Abstract
The maturation and ripening of fleshy fruits is a developmental program that synchronizes seed maturation with metabolism, rendering fruit tissues desirable to seed dispersing organisms. Through RNA interference repression, we show that Tomato AGAMOUS-LIKE1 (TAGL1), the tomato (Solanum lycopersicum) ortholog of the duplicated SHATTERPROOF (SHP) MADS box genes of Arabidopsis thaliana, is necessary for fruit ripening. Tomato plants with reduced TAGL1 mRNA produced yellow-orange fruit with reduced carotenoids and thin pericarps. These fruit are also decreased in ethylene, indicating a comprehensive inhibition of maturation mediated through reduced ACC Synthase 2 expression. Furthermore, ectopic expression of TAGL1 in tomato resulted in expansion of sepals and accumulation of lycopene, supporting the role of TAGL1 in ripening. In Arabidopsis, the duplicate SHP1 and SHP2 MADS box genes regulate the development of separation layers essential for pod shatter. Expression of TAGL1 in Arabidopsis failed to completely rescue the shp1 shp2 mutant phenotypes, indicating that TAGL1 has evolved distinct molecular functions compared with its Arabidopsis counterparts. These analyses demonstrate that TAGL1 plays an important role in regulating both fleshy fruit expansion and the ripening process that together are necessary to promote seed dispersal of fleshy fruit. From this broad perspective, SHP1/2 and TAGL1, while distinct in molecular function, regulate similar activities via their necessity for seed dispersal in Arabidopsis and tomato, respectively.
INTRODUCTION
Angiosperms have evolved many different fruit types, including fleshy berries such as those of tomato (Solanum lycopersicum) and dry and dehiscent fruits exemplified by Arabidopsis thaliana siliques. Fossil records suggest the emergence of fleshy-fruited species from progenitors bearing dry and dehiscent fruit, with examples of conversion between fruit types over evolutionary time (Knapp, 2002; Scutt et al., 2006; Seymour et al., 2008). The existence of closely related species with dry and fleshy fruits, as in the Solanaceae where tomato and pepper (Capsicum annuum) produce fleshy fruits while petunia (Petunia hybrida) and tobacco (Nicotiana tabacum) produce dry capsules, would suggest that the molecular basis of such differences are not necessarily complicated (Knapp, 2002). The association of fleshy fruit development with ripening further suggests that these processes may be related.
Tomato has served as the primary model for fleshy fruit development and ripening (reviewed in Giovannoni, 2007). Relatively few genes involved in fleshy fruit expansion have been described in tomato. Most that have defined functions are associated with cell division (Frary et al., 2000) and the cell cycle (Gonzalez et al., 2007) with no noted downstream impacts on ripening, though quantitative ripening parameters were not always measured. By contrast, fruit ripening has been studied extensively, and a number of important ripening genes have been described. These can be loosely classified as those with functions related to ethylene synthesis and response (reviewed in Barry and Giovannoni, 2007) and those that lie upstream of ethylene regulation and that in some cases have been shown to impact ripening activities beyond ethylene. The latter include the tomato MADS-RIN MADS box (Vrebalov et al., 2002; Ito et al., 2008), Colorless nonripening (Cnr) SPB box (Manning et al., 2006), and HB-1 homeobox (Lin et al., 2008) genes that are all necessary for ethylene induction and ripening in tomato. Though most ripening regulators defined to date have not been demonstrated to impact fruit fleshiness, ectopic expression of homologous and heterologous genes encoding MADS box and HD-ZIP transcription factors in tomato has resulted in fleshy sepal development and pigmentation suggestive of ripening (e.g., lycopene accumulation) in these altered floral organs (Pnueli et al., 1994b; Lin et al., 2008; Tadiello et al., 2009).
MADS box genes have duplicated and diversified extensively in the angiosperms (Becker et al., 2000), and this may have played a central role in the evolution of the great variety of fruit types. In Arabidopsis, several MADS box transcription factors are necessary for different aspects of fruit development, including SHATTERPROOF1 (SHP1) and SHP2, which act redundantly to specify valve margin identity and the dehiscence zones in the fruit (Liljegren et al., 2000). Lignification of valve margin cells and the innermost valve cell layer and subsequent drying of the fruit creates tension that causes fruit shattering (Spence et al., 1996). SHP1 and SHP2 are expressed in the valve margin, and shp1 shp2 double mutants fail to shatter because lignified valve margin layers do not differentiate (Savidge et al., 1995; Flanagan et al., 1996; Liljegren et al., 2000). This appears to be due, at least in part, to a failure in appropriate transcriptional upregulation of the expression of two basic helix-loop-helix genes INDEHISCENT (IND) and ALCATRAZ (ALC) that are also required for fruit dehiscence (Rajani and Sundaresan, 2001; Liljegren et al., 2004).
We have previously mined tomato microarray and digital expression profiling data to identify genes associated with ripening and describe here the functional characterization of the TOMATO AGAMOUS-LIKE (TAGL1) gene and show that it has a role in regulating the ripening process. We specifically targeted TAGL1 because our prior microarray data (available at http://ted.bti.cornell.edu/) showed this gene to be upregulated during both early fruit development and ripening but influenced little by ethylene, suggesting it may represent a regulator prior to the ethylene response. TAGL1 is expressed in ovules, developing carpels, and in the pericarp of developing fruits (Busi et al., 2003; Hileman et al., 2006). Based on phylogenetic analyses, we demonstrate that TAGL1 is orthologous to the duplicate Arabidopsis SHP1 and SHP2 genes. RNA interference (RNAi) repression of TAGL1 in tomato resulted in ripening inhibition and reduced pericarp thickness, suggesting a molecular bridge linking fleshy pericarp development and fruit ripening. Furthermore, overexpression of TAGL1 in tomato fruit induced a ripening-like phenotype in sepals, including the accumulation of carotenoids and increased fleshiness. We also performed heterologous transformation studies to determine the extent to which TAGL1 can replace SHP1/SHP2 function in Arabidopsis. Our results indicate that TAGL1 is not functionally equivalent to SHP1/SHP2 at the molecular level, which in turn suggests that these transcription factors have evolved to regulate different suites of target promoters required for differentiation of distinct fruit types. However, in terms of necessity for normal seed dispersal, the Arabidopsis SHP1/2 and tomato TAGL1 genes retain surprisingly similar roles in plant development.
RESULTS
TAGL1 Is Expressed in Floral Organs, Young Fruit, and during Ripening
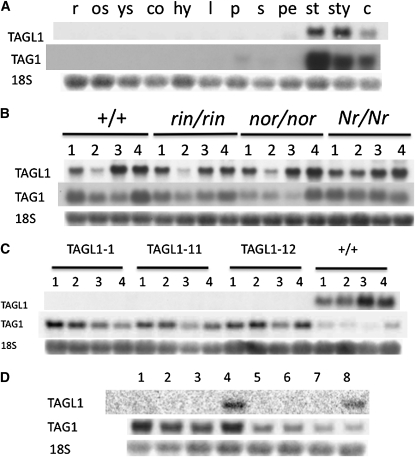
A full-length 1094-bp TAGL1 cDNA cloned into pBluescript was recovered as clone cLEG9L5 from the public tomato cDNA collection (Van der Hoeven et al., 2002; www.sgn.cornell.edu). We termed this clone pBS_SlTAGL1. A DNA sequence corresponding to bases 594 to 1017 of the EST sequence representing the MADS box C domain and 3′ untranslated region (UTR) sequences was shown to be gene specific for TAGL1 via DNA gel blot analysis (see Supplemental Figure 1 online) and was subsequently used for RNA gel blot analysis across a spectrum of tomato tissues, including normal and ripening-impaired mutant fruit. Expression was not detected in roots, seedlings, shoots, leaves, pedicels, or sepals of anthesis stage cv Ailsa Craig flowers but was observed at low levels in petals and with substantial induction in reproductive structures (style, stamen, and carpel) (Figure 1A). This expression is consistent with prior reports (Busi et al., 2003; Hileman et al., 2006) and parallels the expression in these tissues of the related MADS box gene TAG1, which confers AGAMOUS (AG) organ identity function in tomato (Figure 1A; Pnueli et al., 1994b). Maturing fruit show detectable TAGL1 expression in the immature (IM; 28 d postanthesis [DPA]) and mature green stages (MG; 32 DPA, full fruit expansion and mature seeds but preripening), repression in mature green fruit treated with ethylene (MGE), and induction at breaker (BR; initial ripening) and breaker + 7 d red ripe fruit (Figures 1B and 1C). TAGL1 expression was additionally observed in pericarp tissue of 4, 8, and 18 DPA fruit, suggesting continued expression from anthesis throughout carpel development (see Supplemental Figure 2 online).
Figure 1.
Expression of TAGL1 and TAG1 in Tomato Fruit Tissues and in Response to Ethylene.
(A) Total RNA gel blot analysis of TAGL1, TAG1, and 18S rRNA (control). r, roots from 9-d-old seedlings, os, primary stem of 6-week-old plants; ys, primary stem of 3-week-old plants; co, cotyledons of 9-d-old seedlings; h, hypocotyls of 9-d-old seedlings; l, leaves of 6-week-old plants; p, anthesis-stage floral pedicel; s, anthesis sepals; pe, anthesis petals; st, anthesis stamens; sty, anthesis style/stigma; c, anthesis carpels.
(B) Wild-type, rin, nor, and Nr fruit RNAs: 1, MG (mature green); 2, MGE (MG + 10 ppm ethylene for 12 h); 3, BR (breaker; i.e., early ripening); 4, BR +7d (red ripe in the wild type).
(C) RNAs from independently transformed TAGL1 RNAi transgenic lines are designated transgenic lines TAGL1-1, TAGL1-11, and TAGL1-12, respectively. Fruit RNAs: 1, IM (28 DPA); 2, MG; 3, BR; 4, BR +7d (red ripe in the wild type).
(D) Lanes 1 to 4 are combined anthesis-stage stamens and style/stigma from TAGL1-1, 11, 12, and the wild type, respectively. Lanes 5 to 8 are anthesis-stage carpels from TAGL1-1, 11, 12, and the wild type, respectively.
TAGL1 expression in fruit was not appreciably impacted by the single locus rin, nor, or Nr ripening mutations except that it was not repressed in ethylene-treated MGE Nr fruit (Figure 1B), which are insensitive to ethylene (Lanahan et al., 1994). The fact that TAGL1 mRNA accumulation is the same in normal (ethylene producing) and nonripening rin and nor fruit (which do not produce elevated ethylene) indicates that TAGL1 expression is not induced by ethylene during ripening and the observed repression in our ethylene-treated MG fruit is likely transient. To test this hypothesis, we treated wild-type IM, MG, and BR fruit with exogenous ethylene for 12 h and observed TAGL1 downregulation at all three stages, including the ethylene-producing BR fruit (see Supplemental Figure 2 online). This result suggests TAGL1 in fact does respond transiently to increasing ethylene concentrations.
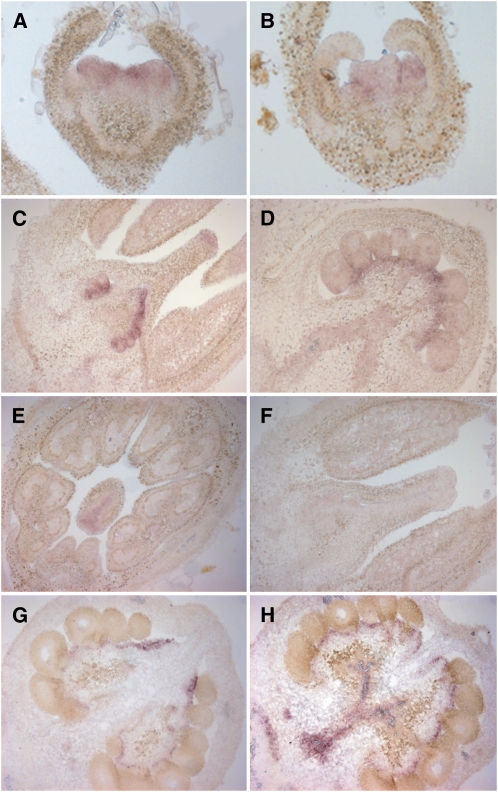
In situ hybridizations demonstrated that TAGL1 expression was predominantly in stamen and carpel primordia at stages 2 and 5 (Figures 2A and 2B) (flower stages according to Brukhin et al., 2003). By stage 9, expression in the stamens was reduced, but expression persisted in carpels, particularly in ovules, placenta, stigmas, and in the transmitting tract compared with a sense control (Figures 2C to 2F). At 0 to 3 DPA, expression was strongest in the placenta and near the vascular bundles with weak expression throughout other fruit tissues (Figures 2G and 2H). These observations differ somewhat from those of Busi et al. (2003), who reported high levels of expression in the tapetal tissues of the stamen at approximately stage 10, in addition to expression in the developing ovules. This reported high level of expression in stamens may reflect cross-hybridization of their probe to other MADS box genes, such as TAG1 (Busi et al., 2003).
