Abstract
The plant hormone abscisic acid (ABA) orchestrates plant adaptive responses to a variety of stresses, including drought. This signaling pathway is regulated by reversible protein phosphorylation, and genetic evidence demonstrated that several related protein phosphatases 2C (PP2Cs) are negative regulators of this pathway in Arabidopsis thaliana. Here, we developed a protein phosphatase profiling strategy to define the substrate preferences of the HAB1 PP2C implicated in ABA signaling and used these data to screen for putative substrates. Interestingly, this analysis designated the activation loop of the ABA activated kinase OST1, related to Snf1 and AMPK kinases, as a putative HAB1 substrate. We experimentally demonstrated that HAB1 dephosphorylates and deactivates OST1 in vitro. Furthermore, HAB1 and the related PP2Cs ABI1 and ABI2 interact with OST1 in vivo, and mutations in the corresponding genes strongly affect OST1 activation by ABA. Our results provide evidence that PP2Cs are directly implicated in the ABA-dependent activation of OST1 and further suggest that the activation mechanism of AMPK/Snf1-related kinases through the inhibition of regulating PP2Cs is conserved from plants to human.
INTRODUCTION
Reversible protein phosphorylation was selected through evolution for the fine-tuning of signaling pathways. The phosphorylation state of Ser, Thr, and Tyr is regulated by the orchestrated activity of protein kinases and phosphatases (Hunter, 1995; Moorhead et al., 2009). However, since the discovery of reversible protein phosphorylation, protein kinases have received more attention, emphasizing their driving role in this regulation mechanism. Consequently, our knowledge of protein phosphatase specificity and regulation is still comparatively limited, thus affecting our general understanding of the fine regulation of signaling pathways.
Ser/Thr phosphatases of the PPP family (PP1, PP2A, and PP2B) are obligate multimeric enzymes. Their distinctive localization, regulation, and substrate specificity are essentially conferred by the large variety of regulatory subunits rather than the promiscuous activity of the few catalytic subunits (Virshup and Shenolikar, 2009). By contrast, Ser/Thr phosphatases of the unrelated PPM family, represented by protein phosphatase 2Cs (PP2Cs), are monomeric proteins. The significantly larger number of genes encoding PP2Cs compared with other phosphatase catalytic subunits in mammals and in plants, suggests that their regulation and substrate specificity might be defined by an increased catalytic specificity and by the presence of regulatory domains frequently associated with the catalytic domain (Schweighofer et al., 2004; Moorhead et al., 2007). PP2Cs act as general negative regulators of stress signaling in yeast and mammals through the regulation of the stress-activated mitogen-activated protein kinase pathway, DNA damage signaling, or the dephosphorylation of AMP-activated kinase (AMPK), the mammalian counterpart of yeast Snf1 kinase (Lammers and Lavi, 2007).
In plants, PP2Cs represent by far the major phosphatase family, with, for example, 76 PP2Cs out of 112 phosphatases encoded in the Arabidopsis thaliana genome (Schweighofer et al., 2004). Genetic evidence revealed that group A PP2Cs, including the related HAB1, ABI1, and ABI2 PP2Cs, are negative regulators of the stress hormone abscisic acid (ABA) signaling. ABA is synthesized in response to drought stress and triggers both the rapid closure of stomatal pores in leaf epidermis to limit water loss through transpiration and the induction of new genetic programs to help the plant withstand this adverse condition (Himmelbach et al., 2003; Hirayama and Shinozaki, 2007; Sirichandra et al., 2009). Loss-of-function and knockout mutants in HAB1, ABI1, and ABI2 are hypersensitive to ABA (Gosti et al., 1999; Merlot et al., 2001; Leonhardt et al., 2004; Saez et al., 2004). Conversely, plants carrying the dominant mutations abi1-1 (abi1G180D) or abi2-1 (abi2G168D) or plants expressing the mutant hab1G246D protein are strongly insensitive to ABA (Leung et al., 1994, 1997; Meyer et al., 1994; Rodriguez et al., 1998; Robert et al., 2006). The distinctive type of mutation in these mutants corresponds to the substitution of a conserved Gly with the negatively charged Asp (G to D) in the phosphatase catalytic center and dramatically reduces the phosphatase activity toward phospho-casein used as heterologous substrate (Leung et al., 1997; Gosti et al., 1999; Robert et al., 2006). However, recent studies revealed that the PP2C G-to-D mutation genetically behaves as a hypermorphic mutation in planta, suggesting that the mutant phosphatase has an exaggerated function compared with the wild-type protein in vivo (Robert et al., 2006; Moes et al., 2008).
Several proteins interacting with the PP2Cs implicated in ABA signaling have been identified. Recently, the Arabidopsis SWI3B subunit of SWI/SNF chromatin-remodeling complex was shown to interact with HAB1 (Saez et al., 2008). ABI1 interacts with the At HB6 transcription factor (Himmelbach et al., 2002), the stress-activated MPK6 kinase (Leung et al., 2006), and the ABA-activated kinase OST1 (Yoshida et al., 2006). OST1, also called SnRK2.6 and SRK2E, belongs to the plant AMPK/Snf1-related kinases family (SnRK). Kinases of the SnRK1, SnRK2, and SnRK3 subfamilies share strong sequence identity in their kinase domains but have divergent C-terminal domains regulating their activity in response to a variety of metabolic and stress signaling pathways (Hrabak et al., 2003; Halford and Hey, 2009). In addition, both ABI2 and ABI1 were shown to interact with the glutathione peroxidase ATGPX3 (Miao et al., 2006) and with the SnRK3 kinases PKS3 and SOS2, although with different apparent affinities (Guo et al., 2002; Ohta et al., 2003). The implication of these PP2C-interacting proteins in ABA signaling was confirmed by phenotypic analysis of the corresponding mutant plants. However, the direct dephosphorylation of these proteins by PP2Cs was not demonstrated; therefore, we do not know whether they are physiological substrates or regulators, or whether, together with the PP2Cs, they form part of signaling complexes that regulate ABA responses.
Here, we developed a phosphatase profiling strategy using a combinatorial phosphopeptide array to precisely characterize the substrate preferences of PP2Cs involved in ABA signaling and used HAB1 preferences in a bioinformatics screen to identify putative substrates. Our results demonstrate that the conserved phosphorylated Ser-175 (pS175) residue located in the activation loop of the SnRK2 kinase OST1 is a target of HAB1. We also provide experimental evidence that OST1 is directly dephosphorylated and regulated by HAB1, ABI1, and ABI2 in vivo. Our results demonstrate that PP2Cs are involved in the ABA-dependant activation of SnRK2s and thus play a critical role at an early step of ABA signaling.
RESULTS
A Protein Phosphatase Profiling Strategy Using a Combinatorial Phosphopeptide Array
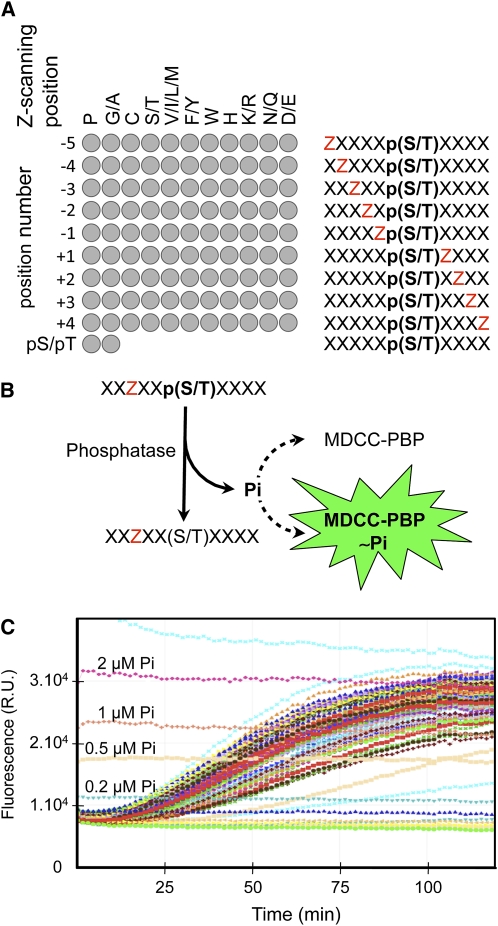
To identify the dephosphorylation preferences of PP2Cs involved in ABA signaling, we developed a combinatorial phosphopeptide array screening strategy using the Escherichia coli phosphate binding protein coupled to the MDCC fluorophore (MDCC-PBP) as a phosphate sensor to continuously measure peptide dephosphorylation (Brune et al., 1994; Pais et al., 2005). Our combinatorial library is composed of semidegenerate phosphopeptides incorporating an equimolar mixture of target pS and pT to report phosphatase activity (Figure 1). Each peptide also contains a Z-scanning residue that is either a single amino acid (P, C, W, or H) or an equimolar mixture of amino acids grouped according to their physicochemical properties ([GA], [ST], [VILM], [FY], [KR], [NQ], or [DE]). The Z position scans the region spanning from positions −5 to +4 around the phosphorylated residues, while degenerate X positions correspond to an equimolar combination of all amino acids except Cys. To these 99 semidegenerate peptides, we included two entirely degenerate peptides containing either a pS or a pT to analyze the preference of the phosphatases toward these phospho-residues (Figure 1A).
Figure 1.
Profiling Strategy to Define Dephosphorylation Preferences of Ser/Thr Phosphatases.
(A) Our phosphatase profiling strategy relies on an array of 101 semidegenerate phosphopeptides (left diagram). Each peptide of the array (right diagram) contains a Z-scanning position (red Z) successively moving from positions −5 to +4 with respect to the fixed target dephosphorylation site (i.e., position 0), which correspond to an equimolar mix of pS and pT. The Z-scanning position is either occupied by a single amino acid (e.g., P) or a limited mixture of amino acids with similar physicochemical properties (e.g., V, I, L, and M). The degenerate X positions consist of an equimolar mix of all amino acids except C. In addition, we used two independant degenerate phosphopeptides, containing either a pS or a pT, to test the selectivity of the phosphatase toward these two phosphorylated amino acids.
