and Co, although the transporters indicated also transport other divalent cations.
Figure 8.
The Loss of Ferroportin Disrupts the Iron Deficiency Response.
(A) Plants were grown on B5 solid medium under constant light for 2 weeks at 21°C and then transferred to either +Fe (black bars) or −Fe (gray bars) minimal medium for 3 d. Ferric chelate reductase activity of a pool of six plants was measured, in triplicate, using the ferrozine assay. Data represent mean ± se. Significant differences from the wild type are indicated by * P < 0.05.
(B) IRT1 protein levels in ferroportin mutants measured by immunoblot using IRT1 antibody. Plants were grown on B5 solid medium under constant light at 21°C for 2 weeks and then transferred to either +Fe or –Fe minimal medium for 3 d. Ten micrograms of total protein were loaded (bottom panel shows Coomassie blue staining).
(C) Plants were grown on B5 solid medium for 2 weeks under constant light at 21°C and then transplanted to –Fe minimal medium. Samples were taken every 24 h. After 72 h on –Fe minimal medium, the plants were transplanted to +Fe minimal medium and samples taken every 4 h. The presence of IRT1 protein was measured by immunoblots using IRT1 antibody.
To further characterize the role of ferroportins in the iron deficiency response, IRT1 protein levels were detected in the mutant backgrounds. IRT1 expression is induced by iron deficiency (Eide et al., 1996; Vert et al., 2002), and immunoblots using an IRT1-specific antibody show IRT1 protein accumulation in roots after 3 d on iron-deficient medium (Figure 8B). In fpn1-2, IRT1 levels are similar to the wild type. In fpn2-2, IRT1 levels are decreased, matching the decreased levels of ferric chelate reductase activity seen. The fpn1 fpn2 double mutant shows greatly increased levels of IRT1; combined with the ferric chelate reductase data, the double mutant clearly has an elevated iron deficiency response. The loss of both ferroportins causes a more extreme iron deficiency response, indicating that iron homeostasis is impaired. This likely exacerbates the increased cobalt and nickel sensitivity seen in the double mutant: increased IRT1 expression would bring more cobalt and nickel into the root, which lacks a means to sequester or efflux the metals. Thus, the changes in the iron deficiency response seen in the fpn2 and fpn1 fpn2 double mutants indicate that ferroportins are an integral part of iron homeostasis in Arabidopsis.
To better characterize how the loss of FPN2 alters the iron deficiency response, a time course comparing IRT1 induction in the wild type and the fpn2 mutant was performed (Figure 8C). Two-week-old plants were transplanted to iron-deficient medium, and roots were harvested every 24 h. IRT1 accumulates more slowly or to lower levels in fpn2-2. At 24 h, IRT1 protein is readily detected in wild-type roots, but much less apparent in fpn2 roots. After 48 h on iron-deficient medium, IRT1 is detected in fpn2 roots, but it never reaches the expression level seen in the wild type. After 72 h, the iron-deprived plants were transferred to iron-sufficient medium and roots harvested every 4 h. After 4 h, IRT1 protein is still present in both wild-type and fpn2 roots but after 8 h, IRT1 protein is no longer detected in fpn2, while it is still present in the wild type. The loss of FPN2 likely mislocalizes iron that would otherwise be sequestered in the vacuoles of the outer root layers, altering either iron-sensing or the speed of iron remobilization during iron deficiency, delaying the induction or the strength of the response and ending it sooner.
Cobalt Accumulation in fpn2 Shoots Is Iron Dependent
To examine the relationship between the iron deficiency response and the fpn2 cobalt accumulation phenotype, we analyzed the shoot metal content of plants supplied with a range of iron supplementation levels (Figure 3B). Wild-type and fpn2-1 plants watered with 30 μM Fe-HBED both accumulate very little cobalt. When plants are grown with decreasing levels of iron supplementation, a clear divergence in cobalt shoot accumulation between fpn2-1 and the wild type emerges. The amount of shoot cobalt increases dramatically in the fpn2-1 mutant with each decrease in iron supplementation: from 5 μM Fe-HBED to 1 μM Fe-HBED, shoot cobalt increases from 2 to 4 μg g−1; when 30 μM Fe-HBED and 1 μM Fe-HBED are compared, the increase in shoot cobalt concentration is fourfold.