Figure 2.
In Situ Expression Analysis of TAGL1 in Wild-Type Floral Buds and Fruit.
(A) TAGL1 expression is first detected in stage 2 to 3 floral buds in presumptive stamen and carpel primordia.
(B) By stage 5, TAGL1 is expressed in stamen and carpel primordia.
(C) At stage 9, expression in stamens is reduced, with TAGL1 expression seen in developing ovules and stigmas of carpels.
(D) Stage 12 to 13 carpel with TAGL1 expression detected in the placenta and weakly in ovules.
(E) Cross section of a flower at stage 9. TAGL1 is expressed in the styles of the carpel and weakly in stamens.
(F) Sense control on stage 9 floral bud.
(G) and (H) In 0 DPA fruit, TAGL1 is expressed in the placenta, areas around vascular bundles, and weakly in the pericarp.
TAGL1 Is the Ortholog of SHP
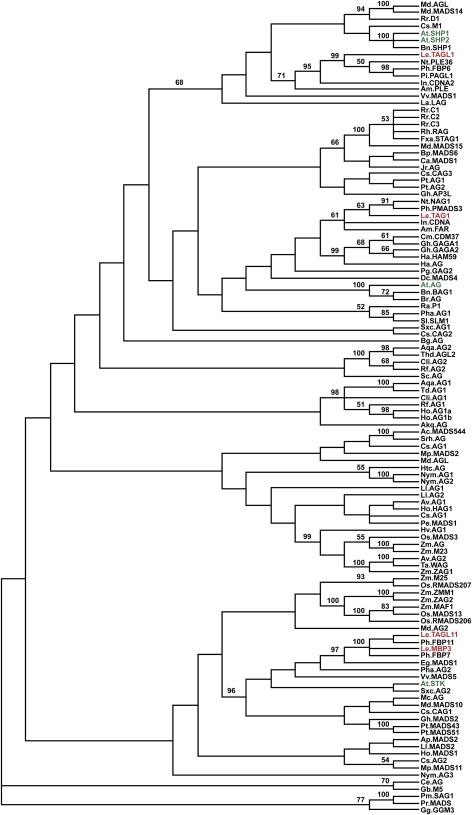
Previous analyses of the large AG clade of MADS box genes have indicated that, prior to the divergence of the rosids and asterids, a duplication event gave rise to the euAG and PLENA (PLE) lineages, which include the Arabidopsis AG and SHP1/2 genes, respectively (Kramer et al., 2004; Zahn et al., 2006). These studies also indicate that the SHP1/SHP2 duplication occurred recently, within the rosids. However, previous analyses either did not include TAGL1 (Kramer et al., 2004) or included relatively few taxa (Hileman et al., 2006; Leseberg et al., 2008), leading in some cases to an equivocal placement of TAGL1 in the AG clade phylogeny. Thus, the relationship of TAGL1 to other AG lineage genes has been unclear, although several recent analyses have suggested that TAGL1 is orthologous to the SHP genes (Zahn et al., 2006; Leseberg et al., 2008).
To define more discretely the orthology of TAGL1, we performed a phylogenetic analysis using maximum parsimony of 144 AG-related sequences from 62 species (Figure 3; see Supplemental Table 1 online). This analysis indicates that of the four identified tomato AG-related genes (Hileman et al., 2006), TAGL1 resides in the well-supported PLE clade, with the most closely related Arabidopsis genes being SHP1/2 (Figure 3). We also identified PLE and euAG clades of genes that are consistent with those defined previously (Zahn et al., 2006), supporting the orthologous relationship of tomato TAGL1 to the Arabidopsis SHP genes.
Figure 3.
Parsimony Analysis of AG Clade MADS Box Proteins.
Consensus tree with bootstrap values greater than or equal to 50% is shown. Tomato proteins are indicated in red and Arabidopsis proteins in green. Gymnosperm sequences were used as outgroups in the analysis. Species names are abbreviated as follows; accession numbers can be found in Supplemental Table 1 online. Asterids: Am, Antirrhinum majus; Ph, Petunia hybrida; Pi, Petunia inflata; Nt, Nicotiana tabacum; Le, Solanum lycopersicum; Dc, Daucus carota; Pg, Panax ginseng; Gh, Gerbera hybrida; Ha, Helianthus annus; Cm, Chrysanthemum × morifolium; In, Ipomoea nil; Eg, Eustoma grandiflorum. Rosids: At, Arabidopsis thaliana; Bn, Brassica napus; Br, Brassica rapa; Cs, Cucumis sativus; Rr, Rosa rugosa; Md, Malus domestica; Jr, Juglans regia; Ca, Corylus pendula; Bp, Betula pendula; Gh, Gossypium hirsutum; Fa, Fragaria × ananassa; Pt, Populus trichocarpa; Mc, Momordica charantia. Vitaceae: Vv, Vitis vinifera. Caryophyllales: Sl, Silene latifolia; Ra, Rumex acetosa; Pha, Phytolacca americana. Saxifragales: Sxc, Saxifraga caryana; La, Liquidamber styraciflua. Sabiaceae: Md, Meliosma dilleniifolia (AG1 and AG2). Ranunculales: Rf, Ranunculales ficaria; Ho, Helleborus orientalis; Cli, Clematis integrifolia; Aqa, Aquilegia alpina; Thd, Thalictrum dioicum; Bg, Berberis gilgiana; Akq, Akebia quinata; Sc, Sanguinaria canadensis. Magnoliales: Mp, Magnolia precossimina. Piperales: Srh, Saruma henryii; Htc, Houttuynia cordata; Ac, Asarum caudigerum. Chloranthaceae: Cs, Chloranthus spicatus (AG1). Liliales: Ll, Lilium longiflorum. Asparagales: Pe, Phalaenopsis equestris; Ho, Hyacinthus orientalis; Ap, Agapanthus praecox; Av, Asparagus virgatus; Cs, Crocus sativus. Poales: Hv, Hordeum vulgare; Ta, Triticum aestivum; Os, Oryza sativa; Zm, Zea mays. Nymphaeales: Nym, Nymphaea sp. Gingkgoales: Gb, Ginkgo biloba. Cycadales: Ce, Cycas edentata. Gnetales: Gg, Gnetum gnemon. Coniferales: Pm, Picea mariana; Pr, Pinus resinosa.
TAGL1 Impacts Fruit Development and Is Necessary for Ripening
To gain insight into TAGL1 function, we created an RNAi construct targeting the C domain and 3′ UTR of TAGL1 in pHELLSGATE2 and transformed wild-type (cv Ailsa Craig) tomato plants via Agrobacterium tumefaciens–mediated T-DNA transfer. Nine independent transgenic RNAi lines confirmed for transgene integration were recovered and the mature fruit of all developed to an orange-yellow color. Three lines (TAGL1-1, -11, and -12) were selected for transgene homozygosity in the T1 generation based on DNA gel blot analysis and phenotypically characterized in the T2 generation. To verify specific repression of TAGL1, total RNA was extracted from IM, MG, BR, and red ripe (BR+7d) stage wild-type and transgenic fruit and hybridized to TAGL1- and TAG1-specific probes. TAG1 was previously shown to be necessary for tomato carpel development (Pnueli et al., 1994b). TAG1 expression was also monitored because it is the most closely related tomato gene to TAGL1 (Figure 3). Tomato gene sequences more closely related to TAGL1 than TAG1 could not be found within the current 40% complete tomato genome sequence (Mueller, 2009) or the over 300,000 tomato ESTs in GenBank nor via low stringency hybridization to over 50,000 ordered cDNA clones from 0 to 7 DPA carpels and ripening fruit (using the TAGL1 RNAi sequence as probe). We have recently generated ∼500,000 EST reads via 454 sequencing from mature green and red ripe Ailsa Craig fruit and using the RNAi construct sequence as a query sequence could find no gene more closely related to TAGL1 than TAG1 in this sequence collection. This same probe sequence gave single copy hybridization in DNA gel blot analysis (see Supplemental Figure 1 online). While not conclusive without the complete tomato genome sequence, together these results strongly suggest that TAG1 is the most closely related gene to TAGL1 and, thus, the best barometer for TAGL1 RNAi specificity.
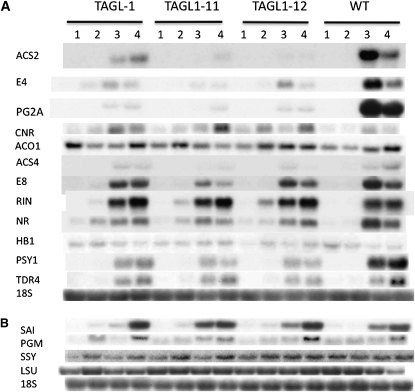
TAGL1 mRNA was detected in the wild type but greatly reduced in transgenic fruit, while TAG1 mRNA was markedly increased in transgenic fruit, suggesting the presence of a regulatory relationship between these genes (Figure 1C). This result also indicated that TAG1 mRNA was not targeted by the TAGL1 RNAi transgene RNA. To ascertain whether or not this relationship is specific to mature fruit tissues, we examined expression in anthesis stage styles, stamen, and carpels. Again, clear repression of TAGL1 was observed in RNAi lines, but little if any impact on TAG1 mRNA accumulation was observed in these tissues (Figure 1D). Together, these results suggest specificity of the TAGL1 RNAi construct for the target gene and the possibility of a regulatory network that induces TAG1 in the mature fruit but not the organs (including carpels) of anthesis stage flowers.
Obvious visual phenotypes of TAGL1 downregulated lines are shown in Figure 4 and include yellow-orange mature fruit, reduced pericarp thickness, and a lack of stylar trichomes. Fruit expansion in tomato can be divided into two general phases. The initial phase of expansion occurs during approximately the first week after pollination and reflects a period of active cell division. The second phase is completed by approximately the MG stage and is characterized by extensive cell expansion and very little cell division (reviewed in Giovannoni, 2004). In tomato, fruit ripening commences after fruit expansion and seed maturation. The fact that TAGL1 impacts both pericarp thickness and ripening phenotypes suggests that this gene has roles both in preripening fruit development impacting fleshiness and in later maturation.
Figure 4.
TAGL1 Repression Phenotypes.
Genotypes are TAGL1 RNAi lines (TAGL-1, 11, and 12) and wild-type Ailsa Craig (AC+/+). TAGL1 RNAi repression alters pericarp (fruit outer wall) thickness, ripening-related fruit pigmentation (A), and style trichome density (B).
TAGL1 Influences Broad Ripening Phenotypes
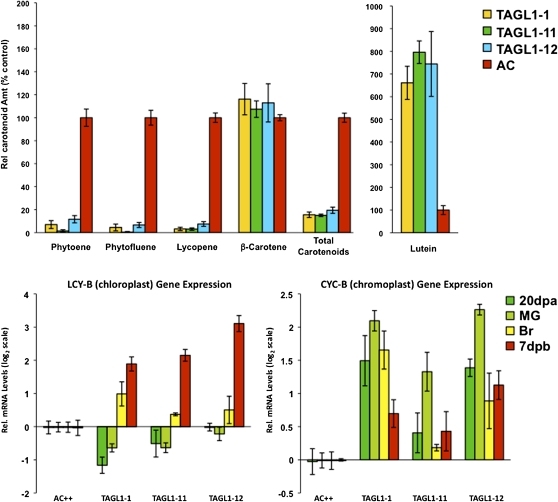
Ripening represents coordinated modification of numerous biochemical pathways associated with pigmentation, cell wall metabolism, pathogen susceptibility, nutrient content, flavor, and aroma. It was therefore important to determine whether TAGL1 repression specifically altered fruit pigmentation or a more comprehensive set of ripening phenomena. The red pigmentation of ripe tomatoes is due to lycopene that accounts for 70 to 90% of the carotenoids in most varieties, while β-carotene accounts for the bulk of the remainder (Burns et al., 2003; Alba et al., 2005). HPLC analyses of mature wild-type and transgenic fruit indicated a dramatic reduction in most carotenoids and a reduction of ∼80% in total carotenoids for the TAGL1 reduced fruit (Figure 5A). Lycopene content was reduced >90% in TAGL1 RNAi lines with the majority of the remaining carotenoid accumulated as β-carotene (and to concentrations typical of wild-type fruit). Lutein is yellow and accounts for a minor fraction of normal ripe tomato carotenoids, though its relative concentration increased six- to eightfold in the transgenics and likely contributes to the coloration of TAGL1 repressed fruit.
Figure 5.
Carotenoid Accumulation Profiles in TAGL1 RNAi Fruit Result in Part from Altered Lycopene-β-Cyclase Expression.