(B) The 101 phosphopeptides were dephosphorylated in solution by the protein phosphatase in the presence of MDCC-PBP, the fluorescent free-phosphate (Pi) sensor.
(C) The dephosphorylation kinetics of the 101 phosphopeptides were recorded in parallel and in real time for 2 h by monitoring the fluorescent signal (R.U., relative unit) increase resulting from the hydrolysis of Pi. The fluorescent signals corresponding to 2, 1, 0.5, and 0.2 μM Pi are indicated on the graph.
The peptides were incubated with protein phosphatases in solution, and the kinetics of dephosphorylation were recorded in real time as an increase of fluorescence emission from the sensor as it complexes with the liberated phosphate (Pi) (Figures 1B and 1C). In our assay conditions, we routinely obtained a logarithmic relation between the fluorescent signal and Pi concentration ranging from 0.2 to 2 μM. In addition, we only observed a weak fluorescent signal loss (<10%) due to the fluorophore bleaching during the 2-h time course of our experiments (80 time points). These characteristics make the MDCC-PBP a very robust and sensitive Pi sensor, allowing the precise calculation of the dephosphorylation speed of the hundred phosphopeptides of the library. The impact or weight of each individual or group of amino acids at each position on dephosphorylation of the target pS/T is calculated as the ratio between the dephosphorylation speed of the corresponding peptide and the mean value of all peptides for a given position. The substrate preference of the phosphatase is illustrated by its position-specific scoring matrix (PSSM) that displays the weight of each amino acid at each position around the phosphorylated site as presented in Figure 2.
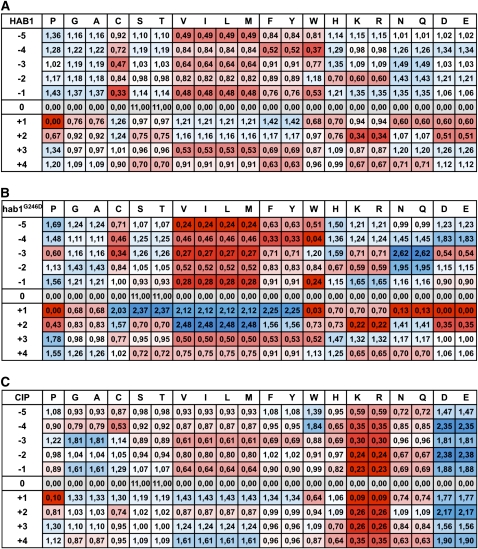
Figure 2.
Substrate Preferences of the Protein Phosphatases HAB1 and hab1G246D.
PSSM of HAB1 (A), hab1G246D (B), and CIP (C). The weight (w) of each amino acid at each position reported in the PSSM corresponds to the ratio between the speed of dephosphorylation of the corresponding peptide and the mean speed of peptide dephosphorylation at a given position. Weights were colored from blue (1<w≤2.5 and over) to red (1 > w ≥ 0.2 and under) using Excel conditional formatting.
The G246-to-D Mutation Narrows the Specificity of HAB1 Phosphatase
The phosphatase profiling revealed that HAB1 has a rather broad preference pattern toward semidegenerate phosphopeptides (Figure 2A) and a slight bias for pT over pS (see Supplemental Figure 1 online). The latter observation contrasts with the usual strong preference observed for other PP2Cs toward pT (Marley et al., 1996; Donella-Deana et al., 2003). The strongest positive effect is observed for [NQ] at position −3, which increases dephosphorylation by 50% above the mean value. By contrast, a P at the +1 position virtually abrogates dephosphorylation, and [VILM] at position −1 and [KR] at position +2 reduces dephosphorylation by 50 and 65%, respectively. We obtained very similar results with wild-type ABI1 and ABI2 PP2Cs (see Supplemental Figure 2 online), which are homologs of HAB1.
As a point of comparison, we analyzed the substrate preferences of calf intestinal phosphatase (CIP), which is considered as a broad specificity protein phosphatase. CIP has a clear preference for peptides containing acidic amino acids but has a strong aversion to basic amino acids containing peptides (Figure 2C). CIP is still able to dephosphorylate peptides containing P at the +1 position and displays a fourfold preference for pS over pT, as previously observed (Donella-Deana et al., 2003; Sun et al., 2008). These results indicate that the profiling strategy we developed is able to reveal subtle substrate preferences between phosphatases.
We then analyzed the effect of the dominant G-to-D mutation on the phosphatase specificity of HAB1 (hab1G246D). In comparison with the crystal structure of human PP2Cα, this mutation is predicted to localize in the close proximity of the catalytic site and would likely affect its activity (Das et al., 1996; Robert et al., 2006). The hab1G246D mutant protein is 10 times less active than HAB1 against the degenerate pT peptide but maintains a preference for pT over pS (see Supplemental Figure 1 online). The comparison of HAB1 and hab1G246D phosphatase profiles revealed that the dominant G-to-D mutation narrows the specificity of the phosphatase (Figure 2B). The presence of [DE] at position +1 of the peptide substrates abrogates its dephosphorylation, indicating that this position is particularly sensitive to the negative charge and/or the steric constraints associated with the G-to-D mutation. The mutation also accentuates the negative effect of [NQ] and W at position +1 and of [KR] and [DE] at position +2. This reduction of activity is compensated by an apparent positive impact of [ST], [VILM], and [FY] at position +1 and [VILM] at position +2. The positive effect of [NQ] at position −3 observed for HAB1 is also amplified by hab1G246D. This analysis suggests that the dominant G246D mutation narrows the intrinsic phosphatase specificity of HAB1. We observed a similar, although less pronounced, effect of the equivalent G180D and G168D mutations on ABI1 and ABI2 activity, respectively (see Supplemental Figure 2 online), indicating that the dominant mutation has a comparable effect on the activity of other group A PP2Cs.
hab1G246D Substrate Preferences Reveal the Activation Loop of the Kinase OST1 as One of Its Putative Targets
We compiled the HAB1 substrate preferences into a PSSM to screen the Arabidopsis protein database using the MAST program. However, because of the broad substrate preferences of HAB1, we were not able to discriminate between putative substrates according to the calculated position P value of the target peptides. On the other hand, while the molecular consequences of the G-to-D mutation on the activity or regulation of group A PP2Cs is still obscure, the genetic nature of the mutation implies that hab1G246D is still able to dephosphorylate at least one of the physiological substrates of HAB1 (Wilkie, 1994). We therefore used the more restrictive hab1G246D preferences to screen for putative HAB1 substrates. To further focus our search toward physiological substrates, we limited our screen to genes that, like HAB1, are strongly expressed in stomata (Leonhardt et al., 2004; Saez et al., 2004). This bioinformatics screen identified several putative hab1G246D substrates, including the AMPK/Snf1-related kinase OST1 (Table 1). OST1 plays an important role in stomatal closure and gene induction in response to ABA (Mustilli et al., 2002; Yoshida et al., 2002). The predicted target site is pS175, located in the activation loop of OST1 that was previously shown to play a critical function in OST1 activation in vitro and in vivo (Belin et al., 2006; Boudsocq et al., 2007). These data suggest that hab1G246D, and consequently HAB1, can dephosphorylate pS175 to directly regulate OST1 activity.
Table 1.
hab1G246D Putative Substrates Expressed in Stomata
| Ranka | AGIb | Description | Peptide Sequence | Position P Valuec |
|---|---|---|---|---|
| #1 | AT4G34110.1 | PAB2, poly(A) binding protein | PEQQRTMLGE | 9.9e-07 |
| #2 | AT3G02360.1 | 6-Phosphogluconate dehydrogenase | ARNGPSMMPG | 4.1e-06 |
| #3 | AT5G56630.1 | Phosphofructo-1-kinase | STNQPSFLGP | 5.4e-06 |
| #4 | AT3G27690.1 AT2G05070.1 | Photosystem II light-harvesting complex gene 2 | SENTPSYLTG | 2.1e-05 |
| #5 | AT4G29220.1 | 6-Phosphofructokinase | STNQPSFMKQ | 2.4e-05 |
| #5 | AT2G22240.1 | Inositol-3-phosphate synthase | GGNNGSTLTA | 2.4e-05 |
| #7 | AT5G13630.1 | GUN5, magnesium chelatase subunit | PENAETLIEE | 2.8e-05 |
| #7 | AT4G26850.1 | VTC2, GDP-l-galactose/GDP-d-glucose phosphorylase | PENSPSVVAI | 2.8e-05 |
| #9 | AT2G25450.1 | 2-Oxoglutarate-dependent dioxygenase | KPNGNSSLDH | 3.2e-05 |
| #10 | AT3G19800.1 | Unknown protein | RENRASTLPE | 3.4e-05 |
| #11 | AT1G78820.1 | Curculin-like lectin family protein | DGQGPTSVND | 3.5e-05 |
| #11 | AT1G15690.1 | AVP1, vacuolar H+-pumping pyrophosphatase | YANARTTLEA | 3.5e-05 |
| #13 | AT4G09320.1 | NDPK1, nucleoside diphosphate kinase | PVNWQSSVHP | 3.8e-05 |
| #14 | AT2G23760.1 | BLH4, homeodomain transcription factor | NNNNNSTLHM | 4.3e-05 |
| #14 | AT1G07720.1 | 3-Ketoacyl-CoA synthase | SGNNRSMILA | 4.3e-05 |
| #16 | AT3G59660.1 | C2 domain-containing protein | DQQGPTIVHQ | 4.7e-05 |
| #17 | AT4G33950.1 | OST1, SnRK2-6, SRK2E, Snf1-related protein kinase | HSQPKSTVGT | 4.8e-05 |
| #18 | AT4G15530.1 | Pyruvate orthophosphate dikinase | QDQGVTVIPE | 4.9e-05 |
| #19 | AT5G54390.1 | HAL2-like phosphatidylinositol phosphatase | GDQNLSIVAE | 5.1e-05 |
| #20 | AT3G21055.1 | Photosystem II subunit T | KEQSSTTMRR | 5.4e-05 |
| #21 | AT4G32940.1 | Vacuolar processing enzyme gamma | ASQACTTLPT | 5.6e-05 |
| #22 | AT3G51370.1 | Protein phosphatase 2C | DTNQVSSVKG | 5.7e-05 |
| #23 | AT3G28710.1 | H+-transporting two-sector ATPase | AQNQKSRIHD | 5.8e-05 |
| #24 | AT1G03550.1 | Secretory carrier membrane protein | PFANHTSVPP | 6.0e-05 |
| #25 | AT5G17170.1 | ENH1, metal ion binding protein | DEQPDTYVCP | 6.2e-05 |
| #26 | AT5G12140.1 | Cys proteinase inhibitor | DQQAGTIVGG | 6.3e-05 |
| #27 | AT2G24520.1 | AHA5, plasma membrane proton ATPase | SEQEASILVP | 6.5e-05 |
| #28 | AT1G50200.1 | Alanyl-tRNA synthetase | PHNDPTLLFA | 6.9e-05 |
| #29 | AT1G27530.1 | Unknown protein | DPNTKSTLTR | 7.3e-05 |
| #30 | AT3G24170.1 | ATGR1, glutathione reductase | GRQEKTLMKL | 8.2e-05 |
| #31 | AT5G12250.1 AT5G44340.1 | β-Tubulin | PNNVKSSVCD | 8.5e-05 |
| #31 | AT5G62690.1 | β-Tubulin | PNNVKSTVCD | 8.5e-05 |
| #33 | AT3G23280.1 | Zinc finger family protein | DAQPRTVMPL | 9.0e-05 |
| #34 | AT2G26080.1 | ATGLDP2, Gly dehydrogenase | HRNGESSLLP | 9.7e-05 |
| #35 |
AT3G26744.1 |
ICE1, MYC-like bHLH transcriptional activator |
SNNNNTMLCG |
1.0e-04 |
The table is sorted by decreasing position P value of peptide match. Only peptide match with position P value ≤0.0001 is shown.