DISCUSSION
Ferroportins Are Required for Normal Cobalt Localization
IRT1 is expressed in response to iron deficiency and transports iron into the root cytoplasm from the rhizosphere, along with zinc, manganese, cadmium, cobalt (Vert et al., 2002), and nickel (Schaaf et al., 2006). FPN2 is also expressed during iron deficiency, localizes to the vacuole in roots (Schaaf et al., 2006), and effluxes metals from the cytoplasm into the vacuole. Without FPN2, cobalt is not sequestered in the root vacuoles; instead, it is able to move to the shoot via FPN1, resulting in an increase in shoot cobalt and cobalt sensitivity (Figure 9). A similar increase in shoot zinc levels is seen in the mtp3 loss-of-function mutant; like FPN2, it is expressed on the vacuolar membrane of iron-deficient roots, sequestering excess incoming zinc in the root (Arrivault et al., 2006). Schaaf et al. (2006) previously reported that FPN2 transported Ni into the vacuole and postulated that FPN2 expression was upregulated during iron deficiency to sequester the nickel that enters the root through IRT1. This explanation can now be extended to cobalt.
Without FPN1, cobalt accumulates in the root and decreases in the shoot. When fpn1-2 is crossed to the cobalt-accumulating frd3-1 mutant, shoot cobalt accumulation is abolished. This confirms that FPN1, despite its low (but constant) expression level, is a primary route for cobalt movement from root to shoot. Cobalt sensitivity does not increase in fpn1 because FPN2 is still able to sequester the increased root cobalt in the vacuole. Thus, in the fpn1 fpn2 double mutant, cobalt is not dispersed into the shoot via FPN1 or sequestered in the root vacuole via FPN2. Instead, cobalt likely accumulates in the cytoplasm of root cells, resulting in a dramatic increase in cobalt sensitivity. Ultimately, it is unclear whether these ferroportin functions are cobalt-specific adaptations or whether they are a side effect of the broad substrate specificity of iron transporters.
Ferroportins Are Required for Iron Homeostasis
FPN2 is an iron effluxer, like its ortholog Hs FPN, but instead of localizing to the plasma membrane, FPN2 is on the vacuolar membrane, moving iron from the cytoplasm into the vacuole. This function is essential for normal iron homeostasis, as fpn2 mutants have a diminished iron deficiency response. Although it seems counterintuitive to sequester iron in the vacuole during iron deficiency, FPN2 may buffer the influx of iron into the cytoplasm of the outer root layers. Expression of FPN2 could serve to sequester excess free iron that would otherwise not be chelated or transported out of the cell quickly enough. This buffering function would be similar to that of yeast zinc transporter ZRC1 that is expressed during zinc deficiency to sequester excess zinc in the vacuole, preventing zinc shock (MacDiarmid et al., 2003).
Iron homeostasis is disrupted in fpn2 because iron that would otherwise be stored in the vacuole is now mislocalized. It seems likely that iron sensing occurs outside the vacuole; thus, it takes the fpn2 mutant longer to perceive deficiency; when iron is restored, the cell is replenished more quickly because FPN2 is not moving iron into the vacuole or because the previously mislocalized iron is remobilized more quickly from nonvacuolar iron pools. The opposite phenotype is seen in the nramp3 loss-of-function mutant: when iron is not released from the vacuole by NRAMP3, the iron deficiency response is elevated (Thomine et al., 2003). While NRAMP3 is expressed in the vasculature, FPN2 is expressed in the root epidermis and cortex. Thus, during iron deficiency, FPN2 buffers the influx of iron into the cytoplasm in the outer tissue layers of the root, while NRAMP3 releases iron in the vascular tissue so that it can immediately move to the shoot.