(A) HPLC analysis of carotenoid accumulation in BR+7 (Breaker plus 7 d) fruit of transgenic TAGL1 RNAi lines presented as percent of control (cv Ailsa Craig [AC]). Standard error is indicated for a minimum of six fruit per sample.
(B) Relative quantitative RT-PCR expression analysis of the chloroplast (LYC-B) and chromoplast (CYC-B) lycopene-β-cyclase genes during fruit development show both are upregulated in ripening stage TAGL1 RNAi fruit, accounting for the metabolism of lycopene to β-carotene and lutein.
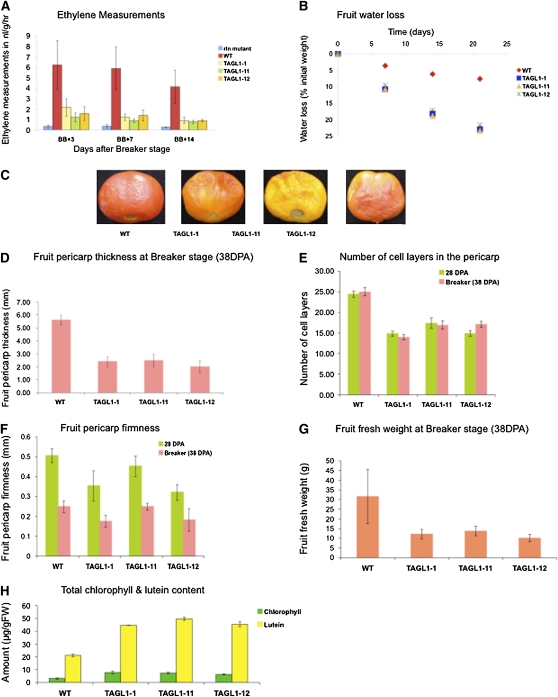
Ethylene is a regulator of carotenoid accumulation during ripening and specifically through upregulation of phytoene synthase (PSY1; Maunders et al., 1987). Ethylene is necessary for many additional ripening activities, and to date relatively few upstream regulators of autocatalytic ethylene synthesis have been defined. One such regulator is encoded by HB1, an HD ZIP homeobox protein shown to positively interact with the ACO1 (ACC OXIDASE1) promoter to foster increased ethylene synthesis and whose repression delays ripening (Lin et al., 2008). Another is the MADS-RIN transcription factor, which is a member of the SEPALLATA (SEP) clade and thus quite distinct from TAGL1 (Hileman et al., 2006). The homozygous rin mutation inhibits ripening and results in green-yellow fruit that produce only basal levels of ethylene (Vrebalov et al., 2002). Measurement of ethylene production from wild-type, nearly isogenic rin/rin, and transgenic lines indicated that TAGL1 repressed fruit were dramatically reduced in ethylene production in a manner much more similar to mutant rin fruit than the wild type (Figure 6A). Characterization of the expression of ethylene biosynthesis genes indicated that ACC SYNTHASE2 (ACS2) mRNA was substantially repressed, ACS4 only slightly repressed if at all in TAGL1 RNAi lines, while ACO1 mRNA accumulation was not altered (Figure 7A). Furthermore, mRNA levels of HB1 were not altered in TAGL1 repressed lines nor were those of MADS-RIN. Expression of COLORLESS NON-RIPENING (CNR), encoding a SQUAMOSA promoter binding protein also known to be necessary for ripening (Manning et al., 2006), varied through development in transgenic lines but was not greatly repressed or induced at any particular fruit development stage, indicating that TAGL1 does not function during ripening by altering expression of these known ripening regulators (Figure 7A). Furthermore, these results indicate that the ethylene reduction in TAGL1 RNAi lines results predominantly through downregulation of ACS2. Whether or not this reflects direct interaction of TAGL1 with the ACS2 promoter remains to be determined. In agreement with reduced ethylene levels in the transgenic fruit, there was reduced accumulation of a number of ethylene-regulated gene mRNAs, including those encoding E4, polygalacturonase (PG), E8, and the carotenoid synthesis enzyme PSY1. PSY1 is a key regulator of flux through the carotenoid pathway (Bird et al., 1991) and its repression is consistent with the reduction of carotenoids observed in TAGL1 repressed lines.
Figure 6.
Physiological, Morphological, and Metabolic Characterization of TAGL1 RNAi Fruit.
(A) Ethylene production (nl/g/h) of transgenic and control fruit at the indicated days after breaker. Standard error is indicated, and a minimum of six fruit per sample were analyzed.
(B) Water loss of transgenic and control fruit over 21 d starting from breaker stage expressed as percentage of original weight.
(C) Ailsa Craig wild-type and TAGL1 RNAi fruit at 30 d after breaker.
(D) Pericarp thickness of breaker (38 DPA) fruit.
(E) Average number of cell layers in immature (28 DPA) and breaker (38 DPA) pericarp.
(F) Pericarp firmness as measured by compression in immature (28 DPA) and breaker (38 DPA) fruit.
(G) Breaker stage fruit fresh weight.
(H) HPLC of immature (28 DPA) fruit total chlorophyll (Total Chl) and lutein concentrations. FW, fresh weight.
Standard error is indicated for a minimum of six fruit per sample in (A) and (H), 10 fruit per sample in (B) and (G), and with a minimum of three fruit per sample in (D) to (F) (though multiple measurements were made on each sample in [D] to [F]. See Methods for details).
Figure 7.
Ripening and Starch Metabolism Gene Expression in TAGL1 RNAi and Control Fruit.
Total fruit RNA was analyzed via gel blot analysis from the indicated genotypes and the following fruit stages: 1, IM (28 DPA); 2, MG; 3, BR; 4, BR + 7d (red ripe in the wild type). Hybridization probes were derived from gene-specific sequences using primers described in Supplemental Table 2 online. Full names and accession numbers for all genes are also listed in Methods.
(A) Ripening-associated genes.
(B) Starch metabolism genes.
The accumulation of TAGL1 mRNA in other Ailsa Craig floral organs suggested the possibility of additional functions in floral development. Stylar trichome development was clearly altered in the TAGL1 repressed lines, though no additional changes in floral organ development or morphology were noted beyond those observed in the carpels. Carotenoid and chlorophyll analyses were also performed on petals, anthers, sepals, and leaves. Leaves and stamens showed no changes in pigment profiles even though the latter accumulate TAGL1 mRNA (Figure 1C). Petals showed altered carotenoid accumulation (see Supplemental Figure 3 online) consistent with the observed expression of TAGL1 in this organ (Figure 1A). While anthesis stage Ailsa Craig sepals showed no detectable TAGL1 mRNA (Figure 1A), they did have altered carotenoids (see Supplemental Figure 3 online), indicating expression either at low levels or at a stage preceding anthesis. Tomato sepals accumulate primarily β-carotene and lutein (similar to leaves) and TAGL1 repression resulted in normal ratios but total elevation of these compounds similar to that observed in immature TAGL1 repressed fruit (Figure 6H). These results suggest TAGL1 influences carotenoid pathway activity primarily in floral organs.
TAGL1 Is Necessary for Fleshy Fruit Development Prior to Ripening
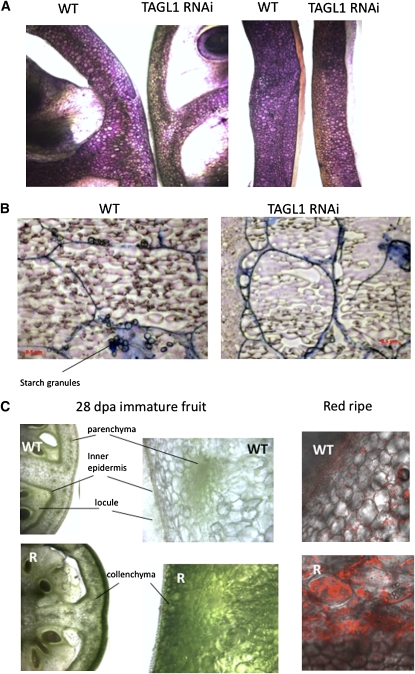
The expression of TAGL1 in styles and carpel tissue through development suggested additional functions prior to ripening. The clear lack of trichomes on the styles of transgenic lines (Figure 4B) represents a function related to normal development of styles in tomato, though this was not studied further. Fruit pericarp thickness was notably reduced in mature TAGL1 RNAi lines (Figure 4A), and this phenotype was also observed in immature fruit (Figure 8A). Indeed, measurement of pericarp thickness at BR stage indicated a reduction in pericarp thickness >50% (Figure 6D) along with a reduction in the number of pericarp cell layers from ∼25 to 15 (Figure 6E). Reduced pericarp thickness correlated with reduced firmness in immature fruit, though as the fruit matured (and ripened in the wild type), softening between control and transgenic lines became indistinguishable (Figure 6F). Reduced pericarp thickness may have also contributed to more rapid dehydration of transgenic fruit than wild-type controls (Figures 6B and 6C). Microscopic examination of the fruit epidermis did not reveal any obvious differences between TAGL1 and control tissues (Figure 8A). To gain insight into the nature of the missing cell layers, we stained BR stage pericarp sections with Nile blue to identify starch granules characteristic of the lower parenchyma cells of the tomato pericarp. While starch-containing cells were observed in control sections, they were absent from the TAGL1 RNAi fruit, suggesting that lower parenchyma cell layers are absent in these lines (Figure 8B). Interestingly, RNA gel blot analysis of a number of available starch metabolism and photosynthesis-related genes did not indicate any notable differences in mRNA accumulation of the wild type versus transgenic pericarp tissues (Figure 7B).
Figure 8.
TAGL1 RNAi Fruit Are Altered in Pericarp Thickness and Starch Accumulation.
Representative section types and stains of the indicated fruit stage and genotype are shown.
(A) Toluidine blue–stained hand-sections for cell counting of wild-type (Ailsa Craig) and TAGL1-1 (TAGL1 RNAi) fruit. The left pair is 28 DPA immature fruit and those on the right are red ripe stage (breaker + 7 d).
(B) Nile blue A stain for starch granules in cryosectioned wild-type (Ailsa Craig) and TAGL1-1 (TAGL1 RNAi) breaker stage fruit.
(C) Optical (left and center) and confocal (right) microscopy of cryosectioned wild-type and TAGL1-1 (R) 28 DPA immature fruit. Confocal microscopy employed chloroplast autofluorescence.
The reduction in pericarp cell layers of TAGL1 RNAi fruit indicates an early role in fruit development, likely manifested during the first week after anthesis when pericarp cell division is the major contributor to fruit expansion. Additional phenotypes were observed later in development, yet prior to ripening, suggesting additional activities during fruit development. For example, transgenic fruit had reduced mass compared with controls by the time they reached BR (Figure 6G) and were darker green when immature (Figure 4A). This later trait is likely due to differential chloroplast distribution and abundance. Specifically, in normal Ailsa Craig immature fruit, chloroplasts are most abundant in the parenchyma cells, near and in the inner epidermis and in the locules. In the TAGL1 RNAi pericarp, plastids were uncharacteristically abundant in the collenchyma cells as shown by optical and confocal microscopy (Figure 8C). The fact that the plastid-rich collenchyma cells are just below the epidermis plausibly contributes to the dark green appearance of immature transgenic fruit. To determine whether or not this phenotype represented a change in total chlorophyll content in addition to altered plastid localization, we measured both chlorophyll and lutein (the predominant carotenoid in immature green tomato fruit) and observed that both compounds were elevated two- to threefold in transgenic lines (Figure 6H). TAGL1 thus influences chloroplast distribution, levels of photosynthetic pigments, and amyloplast-localized starch granule accumulation in immature tomato fruit, in addition to pericarp thickness. Whether or not there is a causal relationship between these phenotypes versus effects of different TAGL1 activities in different cell types could not be assessed in our transgenic system.
Ectopic Expression of TAGL1 Induces Ripening in Sepals
To validate further the role of TAGL1 in regulating fruit ripening, we produced transgenic tomato lines that ectopically expressed TAGL1. We cloned the full-length TAGL1 coding region under the control of the strong constitutive 35S promoter (Benfey and Chua, 1990). Four independent transgenic 35S:TAGL1 lines were generated in the Microtom cultivar and verified for transgene integration; all showed ectopic expression of TAGL1 in leaves and sepals throughout development (see Supplemental Figure 4 online).