AGI, Arabidopsis Genome Initiative.
The position P value of a match is the probability of a single random subsequence of the length of the motif scoring at least as well as the observed match.
hab1G246D and HAB1 Dephosphorylate and Deactivate OST1 in Vitro
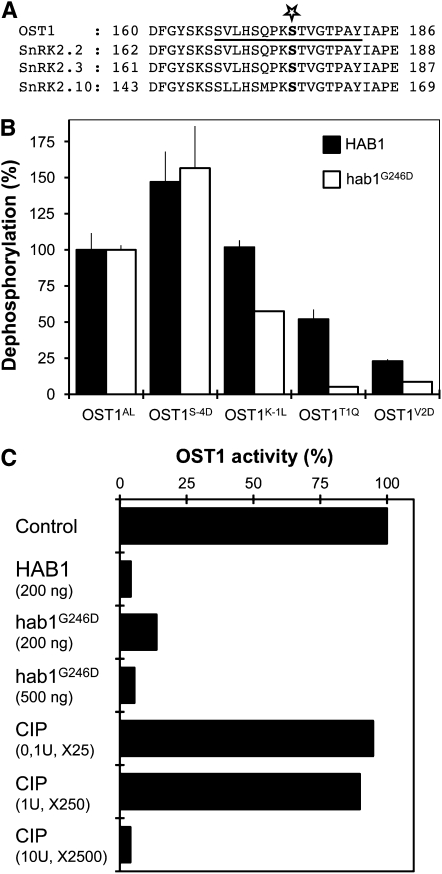
To demonstrate that OST1 is a substrate of hab1G246D and HAB1, we measured their phosphatase activity using the OST1AL peptide, which mimics the portion of the activation loop surrounding pS175, as substrate (Figure 3A). HAB1 dephosphorylates the OST1AL peptide (58.7 ± 6.8 pmol Pi·min−1·μg−1) more efficiently than hab1G246D (11.0 ± 0.4 pmol Pi·min−1·μg−1). We then introduced mutations in OST1AL to confirm the selectivity predictions deduced from the phosphopeptide array screening (Figure 3B). The substitution of K to L at −1 (OST1K-1L) reduces dephosphorylation by hab1G246D by >40%, but, in contrast with predictions, does not reduce dephosphorylation by HAB1. This discrepancy between the predicted and the experimental data for HAB1 may be the consequence of the degenerate nature of the peptide containing [VILM] at this position. The substitutions of T to Q (OST1T1Q) and V to D (OST1V2D) at the +1 and +2 positions reduce the dephosphorylation speed of HAB1 by 50 and 75%, respectively. These mutations have a stronger negative impact on hab1G246D dephosphorylation, resulting, for example, in a 50 times lower activity of hab1G246D against the OST1T1Q peptide (0.56 ± 0.01 pmol Pi·min−1·μg−1), compared with HAB1 (30.5 ± 3.9 pmol Pi·min−1·μg−1). Furthermore, the substitution of S at −4 by D (OST1S-4D) slightly increases the dephosphorylation by both HAB1 and hab1G256D. These results largely confirm the selectivity prediction for HAB1 and hab1G246D and further support the hypothesis that OST1 pS175 is a target of these PP2Cs.
Figure 3.
HAB1 and hab1G246D Dephosphorylate the OST1 Activation Loop in Vitro.
(A) Sequence alignment of OST1 activation loop with two closely related Arabidopsis SnRK2 kinases activated by ABA (SnRK2.2 and SnRK2.3) and the more distant SnRK2.10 kinase, activated by osmotic stress. The star indicates the position of Ser-175 and the underlined sequence corresponds to the OST1AL phosphopeptide.
(B) The phosphatase activity of HAB1 (black bars) and hab1G246D (white bars) was analyzed using OST1AL and variant mutant phosphopeptides. The activity of HAB1 (58.7 ± 6.8 pmol Pi·min−1·μg−1) and hab1G246D (11.0 ± 0.4 pmol Pi·min−1·μg−1) toward OST1AL peptide was normalized to 100%. Experiments were repeated independently two times with similar results. Results from one experiment are presented, and error bars represent se.
(C) The MBP kinase activity of OST1 (100 ng) expressed in E. coli is compared before and after incubation of the kinase with a given amount of HAB1, hab1G246D, and CIP phosphatases. The activity of CIP was normalized to HAB1 using the degenerate YAXXXXXpSXXXXAKKK as standard substrate. Using this peptide, the activity of 1 μg HAB1 is equivalent to 0.02 units of CIP. This experiment was repeated two times independently, with similar results. Data from one representative experiment are presented.
When expressed in E.coli, OST1 autophosphorylates on S175, resulting in an active kinase (Belin et al., 2006), and interacts with purified HAB1 and hab1G246D in vitro (see Supplemental Figure 3 online). Incubation of the active OST1 with both HAB1 and hab1G246D leads to the deactivation of the kinase (Figure 3C). In this assay, hab1G246D is only two to three times less active than HAB1 at deactivating OST1. This result significantly contrasts with the 40 times reduction linked to the G246D mutation observed when using phosphocasein as an artificial substrate of PP2Cs (Robert et al., 2006) and further suggests that hab1G246D might have a significant phosphatase activity against OST1 pS175 in vivo. By contrast, the deactivation of OST1 by CIP requires a considerable excess of alkaline phosphatase activity compared with HAB1, demonstrating the specificity of PP2Cs for OST1.
HAB1 and hab1G246D Interact with OST1 in Vivo
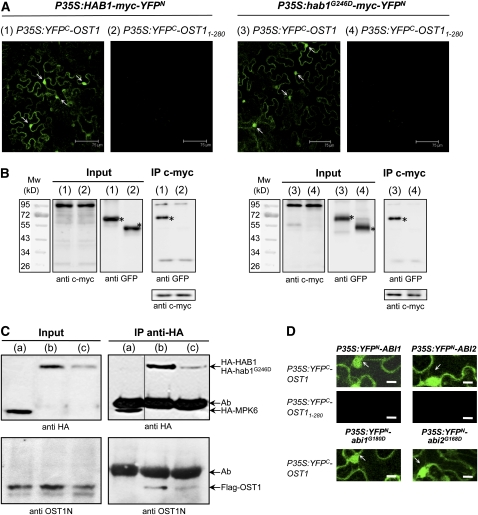
Next, we used bimolecular fluorescence complementation (BiFC) assay to test if HAB1 and hab1G246D interact with OST1 in vivo. HAB1 was translationally fused to the myc epitope and the N-terminal 155–amino acid portion of yellow fluorescent protein (YFPN) to create HAB1-myc-YFPN (Figures 4A and 4B). On the other hand, OST1 was fused to the C-terminal 84–amino acid portion of YFP to generate YFPC-OST1. Transient coexpression of the two proteins in tobacco (Nicotiana benthamiana) cells yielded YFP fluorescence in the nucleus and cytosol (Figure 4A). No fluorescence signal was observed when only one of the two proteins was expressed with the complementary YFP portion or when HAB1-myc-YFPN was expressed together with the truncated YFPC-OST11-280, lacking the C-terminal amino acid residues 281 to 362. These results indicating that HAB1 can interact with OST1 in vivo were confirmed by coimmunoprecipitation experiments using tobacco protein extracts prepared from the BiFC assay (Figure 4B). The expression of hab1G246D-myc-YFPN together with YFPC-OST1 also led to a fluorescent signal qualitatively similar to the one obtained with HAB1-myc-YFPN, indicating that the G246D mutation does not visibly affect the interaction between the phosphatase and the kinase (Figure 4A). Furthermore, BiFC experiments also revealed the interaction of OST1 with the two related group A PP2Cs, ABI1 and ABI2, as well as with their mutant forms, abi1G180D and abi2G168D in plant cells (Figure 4D; see Supplemental Figure 4 online).