The loss of FPN1 results in chlorosis, suggesting that FPN1 loads iron into the vasculature. Yet, fpn1 plants show no change in the iron deficiency response. The loss of both ferroportins, however, results in a greatly increased iron deficiency response, the opposite of the fpn2 phenotype. This could be caused by changes in metal concentration and localization within the root, at both the subcellular and root layer level, altering the kinetics of iron movement to the shoot. While the loss of FPN1 alone is not enough to alter iron sensing, the additional loss of the vacuolar iron buffering of FPN2 adds stress to an impaired system. With iron no longer sequestered in the vacuoles of the outer root layers, more iron likely moves toward the vasculature, which is now lacking FPN1. With increased cytosolic iron levels, competition for access to the remaining transporters and chelators would also increase, perhaps slowing iron movement to the vasculature. This could also be caused, or made worse, by cobalt and nickel accumulating in the root cytoplasm, especially when IRT1 is expressed. Cobalt and nickel are sterically similar to iron, and the presence of cobalt induces the iron deficiency response (see Supplemental Figure 4 online). Their mislocalization to the cytoplasm could competitively inhibit iron transport or bind up the chelators necessary for iron transport (e.g., citrate and NA). This would slow translocation of iron to the shoot, resulting in the more intense iron deficiency response as the shoot becomes iron starved. A similar phenotype was seen in mung bean seedlings treated with cobalt: iron uptake increased, but iron was unable to move from the root to the shoot (Liu et al., 2000).
To confirm these theories, we need to determine how iron and cobalt localization is changing in the ferroportin mutants at both the cellular and subcellular levels. Combining ICP-MS with the recently demonstrated technique of sorting cells by root layer markers (Dinneny et al., 2008) would allow us to determine if root cortex cells contained less iron and cobalt in fpn2 than the wild type. Additionally, improving microscope resolutions and metal binding dyes will eventually allow the real-time visualization of subcellular metal distribution. The ferroportin mutants would be ideal candidates for application of this technology, as we believe that iron and cobalt are mislocalized at the subcellular level.
Cobalt Uptake Is Iron Regulated
The amount of cobalt a plant takes up is dependent on its iron status: as it becomes more iron deficient, more IRT1 is expressed, through which more cobalt is transported. Expression of FPN2 is also iron regulated, increasing simultaneously with IRT1 levels, thus preventing increased cobalt accumulation in the shoot. Without FPN2, less cobalt is sequestered in the root, and each decrease in iron supplementation results in increases in shoot cobalt. Thus, cobalt uptake is iron regulated, while its localization between root and shoot is dependent on FPN2 expression.
The Potential of Cobalt Interference with Iron Sensing
An alternative interpretation of the mutant iron deficiency response phenotypes could center around cobalt localization rather than iron transport. Perhaps the Arabidopsis ferroportins primarily transport cobalt and only minor amounts of iron; the disruption of the iron deficiency response could be caused by changes in cobalt abundance and localization. The trigger of the iron deficiency response in plants is unknown, but if it involves iron binding, cobalt is a likely candidate for inappropriate binding and interference. Exposure to cobalt triggers the iron deficiency response in Arabidopsis, as FIT1 (the transcription factor that controls aspects of the iron deficiency response; Colangelo and Guerinot, 2004) expression increases (J. Morrissey and M.L. Guerinot, unpublished data), as does the amount of ferric iron reduced by the roots (see Supplemental Figure 4 online). This indicates that control of cobalt is essential to maintaining iron homeostasis. It is often speculated that the iron deficiency response has both root and shoot components, and it is possible that the higher shoot cobalt levels in fpn2 could suppress the iron deficiency response, while the concentration of mislocalized cobalt in the roots of fpn1 fpn2 could increase it.
Differences and Similarities to Human Ferroportin
Arabidopsis ferroportins transport cobalt in addition to iron. Our findings raise the obvious question whether the animal FPN proteins also transport cobalt. FPN would be a likely path for cobalt and nickel to move from the digestive track into the bloodstream. Hopefully, the complete range of metals entering into the bloodstream through FPN will be characterized in the future. It would also be interesting to investigate whether there are parallels in regulation of ferroportins in Arabidopsis and humans. Hepcidin is a small peptide that binds to FPN. It is required for normal function of Hs FPN, and the malfunction of either results in iron-related disease (Collins et al., 2008; Muckenthaler et al., 2008). A plant ortholog of hepcidin has not been found, but the number of small plant peptides is very likely underpredicted; in fact, a recent examination of antimicrobial small peptides in Arabidopsis found almost a quarter were not yet annotated (Silverstein et al., 2007). Hepcidin binds a specific domain on an extracellular loop of FPN (De Domenico et al., 2008), leading to phosphorylation of two Tyr residues (De Domenico et al., 2007) and the internalization and degradation of FPN. Both FPN1 and FPN2 have a Tyr and Phe in place of the double Tyr residues (see Supplemental Figure 5 online), and the region corresponding to the hepcidin binding domain lacks the Cys residue essential for binding (which is located in the same position as the Ts-1 frameshift mutation).
