Abstract
A necessary feature of the immune system, TCR (T-cell receptor) cross-reactivity has been implicated in numerous autoimmune pathologies and is an underlying cause of transplant rejection. Early studies of the interactions of αβ TCRs (T-cell receptors) with their peptide–MHC ligands suggested that conformational plasticity in the TCR CDR (complementarity determining region) loops is a dominant contributor to T-cell cross-reactivity. Since these initial studies, the database of TCRs whose structures have been solved both bound and free is now large enough to permit general conclusions to be drawn about the extent of TCR plasticity and the types and locations of motion that occur. In the present paper, we review the conformational differences between free and bound TCRs, quantifying the structural changes that occur and discussing their possible roles in specificity and cross-reactivity. We show that, rather than undergoing major structural alterations or ‘folding’ upon binding, the majority of TCR CDR loops shift by relatively small amounts. The structural changes that do occur are dominated by hinge-bending motions, with loop remodelling usually occurring near loop apexes. As predicted from previous studies, the largest changes are in the hypervariable CDR3α and CDR3β loops, although in some cases the germline-encoded CDR1α and CDR2α loops shift in magnitudes that approximate those of the CDR3 loops. Intriguingly, the smallest shifts are in the germline-encoded loops of the β-chain, consistent with recent suggestions that the TCR β domain may drive ligand recognition.
Keywords: conformational selection, cross-reactivity, crystal structure, induced fit, peptide–MHC (pMHC), T-cell receptor (TCR)
Abbreviations: CDR, complementarity determining region; MBP, myelin basic protein; pMHC, peptide–MHC; TCR, T-cell receptor
INTRODUCTION
αβ TCRs (T-cell receptors) expressed by CD4+ or CD8+ T-cells are responsible for recognizing antigenic peptides bound and presented by class I or class II MHC proteins. Recognition of a pMHC (peptide–MHC) complex by a TCR is required for the initiation and propagation of a cellular immune response, as well as the generation and maintenance of the body's T-cell repertoire. In some regards, TCRs are similar to antibodies. Notably, TCR antigen-binding sites are composed of multiple CDR (complementarity determining region) loops generated via recombination processes similar to those used in antibody generation. However, one of the many differences between antibodies and TCRs is the nature of the ligand recognized. Whereas antibodies recognize linear or non-linear epitopes of seemingly unlimited chemical and structural diversity, TCRs recognize a composite surface consisting of elements of the antigenic peptide as well as the α-helices of the MHC peptide-binding groove. Thus, unlike antibodies, the ligand for the TCR consists of both self (the MHC) and non-self (the peptide) components. TCRs are also cross-reactive, capable of recognizing multiple peptides bound to one or more MHC molecules. TCR cross-reactivity is necessary for T-cell development and maintenance and is crucial given the fixed size of the T-cell repertoire relative to the vast universe of potential antigens [1–3]. Moreover, TCR cross-reactivity has been implicated in numerous autoimmune pathologies and is an underlying cause of transplant rejection.
The structural and physical properties of TCRs and their complexes with pMHC molecules have been reviewed several times, with perspectives provided previously by Garcia and Adams [4] and Rudolph et al. [5]. Yet even since these contributions, the TCR–pMHC structural database has grown considerably. With regards to TCR cross-reactivity, the number of TCRs, both bound and free, for which structural information is available is now large enough to permit general conclusions to be drawn about conformational changes in TCR antigen-binding sites and their roles in receptor specificity and cross-reactivity.
In the present paper, we review the database of TCRs whose structures have been determined both bound and free, focusing on molecular flexibility and dynamics. We primarily discuss TCR CDR loops, initially examining how loop flexibility and dynamics might influence TCR specificity and cross-reactivity, but also comparing loop positions in cases where the same receptor is bound to different ligands. Overall, we conclude that, although TCR conformational changes do broaden TCR reactivity, with few exceptions, the underlying motions do not reflect the large-scale flexibility which is characteristic of disordered regions; rather, the motions tend to be rigid-body shifts facilitated mostly by hinge-bending movements. The largest shifts occur in the randomly generated CDR3α and CDR3β loops, with the germline loops typically making relatively minor rigid-body adjustments. Interestingly, we observe that the germline-encoded CDR1 and CDR2 loops of the TCR β-chain alter their conformations the least upon recognition of pMHC, consistent with a role for the β-chain in driving the recognition of MHC as hypothesized recently [6–8]. Finally, we highlight emerging principles in the basic biophysics of protein–protein recognition, discussing the similarities and differences between induced-fit binding and conformational selection from a pre-existing equilibria, concluding with a call for more sophisticated experiments capable of fully resolving binding mechanisms and characterizing protein conformational dynamics.
SUMMARY OF CONFORMATIONAL SHIFTS IN TCR CDR LOOPS OCCURRING UPON BINDING
In the mid-to-late 1990s, binding studies performed with soluble ectodomains of αβ TCRs and class I or class II pMHC complexes indicated that TCRs bind pMHC weakly with low association rates, typically <105 M−1·s−1 [9]. Rates of this magnitude are lower than expected for a geometrically constrained diffusion-limited interaction, normally expected to be near to 106 M−1·s−1 in the absence of electrostatic steering forces [10,11]. As slow binding kinetics can result from conformational adjustments required for binding, Davis and colleagues suggested that the TCR CDR loops must undergo conformational changes upon recognition of the ligand [12]. This suggestion was supported by crystallographic studies of the murine αβ TCR 2C, for which three out of six CDR loops were shown to display different conformations in the free and bound states [13,14].
Around the time of the first structural and kinetic observations, the inherent cross-reactivity of TCRs was becoming increasingly appreciated [15–17]. This appreciation was highlighted by Mason's [1] estimation that any given T-cell is capable of reacting “productively with approximately 106 different MHC-associated minimal peptide epitopes”. TCR cross-reactivity fits well with the observation that receptor binding occurs with conformational shifts in the CDR loops, as plasticity or adaptability in the binding interface could broaden the reactivity of any given receptor.
The structures of the 2C TCR both free and bound to pMHC were followed by the structures of the human αβ TCR A6, first bound to the native Tax peptide, then to three Tax variants, all presented by the class I MHC molecule HLA-A2 [18,19]. Differences in the positions of the CDR loops were observed when the structures were compared, contributing further to the idea that TCR-binding loops can move upon binding. This study was followed by thermodynamic experiments indicating that TCRs recognize a ligand with unfavourable entropy and large negative heat-capacity changes [20,21], both consistent with a reduction in protein dynamics or conformational changes occurring upon binding [22].
Since these initial studies, many additional TCR crystallographic structures have cemented the notion that CDR loops can adopt different conformations between their bound and free forms. But how extensive are these changes, and what kind of motions do they represent? The most helpful information has come from instances where the structures are available for both bound and free TCRs: at the present time, including structures where different pMHC complexes are bound to the same receptor and excluding structures involving slightly altered peptides, there are 12 such examples with nine different receptors [7,8,13,14,23–33]. Table 1 summarizes the conformational differences in CDR loops seen in these cases, tabulated by reporting the distances between backbone atoms of the loop apexes when the bound and free receptors are superimposed. All of the TCRs have at least one CDR loop that differs in position by 2Å (1Å=0.1 nm) or more, and all but two have at least one CDR loop that differs in position by 5Å or more. There are no distinctions between MHC class I- or MHC class II-specific TCRs. Notably, in all cases, the largest shifts are in the randomly generated CDR3α or CDR3β loops. However, the germline-encoded CDR1 and CDR2 loops also shift, particularly in the α-chains. Indeed, in the 2C, JM22 and LC13 TCRs, shifts in CDR1α and/or CDR2α are of a similar magnitude to those in the CDR3 loops.
Table 1. TCR CDR loop shifts identified through comparison of bound/free structures.
CDR loop shifts (in Å) tabulated by superimposing the backbones of bound and free TCRs and measuring the maximum difference between equivalent backbone atoms at or near the loop apex. For both the unligated and bound structures, the PDB code and the resolution in Å (in parentheses) are stated. Loop shifts greater or equal to 1.5Å were assigned to hinge-bending motions (hb), loop remodelling (rm) and/or rigid-body framework shifts (rb) by visible inspection. Average shifts are the mean ±S. D. of each CDR loop shift.
| CDR loop shifts | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TCR | Unligated structure | pMHC ligand and MHC class | Bound structure | 1α | 2α | 3α | 3β | 2β | 1β | References |
| 1G4* | 2BNU (1.4) | NY-ESO9V/HLA-A2 (class I) | 2BNQ (1.7) | 1.6 (hb) | 1.0 | 2.1 (rm) | 1.5 (hb) | 0.5 | 0.6 | [24] |
| 2C† | 1TCR (2.5) | SIYR/H2-Kb (class I) | 1G6R (2.8) | 3.7 (hb, rb) | 1.2 | 5.6 (hb, rm) | 1.7 (rm) | 1.1 | 0.6 | [14,30] |
| dEV8/H2-Kb (class I) | 2CKB (3.0) | 4.6 (hb, rb) | 0.9 | 6 (hb, rm) | 2.3 (hb, rm) | 1.4 | 0.6 | [13] | ||
| dEV8/H2-Kbm3 (class I) | 1MWA (2.4) | 4.4 (hb, rb) | 0.3 | 5.5 (hb, rm) | 1.3 | 0.9 | 0.6 | [28] | ||
| QL9/H2-Ld (class I) | 2OI9 (2.4) | 4.5 (hb, rb) | 0.6 | 5.5 (hb, rm) | 1.4 | 0.7 | 0.6 | [29] | ||
| LC13 | 1KGC (1.5) | EBV/HLA-B8 (class I) | 1MI5 (2.5) | 3.4 (hb) | 3.6 (rm) | 5.8 (hb, rm) | 3.9 (hb) | 0.9 | 0.5 | [27,32] |
| ELS4 | 2NW2 (1.4) | EPLP/HLA-B3508 (class I) | 2NX5 (2.7) | 1.5 (hb) | 1.0 | 6.1 (hb, rm) | 1.2 | 2.0 (hb, rb) | 1.2 | [26] |
| JM22 | 2VLM (2.0) | FLU/HLA-A2 (class I) | 1OGA (1.4) | 3.4 (rb) | 3.6 (hb, rb) | 5.6 (hb, rm, rb) | 5.0 (hb) | 0.8 | 0.8 | [8,31] |
| KB5-C20‡ | 1KB5 (2.5) | PKB1/H2-Kb (class I) | 1KJ2 (2.7) | 0.7 | 0.6 | 1.6 (hb) | 11.4 (hb, rm) | 2.5 (hb, rb) | 2.8 (hb, rb) | [35] |
| E8§ | 2IAL (1.9) | TPI/HLA-DR1 (class II) | 2IAN (2.6) | 2.0 (rb) | 2.6 (rb) | 1.6 (hb, rb) | 3.4 (hb, rm) | 2.1 (rb) | 1.4 | [25] |
| 1934.4 | 2Z35 (2.2) | MBP/I-Au (class II) | 2PXY (2.2) | 2.0 (rm) | 1.6 (hb) | 4.3 (hb, rm) | 6.6 (hb, rm) | 1.2 | 1.9 (rm) | [23] |
| D10∥ | 1BWM | CA/I-Ak (class II) | 1D9K (3.2) | 1.7 (hb) | 2.8 (hb, rb) | 6.5 (hb) | 5.7 (rb) | 1.2 | 1.5 (hb, rm) | [33,34] |
| Average shifts | 2.8±1.4 | 1.7±1.2 | 4.7±1.8 | 3.8±3.0 | 1.3±0.6 | 1.1±0.7 | ||||
*The structure of 1G4 bound to the very similar NY-ESO9C/HLA-A2 ligand was not included in the analysis.
