Abstract
Balancing selection is potentially an important biological force for maintaining advantageous genetic diversity in populations, including variation that is responsible for long-term adaptation to the environment. By serving as a means to maintain genetic variation, it may be particularly relevant to maintaining phenotypic variation in natural populations. Nevertheless, its prevalence and specific targets in the human genome remain largely unknown. We have analyzed the patterns of diversity and divergence of 13,400 genes in two human populations using an unbiased single-nucleotide polymorphism data set, a genome-wide approach, and a method that incorporates demography in neutrality tests. We identified an unbiased catalog of genes with signatures of long-term balancing selection, which includes immunity genes as well as genes encoding keratins and membrane channels; the catalog also shows enrichment in functional categories involved in cellular structure. Patterns are mostly concordant in the two populations, with a small fraction of genes showing population-specific signatures of selection. Power considerations indicate that our findings represent a subset of all targets in the genome, suggesting that although balancing selection may not have an obvious impact on a large proportion of human genes, it is a key force affecting the evolution of a number of genes in humans.
Keywords: overdominance, frequency-dependent selection, heterosis, human evolution, population genetics, human diversity
Introduction
Balancing selection maintains favorable genetic diversity in populations by a variety of mechanisms, including overdominance and fluctuating selection (e.g., frequency-dependent selection). In the case where one locus with two alleles displays overdominance, the higher fitness of heterozygotes maintains both alleles in the population, eventually leading to an equilibrium allele frequency that maximizes the mean fitness of the population. Under frequency-dependent selection, the fitness associated with an allele varies with its frequency, giving rise to an equilibrium with an enhanced number of alleles at intermediate frequencies (when selection favors intermediate alleles) or low frequencies (in cases of rare allele advantage) (see Richman 2000). Classical examples of balancing selection include the β-globin gene in humans (Pasvol et al. 1978), the major histocompatibility complex (MHC) system in mammals (Hughes and Nei 1988; Takahata and Nei 1990), the disease-response genes (R-genes) in plants (Stahl et al. 1999), the self-incompatibility system in plants (Wright 1939), and the complementary sex determination of haplodiploid species (Yokoyama and Nei 1979; Cho et al. 2006).
By maintaining functional genetic variation in populations, balancing selection is medically relevant. Association between balanced polymorphisms and pathology has been proposed for several human diseases, including the ß-globin gene and sickle cell anemia (Pasvol et al. 1978), CFTR and cystic fibrosis (Gabriel et al. 1994; Pier et al. 1998), and PAH and phenylketonuria (Woolf et al.1967). This is not surprising because, at equilibrium frequencies, a substantial portion of the population is homozygous and carries a deleterious genotype. Balanced polymorphisms present the primary candidates for the common disease–common variant hypothesis because the pattern of natural selection results in elevated frequencies of alleles which, in the homozygous state, may reduce fitness and contribute to disease.
The influence of balancing selection in shaping the levels of diversity in natural populations has long been a subject of debate. Once thought to be the primary driver that maintains the substantial genetic variability observed in populations (Lewontin and Hubby 1966), balancing selection came to be considered rare when polymorphism levels could be explained, without the need of selection, by the neutral theory of evolution (Kimura 1968). It has been proposed that balancing selection cannot be common due to the associated genetic load (the population burden that derives from the reduced fitness of less-favorable homozygotes, maintained by selection than favors advantageous heterozygotes); but the relevance of such arguments in predicting the prevalence of selection has been debated (see Gillespie 1991), and today the debate over the role of selection (and balancing selection) in maintaining polymorphism remains open (Gillespie 1991).
Recent genome-wide scans of selection have dramatically improved our understanding of the influence of purifying and directional selection in shaping the evolution of genes, particularly in humans (Clark et al. 2003; Akey et al. 2004; Bustamante et al. 2005; Chimpanzee Genome and Analysis Consortium 2005; Nielsen, Bustamante, et al. 2005; Sabeti et al. 2006; Voight et al. 2006; Williamson et al. 2007; Barreiro et al. 2008). Such advances have not been applied in a systematic genome-wide fashion to balancing selection, and current biological understanding of balancing selection is mostly limited to a few loci localized by candidate gene approaches (e.g., Hughes and Nei 1988, 1989; Bamshad et al. 2002; Baum et al. 2002; Wooding et al. 2004; Cork and Purugganan 2005; Kroymann and Mitchell-Olds 2005; Tan et al. 2005; Cho et al. 2006; reviewed in Bamshad and Wooding 2003; Mitchell-Olds et al. 2007). This is mainly due to the difficulties associated with the detection of this type of selection at a whole-genome level. The genomic signal of recent balancing selection (extended linkage disequilibrium [LD]) is detectable by LD-based methods (Voight et al. 2006; Wang et al. 2006), but it is indistinguishable from incomplete sweeps of positive selection. The signal of long-term balancing selection is specific (excess of polymorphism) but narrow due to the long-term effects of recombination (Hudson and Kaplan 1988; Charlesworth et al. 1997). Therefore, most available data sets (with low and constant single-nucleotide polymorphism [SNP] density) have little power to detect the localized signals of long-term balancing selection. As a consequence, previous efforts have failed to detect convincing targets in the human genome (Asthana et al. 2005; Bubb et al. 2006).
Here, we present the first concerted effort to detect genes undergoing balancing selection across the genome in human populations. We use a data set of unascertained SNPs and apply a method that contrasts patterns of polymorphism in each gene to the rest of the genome as well as to neutral expectations. Because the timing and type of selection affect its genomic signature, we focus on the identification of genes with strong signals of long-term balancing selection maintaining an excess of intermediate-frequency variants. We find a small but strongly supported set of genes with signatures of selection, providing an unbiased catalog of candidate targets of balancing selection in the human genome.
Materials and Methods
Data
Analyses were performed using polymorphism and divergence data obtained from a complete survey of coding variability in 13,400 human RefSeq genes by direct sequencing of all their well-annotated exons in 39 human subjects (19 African Americans [AA] and 20 European Americans [EA]). The data are described in Bustamante et al. (2005), and a strict bioinformatics pipeline ensured true homology and the use of only well-supported SNPs, as described in Boyko et al. (2008). Substantial effort was taken to avoid biological and technical confounding factors. The original bioinformatics pipeline involved reciprocal Blast searches to avoid misalignments and required a unique high-quality match to the public human chimpanzee sequence PanTro2 (Chimpanzee Sequence and Analysis Consortium 2005) (supplementary Methods, Supplementary Material online). Also, only genes with unique products with in silico polymerase chain reaction (http://genome.ucsc.edu) were used. This process checks for multiple genomic matches of the amplification primers and detects cases of putative nonspecific amplification. Finally, extreme (significant) genes were extensively checked for the presence of close paralogs, including segmental duplications, through BLAT searches of the March 2006 Human Genome Sequence Assembly and test of involvement in segmental duplications (Human Segmental Duplication Database [Cheung et al. 2003]). Fixed differences with respect to chimpanzee and ancestral state of human SNPs were assessed by comparison with PanTro2 chimpanzee reference sequence (Chimpanzee Sequence and Analysis Consortium 2005).