FPN1 and FPN2 likely efflux both iron and cobalt and are essential for cobalt tolerance and iron homeostasis. Cobalt uptake is controlled by iron status, while the tissue-specific accumulation of cobalt is dependent on FPN1 and FPN2. Ferroportin mutants show both increased cobalt sensitivity and shifts in cobalt localization; simultaneously, iron homeostasis in ferroportin mutants is disrupted.
METHODS
Identification of Mutant Lines
The fpn1-1 and fpn2-1 lines were acquired from the Arabidopsis Knockout Facility and screened using FPN1 (5′-GACCACTCTAGAACAAAGTAATACTGAGT-3′) and FPN2-specific primers (5′-GAAATAGAGGTATTTTTACAGTGTGACCA-3′) in combination with a T-DNA–specific primer (5′-CATTTTATAATAACGCTGCGGACATCTAC-3′) (Krysan et al., 1999). The Col lines fpn1-2 (SALK_145458) and fpn2-2 (SALK_074442) were acquired from the Salk Collection via ABRC (Alonso et al., 2003). Homozygous insertion mutants were identified using PCR with primers flanking the insertion site for fpn1 (5′-TGAAATAAATTCTCCATGCGC-3′ and 5′-TGAGGGTTGTTCATCAGGAAG-3′) and fpn2 (5′-GGTACCTGATCATCGTTGACG-3′ and 5′-GATTTGATCAGACCTTGCACC-3′) (see Supplemental Figure 6 online).
Plant Materials and Growth Conditions
Seeds were surface sterilized and stratified for 2 d in the dark at 4°C. B5 medium was supplemented with 1 mM MES and 0.6% agar, pH 5.8. To test for metal sensitivity, B5 medium was amended with either 20 μM CoCl2 or 50 μM NiCl2. Square, gridded Petri dishes were used; six seeds from each line were placed in the center of its own grid box. The plants were grown under a 16/8-h light-dark cycle at 21°C for 3 weeks.
Plants used for ICP-MS of roots and shoots were germinated on B5 and transferred after 2 weeks to quarter-strength Hoagland solution supplemented with 2.5 μM CoCl2. The plants were grown under a 16/8-h light-dark cycle at 21°C, and the hydroponic solution was changed weekly. After 3 weeks, the roots were washed three times in deionized water, and then the roots and shoots were harvested and dried for 48 h.
Ferric Chelate Reductase Assay
Plants were grown under constant light on B5 plates (as described above) for 2 weeks and then transferred to either +Fe or –Fe minimal media. The medium was made with macronutrients and micronutrients (Marschner et al., 1982), 0.6% agar, and 1 mM MES, pH 6.0, then supplemented with either 50 μM Fe(III)-EDTA (+Fe), or 300 μM ferrozine [3- (2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate] (−Fe). After 3 d, pools of six plants were analyzed for ferric chelate reductase activity in triplicate, as previously described (Yi and Guerinot, 1996).
Immunodetection
Plants were grown under constant light on B5 plates for 2 weeks and then transferred to either +Fe or –Fe minimal media for 3 d. The roots were then harvested and prepped, and an immunoblot with IRT1 antibody was performed as previously described (Connolly et al., 2002).
Plasmid Construction and Plant Transformation
The FPN2 promoter was amplified from Col genomic DNA using the primers 5′-TCTGCAGAAATGACTTACGTAAC-3′ and 5′-TGTTCATTGCACGTACCTGGC-3′; these primers added a PstI site at the 5′ end and a NcoI site at the 3′ end. The fragment was subcloned into the pGEM T-easy plasmid (Promega) and sequenced. The FPN2 promoter was then excised by NcoI and PstI digestion and subcloned into pCAMBIA 1302 (GFP) and 1304 (GFP/GUS) (CAMBIA). Agrobacterium tumefaciens strain GV3101 was transformed with these constructs and used to transform wild-type Col-0 using the floral dip method (Clough and Bent, 1998). Transformants were isolated by plating the seeds on B5 with hygromycin (25 mg/mL).
The 35S-FPN2 line was generated by cloning FPN2 from Col genomic DNA using the primers 5′-CATTTGGAGAGAACACGGGGGACTCTTGACATGGAGGAGGAAACAGAAAC-3′ and 5′-CTTCTCCTTTACTAGTCAGATCTACCATGTCATGAAGCAAAAAAGTTGTTC-3′. This was cloned into the NcoI site of pCAMBIA 1302, downstream of the 35S promoter, using the SLIC subcloning method (Li and Elledge, 2007). Agrobacterium and Col-0 were transformed as described above.
