Abstract
The mechanisms involved in sensing oxidative signalling molecules, such as H2O2, in plant and animal cells are not completely understood. In the present study, we tested the postulate that oxidation of Met (methionine) to MetSO (Met sulfoxide) can couple oxidative signals to changes in protein phosphorylation. We demonstrate that when a Met residue functions as a hydrophobic recognition element within a phosphorylation motif, its oxidation can strongly inhibit peptide phosphorylation in vitro. This is shown to occur with recombinant soybean CDPKs (calcium-dependent protein kinases) and human AMPK (AMP-dependent protein kinase). To determine whether this effect may occur in vivo, we monitored the phosphorylation status of Arabidopsis leaf NR (nitrate reductase) on Ser534 using modification-specific antibodies. NR was a candidate protein for this mechanism because Met538, located at the P+4 position, serves as a hydrophobic recognition element for phosphorylation of Ser534 and its oxidation substantially inhibits phosphorylation of Ser534 in vitro. Two lines of evidence suggest that Met oxidation may inhibit phosphorylation of NR-Ser534 in vivo. First, phosphorylation of NR at the Ser534 site was sensitive to exogenous H2O2 and secondly, phosphorylation in normal darkened leaves was increased by overexpression of the cytosolic MetSO-repair enzyme PMSRA3 (peptide MetSO reductase A3). These results are consistent with the notion that oxidation of surface-exposed Met residues in kinase substrate proteins, such as NR, can inhibit the phosphorylation of nearby sites and thereby couple oxidative signals to changes in protein phosphorylation.
Keywords: calcium-dependent protein kinase (CDPK), hydrogen peroxide (H2O2), methionine oxidation, oxidative signalling, phosphorylation motif
Abbreviations: ACS, 1-amino-cyclopropane-1-carboxylate synthase; AMPK, AMP-activated protein kinase; CDPK, calcium-dependent protein kinase; DTT, dithiothreitol; MALDI–TOF-MS, matrix-assisted laser-desorption ionization–time-of-flight MS; MAPK, mitogen-activated protein kinase; Met, methionine; MetSO, methionine sulfoxide; NR, nitrate reductase; PMSR, peptide methionine sulfoxide reductase; PP1α, protein phosphatase 1α; pSer, phosphoserine; ROS, reactive oxygen species; SnRK1, SNF1-related protein kinase; SPS, sucrose-phosphate synthase
INTRODUCTION
ROS (reactive oxygen species) are generated during normal aerobic metabolism by a variety of reactions, including mitochondrial and thylakoid electron transport, and enzymes such as glycolate oxidase and cellular peroxidases [1]. Numerous mechanisms exist to detoxify the most highly reactive ROS, such as superoxide, to form H2O2, which is relatively stable and found at significant levels in plant cells [2]. An emerging concept in both plants and animals is that ROS such as H2O2 play an important role in cellular signalling pathways [3,4]. In plants, H2O2 can alter gene expression [5,6], induce stomatal closure mediated by abscissic acid [7], and at higher concentrations can initiate programmed cell death [8]. However, it is not entirely clear how mild oxidants such as H2O2 are sensed at the molecular level.
Perception of H2O2 could involve chemical modification of amino acid side chains in cellular proteins. Cysteine residues are generally considered to be the archetypal redox-regulatory amino acid as reversible oxidation of the thiol group to form disulfides, or in some cases sulfenic acid (-SOH) or sulfinic acid (-SO2H), is recognized to operate in redox signalling of a variety of proteins [9,10]. However, Met (methionine) residues in proteins can also be highly susceptible to modification by mild oxidants [11]. It has been demonstrated that Met oxidation occurs primarily on the surface of proteins and correlates closely with the solvent-exposed sulfur surface area [12,13]. Consequently, small changes in protein conformation can influence the susceptibility of specific Met residues to oxidation, and static structural analysis can often not accurately predict the degree to which specific residues are solvent exposed [14]. Interestingly, oxidation of Met to the sulfoxide MetSO converts the side chain of this amino acid from hydrophobic to polar and hydrophilic [11,15]. This dramatic change in the chemical nature of the residue, coupled with the fact that oxidation is reversible [16,17], makes this modification of Met of potential regulatory significance. However, whether the rate of Met oxidation is rapid enough to function in redox signalling has been questioned [9]. Although there is some debate about whether the rate of Met oxidation is rapid enough to function in redox signalling, it is clear that MetSO exists in vivo. Thus, MetSO is likely to play a role in ROS responses, but perhaps other mechanisms also play a role in the early and rapid responses. Nonetheless, it is important to understand the impact of MetSO on protein function to develop a fuller understanding of ROS responses.
Much of what is known about the significance of Met oxidation has come from animal studies. While the effect of oxidation is to decrease the hydrophobicity of the residue's side chain [15], structural changes can be induced as a result of Met oxidation that paradoxically result in an overall increase in the surface hydrophobicity of the protein [18]. Regardless of structural changes, Met oxidation can have a large impact on protein functionality, including interactions with other proteins. For example, the oxidation of specific Met residues attenuates binding of calmodulin to clients and is thought to play a role in downregulation of energy metabolism during stress and in altered calcium metabolism in aging animals [19]. Furthermore, a link between Met oxidation and post-translational modification of proteins involving phosphorylation has recently emerged with the demonstration that Met oxidation can alter the activities of protein phosphatases and protein kinases. The activity of calcineurin, a calcium/calmodulin-activated Ser/Thr protein phosphatase, is reduced by oxidation of Met residues in the calmodulin-binding domain of the enzyme [20], whereas CaMKII (Ca2+/calmodulin-dependent protein kinase II), a Ser/Thr protein kinase, is activated by oxidation of Met residues in the autoinhibitory domain of the enzyme [21].
We questioned whether the oxidation of Met residues in the vicinity of phosphorylation sites on the surface of proteins could alter recognition by the requisite protein kinase(s) and thereby serve as an additional mechanism to couple oxidative signals to changes in the post-translational modification of cellular proteins. The rationale behind this notion is that many Ser/Thr protein kinases target primary sequence motifs that involve hydrophobic residues as recognition elements. This includes the basophilic kinases, such as the CDPKs (calcium-dependent protein kinases) and SnRK1s (SNF1-related protein kinases) that are members of the same kinase superfamily [22], where a hydrophobic residue at the P − 5 position (i.e. five residues N-terminal to the phosphorylated residue) is often essential and a hydrophobic residue at P+4 is stimulatory [23–25]. We speculated that when Met is the required hydrophobic residue, its oxidation to MetSO would inhibit phosphorylation of the target Ser/Thr residue because the sequence no longer contains the hydrophobic recognition element. To test this supposition, we initially examined the in vitro phosphorylation of synthetic peptides with Met residues at different positions. Dramatic effects of Met oxidation on phosphorylation in vitro were observed and prompted us to examine whether this might occur in vivo. To do this, we monitored the phosphorylation status of NR (nitrate reductase) at the regulatory Ser534 site, which has a Met residue at the P+4 position that serves as a hydrophobic recognition element. Observed changes in NR phosphorylation in response to: (i) exogenous H2O2 and (ii) overexpression of a cytosolic MetSO-repair enzyme (PMSRA3; peptide MetSO reductase A3) are consistent with the notion that Met oxidation may play a role in coupling oxidative signals to changes in protein phosphorylation in vivo.