All four lines showed dramatic visual phenotypes in flowers and later stage fruits (Figure 9). By stage 7, 35S:TAGL1 flowers show defects in sepal development, with lighter green sepals compared with the equivalent tissues in wild-type plants (Figure 9G). By stage 16, the 35S:TAGL1 sepals fail to open, almost fully enclosing the inner organs; they become fleshier and turn very light green (Figure 9H). When wild-type flowers reach anthesis, 35S:TAGL1 flowers fail to open and sepals continue to swell (Figure 9F). By ∼30 DPA, when both control cv Microtom and 35S:TAGL1 lines have green fruit, the transgenic sepals have a similar color with some dark-green vertical stripes (Figure 9I). At 36 DPA, when control and 35S:TAGL1 lines start to produce breaker fruit, the 35S:TAGL1 sepals also begin to change color, eventually turning to an orange-red or red color (Figure 9J). We also observed varying degrees of transformation of petals into stamenoid structures. In some flowers, the petals are virtually normal, while in other flowers, particularly older flowers, the petals are transformed almost completely into stamens that produce pollen (Figure 9L). Furthermore, the 35S:TAGL1 lines produced pollen but failed to produce any seeds. Crosses of transgenic pollen with wild-type pistils also failed to produce seeds, indicating that overexpression of TAGL1 disrupted normal pollen development.
Figure 9.
Overexpression of TAGL1 Results in Ripening Sepals and Conversion of Petals to Stamens.
(A) to (J) Buds, flowers, and fruit from wild-type ([A] to [E]) and comparably staged 35S:TAGL1 ([F] to [J]) plants. Floral stages are anthesis stage flowers ([A] and [F]), immature inflorescence floral stages 7 to 8 ([B] and [G]), mature inflorescence floral stages 15 to 16 ([C] and [H]), 30 DPA ([D] and [I]), and red ripe ([E] and [J]). Note the following observations.
(F) 35S:TAGL1 flowers fail to open at anthesis.
(G) 35S:TAGL1 buds at stages 7 to 8 have lighter colored sepals than comparably staged wild-type buds (B).
(H) 35S:TAGL1 buds at stages 15 to 16 have sepals that almost fully enclose the inner organs. The sepals show swelling and turn very light green.
(I) 35S:TAGL1 fruit at 30 DPA have mutant sepals that resemble green fruit (D). The mutant sepals also have dark green vertical stripes.
(J) 35S:TAGL1 mature red fruit have sepals that swell and turn red.
(K) Sepal, petal, stamen, and carpel (from left to right) of control flower at anthesis.
(L) Sepal, petal, stamen, and carpel (from left to right) of 35S:TAGL1 flower at anthesis. The sepal is already beginning to turn lighter green. The petal is partially transformed into a stamen. The stamen and carpel both appear normal.
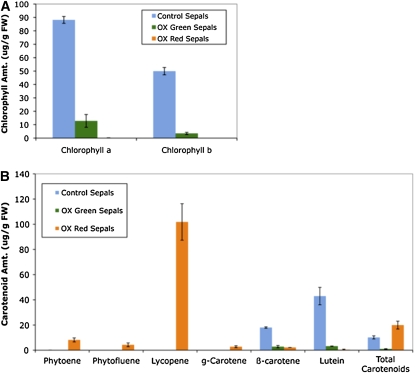
Carotenoid levels were assessed in mature green (30 DPA) and ripe red (45 DPA) sepals dissected from the 35S:TAGL1 lines compared with control cv Microtom mature green sepals at 45 DPA (Figure 10). In comparing 35S:TAGL1 45 DPA sepals to control sepals of the same stage, it is evident that the 35S:TAGL1 sepals accumulated considerably higher amounts of lycopene, g-carotene, phytofluene, and phytoene. In addition, chlorophyll, β-carotene, and lutein levels fall during 35S:TAGL1 sepal ripening, in a manner analogous to that of wild-type fruit.
Figure 10.
Chlorophyll and Carotenoid Levels in 35S:TAGL1 Overexpression Lines.
Chlorophyll (A) and carotenoid (B) levels were analyzed in control (45 DPA) and 35S:TAGL1 green sepals (36 DPA) and red sepals (45 DPA) by HPLC using mean HPLC peak areas (n = 2). Error bars represent se.
A somewhat similar phenotype has been reported for overexpression of TAG1. In 35S:TAG1 transgenic plants, sepals swell, degrade chlorophyll, and express carotenoid biosynthetic genes, but the sepals do not turn red (Pnueli et al., 1994b; Ishida et al., 1998; Bartley and Ishida, 2003). To address whether the sepal ripening phenotype we observed in 35S:TAGL1 plants was at least in part a consequence of inducing expression of TAG1, we examined TAG1 expression levels in 35S:TAGL1 plants (see Supplemental Figure 4 online). We did not observe overexpression of TAG1 in any of the 35S:TAGL1 lines, consistent with our earlier observations (Figure 1C) of the regulatory relationships between TAGL1 and TAG1.
TAGL1 Does Not Completely Rescue Arabidopsis shp Mutants
To examine the extent to which TAGL1 function has been conserved among core eudicots despite considerable diversity in fruit forms, we tested the ability of TAGL1 to rescue shattering in Arabidopsis fruit as well as to produce ectopic phenotypes. We generated a 35S:TAGL1 construct and transformed this into homozygous shp1 shp2 double mutant Arabidopsis plants. shp1 shp2 mutants show defects in siliques, such that the double mutant fruits fail to form dehiscence zones between valves and replums, resulting in a lack of shattering (Liljegren et al., 2000). As the ability of SHP1 or SHP2 transgenes to rescue the shp1 shp2 mutant phenotype has not been reported, we also generated 35S:SHP1 and 35S:SHP2 constructs and transformed these individually into shp1 shp2 double mutants as controls. We produced six 35S:SHP1 lines, eight 35S:SHP2 lines, and eight 35S:TAGL1 lines all in the shp1 shp2 background (Table 1).
Table 1.
Quantification of 35S:TAGL1, 35S:SHP1, and 35S:SHP2 Phenotypes in Transgenic Arabidopsis Plants
| Curly Leaves | Sepals | Petals | No. of Flowers Counted | Transcript Levelsa | Shattering (n)b | |||
|---|---|---|---|---|---|---|---|---|
| Normal | Carpelloid | Normal | Small | |||||
| Wild type | − | 204 | 0 | 204 | 0 | 51 | 0.00 | 60 (60) |
| shp1 shp2 | − | 236 | 0 | 236 | 0 | 59 | 0.00 | 0 (60) |
| 35S:TAGL1-1 | − | 176 | 0 | 176 | 0 | 44 | 0.04 | 0 (50) |
| 35S:TAGL1-2 | − | 164 | 0 | 164 | 0 | 41 | 0.39 | 0 (47) |
| 35S:TAGL1-3 | + | 24 | 28 | 24 | 28 | 13 | 0.63 | 0 (45) |
| 35S:TAGL1-7 | + | 48 | 48 | 12 | 84 | 24 | 0.82 | 0 (40) |
| 35S:TAGL1-8 | + | 132 | 48 | 80 | 100 | 45 | 0.83 | 1 (15) |
| 35S:TAGL1-9 | + | 120 | 0 | 64 | 56 | 30 | 0.50 | 0 (50) |
| 35S:TAGL1-11 | − | 184 | 0 | 184 | 0 | 46 | 0.05 | 0 (50) |
| 35S:TAGL1-12 | − | 200 | 0 | 200 | 0 | 50 | 0.03 | 6 (42) |
| 35S:SHP1-1 | + | 0 | 20 | 0 | 20 | 5 | 1.29 | Reduced siliques |
| 35S:SHP1-4 | + | 0 | 28 | 0 | 28 | 7 | 0.90 | 8 (53) |
| 35S:SHP1-5 | + | 0 | 120 | 0 | 120 | 30 | 1.64 | 24 (57) |
| 35S:SHP1-7 | − | 116 | 0 | 116 | 0 | 29 | 0.46 | 3 (53) |
| 35S:SHP1-8 | + | 0 | 24 | 0 | 24 | 6 | 0.53 | 0 (25) |
| 35S:SHP1-11 | − | 80 | 0 | 80 | 0 | 20 | 0.84 | 3 (40) |
| 35S:SHP2 -1 | − | 148 | 0 | 148 | 0 | 37 | 0.98 | 0 (30) |
| 35S:SHP2 -2 | − | 188 | 0 | 188 | 0 | 47 | 1.45 | 0 (40) |
| 35S:SHP2 -3 | + | 0 | 36 | 0 | 36 | 9 | 0.86 | Reduced siliques |
| 35S:SHP2 -4 | + | 0 | 296 | 0 | 296 | 74 | 1.28 | 5 (58) |
| 35S:SHP2 -7 | + | 0 | 28 | 0 | 28 | 7 | 0.80 | Reduced siliques |
| 35S:SHP2 -8 | + | 0 | 20 | 12 | 8 | 5 | 0.39 | Reduced siliques |
| 35S:SHP2 -9 | − | 120 | 0 | 120 | 0 | 30 | 0.69 | 0 (20) |
|
35S:SHP2 -10 |
− |
168 |
0 |
168 |
0 |
42 |
0.53 |
0 (30) |
All transgenic lines are in the shp1 shp2 double mutant background.
Levels of trangene transcript levels were determined by RT-PCR using transgene specific primers, with the resulting levels normalized to actin controls.
Shattering reflects numbers of siliques that shattered spontaneously when fully dry; shattering was scored as any separation from the apical tip to approximately the midpoint of the silique. “Reduced siliques” indicates the transgenic line was reduced in size.
Although there was some variability in the extent to which different ectopic phenotypes were observed in the transgenic lines, we observed that four of the six 35S:SHP1 and four of the eight 35S:SHP2 lines had curly leaves, reduced petals, and carpelloid sepals that had white stigmatic tissue (Table 1, Figure 11). This phenotype is quite similar to the published phenotypes of 35S:SHP1 and 35S:SHP2 introduced into wild-type Arabidopsis plants (Pinyopich et al., 2003). However, the phenotypes produced by ectopic expression of either SHP1 or SHP2 in the shp1 shp2 background were less severe, in that conversion of petals to stamens or ovules on the margins of sepals was not seen (Figure 11). This could potentially reflect the differing genetic backgrounds in which these transgenes were expressed in the different studies.
Figure 11.
Ectopic Phenotypes Produced by Overexpression of TAGL1, SHP1, and SHP2 in Arabidopsis.
(A) to (H) Individual flower ([A] to [D]) and top view of inflorescence ([E] to [H]) of shp1 shp2 double mutant ([A] and [E]), 35S:SHP1-5 shp1 shp2 ([B] and [F]), 35S:SHP2-7 shp1 shp2 ([C] and [G]), and 35S:TAGL1-7 shp1 shp2 ([D] and [H]) plants.
(I) Leaf phenotypes produced by (from left to right) wild-type (Landsberg erecta), shp1 shp2 double mutant, 35S:SHP1-5 shp1 shp2, 35S:SHP2-4 shp1 shp2, and 35S:TAGL1-8 shp1 shp2 plants.
We also examined the phenotypes produced by heterologous expression of 35S:TAGL1 in the shp1 shp2 background. In four of eight 35S:TAGL1 lines, we observed that the resulting plants had curly leaves, reduced petals, and carpelloid sepals (Table 1, Figure 11). However, this phenotype was weaker than that observed in 35S:SHP1 or 35S:SHP2 lines and only seen in lines that showed higher TAGL1 transcript levels (Table 1). In terms of rescuing the shp1 shp2 silique phenotype, only partial restoration of shattering in 35S:SHP1 or 35S:SHP2 lines was observed (Table 1). Shattering was scored as any separation from the apical tip to approximately the midpoint of the silique. No lines showed full shattering of siliques, where the valve and replum separated completely from each other. 35S:SHP1 and 35S:SHP2 lines with the most severe floral phenotypes also often produced reduced siliques, in which quantifying restoration of shattering was not possible. By contrast, all the 35S:TAGL1 lines showed very little or no restoration of shattering (Table 1), indicating that TAGL1 expression is not as effective as that of either SHP1 or SHP2 in restoring the silique dehiscence zone.
DISCUSSION
Prior efforts by our groups to gain insight into the transcriptional regulation of fruit ripening have focused on positional cloning of known ripening mutants (Vrebalov et al., 2002; Liu et al., 2004; Barry and Giovannoni, 2006; Manning et al., 2006; Barry et al., 2008), expression profiling via microarrays (Alba et al., 2004, 2005), and digital expression analysis (Fei et al., 2004). We selected the MADS box gene TAGL1 for functional analysis because its expression pattern correlates with pericarp expansion and fruit ripening (expression profiling data can be found at http://ted.bti.cornell.edu/). Furthermore, TAGL1 is orthologous to the Arabidopsis SHP1 and SHP2 genes required for fruit dehiscence (Figure 3), suggesting a broadly similar role in fruit maturation or ripening in tomato.