The FPN1 promoter and the coding region for the first 52 amino acids of FPN1 was amplified from Col genomic DNA using primers 5′-CCCGGATCCGTGATTTCTCAGGTTACAAC-3′ and 5′-CCCAAGCTTGGCACTCCATCTTGCAAG-3′, which added BamHI and HindIII sites, respectively. The fragment was subcloned into pSK and sequenced. It was then excised by digestion with BamHI and HindIII and ligated into pCambia1381xA (CAMBIA). The construct was then moved into Agrobacterium strain GV3101 and transformed into Col gl-1 plants using vacuum infiltration (Bent et al., 1994).
GUS Histochemical Staining
FPN2 promoter-GUS T1 lines were germinated on B5 with hygromycin (25 mg/mL). After 14 d, they were transferred to +Fe or –Fe minimal media for 3 d. The plants were incubated with the substrate 5-bromo-4-chloro-3-indolyl β-d-glucuronide as described (Jefferson et al., 1987). FPN1 promoter-GUS T2 lines were germinated on B5 amended with hygromycin (25 mg/mL) and stained after 11 d. To examine the FPN2 expression pattern in detail, roots expressing FPN2-GUS were incubated with a fluorescent GUS substrate, ImaGene Green (Invitrogen), and the cell wall was stained with propidium iodide. The confocal image confirmed that FPN2-GUS expression was predominantly detected in the cortex and epidermis. All fluorescence images were taken with a Nikon Eclipse 80i microscope (Nikon USA) using a green HeNe laser line (543 nm) for the GFP and a red HeNe laser line (633 nm) for the red fluorescence from the propidium iodide staining.
GFP Localization
To check subcellular localization, FPN1-GFP and FPN2-GFP constructs were transformed into Arabidopsis thaliana protoplasts, and then a tonoplast marker dye, FM4-64, was used to stain the protoplasts after 18 h. AHA2-red fluorescent protein was used as a plasma membrane marker (Kim et al., 2001). All fluorescence images were taken with an epifluorescence microscope, Nikon Eclipse 80i (Nikon USA), using the filter sets 31001 (exciter, D480/20x; dichroic, 505DCLP; emitter, D535/40m) for GFP and 31003 (exciter, D546/10x; dichroic, 560DCLP; emitter, D590/30m) from Chroma Technology.
Chlorophyll Assay
Plants were grown on B5 medium for 2 weeks under constant light and then transferred to +Fe or −Fe minimal media for 3 d. The shoots were harvested and assayed for chlorophyll and carotenoid content as previously described (Lichtenthaler, 1987).
Real-Time Quantitative PCR
RNA was prepared from 35S-FPN2 plants (germinated and grown on B5 for 2 weeks under constant light) using the RNeasy kit and protocol (Qiagen) and cDNA synthesized using M-MLV reverse transcriptase and protocol (Invitrogen). Real-time quantitative PCR was performed on an ABI Model 7700 using SYBR Premix ExTaq (Perfect Real Time) reagents and protocol (Takara) and the primers 5′-TTTTGTTAGCGCGTGGAGGAA-3′ and 5′-TAGACCAACACCTGCGCTGATT-3′. Samples were run in quadruplicate, FPN2 levels were normalized to EF1α (5′-CAGTCATTGATGCCCCAGGAC-3′ and 5′-TGTTGTCTCCCTCGAATCCAGAG-3′), and arbitrary transcriptional units calculated.
Plant Growth Conditions of Soil Grown Plants Analyzed by ICP-MS
Plants used for elemental profiling by ICP-MS analysis were grown in a controlled environment, 8 h light/16 h dark (90 μmol·m−2·s−1 light intensity) and 19 to 22°C (Lahner et al., 2003). Briefly, seeds were sown onto moist soil (Sunshine Mix LB2; Carl Brehob and Son) with various elements added at subtoxic concentrations (As, Cd, Co, Li, Ni, Pb, and Se; Lahner et al., 2003) and stratified at 4°C for 3 d. Plants were bottom-watered twice per week with 0.25× Hoagland solution in which iron was replaced with 10 μM Fe-HBED [N,N′-di(2-hydroxybenzyl)ethylenediamine-N,N′-diacetic acid monohydrochloride hydrate; Strem Chemicals). For elemental analysis after 5 weeks, plants were nondestructively sampled by removing one or two leaves. The plant material was rinsed with 18 MΩ water and placed into Pyrex digestion tubes.
