Abstract
The earliest proteolytic event affecting most proteins is the excision of the initiating Met (NME). This is an essential and ubiquitous cotranslational process tightly regulated in all eukaryotes. Currently, the effects of NME on unknown complex cellular networks and the ways in which its inhibition leads to developmental defects and cell growth arrest remain poorly understood. Here, we provide insight into the earliest molecular mechanisms associated with the inhibition of the NME process in Arabidopsis thaliana. We demonstrate that the developmental defects induced by NME inhibition are caused by an increase in cellular proteolytic activity, primarily induced by an increase in the number of proteins targeted for rapid degradation. This deregulation drives, through the increase of the free amino acids pool, a perturbation of the glutathione homeostasis, which corresponds to the earliest limiting, reversible step promoting the phenotype. We demonstrate that these effects are universally conserved and that the reestablishment of the appropriate glutathione status restores growth and proper development in various organisms. Finally, we describe a novel integrated model in which NME, protein N-α-acylation, proteolysis, and glutathione homeostasis operate in a sequentially regulated mechanism that directs both growth and development.
INTRODUCTION
All proteins undergo proteolysis, a major irreversible modification mediated by a large number of proteases. These enzymes are thought to constitute 1 to 5% of eukaryotic proteomes (Lopez-Otin and Overall, 2002). Protease function is essential in numerous physiological processes, including development, blood coagulation, and cell death and in the development of many cancers and infectious diseases (Goldberg, 2003; van der Hoorn, 2008).
N-terminal Met excision (NME) is the earliest proteolytic event affecting proteins that are still bound to the ribosome, before their synthesis has been completed (Giglione et al., 2004, 2009). The NME pathway is ubiquitous, and about two-thirds of the proteins in any organism undergo this type of processing (Meinnel and Giglione, 2008b). Dedicated NME machinery has been found in all cell compartments in which protein synthesis occurs. The NME machinery of higher eukaryotes involves two classes of proteases: peptide deformylases and Met aminopeptidase (MAP; with MAP1D enzymes in the organelles and MAP1A and MAP2 in the cytoplasm) (Giglione et al., 2004; Meinnel et al., 2006). There is evidence that NME is tightly regulated throughout development, tumorigenesis, and during the responses induced by various external signals in both animals and plants (Wang et al., 2000; Cleary et al., 2002; Endo et al., 2002; Florens et al., 2002; Kanno et al., 2002; Selvakumar et al., 2004; Dasgupta et al., 2005; Ross et al., 2005; Selvakumar et al., 2006), corroborating the idea that this mechanism is an important signal to support growth, proliferation, survival, and differentiation. Accordingly, NME enzymes are the targets of various natural compounds with antibiotic, antiparasitic, antifungal, or anticancer effects (for reviews, see Boularot et al., 2004; Giglione et al., 2004). These findings, together with the observation that NME components are overexpressed in many tumor cells, suggest that the targeted inhibition of the corresponding enzymes may provide a means of treating various pathological conditions, including cancer. To date, only a small number of NME inhibitors that are effective in vivo have been discovered. The main difficulty in finding novel potent and effective drugs against NME enzymes is our limited knowledge of the physiological functions of N-terminal Met excision, despite some efforts to characterize the signaling pathways associated with NME (Zhang et al., 2000; Yeh et al., 2006; Zhang et al., 2006). Deciphering the physiological functions of the NME process has been hampered by gene redundancy, lethality upon inhibition, and the fact that NME has more than 15,000 putative distinct targets in higher eukaryotes (see Martinez et al., 2008). One initial basic hypothesis was that NME might facilitate further modifications to the N terminus of the protein, such as N-α-acylation. N-myristoylation (N-Myr) is one such modification. N-Myr involves the addition, by N-myristoyltransferase, of a fatty acid to a set of nuclear-encoded proteins in which the N-terminal Gly is unmasked by NME (Bhatnagar et al., 2001; Resh, 2006; Meinnel and Giglione, 2008a). This lipidation is crucial, as myristoylated proteins are involved in major cell signaling mechanisms. N-Myr has been shown to be necessary, but not sufficient, for the targeting of the protein to the appropriate membrane. However, N-Myr does not occur in bacteria or organelles, and not all proteins with an N-terminal Gly residue undergo N-Myr. This modification alone therefore cannot account for the essential nature of NME in all organisms.
In this study, we used a dedicated inducible system in the model organism Arabidopsis thaliana to investigate the molecular basis of the absolute requirement for cytoplasmic NME (cNME) in higher eukaryotes. Multiple approaches, including proteomics and metabolomics, were used to examine the earliest molecular events associated with the inhibition of this process in Arabidopsis. Our studies demonstrate that developmental defects induced by NME inhibition are associated with an increase in proteolytic activity caused principally by an increase in the size of the pool of proteins available for rapid degradation. This deregulation of proteolysis drives the perturbation of both the gluthatione pool and the redox state. The inhibitory effects of NME can be overcome by supplementation with reduced glutathione in higher plants, fungi, and Archaea, demonstrating the ubiquitous mechanism associated with the NME process. Thus, cNME plays a crucial role in maintaining the appropriate status of the key cellular redox buffer glutathione, and this involves the tight regulation of interconnected processes, including the proteolytic activity of the cell. We also show that this crosstalk between thiol status and proteolysis is essential for normal growth and development.
RESULTS
Validation of a Model System for Studying the Molecular Impact of cNME Inhibition
By combining reverse genetics and reverse chemogenomics in transgenic plant lines, we previously created a specific and reversible switch system for investigating the relevance of cytoplasmic NME in Arabidopsis (Ross et al., 2005). Wild-type seedlings, in which cNME is ensured by two types of MAPs (MAP1 and MAP2s), proved insensitive to micromolar concentrations of Fumagillin, a highly specific, irreversible inhibitor of MAP2s, of which there are two copies in Arabidopsis. By contrast, MAP1A knockout variants (map1A) were unable to germinate under the same conditions. Analysis of the phenotype in the presence of high concentrations (i.e., > 1 μM) of Fumagillin (which blocks all MAP2 activity) in several independent double-stranded RNA interference Arabidopsis lines displaying distinct MAP1A levels demonstrated that the extent of the developmental defect specifically depends on the concentration of MAP1A. We concluded that (1) cNME is an essential pathway in plants, (2) minimum levels of this process are required to ensure correct plant development (Ross et al., 2005), and (3) the phenotype induced by Fumagillin is unambiguously genetically linked to the only MAP1A allele. Fumagillin is therefore a useful tool for inducing reversible cNME deregulation and mimicking the developmental impact of the inhibition of this pathway by comparing the overall effects occurring in the two genetic backgrounds (i.e., the wild type and map1A). We decided to investigate the molecular events associated with cNME by focusing on the primary molecular targets that are sensitive to the inhibition of cytoplasmic NME. To identify the earliest events associated with cNME inhibition, we first established the lowest concentration of the inducer (Fumagillin) required to promote a visible defect in the course of development of the map1A line. The addition of 100 nM Fumagillin to the growth medium of germinating map1A seeds reproducibly triggered a developmental defect that was readily observable at early stages of development, between 2 and 10 d after imbibition (DAI; Figures 1A and 2; see Supplemental Data Sets 1 to 3 online; Ross et al., 2005).
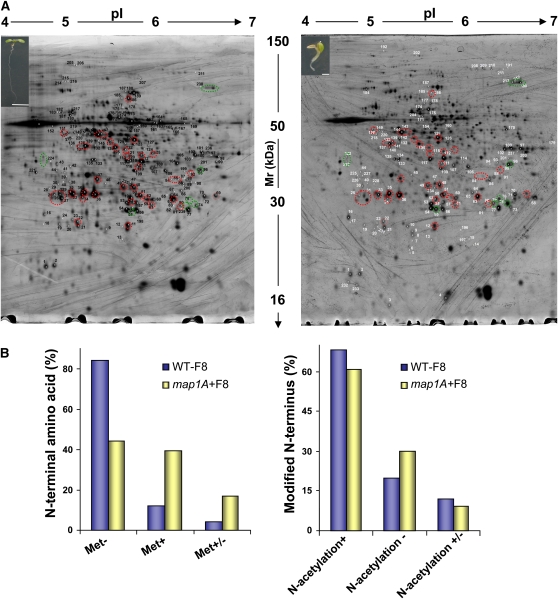
Figure 1.
Proteomics of cNME Inhibition.
(A) Wild-type (left) and map1A (right) Arabidopsis seeds were grown in the presence of 100 nM Fumagillin for 8 DAI. The developmental impact associated with cNME deregulation is illustrated in the insets. Representative differential 2D gel electrophoresis patterns are shown for the two genetic backgrounds. Equal amounts (from 100 to 600 μg) of total proteins were separated on the 18-cm pI 4 to 7 linear gradient IPG strips and by 11% SDS-PAGE. In the gel shown in the figure, 600 μg were used. Proteins displaying differential expression are shown in gels, with their unique sample spot protein numbers. See also Supplemental Tables 1, 3, and 4 online for protein identification. The protein spots outlined with a circle were of significantly different intensities (P ≤ 0.05) for the map1A+F mutant and the wild type. Green circles indicate spots for which abundance systematically increased with cNME inhibition, and red circles indicate spots with systematically lower intensity in map1A+F. We checked that the spots of the reference pattern wild type were also retrieved in both the map1A-F and (WT+F) images. In a previous preliminary analysis of line map1A, we showed that the 2D protein pattern is similar to that of the wild type (Figure 2D in Ross et al., 2005), demonstrating that the knockout of MAP1A has no functional impact at the proteome level.
(B) Comparative analysis of the identified N termini from the wild type and map1A+F at 8 DAI (for more details, see Supplemental Table 2 online). Met-, Met+, and Met+/− mean Met cleavage, Met retaining, and a mixture of the two situations in the sample, respectively.
[See online article for color version of this figure.]
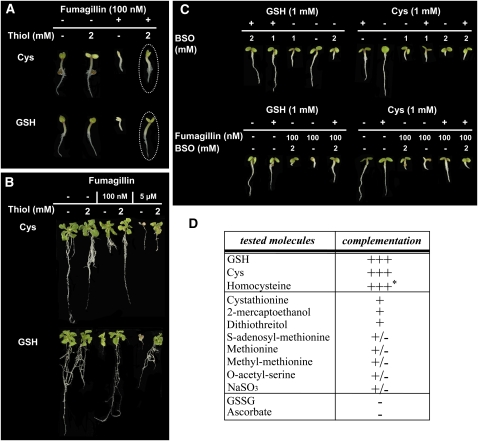
Figure 2.
Rescue of the Developmental Defect Associated with cNME Inhibition in Arabidopsis with GSH and Cys.
(A) map1A line plantlets were vertically grown in the presence or absence of 100 nM Fumagillin and 2 mM of the indicated thiol compound. Dotted ovals show the complementation by Cys and GSH.
(B) map1A line plantlets were vertically grown without any additional compounds. At 8 DAI, plantlets were transferred onto fresh medium containing the indicated concentrations of Fumagillin and 2 mM of the indicated thiol compound.
(C) map1A line plantlets were vertically grown on medium containing GSH or Cys together with the GSH biosynthesis inhibitor BSO or Fumagillin and BSO, at the indicated concentrations.
(D) The table summarizes the effect the various molecules tested on the rescue of the developmental defect associated with cNME inhibition induced in map1A line plantlets in the presence of Fumagillin as in (A). +++ indicates complementation similar to that obtained with GSH or Cys, + indicates very partial complementation, and – indicates no complementation. The asterisk indicates that complementation cannot be obtained with established plants.