EXPERIMENTAL
Materials
Synthetic peptides were produced as amides, and were purchased either from Research Genetics (Huntsville, AL, U.S.A.), Bethyl Laboratories (Montgomery, TX, U.S.A.) or GenScript (Piscataway, NJ, U.S.A.), and were >90% pure. Activated MAPK (mitogen-activated protein kinase; no. 9101) and pSer (phospho-serine)-14-3-3-binding motif antibodies (no. 9606) were purchased from Cell Signaling Technology (Danvers, MA, U.S.A.). Anti-peptide antibodies that recognize an N-terminal sequence of Arabidopsis NR (superscript number indicate residues in Arabidopsis thaliana Nia2: anti-N-terminal, 11RLEPGLNGVVRSYK24) and the regulatory phosphorylation site (anti-pSer534, 528SLKKSVpSTPFMNT540) were produced by Bethyl Laboratories (Montgomery, TX, U.S.A.) and are described in [26]. Heterotrimeric AMPK (AMP-dependent protein kinase; no. PV4672) was purchased from Invitrogen (Carlsbad, CA, U.S.A.); PP1α (protein phosphatase 1α; no. 539493) was purchased from Calbiochem (San Diego, CA, U.S.A.), and soybean CDPKα and CDPKβ were expressed in Escherichia coli as previously described in [27].
Plant growth
A. thaliana (L.) Heynh ecotype Columbia (Col-0) was used as the wild-type. Plants for protein extraction and analysis were grown in a soil mixture with short days (8 h) in growth chambers (Conviron Model PGW36, Winnipeg, Canada) with a photosynthetic photon flux density of 100 μmol·m−2·s−1 at plant level and day/night temperature of 22/18 °C. Fully expanded rosette leaves were harvested into liquid nitrogen from 3- to 4-week-old vegetative plants, as specified in the text.
Peptide kinase assays
The incorporation of radiolabel from [γ-32P]ATP into substrate peptides was measured using the phosphocellulose filter-binding assay. Each 40 μl reaction mixture contained recombinant CDPK in 50 mM Mops/NaOH, pH 7.5, 0.2 mM DTT (dithiothreitol), 0.2 mM CaCl2 and 4 μg of peptide as indicated. Where indicated, CaCl2 was replaced with 2 mM EGTA for determining Ca-independent kinase activity. Reactions were initiated with 0.1 mM [γ-32P]ATP (150 c.p.m.·pmol−1) plus 10 mM MgCl2, and stopped after 10 min at room temperature (22 °C). Peptide assays with AMPK were performed in 25 μl reactions that contained 25 mM Hepes/NaOH, pH 7.4, 2.5 mM DTT, 10 mM MgCl2, 5 mM β-glycerophosphate, 0.5 mM EGTA, 0.01% (v/v) Triton X-100, 0.15 mM AMP, peptide as indicated and 0.1 mM [γ-32P]ATP (150 c.p.m.·pmol−1). As indicated, 1 mg/ml peptide solutions were pretreated with 150 mM H2O2 for 30 min at 25 °C to oxidize Met residue(s) and then taken to dryness under vacuum before resuspending in H2O for use in experiments.
MALDI–TOF (matrix-assisted laser-desorption ionization–time- of-flight)-MS analysis
Peptides (10 μg/ml) were diluted 1:20 into 0.1% TFA (trifluoroacetic acid), and then mixed 1:1 with saturated HCCA (α-cyano-4-hydroxycinnamic acid). An aliquot (usually 0.3 μl) was spotted onto a GE Healthcare probe and allowed to air dry. MS analysis was performed with an Amersham Ettan™ MALDI–TOF/Pro spectrometer, operated in the linear mode.
Immunoblot analysis
Standard SDS/PAGE gels were run using 10 μg of total protein and transferred to PVDF. For the custom antibodies, membranes were blocked with 2% fish gelatin in PBS and then probed with the indicated primary antibody. Detection was completed by using secondary antibodies labelled with IR dyes (Molecular Probes or Rockland Immunochemicals) and scanned and quantitated using a Li-Cor Odyssey scanner and software (Li-Cor Biosciences). Commercial antibodies were used similarly and the recommended protocols were followed.
Feeding exogenous H2O2 to Arabidopsis seedlings in liquid culture
Approx. 50 Arabidopsis seedlings were grown in standard 8 cm plastic Petri plates using half-strength Murashige and Skoog salts supplemented with 0.25% sucrose for 12–14 days in 16 h light, 22 °C. At 2 h into the photoperiod, the seedlings were moved to the dark for 1 h, at which time the indicated concentration of H2O2 was added. Treatments were continued in the dark for an additional 60 min, and then the seedlings were harvested, quickly rinsed in distilled water and frozen in liquid nitrogen. Protein extracts were completed using direct extracts in SDS sample buffer.
CDPK on-blot treatment
Standard SDS/PAGE gels were run using 10 μg of total protein and transferred to PVDF membranes. The membranes were blocked with 2% BSA in TBST (20 mM Tris/HCl, pH 7.5, 140 mM NaCl and 0.1% Tween 20) and then transferred to kinase buffer (50 mM Mops, 0.2 mM CaCl2 and 10 mM MgCl2). Recombinant Soybean CDPKβ was added at 15 μg/ml in kinase buffer containing 50% glycerol, 100 mM NaCl, 2.5 mM DTT and 0.2 mM ATP for 1 h at room temperature. Control reactions were run in parallel, but without ATP. After treatment, the standard procedure for immunoblots was completed.