Figure 4.
Wild-Type and G-to-D Mutant PP2Cs Interact with OST1 in Plant Cells.
(A) The interaction of HAB1 and hab1G246D with OST1 was analyzed by BiFC in Agrobacterium-infiltrated tobacco leaves. Confocal images of epidermal leaf cells expressing HAB1-myc-YFPN with either YFPC-OST1 (1) or YFPC-OST11-280 (2) or expressing hab1G246D-myc-YFPN with YFPC-OST1 (3) or YFPC-OST11-280 (4). The YFP fluorescent signal in the nucleus of epidermal cells is marked with arrows. Bars = 75 μm. All photomicrographs were recorded with the same settings for the microscope and the laser.
(B) The results of the BiFC experiments (1), (2), (3), and (4) presented in (A) were confirmed by coimmunoprecipitation (IP). The expression of the YFPN and YFPC fusion proteins in tobacco leaves was analyzed by immunoblotting using anti-c-myc and anti-GFP antibodies. Proteins immunoprecipitated by the anti-c-myc antibody were probed with anti-GFP antibodies to detect coimmunoprecipitation of YFPC-OST1 and YFPC-OST11-280. The asterisk indicates the size of the expected YFP-OST1 fusion protein.
(C) The molecular interaction between OST1 and both HAB1 and hab1G246D is confirmed by in vivo cross-linking experiments. Arabidopsis protoplasts coexpressing Flag-OST1 with HA-MPK6 (control) (a), HA-HAB1 (b), and HA-hab1G246D (c) were treated with the cross-linker DSP before immunoprecipitation of HA-tagged proteins. Soluble proteins (input) and proteins immunoprecipitated by the anti-HA antibody were analyzed by immunoblotting using anti-HA or anti-OST1N antibodies. Background band (Ab) corresponds to anti-HA immunoglobulin heavy chain.
(D) BiFC experiments as in (A) to analyze the interaction of OST1 with ABI1 and ABI2 and their G-to-D mutant forms. Bars = 25 μm. Larger images are presented in Supplemental Figure 4B online.
We confirmed the interaction between OST1 and both HAB1 and hab1G246D using in vivo cross-linking experiments (Figure 4C). Proteins from protoplasts cotransformed with Flag-OST1 and either HA-HAB1 or HA-hab1G246D were cross-linked in vivo. Our results show that Flag-OST1 coimmunoprecipitated with HA-HAB1 and HA-hab1G246D but not with HA-tagged MPK6, attesting to the specificity of the interaction. The observation that less OST1 coprecipitated with hab1G246D than with HAB1 is likely the consequence of the lower expression of HA-hab1G246D in protoplasts rather than of the reduction of affinity caused by the G246D mutation. Together, these results show that both HAB1 and hab1G246D bind to OST1 in vivo. While the interaction of ABI1 and abi1G180D with OST1 had been previously reported using a yeast two-hybrid assay (Yoshida et al., 2006), our results also reveal the interaction with HAB1, ABI2, and their mutant forms in vivo.
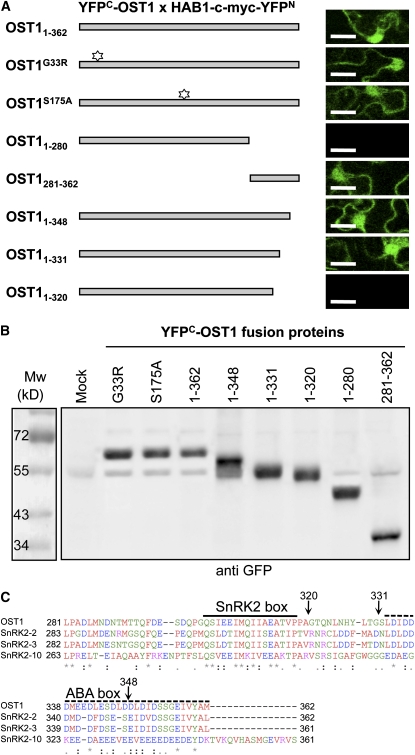
To determine specific regions of OST1 involved in the interaction with HAB1, we repeated BiFC experiments using different OST1 mutant and deleted forms (Figure 5A) and confirmed the expression of these fusion proteins in plant cells by immunoblot analysis (Figure 5B). BiFC experiments using the inactive kinases ost1G33R and ost1S175A indicated that the interaction with HAB1 does not require an active kinase or the target S175. We therefore suspected the presence of a HAB1 docking site in the C-terminal extension of OST1, which was shown to be important for its activation by ABA and for the interaction with ABI1 in yeast (Belin et al., 2006; Yoshida et al., 2006). Accordingly, YFPC-OST1281-362 interacts with HAB1-myc-YFPN, indicating that the OST1 C-terminal extension is necessary and sufficient for the interaction with HAB1 (Figure 5). We then used variant C-terminal deletions of YFPC-OST11-362 to further define the domain of interaction with HAB1. HAB1-myc-YFPN interacts with YFPC-OST11-348 and YFPC-OST11-331 but not with YFPC-OST11-320, further indicating that the OST1 region from amino acids 331 to 362 is not absolutely required for the interaction with HAB1 in vivo.
Figure 5.
The Interaction of OST1 with HAB1 Depends on the Regulatory C-Terminal Domain of OST1.
(A) BiFC analysis of the interaction between HAB1 and different OST1 mutant (G33R and S175A indicated with a star) and deletion forms, as depicted on the left. Protein interaction is revealed by YFP fluorescence analyzed by confocal microscopy (right column). Bars = 25 μm. Larger images are presented in Supplemental Figure 4C online.
(B) The expression in tobacco leaves of the different OST1 mutant and deletion forms used in (A) is verified by immunoblot using the anti-GFP antibody.
(C) Sequence alignment of the OST1 C-terminal regulatory domain with the two closely related Arabidopsis SnRK2 kinases, which are activated by ABA (SnRK2.2 and SnRK2.3), and the more distant SnRK2.10 kinase, which is only activated by osmotic stress. The sequence alignment was realized using ClustalW2. Identical residues (asterisk), conserved substitutions (colon), and semiconserved substitutions (period) are indicated below the alignment. The motifs conserved in all SnRK2s (SnRK2 box) or in ABA-activated SnRK2s (ABA box) are indicated by plain and dotted lines, respectively (Belin et al., 2006). Black arrows indicate the position of the OST1 deletions used in this study.
OST1 Activation by ABA Is Dependent on PP2C Activity in Planta
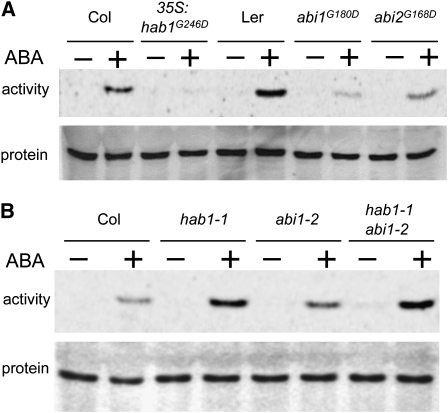
If OST1 is dephosphorylated and negatively regulated by HAB1 in planta, then both loss-of-function and hypermorphic mutations in HAB1 should impact the activation of OST1 in response to ABA. We specifically immunoprecipitated OST1 from hab1-1 knockout plants ectopically expressing hab1G246D treated or not with ABA and measured OST1 activity toward myelin basic protein (MBP) using an in-gel kinase assay (Figure 6A). The results showed that, while OST1 was rapidly and strongly activated in wild-type plants by ABA, this activation was barely detectable in hab1G246D-expressing plants. We also performed this experiment in abi1G180D and abi2G168D mutants and observed that OST1 activation was also strongly reduced. These results confirmed that abi1G180D inhibits ABA-dependent activation of OST1 (Mustilli et al., 2002; Yoshida et al., 2006; Boudsocq et al., 2007) and interestingly revealed a similar effect of abi2G168D, which was not previously observed (Yoshida et al., 2006; see Discussion).
Figure 6.
OST1 Activation by ABA Is Dependent on PP2C Activity in Planta.
(A) Activation of OST1 by ABA in different Arabidopsis mutants expressing G-to-D mutant PP2Cs is analyzed by in-gel kinase assay after endogenous OST1 immunoprecipitation. OST1 activation in plantlets expressing hab1G246D is compared with the corresponding Columbia (Col) wild-type ecotype. OST1 activation in the abi1G180D (abi1-1) and abi2G168D (abi2-1) mutant is compared with that in the Landsberg erecta (Ler) wild-type ecotype. The endogenous OST1 protein level was monitored in parallel by immunoblotting analysis of the soluble protein extract with the anti-OST1N antibody.
(B) Same experiments as in (A) performed in single and double knockout mutants in HAB1 (hab1-1) and ABI1 (abi1-2) genes. These mutants are in the Col ecotype.
By contrast, the ABA-dependent activation of OST1 is increased in the hab1-1 knockout mutant (Figure 6B), but the mutation does not visibly affect the activity of OST1 in the absence of ABA. The ABI1 knockout mutation abi1-2 has a comparable effect on ABA-dependent activation of OST1, but no synergistic or cumulative effect was observed in the double hab1-1 abi1-2 mutant (Figure 6B). These results provide genetic evidence that the PP2Cs HAB1, ABI1, and ABI2 regulate the activation of OST1 in vivo.
DISCUSSION
The primary goal of this work was to identify the substrate(s) of group A PP2Cs, including HAB1, ABI1, and ABI2, which are key elements in the ABA signaling pathway. This pathway plays a crucial role for plant survival under drought stress and coordinates plant growth and development with stress adaptive responses. Using HAB1 and hab1G246D substrate preferences, we were able to identify pS175, located in the activation loop of the AMPK/Snf1-related kinase OST1, as a putative substrate suggesting that HAB1 regulates OST1 activity. This prediction was subsequently supported by in vitro dephosphorylation assay, in vivo interaction assays, and the analysis of OST1 activation in PP2C mutant plants, which provided strong correlative evidence that these PP2Cs play an important and conserved role in the regulation of OST1 activity.