†The structure of the high-affinity 2C variant m6 bound to QL9/H-2Ld was not included in the analysis.
‡The unligated structure of the KB5-C20 TCR was crystallized with a Fab bound to the framework region of the Vβ domain.
§The structure of E8 bound to the very similar TPI-T28I/HLA-DR1 ligand was not included in the analysis.
∥The unligated structure of the D10 TCR was determined via NMR. The co-ordinates used were the best representative conformer of the structural ensemble as identified in the co-ordinate file.
Further insight is gained by examining the means±S.D. of the loop shifts, also reported in Table 1. While still indicating that the greatest shifts occur in the CDR3 loops followed by CDR1α/CDR2α, this analysis also reveals that the shifts for the germline-encoded loops of the β-chains are smaller than those of the α-chains (the average shifts for CDR1β and CDR2β are 1.1±0.7Å and 1.3±0.6Å respectively, compared with 2.8±1.4Å and 1.7±1.2Å for CDR1α and CDR2α respectively).
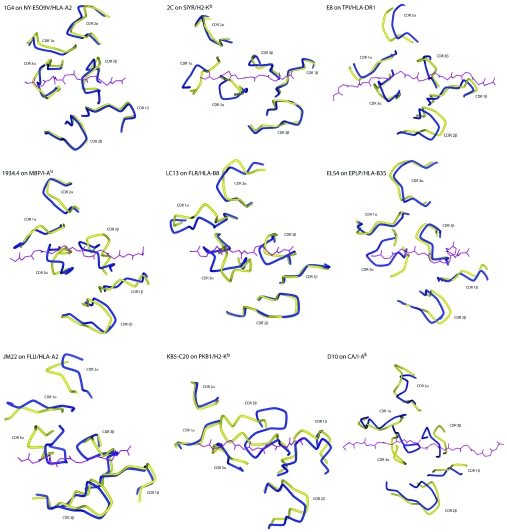
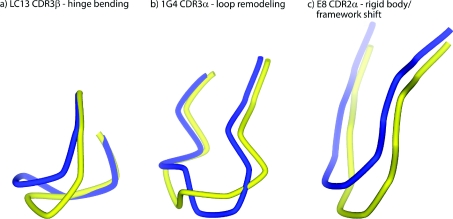
The loop shifts summarized in Table 1 are illustrated in Figure 1, which shows the superimposed bound and free TCRs from the perspective of the pMHC. Through simple visual inspection, the types of loop shift seen in Figure 1 can be placed into three major classes: (i) loop remodelling facilitated by multiple ϕ/ψ bond angle changes, usually near the loop apex, (ii) hinge-bending motions facilitated by correlated bond changes near the beginning and end of the loop, and (iii) rigid-body shifts driven by hinge motions deep within the TCR framework regions or Vα/Vβ domain shifts. Although some loop shifts fit into only one class (e.g. CDR3β of JM22 moves solely by hinge-bending motions), many fit into more than one (e.g. CDR3α of ELS4 moves by hinge bending and loop remodelling). The three general classes of CDR loop movements are illustrated in Figure 2 for LC13 CDR3β (hinge bending), 1G4 CDR3α (loop remodelling) and E8 CDR2α (rigid-body shift due to framework shifts).
Figure 1. Conformational differences in the CDR loops of bound and free TCRs.
The view is of superimposed TCRs from the perspective of the MHC peptide-binding groove, with the peptide from the bound structure shown for reference (purple). Yellow and blue represent the free and bound receptors respectively. For the 2C TCR, only the differences between the free receptor and 2C bound to SIYR/H-2Kb are shown; see Figure 4 for views of loop positions when 2C is bound to other ligands. The discontinuous segment for CDR3α in the unligated ELS4 TCR reflects missing electron density. The image for the unligated D10 TCR, whose structure was determined via NMR, was generated using the best representative conformer of the structural ensemble as identified in the co-ordinate file.
Figure 2. Conformational changes in TCR CDR loops fall into three general classes: hinge-bending movements, loop remodelling or rigid-body movements originating in the TCR framework region.
The three classes are illustrated in (a) by CDR3β of the LC13 TCR (hinge bending), (b) by CDR3α of the 1G4 TCR (loop remodelling) and (c) by CDR3α of the E8 TCR (rigid-body shift). Yellow and blue represent the free and bound receptors respectively.
Each loop shift of 1.5Å or greater is categorized in Table 1. Overall, there are 33 instances of loops that move by hinge-bending motions (with often more than two hinges per loop), 18 instances of loop remodelling and 16 instances of rigid-body shifts. Loop remodelling occurs in CDR3 loops more than three times as often as it does in CDR1/2 loops (14 times for CDR3 compared with four times for CDR1/2), illustrating further the greater plasticity of the randomly generated CDR3 loops. Yet, even with the higher frequency of loop remodelling, the CDR3 loop shifts are still dominated by hinge bending and rigid-body shifts: of all the CDR3 loop shifts greater than 3Å, none of them is facilitated solely by loop remodelling (even CDR3α of the ELS4 TCR, which is missing electron density in the centre of the loop, has clear hinges prior to the region of missing density). Many loop shifts combine all three motional modes, with the largest driven primarily by hinge bends, with loop remodelling at or near the centre of the loop (e.g. CDR3α in LC13 and CDR3β in JM22). The picture that emerges from the data is that, with few exceptions, TCR CDR loops do not wholly reorganize (or fold) upon ligand binding. Rather, motions tend to be block shifts, occasionally enhanced via small amounts of remodelling. Small adaptations dominate for the germline-encoded loops, with the adaptations in Vβ typically less than those in Vα. This picture, which is unaltered if the multiple results for the 2C TCR are excluded from the analysis, is consistent with the prediction of Garcia and Adams [4] that TCR CDR loops are unlikely to be “easily accommodating limp noodles”.
Recent structural studies of the JM22, 1934.4 and 172.10 TCRs suggest that the TCR β-chains may play the dominant role in pMHC recognition [6–8]. As discussed further below, the results in Table 1 and Figure 1 are compatible with this suggestion, evoking a binding mechanism involving rigid-body association using the β-chain germline loops, with structural adaptations occurring in CDR3β and/or one or more of the α-chain loops.
Although the results are not shown, there is not a strong correlation between CDR3 loop length and the magnitude of structural shift. The sole exception is CDR3β of the KB5-C20 TCR, which, at a length of 13 amino acids, has the largest structural shift at 11.4Å [34]. However, the CDR3 loops of the remaining TCRs, which range from seven to eleven amino acids for CDR3α and six to nine amino acids for CDR3β, show no relationship between loop length and structural shift.
Not shown in Figures 1 or 2 are changes in side-chain position coupled to backbone shifts. Usually, changes in side-chain positions are proportional to the magnitude of backbone shifts, with shifts larger than 4Å typically resulting in very large changes in side-chain positions as a result of reorientations of the Cα–Cβ bond vectors. This is particularly so for aromatic side chains which extend far from the peptide backbone. For example, in recognition of the SIYR peptide by the 2C TCR, the 6Å shift in CDR3α translates into a 12Å shift in the side chain of Phe100 at the apex of the loop, moving it to the periphery of the interface out of the way of the pMHC [29]. In recognition of the MBP (myelin basic protein) peptide by the 1934.4 TCR, the 4Å shift in CDR3α results in a very similar 10Å shift in the position of Tyr100 [8]. As expected, smaller backbone shifts typically translate into less dramatic changes in side-chain positions. One exception to this is Tyr95 of CDR3β in the E8 TCR. Despite a moderate loop shift of 3.4Å, the Tyr95 hydroxy group moves by nearly 9Å upon binding, forming a hydrogen bond with the peptide backbone [24]. Another exception is in the CDR2α of the LC13 TCR: despite a loop shift of 3.6Å, the Ser52 hydroxy group moves by 7Å upon binding, interacting with the MHC α2 helix [31].
TCR DOMAIN SHIFTS AND BINDING-SITE STRUCTURAL DIVERSITY
Rigid-body shifts between TCR Vα and Vβ domains have been identified as an additional mechanism in promoting conformational variability in TCR–pMHC interfaces. TCR Vα/Vβ domain shifts were first identified by Reiser et al. [34] in their characterization of the interaction between the KB5-C20 TCR and H-2Kb/pKB1, in which the TCR Vβ domain rotates 8.7° relative to Vα upon binding of pMHC. When the bound and free Vα domains are superimposed, this rotation contributes approx. 1Å to the approx. 3Å displacements seen in the β-chain CDR loops. Gagnon et al. [35] identified a similarly sized Vα/Vβ shift in the A6 TCR upon recognition of the bulky Tax-5K-IBA peptide presented by HLA-A2. More recently, after identifying domain rotations in multiple copies of the free 1.D9.B2 TCR, McBeth et al. [36] performed a rigorous three-dimensional analysis of the relative orientations of TCR Vα/Vβ domains in available TCR–pMHC structures, identifying significant variations in rotation and pitch.