TAGL1 Influences Carotenoid Accumulation during Fruit Ripening
Downregulation of TAGL1 results in substantially reduced total carotenoid levels, reflecting a precipitous decline in the major ripe fruit carotenoid, lycopene (Figure 5A). Furthermore, ectopic overexpression of TAGL1 was sufficient to induce ripening of tomato sepals, including increased fleshiness and accumulation of lycopene, g-carotene, phytofluene, and phytoene, as well as reduced levels of β-carotene and lutein in these tissues (Figure 11). Conversely, the TAGL1 loss-of-function plants had lower levels of lycopene, phytofluene, and phytoene and higher amounts of β-carotene and lutein. PSY1, which is induced by ethylene during ripening and is a major regulator of metabolic flux toward downstream carotenoids during fruit maturation (Fray and Grierson, 1993), was notably reduced in expression in response to reduced TAGL1 (Figure 8). In addition, both the chloroplast and chromoplast lycopene β-cyclases (LYC-B and CYC-B) were upregulated compared with controls in TAGL1 RNAi lines, which would account for the elevated levels of both β-carotene and lutein in the context of a reduced carotenoid pool (Figure 5B). LYC-B is downregulated by ethylene (Alba et al., 2005) consistent with a role of TAGL1 in mediating the ripening shift in carotenoid flux toward lycopene and away from β-carotene via the hormone ethylene in normally ripening fruit and the opposite in TAGL1 repressed fruit.
Interestingly, neither lycopene accumulation nor other ripening phenotypes were observed in 35S:TAGL1 vegetative tissues or immature fruit, implying that specific protein cofactors present in sepals and mature fruit interact with the TAGL1 gene product and are required for ripening. One candidate cofactor is the product of MADS-RIN, a member of the SEP clade of MADS box genes (Vrebalov et al., 2002). The SEP clade genes have duplicated extensively in the eudicots, with four copies identified in Arabidopsis and five in tomato; these five copies appear to have arisen by a duplication event occurring after the diversification of Arabidopsis and tomato (Hileman et al., 2006). Based on limited functional data, it appears as if SEP clade genes have diversified extensively in term of their function as well, having overlapping or distinct roles in regulating floral organ and floral meristem identity, parthenocarpy, or ripening in different species (Pnueli et al., 1994a, 1994b; Pelaz et al., 2000, 2001; Honma and Goto, 2001; Ampomah-Dwamena et al., 2002; Ferrario et al., 2003; Uimari et al., 2004; Malcomber and Kellogg , 2005). MADS-RIN transcripts are localized predominantly in fruit tissue (Vrebalov et al., 2002), and the MADS-RIN protein can heterodimerize with TAGL1 in yeast two-hybrid analyses (Leseberg et al., 2008), suggesting the possibility that TAGL1 and MADS-RIN coordinate their action as part of a protein complex to effect ripening-related processes in tomato.
TAGL1 Broadly Affects Ripening through Regulation of Autocatalytic Ethylene Synthesis
In addition to its role in regulating carotenoid accumulation, RNAi repression of TAGL1 in tomato demonstrates that TAGL1 is necessary for more comprehensive ripening regulation in part through influencing ethylene synthesis. TAGL1 repressed fruit produce substantially less ethylene than do wild-type controls (Figure 6A) and are correspondingly reduced in expression of ethylene-regulated ripening associated genes, including ACS2, E4, E8, PG, PSY1, and NR (Figure 7A), which themselves reflect a range of downstream ripening activities impacting carotenogenesis, cell wall structure, and production of metabolites associated with flavor, aroma, and nutrition (reviewed in Barry and Giovannoni, 2007). It is well documented that repression of ethylene synthesis or perception can retard or arrest all of these processes in climacteric fruits (Oeller et al., 1991; Lanahan et al., 1994; Ayub et al., 1996).
Ethylene synthesis in ripening tomato fruit is regulated by the ACS and ACO gene families (Barry et al., 2000). ACO1 is the predominant member of this family in ripening fruit, and neither its expression nor that of its regulator, HB1, is substantially altered in TAGL1 repressed lines (Figure 7A). A number of ACS genes have been described in tomato, and ACS1A, ACS2, and ACS4 are the predominant ACS transcripts accumulating in ripening fruit. ACS1A and ACS4 are upregulated at ripening initiation and ethylene resulting from their activity induces both ACS2 and ACS4 to mediate the burst of autocatalytic ethylene synthesis characteristic of climacteric ripening (Barry et al., 2000). While ACS2 and ACS4 are both detected, ACS2 is the predominant ACS mRNA in ripening fruit, and repression of ACS2 is sufficient to block ripening (Oeller et al., 1991). In addition to being highly ethylene dependent, ACS2 expression is also regulated by an as yet to be defined ripening-specific factor, in that it is not induced by ethylene in preripening fruit or in other tissues (Barry et al., 2000). This factor may be, or is at least dependent upon, TAGL1. Based on gene expression analysis, the reduction in ethylene synthesis of TAGL1 RNAi fruit is predominantly due to substantially reduced ACS2 gene expression (Figure 7A). Whether or not this reflects direct interaction of TAGL1 with the ACS2 promoter or a downstream effect of TAGL1 is an important question that remains to be answered.
It is interesting that exogenous ethylene treatment (20 ppm at 22°C) for 12 or 96 h did not recover the ripening phenotype of TAGL1 repressed fruit, nor did it promote ethylene synthesis (measured using gas chromatography and a flame ionization detector; data not shown). These observations indicate that while TAGL1 is necessary for induction of the predominant ACS mRNA in ripening tomato fruit (ACS2) leading to autocatalytic ethylene synthesis, TAGL1 has broader functions in ripening regulation (because exogenous ethylene alone could not complement the TAGL1 RNAi phenotype). TAGL1 thus contributes to the fruit-specific mechanism of ACS2 gene expression necessary for autocatalytic ethylene synthesis in addition to ripening activities that are beyond ethylene regulation. MADS-RIN similarly is both necessary for mature fruit ethylene synthesis and ripening activities beyond ethylene regulation (Vrebalov et al., 2002). Analysis of gene expression suggests that while both influence expression of many of the same genes at least in part due to necessity of both for ethylene induction (PSY1, E4, E8, and PG), they also differentially impact other genes. An example is ACO1, which is not altered by TAGL1 RNAi but is repressed in the rin mutant (Figure 7A) (Kitagawa et al., 2006). The fact that TAGL1 and MADS-RIN are capable of interaction in yeast two-hybrid analyses (Leseberg et al., 2008), yet mutation or repression of each affects many similar but some distinct downstream genes, again suggests that they may interact together and/or with additional factors to confer a spectrum of gene target specificities during ripening.
Softening of TAGL1-Downregulated Fruit Is Most Likely due to Altered Pericarp Structure
PG catalyzes the depolymerization of pectins and is one of the most abundant mRNAs during fruit ripening, accounting for as much as 1% of the mRNA in mature tomato fruit (Lincoln and Fischer, 1988). Though PG has been frequently associated with ripening-related textural changes, its repression in the wild type and ectopic expression in unripe rin/rin fruit had minimal impact on softening, suggesting that cell wall metabolism and softening result from more complex changes in gene expression and physiology than a single cell wall hydrolase activity (Smith et al., 1988; Giovannoni et al., 1989). While we did note changes in fruit softening in our transgenic lines, this occurred prior to PG induction and ripening and likely resulted from reductions in the number of pericarp cell layers and corresponding thinner pericarp tissues (Figure 8). While ripe wild-type and TAGL1 RNAi fruit were softer compared with immature wild-type fruit, the alterations in pericarp morphology prior to ripening make it difficult to draw conclusions as to the basis of mature fruit softening phenotypes. TAGL1 repressed fruit also lost considerably more water as they matured than did controls (Figures 6B and 6C). Saladié et al. (2007) have demonstrated that fruit turgor is a major determinant of tomato fruit firmness, and as such, direct changes in pericarp thickness combined with resulting effects on water retention are likely to account for the increased softening of TAGL1 repressed fruit. Because of the effect of TAGL1 on pericarp thickness, it is difficult to separate any ripening-related effects of this gene on softening. Transgenic repression of TAGL1 specific to ripening fruit would be necessary to separate early from late softening determinants.
TAGL1 and TAG1 Expression Appears Compensatory in Fruit Ripening
The paralogous relationship of TAGL1 and TAG1 (Figure 3) suggests the possibility of related or even redundant functions. Both genes are expressed in ripening fruit, and RNAi repression of TAGL1 did not reduce mRNA accumulation of TAG1 (Figures 1 and 2). Indeed, repression of TAGL1 resulted in elevated TAG1 specifically in mature fruit (Figure 2). TAG1 function has been addressed through antisense and ectopic expression, indicating that that this gene is functionally similar to Arabidopsis AG in that TAG1 repression caused inner whorl floral organs to undergo homeotic conversion to petal and sepal-like structures, while ectopic expression caused outer whorls to become carpeloid and fleshy (Pnueli et al., 1994a, 1994b). While many of the resulting organs in TAG1 antisense lines were described as fleshy, they did not undergo a developmental transition analogous to ripening. Because TAGL1 sequences were not available at the time, neither expression compensation, cross-repression, or functional redundancy of TAGL1 in the TAG1 antisense material could be directly addressed. Here, we show that TAGL1 repression clearly stimulated additional TAG1 mRNA accumulation in mature fruit, though the severe impacts of TAGL1 repression on pericarp thickness and ripening indicate that any functional redundancy with TAG1 is not directly related to these aspects of development in normal carpels. However, the fact that ectopic expression of TAG1 (Pnueli et al., 1994b) and TAGL1 (Figure 9) causes similar fleshy phenotypes in sepals suggests they may be capable of eliciting similar responses under specific conditions or tissue types. One plausible scenario would be that TAG1 and TAGL1 proteins, when highly and ectopically overexpressed, can replace each other in transcriptional complexes and regulate a broadly comparable set of target genes.
TAGL1 Plays a Role in Fruit Expansion Contributing to the Fleshiness of Tomato Fruit
Fruits can be generally categorized as either dry or fleshy with the former promoting seed dispersal via wind, water, shattering, or attachment to animal fur and the latter via consumption by seed dispersing organisms. Fleshy fruits appear to have evolved from dry-fruited forms, although there are numerous cases of reversals (Knapp, 2002; Scutt et al., 2006). Our observations that overexpression of TAGL1 can induce fleshy fruit-like development of sepals (Figure 9), coupled with the reduction in pericarp thickness of TAGL1 RNAi fruit (Figures 4A, 6D, and 6E), suggests that changes in TAGL1 expression may be responsible for the evolution of this trait.
TAGL1 clearly has diverged in molecular function compared with the SHP1/2 genes of Arabidopsis, which produce dry dehiscent fruits. TAGL1 expression in Arabidopsis is sufficient to recapitulate the ectopic phenotypes of curled leaves and sepal and petal defects produced by SHP overexpression but is not sufficient to restore normal shattering in shp1 shp2 mutants. Ectopic expression of PLENA in Arabidopsis (Causier et al., 2005) can cause similar sepal and petal defects to what we have observed for ectopic expression of TAGL1 (Figure 11). However, it has not been reported as to whether PLENA can rescue the Arabidopsis shp1 shp2 fruit dehiscence phenotype. Furthermore, overexpression of the peach (Prunus persica) ortholog of TAGL1 in tomato can result in sepal expansion and carotenoid accumulation but does not result in any apparent homeotic conversions of petals (Tadiello et al., 2009). Together, these results argue that there has been considerable plasticity in the ways in which these orthologous proteins from Antirrhinum majus, tomato, peach, and Arabidopsis perform their functions, which may depend in part on their protein interaction partners as well as subtle differences in their expression patterns during flower and fruit development.
The observation that TAGL1 cannot rescue the Arabidopsis shp1 shp2 phenotype also implies that TAGL1 may be transcriptionally regulating a distinct set of downstream genes compared with its Arabidopsis cognates. The pathway through which SHP1 and SHP2 act to specify dehiscence zone formation has been well characterized in Arabidopsis (reviewed in Lewis et al., 2006). REPLUMLESS, encoding a homeodomain transcription factor, and FRUITFULL, encoding a MADS domain transcription factor, have been shown by genetic studies to restrict the domain of SHP1/2 gene expression within the valve margins (Ferrandiz et al., 2000; Roeder et al., 2003). Also, two basic helix-loop-helix genes IND and ALC have been shown to be regulated by SHP1/SHP2 and to act downstream to specify the differentiation of the separation layer in the silique (Rajani and Sundaresan, 2001; Liljegren et al., 2004). One possibility is that TAGL1 regulates a similar suite of genes that in turn have unique roles in regulating aspects of tomato fruit ripening and carotenoid biosynthesis. However, it is not known if orthologs for ALC and IND exist in tomato or if they have roles in fruit ripening. Alternatively, TAGL1 may directly regulate a qualitatively distinct set of target genes compared with SHP1/2 in Arabidopsis.
Given that tomato, Arabidopsis, and Antirrhinum all produce very different fruit types (berries, siliques, and capsules, respectively), it is perhaps surprising that they share components of a conserved regulatory pathway in regulating fruit development and ripening. One possibility is that the tremendous flexibility of MADS box genes to assume new and varied functions and possibly drive the evolution of diverse plant morphology is due to their ability to evolve new protein interaction capabilities and thus modulate the types of higher-order protein complexes that can occur. In summary, transgenic repression demonstrates that TAGL1 participates in normal fleshy fruit development and later ripening, while ectopic expression of TAGL1 in sepals supports the notion that the TAGL1 lineage of MADS box genes may be a key player in evolutionary transitions between dry and fleshy carpel development.