To quantitatively measure the substrate preferences of PP2Cs, we developed an original protein phosphatase profiling strategy using a combinatorial peptide library. Combinatorial peptide array screening strategies are now largely used to reveal the substrate preferences of protein kinases (Manning et al., 2002; Hutti et al., 2004; Doppler et al., 2005; Vlad et al., 2008), but this strategy has only rarely been used with protein phosphatases (Wang et al., 2002). In addition, the analysis of the substrate preferences of Ser/Thr phosphatases using peptide libraries was further limited until recently because of the absence of methods to easily and sensitively detect dephosphorylation of Ser and Thr in high-throughput assays (Sun et al., 2008). Our results show that the simultaneous dephosphorylation of hundreds of peptides can easily and quantitatively be recorded in real time through the release of inorganic phosphate using the MDCC-PBP fluorescent sensor (Brune et al., 1994; Pais et al., 2005). Because of its versatility, sensitivity, and the possibility of measuring phosphate release in real time, we think the MDCC-PBP sensor offers technical advantages over techniques that rely on phosphotyrosine-specific antibody (Kohn et al., 2007) or ProQ-Diamond (Sun et al., 2008) for the calculation of peptide dephosphorylation speed in high-throughput strategies. Furthermore, if necessary, the resolution and versatility of the strategy presented here can be increased using a single amino acid at each Z-scanning position and by designing a similar phosphotyrosine-based peptide library to study phosphotyrosine and dual-specific phosphatases.
These quantitative profiling data can be used to help the identification of phosphatase substrates using a bioinformatic approach. However, we think that bioinformatic screens exclusively based on these profiling data would generate a detrimental amount of false positives in the case of protein phosphatases with low substrate specificity, such as HAB1. We show with this work that this strategy will be valuable when combined with other sets of data, such as gene coexpression and coregulation or protein interactome analysis, to limit the number of potential substrates. In this study, we took advantage of both transcriptomic analysis of guard cells and the specific effect of the hab1G246D mutation to identify putative HAB1 substrates. The hypermorphic G-to-D mutation significantly narrows the specificity of hab1G246D, suggesting that the mutation might restrict the spectra of substrates compared with wild-type HAB1. Nevertheless, the hypermorphic nature of the G-to-D mutation also implies that the mutant phosphatase is still able to dephosphorylate at least one of the wild-type HAB1 substrates in ABA signaling. In addition to the SnRK2 kinase OST1, the bioinformatic screen revealed other putative substrates, including the chloroplastic GUN5 Mg-chelatase H subunit. This protein is, controversially, considered to be an ABA receptor (Shen et al., 2006; Muller and Hansson, 2009); however, because HAB1 does not localize in chloroplasts (Saez et al., 2008), we did not consider this protein as a likely HAB1 substrate and focused on OST1. Further experiments are needed to test if other candidate proteins correspond to physiological HAB1 substrates.
We have subsequently shown that HAB1 directly binds in vivo to the regulatory C-terminal domain of OST1 (Figure 5). We think the presence of a PP2C docking site in the kinase regulatory domain (OST1280-362), outside of the kinase domain, is an important component of the specificity of HAB1 toward OST1, likely bringing the catalytic domain of the PP2C in close proximity to the activation loop to favor the dephosphorylation of pS175.
Our results also reveal that ABI1 and ABI2 play a similar role as HAB1 in ABA signaling. ABI1, ABI2, and their mutant forms display very similar substrate preferences as HAB1 and hab1G246D (Figure 2; see Supplemental Figure 1 online). ABI1 and ABI2 also interact with OST1 in vivo (Figure 4D), and mutations in the corresponding PP2C genes significantly affect ABA-dependent OST1 activation (Figure 6). The direct regulation of OST1 activity by HAB1 and ABI1, but also ABI2, might modify our understanding of ABA signaling. In contrast with the Arabidopsis abi2G168D mutant, abi1G180D and ost1 mutants do not produce reactive oxygen species in response to ABA, suggesting that ABI1 and OST1 act upstream of reactive oxygen species, while ABI2 plays a role downstream of this secondary messenger (Murata et al., 2001; Mustilli et al., 2002). Accordingly, the ABA-dependent activation of OST1 is strongly reduced in the abi1G180D mutant, but abi2G168D does not affect the activation of ectopically expressed OST1 fused to green fluorescent protein (GFP), further suggesting that ABI1, but not ABI2, acts upstream of OST1 (Mustilli et al., 2002; Yoshida et al., 2006). However, we observed that the abi2G168D mutation negatively affects the activation of endogenous OST1 by ABA (Figure 6A). We think that the effect of abi2G168D on ABA-dependent OST1 activation was not previously observed because a significant fraction of the ectopically expressed GFP-OST1 kinase might not have been expressed together with abi2G168D in the same cells and therefore was not affected by the mutant phosphatase. ABI1 and abi1G180D were previously shown to interact with OST1 in yeast (Yoshida et al., 2006). However, the weaker interaction observed with the G-to-D mutant phosphatase led the authors to conclude that OST1 was not a direct substrate of ABI1. By contrast, our results support the idea that these PP2Cs play an important role in an early step of ABA signaling through the dephosphorylation and regulation of OST1 activity. Recently, it was shown that the role of ABI1 and abi1G180D as negative regulators of ABA signaling depends on their nuclear localization (Moes et al., 2008). Our BiFC experiments showing that OST1 interacts with the PP2Cs in the nucleus further suggest that the regulation of OST1 by PP2Cs takes place in this cellular compartment (Figure 4).
Our results also anticipate that these PP2Cs regulate SnRK2.2 and SnRK2.3, two ABA activated kinases homologous to OST1 (Boudsocq et al., 2004; Fujii and Zhu, 2009). The Arabidopsis abi1G180D and abi2G168D mutants are strongly insensitive to ABA for stomatal closure, root growth, and seed germination (Leung et al., 1994, 1997; Meyer et al., 1994), while ost1 only visibly affects ABA responses in stomata (Merlot et al., 2002; Mustilli et al., 2002; Yoshida et al., 2002). The analysis of the snrk2.2 snrk2.3 double knockout mutant revealed that these kinases are required for ABA responses in seeds and roots (Fujii et al., 2007). On the other hand, abi1G180D was shown to affect the activation of all SnRK2s activated by ABA (Boudsocq et al., 2007). Therefore, SnRK2.2 and SnRK2.3, which have the same activation loop as OST1 around S175 (Figure 3A) and share strong homologies in the regulatory C-terminal domain (Figure 5C), are likely targets of the PP2Cs in seeds and roots.
The results presented here do not exclude the possibility that these PP2Cs also perform specific functions at other steps of ABA signaling and therefore may have other physiological substrates, including SWI3B (Saez et al., 2008) and ATHB6 (Himmelbach et al., 2002), that were not revealed in our bioinformatics screen based on the stringent hab1G246D substrate preferences. However, the demonstration that these proteins are genuine substrates of these PP2Cs in ABA signaling further require that they be identified as phosphorylated proteins in vivo that can be directly dephosphorylated by PP2Cs.
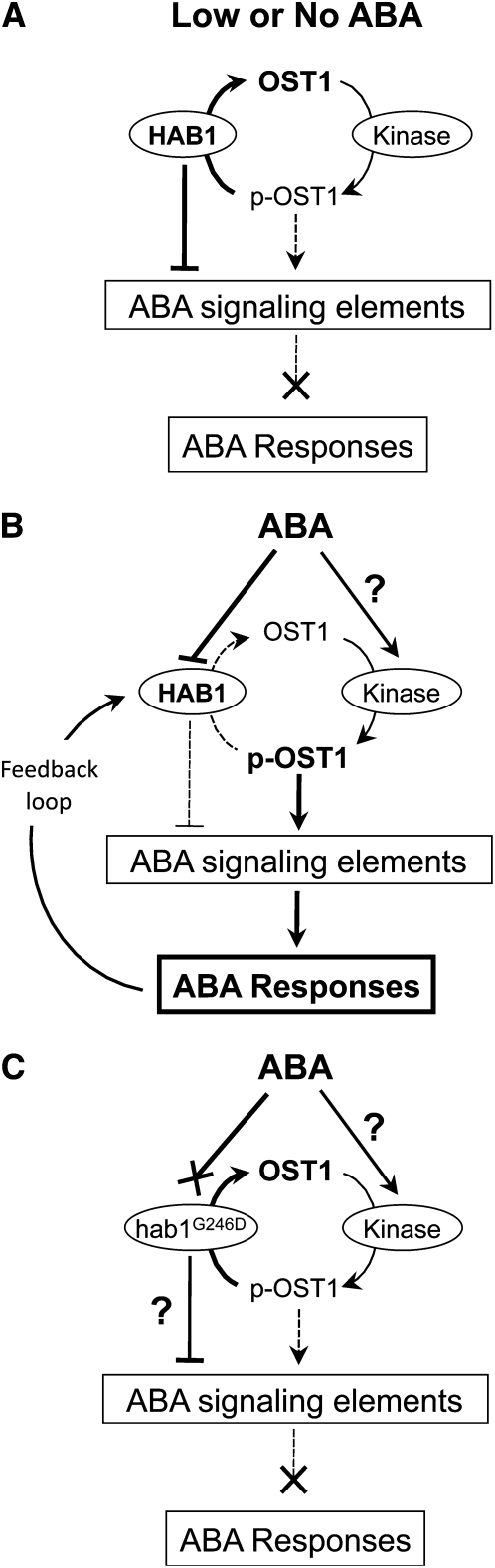
One remaining question is whether PP2Cs play an important role in the activation mechanism of OST1 by ABA. Our results indicate that the ABA-dependent OST1 activation is tightly dependent on PP2C activity in planta. The expression of hab1G246D, abi1G180D, and abi2G168D strongly reduce OST1 activation in response to ABA in plants. By contrast, the ABA-dependent activation of OST1 is significantly increased in the single hab1-1 or abi1-2 knockout mutants. This result is even more surprising if we consider that HAB1, ABI1, and ABI2 play partially redundant roles in ABA signaling in stomata (Merlot et al., 2001; Saez et al., 2004; Rubio et al., 2009), suggesting that even a mild reduction of PP2C activity results in increased OST1 activation. Therefore, we propose that OST1 is activated by ABA at least in part through the inhibition of the dephosphorylation of its activation loop by PP2Cs (Figure 7). While this article was in preparation, two articles reported the identification of a novel class of ABA sensors, called RCAR or PYR/PYL, which specifically binds and inhibits HAB1, ABI1, and ABI2 phosphatases in the presence of ABA (Ma et al., 2009; Park et al., 2009). This new finding provides functional support for our model. In addition to this proposed rapid negative regulation, PP2Cs are also positively regulated by ABA through a feedback loop that involves their transcriptional upregulation (Leung et al., 1997; Merlot et al., 2001; Leonhardt et al., 2004). This suggests that PP2Cs are both regulated negatively by the input ABA and positively by the ABA signaling output to precisely adjust the adaptive response to the strength and duration of the stress the plant is facing (Figure 7). In parallel to the role of PP2Cs, ABA-dependent activation of OST1 might also rely on the activation of upstream kinase(s) that phosphorylate S175 (Boudsocq et al., 2007). We did not observe a constitutive activation of OST1 in the absence of ABA in the single hab1-1 or abi1-2 mutants or in the double hab1-1 abi1-2 knockout mutant, and this suggests that the reduction of PP2C activity is not sufficient to activate OST1. In vitro, OST1 autophosphorylates on S175 leading to its activation (Belin et al., 2006). However, in vivo OST1 activation by ABA is not inhibited by 0.5 μM staurosporine, while OST1 is sensitive to this concentration of kinase inhibitor. This suggests that OST1 is phosphorylated on S175 by a staurosporine-resistant kinase in planta (Boudsocq et al., 2007). The activation of this kinase by ABA might also participate in the activation of OST1. It is now necessary to identify the kinase phosphorylating the OST1 activation loop to evaluate the role of this kinase on the activation of OST1 by ABA.