IMPACT OF TCR LOOP SHIFTS ON THE STRUCTURE AND CHEMISTRY OF THE INTERFACE
If TCR CDR loop shifts are generally not large-scale reorganizations, folding reactions or disorder-to-order transitions, what is the impact of the smaller-scale changes usually observed? With low-molecular-mass antigens, small loop movements can have a dramatic effect on how an antigen ‘sees’ an incoming antibody. Yet when the ligand is a large peptide or protein complex contributing 1000 Å2 or more of surface to the interface, does the capacity for a few loops and their extending side chains to move by a few angstroms make a significant difference? The answer to this question depends on separate but closely related questions: what are the energetic costs of the structural shifts, do they alter the structure and chemistry of the antigen-binding site in a way that influences binding, and to what extent do the new interactions across the interface compensate for the costs associated with the loop and side-chain movements? These questions have been considered in numerous studies of protein–protein interactions, but, unfortunately, although our understanding of the energetics of rigid-body interactions has grown considerably, the situation is greatly complicated when the flexibility of one or more binding partners must be considered. Furthermore, for most of the results in Table 1, we simply do not know whether any of the loop motions are readily sampled by free TCRs or whether they are induced by binding and, if they are induced, we do not have estimates of the associated energetic penalties. An exception to this might the D10 TCR, whose unligated structure was determined using NMR [33]. The 15 lowest energy structures give some indication about the structural ensemble populated by the free receptor, and comparison of the various co-ordinate sets suggests considerable CDR loop mobility: as much as 13Å for the apex of CDR3β. However, although the mobility implied in the D10 structural ensemble is provocative, it is important to keep in mind that protein flexibility is just one of the factors that can influence final NMR models, and the range of motion indicated by an NMR structural ensemble may not directly correlate with the actual extent of mobility [37].
In contemplating the results shown in Table 1, we must also keep in mind that the error associated with crystallographic experiments is exacerbated as the resolution increases. For many of the interactions in Table 1, the quantitative loop shifts reported may be pushing the precision of the structural data, as the shifts represent the differences between two structures, each with their own accuracy and precision. Furthermore, it remains possible that the database of TCRs whose structures have been determined in the unligated state is biased by the inability to crystallize molecules with large amounts of dynamic disorder.
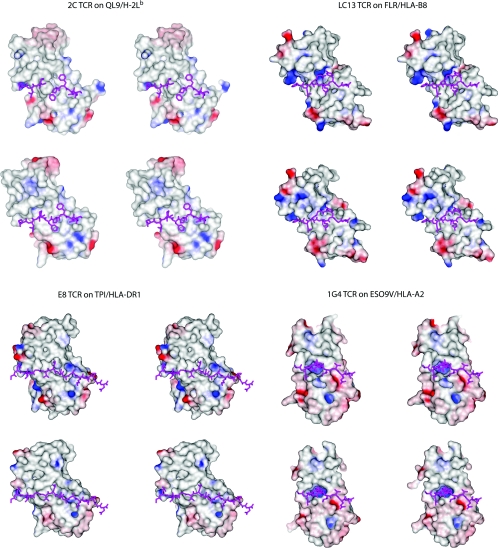
Given the caveats above, how do the loop motions highlighted in Table 1 and Figure 1 influence the structure and chemistry of the antigen-binding sites? A close analysis reveals that, even when TCR conformational changes are small (as in the case of the 1G4 TCR, for which bound and free structures were both solved to <2Å resolution), the interfaces are altered in a fashion that should be expected to influence binding. This is demonstrated in Figure 3, which shows a view of the solvent-accessible surface area of the antigen-binding sites of the 2C, LC13, E8 and 1G4 TCRs from the view of the peptide, coloured by electrostatic potential (for the bound structures, electrostatic potentials were calculated using the co-ordinates of the receptor only). To aid in interpreting how the TCRs adapt to their ligands, the peptides are shown in their correct orientation beneath the receptors (even for the unligated forms). In each case, there are clearly visible differences between the bound and free forms of the TCRs. In the case of the 1G4, the TCR which is most similar in its bound and free states, the small 2Å shift in CDR3α is necessary for opening the central positively charged pocket juxtaposed between the Vα and Vβ domains.
Figure 3. Stereo image of the solvent-accessible surfaces of the 2C, LC13, E8 and 1G4 TCRs from the view of the MHC peptide-binding domains.
Colouring is by electrostatic potential, from −25 kT (red) to +25 kT (blue). For each TCR, the top view is of the unligated TCR superimposed on to the bound receptor, whereas the bottom view is of the ligated TCR. Peptides are shown in both the bound and unbound views for orientation. In each case, there are clear differences in structure and electrostatics between the free and bound receptors. Electrostatic potentials were calculated using the program DelPhi [101].
INFLUENCE OF CDR CONFORMATIONAL CHANGES ON TCR SPECIFICITY AND CROSS-REACTIVITY
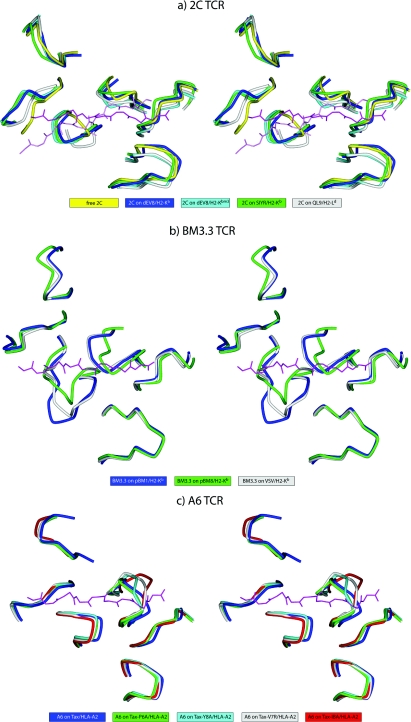
Despite their occurrence, questions remain about the role of TCR conformational changes in binding, particularly in specificity and cross-reactivity. Do CDR loop movements broaden TCR reactivity as originally supposed? Several TCR–pMHC structures with the 2C, BM3.3 and A6 TCRs have allowed this question to be addressed. For the 2C TCR, structures are available for the receptor bound to four different pMHC ligands: dEV8/H-2Kb [13], dEV8/H-2Kbm3 [27], SIYR/H-2Kb [29] and QL9/H-2Ld [28]. These ligands present a range of different structures and chemistries to the TCRs. In the case of the dEV8 peptide, H-2Kb and H-2Kbm3 differ by MHC amino acid substitutions that alter peptide anchoring, causing distortions in the C-terminal end of the dEV8 peptide [27]. For the other 2C structures, although the SIYR and dEV8 peptides share similar charge and aromatic features (SIYRYYGL and EQYKFYSV respectively), the QL9 peptide (QLSPFPFDL) differs substantially from the SIYR and dEV8 peptides. The conformations of the loops for the five 2C TCR structures (one free and four bound) are shown in Figure 4(A). Interestingly, despite many different local interactions in the interfaces, the various loops in the dEV8/Kb, dEV8/Kbm3, SIYR/Kb and QL9/Ld structures are all in similar conformations, each deviating by approximately the same amount from the unligated 2C structure. The differences in the QL9/Ld structure are slightly more dramatic, particularly for CDR3α, which is ‘bent’ more towards the peptide C-terminus than in the other three bound structures, and CDR1α, which deviates less from its unligated conformation. Perhaps most interestingly, the 2C TCR assumes a more diagonal binding orientation on QL9/Ld than on dEV8/Kb or SIYR/Kb, placing the germline-encoded loops over different regions of the MHC α1/α2 helices [28] [this is apparent in Figure 4(A) by the different orientations of the dEV8 and QL9 peptides]. Yet the overall conformations of the various germline loops remain unperturbed, reinforcing the idea that the CDR1 and CDR2 loops undergo relatively minor conformational changes upon recognition of pMHC, and, when they do move, the movements tend to be dominated by rigid-body shifts. Overall, the 2C results indicate that TCR cross-reactivity need not be driven by dramatic alterations in the TCR-binding site, the existence of different interatomic contacts within the interface notwithstanding.
Figure 4. Stereo image showing different CDR loop positions when the same TCR is bound to different pMHC ligands.
Orientation and TCR superimposition is through the MHC peptide-binding groove as in Figure 1. (a) View of the CDR loops of the 2C TCR free (yellow) and bound to dEV8/H-2Kb (blue), dEV8/H-2Kbm3 (cyan), SIYR/H-2Kb (green) and QL9/H-2Ld (grey). The two peptides shown reflect the more orthogonal docking angle of the 2C TCR on the QL9 compared with the dEV8/SIYR ligands. (b) View of the CDR loops of the BM3.3 TCR bound to pBM1/H-2Kb (blue), pBM8/H-2Kb (green) and VSV/H-2Kb (grey). (c) View of the CDR loops of the A6 TCR bound to Tax/HLA-A2 (blue), Tax-P6A/HLA-A2 (green), Tax-Y8A/HLA-A2 (cyan), Tax-V7R/HLA-A (grey) and Tax-5K-IBA/HLA-A2 (red). An interactive three-dimensional version of this Figure can be found at http://www.BiochemJ.org/bj/415/0183/bj4150183add.htm.
For the BM3.3 TCR, although no structure is available for the free receptor, structures are known for the receptor bound to three different pMHC ligands: pBM1/H-2Kb [38], pBM8/H-2Kbm8 [39] and VSV8/H-2Kb [40]. Again, these ligands differ considerably in both sequence and chemistry: the amino acid sequences are INFDFNTI for pBM1, RGYVYQGL for VSV8 and SQYYYNSL for pBM8. The positions of the loops in the three BM3.3 TCR structures are shown in Figure 4(B). The conformation of CDR3α varies dramatically between the three structures, differing by as much as 6.6Å, with the changes driven by hinge bending and loop remodelling. The conformation of CDR3β is the same in the pBM1 and VSV8 structures, but is shifted by as much as 4.4Å in the pBM8 structure as a result of a large wrinkle in the centre of the loop. The positions and conformations of the various germline loops are largely unaltered, with the exception being CDR2α in the pBM8 structure, which is displaced by approx. 2Å owing to hinge bends at the beginning and end of the loop. The affinity of BM3.3 for the pBM8 ligand is substantially weaker than for the pBM1 ligand [39], possibly as a result of the reorganization of CDR3β and displacement of CDR2α, but also possibly from the different interactions present within the interface. Unlike the 2C TCR, BM3.3 binds the three pMHC complexes in the same diagonal orientation.