RESULTS AND DISCUSSION
Methionine oxidation in peptide substrates can block phosphorylation in vitro
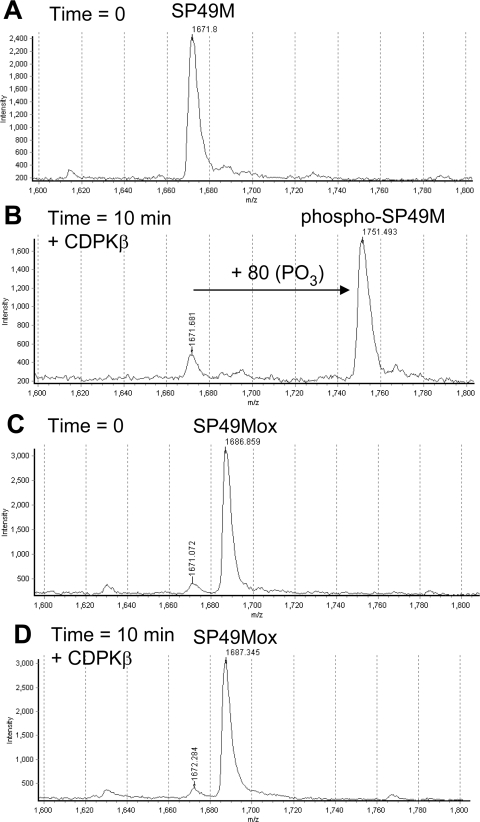
Recombinant CDPKs were tested for the ability to phosphorylate synthetic peptide substrates that in most cases contained a single Met residue that was either reduced (untreated) or oxidized by preincubation with H2O2 prior to the phosphorylation reaction. Initial experiments used the SP49M peptide, which corresponds to the Ser158 regulatory site of spinach SPS (sucrose-phosphate synthase) [28] and fits the canonical motif targeted by CDPKs and SnRK1 with hydrophobic recognition elements at the P−5 and P+4 positions. The sequence surrounding spinach SPS Ser158 contains Met residues at both of these positions as well as the P+3 position. However, the SP49M peptide sequence was modified to contain norleucine (J), a non-oxidizable homologue of Met, at the P+3 and +4 positions. As expected, the reduced form of the SP49M peptide was readily phosphorylated by CDPKβ, as monitored by an increase in the peptide molecular mass of 80 units using linear mode MALDI–TOF-MS analysis (Figures 1A and 1B). Preincubation of the SP49M peptide with 150 mM H2O2 increased the molecular mass by 16 units, corresponding to oxidation of the single Met residue at the P−5 position (designated SP49Mox; Figure 1C). Unlike the reduced peptide, SP49Mox was a poor substrate for CDPKβ, as after 10 min of incubation, there was no increase in peptide mass suggestive of phosphorylation (Figure 1D). It should be noted that H2O2 was effectively removed following the peptide treatment, and thus direct inhibition of CDPK is not the basis for the effect observed. In a separate experiment, the impact of Met oxidation on phosphorylation of the SP49M peptide was confirmed using the more quantitative analysis of peptide kinase activity with [γ-32P]ATP (Table 1, motif A). Thus, oxidation of Met at the P−5 position in a canonical CDPK motif can strongly inhibit phosphorylation by soybean CDPKβ. The SP49 peptide, which contains three Met residues, was a better substrate for CDPKβ than SP49M and, as expected, oxidation of the Met residues resulted in nearly complete inhibition of peptide phosphorylation (97% inhibition; Table 1, motif A). The Ser534 regulatory phosphorylation site of Arabidopsis NADH:NR [23,29,30] also conforms to the canonical motif and has a Met residue at the P+4 position, which is stimulatory but not essential for phosphorylation by CDPK and SnRK1 [23]. As expected, phosphorylation of a synthetic peptide based on the Ser534 site of NR (the S534 peptide, Table 1, motif A) was substantially reduced when the Met residue was oxidized, which is consistent with the known stimulatory role of a hydrophobic residue at this position.
Figure 1. Met oxidation in the synthetic peptide SP49M inhibits phosphorylation by recombinant soybean CDPKβ in vitro.
Peptides were analysed by linear mode MALDI–TOF-MS analysis. (A) The SP49M peptide (KGRMRRISSVEJJK, where J is norleucine and the phosphorylated Ser is in bold) at zero time or (B) after 10 min incubation with CDPKβ and ATP. Note the increase in mass of 80 units corresponding to single site phosphorylation. (C) Oxidation of the SP49M peptide by treatment with 0.5% H2O2 followed by lyophilization to remove the peroxide; note the increase in mass of 16 units corresponding to conversion of Met to MetSO in the SP49Mox peptide. (D) The SP49Mox peptide after incubation with CDPKβ and ATP for 10 min; note the absence of a + 80 peptide.
Table 1. Oxidation of Met residues in substrate peptides inhibits phosphorylation in vitro by soybean CDPKβ.
Residues at essential recognition positions are shown in italics, the phosphorylated residue is underlined, and Met residues are in bold; ϕ is a hydrophobic residue, B is a basic residue and x is any amino acid. Values are means±S.E.M. (n=3) from a representative experiment.
| Kinase activity with peptide (c.p.m.×10−3 incorporated) | |||||
|---|---|---|---|---|---|
| Motif | Peptide | Sequence | Reduced | Oxidized | % Inhibition |
| (A) ϕ-x-B-x-x-[S/T]-x-x-x-ϕ | |||||
| SP49 | KGRMRRISSVEMMK | 51.3±3.4 | 1.4±0.1 | 97 | |
| SP49M | KGRMRRISSVEJJK | 21.9±0.6 | 2.2±0.3 | 90 | |
| NR-S534 | SLKKSVSTPFMNT | 21.2±2.0 | 8.5±0.4 | 60 | |
| (B) B-B-x-B-ϕ-x(4)-[S/T]-x-B | |||||
| ACA2M-5 | RRFRMTANLSKRY | 161±8.1 | 9.6±1.4 | 94 | |
| (C) ϕ-B-ϕ-[S/T]-ϕ-x-B-B | |||||
| ACS-3 | KKNNLRLSFGKRMY | 39.1±0.3 | 40.4±0.7 | (3) | |
| ACSM−1 | NNLRMSFGKR | 106.4±4.6 | 68.2±2.8 | 36 | |
| ACSM+1 | NNLRLSMGKR | 47.0±2.5 | 5.3±0.1 | 89 | |
| ACSM-3 | NNMRLSFGKR | 96.3±3.1 | 33.8±0.7 | 65 | |
The recombinant CDPKβ will also readily phosphorylate certain peptides that do not match the canonical motif. For example, a peptide based on Ser45 of the Ca+2-ATPase, ACA2 [31], can be phosphorylated by CDPKs in vitro but lacks a basic residue at the P − 3 position (See Table 1, motif B). The ACA2 motif is defined by basic residues at P − 6 (and beyond) and P+2 and with a hydrophobic residue at P−5 [32]. Similar to the peptides conforming to the canonical motif, the ACA2M-5 peptide was a good substrate when the Met residue was reduced, but a very poor substrate when the Met at P−5 was oxidized (Table 1, motif B).