Figure 7.
Proposed Model for the Negative Regulation of OST1 Activity and ABA Responses by HAB1.
(A) In the absence of ABA, HAB1 dephosphorylates pS175, located in the OST1 activation loop, leading to the accumulation of the dephosphorylated inactive form of the kinase.
(B) Perception of ABA leads to the inhibition of HAB1 activity toward the OST1 activation loop. The underlying mechanism likely involves the RCARs/PYR/PYLs inhibitor proteins. The phosphorylated pS175 OST1 active form accumulates, triggering downstream ABA responses.
(C) hab1G246D PP2C activity toward the OST1 activation loop is proposed to be insensitive to the inhibition by ABA, leading to a constitutive dephosphorylation and inhibition of OST1 and therefore of ABA signaling.
Our results enable us to better understand the hypermorphic nature of the G-to-D mutation that was recently proposed to cause an exaggerated activity of the clade A PP2Cs in ABA signaling (Robert et al., 2006; Moes et al., 2008). So far, the genetic behavior of the G-to-D mutation was difficult to reconcile with its strong deleterious effect on both the PP2C activity when assayed against phosphocasein as heterologous substrate (Leung et al., 1997; Gosti et al., 1999; Robert et al., 2006) and the interaction with putative substrates (Guo et al., 2002; Himmelbach et al., 2002; Saez et al., 2008). However, it was recently shown that the abi1G180D mutation causes a preferential accumulation of the mutant PP2C in the nucleus, where it acts as a negative regulator of ABA signaling (Moes et al., 2008). The G-to-D mutation also disrupts the interaction between PP2Cs and their inhibitory proteins RCAR/PYR/PYL, suggesting that the mutant PP2Cs escape the negative regulation by ABA (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009). Furthermore, we show here that the G-to-D mutation only weakly reduces the in vitro activity of hab1G246D toward the OST1 activation loop (Figure 3C) and does not abrogate or visibly affect the interaction between the PP2Cs and OST1 in vivo (Figure 4). Together, these results suggest that the G-to-D mutant forms of PP2Cs dephosphorylate and deactivate the kinase OST1 in vivo, whether or not ABA is present, providing a tangible molecular explanation for the genetic hypermorphic nature of the G-to-D mutation in ABA signaling.
Mammalian PP2Cα dephosphorylates pT172 in the activation loop of AMPK in vitro, resulting in the deactivation of the kinase (Moore et al., 1991; Davies et al., 1995; Marley et al., 1996). More recently, it was proposed that the negative regulation of PP2C activity mainly participates in AMPK activation in response to an elevated AMP/ATP ratio following metabolic stress (Suter et al., 2006; Sanders et al., 2007). Our work supports that the activation mechanism of certain members of the AMPK/Snf1/SnRKs kinase family by the inhibition of regulating PP2Cs is conserved from plants to humans and therefore designates PP2Cs as key players in the regulation of stress signaling in eukaryotes.
METHODS
Protein Phosphatase Profiling and Bioinformatic Screen
The 101 semidegenerate phosphopeptides of the array were synthesized by Intavis AG as crude peptides at a 5-mg scale. The sequence of the peptides in this set have the common YAZXXXXp(S/T)XXXXAKKK structure. The fixed central position p(S/T), or position 0, is an equimolar mix of phosphorylated Ser and Thr. The scanning Z position, which moves from position −5 to +4, is successively occupied by single amino acids (P, C, W, and H) or an equimolar mix of amino acids grouped according to their physicochemical properties ([GA], [ST], [VILM], [FY], [KR], [NQ], and [DE]). The X positions represent an equimolar mix of all natural amino acids, except Cys, to avoid problems with oxidation. In addition to these 99 phosphopeptides, two degenerate phospho-S and phospho-T peptides (YAXXXXXpSXXXXAKKK and YAXXXXXpTXXXXAKKK) were added in this set to measure the preference of the phosphatases for pS and pT. The 101 phosphopeptides were first dissolved as 20 mM stock in DMSO and then diluted to 200 μM in 5 mM Tris-HCl, pH 7.4. The dephosphorylation of the peptides was analyzed in 384-well black plates (Greiner Bio-One; 781076) containing 5 μL of the 200 μM phosphopeptide solutions. In addition, inorganic phosphate (Pi) standards (from 8 to 0.002 μM) were added in different wells for absolute quantification of Pi. Phosphopeptides (50 μM final concentration) were simultaneously dephosphorylated at 25°C in 20 μL of reaction solution (50 mM Tris-HCl, pH 7.8, 20 mM magnesium acetate, 1 mM DTT, and 0.05% Tween 20), containing 0.5 μM phosphate sensor (Invitrogen; PV4406) and the protein phosphatase (0.15 to 7.5 ng/μL). In a preliminary study, we verified that we did not detect fluorescent background coming from contaminating phosphate up to a final phosphopeptide concentration of 50 μM. The dephosphorylation of peptides was recorded in real time for 2 h (at 90-s time points) as the increase of fluorescence of the phosphate sensor using a Tecan Infinite M200 (excitation 415 nm/emission 450 nm). The fluorescent signal was converted to an amount of free phosphate using a logarithmic Pi standard curve, and dephosphorylation speed (Vi) for each of the 101 phosphopeptides was calculated during the linear phase of the curve.
The weight of individual or a group of amino acids at each position from −5 to +4 corresponds to the ratio between Vi and the mean dephosphorylation speed at this position, called Vposition (Vposition= ∑Vi/11). At position 0, a weight of 11 is given to Ser and Thr. The weight of each amino acid at each position is reported in a PSSM. For in silico search of putative hab1G246D substrates, the MAST program (Bailey and Gribskov, 1998; Bailey et al., 2006) was used, as previously described (Vlad et al., 2008), using a protein database of ∼1300 Arabidopsis thaliana annotated genes that are the most expressed in guard cells (Yang et al., 2008; signal ≥2500 from experiment 1).
PP2C Activity Assay
The PP2Cs ABI1, ABI2, and HAB1 and their mutant forms used in this study were produced as glutathione S-transferase fusion proteins in Escherichia coli and purified using a standard protocol (Leung et al., 1997; Gosti et al., 1999; Robert et al., 2006). CIP was purchased from New England Biolabs. Defined sequence phosphopeptides were synthesized by Eurogentec as crude peptides as follows: OST1AL, SVLHSQPKpSTVGTPAY; OST1S-4D, SVLHDQPKpSTVGTPAY; OST1K-1L, SVLHSQPLpSTVGTPAY; OST1T1Q, SVLHSQPKpSQVGTPAY; and OST1V2D, SVLHSQPKpSTDGTPAY. Reaction conditions, data processing, and dephosphorylation speed calculation were the same as for the phosphopeptide array. All samples were duplicated.
OST1 Deactivation Assay
Active His-tagged OST1 protein kinase (100 ng) produced in E. coli (Belin et al., 2006) was incubated with protein phosphatase for 30 min at 25°C in 20 mM Tris-HCl, pH 7.8, 20 mM MgCl2, and 1 mM DTT. We then added NaF (10 mM), β-glycerophosphate (25 mM), and ATPγ32P (20 μM, 1 μCi·nmol−1) to a final volume of 30 μL and incubated the reaction for 90 min at 25°C in the presence of 5 μg MBP. The reaction was then analyzed by SDS-PAGE, and MBP phosphorylation was quantified by phosphor imaging using a Storm 840 and ImageQuant 5.2 software (Molecular Dynamics).