Lastly, although no structure is available for the free receptor, there are five structures of the A6 TCR bound to peptides presented by HLA-A2: the native Tax peptide and four single amino acid variants [18,19,35]. Three of the altered peptides result in large alterations in the interface: the Y8A variant removes a bulky tyrosine side chain, the V7R variant replaces a hydrophobic valine residue with a large positively charged arginine residue, and the Y5K-IBA variant replaces the central tyrosine residue with a bulkier and more flexible hapten. The positions of the loops in the five A6 TCR structures are shown in Figure 4(C). Structural variation is seen mostly in CDR3β, which moves by an amount that correlates directly with the size of the perturbation in the interface, culminating in a 5.5Å shift with the Y5K-IBA peptide. As with the BM3.3 TCR, the affinity of the A6 TCR weakens with greater shifts in CDR3β (from 1 μM with native Tax to 7 μM with V7R and >160 μM with Y5K-IBA), but, as noted above, we cannot easily separate the energy associated with the loop shift from other structural and chemical changes in the interface.
BIOPHYSICAL CORRELATES WITH CONFORMATIONAL CHANGES AND DYNAMICS
As noted above, early evidence for CDR loop mobility in TCRs came from kinetic and thermodynamic measurements of TCR–pMHC interactions. Although any one set of results is useful in examining a particular interaction, with the multiple bound/free TCR structures now available, can quantitative or predictive relationships be drawn between the aggregated biophysical and structural data?
Unfortunately, equivalent binding data are not available for all of the TCR–pMHC interactions for which both bound and free TCR structures are available. The results that are available are listed in Table 2. Notably, all of the interactions tabulated have association rates of less than 105 M−1·s−1 [8,20,23,29,41,42], less than the rate of nearly 106 M−1·s−1 expected for a rigid-body interaction in the absence of electrostatic steering [10,11]. As mentioned above, this is consistent with the structural differences that are observed between bound and free receptors: if conformational changes need to occur for binding, then binding will be slowed by an amount proportional to the rate at which these changes proceed. However, there is no correlation between the extent of loop shifts and association rate, nor is there any correlation with affinity. Neither of these findings is surprising, as factors other than conformational changes and their associated exchange rates will influence binding kinetics and affinity.
Table 2. Biophysical for TCR–pMHC interactions with structures available in the free and bound states.
1 kcal≈4.184 kJ. N/A, not applicable.
| TCR–pMHC interaction | Maximum loop shift (Å)* | Affinity (μM)† | kon (×104 M−1·s−1)† | ΔS° (cal/mol/K)† | ΔCp experimental/predicted assuming rigid body (cal/mol/K)‡ | SASA buried in TCR transition from unbound to bound conformation (Å2)§ | Predicted ΔCp for TCR structural transition (cal/mol/K)∥ |
|---|---|---|---|---|---|---|---|
| 1G4–NY-ESO9V/HLA-A2 | 2.1 (3α) | 5.7 | 1.2 | N/A | N/A | N/A | N/A |
| 2C–SIYR/H2-Kb | 5.6 (3α) | 32 | 0.2 | −4 | −1100/−260 | 60 apolar, −262 polar | −95 |
| 2C–dEV8/H2-Kb | 6.0 (3α) | 84 | 0.2 | −54 | −1100/−270 | 73 apolar, 16 apolar | −28 |
| 2C–dEV8/H2-Kbm3 | 5.5 (3α) | 56 | 0.09 | N/A | N/A | N/A | N/A |
| 2C–QL9/H2-Ld | 5.5 (3α) | 3.9 | 0.6 | 12 | N/A | N/A | N/A |
| 1934.4–MBP/I-Au | 6.6 (3β) | 31 | 0.5 | −32 | −1250/−230 | 127 apolar, −255 polar | −124 |
| LC13–EBV/HLA-B8 | 5.8 (3α) | 8.1 | N/A | 14 | −620/−270 | 143 apolar, 81 polar | −40 |
| JM22–FLU/HLA-A2 | 5.6 (3α) | 6.6 | 3.0 | −50 | −640/−250 | −107 apolar, 80 polar | +69 |
*Maximum loop shift from Table 1, with the shifting loop in parentheses.
†Reported affinity, on-rate and enthalpy change at 25 °C.
‡Experimental heat-capacity change determined by van 't Hoff analysis, followed by the value predicted from the structure of the complex assuming a rigid-body interaction.
§Solvent-accessible surface area buried upon transition from the unligated TCR conformation to the conformation found in the bound state, calculated using co-ordinates for TCR variable domains only. Negative values reflect exposure, rather than burial, of additional surface area. Surface-area calculations performed with naccess [100], using a probe radius of 1.4Å and a slice width of 0.05Å.
∥Predicted heat-capacity change for the unbound-to-bound structural transition in the TCR, calculated using empirical relationships between changes in solvent-accessible surface area and heat capacity [22].
Unfavourable binding entropy changes have also been used to support the presence of conformational changes (or reductions in flexibility) occurring upon TCR recognition of pMHC. The first several TCR–pMHC interactions studied thermodynamically all bound with unfavourable entropy changes at 25 °C [20,43,44], and, for a brief period, unfavourable binding entropy changes were presumed to serve as a ‘thermodynamic signature’ for TCR binding. Subsequently, however, several TCRs were shown to bind with favourable entropy changes [28,39,45–48], demonstrating that there is no clear entropic signature for TCR recognition of pMHC [49]. Indeed, the interactions shown in Table 2 include some that proceed with favourable and some that proceed with unfavourable entropy changes, yet all involve similarly large changes in CDR loop positions. This result is also not surprising, as conformational dynamics are only one of many contributors to binding entropy changes (others include changes in solvation of polar and apolar surfaces and water/ion release/incorporation). As every TCR–pMHC interface in Table 2 is different, it is not unexpected that the various contributions are present to different extents, yielding positive or negative entropy changes independently of whether or not reductions in protein flexibility occur upon binding.
Changes in TCR–pMHC binding heat capacity (ΔCp) have been used in a number of cases as supporting evidence for conformational differences between bound and free TCRs [21,41]. Such evidence stems from the relationship between heat capacity and the hydration of protein surface: changes in hydration that occur upon protein binding give rise to changes in heat capacity [50]. Empirical relationships between polar and apolar solvent-accessible surface area are frequently used to predict ΔCp values from the structures of protein–protein complexes [22,51], with the differences between predicted and experimental values usually attributed to conformational differences between bound and free proteins, i.e. if treating the complex as a rigid-body interaction does not correctly predict ΔCp, the interaction must not be rigid body.
Experimentally determined heat-capacity changes (ΔCp) are available for five of the interactions listed in Table 2 [7,20,41,43,46]. In each case, the experimental ΔCp was determined by van 't Hoff analysis (measurements of affinity as a function of temperature), and each is significantly more negative than the ΔCp predicted by treating the interaction as rigid body, consistent with the existence of structural differences between bound and free receptors. Yet do the actual structural differences between the bound and free receptors account for the deviations between experimental and predicted ΔCp values? The question is pertinent, as ΔCp values are traditionally difficult to measure via van 't Hoff analysis, particularly for interactions with weak to moderate affinities [52]. Notably, some of the ΔCp values in Table 2 approach or exceed heat-capacity changes for the unfolding of small proteins (for an example, see [53]).
To address this question, for each of the interactions with ΔCp measurements available, Table 2 also lists the changes in polar and apolar solvent-accessible surface area associated with TCR movement from the unligated to the ligated conformation. These values were calculated using the co-ordinates of the unligated TCR variable domains and the co-ordinates of the bound TCR variable domains extracted from the complex. Also shown is the predicted ΔCp for this transition, calculated using the same empirical relationships used to estimate the binding ΔCp (this value has been referred to in previous publications as the ‘conformational ΔCp’). In no case does the structural transition from the unligated to the ligated conformation account for the discrepancies between experimental and predicted binding heat-capacity changes, as the changes in the solvent-accessible surface area are simply too small, and the ‘conformational ΔCp’ values never reach more than 10% of the experimental value. This result is entirely consistent with the results in Table 1, showing that the overall magnitude of CDR loop conformational changes occurring upon TCR binding is relatively small. The conclusion from this analysis is that, although TCR–pMHC binding heat-capacity changes are suggestive of conformational differences between free and bound TCRs, they do not quantitatively report on these structural differences. In many cases, the discrepancy is likely to be the result of experimental difficulties in measuring accurate heat-capacity changes for weak protein–protein interactions [52].
CONFORMATIONAL CHANGES OCCURRING UPON TCR–pMHC BINDING: INDUCED FIT OR CONFORMATIONAL SELECTION FROM A PRE-EXISTING EQUILIBRIUM?
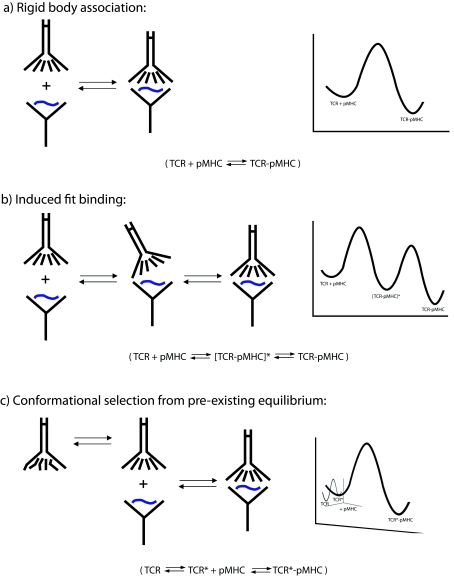
The structural results indicate that, although in most cases they are not dramatic reorganizations, conformational shifts do facilitate TCR recognition of pMHC ligands and contribute to TCR cross-reactivity. The largest conformational shifts tend to be in the CDR3 loops, with smaller rigid-body shifts occurring in the germline-encoded CDR1/CDR2 loops. But what is the origin of the underlying motions? Are they induced upon binding or do they reflect pre-existing equilibria, where binding occurs only to a compatible conformation present in a larger structural ensemble? The differences between these two modes of recognition are illustrated in Figure 5. The extent to which TCRs use one or both of these mechanisms and whether they arise due to biological or chemical/structural necessity has implications for the molecular-recognition properties of TCRs, and, by extension, the nature of T-cell specificity and cross-reactivity.