[See online article for color version of this figure.]
We previously reported that the protein profiles of both the wild type grown in the presence of Fumagillin and of map1A were identical to that of the wild type grown in the absence of the drug (Ross et al., 2005). We investigated the global effects on the cellular proteome of early cNME inhibition and tried to identify the proteins involved by carrying out differential two-dimensional (2D) gel electrophoresis followed by tandem mass spectrometry (MS/MS) for protein identification on wild-type and map1A seedlings grown in the presence of 100 nM Fumagillin (map1A+F, insets in Figure 1A) 8-DAI. The MS/MS analysis of 441 spots (see Supplemental Data Set 1 online) led to the identification of several independent cytosolic N-terminal peptides (see Supplemental Data Set 2 online) from the wild type (42) and map1A+F (42). The nature of the N termini identified in the wild-type background confirmed the high degree of accuracy of the bioinformatics tool recently made available for the species-specific prediction of NME and related modifications (Martinez et al., 2008). Complete or partial retention of the N-terminal Met was observed for several proteins identified in the map1A+F context. Retention of the N-terminal Met was observed for 40% of the N termini, confirming effective and partial inhibition of the cNME in vivo, as expected from our strategy of focusing on the most sensitive targets (Figure 1B). Moreover, 68% of wild-type proteins for which the peptide N termini were identified were N-acetylated (the largest population of stable proteins), 20% were not N-acetylated, and the remaining proteins displayed mixed modifications. By contrast, in the map1A+F context, a smaller proportion of proteins (60%) were N-acetylated, and a significantly higher proportion of N termini were not N-acetylated (30%) (Figure 1B). This difference probably resulted from the N termini with a retained Met not being recognized by the enzymes responsible for N-α-acetylation. Thus, at the molecular level, cNME inhibition essentially induces a significant increase in the pool of proteins with both an N-terminal Met and a free αNH2 moiety.
Proteomic Analysis of the Effects of Inhibiting cNME
With the above system, cNME inhibition in the map1A background appeared to be associated with only slight changes of the 2D electrophoresis profile (<5% of all spots; Figure 1A; see Supplemental Table 1 and Supplemental Data Set 3 online). The differential characterization of the 441 spots (see Supplemental Data Set 1 online) by MS/MS revealed that, in contrast with the small number of spots for which protein abundance increased (only 11 spots, corresponding to 11 different proteins), 100 spots, corresponding to >150 different proteins, decreased in intensity following partial cytosolic NME inhibition. Highly reproducible results in various biological replicates were obtained for 40 spots, corresponding to >60 different proteins (Figure 1A, red circles; see Supplemental Data Set 3 online). An interesting subclass of proteins, the accumulation of which was reduced by cNME inhibition, corresponded to the SnRK2.8 (for SNF1-related kinase) kinase group identified on the phosphoproteomic profile (Shin et al., 2007). These proteins included a non-ε 14-3-3-like protein, an adenosine kinase, and glyoxalase I. As these kinases regulate plant growth and development and we have recently shown that N-Myr, a cNME-dependent modification, regulates the SnRK pathway (Pierre et al., 2007), we checked whether (1) general phosphorylation and (2) SnRK1 activity were affected by the inhibition of cytoplasmic NME. Using Pro-Q Diamond staining to detect phosphorylated proteins on one-dimensional (1D) gels, we observed no significant change in the phosphoproteome of plants in which cytoplasmic NME was inhibited, at any development stage (see Supplemental Figure 1A online). Accordingly, no significant changes in SnRK1 enzyme activity were observed upon NME inhibition.
The most intriguing result of our proteomic characterization of cNME was the finding that a striking number of the modified spots corresponded to pools of thiol proteins (see Supplemental Table 1 and Supplemental Data Set 3 online), for which strong dependence on glutathione homeostasis and/or glutathionylation (i.e., the formation of protein-glutathione mixed disulfides) has already been demonstrated (Ito et al., 2003; Ball et al., 2004; Dixon et al., 2005). Interestingly, the phenotype of plants with inhibited cNME (Figure 2) was very similar to that of several mutants of γ-glutamyl-Cys synthetase. This protein is the first dedicated enzyme of the glutathione biosynthesis pathway (γ-ECS or GSH1). The phenotype induced by NME inhibition resembled those observed in the root meristemless1 mutant line and in wild-type plants grown in the presence of l-buthionine-S-sulfoximine (BSO), a specific inhibitor of γ-ECS (Sanchez-Fernandez et al., 1997; Vernoux et al., 2000). From the similarities of phenotypes together with the proteomic analysis (see above), we hypothesized that the abnormal development induced by NME inhibition could be due to a deregulation of the glutathione pool.
Modulated Glutathione Status Is a Ubiquitous Feature Associated with cNME Inhibition
We tested the above hypothesis by supplementing the growth medium with the reduced form of glutathione, which is known to rescue the phenotype of GSH1 mutants (Cairns et al., 2006), or Cys, an amino acid precursor of glutathione, to determine whether these compounds could rescue the abnormal phenotype induced by low levels of cytoplasmic NME. The abnormal phenotype was complemented when the map1A+F mutant was grown in the presence of the reduced form of glutathione (GSH) or Cys (Figure 2A). This complementation was dependent on the GSH and Cys concentration, complete with 2 mM Cys or GSH and also observed when cNME was inhibited at later stages of development (Figure 2B). Full complementation, with reversion of the abnormal developmental phenotype induced by cNME inhibition, was also observed if homocysteine was added to the medium before germination. In contrast with our findings for Cys and GSH, no complementation was detected if homocysteine was supplied to established plants (Figure 2D).
We then performed various complementation experiments on plants with reduced cNME in the presence of BSO. Complementation was observed with GSH but not with its precursor, Cys (Figure 2C). Thus, complementation was primarily supported by GSH through its biosynthesis from free Cys and was independent of any direct interference from added thiols and Fumagillin. To explore whether the GSH molecule was the key element responsible for cNME complementation, we further assessed the effects of (1) the principal oxidized form of glutathione, GSSG; (2) another key redox buffer, ascorbate; (3) two general thiol reductants (2-mercaptoethanol and DTT); and (4) several intermediates of Cys and Met biosynthesis. With its limiting precursor Cys, GSH was the only factor, which we have identified so far to be able to completely cure the phenotype associated with the map1A+ line. Only the thiol reductants and cystathionine gave very partial complementation, which could correspond to complementation of one of the defects induced by cNME inactivation (i.e., the general effect on cellular redox status; Figure 2D). Finally, complementation experiments with GSH or Cys were also successful if a MAP2 inhibitor other than Fumagillin was used and with triple MAP RNA interference mutants displaying weak and intermediate developmental defects (ψMAP1A2, see phenotype shown in Figure 5E in Ross et al., 2005; see Supplemental Figure 2A online).
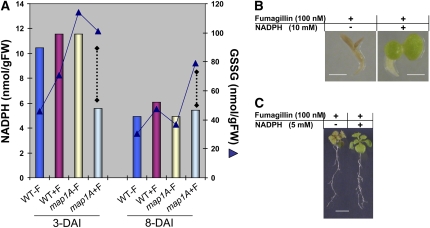
Figure 5.
The Low Concentration of NADPH in map1A+F Plantlets Limits GR Activity.
(A) The total amount of NADPH was measured in wild-type and map1A plantlets grown in the presence or absence of 100 nM Fumagillin during early stages of development. The amount of NADPH was compared with that of GSSG detected in each set of conditions. Dotted lines accentuate the loss of correlation between NADPH and GSSG levels in the map1A+F mutant. se has been reported in Figure 2 for GSSG and Supplemental Figure 3 online for NADPH.
(B) map1A line plantlets were grown in the presence of 100 nM Fumagillin and in the presence or absence of 10 mM NADPH. Bar = 1 mm.
(C) map1A line plantlets were grown for 8 DAI with no additional compounds and then transferred to new medium containing the indicated concentrations of Fumagillin and NADPH. Bar = 1 cm.
[See online article for color version of this figure.]
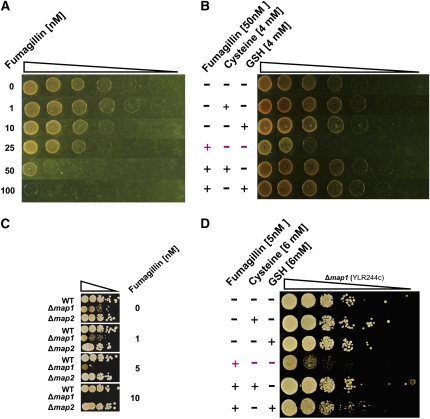
We then investigated whether the significance of the link between cNME inhibition and GSH pathway deregulation was restricted to the plant kingdom or whether it was of broader biological relevance. Interestingly, all archaeal members possess only one active MAP2, facilitating studies of the effects of MAP2 inhibitors, such as Fumagillin. Using the conditions recently established for assays of MAP activity in vitro (Frottin et al., 2006), we first checked that the archaeon MAP2 (Pyrococcus furiosus) was specifically inhibited by Fumagillin. An IC50 value of 25 nM was obtained at concentrations of 50 nM MAP2, indicating stoichiometric binding. This result is consistent with the expected covalent binding of the drug to the enzyme target (i.e., IC50 = [MAP2]/2). We then showed that micromolar amounts of the drug fully inhibited the growth of the archaeon Haloferax volcanii, demonstrating the absolute requirement for cNME in archaea. We then performed complementation experiments with GSH or Cys in the growth medium. Increasing Fumagillin concentrations inhibited cell growth, and this inhibition was reversed by supplementation with Cys or GSH (Figures 3A and 3B). Low concentrations of Fumagillin had cytostatic effects, whereas high concentrations were cytotoxic to H. volcanii, as also observed with Arabidopsis (Figure 2B). Remarkably, the effects of Fumagillin on wild-type Saccharomyces cerevisiae map1 and map2 mutant lines were also very similar to those observed in Arabidopsis (Figures 3C and 3D). As previously described (Li and Chang, 1995; Griffith et al., 1997), the map1 null strain (Δmap1) grew more slowly and was more sensitive to Fumagillin than the wild-type and map2-null (Δmap2) strains. Using the Δmap1 variant, we showed that either Cys or GSH restored normal growth in Fumagillin-treated yeast.
Figure 3.
Ubiquitous Mechanism Induced by cNME Blockade as Evidenced by Characterization of the Fumagillin Effects on the Haloarchaeon H. volcanii and Several Yeast Mutants.
The triangles above each panel represent the use of serial dilutions by 10-fold of the strain considered.
(A) The wild-type strain haloarchaeon H. volcanii was spotted onto rich medium plates containing the indicated concentrations of Fumagillin.
(B) For complementation experiments, H. volcanii was cultured in the presence (+) or absence (−) of 50 nM Fumagillin and 4 mM Cys or 4 mM GSH.
(C) The effect of Fumagillin was investigated in the yeast S. cerevisiae with two KO mutants: Δmap1 and Δmap2. Wild-type and KO lines were spotted on rich medium containing a range of Fumagillin concentrations.
(D) Complementation experiments with the line expressing only MAP2 (Δmap1) were performed on rich medium with or without 5 nM Fumagillin and supplemented with 6 mM Cys or 6 mM GSH.
[See online article for color version of this figure.]
How Does GSH Rescue the Abnormal Development Induced by cNME Inhibition in Arabidopsis?