Consideration of TAGL1 in the Context of Fruit Development and Ripening Transcription Factors
Given the recent description of a number of ripening regulators, it is important to consider how TAGL1 participates in ripening in the context of said regulators and especially previously described ripening transcription factors. TAGL1, MADS-RIN (Vrebalov et al., 2002), CNR-SBP (Manning et al., 2006), and the HB-1 HD-ZIP homeobox protein (Lin et al., 2008) have all been shown to be necessary for ripening. Tomato EIN3-like (EIL) transcription factors have redundant functions associated with ethylene response and ripening (Tieman et al., 2001; Chen et al., 2004), and additional transcription factors such as AUX/IAA gene IAA9 impact fruit development when repressed (resulting in early and parthenocarpic fruit set) but not ripening (Wang et al., 2005). The specific nature of the regulatory hierarchy and interactions among these regulators remains unclear, but some general conclusions can be drawn. Repression of EIL genes inhibits ripening as part of a general reduction in plant ethylene sensitivity, and overexpression of EIL1 restores ripening in the Nr ethylene receptor mutant, confirming activity downstream of ethylene synthesis and the receptors in the ethylene signal transduction network. Repression of TAGL1, MADS-RIN, CNR-SBP, or HB-1 is similar in that all result in low ethylene, nonripening fruit, suggesting that all four lie upstream of ethylene synthesis control. HB1 regulates climacteric ethylene through direct regulation of ACO1, while TAGL1 regulates ethylene (either directly or indirectly) through regulation of ACS2 with no effect on ACO1. CNR-SBP mRNA accumulation is reduced in the rin mutant, suggesting that MADS-RIN has a positive regulatory role on CNR-SBP expression. TAGL1 expression is not altered in the rin mutation (Figure 1), and neither MADS-RIN nor CNR-SBP mRNA accumulation is substantially altered in transgenic fruit repressed for TAGL1 except that both appear to be induced early in TAGL1 repressed fruit (Figure 8A). Together, these results indicate that the pathways influencing TAGL1 versus MADS-RIN and CNR-SPB are distinct, suggesting that additional regulators remain to be discovered for regulation of these key ripening genes. TAGL1 adds a new component to the emerging networks regulating fleshy fruit expansion and ripening and uniquely links these processes to promote seed dispersal.
METHODS
Plant Materials and Growth Conditions
All tomato cultivars (Solanum lycopersicum cv Ailsa Craig and Microtom) were kindly provided by the Tomato Genetics Resource Center in Davis, CA (http://tgrc.ucdavis.edu/). Plants were either grown under field conditions in Freeville, NY or in greenhouses under sodium lights timed for 16-h days (27°C) and 8-h nights (19°C). TAGL1 RNAi transgenic lines were advanced to the T2 generation, and only plants homozygous for the transgene were used for quantitative analyses. Fruit of normal and transgenic lines reached the 1 cm stage uniformly at 7 to 8 DPA, at which point they were tagged to synchronize developmental comparisons. Fruit of the normal and transgenic lines analyzed reached breaker stage uniformly at 37 to 38 DPA.
Transgenic and control Arabidopsis thaliana plants (ecotype Columbia-0) were grown in growth chambers at 22°C with a 16-h-light and 8-h-dark cycle. Transgenic lines were assayed as homozygotes in the T2 generation.
Ethylene Measurement
Ethylene was measured from fruits by sealing whole fruits in airtight jars for 2 h at 22°C, after which a 1-mL sample of the headspace was taken and injected on to a Hewlett-Packard 5890 series II gas chromatograph equipped with a flame ionization detector. Samples were compared with a standard of known concentration and normalized for fruit mass.
Phylogenetic Analyses
AG-like sequences were identified based on BLAST searches and previously published data (Kramer et al., 2004; Zahn et al., 2006). Full-length amino acid sequences were aligned using ClustalW using pairwise parameters: gap opening 45, gap extend 2; multiple alignment parameters: gap opening 25, gap extend 2, and both using Gonnet protein weight matrix. Alignments were refined by hand using MacClade 4.03. The alignment is available as Supplemental Data Set 1 online. Maximum parsimony trees were generated using PAUP 4.0 through heuristic searches of 10 random stepwise additions; tree support was assessed with bootstrap analysis. Gymnosperm sequences were used as outgroups.
DNA Isolation and DNA Gel Blot Analysis
Genomic DNA was isolated, digested with restriction enzymes, transferred to hybridization membranes, hybridized to radioactive DNA probes, and visualized as described previously by Vrebalov et al. (2002). Bases 594 to 1017 of the TAGL1 cDNA were used as a hybridization probe.
RNAi and Overexpression Constructs and Transformation
The TAGL1 RNAi construct was made using the pHELLSGATE 2 vector (kindly provided by CSIRO, Plant Industry, Canberra, Australia). TAGL1 cDNA sequences used in the haripin included 252 bp from the C domain and the subsequent 172 bp of 3′ UTR (i.e., bases 594 to 1017 of the full-length cDNA). The target sequences were PCR amplified from EST clone cLEG9L5 using gene-specific primers cLEG9L5For and cLEG9L5FRev with addition of the corresponding recombination sequences as defined in the kit for site-specific recombination used (Gateway BP Clonase enzyme mix; Invitrogen). The resulting PCR product was gel purified and cloned into pHELLSGATE 2 via homologous recombination using the kit above. The resulting construct was sequence confirmed and transformed into tomato cv Ailsa Craig by Agrobacterium tumefaciens (strain ABA4404) as described previously (Vrebalov et al., 2002).
Overexpression constructs were generated using the Gateway system (Invitrogen). For the tomato overexpression construct, the complete open reading frame of TAGL1 was amplified using the TAGL1FLFB1 primer containing an attB1 site, and the TAGL1FLR-B2 primer containing an attB2 site DNA product was amplified using the following program: 95° for 10 min, 40 cycles of 95°C for 30 s, 50° for 1 min and 30 s, 72° for 1 min, followed by 72° for 7 min. These products were cloned into the pH7WG2 destination vector (Karimi et al., 2002). All constructs were transformed into A. tumefaciens strain LBA 4404 by electroporation.
Transformation of tomato (S. lycopersicum cv Micro-Tom) wild-type cotyledon explants was performed as previously described (McCormick, 1991). The presence of the transgene was verified in the T0 and T1 generations by PCR using two sets of primers. HYG1-F and HYG1-R amplified the Hygromycin resistance gene. The 35S2 primer and the gene-specific reverse primers with an attB2 site (TAGL1RB2 or TAGL1FLR-B2) amplified the region encompassing the end of the 35S promoter and the transgene. The PCR program for Hygromycin was 95°C for 10 min, 40 cycles of 95°C for 30 s, 60°C for 1 min, 72°C for 1 min, followed by 72°C for 7 min. The PCR program for the 35S2 and gene-specific product was 95°C for 10 min, 40 cycles of 95°C for 30 s, 50°C for 45 s, 72°C for 1 min, followed by 72°C for 7 min.
For Arabidopsis transformation experiments, the complete open reading frames of TAGL1, SHP1, and SHP2 were amplified with the following primers containing an XbaI site in the F primer and a BamHI site in the R primer: TAGL1FL-F and TAGL1FL-R for TAGL1, SHP1FL-F and SHP1FL-R for SHP1, and SHP2FL-F and SHP2FL-R for SHP2 (see Supplemental Table 2 online). DNA products were amplified using the following program: 95°C for 10 min, 40 cycles of 95°C for 30 s, 50°C for 45 s, 72°C for 1 min, followed by 72°C for 7 min.
These products were cloned into a Topo 4.0 vector (Invitrogen), cut with BamHI and XbaI restriction enzymes, and ligated into a p235 binary vector (pPZP221; Hajdukiewicz et al., 1994) containing the 35S promoter from Cauliflower mosaic virus. Plants were transformed using the floral dip method (Clough and Bent, 1998). The presence of the transgenes was verified in the T0 and T1 generations using PCR with the 35S2 primer and the gene-specific reverse primers (TAGL1FL-R, SHP1FL-R, and SHP2FL-R).
RNA Isolation, Gel Blot, RT-PCR, and Quantitative RT-PCR Analysis
Harvested tomato tissue was immediately frozen in liquid nitrogen and stored at −80 C. Total RNA from tomato vegetative tissue, flowers, and fruit was isolated using procedures and reagents described by Chang et al. (1993). RNA gel blot analyses used 30 μg of total RNA per lane. RNA was loaded onto 1.2% formaldehyde agarose gels, transferred on Hybond-N membrane, and cross-linked by baking as per the protocol of the membrane supplier (Amersham). Radiolabeled probe preparation and hybridizations were as described by Vrebalov et al. (2002) with visualization via audioradiography also as described therein. Gene-specific probes were generated using PCR using primers for the following genes (all primers are listed in Supplemental Table 2 online): ACC synthase 4 (ACS4F/R), ACC synthase 2 (ACS2F/R), ACC oxidase 1 (ACO1F/R), E4 (E4F/R), Polygalacturonase 2A (PG2AF/R), Never-Ripe (NRF/R), E8 (E8F/R), Phytoene synthase 1 (PSY1F/R), CNR-SPB (CNRF/R), HB1 (HB1F/R), MADS-RIN (RINF/R), Soluble acid invertase (SAIF/R), Phosphoglucomutase (PGMF/R), Soluble starch synthase (SSYF/R), ADP Glucose Pyrophosporilase Large subunit (LSUF/R), TAG (TAG1-F/R), TAGL1 (TAGL1-F/R), TDR4 (TDR4F/R), and 18S rDNA (18SF/R).
For quantitative RT-PCR analysis, total RNA as isolated above was DNase treated, phenol:chloroform (1:1, v/v) extracted, and purified using the RNA clean-up protocol from the RNeasy Mini kit (Qiagen Sciences). Final concentration was assessed using the ND-1000 v3.1.0 (NanoDrop Technologies). Quantitative real-time PCR was performed using the Power SYBR Green RNA-to-CT 1-step kit (Applied Biosystems) in a 10-μL total sample volume (1× RT-PCR buffer, 1× final of 125X RT enzyme mix, 100 nM of each primer, and 50 and 0.005 ng total RNA for gene of interest and internal control). The wild-type and transgenic lines were represented by three biological replicates (each with three technical replicates) for each stage. Gene-specific primer concentrations were optimized using wild-type RNA. To be able to apply the standard curve method described in User Bulletin #2 (Applied Biosystems), a standard curve was included on each plate for the specific gene being analyzed using wild-type RNA (serial dilutions: 500, 50, 5, 0.5, 0.05, and 0.005 ng) in triplicate. For each gene analysis, template-free and negative-RT controls were included. Real-time PCR reactions were performed using an ABI PRISM 7900HT sequence detection system (Applied Biosystems) under the following reaction conditions: reverse transcription at 48°C for 30 min, AmpliTaq Gold Activation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The PCR reaction was followed by a dissociation stage composed of 95°C for 15 s and then 60°C for 15 s and 95°C for 15 s. ABI PRISM SDS version 2.1 software (Applied Biosystems) was used to determine gene-specific threshold cycles (CT) using the endogenous reference (18S rRNA) for every sample. CT's were extracted and relative quantification was performed using the standard curve method (Applied Biosystems) and applied to calculate relative mRNA levels in comparison to the wild-type control. Quantitative RT-PCR primer sequences were chloroplast lycopene-β-cyclase (LYC-BF/R) and chromoplast lycopene-β-cyclase (CYC-BF/R).
For RT-PCR, total RNA from tomato and Arabidopsis tissue were isolated using Trizol (Invitrogen) according to the manufacturer's instructions. For tomato seeds, total RNA was isolated using the Invisorb Spin Plant RNA Mini Kit (Invitek). For cDNA synthesis, 2.5 μg were used using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The cDNA was diluted 1:10 and 1 to 2 μL were used for PCR. The following primers were used: ACT1, ACT2, TAG1F, TAG1R, TAGL1-F, and TAGL1-R.
The PCR program for TAGL1 and ACTIN was 94°C for 5 min, 28 cycles of 94°C for 30 s, 59°C for 45 s, 72°C for 1 min, followed by 72°C for 10 min. The PCR program for TAG1 was 94°C for 5 min, 29 cycles of 94°C for 30 s, 59°C for 45 s, 72°C for 1 min, followed by 72°C for 10 min. Gel images were scanned, and band intensities were normalized to ACTIN and quantified using NIH Image J (http://rsb.info.nih.gov/ij/).