The second non-canonical motif identified for CDPKs was based on the Ser460 regulatory phosphorylation site of tomato ACS (1-amino-cyclopropane-1-carboxylate synthase) 2 [33]. The sequence around Ser460 does not conform to the canonical motif for CDPK substrates, because it lacks both the basic residue at P−3 and the hydrophobic residue at P−5, but nonetheless was shown to be efficiently phosphorylated by crude tomato extracts [33] and also recombinant CDPKs [27]. Analysis of peptide variants defined the ACS motif as Φ-Basic-Φ-Ser/Thr-Φ-X-Basic-Basic, where Φ is a hydrophobic residue and X is any amino acid [27]. The ACS3 peptide (KKNNLRLSFGKRMY) is based on the tomato ACS2 protein sequence and contains a single Met residue at the P+5 position (underlined); however, there is no recognized requirement for a hydrophobic residue at this position. Accordingly, treatment of the ACS3 peptide with H2O2 oxidized the single Met residue and increased the molecular mass by 16 Da to form the ACS3ox peptide, which was readily phosphorylated as monitored qualitatively by MALDI–TOF-MS (results not shown) or quantitatively by radiolabel incorporation (Table 1, motif C). Thus oxidation of Met residues at positions that are not essential as hydrophobic recognition elements does not inhibit phosphorylation. These results also indicate that the mild oxidant was effectively removed from the peptide preparations during processing, so that there was no H2O2 remaining that could directly inhibit the protein kinase.
CDPKs will also phosphorylate a shorter 10-residue peptide based on the non-canonical ACS motif [27]. Variants of the native peptide sequence (NNLRLSFGKR) were produced with a single Met substituted at each of the three positions (P−3, −1 and +1; underlined in the sequence) thought to be important for hydrophobic interactions with the protein kinase. All three of the Met-containing peptides were readily phosphorylated by CDPKβ when the Met was reduced, and in each case, oxidation of the Met residue resulted in substantial inhibition of phosphorylation (Table 1, motif C). Complete conversion of Met to MetSO was verified for each peptide by MALDI–TOF-MS analysis (results for the ACSM-1 peptide are shown in Supplementary Figure S1 at http://www.BiochemJ.org/bj/422/bj4220305add.htm), but the impact on peptide phosphorylation clearly varied with position, with MetSO at the P+1 position having the greatest effect and the P−1 position having the least effect. It is important to note that the relative importance of the hydrophobic residues at the three different positions was not established in the original report. Rather, it was simply determined that combined substitution of alanine residues at all three positions prevented phosphorylation [27]. Consistently, oxidation of a single Met residue at each of the three positions had a significant effect on peptide phosphorylation.
Collectively, the results suggest that Met residues must be in the reduced form in order to serve as hydrophobic recognition elements in phosphorylation motifs. This requirement is documented with three distinct motifs (canonical and non-canonical) and two protein kinases: CDPKβ (Table 1) and CDPKα (Supplementary Table S1 at http://www.BiochemJ.org/bj/422/bj4220305add.htm). This effect can also be extended to positions not recognized as hydrophobic recognition elements and to another distinct protein kinase, AMPK. One well known physiological target of human AMPK is Ser80 of ACC1 (acetyl-CoA carboxylase 1) [34], which contains a Met residue at the P−1 position. A 15-residue synthetic peptide based on this site (LHIRSSMS80GLHLY) conforms to the canonical motif and is a good substrate for human AMPK and soybean CDPKβ (Supplementary Figure S2 at http://www.BiochemJ.org/bj/422/bj4220305add.htm). Oxidation of the Met at the P−1 position strongly inhibited phosphorylation by both kinases, suggesting that while a hydrophobic residue at this position is not essential, a large polar residue is not well tolerated. Hence, the impact of Met oxidation at a specified position is motif-specific and can have a significant effect even at positions not thought to serve as hydrophobic recognition elements.
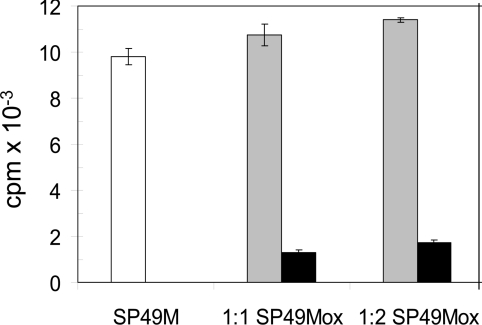
In principle, failure to phosphorylate peptides containing oxidized Met residues could be the result of a lack of peptide binding to the protein kinase or an inability of the kinase to phosphorylate the oxidized peptide. In order to distinguish between these two possibilities, increasing amounts of the SP49Mox peptide were added to reaction mixtures containing the untreated (reduced) SP49M peptide as substrate. As shown in Figure 2, addition of the SP49Mox peptide to reaction mixtures containing the SP49M peptide resulted in a slight increase in total [32P] incorporation equivalent to the activity supported by the oxidized peptide alone. The clear lack of competition suggests that the oxidized peptide is not recognized and does not bind to the kinase active site.
Figure 2. The oxidized peptide does not inhibit phosphorylation of the reduced SP49 peptide by CDPKβ.
Increasing amounts of the SP49Mox peptide [KGR(MetSO)RRISSVEJJK] were added to complete phosphorylation reaction mixtures containing the SP49M peptide (KGRMRRISSVEJJK). Total incorporation of [32P] was monitored with the SP49M peptide alone (white bar) or with an equivalent amount or 2-fold excess of the SP49Mox peptide (grey bars), or with the SP49Mox peptide alone (black bars). Values are means±S.E.M. (n=3) from a representative experiment.
In contrast, dephosphorylation of phosphorylated SP49M by lambda protein phosphatase or recombinant PP1α of rabbit skeletal muscle was relatively unaffected by oxidation of the Met residue at the P − 5 position. In these experiments, MALDI–TOF-MS was used to monitor the dephosphorylation of the synthetic phosphopeptides (designated phospho-SP49M or phospho-SP49Mox) during a 30 min incubation with the recombinant protein phosphatases. As shown in Supplementary Figure S3 (at http://www.BiochemJ.org/bj/422/bj4220305add.htm), the progress curves for dephosphorylation catalysed by PP1α were generally similar for the phosphorylated reduced and oxidized peptides, and quite distinct from the large effect Met oxidation had on peptide phosphorylation. Similar results were obtained with lambda protein phosphatase (results not shown). Collectively, these results suggest that peptidyl Met oxidation may potentially have more impact on phosphorylation rather than dephosphorylation, but this certainly needs to be studied further.