Tissue Elemental Analysis
Tissue samples were dried at 92°C for 20 h in Pyrex tubes (16 × 100 mm) to yield ∼2 to 4 mg of tissue for elemental analysis. After cooling, seven of ∼100 samples from each sample set were weighed. All samples were digested with 0.7 mL of concentrated nitric acid (OmniTrace; VWR Scientific Products) and diluted to 6.0 mL with 18 MΩ water. Elemental analysis was performed with an ICP-MS (Elan DRCe; Perkin-Elmer) for Li, B, Na, Mg, P, S,K, Ca, Mn, Fe, Co, Ni, Cu, Zn, As, Se, Mo, and Cd. All samples were normalized to calculated weights, as determined with an iterative algorithm using the best-measured elements, the weights of the seven weighed samples, and the solution concentrations, implemented in the PiiMS database (Baxter et al., 2007).
DNA Microarray-Based Bulk Segregant Analysis
DNA microarray-based bulk segregant analysis was performed as previously described (Borevitz et al., 2003; Hazen et al., 2005). Briefly, SFPs were identified between Col-0 and Ts-1 by hybridizing labeled genomic DNA from each one of the accessions to Affymetrix ATH1 microarrays and comparing them to Col-0 hybridizations downloaded from http://www.naturalvariation.org/xam. Two genomic DNA pools from an F2 population of a cross between Ts-1 and Col-0 were created and hybridized to separate DNA microarrays. Each one of the pools contained plants with either shoot Co contents similar to Col-0 (control pool) or high shoot Co contents similar to Ts-1 (High Co pool). At loci unlinked to the high Co phenotype, the pools should have equivalent amounts of each genotype, and the hybridization signal at each SFP should be intermediate between the two parent accessions, for an average difference between the two DNA microarrays of zero. At linked loci, the difference between the two DNA pools should be approximately two-thirds the difference between the parent accessions. By smoothing the signal across multiple SFPs, noise is reduced and the peak of the differences in hybridization signal will correspond to the chromosomal region of the loci controlling the high Co trait. Raw hybridization data (.CEL files) for each probe on the ATH1 DNA microarrays used in these experiments have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) for public distribution (Barrett et al., 2007).
Yeast Strains, Plasmids, and Growth Conditions
Δccc1∷HIS3, Δzrc1∷HIS3, and their wild-type strains were in the W303 background (Li et al., 2001). Δcot1∷KanMX and its wild-type strain were in the BY4743 background (Conklin et al., 1992). Yeast strains were grown in CM medium (yeast nitrogen base, amino acids, and dextrose). For assay of metal toxicity, different concentrations of metals were added to the medium and serial dilutions of cells were spotted. Plant FPN1 or FPN2 genes were cloned into the yeast expression vector M3385 reference with a MET25 promoter and a C-terminal GFP epitope tag. To induce expression, cells were grown in medium lacking Met.
GFP Protein Localization in Yeast
Cells with plant FPN1/2-GFP expression plasmids were grown overnight in medium lacking Met. Cells were plated on 1 mg/mL Conconavalin A–coated glass slides. GFP images were captured by an Olympus BX051 epifluorescence microscope using a ×100 1.4 aperture.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or the EMBL/GenBank databases under accession numbers At2g38460 (FPN1) and At5g03570 (FPN2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Some Arabidopsis Accessions Contain a Frameshift Mutation in FPN2.
Supplemental Figure 2. fpn2-1 Roots Have Less Ferric Chelate Reductase Activity Than Wild-Type Ws.
Supplemental Figure 3. Overexpression of FPN2 Induces Iron Deficiency.
Supplemental Figure 4. Cobalt Triggers the Iron Deficiency Response.
Supplemental Figure 5. Alignment of FPN Domains from Humans and Arabidopsis.
Supplemental Figure 6. Ferroportin Alleles.
Supplemental Table 1. Fe, Co, and Ni Levels in the 10 Accessions with the Highest Levels of Co.
Supplemental Table 2. ICP-MS Data from Hydroponically Grown Plants.