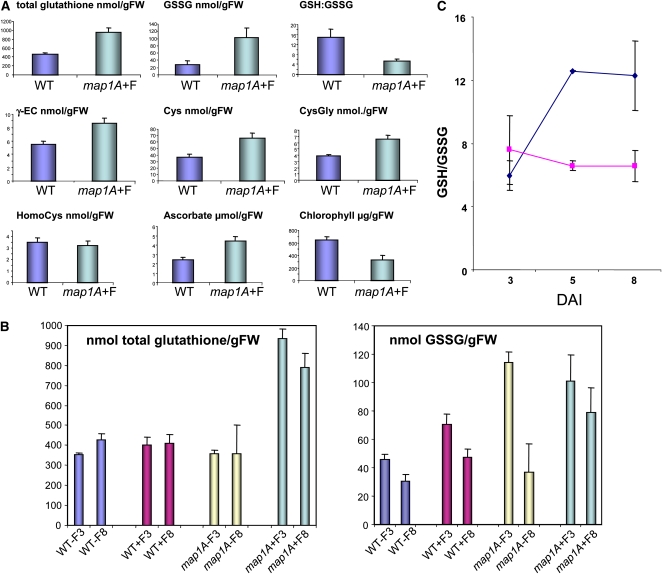
The data shown in Figure 2 suggest that glutathione is not only a key molecule responsible for the abnormal phenotype induced by cNME inhibition but also that reversion is partially dependent on the redox state of glutathione. Indeed, thiol-reducing agents, such as 2-mercaptoethanol or DTT, also attenuated the phenotype induced by low-level cNME activity, albeit to a lesser extent than reduced GSH or Cys. We therefore investigated the thiol profiles in both backgrounds: wild-type plants and plants with reduced cNME. Unexpectedly, determinations of the amounts of the various thiol-containing metabolites (Figure 4A) revealed that 8-DAI plants with low levels of cNME activity exhibited higher levels of most thiol compounds, including Cys and glutathione. The increase observed for all the compounds analyzed in Figure 4A involved both the reduced and oxidized forms of the molecule.
Figure 4.
Glutathione Redox Potential Remains Constant during Development in map1A+F Lines and Lower Than That in Wild-Type Plants.
(A) Endogenous levels of the indicated thiols, ascorbate, and chlorophyll were measured in wild-type and map1A+F plantlets at 8 DAI as described in Methods. Thiol concentrations are expressed in nmol/g fresh weight (FW), ascorbate in μmol/gFW, and chlorophyll in μg/gFW.
(B) Endogenous levels of total glutathione and oxidized glutathione were analyzed at 3 or 8 DAI in both the wild type and map1A grown in the presence or absence of 100 nM Fumagillin.
(C) During early plant development, the GSH/GSSG ratio, which is proportional to the glutathione redox potential, was calculated as follows: (total glutathione − 2GSSG)/2GSSG. Blue dots indicate the mean GSH/GSSG ratio for the wild type, WT+F, and map1A; pink dots indicate the mean ratio for map1A+F.
[See online article for color version of this figure.]
We observed high levels of GSSG during the early stages of development (Figure 4B), as reported in other studies (Tommasi et al., 2001; Henmi et al., 2005). Interestingly, this significant increase in GSSG levels was observed in both the map1A background alone and in the same plants challenged with Fumagillin to inhibit cNME. This observation thus identifies the first marker unambiguously associated with MAP1A gene inactivation. However, Fumagillin treatment modulated the age-dependent pattern of GSSG accumulation. GSSG contents decreased between 3 and 8 DAI in the wild type and in untreated map1A plants but remained high in map1A plants in which NME was further repressed by Fumagillin treatment (see map1A+F8 in Figure 4B). In this background, the GSSG levels remained constantly high throughout development.
Together with the partial complementation, by general reductants, of the phenotype produced by NME inhibition (Figure 2D), the relationship between NME inhibition, GSH, and GSSG contents suggests that one of the effectors in the system is the intracellular glutathione redox potential. This factor is determined by [GSH]2/[GSSG] and thus depends both on the GSH/GSSG ratio and the total glutathione concentration. In plants with reduced cNME, the redox potential of the glutathione system was kept high (i.e., more positive or less reducing) throughout seedling development (Figure 4C). By contrast, in wild-type plants and in the map1A mutants grown in the absence of Fumagillin, the GSH/GSSG ratio was low due to the presence of large amounts of GSSG at early stages of seedling development. Greater reducing potential was then acquired during development (Figures 4B and 4C) as GSSG decreased, probably due to an increase in glutathione reductase activity (Henmi et al., 2005). Our data are consistent with previous studies showing remarkable changes in the content and redox balance of glutathione throughout development, particularly during germination (Tommasi et al., 2001; Foyer and Noctor, 2005; Cairns et al., 2006) and complementation experiments in which supplementation with reduced glutathione, but not the oxidized form GSSG, rescued the abnormal phenotype associated with decreased NME. These observations suggest that NME inhibition prevents the adjustment of the glutathione redox potential to the reducing values required to promote correct development.
A Decreased Pool of NADPH Participates in Keeping the Level of GSSG Elevated When NME Is Inhibited
As the GSH/GSSG ratio is known to be altered by ROS production (Foyer and Noctor, 2005), we checked whether cNME inhibition induced generalized oxidative stress conditions. We controlled overall oxidative state by assessing protein carbonylation in crude plant homogenates. No global variation of carbonyl content during the first 8 d of development was detected between plants with reduced cNME and the wild type (see Supplemental Figure 1B online). We also measured the antioxidative enzyme activities of both catalase and ascorbate peroxidase, the two major H2O2 scavengers of leaves. The corresponding values remained unaffected in both backgrounds. Finally, our proteomic analysis was inconsistent with an oxidative stress because it revealed a reduced accumulation of proteins usually increased and abundant under oxidative stress conditions (see Supplemental Data Set 2 online and Chew et al., 2003; Foyer and Noctor, 2005). These data indicate that the effects of NME inhibition on glutathione are specific and do not result from the induction of generalized oxidative stress.
As GSSG is mostly recycled into GSH by glutathione reductase (GR), we investigated the expression of the two plant GR genes (the cytosolic GR1 gene and the plastid GR2 gene). We compared the total enzyme activity in plants with reduced cNME with that in the wild type (see Supplemental Figure 3 online). The data excluded the possibility that the high levels of GSSG induced by lower levels of cNME in the mutant were caused by compromised GR capacity, measured as extractable activity at optimal substrate concentrations. The activity of the enzyme is highly dependent on the presence of its cofactor, NADPH. As NADPH is added exogenously in experiments in vitro, extractable GR activities essentially reflect the concentration of the enzyme rather than its actual activity in vivo. In agreement with this, the map1A+F line did not exhibit insufficient GR amounts, as at 3 DAI both the mRNA and the protein levels were doubled compared with in wild-type conditions (see Supplemental Figure 3 online). In this context, it should be stressed that a GR1 knockout (KO) would not reproduce the map1A+F phenotype. Indeed, gr1 KO mutants, which have no visible aberrant phenotype, show an increase of GSSG but only a minor increase in the total glutathione pool (Henmi et al., 2005; Marty et al., 2009). This is in agreement with our data on line map1A alone, for which we observe no visible phenotype, except an increase of GSSG in the very early development stages that was not accompanied by an increase in the total glutathione pool (Figure 4B). We tried to overproduce GR activity from the GR1 gene. We hypothesized that, if NADPH cofactor was not limiting in the cell, the increase in GR activity would decrease the extent of the phenotype or increase resistance to Fumagillin. Unfortunately, our several attempts to increase GR concentration were not successful, most likely because of the tight regulation required for GR activity. Finally, it has recently been shown that GR1 activity could be compensated for by the NADPH-dependent thioredoxin system and that there is gene redundancy to regulate the glutathione redox state in a GR1 KO context (Marty et al., 2009).
We finally investigated whether the accumulation of GSSG resulted from a decrease in the in vivo availability of its cosubstrate, NADPH, by determining the pyridine nucleotide contents (i.e., NADPH, NADH, NADP, and NAD) of total extracts from wild-type and NME-deficient plants at various stages of development (see Supplemental Figure 4 online). The measurement of the single pyridine nucleotides (total and ratios) shows that the NADH level was unchanged in any tested condition, unlike NADPH and to a lesser extent NADP, which causes reduction of the total pyridine pool and redox at very early stages. This analysis revealed that an early effect of NME inhibition was a decrease in NADPH levels below those in the wild type at 3 DAI. NADPH levels then remained constant throughout development in the map1A+F seedlings. Wild-type plants grown in the presence of 100 nM Fumagillin or map1A seedlings contained similar amounts of NADPH to the untreated wild type at the corresponding developmental stage but had a lower concentration of NADPH at later developmental stages, with smaller amounts of GSSG (Figure 5A). At later development stages (i.e., 8 DAI), the NADPH levels were comparable in both wild-type and NME-deficient plants (see Supplemental Figure 4 online; Figure 5A). Thus, given both the amount of GSSG present in the different conditions and the corresponding amounts of NADPH detected (Figures 4B and 5A; see Supplemental Figure 4 online), GR activity is probably limited by the small amount of NADPH available when NME is inhibited, causing an increase in GSSG levels and decrease in the reducing glutathione redox potential. This hypothesis was tested directly by supplementing the growth medium of plants of various backgrounds, including wild-type and cNME-deficient seedlings, with NADPH. Unambiguous and reproducible complementation was observed when the map1A+F mutants were treated with NADPH, but not with other pyridine nucleotides, at a concentration exceeding 5 mM (Figures 5B and 5C), and this complementation was particularly apparent for leaf development. Complementation by NADPH was less pronounced in the roots, but its effects may have been masked, as the NADPH concentrations required to complement NME inhibition also strongly inhibited the growth of both wild-type and map1A mutant roots grown in the absence of Fumagillin. Although GR has a high affinity for NADPH, decreases in NADPH/NADP+ ratios could nonetheless cause adjustments in the GSH/GSSG ratio. This interpretation is supported by our observations that the map1A+F phenotype can be complemented by GSH (which restores the glutathione redox balance directly) or by NADPH, which restores it through GSSG reduction.
Maintenance of a low level of NADPH in map1A+F mutants was supported by the proteomic analysis (see Supplemental Data Set 3 online), which revealed a strongly diminished pool of enzymes involved in the glycolytic and oxidative pentose phosphate pathway, two pathways that operate together to produce NADPH (Schwender et al., 2003).
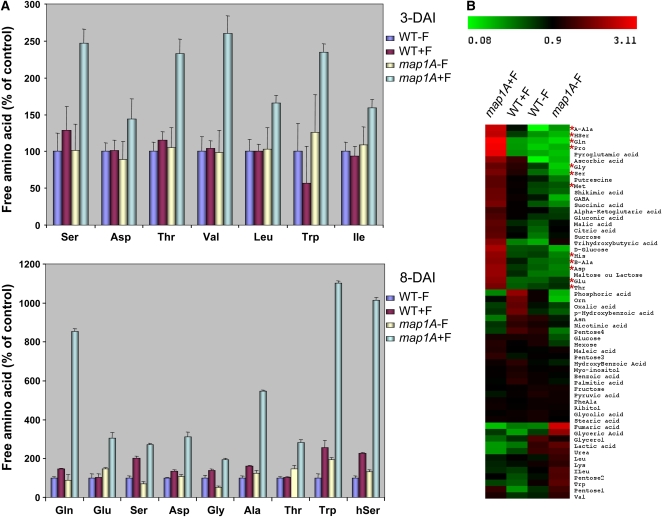
Metabolic Profiling Reveals That cNME Inhibition Dramatically Increases the Free Amino Acid Pool
The data presented above suggest that NME inhibition leads to changes in enzyme profiles, resulting in the production of smaller amounts of NADPH, which could be considered to be one of the factors responsible for the maintenance of the elevated oxidized glutathione pool. Since elevated GSSG is not the major problem of map1A+F plants, we next investigated the way in which NME inhibition led also to an increase in total glutathione content (Figure 4B). We investigated whether Cys was the only amino acid for which concentration was induced by cNME inhibition or whether other amino acids were affected by performing HPLC profiling of amino acids in the two backgrounds at various developmental stages. Most amino acids were found to be present in larger amounts in the NME-inhibited background, and this trend became increasingly pronounced as development progressed (Figure 6A). At 8 DAI, when cNME was inhibited, the total free amino acid concentration was five times higher than that of the wild type. HPLC analyses were extended by nonspecific metabolic profiling based on gas chromatography–coupled time-of-flight mass spectrometry (GC-TOF-MS), which confirmed the observed increases in free amino acid levels (Figure 6B, asterisks). Surprisingly, the levels of enzymes involved in amino acid biosynthesis were not higher in the cNME-inhibited background. Instead, some of these enzymes, including both Gln synthetase and Cys synthase, were less abundant (Figure 1; see Supplemental Data Set 3 online). As proteins are the main reservoir of amino acids in the cell, this finding strongly indicated that the increase in free amino acids, including Cys, was probably linked to overall cellular proteolysis rather than to specific increases in amino acid biosynthesis.