Effect of oxidative signals on phosphorylation of NR in vivo
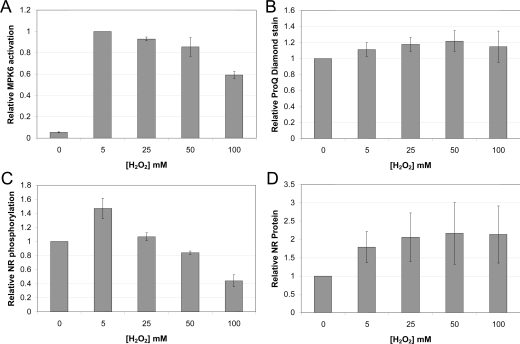
In order to determine whether Met oxidation affects protein phosphorylation in vivo, we focused on NR, which has a Met residue at the P+4 position that, when oxidized, can reduce phosphorylation of the regulatory Ser534 site (Table 1, motif A). The phosphorylation status of NR at the regulatory site can be monitored using modification-specific antibodies (anti-pSer534 antibodies), and total NR protein can be monitored using antibodies directed against an N-terminal sequence (anti-NT antibodies) [26]. Initially, we examined the effect of exogenous H2O2 on NR phosphorylation in Arabidopsis seedlings grown in liquid culture [35]. As a marker for ROS signalling, we used the activation of MAPKs, which is well known to respond to exogenous H2O2 [36–38]. In our system, MPK6 [38] activation was the most pronounced and was maximal with 5 mM H2O2. At higher concentrations of H2O2, MPK6 activation was reduced slightly and only decreased appreciably at 100 mM H2O2 (Figure 3A and Supplementary Figure S4A at http://www.BiochemJ.org/bj/422/bj4220305add.htm). Because the exposure time to H2O2 was relatively short (60 min), there was no apparent phytotoxicity or cell death in our seedlings, which can occur in response to high concentrations of H2O2 (50 to 100 mM) but requires longer periods of exposure. For example, infiltration of 70 mM H2O2 into Arabidopsis leaves resulted in cell death after 7 days [39], and treatment of the more sensitive suspension cells with 50 mM H2O2 produced little cell death within the first 3 h [40]. Accordingly, the seedlings in our system remained fully viable as evidenced by maintenance of phosphorylation of a subset of the plant proteome (arbitrarily set as proteins greater than 50 kDa; Figure 3B and Supplementary Figure S4D). In marked contrast, NR-Ser534 phosphorylation was increased by 5 mM H2O2 and then progressively decreased at higher concentrations (Figure 3C and Supplementary Figure S4B). Undoubtedly, many factors contribute to these changes in NR phosphorylation, with increased cytosolic [Ca2+] in response to exogenous H2O2 [41], potentially increasing phosphorylation of NR by CDPKs, whereas oxidation of NR-Met538 would be expected to cause inhibition of phosphorylation. The observed changes in relative NR phosphorylation (Figure 3C) are consistent with the notion that low concentrations of H2O2 are sufficient to trigger a rise in cytosolic [Ca2+], whereas higher concentrations are required for protein oxidation. It is also interesting to note that exogenous H2O2 increased total NR protein in the seedlings (Figure 3D and Supplementary Figure S4C). The steady-state level of Nia2 transcript is only increased 1.11-fold by H2O2 (20 mM for 1 h; Genevestigator, http://www.genevestigator.ethz.ch/gv), and so increased translation and/or decreased proteolytic degradation of NR protein may be responsible for the rapid changes in content observed. The upregulation of NR protein even at the highest concentration of H2O2 is also consistent with the notion that the seedlings were completely viable under the conditions tested.
Figure 3. Effect of exogenous H2O2 on the phosphorylation status of cellular proteins.
Liquid cultured Arabidopsis seedlings were treated with various concentrations of H2O2 for 1 h in the dark. (A) MAPK activation as monitored by immunoblotting with activation loop antibodies. (B) Integrated signal for ProQ Diamond phosphoprotein staining of cellular proteins greater than 50 kDa. (C) Relative phosphorylation of NR at the Ser534 site, expressed as the pSer534/NR ratio, where NR is total enzyme protein detected with an antibody directed against the N-terminus of NR. (D) NR protein content by immunoblotting. At 2 h into the photoperiod, the seedlings were moved to the dark for 1 h, at which time the indicated concentration of H2O2 was added. Treatments were completed in the dark, and after 1 h the seedlings were harvested. Values shown are means±S.E.M of three biological replicates, and within each panel values are normalized to the 0 mM H2O2 control, except in (A), which was normalized to the 5 mM H2O2 treatment.
The concentrations of exogenous H2O2 supplied to intact seedlings in the present study (up to 100 mM) are certainly in excess of reported values for the leaf content of H2O2, which range from roughly 0.5 to 5 μmol·g−1 fresh weight for different species (corresponding to 0.5 to 5 mM if distributed uniformly) [2,42]. The extent to which the internal H2O2 concentration was increased in our experiments is not known, but tissue barriers to uptake coupled with various mechanisms to remove H2O2 almost certainly reduced the internal concentration to well below the external concentration. Consequently, we applied exogenous H2O2 over a concentration range that was empirically determined to enhance ROS signalling but without loss of tissue viability. Hence, the results obtained should be physiologically relevant.
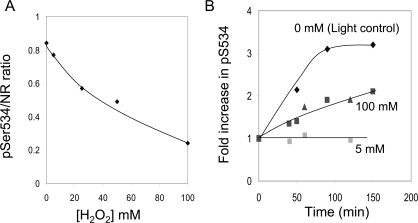
We developed an on-blot CDPK phosphorylation assay to determine whether Met538 oxidation might contribute to the reduced phosphorylation of NR-Ser534 in vivo in response to exogenous H2O2. In the experiment presented in Figure 4, exogenous H2O2 resulted in a progressive inhibition of NR phosphorylation (Figure 4A). Three samples of NR, differing in phosphorylation status at the Ser534 site, were then compared as substrates for CDPK phosphorylation on the PVDF membrane. The results are presented as the relative increase in the pSer534 signal in response to incubation with recombinant CDPKβ (Figure 4B). The sample prepared from seedlings exposed to 5 mM H2O2 had an initially high phosphorylation status and, as expected, no increase in phosphorylation was observed when incubated with CDPKβ. In contrast, the light control sample (obtained before seedlings were darkened) had an initial pSer534/NR ratio of approx. 0.35 and was rapidly phosphorylated in the on-blot assay. However, the extract from seedlings treated with 100 mM H2O2 also had a low pSer534/NR ratio and was phosphorylated on the blot, but at a lower rate compared with the light control sample. The difference in rate of on-blot phosphorylation of these two samples is consistent with the notion that oxidation of NR-Met538 occurred in vivo in response to exogenous H2O2 and contributed to the reduction in phosphorylation at the Ser534 site.
Figure 4. On-blot phosphorylation of NR at the Ser534 site.
(A) Arabidopsis seedlings were treated with exogenous H2O2 to generate samples differing in NR-Ser534 phosphorylation status as described in the legend of Figure 3. (B) On-blot phosphorylation of NR samples differing in phosphorylation stoichiometry. Samples from the 0 mM H2O2 (light control harvested before transferring plants to the dark) and 100 mM H2O2 treatment (dark sample) had low initial phosphorylation, whereas the 5 mM H2O2 treatment (dark sample) had a high initial phosphorylation status. The blotted proteins were incubated with CDPK plus ATP for the indicated time before probing with anti-NR-pSer534 antibodies. The relative increase in immunoblot signal is plotted. Results for the 100 mM H2O2 sample were obtained from two separate experiments.