A total of 4,877 genes had at least ten informative sites (polymorphic or fixed relative to chimpanzee) and were further considered. This condition filtered out genes that lacked sufficient information for a valid test without biasing the data set. Also, all genes had to contain at least one polymorphic site for neutrality tests to be performed. These data do not suffer from ascertainment bias, have power to detect the localized signals of long-term balancing selection, and are expected to contain the majority of common variants in these populations. In short, this is a particularly well-suited data set for the detection of balancing selection.
Null Model
The choice of an adequate null model is crucial for detection of selection because some demographic scenarios can mimic the effects of selection on diversity. To avoid such confounding effects, we applied a method designed to minimize the effects of demography in neutrality tests (Nielsen et al. 2009). In essence, the method uses the complete data set to estimate parameters of the past demographic history that best fit the data and considers such estimates as the null (neutral) demographic model against which neutrality is tested.
All demographic inferences were based on the complete data set (13,400 genes). Briefly, the method infers admixture proportions of individuals using a maximum likelihood (ML) method. The demographic parameters that best fit the data are estimated using an (composite) ML approach through coalescent simulations and considering the estimated admixture proportions. Demography was inferred separately for the X and autosomes due to the possibility of sex-specific differential migration. The best-fit demographic model allows for a bottleneck in Europeans upon emergence from Africa and exponential growth in both populations (fig. 2 legend and supplementary Methods, Supplementary Material online). It provides a very good fit of the data, indicating that the demographic scenario explains most of the patterns observed in the data (Nielsen et al. 2009).
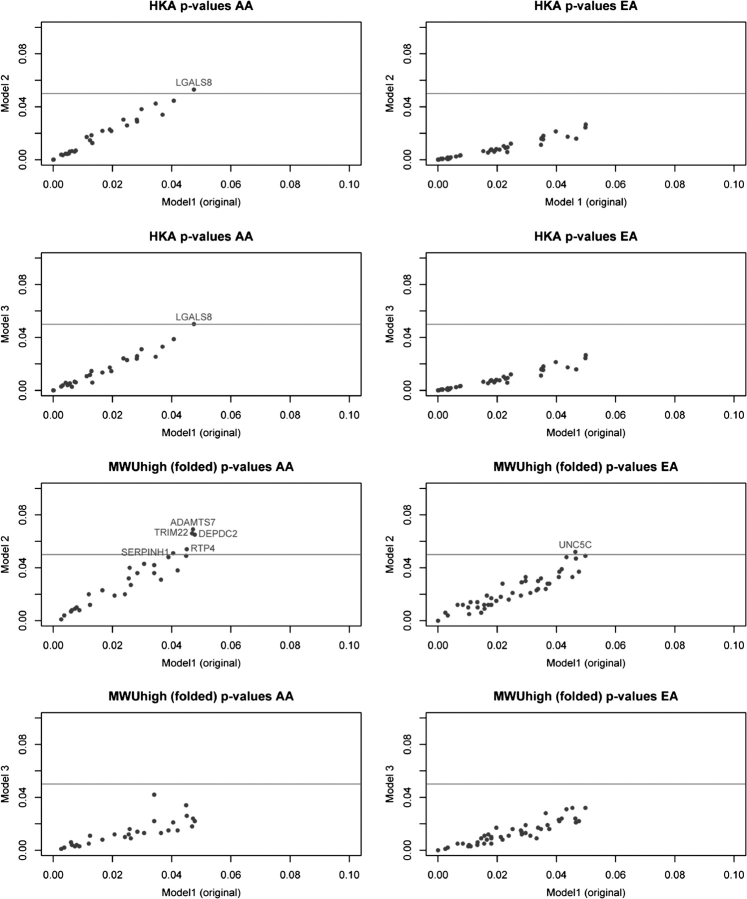
FIG. 2.—
Correlation of results under different demographic scenarios. Correlation between the P values under the originally estimated demographic scenario (Model 1) and the two alternative demographic models (Model 2 and Model 3). Extreme genes showing P > 0.05 for the alternative demographic scenario are annotated. All correlations (Spearman) are rho > 0.9 and P < 4 × 10−11. Model 1: T = 0.099 (divergence time between the two populations); αA = 9.5, αE = 21.1 (rate of expansion of African and European populations since divergence time); m = 6.67 (gene flow rate of migrants per generation between the two populations); bottleneck in European population 0.1 generations ago lasting 0.01 generations, with a reduction in population size (β) = 0.018; γ = 1.82 (ratio of the current African to European population size). Model 2: same model, but with all genetic admixture explained by recent admixture rather than migration; f (admixture proportion EA to AA) = 0.20, m = 0. Model 3: best demographic model inferred from this data using an independent method, δaδi (Gutenkunst R, in preparation): T = 0.142, αA = 10.2, αB = 14.3, γ = 1.95, β = 0.021, m = 4.6, f = 0.18.
For each gene, neutrality tests are then performed and their statistical significance is assessed by extensive neutral coalescent simulations under the inferred demographic scenario, with the number of segregating sites and missing data of the gene, and a recombination rate of 7.5 × 10−4 per base pair (Nielsen, Williamson, et al. 2005). Further details can be found in supplementary Methods (Supplementary Material online); for a formal description of the method, statistical details, and discussion of the demographic inference, readers are referred to Nielsen et al. (2009). Genes showing the most unusual patterns of variability considering the demographic history of the populations are identified based on the P values from these neutrality tests. Although the demographic model inferred does not necessarily represent the exact demographic history of the populations, its application as the null model in neutrality tests represents a conservative approach: The tests will only identify genes with a sufficiently extreme departure from the overall patterns observed in the genome, according to the demographic history of the sample (assessed by neutral simulations). The influence of the demographic model was assessed by comparing the probability of the tests under the original and two alternative demographic scenarios (described in fig. 2 legend and supplementary Methods, Supplementary Material online).
Neutrality Tests
Balancing selection may vary in timescale, strength, type (e.g., overdominance vs. frequency-dependent selection), and target (e.g., single locus vs. multiple loci). Such parameters influence the expected effect of selection in linked variation and therefore the strategies for their detection. We aim at detecting long-term balancing selection toward intermediate-frequency alleles, either due to overdominance or frequency-dependent selection, and either targeting single sites or combinations of variants in an epistatic way.