Supplementary Material
Acknowledgments
We thank Marina Tikhonova and Elena Yakubova for plant cultivation and harvesting and Balasubramaniam Muthukumar for performing crosses of various Arabidopsis accessions. This research was supported by the National Institutes of Health, the National Institute of General Medicine (R01 GM78536-01A1), and the National Science Foundation Arabidopsis 2010 program (I OB 0419695).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Mary Lou Guerinot (guerinot@dartmouth.edu).
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Adrait, A., Jacquamet, L., Le Pape, L., de Peredo, A.G., Aberdam, D., Hazemann, J.L., Latour, J.M., and Michaud-Soret, I. (1999). Spectroscopic and saturation magnetization properties of the manganese- and cobalt-substituted Fur (ferric uptake regulation) protein from Escherichia coli. Biochemistry. 38 6248–6260. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Arrivault, S., Senger, T., and Kramer, U. (2006). The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 46 861–879. [DOI] [PubMed] [Google Scholar]
- Barrett, T., Troup, D.B., Wilhite, S.E., Ledoux, P., Rudnev, D., Evangelista, C., Kim, I.F., Soboleva, A., Tomashevsky, M., and Edgar, R. (2007). NCBI GEO: Mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 35 D760–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter, I., Ouzzani, M., Orcun, S., Kennedy, B., Jandhyala, S.S., and Salt, D.E. (2007). Purdue Ionomics Information Management System. An integrated functional genomics platform. Plant Physiol. 143 600–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265 1856–1860. [DOI] [PubMed] [Google Scholar]
- Borevitz, J.O., Liang, D., Plouffe, D., Chang, H.S., Zhu, T., Weigel, D., Berry, C.C., Winzeler, E., and Chory, J. (2003). Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res. 13 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Colangelo, E.P., and Guerinot, M.L. (2004). The essential bHLH protein FIT1 is required for the iron deficiency response. Plant Cell 16 3400–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, J., Wessling-Resnick, M., and Knutson, M. (2008). Hepcidin regulation of iron transport. J. Nutr. 138 2284–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin, D.S., McMaster, J.A., Culbertson, M.R., and Kung, C. (1992). COT1, a gene involved in cobalt accumulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 12 3678–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, E.L., Fett, J.P., and Guerinot, M.L. (2002). Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Domenico, I., Nemeth, E., Nelson, J., Phillips, J., Ajioka, R., Kay, M., Kushner, J., Ganz, T., Ward, D., and Kaplan, J. (2008). The hepcidin-binding site on ferroportin is evolutionarily conserved. Cell Metab. 8 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- De Domenico, I., Ward, D., Langelier, C., Vaughn, M., Nemeth, E., Sundquist, W., Ganz, T., Musci, G., and Kaplan, J. (2007). The molecular mechanism of hepcidin-mediated ferroportin down-regulation. Mol. Biol. Cell 18 2569–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny, J.R., Long, T.A., Wang, J.Y., Jung, J.W., Mace, D., Pointer, S., Barron, C., Brady, S.M., Schiefelbein, J., and Benfey, P.N. (2008). Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320 942–945. [DOI] [PubMed] [Google Scholar]
- Durrett, T.P., Gassmann, W., and Rogers, E.E. (2007). The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 144 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide, D., Broderius, M., Fett, J., and Guerinot, M.L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 93 5624–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen, S.P., Borevitz, J.O., Harmon, F.G., Pruneda-Paz, J.L., Schultz, T.F., Yanovsky, M.J., Liljegren, L.J., Ecker, J.R., and Kay, S.A. (2005). Rapid array mapping of circadian clock and developmental mutations in Arabidopsis. Plant Physiol. 138 990–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.H., Eu, Y.J., Yoo, C.M., Kim, Y.W., Pih, K.T., Jin, J.B., Kim, S.J., Stenmark, H., and Hwang, I. (2001). Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunova, Y.O., Eide, D., Clark, W.G., Guerinot, M.L., and Pakrasi, H.B. (1999). The IRT1 protein from Arabidopsis thaliana is a metal transporter with broad specificity. Plant Mol. Biol. 40 37–44. [DOI] [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahner, B., Gong, J., Mahmoudian, M., Smith, E.L., Abid, K.B., Rogers, E.E., Guerinot, M.L., Harper, J.F., Ward, J.M., McIntyre, L., Schroeder, J.I., and Salt, D.E. (2003). Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotechnol. 21 1215–1221. [DOI] [PubMed] [Google Scholar]