Figure 5. Schematic diagrams indicating three possible TCR–pMHC binding mechanisms.
The left-hand side of each panel illustrates protein association, whereas the right-hand side shows traditional free-energy diagrams showing energy as a function of reaction progress. (a) Rigid-body docking, where the structures of the TCR and pMHC are identical in the bound and free states. (b) Induced–fit binding, where a loose intermediate energy complex ([TCR-pMHC]*) is initially formed, followed by rearrangements in the TCR giving rise to the final lowest-energy-bound state. (c) Conformational selection from a pre-existing equilibrium, in which the free TCR samples multiple conformations (two are illustrated here), with only one being binding competent. As shown, binding to the competent TCR conformation follows a rigid-body mechanism, although, as discussed in the text, the association phase could also follow an induced-fit mechanism.
Induced-fit interactions
The notion of induced-fit conformational changes in protein–ligand interactions began with Koshland's [54] extension of the ‘lock and key’ model for enzyme–substrate interactions. Accordingly, the adaptability of substrates and active-site side chains allows for the best energetic fit within enzyme active sites; the same logic is implied when induced-fit motions are used to describe larger protein–protein interactions. A key requirement for induced-fit binding is an initial ‘loose’ contact between the two molecules, followed by structural adaptations to form a higheraffinity complex. If the crystallographically observed TCR conformational changes occurring upon binding do represent induced-fit motions, the requisite weak initial contact may provide a mechanism for TCR ‘scanning’ of different peptides, as originally proposed by Boniface et al. [21]. In this model, initial weak binding to MHC molecules would allow higher-affinity binding only to those ligands to which the best fit could be obtained before the encounter complex dissociates. Implied in this model is a physical ‘bias’ of TCRs towards the peptide-binding domains of MHC molecules. A bias towards TCR recognition of MHC could have resulted from the co-evolution of immune receptor gene segments and MHC molecules, as suggested by Jerne [55], and more recently discussed by Kim et al. [56].
Evidence for an inherent bias for TCR recognition of MHC was presented in the influential work of Zerrahn et al. [57], who characterized the MHC reactivity of the T-cell repertoire prior to positive and negative selection and concluded that TCRs are inherently MHC reactive. Their results built on work from Gascoigne and colleagues, who demonstrated a role for germline-encoded CDR loops in directing restriction towards class I or class II MHC [58]. This was recently followed by data from Kappler and colleagues, who found that, after relaxing negative selection in a mouse model, the resultant T-cell repertoire was substantially more cross-reactive than normal [59]. Biophysical evidence for biased TCR recognition of MHC has also been presented: using ϕ analysis [60], and Wu et al. [61] showed that many amino acid substitutions in the MHC molecule weakened receptor binding by lowering association rates, whereas mutations in the peptide weakened binding by increasing dissociation rates. The results were interpreted as resulting from a ‘two-step’ binding mechanism: an initial TCR contact to the MHC, followed by induced-fit adjustments as the receptor ‘settled down’ on the peptide. More recent biophysical evidence in support of this model has come from the rapid kinetic measurements of Gakamsky et al. [48], who found that a TCR specific for the CMV pp65 peptide presented by HLA-A2 bound with a kinetic mechanism consistent with an induced-fit reaction. Finally, recent structural studies of the JM22, 1934.4 and 172.10 TCRs suggest that the TCR β-chains may play the dominant role in pMHC recognition [7,8]. The results in Table 1 showing that the germline-encoded CDR1β and CDR2β loops which shift the least upon binding pMHC are compatible with this suggestion, evoking a binding mechanism involving rigid-body association using the β-chain germline loops, with structural adaptations occurring in CDR3β and/or one or more of the α-chain loops.
Undoubtedly, the presence of induced-fit motions coupled with an inherent TCR bias for recognition of MHCs provides an attractive way to achieve TCR cross-reactivity. Although sometimes controversial owing to the difficulty of distinguishing between induced-fit and pre-existing conformational equilibria [62,63], the presence of induced-fit motions has been firmly established in other systems (for a review, see [64]). Experimental and theoretical investigations have begun to elucidate what initial ‘loose’ encounter complexes could look like [65,66]. Of particular interest is the use of paramagnetic relaxation measurements in elucidating these structures. Tang et al. [65] recently used this approach to characterize the transient interaction between enzyme I and the phosphocarrier protein HPr, showing that HPr can initially bind over a large portion of the surface of enzyme I, followed by electrostatically driven diffusion into the high-affinity-binding site. Although this interaction is technically not an ‘induced-fit’ binding reaction, one could envision a more refined mechanism for TCR recognition of pMHC, where the initial binding and subsequent isomerization of the TCR is limited to the ‘business ends’ of the TCR and MHC.
Despite the appeal of TCR conformational shifts resulting from induced-fit motions occurring upon binding, the corollary of a transient TCR–MHC association arising from the co-evolution of TCR and MHC genes has been difficult to establish. Mutagenesis experiments have not identified binding hot spots that are well conserved across TCR–pMHC interactions [67,68]. The ‘two-step’ binding model of Wu et al. [61] as originally formulated has been questioned both biophysically and structurally [40,69,70]. Structures of TCR–pMHC complexes have not shown conserved contacts that could mediate an MHC bias, although structures of different receptors sharing common variable domains suggest types of interaction that may be conserved between particular gene segment–MHC pairs [8,71], and, as noted above, recent results are compatible with the germline β-chain loops driving the initial pMHC contact [7]. Finally, Buslepp et al. [72] have suggested that an inherent bias towards the recognition of MHC is imparted on the T-cell repertoire not by the co-evolution of TCR and MHC genes, but by the need for co-receptor during thymic selection. Thus, although induced-fit motions may characterize many TCR–pMHC interactions, the extent to which T-cells take advantage of this for ‘scanning’ peptides still remains unclear.
Complicating the role of induced fit in TCR recognition of pMHC are observations that peptides occasionally alter their conformations upon TCR binding. This has been seen most dramatically for the extensively bulged EPLP (endorphin-like peptide) presented by HLA-B35, which, as a 13-mer, bulges up and above the plane of the peptide-binding groove. Binding of the ELS4 TCR dramatically ‘squishes’ the peptide backbone as described by Tynan et al. [25], with a large shift of 5Å occurring in the centre of the peptide. Smaller backbone shifts in the range 2–3Å have been seen upon TCR binding to the Tax and NY-ESO peptides presented by HLA-A2 [18,23,73–75] and the dEV8 peptide presented by H-2Kb and H-2Kbm8 [13,27]. The α-helices flanking the peptide-binding groove can also shift upon TCR binding, as seen in the recognition of pBM8/H-2Kbm8 (class I MHC) by the BM3.3 TCR [39,76] and MBP/HLA-DR2a (class II MHC) by the 3A6 TCR [77], where, in both cases, rigid-body shifts of approx. 1Å are distributed across both the α1 and α2 helices. Overall, most of the changes in pMHC are small, but they are not much smaller than the magnitude of changes seen in the TCR CDR loops. Results such as these highlight the danger, in the absence of supporting structures, of interpreting TCR–pMHC biophysical data in terms of induced-fit conformational changes occurring only within the TCR CDR loops.
Conformational selection from a pre-existing equilibrium
An alternative mechanism for achieving the conformational changes observed in TCR–pMHC interactions is selection from a pre-existing conformational equilibrium. This mechanism is related to the energy landscape treatment of protein conformational dynamics [78], which accounts for the intrinsic flexibility of proteins and how the free energies of different structural states dictate the extent to which they are populated. Although commonly used in discussions of protein folding, energy landscapes are also applicable to protein binding, particularly when conformational changes or alterations in dynamics are involved [79–82]. Holler and Kranz [83] discussed how pre-existing equilibria could promote TCR cross-reactivity in their ‘conformer’ model of TCR recognition of pMHC. Briefly, different conformers (or structural substates) could recognize different pMHC ligands, with the potential for each conformer to have different degrees of fine specificity. TCR cross-reactivity would then be driven not only by the intrinsic affinity of a conformer for a given pMHC, but also by the relative populations of substates within the overall structural ensemble. In TCRs, the conformational ensemble would represent alternative loop configurations (based on the results in Table 1, mostly limited to the CDR3 loops) and possibly different Vα/Vβ orientations.
Although evidence for pre-existing conformational equilibria in TCR CDR loops as a means to promote T-cell cross-reactivity has not yet been presented, this mechanism has been demonstrated in antibodies. In their classic paper, Foote and Milstein [84] identified antibodies to the hapten 2-phenyl-5-oxazolone that bound with complex kinetics best described not by an induced-fit mechanism, but by a mechanism in which the antibodies populated multiple conformations in the unbound state, of which only a subset was binding competent. This work built on earlier studies by Lancet and Pecht [85], and was related to the hypothesis for antibody cross-reactivity initially formulated by Linus Pauling in 1940 [86]. However, the model was not widely accepted until 2003, when Tawfik and colleagues showed that the antibody SPE7 uses this mechanism to cross-react with the hapten 2-nitro-4-iodophenol and the protein Trx-Shear3 [87,88]. Key to the acceptance of the SPE7 results was the availability of not only rapid kinetic data consistent with a pre-existing conformational equilibrium, but also crystallographic structures of the bound and free antibody, demonstrating that conformations seen in different bound states were sampled in the unligated antibody.