Figure 6.
cNME Inhibition Dramatically Increases the Size of the Free Amino Acid Pool.
(A) All free amino acids were determined using two independent HPLC methods (see Methods) in all the conditions indicated in the figure (i.e., wild type, WT+F, map1A+F, map1A). Amino acid concentrations are expressed in nmol/g FW, with the values obtained for the wild-type plantlets taken as 100%. Only amino acids for which a significant difference in content was observed are reported.
(B) Global metabolic footprinting was performed for wild-type and map1A plantlets in the presence and absence of 100 nM Fumagillin at 8 DAI with the GC-TOF-MS system (see Methods). Red asterisks indicate the amino acids that accumulated in the map1A+F sample.
[See online article for color version of this figure.]
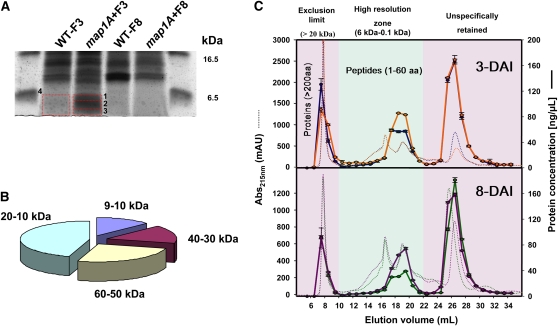
We assessed overall proteolytic activity when cNME is inhibited using high-resolution denaturing 1D gel electrophoresis followed by MS/MS analysis and size-exclusion chromatography independently, to separate, compare, and characterize the pool of small proteins/peptides detectable in both backgrounds (i.e., wild-type and cNME inhibited). At 3 DAI, the map1A+F plants were significantly enriched in polypeptides of a molecular mass of 4 to 6 kD (Figure 7A). MS/MS analysis of several of the species with the highest levels of accumulation showed that most (>80%) were the degradation products of proteins with significantly higher molecular masses (Figure 7B; see Supplemental Table 2 online). Similarly, at all stages, cNME inhibition resulted in larger numbers of peptide fragments with molecular masses ranging from 60 to 640 D (i.e., ∼1 to 10 amino acids in length) (Figure 7C). Finally, we used a peptidase/protease assay to quantify accurately the widest range of protease activities. The inhibition of cNME induced no global change in protease activity (see Supplemental Figure 5 online). However, at later stages of cNME inhibition, a slight but significant induction of protease activities, such as those related to the proteasome, was observed (see Supplemental Figure 5 online). This suggests that proteasome activity may increase due to an increase in the abundance of target proteins.
Figure 7.
Dynamic Accumulation of Protein Degradation Products in the map1A+F Plantlets.
(A) The accumulation of low molecular mass peptides (<6 kD) in the map1A+F line was evidenced in 1D gel electrophoresis (16%), and the band annotations correspond to the identified proteins reported in Supplemental Table 5 online.
(B) The exploded pie chart displays the relative amounts of the peptides identified in spots 1 to 3 (Figure 6A) as a function of the calculated molecular weight of the corresponding protein (for protein identifications, see Supplemental Table 5 online).
(C) Gel filtration chromatography was performed with map1A + 100 nM Fumagillin (orange and purple) and the wild type (blue and green) at 3 DAI (top panel) and 8 DAI (bottom panel). Chromatograms at 215 nm are shown as dotted lines. The total elution volume was collected in 1-mL fractions, and peptides/small proteins were quantified in each fraction (solid line). The green zone defines the resolution volume efficiency of the column, the pink zone on the left depicts the size-exclusion limit (20 kD), and the pink zone on the right shows nonspecific components retained on the column.
[See online article for color version of this figure.]
In conclusion, our data support the idea that the increase in proteolytic activity associated with cNME inhibition is caused principally by an increase in the size of the pool of proteins available for rapid degradation. This degradation process probably involves the proteasome machinery.
DISCUSSION
Partial Retention of the N-Terminal Met of Cytosolic Proteins Increases General Cellular Proteolysis
The proteolytic process of NME was discovered several decades ago (Waller, 1963; Capecchi, 1966). Our current knowledge of this essential mechanism, particularly in higher eukaryotes, has significantly advanced in recent years. This is probably because NME is recognized as an excellent therapeutic target, with several natural and synthetic inhibitors (Boularot et al., 2004). These inhibitors have considerable potential for use in the treatment of various human diseases, from cancer to bacterial and parasitic infections. The inhibition of NME causes cell cycle arrest in tumor cells (Yeh et al., 2000; Zhang et al., 2000), developmental defects (Boxem et al., 2004; Yeh et al., 2006), and early senescence in plants (Ross et al., 2005). Accordingly, recent data have also suggested that the expression of NME enzymes is tightly regulated throughout development, in response to abiotic stress and tumorigenesis (Selvakumar et al., 2006; Dill et al., 2009), suggesting that the fraction of proteins undergoing NME may vary over space and time. Despite the demonstration that this process is essential in all organisms and its identification as a valuable target for the design of new antibacterial and anticancer agents, the molecular mechanism underlying the physiological requirement for NME function in living cells remains largely unknown.
The analysis presented here deals with the >15,000 nuclear-encoded proteins that are the substrates of NME in the cytosol. Our global study provides evidence that the in vivo inhibition of NME activity induces the retention of N-Met in proteins that normally undergo this process (see Supplemental Data Set 1 online), leading to an increase in the pool of proteins available for rapid degradation. Indeed, our proteomic, metabolic, and peptide profiling studies highlight an increase in proteolysis in vivo directly associated with NME inhibition.
The Proteolysis Triggered by cNME Inhibition Illustrates the Importance of Both the Nature and Size of the Protein Substrate Pool for Regulating Protein Degradation Rates
Proteolysis plays a crucial role in many vital processes (Vierstra, 2003; Doucet et al., 2008). In each of these processes, proteolysis arbitrates protein lifespan, eliminates misfolded or damaged proteins, and maintains free amino acid pools. As expected for a fundamental process, proteolysis is tightly regulated, and the uncontrolled degradation of intracellular proteins has been systematically linked with major effects in many organisms (Marino et al., 2008; Mizushima et al., 2008; Taylor et al., 2008; van der Hoorn, 2008). Efforts have been made to characterize the upstream mechanisms operating in the control of proteolytic activities, but much less is known about the precise cascade of events following the deregulation of this process. Dynamic increases or decreases in protein degradation may be accomplished in many ways: by altering protease pools, modulating protease activity, and/or controlling protein turnover (Hanna and Finley, 2007; Kurepa and Smalle, 2008; Ravid and Hochstrasser, 2008; Yen et al., 2008). In various physiological processes and under changing environmental and physiopathological conditions, the transcriptional and posttranslational regulation of most proteases has been shown to be strong and highly structured (Montuori et al., 2002; Yan and Boyd, 2007; Kurepa and Smalle, 2008; Wladyka and Pustelny, 2008). For instance, a combination of gene expression analyses and proteasome suppression studies has unraveled the coordinated regulation of 26S proteasome genes during plant development (Kurepa and Smalle, 2008). Caspases, the major effectors of apoptosis, constitute a family of Asp-specific proteases normally synthesized as inactive precursors but activated at the onset of apoptosis after proteolytic cleavage (Logue and Martin, 2008). The calcium-dependent proteolytic system involves calpains, a group of Cys proteases. In addition to being regulated by calcium, calpain activity is tightly regulated by the specific endogenous inhibitor, calpastatin, and by binding to phospholipids, autoproteolysis, and phosphorylation (Dargelos et al., 2008).
Diverse protease activities were measured to obtain a general picture of changes in protease pools and protease activity (see Supplemental Figure 5 online). No significant differences were found between the wild type and the map1A+F mutant, consistent with the notion that the increase in proteolytic activity associated with cNME inhibition resulted principally from an increase in the pool of proteins available for rapid degradation. Thus, cNME activity, by modifying the availability of several proteins for degradation by proteolytic systems, is an integral and fundamental element of the regulation of protein turnover.
Redox State of Glutathione and Proteolysis: Who's Controlling Who?
Hundreds of cell proteins, including many proteases, undergo reversible transitions between redox states. It is unclear whether redox-modified proteins/proteases have common redox controls and serve coordinate functions. The putative redox code integrating the metabolome, proteome, and genome remains largely undefined. By contrast, in complex biological mixtures, the redox state of a protein site is the net result of its many interactions with sulfur, metal, oxygen, and other redox factors. Cathepsin B is a model, redox-responsive, catabolic protein involved in turnover. The Cys/His pair within this protein acts simultaneously as a redox-responsive site and an inhibitory metal binding site and mediates a peptidolytic reaction mechanism. Mature cathepsin B can be inactivated by partitioning into multiple oxidation states. Cathepsin B can be reductively activated by GSH or disulfhydryl reductases and redox-buffered by GSH/GSSG. The interaction of the Cys/His site with iron provides a sensor, integrator, and effector switch coupling cathepsin B to metal-sulfuroxygen redox (Lockwood, 2005). It has also been shown that the proteasome is affected by oxidative stress to various degrees. Mild oxidation increases proteasomal degradation, whereas higher oxidant levels decrease proteasomal degradation. Moreover, the ATP-stimulated 26S proteasome is sensitive to oxidative stress, whereas the 20S form seems to be resistant (Breusing and Grune, 2008). As the downregulation of cNME generates a sustained low GSH/GSSG ratio, we checked whether this downregulation induced an oxidative stress that might affect proteolytic activity in map1A+F conditions. A slight increase in the production of intracellular reactive oxygen species was observed when human hepatoma cells were cultured with a high concentration of TNP470, a derivative of Fumagillin (Yoshida et al., 1998). However, we observed no difference in the accumulation of cellular proteins irreversibly oxidized during development between the wild type and map1A+F, both of which behaved as previously described in other model systems (Johansson et al., 2004). Thus, the low concentration of Fumagillin used in this study was insufficient to induce generalized cellular oxidation. Moreover, the lack of complementation of the map1A+F mutant phenotype under non-iron-limiting conditions also rules out the possibility that changes in intracellular glutathione levels reflect an adaptation of cells to iron stress (see Supplemental Figure 2B online). Finally, as cNME inhibition was always accompanied by a substantial increase in the total pool of amino acids, including the limiting precursors of glutathione, it is likely that also the increases in the levels of the reduced and oxidized forms of glutathione were reinforced by the increase in proteolytic activity. Accordingly, blocking the later step of glutathione biosynthesis with BSO prevented any complementation of the map1A+F mutant phenotype by amino acid precursors of the tripeptide thiol. Although no more than 50% of all protein species are thought to be sensitive to cNME inhibition (i.e., those losing their N-Met), these proteins are known to be the most abundant proteins in the cell (i.e., accounting for 80% of cellular proteins) (Martinez et al., 2008). They include key enzymes involved in energy metabolism and redox homeostasis (Boucherie et al., 1996). In the map1A+F mutant, increased degradation of proteins within this group most likely explains why NADPH concentration is depleted consequently. Together with the overall increase in glutathione linked to an increase in the levels of free amino acids, including Cys, NADPH depletion would further favor accumulation of GSSG.