Signatures of balancing selection were detected based on two different properties of sequence variation, as the use of different attributes of the data can be more powerful than the consideration of single neutrality tests (Innan 2006). The main effect in genealogies of long-term balancing selection is an increased coalescence time when compared with neutral expectations. This leads to an excess of polymorphism in the genomic region linked to the selected variant(s) (Hudson and Kaplan 1988; Takahata and Nei 1990; Nordborg 1997; Barton and Etheridge 2004; Williamson et al. 2004). A modified HKA test (Hudson et al. 1987) was applied to detect such excess of diversity. Whereas the original HKA test rejects neutrality with both excess of polymorphism and divergence, our “HKAlow” test is a one-sided HKA test that rejects neutrality only with excess polymorphism. Besides affecting the time to coalescence, balancing selection also affects allele frequencies. Both overdominance (with similar fitness of both homozygotes) and frequency-dependent selection (with optimum at frequencies ∼0.5) can produce an excess of intermediate-frequency alleles. This yields a local site frequency spectrum (SFS) skewed toward intermediate-frequency alleles with respect to the genome as a whole (global SFS). Such a difference between the local and global SFS was tested with a one-sided Mann–Whitney U (MWU) test on the “folded” SFSs. This test, which we call “MWUhigh,” rejects neutrality only in the presence of excess of intermediate-frequency alleles.
The signature of balancing selection is defined by the intersection of the two tests. Genes with signatures of balancing selection (here referred to as extreme genes) are selected as those with significant departures from the neutral model both for HKAlow and MWUhigh tests (5% significance level). The intersection defines genes with both a significant excess of polymorphism and a significant excess of intermediate-frequency alleles. The two tests are sensitive to additional selective forces, but their combination is expected to specifically detect the effects of long-term balancing selection maintaining intermediate frequencies.
The limited number of variants per gene prevents the separate analysis of synonymous and nonsynonymous sites, as well as sliding window type of approaches. For test of gene categories, all genes (irrespective of the number of informative sites) were divided into biological process and molecular function categories according to Panther (http://www.pantherdb.org/), and the distribution of P values of each category was compared with the rest of the data set with a Mann–Whitney U test.
Linkage, Haplotypes, Genealogies, and PolyPhen
Because linkage phase of haplotypes in this data is unknown, LD was measured by the composite LD (Weir 1996), which does not require phase information and avoids introducing uncertainty during haplotype inference (Andrés et al. 2007). We used composite_LD, a Bioperl package from Matthew Hahn and Jason Stajich (http://www.bioperl.org). LD was computed for all SNP pairs in the gene, and the percentage of unmatched, frequency-matched or distance-matched SNP pairs showing significant LD was compared between extreme and nonextreme genes through 10,000 permutations. Complete haplotypes were inferred with PHASE 2.0 (Stephens et al. 2001), and haplotype networks constructed using Network 4.1.1.2 (Bandelt and Dress 1992). When necessary for comparison, HapMap SNP frequency and LD information were obtained from the HapMap database (http://www.hapmap.org) for the Yoruba from Nigeria (YRI) and western Europeans (CEU). The potential functional consequences of nonsynonymous SNPs were predicted with PolyPhen (Sunyaev et al. 2001) as described in Lohmueller et al. (2008).
Results
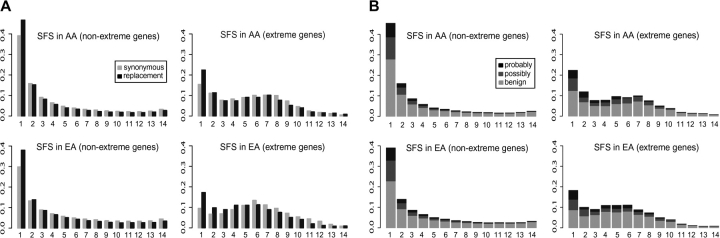
We detect 60 genes with significant signatures of long-term balancing selection (table 1) as shown by their excess of polymorphism (significant HKAlow test) and excess of intermediate-frequency alleles (significant MWUhigh test). We refer to these genes as extreme genes. The average ratio of counts of polymorphic to divergent sites in nonextreme genes is 0.6, whereas the ratio is 1.9 for extreme genes in both populations. This represents a 3-fold increase in the number of polymorphic nucleotides in extreme genes. Allele frequencies also show substantial differences between extreme and nonextreme genes (fig. 1A). The SFS of nonextreme genes has the expected skew toward low-frequency alleles, slight differences between populations due to demographic differences, and a relative enrichment of replacement sites at very low frequencies due to purifying selection against deleterious alleles. As expected by their significant MWUhigh test, extreme genes have a considerable skew toward intermediate-frequency alleles (fig. 1A). The bimodal SFS may reflect a combination of selective forces, with purifying selection keeping deleterious variants at low frequencies and balancing selection maintaining alleles at intermediate frequencies. Note that the contribution of both synonymous and replacement sites is similar at intermediate frequencies, indicating that the excess is not only due to silent (neutral) alleles but also to putatively functional replacement variants. The highly similar SFS of synonymous and replacement sites can be explained simply by linkage.
Table 1.