BiFC Interaction Experiments
BiFC experiments were performed using transient transfection of Nicotiana benthamiana leaves with Agrobacterium tumefaciens as previously described (Voinnet et al., 2003; Saez et al., 2008). The entire coding sequence of OST11-362 was amplified by RT-PCR and cloned into the pCR8/GW/TOPO entry vector (Invitrogen). The coding sequence of the OST1 deletions (OST11-348, OST11-331, OST11-320, and OST11-280) and the inactive kinases ost1G33R and ost1S175A were cloned into the pENTR201 entry vector (Belin et al., 2006). OST1281-362 was amplified with primers 5′-CTACCGGCAGATCTAATGAAC-3′ and 5′-TTTGTCGACTCACATTGCGTACACAATCT-3′ and cloned in pCR8/GW/TOPO. All these clones were recombined into the pYFPC43 vector (kindly provided by A. Ferrando, Universidad de Valencia, Spain), a derivative of pMDC43 (Curtis and Grossniklaus, 2003), to express YFPC-OST1 fusion proteins. HAB1-c-myc-YFPN and hab1G246D-c-myc-YFPN fusion proteins were expressed using pSPYNE-35S-HAB1 and pSPYNE-35S-hab1G246D constructs (Saez et al., 2008). The coding sequence of ABI1 and abi1G180D was amplified by RT-PCR with primers 5′-CCGGCCCTCGAGATGGAGGAAGTATCTCCGGC-3′ and 5′-CCGGCCCTCGAGTCAGTTCAAGGGTTTGCT-3′ using mRNA extracted from Ler and the abi1-1 mutant, respectively. The same strategy was used for ABI2 and abi2G168D using primers 5′-ATGGACGAAGTTTCTCCTGCAG-3′ and 5-TCAATTCAAGGATTTGCTCTT-3′. PCR fragments were cloned into pCR8/GW/TOPO and recombined by LR reaction into the pYFPN43 destination vector. A. tumefaciens C58C1 (pGV2260) transformed with the given constructs was injected into young, fully expanded leaves of N. benthamiana plants together with the silencing suppressor p19. YFP fluorescence of infected leaves was examined after 3 to 4 d with a Leica TCS-SL confocal microscope. For analysis of fusion protein expression, total proteins were extracted in 2× Laemmli buffer (125 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 2% mercaptoethanol, and 0.001% bromophenol blue) and analyzed by immunoblotting using a standard protocol with monoclonal anti-c-myc (clone 9E10; Roche) or anti-GFPC (clone JL-8; Clontech). Detection of primary antibody was performed using the ECL Advance Western Blotting Detection Kit (GE Healthcare). For immunoprecipitation experiments, soluble proteins were extracted in PBS containing 1 mM EDTA, 0.05% Triton X-100, and protease inhibitor cocktail (Roche). C-myc-tagged proteins were immunoprecipitated and purified using superparamagnetic micro MACS beads coupled to monoclonal anti c-myc antibody according to the manufacturer's instructions (Miltenyi Biotec). Purified immunocomplexes were eluted with hot Laemmli buffer and analyzed by immunoblotting as previously described.
OST1 in-Gel Kinase Assay
ABA-dependent activation of OST1 was analyzed in Arabidopsis mutant lines hab1-1, abi1-2, hab1-1 abi1-2 (Saez et al., 2006), abi1-1 and abi2-1 (Leung et al., 1997; abi1G180D and abi2G168D), hab1-1 transgenic plants expressing the mutant hab1G246D protein (Robert et al., 2006), and the corresponding Col and Ler wild-type ecotypes. Ten- to twelve-day-old plantlets grown in vitro were transferred to liquid Murashige and Skoog medium for 3 h before a 30-min treatment with 30 μM ABA, or ethanol as control, and then rapidly frozen in liquid nitrogen.
Soluble proteins were extracted and an in-gel kinase assay was performed as previously described (Boudsocq et al., 2007). Endogenous OST1 was immunoprecipitated at 4°C from 300 μg soluble protein extract using 5 μL of anti-OST1N serum (Belin et al., 2006) for 12 h followed by incubation with protein A-Sepharose CL-4B (Sigma-Aldrich; P3391) for 2 h. After three washes in immunoprecipitation buffer, the kinase activity of immunoprecipitated OST1 was revealed by an in-gel kinase assay and quantified by phosphor imaging. Endogenous OST1 expression in different mutant plant backgrounds was analyzed by immunoblotting using purified anti-OST1N antibody and horseradish peroxidase–conjugated protein A (Sigma-Aldrich; P8651) with ECL chemiluminescence substrate (GE Healthcare).
In Vivo Protein Cross-Linking and Coimmunoprecipitation Assays
Constructs to express tagged HA-HAB1, HA-hab1G246D (Robert et al., 2006), or HA-MPK6 were transfected together with the Flag-OST1 construct (the latter two constructs were kindly provided by M. Boudsocq, Institut des Sciences du Végétal) in Arabidopsis cell suspension protoplasts for transient protein expression as previously described (Boudsocq et al., 2004). Fifteen hours after transfection, protoplasts were treated for 30 min at room temperature with 2 mM DSP (Sigma-Aldrich; D3669). The in vivo protein cross-linking reaction was quenched by the addition of Tris-HCl, pH 7.5, to a final concentration of 25 mM for 15 min on ice. Proteins were extracted in lysis buffer (40 mM Tris-HCl, pH 7.5, 75 mM NaCl, 5 mM EDTA, 5 mM EGTA, 10 mM orthovanadate, 10 mM NaF, 60 mM β-glycerophosphate, 1 mM PMSF, 5 μg/mL leupeptin, 5 μg/mL antipain, and 1% Triton X-100) and immunoprecipitated with 20 μL of 50% slurry agarose conjugated anti-HA antibody (Clone HA-7; Sigma-Aldrich). Extracted proteins and immunocomplexes were solubilized in Laemmli buffer containing DTT and analyzed by immunoblotting using purified anti-OST1N antibody or anti-HA monoclonal antibody (Clone HA-7; Sigma-Aldrich).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At1g72770 (HAB1), At4g33950 (OST1), At4g26080 (ABI1), and At5g57050 (ABI2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. HAB1 and hab1G246D Show a Slight Preference for Phospho-Thr (pT).
Supplemental Figure 2. Position-Specific Scoring Matrix of ABI1, abi1G180D, ABI2, and abi2G168D.
Supplemental Figure 3. OST1 Interacts with HAB1 and ABI1 in Vitro.
Supplemental Figure 4. Wild-Type and G-to-D Mutant PP2Cs Interact with OST1 in Plant Cells.
Supplementary Material
Acknowledgments
We thank A. Ferrando for kindly providing the pYFPC43 vector and M. Boudsocq for kindly providing HA-MPK6 and Flag-OST1 constructs used in the cross-linking assays, as well as for precious advice on protoplast transformation. This work was supported in part by the Centre National de la Recherche Scientifique, Genoplante (ABRUPT/GNP05037G to J.L.) and the Ministerio de Educación y Ciencia and Fondo Europeo de Desarrollo Regional (BIO2008-00221 to P.L.R.). F.V. is supported by a Marie-Curie early-stage training fellowship ADONIS (MEST-CT-2005-020232 to J.L.), S.R. by a fellowship from the Consejo Superior de Investigaciones Científicas, and C.S. by a doctoral fellowship from the french Ministère de l'Enseignement Supérieur et de la Recherche.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Pedro L. Rodriguez (prodriguez@ibmcp.upv.es), Christiane Laurière (christiane.lauriere@isv.cnrs-gif.fr), and Sylvain Merlot (smerlot@noumea.ird.nc).
Online version contains Web-only data.