As glutathione is a major cellular antioxidant and thiol/disulfide buffer, it is of considerable importance for cellular redox homeostasis and for diverse fundamental cellular functions. Several lines of evidence suggest that changes in the redox state of glutathione regulate one or more phases in the cell cycle closely linked to cell differentiation and programmed cell death (Cai and Jones, 1998; Henmi et al., 2005). Consistent with this, our in vivo data highlight the importance of changes in glutathione redox state during plant development, and the results presented here reveal that the cellular glutathione redox network has evolved to be closely intertwined with the machinery of protein turnover controlled by the cNME process.
A Working Model Depicting the Essential Role of cNME in the Crosstalk between the Cellular Proteolytic Machinery and Glutathione Homeostasis
Our global comparative analysis pointed out that the cNME developmental defect was primarily due to the maintenance of the glutathione redox state at an inappropriately oxidized level throughout plant development. We showed that the abnormal development induced by insufficient cNME pathway activity was linked to both the presence of large amounts of glutathione and a lower GSH/GSSG ratio in the map1A+F mutants. Based on our results, this disturbance of glutathione levels seems to be caused by two effects of cNME inhibition acting in synergy. Comparative proteomic studies, peptide profile analysis, protease activities, and HPLC analysis of free amino acids strongly suggested that retention of the first Met residue increased cellular proteolytic activity by incrementing the size of the pool of proteins suitable for processing. The large increase in amino acid content, including all the constituent amino acids of glutathione (Glu, Cys, and Gly), detected when cNME was inhibited is a cellular marker of this situation. Thus, the concomitant increases in Glu, Cys, Gly, and glutathione concentrations are consistent with (though do not prove) activation of glutathione synthesis rates. Finally, the identification of otherwise stable proteins that become unstable when NME is inhibited and an analysis of GSH/GSSG ratios and NADPH provided evidence that the enzymes involved in the reductant generation are targeted for degradation, preventing the efficient reduction of GSSG to GSH. The crucial role of glutathione redox status as a redox stabilizer preventing the stimulation of proteolysis by NME inhibition is demonstrated by complementation of the developmental defect associated with cNME inhibition by reduced glutathione and the partial complementation of this defect observed with NADPH. Taken together, our data demonstrate that cNME plays a key role in maintaining cellular glutathione content and redox state during development via a tight control of interconnected processes encompassing the proteolytic activity of the cell.
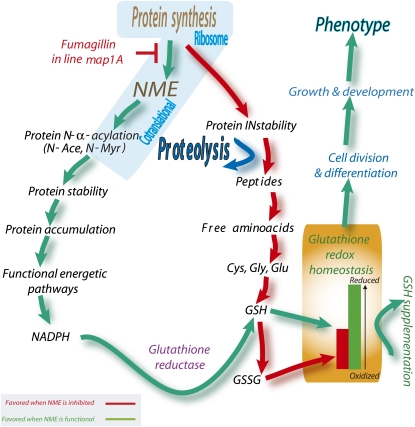
Our observations are consistent with a model in which, during rapid cell division, such as occurs in the early growth of plantlets after germination, cNME is an essential player in the dialogue between proteolytic activity and glutathione redox state that functions as part of a cellular checklist for continued development (Figure 8). It should be remembered that in the course of protein synthesis, the first Met residue of most accumulating proteins (∼80%) is removed by cNME. The released Met residues join the cellular free Met pool, whereas the new nascent chain becomes part of the largest population of stable proteins. At steady state, a small but non-negligible percentage of proteins do not undergo N-Met excision (∼20%). As a result, two distinct pools of Met polypeptide chains can be distinguished in the cell: one half with an N-α-acetylated Met residue (these proteins join the pool of stable cellular proteins) and the other with free Met residue. This second group of proteins establishes a pool of highly unstable proteins, the degradation of which supplies amino acids, including Cys, the key precursor of GSH, to free amino acid pools. In the model shown in Figure 8, fine adjustments in cNME regulate glutathione homeostasis by modulating the pool of unstable proteins with an N-terminal free Met. Thus, cNME inhibition is likely to increase the pool of proteins with a free Met residue, probably because the N-α-acetylation of this residue is much less frequent than that of other residues, such as Ala or Ser, which are unmasked by the removal of Met. Indeed, N-terminal Ala or Ser residues are substrates of NatA, which cannot acylate N-terminal Met. The narrow substrate specificity of the other two Nat enzymes (NatB and NatC) cannot compensate for the increase in the amount of substrates with an N-terminal Met.
Figure 8.
A Crucial Role for NME in the Glutathione-Proteolysis Crosstalk Necessary for Appropriate Growth and Development.
NME downregulation (red path) triggers an increase of cellular proteolytic activity by incrementing the size of the pool of proteins suitable for processing (Proteolysis). The large increase in amino acid content, including all precursors of glutathione (Glu, Cys, and Gly), detected when cNME was inhibited, is a cellular marker of this situation. Thus, the increase in Glu, Cys, and Gly concentrations probably drives glutathione synthesis rates up. Finally, the identification of otherwise stable proteins that become unstable when NME is inhibited and an analysis of GSH/GSSG ratios and NADPH provided evidence that the enzymes involved in reductive power generation are targeted for degradation, preventing the efficient reduction of GSSG to GSH. In cells undergoing appropriate development, this loop is broken by NME, which thus functions as a cycle interrupter (green path). As we have shown here, the loop can also be broken by establishing a more appropriate GSH/GSSG ratio (GSH supplementation). Glutathione reductase activity might correspond to GR1 and to the NADPH-dependent thioredoxin system, as recently proposed (Marty et al., 2009).
[See online article for color version of this figure.]
Conclusion
While important crosstalk between cNME and the NADPH-glutathione system has been demonstrated here, it cannot be ruled out that other crucial cellular regulatory mechanisms can be controlled by cNME. In our tightly controlled system in which everything is regulated to depict the earliest molecular mechanistic insight associated with the deregulation of the NME process, the perturbation of glutathione redox state was found to be the first limiting reversible step promoting the phenotype associated with map1A+F mutants. Thus, we suggest that the regulation of other mechanisms by NME will be influenced by the crucial crosstalk identified between NME, general proteolysis, and thiol status. We expect these new mechanisms will be described in the future, for instance, by studying the NME process and its deregulation in other specific conditions.
METHODS
All data presented are the means of two to four technical replicates for each of at least three biological replicates (i.e., at least six distinct measurements). All vertical bars in the figures indicate the se.
Materials
All chemicals were purchased from Sigma-Aldrich Chimie. All compounds and inhibitors were dissolved in water, with the exception of Fumagillin, which was dissolved in methanol.
Plant Material and Growth Conditions
The material used was sampled from Arabidopsis thaliana ecotype Columbia. The SALK_021985 (map1A) Salk T-DNA insertion line was obtained from the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk) as previously described (Ross et al., 2005). Seeds were sterilized and sown on 0.5× Murashige and Skoog (Sigma-Aldrich) medium supplemented with 0.8% agarose (Difco) and 1% sucrose (Sigma-Aldrich). Petri dishes were incubated in a growth chamber (22°C, 16 h of daylight; light intensity 100 μE·m−2·s−1) for up to 2 weeks. Plant material samples were collected in 2-mL Eppendorf tubes, weighed, rapidly frozen in liquid nitrogen, and stored at −80°C or in liquid nitrogen until protein or metabolite extractions. For the complementation experiments, Arabidopsis seeds or seedling were grown in plates containing the indicated molecules.
Yeast Strains, Growth Conditions, and Recovery Experiments
Experiments were performed with Saccharomyces cerevisiae wild-type, Δmap1 (BY4741; MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ; YLR244c∷kanMX4), and Δmap2 (BY4741; MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ; YBL091c∷kanMX4) strains from Euroscarf. This bank provided deletion strains in which the gene of interest was replaced by a KanMX module and a gene-specific tag. The deletion was confirmed by PCR, according to the procedure described and with the primers indicated on the Saccharomyces Genome Deletion Project website (http://www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html). Yeast strains were usually propagated on rich medium (2% [w/v] peptone, 1% [w/v] yeast extract, and 2% [w/v] glucose) at 30°C. For recovery experiments, the phenotype was investigated on plates containing the indicated molecules by spotting 1:10 serial dilutions of exponential cultures and recording growth after 3 or 6 d of incubation at 30°C.
Haloferax volcanii Growth Conditions
The H. volcanii wild-type (DS2) strain was grown at 45°C in a rich medium containing yeast extract (5.2 g/L final concentration), peptone (1 g/L), casamino acid (1 g/L), NaCl (2.46 M), MgCl2 (88 mM), MgSO4 (89 mM), KCl (54 mM), CaCl2 (3 mM), Tris-HCl, pH 7.5 (12 mM), and KOH (2 mM). For rich solid medium, 15 g/L Bacto agar was added to the solution. Colonies were harvested from a fresh culture and resuspended in rich liquid medium and grown to an OD650 of 0.5 to 0.8. Then, 10 μL of a 10-fold serial dilution was spotted in triplicate onto plates containing rich medium supplemented with the indicated compound(s). Photographs were taken after 3 to 7 d of incubation.
Protein Extraction and Analysis
Plant tissues (10 to 400 mg) were frozen in liquid nitrogen and ground in a 2-mL microcentrifuge tube containing both 3 and 5 mm iron beads for 1 min each, using a MM 300 mixer mill at 30 Hz (Qiagen). The resulting fine powder was dissolved in 1 mL of lysis buffer as previously described (Espagne et al., 2007). The homogenates were incubated at 4°C for 20 to 45 min with shaking. The supernatants were separated from the insoluble fraction by centrifugation at 15,000g at 4°C for 0.5 to 1 h. The resulting supernatants were used to measure protein concentration by the Bradford protocol (Bio-Rad) and were stored at −80°C unless used immediately. Protein gel blotting was performed as previously described (Ross et al., 2005), and antibody against both GR1 and GR2 was provided by Agrisera. Before 2D electrophoresis analysis, samples were subjected to a modified TCA/acetone precipitation (Espagne et al., 2007). The precipitated pellets were resuspended in 50 μL of rehydration buffer E (8.75 M urea, 2.5 M thiourea, 1.25% [v/v] IPG buffer containing 0.5% ampholine, pH 4 to 7 [GE Healthcare Life Sciences], 0.25% Triton X-100, 2.5% CHAPS, and 25 mM DTT) and stored at −80°C until use.