Sequence Traits and Neutrality Tests of Extreme Genes
| Gene | chromosome | ns | nf | pMWUhigh |
pHKAlow |
||
| AA | EA | AA | EA | ||||
| AA and EA | |||||||
| ADAM11 | 17 | 7 | 4 | 0.006 | 0.043 | 0.012 | 0.050 |
| ALPK2 | 18 | 39 | 31 | 0.026 | 0.028 | 0.000 | 0.000 |
| BTN1A1 | 6 | 7 | 5 | 0.012 | 0.030 | 0.028 | 0.036 |
| DEPDC2 | 8 | 7 | 4 | 0.048 | 0.028 | 0.025 | 0.018 |
| KRT14 [] | 17 | 10 | 6 | 0.004 | 0.007 | 0.005 | 0.003 |
| LGALS8 | 1 | 9 | 10 | 0.017 | 0.034 | 0.048 | 0.022 |
| LILRB4 [] | 19 | 8 | 4 | 0.009 | 0.000 | 0.006 | 0.002 |
| LINS1 | 15 | 15 | 14 | 0.031 | 0.008 | 0.020 | 0.019 |
| RCBTB1 | 13 | 11 | 6 | 0.007 | 0.017 | 0.003 | 0.001 |
| RPS7 | 2 | 10 | 1 | 0.003 | 0.003 | 0.000 | 0.000 |
| RTP4 | 3 | 7 | 5 | 0.045 | 0.013 | 0.041 | 0.007 |
| TRIM22 | 11 | 9 | 5 | 0.047 | 0.038 | 0.004 | 0.008 |
| WDR40C [] | X | 7 | 3 | 0.034 | 0.036 | 0.006 | 0.003 |
| AA | |||||||
| ADAMTS7 | 15 | 7 | 3 | 0.047 | 0.107 | 0.007 | 0.025 |
| C14orf124 | 14 | 8 | 2 | 0.034 | 0.673 | 0.003 | 0.000 |
| CLCNKB [] | 1 | 16 | 18 | 0.024 | 0.407 | 0.011 | 0.205 |
| COL27A1 | 9 | 18 | 19 | 0.026 | 0.107 | 0.017 | 0.017 |
| COPE | 19 | 7 | 5 | 0.036 | 0.158 | 0.024 | 0.110 |
| FGF6 | 12 | 6 | 4 | 0.012 | 0.066 | 0.030 | 0.080 |
| FLJ40243 | 5 | 10 | 9 | 0.042 | 0.061 | 0.019 | 0.014 |
| KRT6B [] | 12 | 8 | 6 | 0.045 | 0.120 | 0.037 | 0.027 |
| KRT84 | 12 | 10 | 6 | 0.006 | 0.055 | 0.005 | 0.008 |
| LRRN6A | 15 | 8 | 3 | 0.028 | 0.101 | 0.008 | 0.003 |
| PPP1R15A | 19 | 15 | 14 | 0.008 | 0.145 | 0.028 | 0.003 |
| SERPINH1 [] | 11 | 7 | 3 | 0.041 | 0.106 | 0.013 | 0.003 |
| TARBP1 | 1 | 15 | 15 | 0.026 | 0.266 | 0.013 | 0.025 |
| TNS1 | 2 | 32 | 25 | 0.039 | 0.115 | 0.000 | 0.002 |
| TRPV6 [] | 7 | 10 | 11 | 0.021 | 0.410 | 0.035 | 0.030 |
| EA | |||||||
| ALDH4A1 | 1 | 10 | 5 | 0.141 | 0.035 | 0.015 | 0.002 |
| ARHGEF3 | 3 | 8 | 3 | 0.091 | 0.041 | 0.002 | 0.001 |
| C20orf186 | 20 | 12 | 9 | 0.072 | 0.025 | 0.009 | 0.003 |
| CAMK2B | 7 | 6 | 6 | 0.154 | 0.011 | 0.123 | 0.035 |
| CD200R1 | 3 | 5 | 5 | 0.016 | 0.015 | 0.159 | 0.050 |
| CDSN | 6 | 20 | 8 | 0.130 | 0.018 | 0.000 | 0.000 |
| FLJ90650 | 5 | 6 | 5 | 0.484 | 0.047 | 0.080 | 0.040 |
| FUT2 [] | 19 | 11 | 7 | 0.051 | 0.041 | 0.004 | 0.021 |
| GM632 | 20 | 9 | 12 | 0.407 | 0.037 | 0.334 | 0.044 |
| GPR111 | 6 | 6 | 5 | 0.009 | 0.018 | 0.080 | 0.020 |
| GRIN3A | 9 | 13 | 19 | 0.357 | 0.050 | 0.238 | 0.019 |
| HLA-B [] | 6 | 13 | 1 | 0.123 | 0.024 | 0.000 | 0.000 |
| KIAA0753 | 17 | 11 | 11 | 0.072 | 0.028 | 0.098 | 0.017 |
| KIAA1303 | 17 | 14 | 19 | 0.247 | 0.034 | 0.110 | 0.023 |
| KRT6E [] | 12 | 8 | 8 | 0.828 | 0.003 | 0.355 | 0.023 |
| LHB [] | 19 | 6 | 4 | 0.073 | 0.020 | 0.031 | 0.025 |
| LOC197322 | 16 | 10 | 14 | 0.004 | 0.016 | 0.095 | 0.047 |
| LRAP | 5 | 12 | 10 | 0.022 | 0.011 | 0.068 | 0.004 |
| MYO1G | 7 | 12 | 14 | 0.486 | 0.033 | 0.092 | 0.023 |
| NALP13 | 19 | 20 | 12 | 0.144 | 0.031 | 0.000 | 0.000 |
| PCDHB16 [] | 5 | 21 | 26 | 0.115 | 0.010 | 0.042 | 0.003 |
| RABEP1 | 17 | 9 | 9 | 0.126 | 0.030 | 0.061 | 0.036 |
| RIOK2 | 5 | 7 | 9 | 0.136 | 0.018 | 0.445 | 0.035 |
| SAMM50 | 22 | 9 | 5 | 0.118 | 0.045 | 0.009 | 0.004 |
| SERPINB5 | 18 | 8 | 4 | 0.130 | 0.042 | 0.010 | 0.015 |
| SLC2A9 | 4 | 7 | 3 | 0.068 | 0.017 | 0.006 | 0.003 |
| SMARCAD1 | 4 | 8 | 4 | 0.662 | 0.048 | 0.013 | 0.006 |
| TMEM171 | 5 | 6 | 4 | 0.096 | 0.016 | 0.118 | 0.024 |
| TSPAN10 | 17 | 10 | 4 | 0.106 | 0.021 | 0.011 | 0.000 |
| UNC5C | 4 | 10 | 13 | 0.419 | 0.046 | 0.240 | 0.035 |
| VARSL | 6 | 14 | 6 | 0.055 | 0.013 | 0.000 | 0.000 |
| ZNF415 [] | 19 | 10 | 6 | 0.088 | 0.022 | 0.008 | 0.018 |
NOTE.—ns: number of segregating sites; and nf: number of fixed divergent sites (human vs. chimpanzee). pMWUhigh: P value of MWUhigh test in AA or EA. pHKAlow: P value of HKAlow test in AA or EA. Bolded genes are considered population-specific because they show P value >0.2 for at least one test in the other population. Genes marked with [] represent cases where events of gene conversion could not be completely discarded. Full names of all genes are shown in supplementary table 1 (Supplementary Material online).
FIG. 1.—
Allele SFS of all segregating sites in extreme and nonextreme genes. The x axis represents the absolute allele frequency of SNPs in the sample; to account for missing data, all SFSs were projected into a sample of 15 chromosomes (Nielsen, Williamson, et al. 2005). The y axis represents the frequency in the data of each respective allele frequency bin. (A) SFS by mutation type: synonymous sites and replacement sites shown for nonextreme genes (left) and extreme genes (right), as shown for each population. (B) SFS by PolyPhen category: benign sites, sites with possible phenotypic effect, and sites with probable phenotypic effect. Color figure in supplementary fig. 1 (Supplementary Material online).