The discussion of pre-existing conformational equilibrium compared with induced fit in protein binding has a parallel in allostery, in which a protein adopts a different functional (and thus structural) state in response to the binding of an allosteric effector. Although the traditional explanation has been one of induced fit, the notion of pre-existing conformational equilibria as an explanatory mechanism for protein allostery has been gaining ground [89,90]. For example, the NtrC (nitrogen regulatory protein C) signalling protein exists in two conformational states depending on its phosphorylation state, yet NMR studies indicate that it samples the phosphorylated conformation even when unphosphorylated [91]. The more traditional view of allostery has been questioned further with observations that allostery can result purely from dynamical changes: Popovych et al. [92] recently showed that the binding of one molecule of cAMP to the catabolic activator protein changed the dynamics of the protein but not the average structure, strongly influencing the affinity of a second molecule of cAMP. Similar discussions of induced fit compared with conformational dynamics have been ongoing in the field of enzyme catalysis, in which movements necessary for catalysis are not seen as being induced by substrate binding, but instead as reflecting an existing flexibility within an active site (for an example, see [93]).
One issue with pre-existing conformational equilibria in TCR cross-reactivity concerns the relative populations of differing conformations. If a conformation capable of recognizing a specific ligand is only populated a fraction of the time, unless that conformation has a high intrinsic affinity for the ligand, the overall binding affinity will be very weak, as the effective concentration of binding-competent receptor is reduced. Yet, by and large, crystallographic structures of TCR–pMHC interfaces do not have hallmarks of high-affinity complexes: the interfaces tend to be relatively flat, with poor packing, suboptimal surface complementarity and buried water molecules [5]. Although quantitative predictions of binding affinity from structure are notoriously inaccurate, in general, the TCR–pMHC complexes whose structures have been solved are consistent with the weak to moderate affinities that characterize them. We might conclude from this that if unligated TCR CDR loops do sample conformational states that promote cross-reactivity, the different structural substates sampled will be likely to be of similar energies (and thus populations), as otherwise the large energetic penalties for shifting equilibria towards minor poorly populated states would need to be compensated by extremely well optimized protein–protein interfaces, in contrast with what is seen in most TCR–pMHC structures. Consequently, TCR structural ensembles are not likely to be described by large amounts of disorder. Rather, the conformational ensembles should be expected to be dominated by local backbone/side-chain dynamics or block shifts [4], as is observed in the structural results summarized in Table 1 and Figure 1. Consistent with this conclusion, of the nine structures of unligated TCRs currently available, only two (ELS4 and 1.D9.B2) have missing electron density, suggesting the dynamic disorder of a CDR loop [25,36]. An important caveat to this conclusion is that the database of unligated TCR structures may be biased by those that are crystallizable, as discussed above.
As noted above, peptides and MHC molecules can undergo conformational changes upon TCR recognition, and the discussion of a pre-existing conformational equilibria applies equally to the pMHC complex. Certainly, cases where the peptide has been shown to be disordered or adopt multiple structures in the MHC-binding groove are candidates for peptide pre-existing equilibria having influence on receptor recognition [35,94–97]. However, if both the receptor and the ligand populate broad conformational ensembles, statistically it seems less probable that a TCR–pMHC complex would assemble at a reasonable rate if both molecules need to shift into a binding competent conformation.
Finally, it is possible, if not likely, that different TCRs and pMHC complexes interact via a combination of induced fit and conformational selection in ways that are ultimately dictated by the structure, chemistry and dynamics of each component. Combinations of these binding modes have been seen in other protein–ligand interactions, most notably in work with the SPE7 antibody [98]. Further evidence for such a combination comes from the computational studies of Grunberg et al. [99], who studied 17 protein–protein interactions and elegantly combined conformational selection and induced fit into a more general model for protein–protein recognition. In other computational studies, regions of proteins that undergo induced-fit motions have been shown to possess high intrinsic flexibility [62,63], blurring the distinction between induced fit and conformational equilibrium.
However, the concept that, biophysically, every TCR–pMHC interaction is different must be reconciled with the evidence for a TCR bias towards MHC. The possibility of germline-encoded β-chains driving initial TCR binding followed by conformational section from a limited ensemble or moderate induced fit elsewhere is an attractive means to achieve this reconciliation. What is needed to fully address these issues are detailed binding investigations that go beyond structural studies and the traditional surface plasmon resonance experiments commonly performed in studies of TCR–pMHC interactions. Attractive experiments include solution kinetic measurements capable of identifying induced fit and conformational selection (e.g. [48]), as well as studies of the intrinsic flexibility of TCR CDR loops and peptides bound to MHC molecules (e.g. [33,100]). Correlation of the resulting data with loop and peptide sequences and the expanding database of structural and mutational studies should shed further light on how TCRs use conformational dynamics to achieve their remarkable molecular recognition properties.
Acknowledgments
We thank Dr Alex Toker for the invitation, members of the Baker research group for discussion and an anonymous reviewer for helpful comments.
FUNDING
Supported by grant R01GM067097 from the NIGMS (National Institute of General Medical Sciences), NIH (National Institutes for Health). K. H. P. was supported by the Notre Dame Chemistry–Biochemistry–Biology Interface training program, funded by grant T32GM075762 from the NIGMS, NIH.
References
- 1.Mason D. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol. Today. 1998;19:395–404. doi: 10.1016/s0167-5699(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 2.Wilson D. B., Wilson D. H., Schroder K., Pinilla C., Blondelle S., Houghten R. A., Garcia K. C. Specificity and degeneracy of T cells. Mol. Immunol. 2004;40:1047–1055. doi: 10.1016/j.molimm.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 3.Maverakis E., van den Elzen P., Sercarz E. E. Self-reactive T cells and degeneracy of T cell recognition: evolving concepts–from sequence homology to shape mimicry and TCR flexibility. J. Autoimmun. 2001;16:201–209. doi: 10.1006/jaut.2000.0493. [DOI] [PubMed] [Google Scholar]
- 4.Garcia K. C., Adams E. J. How the T cell receptor sees antigen – a structural view. Cell. 2005;122:333–336. doi: 10.1016/j.cell.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 5.Rudolph M. G., Stanfield R. L., Wilson I. A. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 6.Wang J. H., Mallis R. J., Reinherz E. L. Immunodominant-peptide recognition: beta testing TCRαβ. Immunity. 2008;28:139–141. doi: 10.1016/j.immuni.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Ishizuka J., Stewart-Jones G. B., van der Merwe A., Bell J. I., McMichael A. J., Jones E. Y. The structural dynamics and energetics of an immunodominant T cell receptor are programmed by its V[β] domain. Immunity. 2008;28:171–182. doi: 10.1016/j.immuni.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 8.Feng D., Bond C. J., Ely L. K., Maynard J., Garcia K. C. Structural evidence for a germline-encoded T cell receptor–major histocompatibility complex interaction ‘codon’. Nat. Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 9.Davis M. M., Boniface J. J., Reich Z., Lyons D., Hampl J., Arden B., Chien Y. Ligand recognition by αβ T cell receptors. Annu. Rev. Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 10.Vijayakumar M., Wong K.-Y., Schreiber G., Fersht A. R., Szabo A., Zhou H.-X. Electrostatic enhancement of diffusion-controlled protein–protein association: comparison of theory and experiment on barnase and barstar1. J. Mol. Biol. 1998;278:1015–1024. doi: 10.1006/jmbi.1998.1747. [DOI] [PubMed] [Google Scholar]
- 11.Janin J. The kinetics of protein–protein recognition. Proteins. 1997;28:153–161. doi: 10.1002/(sici)1097-0134(199706)28:2<153::aid-prot4>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 12.Matsui K., Boniface J. J., Steffner P., Reay P. A., Davis M. M. Kinetics of T-cell receptor binding to peptide/I-Ek complexes: correlation of the dissociation rate with T-cell responsiveness. Proc. Natl. Acad. Sci. U.S.A. 1994;91:12862–12866. doi: 10.1073/pnas.91.26.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia K. C., Degano M., Pease L. R., Huang M., Peterson P. A., Teyton L., Wilson I. A. Structural basis of plasticity in T cell receptor recognition of a self peptide–MHC antigen. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- 14.Garcia K. C., Degano M., Stanfield R. L., Brunmark A., Jackson M. R., Peterson P. A., Teyton L., Wilson I. A. An αβ T cell receptor structure at 2.5Å and its orientation in the TCR–MHC complex. Science. 1996;274:209–219. [Google Scholar]
- 15.Bhardwaj V., Kumar V., Geysen H. M., Sercarz E. E. Degenerate recognition of a dissimilar antigenic peptide by myelin basic protein-reactive T cells. Implications for thymic education and autoimmunity. J. Immunol. 1993;151:5000–5010. [PubMed] [Google Scholar]
- 16.Evavold B. D., Sloan-Lancaster J., Wilson K. J., Rothbard J. B., Allen P. M. Specific T cell recognition of minimally homologous peptides: evidence for multiple endogenous ligands. Immunity. 1995;2:655–663. doi: 10.1016/1074-7613(95)90010-1. [DOI] [PubMed] [Google Scholar]