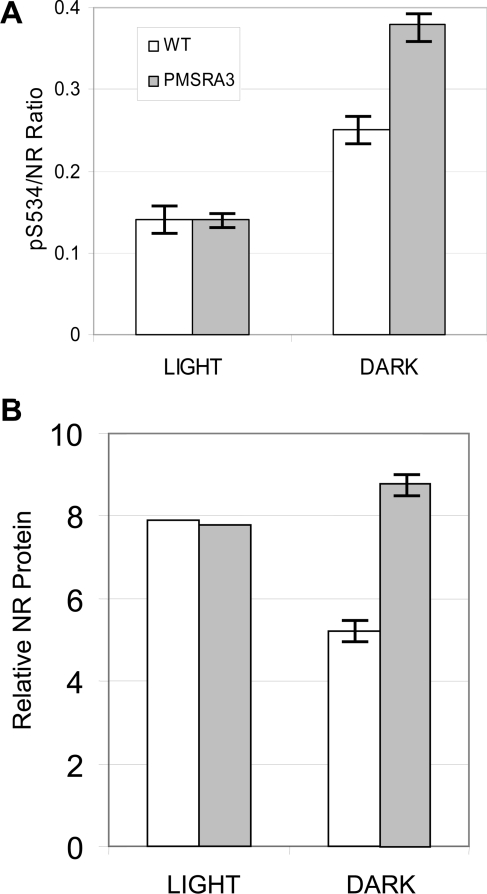
Another approach to determine whether Met oxidation affects NR phosphorylation in vivo was undertaken with plants overexpressing the MetSO-repair enzyme PMSRA3. A. thaliana MSRA3 encodes a cytosolic enzyme involved in the repair of S-MetSO diastereoisomers, and plants overexpressing this gene have increased tolerance to acute oxidative stress (treatment with methyl viologen) [43]. As shown in Figure 5(A), the relative phosphorylation status of NR at the Ser534 site was greater in darkened leaves compared with illuminated leaves [26,44], but the striking result was that overexpression of PMSRA3 increased NR-Ser534 phosphorylation in the dark, which is consistent with the notion that Met oxidation may inhibit NR phosphorylation in vivo. The increased relative phosphorylation of NR was also associated with increased absolute levels of NR protein in the transgenic plants relative to wild-type (Figure 5B). These results suggest that under normal dark conditions Met oxidation can attenuate NR phosphorylation.
Figure 5. Overexpression of PMSRA3 affects NR protein content and phosphorylation status in leaves of Arabidopsis plants grown in soil.
(A) Relative phosphorylation of NR at the Ser534 site, expressed as the pSer534/NR ratio. (B) NR protein content by immunoblotting. Light samples were collected from plants 2 h into the photoperiod, at which time lights were turned off and dark samples were collected after 2 and 12 h; values shown are means±S.E.M (n=3 for light samples and n=6 for dark samples).
The anti-pSer534 antibodies used in the experiments presented in Figures 4 and 5 were produced using the phosphopeptide SLKKSVpSTPFMNT as the antigen [26], raising the possibility that oxidation of Met538 may directly interfere with antibody cross- reactivity. In that case, a reduced immunoblot signal may not be the result of decreased phosphorylation but rather oxidation of the nearby Met residue. However, this seems unlikely for several reasons. First, pre-absorption of antibodies on a column with an immunobilized phosphopeptide corresponding to the C-terminal portion of the antigen peptide (SVpSTPFMNT) did not reduce the intensity of immunoblot signals, suggesting that the Met residue lies outside of the epitope (results not shown). Secondly, treatment of a blot with 1% H2O2 to oxidize Met residues on immobilized proteins had no effect on detection of total NR protein on the blot (using the anti-NT antibodies), whereas the anti-pSer534 immunoblot signal was increased slightly (results not shown). That inhibition was not observed strongly suggests that Met oxidation is not affecting our ability to monitor phosphorylation of Ser534. Finally, equivalent results for phosphorylation of NR at the Ser534 site were obtained with our antibodies compared with commercial antibodies against the mode 1 14-3-3 binding motif ([KR]-x-x-pSer-x-P; where x is any amino acid) that does not include the Met residue (Supplementary Figure S5 at http://www.BiochemJ.org/bj/422/bj4220305add.htm). Hence, reduced phosphorylation of NR-Ser534 at high exogenous H2O2 concentrations cannot be attributed to failure of the antibodies to bind to the phosphorylated Ser534 sequence when the nearby Met is oxidized.
Concluding remarks
The most important result of the present study is the demonstration that Met oxidation can act as a redox switch coupling oxidative signals to protein phosphorylation. This is shown to occur with synthetic peptides phosphorylated in vitro with CDPKs and human AMPK, and we suggest may apply broadly to protein kinases that target motifs with essential hydrophobic recognition elements. Identifying proteins where this mechanism may be important in vivo is a challenge for future research. Because phosphorylation sites are typically on the surface of proteins, Met residues in the vicinity are also likely to be solvent accessible and therefore susceptible to oxidation. We are speculating that the propensity for oxidation of Met residues has been exploited in Nature to serve as a redox switch to regulate phosphorylation of nearby sites. Identifying proteins with conserved Met residues in the vicinity of known phosphorylation sites is certainly one approach to catalogue possible candidate proteins that might be regulated in this manner.
We provide evidence in the present study that phosphorylation of NR may be regulated by oxidative signals involving the Met redox switch. The regulatory Ser534 phosphorylation site on NR controls binding of a 14-3-3 protein, to form a catalytically inactive complex (NR-pSer534–14-3-3) that modulates NR activity in response to light and other environmental signals [45,46]. The phosphorylation site occurs in a region of the NR molecule known as hinge 1, which contains ∼60 amino acids that connect the molybdenum cofactor domain and cytochrome b5 domain [47]. Based on limited structural information [48], this region is suggested to be surface exposed, which would certainly be necessary to allow phosphorylation and binding of a 14-3-3 protein, and also suggests that Met538 would be solvent exposed and therefore susceptible to oxidation. NR phosphorylation is sensitive to oxidative signals and two lines of evidence are consistent with the notion that Met538 oxidation may play a role. Certainly more work needs to be done to confirm and extend these results, but NR appears to be a good candidate protein to examine the role of Met oxidation as a redox switch controlling phosphorylation. It is possible that this mechanism may be involved in the activation of NR in vivo in response to hypoxia/anoxia [49], which paradoxically involves increased ROS production [50] and is beneficial for plant tolerance to root flooding [51]. This work just begins to explore the role Met oxidation plays in modulating protein phosphorylation of both plant and animal systems and future work will probably uncover many additional aspects in which the Met oxidation redox switch plays critical roles.
Online data
AUTHOR CONTRIBUTION
Shane Hardin, Clayton Larue, Man-Ho Oh and Vanita Jain performed research and analysed data. Steven Huber analysed data and wrote the paper.
ACKNOWLEDGEMENTS
We thank Professor Ming Tien (Department of Biochemistry and Molecular Biology, Pennsylvania State University, State College, PA, U.S.A.) for providing seeds of the PMSRA3-overexpressing plants. Mention of a trademark of proprietary product does not constitute a guarantee or warranty by the USDA-ARS and does not imply its approval to the exclusion of other products that might also be suitable. Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the National Research Initiative or the USDA-ARS.