The effect on fitness of replacement mutations ranges from mild to severe. Although the phenotypic consequences of most mutations are unknown, inferences can be made based on the physicochemical properties of the change and the evolutionary conservation at the site. For example, because most mutations are expected to be deleterious, Polyphen (Sunyaev et al. 2001) classifies mutations in increasing order of expected phenotypic effect as benign, possibly damaging, or probably damaging. In nonextreme genes, the majority of mutations with possible and probable phenotypic effect are likely deleterious and maintained at low frequencies (fig. 1B). In extreme genes, many such variants are also at low frequencies, probably due to purifying selection against deleterious alleles. Nevertheless, a considerable proportion of possibly and probably functional variants are present at intermediate frequencies in extreme genes (fig. 1B), presumably maintained by balancing selection. This proportional enrichment for likely functional variants at very low frequencies and intermediate frequencies in extreme genes (supplementary fig. 2, Supplementary Material online) is not significant. Still, the trend again illustrates the combination of purifying selection (maintaining the functionality of genes) and balancing selection (maintaining functional variants in the population) in extreme genes.
Our multistep process involves the inference of the demographic scenario that best fits the data and the use of this model as the null against which neutrality is tested. Although the demographic inference is not intended to disentangle the exact demographic history of human populations (no genetic inference can), this strategy is a conservative one (supplementary Methods, Supplementary Material online). Other demographic scenarios are compatible with the data, though, and their use could, in theory, affect neutrality tests. We investigated the influence of the underlying demographic scenario by assessing the probability of the two tests under two additional demographic scenarios in extreme genes. The P values show a high correlation between the original and the two alternative demographic models (fig. 2). Only six genes in AA and one gene in EA found to be significant in the original analysis do not reach significance under the alternative scenarios, in all cases with P < 0.07 (fig. 2). These results show a modest influence of the demographic scenario and suggest that our results will be largely robust regardless of the demographic model assumed, as long as the model is a realistic one.
Extreme Genes
An advantage of the gene-centric nature of the data set is that, rather than detecting long genomic regions containing several genes, we identify the specific gene under selection. This makes the interpretation of selective signatures considerably easier and more precise than other genome-scan methods. A total of 28 genes show signals of selection in AA and 45 genes show signals of selection in EA, with 13 showing consistent signatures in both populations (table 1). Although selective differences between the two populations cannot be discarded, the asymmetry is likely due to differences in power between the two populations because of their dissimilar neutral and genomic distributions. Assessing the false discovery rate is not trivial because the criterion to select extreme genes integrates information from two nonindependent tests. If the tests were independent, we would expect 12 extreme genes in each population just by chance (at the 5% significance level for each test). We observe an excess of 16 extreme genes for AA and 33 extreme genes for EA, indicating that there are real signatures of selection in the data.
Most genes with significant signals of selection in only one population show similar patterns in the second population (although not reaching statistical significance, table 1) and cannot be considered population specific. This is consistent with selection predating the relatively recent separation of the two populations, as expected with long-term balancing selection. Some genes, though, show unexpected population-specific patterns. Specifically, four genes show AA-only signatures (with P values >0.2 in EA) and nine genes show EA-only signatures (table 1). Those patterns likely result from recent demographic or selective population-specific factors contributing to the loss of the balanced equilibrium in one of the populations and may represent interesting cases of population-specific loss of an advantageous functional variant.
Whenever possible, results of large scans should be compared with examples of genes known to be undergoing balancing selection, which serve as internal positive controls. In the case of long-term balancing selection, this represents a challenge due to the scarcity of known examples. The best-characterized case in humans is the MHC, with several human leukocyte antigen (HLA) loci showing excess of polymorphism, complex haplotype structures, and trans-specific polymorphism (Hughes and Yeager 1998). Of the five HLA genes analyzed, the only one for which signatures of balancing selection have been previously reported is HLA-B (Hedrick et al. 1991; Sánchez-Mazas 2007), which shows signatures of selection in our data set. We also detect FUT2/Secretor factor (Se), an ABO-secretor gene considered an “honorary blood group” and associated with signatures of balancing selection in humans (Koda et al. 2000; Soejima et al. 2007; Ferrer-Admetlla et al. 2009) (supplementary table 2A, Supplementary Material online). Other historically proposed targets of balancing selection are either cases of recent selection (β-globin, CFTR, G6PD)—not targeted or detected by our method—or genes (like ABO) that show incomplete signatures of selection according to our strict criterion (see supplementary table 2B, Supplementary Material online). Overall, the comparison of extreme genes with previously reported targets confirms the detection of strong signatures of selection (like those in HLA-B and FUT2) and the specificity of the method to detect only genes under strong, long-term selection maintaining intermediate-frequency alleles.
An excess of heterozygotes (one of the signatures of present-day overdominance) has been reported for olfactory receptors (Alonso et al. 2008), but we find no evidence of increased selection in this functional category. The molecular function categories showing the strongest excess of low P values for the two tests and in the two populations are extracellular matrix, extracellular matrix structural protein, structural protein, intermediate filament, and serine protease inhibitor. The extracellular matrix comprise a large variety of proteins, including diverse structural molecules; high genetic variability in these proteins might contribute to the diversity and complexity of the matrix. We observe fewer signals in biological process categories, with no category showing consistent excess of low P values in both populations (supplementary table 3, Supplementary Material online).