- 17.Wucherpfennig K. W., Strominger J. L. Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garboczi D. N., Ghosh P., Utz U., Fan Q. R., Biddison W. E., Wiley D. C. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 19.Ding Y. H., Baker B. M., Garboczi D. N., Biddison W. E., Wiley D. C. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity. 1999;11:45–56. doi: 10.1016/s1074-7613(00)80080-1. [DOI] [PubMed] [Google Scholar]
- 20.Willcox B. E., Gao G. F., Wyer J. R., Ladbury J. E., Bell J. I., Jakobsen B. K., van der Merwe P. A. TCR binding to peptide–MHC stabilizes a flexible recognition interface. Immunity. 1999;10:357–365. doi: 10.1016/s1074-7613(00)80035-7. [DOI] [PubMed] [Google Scholar]
- 21.Boniface J. J., Reich Z., Lyons D. S., Davis M. M. Thermodynamics of T cell receptor binding to peptide–MHC: evidence for a general mechanism of molecular scanning. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11446–11451. doi: 10.1073/pnas.96.20.11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker B. M., Murphy K. P. Prediction of binding energetics from structure using empirical parameterization. Methods Enzymol. 1998;295:294–315. doi: 10.1016/s0076-6879(98)95045-5. [DOI] [PubMed] [Google Scholar]
- 23.Chen J.-L., Stewart-Jones G., Bossi G., Lissin N. M., Wooldridge L., Choi E. M. L., Held G., Dunbar P. R., Esnouf R. M., Sami M., et al. Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J. Exp. Med. 2005;201:1243–1255. doi: 10.1084/jem.20042323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng L., Langley R. J., Brown P. H., Xu G., Teng L., Wang Q., Gonzales M. I., Callender G. G., Nishimura M. I., Topalian S. L., Mariuzza R. A. Structural basis for the recognition of mutant self by a tumor-specific, MHC class II-restricted T cell receptor. Nat. Immunol. 2007;8:398–408. doi: 10.1038/ni1447. [DOI] [PubMed] [Google Scholar]
- 25.Tynan F. E., Reid H. H., Kjer-Nielsen L., Miles J. J., Wilce M. C., Kostenko L., Borg N. A., Williamson N. A., Beddoe T., Purcell A. W., et al. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat. Immunol. 2007;8:268–276. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]
- 26.Kjer-Nielsen L., Clements C. S., Brooks A. G., Purcell A. W., McCluskey J., Rossjohn J. The 1.5Å crystal structure of a highly selected antiviral T cell receptor provides evidence for a structural basis of immunodominance. Structure. 2002;10:1521–1532. doi: 10.1016/s0969-2126(02)00878-x. [DOI] [PubMed] [Google Scholar]
- 27.Luz J. G., Huang M., Garcia K. C., Rudolph M. G., Apostolopoulos V., Teyton L., Wilson I. A. Structural comparison of allogeneic and syngeneic T cell receptor–peptide–major histocompatibility complex complexes: a buried alloreactive mutation subtly alters peptide presentation substantially increasing V{β} interactions. J. Exp. Med. 2002;195:1175–1186. doi: 10.1084/jem.20011644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colf L. A., Bankovich A. J., Hanick N. A., Bowerman N. A., Jones L. L., Kranz D. M., Garcia K. C. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 29.Degano M., Garcia K. C., Apostolopoulos V., Rudolph M. G., Teyton L., Wilson I. A. A functional hot spot for antigen recognition in a superagonist TCR/MHC complex. Immunity. 2000;12:251–261. doi: 10.1016/s1074-7613(00)80178-8. [DOI] [PubMed] [Google Scholar]
- 30.Stewart-Jones G. B., McMichael A. J., Bell J. I., Stuart D. I., Jones E. Y. A structural basis for immunodominant human T cell receptor recognition. Nat. Immunol. 2003;4:657–663. doi: 10.1038/ni942. [DOI] [PubMed] [Google Scholar]
- 31.Kjer-Nielsen L., Clements C. S., Purcell A. W., Brooks A. G., Whisstock J. C., Burrows S. R., McCluskey J., Rossjohn J. A structural basis for the selection of dominant αβ T cell receptors in antiviral immunity. Immunity. 2003;18:53–64. doi: 10.1016/s1074-7613(02)00513-7. [DOI] [PubMed] [Google Scholar]
- 32.Reinherz E. L., Tan K., Tang L., Kern P., Liu J., Xiong Y., Hussey R. E., Smolyar A., Hare B., Zhang R., et al. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science. 1999;286:1913–1921. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- 33.Hare B. J., Wyss D. F., Osburne M. S., Kern P. S., Reinherz E. L., Wagner G. Structure, specificity and CDR mobility of a class II restricted single-chain T-cell receptor. Nat. Struct. Biol. 1999;6:574–581. doi: 10.1038/9359. [DOI] [PubMed] [Google Scholar]
- 34.Reiser J. B., Gregoire C., Darnault C., Mosser T., Guimezanes A., Schmitt-Verhulst A. M., Fontecilla-Camps J. C., Mazza G., Malissen B., Housset D. A T cell receptor CDR3β loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex. Immunity. 2002;16:345–354. doi: 10.1016/s1074-7613(02)00288-1. [DOI] [PubMed] [Google Scholar]
- 35.Gagnon S. J., Borbulevych O. Y., Davis-Harrison R. L., Turner R. V., Damirjian M., Wojnarowicz A., Biddison W. E., Baker B. M. T cell receptor recognition via cooperative conformational plasticity. J. Mol. Biol. 2006;363:228–243. doi: 10.1016/j.jmb.2006.08.045. [DOI] [PubMed] [Google Scholar]
- 36.McBeth C., Seamons A., Pizarro J. C., Fleishman S. J., Baker D., Kortemme T., Goverman J. M., Strong R. K. A new twist in TCR diversity revealed by a forbidden [α][β] TCR. J. Mol. Biol. 2008;375:1306–1319. doi: 10.1016/j.jmb.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Philippopoulos M., Lim C. Exploring the dynamic information content of a protein NMR structure: comparison of a molecular dynamics simulation with the NMR and X-ray structures of Escherichia coli ribonuclease HI. Proteins. 1999;36:87–110. doi: 10.1002/(sici)1097-0134(19990701)36:1<87::aid-prot8>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 38.Reiser J.-B., Darnault C., Guimezanes A., Grégoire C., Mosser T., Schmitt-Verhulst A.-M., Fontecilla-Camps J. C., Malissen B., Housset D., Mazza G. Crystal structure of a T cell receptor bound to an allogeneic MHC molecule. Nat. Immunol. 2000;1:291–297. doi: 10.1038/79728. [DOI] [PubMed] [Google Scholar]
- 39.Mazza C., Auphan-Anezin N., Gregoire C., Guimezanes A., Kellenberger C., Roussel A., Kearney A., van der Merwe P. A., Schmitt-Verhulst A. M., Malissen B. How much can a T-cell antigen receptor adapt to structurally distinct antigenic peptides? EMBO J. 2007;26:1972–1983. doi: 10.1038/sj.emboj.7601605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reiser J. B., Darnault C., Gregoire C., Mosser T., Mazza G., Kearney A., van der Merwe P. A., Fontecilla-Camps J. C., Housset D., Malissen B. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat. Immunol. 2003;4:241–247. doi: 10.1038/ni891. [DOI] [PubMed] [Google Scholar]
- 41.Krogsgaard M., Prado N., Adams E. J., He X., Chow D. C., Wilson D. B., Garcia K. C., Davis M. M. Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T cell activation. Mol. Cell. 2003;12:1367–1378. doi: 10.1016/s1097-2765(03)00474-x. [DOI] [PubMed] [Google Scholar]
- 42.Garcia K. C., Tallquist M. D., Pease L. R., Brunmark A., Scott C. A., Degano M., Stura E. A., Peterson P. A., Wilson I. A., Teyton L. αβ T cell receptor interactions with syngeneic and allogeneic ligands: affinity measurements and crystallization. Proc. Natl. Acad. Sci. U.S.A. 1997;94:13838–13843. doi: 10.1073/pnas.94.25.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia K. C., Radu C. G., Ho J., Ober R. J., Ward E. S. Kinetics and thermodynamics of T cell receptor–autoantigen interactions in murine experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6818–6823. doi: 10.1073/pnas.111161198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anikeeva N., Lebedeva T., Krogsgaard M., Tetin S. Y., Martinez-Hackert E., Kalams S. A., Davis M. M., Sykulev Y. Distinct molecular mechanisms account for the specificity of two different T-cell receptors. Biochemistry. 2003;42:4709–4716. doi: 10.1021/bi026864+. [DOI] [PubMed] [Google Scholar]
- 45.Davis-Harrison R. L., Armstrong K. M., Baker B. M. Two different T cell receptors use different thermodynamic strategies to recognize the same peptide/MHC ligand. J. Mol. Biol. 2005;346:533–550. doi: 10.1016/j.jmb.2004.11.063. [DOI] [PubMed] [Google Scholar]
- 46.Ely L. K., Beddoe T., Clements C. S., Matthews J. M., Purcell A. W., Kjer-Nielsen L., McCluskey J., Rossjohn J. Disparate thermodynamics governing T cell receptor–MHC-I interactions implicate extrinsic factors in guiding MHC restriction. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6641–6646. doi: 10.1073/pnas.0600743103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller P. J., Pazy Y., Conti B., Riddle D., Appella E., Collins E. J. Single MHC mutation eliminates enthalpy associated with T cell receptor binding. J. Mol. Biol. 2007;373:315–327. doi: 10.1016/j.jmb.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gakamsky D. M., Lewitzki E., Grell E., Saulquin X., Malissen B., Montero-Julian F., Bonneville M., Pecht I. Kinetic evidence for a ligand-binding-induced conformational transition in the T cell receptor. Proc. Natl. Acad. Sci. U.S.A. 2007;104:16639–16644. doi: 10.1073/pnas.0707061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armstrong K. M., Insaidoo F. K., Baker B. M. Thermodynamics of T-cell receptor–peptide/MHC interactions: progress and opportunities. J. Mol. Recognit. 2008;21:275–287. doi: 10.1002/jmr.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prabhu N. V., Sharp K. A. Heat capacity in proteins. Annu. Rev. Phys. Chem. 2005;56:521–548. doi: 10.1146/annurev.physchem.56.092503.141202. [DOI] [PubMed] [Google Scholar]
- 51.Spolar R. S., Record M. T., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 52.Zhukov A., Karlsson R. Statistical aspects of van't Hoff analysis: a simulation study. J. Mol. Recognit. 2007;20:379–385. doi: 10.1002/jmr.845. [DOI] [PubMed] [Google Scholar]
- 53.Murphy K. P., Freire E. Thermodynamics of structural stability and cooperative folding behavior in proteins. Adv. Protein Chem. 1992;43:313–361. doi: 10.1016/s0065-3233(08)60556-2. [DOI] [PubMed] [Google Scholar]
- 54.Koshland D. E. Application of a theory of enzyme specificity to protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jerne N. K. The somatic generation of immune recognition. Eur. J. Immunol. 1971;1:1–9. doi: 10.1002/eji.1830010102. [DOI] [PubMed] [Google Scholar]