2D Gel Electrophoresis
Protein concentration was determined using the 2-D Quant Kit (GE Healthcare Life Sciences). Sample preparation and gel electrophoresis were essentially as described elsewhere (Espagne et al., 2007). The Ettan DALT System (GE-Healthcare Life Sciences) and 1-mm-thick SDS-denaturing polyacrylamide gels (11%) were used for the second dimension. Gels were run at 50 V for 1 h and 100 V for 16 h at 10°C using a cathodic solution (384 mM glycine, 25 mM Trizma base, and 0.2% [w/v] SDS) and an anodic solution (192 mM glycine, 25 mM Trizma base, and 0.15% [w/v] SDS). Gels were stained with colloidal Coomassie blue, and spots were excised for MS. The gels were scanned with a GS800 imaging densitometer (Bio-Rad) and analyzed with PD-Quest 7.1 software (Bio-Rad). Each analysis reported is the result of six replicates. Selected spots were picked with a sterile 1-mL Eppendorf tip. The complete data set is shown in Supplemental Table 1 online.
LC-MS and Protein Identification
The 2D gel spots were picked, digested, and analyzed essentially as described elsewhere (Espagne et al., 2007) and as follows. In-gel digestion was performed with the Progest system (Genomic Solution) using a standard trypsin protocol. Gel pieces were washed twice in 10% acetic acid, 40% ethanol, and acetonitrile (ACN) (in two separate baths). They were then washed twice in 25 mM NH4CO3 and ACN (again, two separate baths). Digestion was then performed for 6 h at 37°C with 125 ng of modified trypsin (Promega) dissolved in 20% methanol and 20 mM NH4CO3. The peptides were extracted with 2% trifluoroacetic acid (TFA) and 50% ACN and then with pure ACN alone. Peptide extracts were dried in a vacuum centrifuge, and the resulting powder was suspended in 20 μL of 0.05% TFA, 0.05% HCOOH, and 2% ACN. HPLC was performed on an Ultimate LC system combined with a Famos autosampler and a Switchos II microcolumn switch system (Dionex). The sample (4 μL) was loaded at a flow rate of 5 μL/min−1 onto precolumn cartridge (stationary phase: C18 PepMap 100, 5 μm; column: 300 μm i.d., 5 mm; Dionex) and desalted with 0.05% TFA, 0.05% HCOOH, and 2% ACN. After 2.5 min, the precolumn cartridge was connected to the separating PepMap C18 column (stationary phase: C18 PepMap 100, 3 μm; column: 75 μm i.d., 150 mm; Dionex). The buffers used were 0.1% acetic acid (HCOOH), 3% ACN (buffer A), and 0.1% HCOOH, 95% ACN (buffer B). Peptides were separated using a linear gradient from 5 to 30% B for 25 min at 200 nL/min−1. A single run took 45 min, including the regeneration step in 100% buffer B and the equilibration step in 100% buffer A. Eluted peptides were analyzed online with a LCQ Deca XP+ ion trap (Thermo Electron), using a nanoelectrospray interface. Ionization (1.2 to 1.5 kV ionization potential) was achieved with a liquid junction and an uncoated capillary probe (10 mm i.d.; New Objective). Peptide ions were analyzed with Xcalibur 1.4, with the following data-dependent acquisition steps: (1) full MS scan (mass-to-charge ratio [m/z] 400 to 1900, centroid mode), (2) ZoomScan on a selected precursor (scan at high resolution in profile mode over an m/z window of 4), and (3) MS/MS (qz = 0.22, activation time = 50 ms, and collision energy = 40%; centroid mode). Steps 2 and 3 were repeated for the two major ions detected in step 1. Dynamic exclusion was set to 30 s. A database search was performed with Bioworks 3.2 (Thermo Electron). We looked for trypsin digestion (enzymatic cleavage), Cys carboxyamidomethylation (static), and Met oxidation (possible modifications). Precursor mass and fragment mass tolerance were 1.4 and 1 D, respectively. The Arabidopsis protein database (30,692 entries, 08/11/2005 update) from The Arabidopsis Information Resource (http://www.Arabidopsis.org/) was used. The tryptic peptides identified were filtered according to (1) cross-correlation score (Xcorr) >1.7, 2.2, and 3.3 for peptides with one, two, and three charges, respectively, and (2) a probability of <0.05. At least two different peptides were required. In cases of identification with only two or three MS/MS spectra, similarity between the experimental and theoretical MS/MS spectra was confirmed by eye.
For the identification of small polypeptides separated by 1D gels, the eluted peptides were analyzed online with a LTQ XL ion trap (Thermo Electron) using a nanoelectrospray interface. Ionization (1.5 kV ionization potential) was performed with a liquid junction and a noncoated capillary probe (10 μm i.d.; New Objective). Peptide ions were analyzed using Xcalibur 2.07 and the following data-dependent acquisition steps: (1) full MS scan (m/z 400 to 1900, centroid mode), (2) ZoomScan on a selected precursor (scan at high resolution in profile mode on a m/z window of 4), and (3) MS/MS (qz = 0.25, activation time = 30 ms, and collision energy = 35%; centroid mode). Steps 2 and 3 were repeated for the two major ions detected in step 1. Dynamic exclusion was set to 30 s. A database search was performed with XTandem 2008.02.01 (http://www.thegpm.org/TANDEM/). Enzymatic cleavage was declared as a trypsin digestion with one possible miscleavage event. Cys carboxyamidomethylation and Met oxidation were set to static and possible modifications, respectively. Precursor mass and fragment mass tolerance were 2.0 and 0.8 D, respectively. A refinement search was added with similar parameters except that semitrypsic peptide and possible N-terminal protein acetylation was sought. The Arabidopsis protein database (32,825 entries, version 8) from The Arabidopsis Information Resource (http://www.Arabidopsis.org/) and a contaminant database (trypsin, keratins, etc.) were used. Only peptides with an E value below 0.1 are reported. Identified proteins were filtered according to the following specifications: (1) at least two different peptides with an E value below 0.05 and (2) a protein E value below 10−4. In the case of identification on the basis of only two or three MS/MS spectra, similarity between the experimental and the theoretical MS/MS spectra was visually checked.
Amino Acid Analysis
Arabidopsis plantlets were grown as described above. For HPLC analyses, young plants were harvested at 3 and 8 DAI. Frozen plants were ground with a pestle in a mortar with liquid nitrogen to obtain a fine powder. Two different methods were used to analyze and confirm amino acid content. For the first method, the powder was further homogenized with 1 mL of 20 mM HCl. Plantlet extracts were centrifuged at 14,000g for 10 min at 4°C. Ten microliters of the supernatant was used for amino acid derivatization with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, using an AccQ-Fluor Reagent Kit (Waters). Derivatization was performed according to Waters' recommendation and 10 μL was injected onto the column. HPLC was performed with a Waters model 2695 separation module. Separations were performed on a 3.9 × 150-mm Waters Nova-Pak C18 column equipped with a Nova-Pak C18 Guard-Pak insert. Eluted amino acid derivatives were detected using a Waters model 2475 multi λ fluorescence detector with an excitation wavelength of 250 nm and a detection wavelength of 395 nm. Eluent A was 140 mM sodium acetate and 6.9 mM triethylamine at pH 5.05; eluent B was acetonitrile:water (60:40). The elution protocol was 0 to 0.5 min, 100% A; 0.5 to 5 min, linear gradient to 5% B; 5 to 35 min, linear gradient to 7.5% B; 35 to 41 min, linear gradient to 10% B; 41 to 44 min, 10% B; 44 to 54 min, linear gradient to 20% B; 54 to 61 min, 20% B; 61 to 71 min, linear gradient to 30% B; 71 to 86 min, linear gradient to 100% B as previously described (Kim et al., 2002). The flow rate was 1.0 mL/min. All standard solutions of amino acids (Sigma-Aldrich Chimie) were dissolved in and diluted with 20 mM HCl and analyzed at different concentrations to calculate a detector response factor. The second methods used for amino acid content determination involved GC-TOF-MS technology and derivatization with methoxyamine and N-methyl-N-(trimethylsilyl)trifluoroacetamide as previously described (Noctor et al., 2007).
Separation and Quantification of Peptides
One hundred milligrams of plantlets samples were mechanically crushed in liquid nitrogen, and proteins were extracted with 1 mL of protein extraction buffer as described above. After incubation and centrifugation at 4°C, the supernatant was recovered and 50 μL was separated by fast protein liquid chromatography (AKTA Explorer 10S; GE Healthcare) on a gel filtration column optimized for peptides analysis (Superdex peptide 10/300 GL; GE Healthcare). Elution was performed with a buffer at pH 7.0 containing 50 mM sodium phosphate and 150 mM NaCl at a flow rate of 0.5 mL·min−1 and recorded at 215 nm. Then, 1-mL fractions were collected and the peptide content of each fraction was quantified using the LavaPep Peptide and Protein Assay Kit (FluoProbes; Interchim) according to the procedure provided by the manufacturer. Fluorescence (excitation 540 nm; emission 630 nm) was recorded by a fluorescence microplate reader (Infinite M200; Tecan), and the peptide concentration was deduced from a standard curve. Each measurement is the average of three independent biological replicates each assessed twice.
Analysis of Small Metabolites
Analysis of small metabolic compounds was as previously described (Queval and Noctor, 2007). Oxidized and reduced forms of glutathione, ascorbate, and NADP(+) were quantified in HCl extracts. NADH and NADPH were quantified in alkaline extracts using spectrophotometric cycling assays. Thiol analyses were validated by HPLC after a derivation with monobromobimane as described (Queval and Noctor, 2007). GC-TOF-MS was performed as described elsewhere (Noctor et al., 2007).
Staining of Specific Protein Modifications
To detect the phosphorylated proteins, 1D gels were stained with ProQ-Diamond Phosphoprotein (Molecular Probes). Gels were stained according to the manufacturer's protocol. Carbonylated proteins were detected with the chemical and immunological reagents of the OxyBlot Oxidized Protein Detection Kit (Chemicon). The carbonyl groups in the protein side chains were derivatized to 2,4-dinitrophenylhydrazone (DNP) by reaction with 2,4-dinitrophenylhydrazine (DNPH). The chemiluminescence blotting substrate (ECLplus) was obtained from GE Healthcare Life Sciences and used according to the instructions provided by the manufacturer. Immobilon-P polyvinylidene diflouride membrane was from GE Healthcare Life Sciences. As described above, crude protein extracts were obtained at various developmental stages, the extracts were reacted with the carbonyl reagent, DNPH, dot-blotted onto polyvinylidene diflouride membranes, and oxidatively modified proteins were detected with anti-DNP antibodies. In general, 10 to 15 μg of protein were loaded into the slot-blot apparatus.
Antioxidant Enzyme Activity Measurements
Plantlet material (100 mg) was ground in liquid nitrogen, 100 mg insoluble polyvinylpyrrolidone was added, and the powder was extracted into 1.5 mL 0.1 m NaH2PO4, pH 7.5, and 1 mm EDTA. Samples were taken for chlorophyll analysis and the remainder of the extract was centrifuged for 10 min. An aliquot of the supernatant was loaded onto NAP-5 columns and the desalted eluant used for enzyme assays. Catalase was assayed as previously described (Veljovic-Jovanovic et al., 2001). Protein was assayed using the Bio-Rad kit. Chlorophyll was measured in 80% acetone at 663 and 645 nm. GR activity was determined by monitoring the oxidation of NADPH at 340 nm (ε = 6.22 M−1 cm−1) as described by Donahue et al. (1997). Ascorbate peroxidase activity was determined by monitoring the oxidation of ascorbate at 290 nm (ε = 2.88 mM−1 cm−1) as described elsewhere (Dutilleul et al., 2003).