FUNDING
This work was supported in part by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service [grant numbers 2007-35318-17801 and 2008-35318-18650] and by the US Department of Agriculture (USDA)-Agricultural Research Service (ARS). V. J. thanks the Department of Science and Technology for providing support in the form of a BOYSCAST fellowship.
References
- 1.Foyer C. H., Noctor G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol. Plant. 2003;119:355–364. [Google Scholar]
- 2.Cheeseman J. M. Hydrogen peroxide concentrations in leaves under natural conditions. J. Exp. Bot. 2006;57:2435–2444. doi: 10.1093/jxb/erl004. [DOI] [PubMed] [Google Scholar]
- 3.Foyer C. H., Noctor G. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neill S. J., Desikan R., Clarke A., Hurst R. D., Hancock J. T. Hydrogen peroxide and nitric oxide as signalling molecules in plants. J. Exp. Bot. 2002;53:1237–1247. [PubMed] [Google Scholar]
- 5.Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. Reactive oxygen gene networks of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Vranová E., Inzé D., Van Breusegem F. Signal transduction during oxidative stress. J. Exp. Bot. 2002;53:1227–1236. [PubMed] [Google Scholar]
- 7.Pei Z.-M., Murata Y., Benning G., Thomine S., Klusener B., Allen G. J., Grill E., Schroeder J. L. Calcium channels activated by hydrogen peroxide mediate abscisic acid signaling in guard cells. Nature. 2000;406:731–734. doi: 10.1038/35021067. [DOI] [PubMed] [Google Scholar]
- 8.Desikan R., Cheung M.-K., Bright J., Henson D., Hancock J. T., Neill S. J. ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J. Exp. Bot. 2004;395:205–212. doi: 10.1093/jxb/erh033. [DOI] [PubMed] [Google Scholar]
- 9.D'Autréaux B., Toledano M. B. ROS as signaling molecules: mechanisms that generate specificity in ROS homeostatis. Nat. Rev. Mol. Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 10.Fourquet S., Huang M.-E., D'Autréaux B., Toledano M. B. The dual functions of thiol-based peroxidases in H2O2 scavenging and signaling. Antiox. Redox Signal. 2008;10:1565–1576.. doi: 10.1089/ars.2008.2049. [DOI] [PubMed] [Google Scholar]
- 11.Vogt W. Oxidation of methionyl residues in proteins: tools, targets and reversal. Free Radical Biol. Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths S. W., Cooney C. L. Relationship between protein structure and methionine oxidation in recombinant human α1-antitrypsin. Biochemistry. 2002;41:6245–6252. doi: 10.1021/bi025599p. [DOI] [PubMed] [Google Scholar]
- 13.Yin D., Kuczera K., Squier T. C. The sensitivity of carboxyl-terminal methionines in calmodulin isoforms to oxidation by H2O2 modulates the ability to activate the plasma membrane Ca-ATPase. Chem. Res. Toxicol. 2000;13:103–110. doi: 10.1021/tx990142a. [DOI] [PubMed] [Google Scholar]
- 14.Chu J.-W., Yin J., Wang D. I. C., Trout B. L. Molecular dynamics simulations and oxidation rates of methionine residues of granulocyte colony-stimulating factor at different pH values. Biochemistry. 2004;43:1019–1029. doi: 10.1021/bi0356000. [DOI] [PubMed] [Google Scholar]
- 15.Black S. D., Mould D. R. Development of hydrophobicity parameters to analyze proteins which bear post- or cotranslational modifications. Anal. Biochem. 1991;193:72–82. doi: 10.1016/0003-2697(91)90045-u. [DOI] [PubMed] [Google Scholar]
- 16.Kim H. Y., Gladyshev V. N. Methionine sulfoxide reductases: selenoprotein forms and roles in antioxidant protein repair in mammals. Biochem. J. 2007;407:321–329. doi: 10.1042/BJ20070929. [DOI] [PubMed] [Google Scholar]
- 17.Tarrago L., Laugier E., Rey P. Protein-repairing methionine sulfoxide reductases in photosynthetic organisms: gene organization, reduction mechanisms, and physiological roles. Mol. Plant. 2009;2:202–217. doi: 10.1093/mp/ssn067. [DOI] [PubMed] [Google Scholar]
- 18.Chao C.-C., Ma Y.-S., Stadtman E. R. Modification of protein surface hydrophobicity and methionine oxidation by oxidative systems. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2969–2974. doi: 10.1073/pnas.94.7.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bigelow D. J., Squier T. C. Redox modulation of cellular signaling and metabolism through reversible oxidation of methionine sensors in calcium regulatory proteins. Biochim. Biophys. Acta. 2005;1703:121–134. doi: 10.1016/j.bbapap.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 20.Carruthers N. J., Stemmer P. M. Methionine oxidation in the calmodulin-binding domain of calcineurin disrupts calmodulin binding and calcineurin activation. Biochemistry. 2008;47:3085–3095. doi: 10.1021/bi702044x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erickson J. R., Joiner M. A., Guan X., Kutschke W., Yang J., Oddis C. V., Bartlett R. K., Lowe J. S., O'Donnell S. E., Aykin-Burns N., et al. A dynamic pathway for calcium-independent activation of CAMKII by methionine oxidation. Cell. 2008;133:462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hrabak E. M., Chan C. W. M., Gribskov M., Harper J. F., Choi J. H., Halford N., Kudla J., Luan S., Nimmo H. G., Sussmann M. R., et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachmann M., Shiraishi N., Campbell W. H., Yoo B.-C., Harmon A. C., Huber S. C. Identification of Ser-543 as the major regulatory phosphorylation site in spinach leaf nitrate reductase. Plant Cell. 1996;8:505–517. doi: 10.1105/tpc.8.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMichael R. W., Jr, Kochansky J., Klein R. R., Huber S. C. Characterization of the substrate specificity of sucrose-phosphate synthase protein kinase. Arch. Biochem. Biophys. 1995;321:71–75. doi: 10.1006/abbi.1995.1369. [DOI] [PubMed] [Google Scholar]
- 25.Weekes J., Ball K. L., Caudwell F. B., Hardie D. G. Specificity determinants for the AMP-activated protein kinase and its plant homologue analyzed using synthetic peptides. FEBS Lett. 1993;334:335–339. doi: 10.1016/0014-5793(93)80706-z. [DOI] [PubMed] [Google Scholar]
- 26.Oh M.-H., Huber J. L., Shen W., Athwal G. S., Wu X., Huber S. C. Overexpression of a directed mutant of 14-3-3ω in Arabidopsis leaves affects phosphorylation and protein content of nitrate reductase. Can. J. Bot. 2009;87:691–701. [Google Scholar]