Characteristics of Extreme Genes
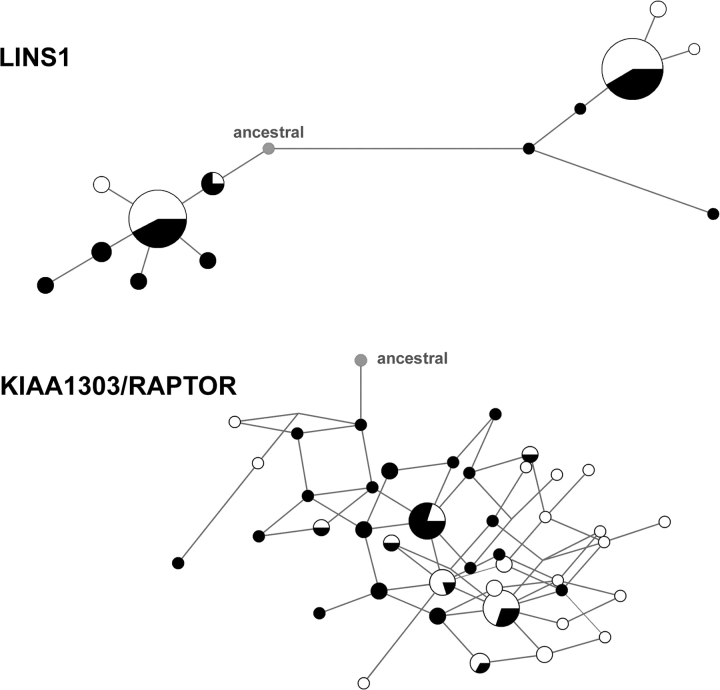
The genome-wide scale of the project allowed us to analyze the specific characteristics of the selection targets, rather than focusing on the particulars of one or two candidate genes. For example, balancing selection can alter the haplotype and LD structure of a gene, either by reducing the association between sites due to increased coalescent time and recombination (Charlesworth et al. 1997) or some types of epistasis (Navarro and Barton 2002), or by raising it due to positive epistasis between selected sites. Extreme genes show significantly higher LD (MWU P [AA] = 4.16 × 10−6, P [EA] = 6.01 × 10−10). They also show an excess of SNP pairs in significant LD (in both populations, 1-tailed permutation test P < 10−4). This is true even after correcting for the intrinsically higher SNP density (which may increase LD) or higher average allele frequency (which may increase the power to detect significance) of extreme genes (in all cases, one-tailed permutation test P < 10−4). Unfortunately, simultaneously controlling for these two factors is not feasible because the combination of high SNP density and allele frequency is an intrinsic trait of extreme genes. The elevated LD within genes cannot be explained by reduced recombination rate in extreme genes (data not shown), and the increased LD does not extend over the limits of the genes: The average LD (r2) in HapMap SNPs (CEU and YRI) for regions of 20 kb centered on every gene is not unusual in extreme genes (t-test P [AA] = 0.1497, P [EA] = 0.1604). Similar results were obtained for regions of 50 kb (t-test P [AA] = 0.9942, P [EA] = 0.3751). This confirms that the signal is specific to extreme genes and not to the genomic regions in which they reside, and that balancing selection may favor mostly specific haplotypes, rather than individual SNPs, in this set of genes. In any case, the pattern is by no means universal, with LD and haplotype structure varying substantially among genes, from genes with two distant and rarely recombining haplogroups to genes with pervasive signals of recombination and/or gene conversion (fig. 3).
FIG. 3.—
Network of the inferred haplotypes for LINS1 and KIAA1303 (also known as RAPTOR) genes. Circles represent haplotypes (size proportional to frequency), black for haplotypes in AA and light grey in EA. Length of the branches between haplotypes is proportional to the number of SNPs present in that branch. The ancestral haplotype corresponds to the ancestral allele for all SNPs, as inferred by comparison with chimpanzee. Reticulations denote unresolved paths due to recurrent mutation or recombination. In LINS1, manual inspection revealed three possible additional singleton reticulations (supplementary fig. 4, Supplementary Material online).
Nonhomologous gene conversion has a recognized influence in the high levels of variability of the MHC complex by introducing new variants from paralogous sequences. In the absence of selection, such variants, at low frequency, mimic the patterns of purifying selection rather than those of balancing selection. Still, nonhomologous gene conversion could not be completely discounted for 13 extreme genes (including HLA-B and FUT2), where more than one SNP could be mapped to a paralogous sequence on the same chromosome (table 1). Most of these SNPs are transitions, and therefore, independent mutations in the two copies can account for some of the cases. Nevertheless, our results suggest that gene conversion may be a mechanism for introducing variants to genes evolving under balancing selection outside the HLA complex.
Discussion
Here, we report the results of a systematic genome-wide scan of balancing selection that, in contrast to previous studies, reveals a number of candidate target of balancing selection in the human genome. Asthana et al. (2005) reported that transspecific polymorphism between humans and chimpanzees is rare, suggesting a limited role of long-term balancing selection in the two species. The power to detect events of transspecific polymorphism is small (Clark 1997; Wiuf et al. 2004) and still limited by the data, but those results suggest that transspecific patterns like those found in the MHC locus are most likely an exception. Focusing on variants recovered from genomic sequence reads, Bubb et al. (2006) also failed to detect convincing targets of long-term balancing selection in humans. They focused on large genomic regions with high SNP density and high LD. Although some targets of balancing selection fulfill those requirements (HLA is the most prominent example), neither high LD nor extension of the highly polymorphic region are necessary predictions of the action of balancing selection. In nonselfing species and in the absence of epistasis, the signal of balancing selection is expected to be broken by recombination and affect only narrow genomic regions (Charlesworth et al. 1997). Bubb et al. (2006) clearly established that the complex patterns seen in the MHC locus are unusual, possibly resulting from a combination of directional and balancing selection, recombination/gene conversion, and epistasis between distant sites (Hughes and Yeager 1998). But until now, it had remained unclear whether other, more typical cases of balancing selection exist in the human genome.
New genomic data sets allow us to tackle this question now. We have identified genes experiencing effects of balancing selection, showing that these cases do exist. The double signature of excess of polymorphism and intermediate-frequency alleles is difficult to reconcile with forces other than balancing selection, including purifying selection and positive selection, from new or standing variation (supplementary Discussion, Supplementary Material online). Because weak overdominance does not increase polymorphism (Williamson et al. 2004), selection must be strong to lead to the patterns that we observe here. Likewise, the genetic signals observed do not agree with the expected patterns produced by other possible causes of deep coalescence, like ancestral admixture, ancient population structure, or putative hybridization between ancestral humans and chimpanzees (supplementary Discussion, Supplementary Material online).
Other demographic factors are also an unlikely cause for the patterns observed. First, populations were analyzed separately, and population history (including admixture) was intrinsically accounted for in the statistical test. Second, demographic effects are expected to affect the whole genome, not a small number of genes, although some demographic models may increase the variance among genes. Third, we have shown that our results are largely robust to the demographic model used. Fourth, admixture is not expected to increase the coalescence time of the gene, and in humans, with little population stratification, population structure increases the proportion of low-frequency alleles, and not intermediate-frequency alleles, with increasing numbers of populations (Ptak and Przeworski 2002). This is the opposite pattern to balancing selection. Finally, to ensure that the use of potentially admixed American populations does not affect our results, we compared the frequency in our samples (AA and EA) and potentially nonadmixed HapMap populations (Yoruba and CEU) for SNPs present in both data sets. Only three genes (LRRN6A [also known as LINGO], LINS1, and TARBP1) had two or more SNPs with more intermediate frequency in this data set than in HapMap samples (allele frequency difference ≥0.2). This is a very small proportion of extreme genes, and some variance in allele frequencies between data sets is expected. So, even if the influence of potential admixture cannot be completely discarded for these three genes, it should not be a concern to the overall results.
An additional element that requires attention is gene duplication because inadvertent confounding of the sequences of two distinct copies of a gene would alter the patterns of variation. Nevertheless, this is an unlikely source of error in our analysis. Only old events (with fixed differences among copies) would produce false high-frequency variants; such events are likely well annotated in genome assemblies and would be detected by our strict bioinformatics pipeline, which was designed to remove such regions from the analysis (see Materials and Methods). To further discard the influence of copy number variation (CNV), we calculated the fraction of extreme genes overlapping CNVs according to Redon et al. (2006). Extreme genes do not have a greater likelihood to overlap CNVs than other genes in our data set (P value = 0.209), and only two genes (PCDHB16 and ZNF512b) are present in CNVs reported in more than one study. Identifying CNVs is an error-prone task and their annotation at the genome scale might still be incomplete, but this analysis suggests that duplications do not significantly impact our results.