Carotenoid and Chlorophyll Extraction and Analysis
Carotenoids for mature green and red ripe fruit were extracted from 200 mg of frozen tomato pericarp using a modified protocol from Alba et al. (2005). The frozen tissue was homogenized in a Savant FP120 Fast Prep machine with 15 mg of Mg-carbonate and 450 mL of tetrahydrofuran twice and then a third time with 450 mL of methanol containing 2,6-Di-tert-butyl-4-methylphenol. The homogenate was filtered through Spin-X centrifuge filters (0.45-mm nylon filter; Corning/Costar 8170), and tissue debris was reextracted with an additional 500 mL of tetrahydrofuran to ensure complete extraction of carotenoids. The carotenoid/nonpolar phase was separated from the aqueous phase through two separation steps, first with 375 mL of petroleum ether and 150 mL of 25% NaCl and next with 500 mL of petroleum ether. The two upper phase aliquots were combined and dried down in a vacufuge (Eppendorf). For carotenoids, the dried extract was resuspended in 1 mL of methyl t-butyl ether and 970 mL of 2,6-Di-tert-butyl-4-methylphenol. All solvents used were HPLC grade. Carotenoid analysis was performed using a Dionex HPLC (P680 HPLC pump, ASI-100 automated sample injector, and PDA-100 photodiode array detector) and the Chromeleon (v6.40) software package. Separation of carotenoids was achieved under a polar to nonpolar gradient (0 to 5 min 100% methanol:0.1% ammonium acetate; 6 to 25 min ramp to 4% methanol:ammonium acetate and 96% methyl t-butyl ether; 26 to 30 min ramp to 100% methanol:ammonium acetate; 31 to 35 min 100% methanol:ammonium acetate) through a guard cartridge (YMC Carotenoid S-5, 4.0 × 20 mm DC guard; Waters), C30 column (YMC Carotenoid S-5, 4.6 × 250 mm; Waters) assembly. Five channels were used for data acquisition: channel 1 (286 nm), channel 2 (348 nm), channel 3 (434 nm), channel 4 (450 nm), and channel 5 (471 nm). Peak identification was performed as described by Alba et al. (2005). For chlorophyll, the dried down extracts were resuspended in 1 mL of ethyl acetate, and 25 μL of sample was applied to a modified gradient system described by Fraser et al. (2000) (0 to 8 min 5 of 80% methanol:20% water:0.2% ammonium acetate, and 95 of 100% methanol; 8 min step to 5% of 80/20 methanol:0.2% ammonium acetate, 80% methanol, and 15% methyl t-butyl ether; 8 to 40 min ramp to 5% 80/20 methanol:0.2% ammonium acetate, 10% of methanol, and 85% methyl t-butyl ether; 40 to 50 min ramp to 5% of 80/20 methanol:0.2% ammonium acetate, and 95% methanol; 50 to 60 min 5% of 80/20 methanol:0.2% ammonium acetate, and 95% methanol). HPLC peak identification and data analysis were performed as described previously for fruit tissue.
Fruit Pericarp Thickness, Firmness, and Water Loss
Tomato fruits were cut in half, and pericarp thickness was measured at three different points for each locule using a digital caliper for a total of six measurements per fruit performed on a minimum of three fruits per genotype for a total minimum of 18 measurements per genotype/stage. The final thickness represents an average of these measurements. Firmness measurements were made by recording a force-deformation curve using an Instron Materials Tester (model 3342) with a 100 N load cell following Wu and Abbott (2002). A flat probe was used at a displacement rate of 1 mm s−1 to compress an intact tomato fruit a total distance of 3 mm. The maximum force recorded at 3 mm of compression was used as an estimation of the fruit firmness from the averaged value of at least three tested fruits with a minimum of three compressions per fruit. Water loss analysis was performed using 10 fruit from the wild type and each transgenic line harvested at the breaker stage (BR). The fresh weight was recorded as a starting point. Fruit was kept at room temperature for 3 weeks, and a fresh weight was recorded every 7 d. Water loss was calculated as a percentage in fresh weight difference between the starting weight and each individual measurement.
Quantitation of Cell Layers
Toluidine blue–stained sections were used to estimate the number of cell layers that composes the wild type and transgenic fruit pericarp. At least 10 transects were drawn from the epidermis to the endodermis of the pericarp for each section and the number of cells counted manually. The average value of at least four different fruit samples was presented, reflecting a minimum of 40 measurements per genotype at each developmental stage measured.
Tissue Preparation for Microscopy
Fresh tomato fruit pericarp tissue was sectioned as follows. Tissue was cut into small pieces and fixed in FAA (5% formaldehyde, 5% glacial acetic acid, and 45% ethanol). This fixative was infiltrated into the sections under vacuum (400 mm of Hg) for 15 min on ice. Fresh FAA was added to the samples and incubated overnight at 4°C. Tissue blocks were then cryoprotected by immersion in a 10 and 20% (w/v) sucrose solution dissolved in 100 mM sodium phosphate buffer, pH 7.2. Successive sucrose solutions were infiltrated into the sections under vacuum (400 mm of Hg) on ice for 15 min and then held at 4°C for 2 h. The tissue pieces were embedded in TissueTek OCT medium (Sakura Finetek), frozen in liquid nitrogen, and stored at −80°C. Tomato sepals and fruit pericarp were sectioned at 6 and 10 mm, respectively, in a cryostat (HM500; Microm), and tissue was mounted on a 1× adhesive-coated slide using the CryoJane Tape-Transfer System (Instrumedics) at −26°C following the system instructions. Briefly, tissue sections were captured on an adhesive tape window (Instrumedics) as they were being cut. The sections then were laminated to an adhesive-coated slide, anchored tightly on the slide by a flash of UV light (360 nm), and the tape was removed leaving the sections on the slide. Each slide was postfixed in room temperature CryoJane Aqueous Slide Fixative (40% glutaraldehyde [25% aqueous] and 60% CryoJane Salt Buffer) for 45 s and rinsed gently with distilled water immediately prior to staining. For chloroplast distribution and fruit pericarp thickness microscopy images, fresh tomato fruits from 28 DPA and BB+7 were hand-sectioned to ∼1-mm thickness.
Tissue Staining
Two different staining procedures were applied to the tomato pericarp sections. One was toluidine blue O (Sigma-Aldrich; 0.05%, w/v, in distilled water) incubation for ∼30 s, followed by rinsing with distilled water, and mounting in water with a cover slip. Sections stained this way were used to determine the number of cells layers that compose the pericarp and to stain hand-cut fruit samples in wild-type and mutant lines. For starch granule detection, a Nile blue A stain (Sigma-Aldrich; 1%, w/v, in distilled water) was added to the sections for 30 s. The stain was poured off, 1% acetic acid was added to the slides for another 30 s, and the slides were rinsed with distilled water as described (Gahan, 1984). Starch granules of the pericarp parenchyma cells were identified by their starch-associated birefringence under polarized light.
Microscopy and Image Processing
Stained sections were observed using a Zeiss AxioImager A1 microscope equipped with a Zeiss AxioCam MRc color video camera and Zeiss AxioVs40 4.6.3.0 software. When necessary, several images of the same sample were collated using the software hugin Panorama photo stitcher (http://hugin.sourceforge.net/). Chloroplast images were collected on a Leica TCS-SP5 confocal microscope (Leica Microsystems) using a ×63 water immersion objective with a numerical aperture of 1.2 and zoom 1.6. For autofluorescence, chloroplasts were excited with the blue argon laser (488 nm), and emitted light was collected from 626 to 731 nm. Images were processed using Leica LAS-AF software (versions 1.6.3 and 1.7.0) and Adobe Photoshop CS2 version 9.0.2 (Adobe Systems).
In Situ Hybridizations
In situ hybridizations were conducted using published protocols (Jackson, 1991; de Martino et al., 2006). Tomato (S. lycopersicum cv Micro-Tom) wild-type floral buds of varying stages and young fruit (0 to 3 DPA) were fixed in 4% paraformaldehyde in PBS and embedded in Paraplast Plus tissue embedding medium (Tyco Healthcare).
For in situ on floral buds, a 200-bp TAGL1 antisense probe was made using the TAGL1F primer and the TAGL1R7 primer, which adds the T7 promoter For in situ on young fruit, a 1048-BP TAGL1 antisense probe targeting the full-length coding region, and the 3′ UTR was made using the TAGLIFLF0 primer and the TAGL1UR7 primer, which adds the T7 promoter. DNA products were amplified using the following program: 95°C for 10 min, 40 cycles of 95°C for 30 s, 50°C for 1 min 30 s, 72°C for 1 min, followed by 72°C for 7 min. Hybridizations were performed at 52°C and washes were performed at 55°C.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ACO1 (XO4792), ACS2 (AY326958), ACS4 (M63490), CNR (DQ672601), CYC-B (AF254793), E4 (S44898), E8 (X13437), HB1 (BTO14213), LCY-B (X86452), LSU (U85496), MADS-RIN (AF448522), NR (U38666), PG2A (XD4583), PGM (BTO14628), PSY1 (EF157836), SAI (S70040), SSY (BTO12843), TAG1 (L26295), TAGL1/cLEG9L5 (AY098735.2), TDR4 (X60757.1), and 18S rDNA (X51576.1). The accession numbers for coding regions used in the phylogenetic analysis can be found in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. TAGL1 Hybridization to Tomato Genomic DNA.
Supplemental Figure 2. Total RNA Gel Blot Analysis of TAGL1 and 18S rRNA (Control).
Supplemental Figure 3. Chlorophyll and Carotenoid Accumulation in Sepal and Petals of TAGL1 Repressed Lines.
Supplemental Figure 4. TAGL1 Expression in TAGL1 Overexpression Lines.
Supplemental Table 1. List of Species and Accession Numbers of Coding Regions Used for Phylogenetic Analysis.
Supplemental Table 2. DNA Primer Sequences.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis in Figure 3.
Supplementary Material
Acknowledgments
We acknowledge the Tomato Genetics Resource Center at UC Davis for kindly providing tomato seed. This work was supported by the USDA-Agricultural Research Service, National Science Foundation Plant Genome Grants 05-01778 and 06-06595, United States-Israel Binational Agriculture Research and Development Fund Grant IS-3803-05, USDA-National Research Initiative Grants 2007-02773 and 2006-35304-17323, and National Science Foundation Grant DBI-0411960.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: James Giovannoni (jjg33@cornell.edu) and Vivian F. Irish (vivian.irish@yale.edu).