Endoproteolytic Activities
Endoproteolytic activities were investigated using the EnzChek peptidase/protease assay kit (Invitrogen). Proteins were obtained using the extraction buffer described above without EDTA, PMSF, and protease cocktail inhibitors. After cell breaking and 30 min incubation at 4°C with extraction buffer, crude extracts were collected by centrifugation. One microliter was used in the test. The final volume was 100 μL with 90 μL of the manufacturer's buffer, 4 μL water, and 5 μL of the substrate solution. Time-course fluorescence intensity studies of the product released were performed with a Tecan Infinite M200 microplate fluorescence reader (excitation wavelength 490 nm and emission 520 nm). Slopes were determined during the first 20 min. Results were normalized for 1 μg of proteins in crude extract.
Proteasome Assay
Protein extracts were obtained using the procedure as described above. The extraction buffer contained no EDTA, PMSF, or protease cocktail inhibitor. Protein quantification was performed using the Bradford principle (Bio-Rad) with BSA as the standard. Proteasome assays were performed with the commercially available 20S Proteasome Activity Assay Kit (Chemicon International) and the fluorogenic substrates pack provided by Biomol. Suc-LLVY-AMC, Boc-LRR-AMC, or Z-LLE-AMC was used for assays of the chymotrypsin-like, trypsin-like, and caspase-like activity, respectively. Proteasome assays were performed at 37°C in 100-μL reaction volume containing 50 μM fluorogenic substrate and protein sample. The same amount of protein was used in each test (10 μg for 3-DAI samples and 15 μg for 8-DAI samples). Fluorescence intensity (λex. = 380 nm; λem. = 460 nm) was monitored with a Tecan Infinite microplate fluorescence reader for 2 h, with measurements taken every 3 min. Initial velocities were determined from the first linear part of the curves, and data were normalized per μg proteins. Specific activity, in pmol/min/μg of protein, was obtained with AMC standard curve. Statistical analysis was performed with R software (http://cran.cict.fr/).
Real-Time Quantitative RT-PCR
Total RNA extraction was performed from Arabidopsis tissues as previously described (Kay et al., 1987). Poly(A)+ RNA were purified using oligo(dT) 25 Dynabeads (Dynal). mRNA (200 ng) was reverse-transcribed and the obtained cDNA was then diluted 20 times. The PCR amplification was performed using the following specific oligonucleotides: for the cytosolic glutathione reductase (GR1; At3g24170), 5′-TGGATTAAAGCCTGAGGTGAAGACCAGAAT-3′ and 5′- CAGCAATGTCGCCTGTGCTGTATTTTGCA-3′; for the chloroplastic glutathione reductase (GR2; At3g54660), 5′-GCCCTCATAGTGACAAACTCCTCAGCT-3′ and 5-GGGGATGTTACTGACCGAATCAATTTGACT-3′. Real-time PCR mix was optimized with the LightCycler Faststart DNA Master SYBER Green Kit (Roche) for each primer pair using a standard cDNA. After the initial activation step (95°C, 10 min), amplification and quantification were performed as follows: denaturation for 5 s at 95°C, annealing for 5 s at 60°C, and extension for 10 s (for GR1 and GR2) or 20 s (for EF1α) at 72°C. The absence of nonspecific products was checked by melting curve analysis (95°C, 5 s and 60°C, 15 s, 95°C, heating rate 0.1°C/s). Each cDNA sample was quantified at least twice, using as reference a standard cDNA, studied in the same conditions. All experiments were performed with a LightCycler (Roche) and the data were processed with the LightCycler quantitative software 1.0 (Roche).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MAP2A (At2g44180), MAP2B (At3g59990), MAP1A (At2g45240), the cytosolyc MAPs; At3g24170, the cytosolic GR1 gene; and At3g54660, the plastid GR2 gene.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. cNME Inhibition Does Not Modify the Phosphoproteome or Protein Oxidation Levels.
Supplemental Figure 2. Rescue of the Defect Associated with cNME Inhibition in dsRNA Interference Arabidopsis Lines with Cys and Impact of Iron Cations on Growth.
Supplemental Figure 3. The High Level of GSSG in cNME-Deficient Plants Is Not Due to a Decrease in Glutathione Reductase Concentration.
Supplemental Figure 4. cNME Inhibition Rapidly Induces a Decrease in NADPH Nucleotide Levels.
Supplemental Figure 5. NME Perturbation Induced a Slight Adjustment of Proteolytic Activities.
Supplemental Table 1. Identities of Protein Spots with a Higher Intensity in NME-Inhibited Conditions.
Supplemental Table 2. Low Molecular Weight Proteins Are More Abundant in map1A+F 3 DAI Than in the Equivalent Fractions from the Wild Type.
Supplemental Data Set 1. A Complete List of the 441 Analyzed Spots (Giving a Total of 821 Expressed Protein Identifications) for Wild-Type and map1A+F Plantlets at 8 DAI.
Supplemental Data Set 2. Mass Spectrometry–Based Analysis of the Identified N Termini from Supplemental Table 1.
Supplemental Data Set 3. Identities of Protein Spots with a Lower Intensity in NME-Inhibited Conditions.
Supplementary Material
Acknowledgments
F.F. was supported by a PhD grant from the French Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche (MENRT) as part of Ecole Doctorale ED145 (Université Paris-Sud). We thank H. Millikallio (IGM, Orsay, France) and Guillaume Queval (IBP, Orsay, France) for technical advice and E. Espagne (IGM), D. Libri (CGM, Gif/Yvette, France), Z. Adam (Rehovot, Israel), and C. Foyer for critical reading of the manuscript. We also thank B. Franzetti (IBS, Grenoble, France) for providing H. volcanii wild-type strain and M. Toledano (CEA, Gif/Yvette, France) for providing the S. cerevisiae strains and helpful comments. This work was supported by Grant PGP04-11 from Centre National de la Recherche Scientifique, Grant BCMS-275 (MENRT, France), by Projets transversaux Institut Fédératif 87, and by Grants 3603 and 4920 from the Association pour la Recherche sur le Cancer (Villejuif, France).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Carmela Giglione (carmela.giglione@isv.cnrs-gif.fr).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Ball, L., Accotto, G.P., Bechtold, U., Creissen, G., Funck, D., Jimenez, A., Kular, B., Leyland, N., Mejia-Carranza, J., Reynolds, H., Karpinski, S., and Mullineaux, P.M. (2004). Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 16 2448–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar, R.S., Ashrafi, K., Futterer, K., Waksman, G., and Gordon, J.I. (2001). Biology and enzymology of protein N-myristoylation. In The Enzymes, F. Tamanoi and D.S. Sigman, eds (San Diego, CA: Academic Press), pp. 241–286.
- Boucherie, H., Sagliocco, F., Joubert, R., Maillet, I., Labarre, J., and Perrot, M. (1996). Two-dimensional gel protein database of Saccharomyces cerevisiae. Electrophoresis 17 1683–1699. [DOI] [PubMed] [Google Scholar]
- Boularot, A., Giglione, C., Artaud, I., and Meinnel, T. (2004). Structure-activity relationship and therapeutic potential of peptide deformylase inhibitors. Curr. Opin. Investig. Drugs 5 809–822. [PubMed] [Google Scholar]
- Boxem, M., Tsai, C.W., Zhang, Y., Saito, R.M., and Liu, J.O. (2004). The C. elegans methionine aminopeptidase 2 analog map-2 is required for germ cell proliferation. FEBS Lett. 576 245–250. [DOI] [PubMed] [Google Scholar]
- Breusing, N., and Grune, T. (2008). Regulation of proteasome-mediated protein degradation during oxidative stress and aging. Biol. Chem. 389 203–209. [DOI] [PubMed] [Google Scholar]
- Cai, J., and Jones, D.P. (1998). Superoxide in apoptosis. Mitochondrial generation triggered by cytochrome c loss. J. Biol. Chem. 273 11401–11404. [DOI] [PubMed] [Google Scholar]
- Cairns, N.G., Pasternak, M., Wachter, A., Cobbett, C.S., and Meyer, A.J. (2006). Maturation of arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol. 141 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi, M.R. (1966). Initiation of E. coli proteins. Proc. Natl. Acad. Sci. USA 55 1517–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew, O., Whelan, J., and Millar, A.H. (2003). Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 278 46869–46877. [DOI] [PubMed] [Google Scholar]
- Cleary, M.D., Singh, U., Blader, I.J., Brewer, J.L., and Boothroyd, J.C. (2002). Toxoplasma gondii asexual development: identification of developmentally regulated genes and distinct patterns of gene expression. Eukaryot. Cell 1 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dargelos, E., Poussard, S., Brule, C., Daury, L., and Cottin, P. (2008). Calcium-dependent proteolytic system and muscle dysfunctions: a possible role of calpains in sarcopenia. Biochimie 90 359–368. [DOI] [PubMed] [Google Scholar]
- Dasgupta, B., Yi, Y., Hegedus, B., Weber, J.D., and Gutmann, D.H. (2005). Cerebrospinal fluid proteomic analysis reveals dysregulation of methionine aminopeptidase-2 expression in human and mouse neurofibromatosis 1-associated glioma. Cancer Res. 65 9843–9850. [DOI] [PubMed] [Google Scholar]
- Dill, B.D., Dessus-Babus, S., and Raulston, J.E. (2009). Identification of iron-responsive proteins expressed by Chlamydia trachomatis reticulate bodies during intracellular growth. Microbiology 155 210–219. [DOI] [PubMed] [Google Scholar]
- Dixon, D.P., Skipsey, M., Grundy, N.M., and Edwards, R. (2005). Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 138 2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue, J.L., Okpodu, C.M., Cramer, C.L., Grabau, E.A., and Alscher, R.G. (1997). Responses of antioxidants to paraquat in pea leaves (relationships to resistance). Plant Physiol. 113 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet, A., Butler, G.S., Rodriguez, D., Prudova, A., and Overall, C.M. (2008). Metadegradomics: Toward in vivo quantitative degradomics of proteolytic post-translational modifications of the cancer proteome. Mol. Cell. Proteomics 7 1925–1951. [DOI] [PubMed] [Google Scholar]
- Dutilleul, C., Garmier, M., Noctor, G., Mathieu, C., Chetrit, P., Foyer, C.H., and de Paepe, R. (2003). Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, H., Takenaga, K., Kanno, T., Satoh, H., and Mori, S. (2002). Methionine aminopeptidase 2 is a new target for the metastasis-associated protein, S100A4. J. Biol. Chem. 277 26396–26402. [DOI] [PubMed] [Google Scholar]
- Espagne, C., Martinez, A., Valot, B., Meinnel, T., and Giglione, C. (2007). Alternative and effective proteomic analysis in Arabidopsis. Proteomics 7 3788–3799. [DOI] [PubMed] [Google Scholar]
- Florens, L., et al. (2002). A proteomic view of the Plasmodium falciparum life cycle. Nature 419 520–526. [DOI] [PubMed] [Google Scholar]
- Foyer, C.H., and Noctor, G. (2005). Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17 1866–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frottin, F., Martinez, A., Peynot, P., Mitra, S., Holz, R.C., Giglione, C., and Meinnel, T. (2006). The proteomics of N-terminal methionine cleavage. Mol. Cell. Proteomics 5 2336–2349. [DOI] [PubMed] [Google Scholar]