The signature of balancing selection affects extremely narrow genomic regions, as predicted by theory (Hudson and Kaplan 1988; Charlesworth et al. 1997). Note, for example, that the signal in one gene does not extend to neighboring genes (supplementary fig. 3, Supplementary Material online). One unique large cluster of genes is evident, on chromosome 19 (a gene-rich chromosome), with extreme genes separated by many nonextreme genes, indicating that their signals are most likely independent. Only the signals of VARSL and CDSN should be interpreted with caution due to their close proximity to the HLA loci, and the double signature in KRT84 and KRT6B (adjacent in the genome) in AA could be caused by strong selection in one of the two genes or an intermediate region. The remarkably tight localization of signals confirms that only data sets with a high density of SNPs (i.e., resequencing data) will have the power to detect balancing selection.
The 60 candidate targets of balancing selection were identified after careful efforts to remove or account for possible confounding factors and using very stringent criteria. Still, confirmation of selective signatures detected by genome scans, such as this one, will best be performed on a gene-by-gene basis. Nevertheless, an inspection of genes in table 1 already reveals interesting patterns. For example, a disproportionate number of extreme genes are involved in immunology and response to pathogens. In addition to HLA-B, LRAP and LILRB4 are directly involved in MHC function; BTN1A1 is a member of the immunoglobulin superfamily; LRRN6A (LINGO1) contains an immunoglobulin domain; C20orf186 codes for the antimicrobial peptide RY2G5; CD200R1 is an important immune regulator; TARBP1 and TRIM22 are involved in HIV infection; and FUT2 determines blood group and its variants modulate susceptibility to Norwalk virus and HIV-1 infection (supplementary table 4, Supplementary Material online). This is expected if, as predicted based on the MHC loci, the maintenance of genetic diversity is selectively advantageous in response to pathogens (Hughes 2002). The signatures of balancing selection in a variety of immune genes illustrate the beneficial role of genetic variability in diverse steps of the immunological process (e.g., Ferrer-Admetlla et al. 2008).
A number of keratin genes (KRT14, KRT6B, KRT6C [KRT6E], KRT84) show signals of balancing selection, as does CDSN, a protein expressed specifically in corneocytes (keratinocyte-derived cells). Interestingly, other identified genes include those encoding a glucose solute carrier (SLC2A9) and three ion channels (CLCNKB, GRIN3A, and TRPV6). The pattern of variation in the gene encoding a prominent chloride channel, CFTR, has been proposed to reflect recent balancing selection in humans (Quinton 1994), consistent with reports suggesting improved survival of heterozygotes to certain infections (Gabriel et al. 1994). Like CFTR, the genes encoding other membrane channels may work as a gateway for entrance of pathogens into cells or may be important in controlling the response to infection, in addition to their primary role in cellular transport.
Many extreme genes are disease-causing or have association with disease (supplementary table 5, Supplementary Material online). Still, we find no overrepresentation of targets of balancing selection in OMIM. If SNPs in genes under balancing selection are associated with disease (and the disease is not the direct selective force), their common variants will most likely have a role in common, complex diseases, where identification of causal mutations is a challenge (Reich and Lander 2001). For example, immune-related genes, as well as those involved in inflammation (ADAMST7, NALP13, and PPP1R15A), are candidates of this class. The possibility remains that balancing selection has a modest influence on human disease, but we believe that these genes should be considered candidates for the genetic basis of common, complex diseases of unclear etiology, due to their excess of putatively functional common variants.
To our knowledge, this study provides the first unbiased set of candidate targets of balancing selection in humans. Our method was designed for the detection of a specific type of selection and, consequently, has little or no power to detect other classes of selection. We will not detect selection if the bouts are particularly recent or if the form of balancing selection yields no excess of intermediate-allele frequency at equilibrium. Likewise, we have reduced power to identify very short genes and no coverage in nongenic genomic regions. Because, in addition, our data set contains about one-third of the estimated number of genes in the genome, it is likely that we have identified only a portion of the genes that may have evolved under balancing selection in the human genome. The identification of such elements is important not only for understanding their evolutionary history but also for finding functional variants of potential phenotypic and medical relevance. In this respect, this study represents a step forward in the evolutionary annotation of the human genome.
Supplementary Material
Supplementary Methods, Discussion, tables 1–5, and figures 1–4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
The authors would like to thank Scott Williamson for his constant support and for always-insightful comments, discussions, and suggestions. We thank Sergio Castellano for helpful discussions and Kirk Lohmueller for useful suggestions and help with PolyPhen analysis. We thank John Sninsky (Celera Diagnostics) for his active role in the Applera Genome Initiative project, which generated the data for this study, and for stimulating discussion and support. This work was supported by the National Institutes of Health (grants HL072904 and GM065509 to A.M.A. and A.G.C.) This work was supported in part by the Intramural Program of the National Human Genome Research Institute, National Institutes of Health.