- Latunde-Dada, G.O., Shirali, S., McKie, A.T., Simpson, R.J., and Peters, T.J. (2004). Effect of transition metal ions (cobalt and nickel chlorides) on intestinal iron absorption. Eur. J. Clin. Invest. 34 626–630. [DOI] [PubMed] [Google Scholar]
- Li, L., Chen, O.S., Ward, D.M., and Kaplan, J. (2001). CCC1 is a transporter that mediates vacuolar iron storage in yeast. J. Biol. Chem. 276 29515–29519. [DOI] [PubMed] [Google Scholar]
- Li, M.Z., and Elledge, S.J. (2007). Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4 251–256. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler, H.K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 148 350–382. [Google Scholar]
- Liu, J., Reid, R.J., and Smith, F.A. (2000). The mechanism of cobalt toxicity in mung beans. Physiol. Plant. 110 104–110. [Google Scholar]
- MacDiarmid, C.W., Milanick, M.A., and Eide, D.J. (2003). Induction of the ZRC1 metal tolerance gene in zinc-limited yeast confers resistance to zinc shock. J. Biol. Chem. 278 15065–15072. [DOI] [PubMed] [Google Scholar]
- Marschner, H., Römheld, V., and Ossenberg-Neuhaus, H. (1982). Rapid method for measuring changes in pH and reducing processes along roots of intact plants. Z. Pflanzenphysiol. 105 407–416. [Google Scholar]
- Muckenthaler, M.U., Galy, B., and Hentze, M.W. (2008). Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu. Rev. Nutr. 28 197–213. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., Hu, T.T., Ishimo, Y., Toomajian, C., Zheng, H.G., Bakker, E., and Calabrese, P. (2005). The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3 e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranquet, C., Ollagnier de Choudens, S., Loiseau, L., Barras, F., and Fontecave, M. (2007). Cobalt stress in Escherichia coli: The effect on the iron-sulfur proteins. J. Biol. Chem. 282 30442–30451. [DOI] [PubMed] [Google Scholar]
- Robinson, N.J., Proctor, C.M., Connolly, E.L., and Guerinot, M.L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397 694–697. [DOI] [PubMed] [Google Scholar]
- Salnikow, K., Su, W., Blagosklonny, M.V., and Costa, M. (2000). Carcinogenic metals induce hypoxia-inducible factor-stimulated transcription by reactive oxygen species-independent mechanism. Cancer Res. 60 3375–3378. [PubMed] [Google Scholar]
- Schaaf, G., Honsbein, A., Meda, A.R., Kirchner, S., Wipf, D., and von Wiren, N. (2006). AtIREG2 encodes a tonoplast transport protein involved in iron-dependent nickel detoxification in Arabidopsis thaliana roots. J. Biol. Chem. 281 25532–25540. [DOI] [PubMed] [Google Scholar]
- Silverstein, K.A., Moskal, W.A.J., Wu, H.C., Underwood, B.A., Graham, M.A., Town, C.D., and VandenBosch, K.A. (2007). Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 51 262–280. [DOI] [PubMed] [Google Scholar]
- Stadler, J., and Schweyen, R. (2002). The yeast iron regulon is induced upon cobalt stress and crucial for cobalt tolerance. J. Biol. Chem. 277 39649–39654. [DOI] [PubMed] [Google Scholar]
- Thomine, S., Lelievre, F., Debarbieux, E., Schroeder, J.I., and Barbier-Brygoo, H. (2003). AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant J. 34 685–695. [DOI] [PubMed] [Google Scholar]
- Thorgersen, M.P., and Downs, D.M. (2007). Cobalt targets multiple metabolic processes in Salmonella enterica. J. Bacteriol. 189 7774–7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert, G., Grotz, N., Dedaldechamp, F., Gaymard, F., Guerinot, M.L., Briat, J.-F., and Curie, C. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and plant growth. Plant Cell 14 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, S., Baron, J., and Tephly, T.R. (1980). Identification of cobalt protoporphyrin IX formation in vivo following cobalt administration to rats. Biochem. Pharmacol. 29 2319–2323. [DOI] [PubMed] [Google Scholar]
- Yi, Y., and Guerinot, M.L. (1996). Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 10 835–844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.