- 56.Kim H.-J., Guo D., Sant'Angelo D. B. Coevolution of TCR-MHC interactions: conserved MHC tertiary structure is not sufficient for interactions with the TCR. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7263–7267. doi: 10.1073/pnas.0502751102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zerrahn J., Held W., Raulet D. H. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- 58.Sim B.-C., Zerva L., Greene M. I., Gascoigne N. R. Control of MHC restriction by TCR Vα CDR1 and CDR2. Science. 1996;273:963–966. doi: 10.1126/science.273.5277.963. [DOI] [PubMed] [Google Scholar]
- 59.Huseby E. S., White J., Crawford F., Vass T., Becker D., Pinilla C., Marrack P., Kappler J. W. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 60.Fersht A. R., Sato S. Φ-Value analysis and the nature of protein-folding transition states. Proc. Natl. Acad. Sci. U.S.A. 2004;101:7976–7981. doi: 10.1073/pnas.0402684101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu L. C., Tuot D. S., Lyons D. S., Garcia K. C., Davis M. M. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature. 2002;418:552–556. doi: 10.1038/nature00920. [DOI] [PubMed] [Google Scholar]
- 62.Tobi D., Bahar I. Structural changes involved in protein binding correlate with intrinsic motions of proteins in the unbound state. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18908–18913. doi: 10.1073/pnas.0507603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keskin O. Binding induced conformational changes of proteins correlate with their intrinsic fluctuations: a case study of antibodies. BMC Struct. Biol. 2007;7:1–11. doi: 10.1186/1472-6807-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goh C.-S., Milburn D., Gerstein M. Conformational changes associated with protein–protein interactions. Curr. Opin. Struct. Biol. 2004;14:104–109. doi: 10.1016/j.sbi.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 65.Tang C., Iwahara J., Clore G. M. Visualization of transient encounter complexes in protein–protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 66.Tang C., Ghirlando R., Clore G. M. Visualization of transient ultra-weak protein self-association in solution using paramagnetic relaxation enhancement. J. Am. Chem. Soc. 2008;130:4048–4056. doi: 10.1021/ja710493m. [DOI] [PubMed] [Google Scholar]
- 67.Wang Z., Turner R., Baker B. M., Biddison W. E. MHC allele-specific molecular features determine peptide/HLA-A2 conformations that are recognized by HLA-A2-restricted T cell receptors. J. Immunol. 2002;169:3146–3154. doi: 10.4049/jimmunol.169.6.3146. [DOI] [PubMed] [Google Scholar]
- 68.Gagnon S. J., Borbulevych O. Y., Davis-Harrison R. L., Baxter T. K., Clemens J. R., Armstrong K. M., Turner R. V., Damirjian M., Biddison W. E., Baker B. M. Unraveling a hotspot for TCR recognition on HLA-A2: evidence against the existence of peptide-independent TCR binding determinants. J. Mol. Biol. 2005;353:556–573. doi: 10.1016/j.jmb.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 69.Davis-Harrison R. L., Insaidoo F. K., Baker B. M. T cell receptor binding transition states and recognition of peptide/MHC. Biochemistry. 2007;46:1840–1850. doi: 10.1021/bi061702p. [DOI] [PubMed] [Google Scholar]
- 70.Housset D., Malissen B. What do TCR–pMHC crystal structures teach us about MHC restriction and alloreactivity? Trends Immunol. 2003;24:429–437. doi: 10.1016/s1471-4906(03)00180-7. [DOI] [PubMed] [Google Scholar]
- 71.Dai S., Huseby E. S., Rubtsova K., Scott-Browne J., Crawford F., Macdonald W. A., Marrack P., Kappler J. W. Crossreactive T cells spotlight the germline rules for [α][β] T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buslepp J., Wang H., Biddison W. E., Appella E., Collins E. J. A correlation between TCR Vα docking on MHC and CD8 dependence: implications for T cell selection. Immunity. 2003;19:595–606. doi: 10.1016/s1074-7613(03)00269-3. [DOI] [PubMed] [Google Scholar]
- 73.Khan A. R., Baker B. M., Ghosh P., Biddison W. E., Wiley D. C. The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J. Immunol. 2000;164:6398–6405. doi: 10.4049/jimmunol.164.12.6398. [DOI] [PubMed] [Google Scholar]
- 74.Ding Y. H., Smith K. J., Garboczi D. N., Utz U., Biddison W. E., Wiley D. C. Two human T cell receptors bind in a similar diagonal mode to the HLA- A2/Tax peptide complex using different TCR amino acids. Immunity. 1998;8:403–411. doi: 10.1016/s1074-7613(00)80546-4. [DOI] [PubMed] [Google Scholar]
- 75.Webb A. I., Dunstone M. A., Chen W., Aguilar M.-I., Chen Q., Jackson H., Chang L., Kjer-Nielsen L., Beddoe T., McCluskey J., et al. Functional and Structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue. J. Biol. Chem. 2004;279:23438–23446. doi: 10.1074/jbc.M314066200. [DOI] [PubMed] [Google Scholar]
- 76.Auphan-Anezin N., Mazza C., Guimezanes A., Barrett-Wilt Gregory A., Montero-Julian F., Roussel A., Hunt Donald F., Malissen B., Schmitt-Verhulst A.-M. Distinct orientation of the alloreactive monoclonal CD8 T cell activation program by three different peptide/MHC complexes. Eur. J. Immunol. 2006;36:1856–1866. doi: 10.1002/eji.200635895. [DOI] [PubMed] [Google Scholar]
- 77.Li Y., Huang Y., Lue J., Quandt J. A., Martin R., Mariuzza R. A. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 2005;24:2968–2979. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dill K. A., Chan H. S. From Levinthal to pathways to funnels. Nat. Struct. Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 79.Miller D. W., Dill K. A. Ligand binding to proteins: the binding landscape model. Protein Sci. 1997;6:2166–2179. doi: 10.1002/pro.5560061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsai C., Kumar S., Ma B., Nussinov R. Folding funnels, binding funnels, and protein function. Protein Sci. 1999;8:1181–1190. doi: 10.1110/ps.8.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Levy Y., Cho S. S., Onuchic J. N., Wolynes P. G. A survey of flexible protein binding mechanisms and their transition states using native topology based energy landscapes. J. Mol. Biol. 2005;346:1121–1145. doi: 10.1016/j.jmb.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 82.Kumar S., Ma B., Tsai C. J., Sinha N., Nussinov R. Folding and binding cascades: dynamic landscapes and population shifts. Protein Sci. 2000;9:10–19. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Holler P. D., Kranz D. M. T cell receptors: affinities, cross-reactivities, and a conformer model. Mol. Immunol. 2004;40:1027–1031. doi: 10.1016/j.molimm.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 84.Foote J., Milstein C. Conformational isomerism and the diversity of antibodies. Proc. Natl. Acad. Sci. U.S.A. 1994;91:10370–10374. doi: 10.1073/pnas.91.22.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lancet D., Pecht I. Kinetic evidence for hapten-induced conformational transition in immunoglobulin MOPC 460. Proc. Natl. Acad. Sci. U.S.A. 1976;73:3549–3553. doi: 10.1073/pnas.73.10.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pauling L. A theory of the structure and process of formation of antibodies. J. Am. Chem. Soc. 1940;62:2643–2657. [Google Scholar]
- 87.James L. C., Roversi P., Tawfik D. S. Antibody multispecificity mediated by conformational diversity. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- 88.James L. C., Tawfik D. S. Conformational diversity and protein evolution – a 60-year-old hypothesis revisited. Trends Biochem. Sci. 2003;28:361–368. doi: 10.1016/S0968-0004(03)00135-X. [DOI] [PubMed] [Google Scholar]
- 89.Swain J. F., Gierasch L. M. The changing landscape of protein allostery. Curr. Opin. Struct. Biol. 2006;16:102–108. doi: 10.1016/j.sbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 90.Kern D., Zuiderweg E. R. The role of dynamics in allosteric regulation. Curr. Opin. Struct. Biol. 2003;13:748–757. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 91.Volkman B. F., Lipson D., Wemmer D. E., Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 92.Popovych N., Sun S., Ebright R. H., Kalodimos C. G. Dynamically driven protein allostery. Nat. Struct. Mol. Biol. 2006;13:831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Benkovic S. J., Hammes-Schiffer S. A perspective on enzyme catalysis. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 94.Sharma A. K., Kuhns J. J., Yan S., Friedline R. H., Long B., Tisch R., Collins E. J. Class I major histocompatibility complex anchor substitutions alter the conformation of T cell receptor contacts. J. Biol. Chem. 2001;276:21443–21449. doi: 10.1074/jbc.M010791200. [DOI] [PubMed] [Google Scholar]
- 95.Kuhns J. J., Batalia M. A., Yan S., Collins E. J. Poor binding of a HER-2/neu epitope (GP2) to HLA-A2.1 is due to a lack of interactions with the center of the peptide. J. Biol. Chem. 1999;274:36422–36427. doi: 10.1074/jbc.274.51.36422. [DOI] [PubMed] [Google Scholar]
- 96.Speir J. A., Stevens J., Joly E., Butcher G. W., Wilson I. A. Two different, highly exposed, bulged structures for an unusually long peptide bound to rat MHC class I RT1-Aa. Immunity. 2001;14:81–92. doi: 10.1016/s1074-7613(01)00091-7. [DOI] [PubMed] [Google Scholar]
- 97.Borbulevych O. Y., Insaidoo F. K., Baxter T. K., Powell D. J., Jr, Johnson L. A., Restifo N. P., Baker B. M. Structures of MART-1(26/27–35) peptide/HLA-A2 complexes reveal a remarkable disconnect between antigen structural homology and T cell recognition. J. Mol. Biol. 2007;372:1123–1136. doi: 10.1016/j.jmb.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.James L. C., Tawfik D. S. Structure and kinetics of a transient antibody binding intermediate reveal a kinetic discrimination mechanism in antigen recognition. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12730–12735. doi: 10.1073/pnas.0500909102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grunberg R., Leckner J., Nilges M. Complementarity of structure ensembles in protein–protein binding. Structure. 2004;12:2125–2136. doi: 10.1016/j.str.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 100.Pohlmann T., Bockmann R. A., Grubmuller H., Uchanska-Ziegler B., Ziegler A., Alexiev U. Differential peptide dynamics is linked to MHC polymorphism. J. Biol. Chem. 2004;279:28197–28201. doi: 10.1074/jbc.C400128200. [DOI] [PubMed] [Google Scholar]
- 101.Rocchia A., Alexov E., Honig B. Extending the applicability of the nonlinear Poisson–Boltzmann equation: multiple dielectric constants and multivalent ions. J. Phys. Chem. B. 2001;105:6507–6514. [Google Scholar]