References
- Bailey, T.L., and Gribskov, M. (1998). Methods and statistics for combining motif match scores. J. Comput. Biol. 5 211–221. [DOI] [PubMed] [Google Scholar]
- Bailey, T.L., Williams, N., Misleh, C., and Li, W.W. (2006). MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34 W369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin, C., de Franco, P.O., Bourbousse, C., Chaignepain, S., Schmitter, J.M., Vavasseur, A., Giraudat, J., Barbier-Brygoo, H., and Thomine, S. (2006). Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 141 1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq, M., Barbier-Brygoo, H., and Lauriere, C. (2004). Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279 41758–41766. [DOI] [PubMed] [Google Scholar]
- Boudsocq, M., Droillard, M.J., Barbier-Brygoo, H., and Lauriere, C. (2007). Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol. Biol. 63 491–503. [DOI] [PubMed] [Google Scholar]
- Brune, M., Hunter, J.L., Corrie, J.E., and Webb, M.R. (1994). Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry 33 8262–8271. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, A.K., Helps, N.R., Cohen, P.T., and Barford, D. (1996). Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J. 15 6798–6809. [PMC free article] [PubMed] [Google Scholar]
- Davies, S.P., Helps, N.R., Cohen, P.T., and Hardie, D.G. (1995). 5′-AMP inhibits dephosphorylation, as well as promoting phosphorylation, of the AMP-activated protein kinase. Studies using bacterially expressed human protein phosphatase-2C alpha and native bovine protein phosphatase-2AC. FEBS Lett. 377 421–425. [DOI] [PubMed] [Google Scholar]
- Donella-Deana, A., Boschetti, M., and Pinna, L.A. (2003). Monitoring of PP2A and PP2C by phosphothreonyl peptide substrates. Methods Enzymol. 366 3–17. [PubMed] [Google Scholar]
- Doppler, H., Storz, P., Li, J., Comb, M.J., and Toker, A. (2005). A phosphorylation state-specific antibody recognizes Hsp27, a novel substrate of protein kinase D. J. Biol. Chem. 280 15013–15019. [DOI] [PubMed] [Google Scholar]
- Fujii, H., Verslues, P.E., and Zhu, J.K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, H., and Zhu, J.K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 106 8380–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti, F., Beaudoin, N., Serizet, C., Webb, A.A., Vartanian, N., and Giraudat, J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y., Xiong, L., Song, C.P., Gong, D., Halfter, U., and Zhu, J.K. (2002). A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev. Cell 3 233–244. [DOI] [PubMed] [Google Scholar]
- Halford, N.G., and Hey, S.J. (2009). Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem. J. 419 247–259. [DOI] [PubMed] [Google Scholar]
- Himmelbach, A., Hoffmann, T., Leube, M., Hohener, B., and Grill, E. (2002). Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 21 3029–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach, A., Yang, Y., and Grill, E. (2003). Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol. 6 470–479. [DOI] [PubMed] [Google Scholar]
- Hirayama, T., and Shinozaki, K. (2007). Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 12 343–351. [DOI] [PubMed] [Google Scholar]
- Hrabak, E.M., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, T. (1995). Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80 225–236. [DOI] [PubMed] [Google Scholar]
- Hutti, J.E., Jarrell, E.T., Chang, J.D., Abbott, D.W., Storz, P., Toker, A., Cantley, L.C., and Turk, B.E. (2004). A rapid method for determining protein kinase phosphorylation specificity. Nat. Methods 1 27–29. [DOI] [PubMed] [Google Scholar]
- Kohn, M., Gutierrez-Rodriguez, M., Jonkheijm, P., Wetzel, S., Wacker, R., Schroeder, H., Prinz, H., Niemeyer, C.M., Breinbauer, R., Szedlacsek, S.E., and Waldmann, H. (2007). A microarray strategy for mapping the substrate specificity of protein tyrosine phosphatase. Angew. Chem. Int. Ed. Engl. 46 7700–7703. [DOI] [PubMed] [Google Scholar]
- Lammers, T., and Lavi, S. (2007). Role of type 2C protein phosphatases in growth regulation and in cellular stress signaling. Crit. Rev. Biochem. Mol. Biol. 42 437–461. [DOI] [PubMed] [Google Scholar]
- Leonhardt, N., Kwak, J.M., Robert, N., Waner, D., Leonhardt, G., and Schroeder, J.I. (2004). Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16 596–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, J., Bouvier-Durand, M., Morris, P.C., Guerrier, D., Chefdor, F., and Giraudat, J. (1994). Arabidopsis ABA response gene ABI1: Features of a calcium-modulated protein phosphatase. Science 264 1448–1452. [DOI] [PubMed] [Google Scholar]
- Leung, J., Merlot, S., and Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, J., Orfanidi, S., Chefdor, F., Meszaros, T., Bolte, S., Mizoguchi, T., Shinozaki, K., Giraudat, J., and Bogre, L. (2006). Antagonistic interaction between MAP kinase and protein phosphatase 2C in stress recovery. Plant Sci. 171 596–606. [Google Scholar]
- Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., and Grill, E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324 1064–1068. [DOI] [PubMed] [Google Scholar]
- Manning, B.D., Tee, A.R., Logsdon, M.N., Blenis, J., and Cantley, L.C. (2002). Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell 10 151–162. [DOI] [PubMed] [Google Scholar]
- Marley, A.E., Sullivan, J.E., Carling, D., Abbott, W.M., Smith, G.J., Taylor, I.W., Carey, F., and Beri, R.K. (1996). Biochemical characterization and deletion analysis of recombinant human protein phosphatase 2C alpha. Biochem. J. 320 801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25 295–303. [DOI] [PubMed] [Google Scholar]
- Merlot, S., Mustilli, A.C., Genty, B., North, H., Lefebvre, V., Sotta, B., Vavasseur, A., and Giraudat, J. (2002). Use of infrared thermal imaging to isolate Arabidopsis mutants defective in stomatal regulation. Plant J. 30 601–609. [DOI] [PubMed] [Google Scholar]
- Meyer, K., Leube, M.P., and Grill, E. (1994). A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264 1452–1455. [DOI] [PubMed] [Google Scholar]
- Miao, Y., Lv, D., Wang, P., Wang, X.C., Chen, J., Miao, C., and Song, C.P. (2006). An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18 2749–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moes, D., Himmelbach, A., Korte, A., Haberer, G., and Grill, E. (2008). Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. Plant J. 54 806–819. [DOI] [PubMed] [Google Scholar]
- Moore, F., Weekes, J., and Hardie, D.G. (1991). Evidence that AMP triggers phosphorylation as well as direct allosteric activation of rat liver AMP-activated protein kinase. A sensitive mechanism to protect the cell against ATP depletion. Eur. J. Biochem. 199 691–697. [DOI] [PubMed] [Google Scholar]
- Moorhead, G.B., De Wever, V., Templeton, G., and Kerk, D. (2009). Evolution of protein phosphatases in plants and animals. Biochem. J. 417 401–409. [DOI] [PubMed] [Google Scholar]
- Moorhead, G.B., Trinkle-Mulcahy, L., and Ulke-Lemee, A. (2007). Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell Biol. 8 234–244. [DOI] [PubMed] [Google Scholar]
- Muller, A.H., and Hansson, M. (2009). The barley magnesium chelatase 150-kd subunit is not an abscisic acid receptor. Plant Physiol. 150 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata, Y., Pei, Z.M., Mori, I.C., and Schroeder, J. (2001). Abscisic acid activation of plasma membrane Ca(2+) channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell 13 2513–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli, A.C., Merlot, S., Vavasseur, A., Fenzi, F., and Giraudat, J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, M., Guo, Y., Halfter, U., and Zhu, J.K. (2003). A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. USA 100 11771–11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais, J.E., Bowers, K.E., Stoddard, A.K., and Fierke, C.A. (2005). A continuous fluorescent assay for protein prenyltransferases measuring diphosphate release. Anal. Biochem. 345 302–311. [DOI] [PubMed] [Google Scholar]
- Park, S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, N., Merlot, S., N'Guyen, V., Boisson-Dernier, A., and Schroeder, J.I. (2006). A hypermorphic mutation in the protein phosphatase 2C HAB1 strongly affects ABA signaling in Arabidopsis. FEBS Lett. 580 4691–4696. [DOI] [PubMed] [Google Scholar]
- Rodriguez, P.L., Benning, G., and Grill, E. (1998). ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett. 421 185–190. [DOI] [PubMed] [Google Scholar]
- Rubio, S., Rodrigues, A., Saez, A., Dizon, M.B., Galle, A., Kim, T.H., Santiago, J., Flexas, J., Schroeder, J.I., and Rodriguez, P.L. (2009). Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 150 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez, A., Apostolova, N., Gonzalez-Guzman, M., Gonzalez-Garcia, M.P., Nicolas, C., Lorenzo, O., and Rodriguez, P.L. (2004). Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 37 354–369. [DOI] [PubMed] [Google Scholar]
- Saez, A., Robert, N., Maktabi, M.H., Schroeder, J.I., Serrano, R., and Rodriguez, P.L. (2006). Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol. 141 1389–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez, A., Rodrigues, A., Santiago, J., Rubio, S., and Rodriguez, P.L. (2008). HAB1–SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell 20 2972–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, M.J., Grondin, P.O., Hegarty, B.D., Snowden, M.A., and Carling, D. (2007). Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. Biochem. J. 403 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago, J., Rodrigues, A., Saez, A., Rubio, S., Antoni, R., Dupeux, F., Park, S.Y., Marquez, J.A., Cutler, S.R., and Rodriguez, P.L. (2009). Modulation of drought resistance by the abscisic acid-receptor PYL5 through inhibition of clade A PP2Cs. Plant J., in press. [DOI] [PubMed]
- Schweighofer, A., Hirt, H., and Meskiene, I. (2004). Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 9 236–243. [DOI] [PubMed] [Google Scholar]
- Shen, Y.Y., et al. (2006). The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443 823–826. [DOI] [PubMed] [Google Scholar]
- Sirichandra, C., Wasilewska, A., Vlad, F., Valon, C., and Leung, J. (2009). The guard cell as a single-cell model towards understanding drought tolerance and abscisic acid action. J. Exp. Bot. 60 1439–1463. [DOI] [PubMed] [Google Scholar]
- Sun, H., Lu, C.H., Uttamchandani, M., Xia, Y., Liou, Y.C., and Yao, S.Q. (2008). Peptide microarray for high-throughput determination of phosphatase specificity and biology. Angew. Chem. Int. Ed. Engl. 47 1698–1702. [DOI] [PubMed] [Google Scholar]
- Suter, M., Riek, U., Tuerk, R., Schlattner, U., Wallimann, T., and Neumann, D. (2006). Dissecting the role of 5′-AMP for allosteric stimulation, activation, and deactivation of AMP-activated protein kinase. J. Biol. Chem. 281 32207–32216. [DOI] [PubMed] [Google Scholar]
- Virshup, D.M., and Shenolikar, S. (2009). From promiscuity to precision: Protein phosphatases get a makeover. Mol. Cell 33 537–545. [DOI] [PubMed] [Google Scholar]
- Vlad, F., Turk, B.E., Peynot, P., Leung, J., and Merlot, S. (2008). A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 55 104–117. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33 949–956. [DOI] [PubMed] [Google Scholar]
- Wang, P., Fu, H., Snavley, D.F., Freitas, M.A., and Pei, D. (2002). Screening combinatorial libraries by mass spectrometry. 2. Identification of optimal substrates of protein tyrosine phosphatase SHP-1. Biochemistry 41 6202–6210. [DOI] [PubMed] [Google Scholar]
- Wilkie, A.O. (1994). The molecular basis of genetic dominance. J. Med. Genet. 31 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Costa, A., Leonhardt, N., Siegel, R.S., and Schroeder, J.I. (2008). Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, R., Hobo, T., Ichimura, K., Mizoguchi, T., Takahashi, F., Aronso, J., Ecker, J.R., and Shinozaki, K. (2002). ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 43 1473–1483. [DOI] [PubMed] [Google Scholar]
- Yoshida, R., Umezawa, T., Mizoguchi, T., Takahashi, S., Takahashi, F., and Shinozaki, K. (2006). The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281 5310–5318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.