- Giglione, C., Boularot, A., and Meinnel, T. (2004). Protein N-terminal methionine excision. Cell. Mol. Life Sci. 61 1455–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglione, C., Fieulaine, S., and Meinnel, T. (2009). Cotranslational processing mechanisms: Towards a dynamic 3D model. Trends Biochem. Sci. 34 417–426. [DOI] [PubMed] [Google Scholar]
- Goldberg, A.L. (2003). Protein degradation and protection against misfolded or damaged proteins. Nature 426 895–899. [DOI] [PubMed] [Google Scholar]
- Griffith, E.C., Su, Z., Turk, B.E., Chen, S., Chang, Y.-H., Wu, Z., Biemann, K., and Liu, J. (1997). Methionine aminopeptidase (type 2) is the common target for angiogenesis AGM-1470 and ovalicin. Chem. Biol. 4 461–471. [DOI] [PubMed] [Google Scholar]
- Hanna, J., and Finley, D. (2007). A proteasome for all occasions. FEBS Lett. 581 2854–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henmi, K., Demura, T., Tsuboi, S., Fukuda, H., Iwabuchi, M., and Ogawa, K. (2005). Change in the redox state of glutathione regulates differentiation of tracheary elements in Zinnia cells and Arabidopsis roots. Plant Cell Physiol. 46 1757–1765. [DOI] [PubMed] [Google Scholar]
- Ito, H., Iwabuchi, M., and Ogawa, K. (2003). The sugar-metabolic enzymes aldolase and triose-phosphate isomerase are targets of glutathionylation in Arabidopsis thaliana: Detection using biotinylated glutathione. Plant Cell Physiol. 44 655–660. [DOI] [PubMed] [Google Scholar]
- Johansson, E., Olsson, O., and Nystrom, T. (2004). Progression and specificity of protein oxidation in the life cycle of Arabidopsis thaliana. J. Biol. Chem. 279 22204–22208. [DOI] [PubMed] [Google Scholar]
- Kanno, T., Endo, H., Takeuchi, K., Morishita, Y., Fukayama, M., and Mori, S. (2002). High expression of methionine aminopeptidase type 2 in germinal center B cells and their neoplastic counterparts. Lab. Invest. 82 893–901. [DOI] [PubMed] [Google Scholar]
- Kay, R., Chan, A., Daly, M., and McPherson, J. (1987). Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 236 1299–1302. [DOI] [PubMed] [Google Scholar]
- Kim, J., Lee, M., Chalam, R., Martin, M.N., Leustek, T., and Boerjan, W. (2002). Constitutive overexpression of cystathionine gamma-synthase in Arabidopsis leads to accumulation of soluble methionine and S-methylmethionine. Plant Physiol. 128 95–107. [PMC free article] [PubMed] [Google Scholar]
- Kurepa, J., and Smalle, J.A. (2008). Structure, function and regulation of plant proteasomes. Biochimie 90 324–335. [DOI] [PubMed] [Google Scholar]
- Li, X., and Chang, Y.H. (1995). Amino-terminal protein processing in Saccharomyces cerevisiae is an essential function that requires two distinct methionine aminopeptidases. Proc. Natl. Acad. Sci. USA 92 12357–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood, T.D. (2005). The transfer of reductive energy and pace of proteome turnover: A theory of integrated catabolic control. Antioxid. Redox Signal. 7 982–998. [DOI] [PubMed] [Google Scholar]
- Logue, S.E., and Martin, S.J. (2008). Caspase activation cascades in apoptosis. Biochem. Soc. Trans. 36 1–9. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin, C., and Overall, C.M. (2002). Protease degradomics: A new challenge for proteomics. Nat. Rev. Mol. Cell Biol. 3 509–519. [DOI] [PubMed] [Google Scholar]
- Marino, G., Ugalde, A.P., Salvador-Montoliu, N., Varela, I., Quiros, P.M., Cadinanos, J., van der Pluijm, I., Freije, J.M., and Lopez-Otin, C. (2008). Premature aging in mice activates a systemic metabolic response involving autophagy induction. Hum. Mol. Genet. 17 2196–2211. [DOI] [PubMed] [Google Scholar]
- Martinez, A., Traverso, J.A., Valot, B., Ferro, M., Espagne, C., Ephritikhine, G., Zivy, M., Giglione, C., and Meinnel, T. (2008). Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics 8 2809–2831. [DOI] [PubMed] [Google Scholar]
- Marty, L., Siala, W., Schwarzlander, M., Fricker, M.D., Wirtz, M., Sweetlove, L.J., Meyer, Y., Meyer, A.J., Reichheld, J.P., and Hell, R. (2009). The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc. Natl. Acad. Sci. USA 106 9109–9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel, T., and Giglione, C. (2008. a). Protein lipidation meets proteomics. Front. Biosci. 13 6326–6340. [DOI] [PubMed] [Google Scholar]
- Meinnel, T., and Giglione, C. (2008. b). Tools for analyzing and predicting N-terminal protein modifications. Proteomics 8 626–649. [DOI] [PubMed] [Google Scholar]
- Meinnel, T., Serero, A., and Giglione, C. (2006). Impact of the N-terminal amino acid on targeted protein degradation. Biol. Chem. 87 839–851. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., Levine, B., Cuervo, A.M., and Klionsky, D.J. (2008). Autophagy fights disease through cellular self-digestion. Nature 451 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montuori, N., Carriero, M.V., Salzano, S., Rossi, G., and Ragno, P. (2002). The cleavage of the urokinase receptor regulates its multiple functions. J. Biol. Chem. 277 46932–46939. [DOI] [PubMed] [Google Scholar]
- Noctor, G., Bergot, G., Mauve, C., Thominet, D., Lelarge-Trouverie, C., and Prioul, J.-L. (2007). A comparative study of amino acid measurement in leaf extracts by gas chromatography-time of flight-mass spectrometry and high performance liquid chromatography with fluorescence detection. Metabolomics 3 161–174. [Google Scholar]
- Pierre, M., Traverso, J.A., Boisson, B., Domenichini, S., Bouchez, D., Giglione, C., and Meinnel, T. (2007). N-Myristoylation regulates the SnRK1 pathway in Arabidopsis. Plant Cell 19 2804–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval, G., and Noctor, G. (2007). A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: Application to redox profiling during Arabidopsis rosette development. Anal. Biochem. 363 58–69. [DOI] [PubMed] [Google Scholar]
- Ravid, T., and Hochstrasser, M. (2008). Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh, M.D. (2006). Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2 584–590. [DOI] [PubMed] [Google Scholar]
- Ross, S., Giglione, C., Pierre, M., Espagne, C., and Meinnel, T. (2005). Functional and developmental impact of cytosolic protein N-terminal methionine excision in Arabidopsis. Plant Physiol. 137 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Fernandez, R., Fricker, M., Corben, L.B., White, N.S., Sheard, N., Leaver, C.J., Van Montagu, M., Inze, D., and May, M.J. (1997). Cell proliferation and hair tip growth in the Arabidopsis root are under mechanistically different forms of redox control. Proc. Natl. Acad. Sci. USA 94 2745–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender, J., Ohlrogge, J.B., and Shachar-Hill, Y. (2003). A flux model of glycolysis and the oxidative pentosephosphate pathway in developing Brassica napus embryos. J. Biol. Chem. 278 29442–29453. [DOI] [PubMed] [Google Scholar]
- Selvakumar, P., Lakshmikuttyamma, A., Dimmock, J.R., and Sharma, R.K. (2006). Methionine aminopeptidase 2 and cancer. Biochim. Biophys. Acta 1765 148–154. [DOI] [PubMed] [Google Scholar]
- Selvakumar, P., Lakshmikuttyamma, A., Lawman, Z., Bonham, K., Dimmock, J.R., and Sharma, R.K. (2004). Expression of methionine aminopeptidase 2, N-myristoyltransferase, and N-myristoyltransferase inhibitor protein 71 in HT29. Biochem. Biophys. Res. Commun. 322 1012–1017. [DOI] [PubMed] [Google Scholar]
- Shin, R., Alvarez, S., Burch, A.Y., Jez, J.M., and Schachtman, D.P. (2007). Phosphoproteomic identification of targets of the Arabidopsis sucrose nonfermenting-like kinase SnRK2.8 reveals a connection to metabolic processes. Proc. Natl. Acad. Sci. USA 104 6460–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, R.C., Cullen, S.P., and Martin, S.J. (2008). Apoptosis: Controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 9 231–241. [DOI] [PubMed] [Google Scholar]
- Tommasi, F., Paciolla, C., de Pinto, M.C., and De Gara, L. (2001). A comparative study of glutathione and ascorbate metabolism during germination of Pinus pinea L. seeds. J. Exp. Bot. 52 1647–1654. [PubMed] [Google Scholar]
- van der Hoorn, R.A. (2008). Plant proteases: From phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 59 191–223. [DOI] [PubMed] [Google Scholar]
- Veljovic-Jovanovic, S.D., Pignocchi, C., Noctor, G., and Foyer, C.H. (2001). Low ascorbic acid in the vtc-1 mutant of Arabidopsis is associated with decreased growth and intracellular redistribution of the antioxidant system. Plant Physiol. 127 426–435. [PMC free article] [PubMed] [Google Scholar]
- Vernoux, T., Wilson, R.C., Seeley, K.A., Reichheld, J.P., Muroy, S., Brown, S., Maughan, S.C., Cobbett, C.S., Van Montagu, M., Inze, D., May, M.J., and Sung, Z.R. (2000). The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra, R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8 135–142. [DOI] [PubMed] [Google Scholar]
- Waller, J.-P. (1963). The NH2-terminal residue of the proteins from cell-free extract of E. coli. J. Mol. Biol. 7 483–496. [DOI] [PubMed] [Google Scholar]
- Wang, J., Lou, P., and Henkin, J. (2000). Selective inhibition of endothelial cell proliferation by fumagillin is not due to differential expression of methionine aminopeptidases. J. Cell. Biochem. 77 465–473. [PubMed] [Google Scholar]
- Wladyka, B., and Pustelny, K. (2008). Regulation of bacterial protease activity. Cell. Mol. Biol. Lett. 13 212–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, C., and Boyd, D.D. (2007). Regulation of matrix metalloproteinase gene expression. J. Cell. Physiol. 211 19–26. [DOI] [PubMed] [Google Scholar]
- Yeh, J.R., Ju, R., Brdlik, C.M., Zhang, W., Zhang, Y., Matyskiela, M.E., Shotwell, J.D., and Crews, C.M. (2006). Targeted gene disruption of methionine aminopeptidase 2 results in an embryonic gastrulation defect and endothelial cell growth arrest. Proc. Natl. Acad. Sci. USA 103 10379–10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, J.R., Mohan, R., and Crews, C.M. (2000). The antiangiogenic agent TNP-470 requires p53 and p21CIP/WAF for endothelial cell growth arrest. Proc. Natl. Acad. Sci. USA 97 12782–12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, H.C., Xu, Q., Chou, D.M., Zhao, Z., and Elledge, S.J. (2008). Global protein stability profiling in mammalian cells. Science 322 918–923. [DOI] [PubMed] [Google Scholar]
- Yoshida, T., Kaneko, Y., Tsukamoto, A., Han, K., Ichinose, M., and Kimura, S. (1998). Suppression of hepatoma growth and angiogenesis by a fumagillin derivative TNP470: possible involvement of nitric oxide synthase. Cancer Res. 58 3751–3756. [PubMed] [Google Scholar]
- Zhang, Y., Griffith, E.C., Sage, J., Jacks, T., and Liu, J.O. (2000). Cell cycle inhibition by the anti-angiogenic agent TNP-470 is mediated by p53 and p21WAF1/CIP1. Proc. Natl. Acad. Sci. USA 97 6427–6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Yeh, J.R., Mara, A., Ju, R., Hines, J.F., Cirone, P., Griesbach, H.L., Schneider, I., Slusarski, D.C., Holley, S.A., and Crews, C.M. (2006). A chemical and genetic approach to the mode of action of fumagillin. Chem. Biol. 13 1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.