- 27.Sebastia C. H., Hardin S. C., Clouse S. D., Kieber J. J., Huber S. C. Identification of a new motif for CDPK phosphorylation in vitro that suggests ACC synthase may be a CDPK substrate. Arch. Biochem. Biophys. 2004;428:81–91. doi: 10.1016/j.abb.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 28.McMichael R. W., Jr, Klein R. R., Salvucci M. E., Huber S. C. Identification of the major regulatory phosphorylation site in sucrose-phosphate synthase. Arch. Biochem. Biophys. 1993;307:248–252. doi: 10.1006/abbi.1993.1586. [DOI] [PubMed] [Google Scholar]
- 29.Douglas P., Morrice N., MacKintosh C. Identification of a regulatory phosphorylation site in the hinge 1 region of nitrate reductase from spinach (Spinacea oleracea) leaves. FEBS Lett. 1995;377:113–117. doi: 10.1016/0014-5793(95)01300-8. [DOI] [PubMed] [Google Scholar]
- 30.Su W., Huber S. C., Crawford N. M. Identification in vitro of a post-translational regulatory site in the hinge 1 region of Arabidopsis nitrate reductase. Plant Cell. 1996;8:519–527. doi: 10.1105/tpc.8.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang I., Sze H., Harper J. F. A calcium-dependent protein kinase can inhibit a calmodulin-stimulated Ca2+ pump (ACA2) located in the endoplasmic reticulum of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6224–6229. doi: 10.1073/pnas.97.11.6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang J.-Z., Hardin S. C., Huber S. C. Identification of a novel phosphorylation motif for CDPKs: phosphorylation of synthetic peptides lacking basic residues at P−3/P−4. Arch. Biochem. Biophys. 2001;393:61–66. doi: 10.1006/abbi.2001.2476. [DOI] [PubMed] [Google Scholar]
- 33.Tatsuki M., Mori H. Phosphorylation of tomato 1-aminocyclopropane- 1-carboxylic acid synthase, LE-ACS2, at the C-terminal region. J. Biol. Chem. 2001;276:28051–28057. doi: 10.1074/jbc.M101543200. [DOI] [PubMed] [Google Scholar]
- 34.Hardie D. G., Scott J. W., Pan D. A., Hudson E. R. Management of cellular energy by the AMP-activated protein kinase system. FEBS Lett. 2003;546:113–120. doi: 10.1016/s0014-5793(03)00560-x. [DOI] [PubMed] [Google Scholar]
- 35.Wang X., Goshe M. B., Soderblom E. J., Phinney B. S., Kuchar J. A., Li J., Asami T., Yoshida S., Huber S. C., Clouse S. D. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell. 2005;17:1685–1703. doi: 10.1105/tpc.105.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagami H., Soukupová H., Schikora A., Žárský V., Hirt H. A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 2006;281:38697–38704. doi: 10.1074/jbc.M605293200. [DOI] [PubMed] [Google Scholar]
- 37.Pitzschke A., Hirt H. Mitogen-activated protein kinases and reactive oxygen species signaling in plants. Plant Physiol. 2006;141:351–356. doi: 10.1104/pp.106.079160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuasa T., Ichimura K., Mizoguchi T., Shinozaki K. Oxidative stress activates ATMPK6, an Arabidopsis homologue of MAP kinase. Plant Cell Physiol. 2001;42:1012–1016. doi: 10.1093/pcp/pce123. [DOI] [PubMed] [Google Scholar]
- 39.Yang L.-X., Wang R.-Y., Ren F., Liu J., Cheng J., Lu Y.-T. AtGLB1 enhances the tolerance of Arabidopsis to hydrogen peroxide stress. Plant Cell Physiol. 2005;46:1309–1316. doi: 10.1093/pcp/pci140. [DOI] [PubMed] [Google Scholar]
- 40.Desikan R., Reynolds A., Hancock J. T., Neill S. J. Harpin and hydrogen peroxide both initiate programmed cell death but have differential effects on defence gene expression in Arabidopsis suspension cultures. Biochem. J. 1998;330:115–220. doi: 10.1042/bj3300115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rentel M. C., Knight M. R. Oxidative stress-induced calcium signaling in Arabidopsis. Plant Physiol. 2004;135:1471–1479. doi: 10.1104/pp.104.042663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Queval G., Hager J., Gakière B., Noctor G. Why are literature data for H2O2 contents so variable? A discussion of potential difficulties in the quantitative assay of leaf extracts. J. Exp. Bot. 2008;59:135–146. doi: 10.1093/jxb/erm193. [DOI] [PubMed] [Google Scholar]
- 43.Romero-Angulo H. M. On the role of the enzyme peptide methionine sulfoxide reductase in the response of Arabidopsis plants to oxidative stress [Ph.D. Thesis] State College, PA, U.S.A.: Pennsylvania State University; 2005. [Google Scholar]
- 44.Weiner H., Kaiser W. M. Antibodies to assess phosphorylation of spinach leaf nitrate reductase on serine 543 and its binding to 14-3-3 proteins. J. Exp. Bot. 2001;52:1165–1172. [PubMed] [Google Scholar]
- 45.Huber S. C., Bachmann M., Huber J. L. Post-translational regulation of nitrate reductase activity in higher plants: a role for Ca2+ and 14-3-3 proteins. Trends Plant Sci. 1996;1:432–438. [Google Scholar]
- 46.Moorhead G., Douglas P., Morrice N., Scarabel M., Aitken A., MacKintosh C. Phosphorylated nitrate reductase from spinach leaves is inhibited by 14-3-3 proteins and activated by fusicoccin. Curr. Biol. 1996;6:1104–1113. doi: 10.1016/s0960-9822(02)70677-5. [DOI] [PubMed] [Google Scholar]
- 47.Campbell W. H. Structure and function of eukaryotic NAD(P)H:nitrate reductase. Cell. Mol. Life Sci. 2001;58:194–204. doi: 10.1007/PL00000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer K., Barbier G. G., Hecht H.-J., Mendel R. R., Campbell W. H., Schwarz G. Structural basis of eukaryotic nitrate reduction: crystal structures of the nitrate reductase active site. Plant Cell. 2005;17:1167–1179. doi: 10.1105/tpc.104.029694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaiser W. M., Huber S. C. Post-translational regulation of nitrate reductase: mechanism, physiological relevance and environmental triggers. J. Exp. Bot. 2001;52:1981–1989. doi: 10.1093/jexbot/52.363.1981. [DOI] [PubMed] [Google Scholar]
- 50.Fukao T., Bailey-Seres J. Plant responses to anoxia – is survival a balancing act? Trends Plant Sci. 2004;9:449–456. doi: 10.1016/j.tplants.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 51.Stoimenova M., Libourel I. G. L., Ratcliffe R. G., Kaiser W. M. The role of nitrate reduction in the anoxic metabolism of roots II. Anoxic metabolism of tobacco roots with or without nitrate reductase activity. Plant Soil. 2003;253:155–167. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.