Online version contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Alba, R., et al. (2004). ESTs, cDNA microarrays, and gene expression profiling: Tools for dissecting plant physiology and development. Plant J. 39 697–714. [DOI] [PubMed] [Google Scholar]
- Alba, R., Payton, P., Fei, Z., McQuinn, R., Debbie, P., Martin, G.B., Tanksley, S.D., and Giovannoni, J.J. (2005). Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17 2954–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampomah-Dwamena, C., Morris, B.A., Sutherland, P., Veit, B., and Yao, J.L. (2002). Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 130 605–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayub, R., Guis, M., Ben Amor, M., Gillot, L., Roustan, J.P., Latche, A., Bouzayen, M., and Pech, J.C. (1996). Expression of ACC oxidase antisense gene inhibits ripening of cantaloupe melon fruits. Nat. Biotechnol. 14 862–866. [DOI] [PubMed] [Google Scholar]
- Barry, C., and Giovannoni, J. (2007). Ethylene and fruit ripening. J. Plant Growth Regul. 26 143–159. [Google Scholar]
- Barry, C.S., and Giovannoni, J.J. (2006). Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proc. Natl. Acad. Sci. USA 103 7923–7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, C.S., Llop-Tous, M.I., and Grierson, D. (2000). The regulation of 1-aminocyclopropane-1-carboxylic acid synthase gene expression during the transition from system-1 to system-2 ethylene synthesis in tomato. Plant Physiol. 123 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, C.S., McQuinn, R.P., Chung, M.Y., Besuden, A., and Giovannoni, J.J. (2008). Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 147 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartley, G.E., and Ishida, B.K. (2003). Developmental gene regulation during tomato fruit ripening and in vitro sepal morphogenesis. BMC Plant Biol. 3 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, A., Winter, K.-U., Saedler, H., and Theissen, G. (2000). MADS-box gene diversity in seed plants 300 million years ago. Mol. Biol. Evol. 17 1425–1434. [DOI] [PubMed] [Google Scholar]
- Benfey, P.N., and Chua, N.-H. (1990). The Cauliflower Mosaic virus 35S promoter: Combinatorial regulation of transcription in plants. Science 250 959–966. [DOI] [PubMed] [Google Scholar]
- Bird, C., Ray, J., Fletcher, J.C., Boniwell, J., Bird, A., Teulieres, C., Blain, I., Bramley, M., and Schuch, W. (1991). Using antisense RNA to study gene function: Inhibition of carotenoid biosynthesis in transgenic tomatoes. Biotechnology 9 635–639. [Google Scholar]
- Brukhin, V., Hernould, M., Gonzalez, N., Chevalier, C., and Mouras, A. (2003). Flower development schedule in tomato Lycopersicon esculentum cv. sweet cherry. Sex. Plant Reprod. 15 311–320. [Google Scholar]
- Burns, J., Fraser, P.D., and Bramley, P.M. (2003). Identification and quantification of carotenoids, tocopherols and chlorophylls in commonly consumed fruits and vegetables. Phytochemistry 62 939–947. [DOI] [PubMed] [Google Scholar]
- Busi, M.V., Bustamante, C., D'Angelo, C., Hidalgo-Cuevas, M., Boggio, S.B., Valle, E.M., and Zabaleta, E. (2003). MADS-box genes expressed during tomato seed and fruit development. Plant Mol. Biol. 52 801–815. [DOI] [PubMed] [Google Scholar]
- Causier, B., Castillo, R., Zhou, J., Ingram, R., Xue, Y., Schwarz-Sommer, Z., and Davies, B. (2005). Evolution in action: Following function in duplicated floral homeotic genes. Curr. Biol. 15 1508–1512. [DOI] [PubMed] [Google Scholar]
- Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11 113–116. [Google Scholar]
- Chen, G., Alexander, L., and Grierson, D. (2004). Constitutive expression of EIL-like transcription factor partially restores ripening in the ethylene-insensitive Nr tomato mutant. J. Exp. Bot. 55 1491–1497. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- de Martino, G., Pan, I., Emmanuel, E., Levy, A., and Irish, V.F. (2006). Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell 18 1833–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei, Z., Tang, X., Alba, R.M., White, J.A., Ronning, C.M., Martin, G.B., Tanksley, S.D., and Giovannoni, J.J. (2004). Comprehensive EST analysis of tomato and comparative genomics of fruit ripening. Plant J. 40 47–59. [DOI] [PubMed] [Google Scholar]
- Ferrandiz, C., Liljegren, S.J., and Yanofsky, M.F. (2000). Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289 436–438. [DOI] [PubMed] [Google Scholar]
- Ferrario, S., Immink, R.G., Shchennikova, A., Busscher-Lange, J., and Angenent, G.C. (2003). The MADS box gene FBP2 is required for SEPALLATA function in petunia. Plant Cell 15 914–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan, C.A., Hu, Y., and Ma, H. (1996). Specific expression of the AGL1 MADS-box gene suggests regulatory functions in Arabidopsis gynoecium and ovule development. Plant J. 10 343–353. [DOI] [PubMed] [Google Scholar]
- Frary, A., Nesbitt, T.C., Grandillo, S., Knaap, E., Cong, B., Liu, J., Meller, J., Elber, R., Alpert, K.B., and Tanksley, S.D. (2000). fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289 85–88. [DOI] [PubMed] [Google Scholar]
- Fraser, P.D., Pinto, M.E.S., Holloway, D.E., and Bramley, P.M. (2000). Application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J. 24 551–558. [DOI] [PubMed] [Google Scholar]
- Fray, R.G., and Grierson, D. (1993). Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol. Biol. 22 589–602. [DOI] [PubMed] [Google Scholar]
- Gahan, P. (1984). Experimental Botany: An International Series of Monographs: Plant Histochemistry and Cytochemistry: An Introduction, Vol. 18. (Orlando, FL: Academic Press, Inc.).
- Giovannoni, J.J. (2004). Genetic regulation of fruit development and ripening. Plant Cell 16 (suppl. 1): S170–S180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni, J.J. (2007). Fruit ripening mutants yield insights into ripening control. Curr. Opin. Plant Biol. 10 283–289. [DOI] [PubMed] [Google Scholar]
- Giovannoni, J.J., DellaPenna, D., Bennett, A.B., and Fischer, R.L. (1989). Expression of a chimeric polygalacturonase gene in transgenic rin (ripening inhibitor) tomato fruit results in polyuronide degradation but not fruit softening. Plant Cell 1 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, N., Gevaudant, F., Hernould, M., Chevalier, C., and Mouras, A. (2007). The cell cycle-associated protein kinase WEE1 regulates cell size in relation to endoreduplication in developing tomato fruit. Plant J. 51 642–655. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25 989–994. [DOI] [PubMed] [Google Scholar]
- Hileman, L.C., Sundstrom, J.F., Litt, A., Chen, M., Shumba, T., and Irish, V.F. (2006). Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol. Biol. Evol. 23 2245–2258. [DOI] [PubMed] [Google Scholar]
- Honma, T., and Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 469–471. [DOI] [PubMed] [Google Scholar]
- Ishida, B.K., Jenkins, S.M., and Say, B. (1998). Induction of AGAMOUS gene expression plays a key role in ripening of tomato sepals in vitro. Plant Mol. Biol. 36 733–739. [DOI] [PubMed] [Google Scholar]
- Ito, Y., Kitagawa, M., Ihashi, N., Yabe, K., Kimbara, J., Yasuda, J., Ito, H., Inakuma, T., Hiroi, S., and Kasumi, T. (2008). DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 55 212–223. [DOI] [PubMed] [Google Scholar]
- Jackson, D. (1991). In situ hybridisation in plants. In Molecular Plant Pathology: A Practical Approach, D.J. Bowles, S.J. Gurr, and P. McPherson, eds (Oxford, UK: Oxford University Press), pp. 163–174.
- Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kitagawa, M., Nakamura, N., Usuda, H., Shiina, T., Ito, H., Yasuda, J., Inakuma, T., Ishiguro, Y., Kasumi, T., and Ito, Y. (2006). Ethylene biosynthesis regulation in tomato fruit from the F1 hybrid of the ripening inhibitor (rin) mutant. Biosci. Biotechnol. Biochem. 70 1769–1772. [DOI] [PubMed] [Google Scholar]
- Knapp, S. (2002). Tobacco to tomatoes: A phylogenetic perspective on fruit diversity in the Solanaceae. J. Exp. Bot. 53 2001–2022. [DOI] [PubMed] [Google Scholar]
- Kramer, E.M., Jaramillo, M.A., and Di Stilio, V.S. (2004). Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan, M.B., Yen, H.C., Giovannoni, J.J., and Klee, H.J. (1994). The never ripe mutation blocks ethylene perception in tomato. Plant Cell 6 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leseberg, C.H., Eissler, C.L., Wang, X., Johns, M.A., Duvall, M.R., and Mao, L. (2008). Interaction study of MADS-domain proteins in tomato. J. Exp. Bot. 59 2253–2265. [DOI] [PubMed] [Google Scholar]
- Lewis, M.W., Leslie, M.E., and Liljegren, S.J. (2006). Plant separation: 50 ways to leave your mother. Curr. Opin. Plant Biol. 9 59–65. [DOI] [PubMed] [Google Scholar]
- Liljegren, S.J., Ditta, G.S., Eshed, Y., Savidge, B., Bowman, J.L., and Yanofsky, M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404 766–770. [DOI] [PubMed] [Google Scholar]
- Liljegren, S.J., Roeder, A.H., Kempin, S.A., Gremski, K., Ostergaard, L., Guimil, S., Reyes, D.K., and Yanofsky, M.F. (2004). Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116 843–853. [DOI] [PubMed] [Google Scholar]
- Lin, Z., Hong, Y., Yin, M., Li, C., Zhang, K., and Grierson, D. (2008). A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 55 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, J.E., and Fischer, R.L. (1988). Regulation of gene expression by ethylene in wild-type and rin tomato (Lycopersicon esculentum) fruit. Plant Physiol. 88 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Roof, S., Ye, Z., Barry, C., van Tuinen, A., Vrebalov, J., Bowler, C., and Giovannoni, J. (2004). Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc. Natl. Acad. Sci. USA 101 9897–9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber, S.T., and Kellogg, E.A. (2005). SEPALLATA gene diversification: brave new whorls. Trends Plant Sci. 10: 427–435. [DOI] [PubMed]
- Manning, K., Tor, M., Poole, M., Hong, Y., Thompson, A.J., King, G.J., Giovannoni, J.J., and Seymour, G.B. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38 948–952. [DOI] [PubMed] [Google Scholar]
- Maunders, M., Holdsworth, M., Slater, A., Knapp, J., Bird, C., Schuch, W., and Grierson, D. (1987). Ethylene stimulated the accumulation of ripening-related mRNAs in tomatoes. Plant Cell Environ. 10 177–184. [Google Scholar]
- McCormick, S. (1991). Transformation of tomato with Agrobacterium tumefaciens. In Plant Tissue Culture Manual, K. Lindsey, ed (Dortrecht, The Netherlands: Kluwer), pp. 1–9.
- Mueller, L.A. (2009). A snapshot of the emerging tomato genome sequence. Plant Genome 2 78–92. [Google Scholar]
- Oeller, P.W., Lu, M.W., Taylor, L.P., Pike, D.A., and Theologis, A. (1991). Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254 437–439. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Ditta, G.S., Baumann, E., Wisman, E., and Yanofsky, M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 200–203. [DOI] [PubMed] [Google Scholar]
- Pelaz, S., Gustafson-Brown, C., Kohalmi, S.E., Crosby, W.L., and Yanofsky, M.F. (2001). APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 26 385–394. [DOI] [PubMed] [Google Scholar]
- Pinyopich, A., Ditta, G.S., Savidge, B., Liljegren, S.J., Baumann, E., Wisman, E., and Yanofsky, M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424 85–88. [DOI] [PubMed] [Google Scholar]
- Pnueli, L., Hareven, D., Broday, L., Hurwitz, C., and Lifshitz, E. (1994. a). The TM5 MADS box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell 6 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli, L., Hareven, D., Rounsley, S.D., Yanofsky, M.F., and Lifschitz, E. (1994. b). Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 6 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajani, S., and Sundaresan, V. (2001). The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr. Biol. 11 1914–1922. [DOI] [PubMed] [Google Scholar]
- Roeder, A.H., Ferrandiz, C., and Yanofsky, M.F. (2003). The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 13 1630–1635. [DOI] [PubMed] [Google Scholar]
- Saladié, M., et al. (2007). A reevaluation of the key factors that influence tomato fruit softening and integrity. Plant Physiol. 144 1012–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidge, B., Rounsley, S.D., and Yanofsky, M.F. (1995). Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral organ identity genes. Plant Cell 7 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scutt, C.P., Vinauger-Douard, M., Fourquin, C., Finet, C., and Dumas, C. (2006). An evolutionary perspective on the regulation of carpel development. J. Exp. Bot. 57 2143–2152. [DOI] [PubMed] [Google Scholar]
- Seymour, G., Poole, M., Manning, K., and King, G.J. (2008). Genetics and epigenetics of fruit development and ripening. Curr. Opin. Plant Biol. 11 58–63. [DOI] [PubMed] [Google Scholar]
- Smith, C., Watson, C.F., Ray, J., Bird, C., Morris, P., Schuch, W., and Grierson, D. (1988). Antisense RNA inhibition of polygalacturonase gene expression in transgenic tomatoes. Nature 334 724–726. [Google Scholar]
- Spence, J., Vercher, Y., Gates, P., and Harris, N. (1996). Pod shatter in Arabidopsis thaliana, Brassica napus and B. juncea. J. Microsc. 181 195–203. [Google Scholar]
- Tadiello, A., Pavanello, A., Zanin, D., Caporali, E., Colombo, L., Rotino, G.L., Trainotti, L., and Casadoro, G. (2009). A PLENA-like gene of peach is involved in carpel formation and subsequent transformation into a fleshy fruit. J. Exp. Bot. 60 651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman, D.M., Ciardi, J.A., Taylor, M.G., and Klee, H.J. (2001). Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J. 26 47–58. [DOI] [PubMed] [Google Scholar]
- Uimari, A., Kotilainen, M., Elomaa, P., Yu, D., Albert, V.A., and Teeri, T.H. (2004). Integration of reproductive meristem fates by a SEPALLATA-like MADS-box gene. Proc. Natl. Acad. Sci. USA 101 15817–15822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoeven, R., Ronning, C., Giovannoni, J., Martin, G., and Tanksley, S. (2002). Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14 1441–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., Schuch, W., and Giovannoni, J. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296 343–346. [DOI] [PubMed] [Google Scholar]
- Wang, H., Jones, B., Li, Z., Frasse, P., Delalande, C., Regad, F., Chaabouni, S., Latche, A., Pech, J.C., and Bouzayen, M. (2005). The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17 2676–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T., and Abbott, J. (2002). Firmness and force relaxation characteristics of tomatoes stored intact or as slices. Postharvest Biol. Technol. 24 59–68. [Google Scholar]
- Zahn, L.M., Leebens-Mack, J.H., Arrington, J.M., Hu, Y., Landherr, L.L., dePamphilis, C.W., Becker, A., Theissen, G., and Ma, H. (2006). Conservation and divergence in the AGAMOUS subfamily of MADS-box genes: Evidence of independent sub- and neofunctionalization events. Evol. Dev. 8 30–45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.