References
- Akey JM, Eberle MA, Rieder MJ, Carlson CS, Shriver MD, Nickerson DA, Kruglyak L. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2004;2:e286. doi: 10.1371/journal.pbio.0020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso S, López S, Izagirre N, de la Rúa C. Overdominance in the human genome and olfactory receptor activity. Mol Biol Evol. 2008;25:997–1001. doi: 10.1093/molbev/msn049. [DOI] [PubMed] [Google Scholar]
- Andrés AM, Clark AG, Shimmin L, Boerwinkle E, Sing CF, Hixson JE. Understanding the accuracy of statistical haplotype inference with sequence data of known phase. Genet Epidemiol. 2007;31:659–671. doi: 10.1002/gepi.20185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asthana S, Schmidt S, Sunyaev S. A limited role for balancing selection. Trends Genet. 2005;21:30–32. doi: 10.1016/j.tig.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Bamshad M, Wooding SP. Signatures of natural selection in the human genome. Nat Rev Genet. 2003;4:99–111. doi: 10.1038/nrg999. [DOI] [PubMed] [Google Scholar]
- Bamshad MJ, Mummidi S, Gonzalez E, et al. (11 co-authors) A strong signature of balancing selection in the 5’ cis-regulatory region of CCR5. Proc Natl Acad Sci USA. 2002;99:10539–10544. doi: 10.1073/pnas.162046399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Dress AWM. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol Phylogenet Evol. 1992;1:242–252. doi: 10.1016/1055-7903(92)90021-8. [DOI] [PubMed] [Google Scholar]
- Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40:340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- Barton NH, Etheridge AM. The effect of selection on genealogies. Genetics. 2004;166:1115–1131. doi: 10.1534/genetics.166.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum J, Ward RH, Conway DJ. Natural selection on the erythrocyte surface. Mol Biol Evol. 2002;19:223–229. doi: 10.1093/oxfordjournals.molbev.a004075. [DOI] [PubMed] [Google Scholar]
- Boyko AR, Williamson SH, Indap AR, et al. (14 co-authors) Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;45:e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb KL, Bovee D, Buckley D, et al. (12 co-authors) Scan of human genome reveals no new loci under ancient balancing selection. Genetics. 2006;173:2165–2177. doi: 10.1534/genetics.106.055715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, et al. (14 co-authors) Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Nordborg M, Charlesworth D. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet Res. 1997;70:155–174. doi: 10.1017/s0016672397002954. [DOI] [PubMed] [Google Scholar]
- Cheung J, Estivill X, Khaja R, MacDonald JR, Lau K, Tsui LC, Scherer SW. Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol. 2003;4:R25. doi: 10.1186/gb-2003-4-4-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Cho S, Huang ZY, Green DR, Smith DR, Zhang J. Evolution of the complementary sex-determination gene of honey bees: balancing selection and trans-species polymorphisms. Genome Res. 2006;16:1366–1375. doi: 10.1101/gr.4695306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG. Neutral behavior of shared polymorphism. Proc Natl Acad Sci USA. 1997;94:7730–7734. doi: 10.1073/pnas.94.15.7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Glanowski S, Nielsen R, et al. (17 co-authors) Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Cork JM, Purugganan MD. High-diversity genes in the Arabidopsis genome. Genetics. 2005;170:1897–1911. doi: 10.1534/genetics.104.036152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Admetlla A, Bosch E, Sikora M, et al. (11 co-authors) Balancing selection is the main force shaping the evolution of innate immunity genes. J Immunol. 2008;181:1315–1322. doi: 10.4049/jimmunol.181.2.1315. [DOI] [PubMed] [Google Scholar]
- Ferrer-Admetlla A, Sikora M, Laayouni H, Esteve A, Roubinet F, Blancher A, Calafell F, Bertranpetit J, Casals F. A natural history of FUT2 polymorphism in humans. Mol Biol Evol. 2009 doi: 10.1093/molbev/msp108. 26:1993-2003. [DOI] [PubMed] [Google Scholar]
- Gabriel SE, Brigman KN, Koller BH, Boucher RC, Stutts MJ. Cystic fibrosis heterozygote resistance to cholera toxin in the cystic fibrosis mouse model. Science. 1994;266:107–109. doi: 10.1126/science.7524148. [DOI] [PubMed] [Google Scholar]
- Gillespie JH. The causes of molecular evolution. Oxford: Oxford University Press; 1991. [Google Scholar]
- Hedrick PW, Whittam TS, Parham P. Heterozygosity at individual amino acid sites: extremely high levels for HLA-A and -B genes. Proc Natl Acad Sci USA. 1991;88:5897–5901. doi: 10.1073/pnas.88.13.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. The coalescent process in models with selection and recombination. Genetics. 1988;120:831–840. doi: 10.1093/genetics/120.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M. A test of neutral molecular evolution base on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. Natural selection and the diversification of vertebrate immune effectors. Immunol Rev. 2002;190:161–168. doi: 10.1034/j.1600-065x.2002.19012.x. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc Natl Acad Sci USA. 1989;86:958–962. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Yeager M. Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet. 1998;32:415–435. doi: 10.1146/annurev.genet.32.1.415. [DOI] [PubMed] [Google Scholar]
- Innan H. Modified Hudson-Kreitman-Aguade test and two-dimensional evaluation of neutrality tests. Genetics. 2006;173:1725–1733. doi: 10.1534/genetics.106.056242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Koda Y, Tachida H, Soejima M, Takenaka O, Kimura H. Ancient origin of the null allele se428. of the human ABO-secretor locus FUT2. J Mol Evol. 2000;50:243–248. doi: 10.1007/s002399910028. [DOI] [PubMed] [Google Scholar]
- Kroymann J, Mitchell-Olds T. Epistasis and balanced polymorphism influencing complex trait variation. Nature. 2005;435:95–98. doi: 10.1038/nature03480. [DOI] [PubMed] [Google Scholar]
- Lewontin RC, Hubby JL. A molecular approach to the study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics. 1966;54:595–609. doi: 10.1093/genetics/54.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Indap AR, Schmidt S, et al. (12 co-authors) Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Willis JH, Goldstein DB. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat Rev Genet. 2007;8:845–856. doi: 10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- Navarro A, Barton NH. The effects of multilocus balancing selection on neutral variability. Genetics. 2002;161:849–863. doi: 10.1093/genetics/161.2.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, et al. (13 co-authors) A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, Bustamante C. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Hubisz MJ, Torgerson D, et al. (13 co-authors) Darwinian and demographic forces affecting human protein coding genes. Genome Res. 2009;19:838–849. doi: 10.1101/gr.088336.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M. Structured coalescent processes on different time scales. Genetics. 1997;146:1501–1514. doi: 10.1093/genetics/146.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasvol G, Weatherall DJ, Wilson RJ. Cellular mechanism for the protective effect of haemoglobin S against P. falciparum malaria. Nature. 1978;274:701–703. doi: 10.1038/274701a0. [DOI] [PubMed] [Google Scholar]
- Pier GB, Grout M, Zaidi T, Meluleni G, Mueschenborn SS, Banting G, Ratcliff R, Evans MJ, Colledge WH. Salmonella typhi uses CFTR to enter intestinal epithelial cells. Nature. 1998;393:79–82. doi: 10.1038/30006. [DOI] [PubMed] [Google Scholar]
- Ptak SE, Przeworski M. Evidence for population growth in humans is confounded by fine-scale population structure. Trends Genet. 2002;18:559–563. doi: 10.1016/s0168-9525(02)02781-6. [DOI] [PubMed] [Google Scholar]
- Quinton PM. Human genetics. What is good about cystic fibrosis? Curr Biol. 1994;4:742–743. doi: 10.1016/s0960-9822(00)00166-4. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, et al. (43 co-authors) Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- Richman A. Evolution of balanced genetic polymorphism. Mol Ecol. 2000;9:1953–1963. doi: 10.1046/j.1365-294x.2000.01125.x. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES. Positive natural selection in the human genome. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- Sánchez-Mazas A. An apportionment of human HLA diversity. Tissue Antigens. 2007;69(Suppl 1):198–202. doi: 10.1111/j.1399-0039.2006.00802.x. [DOI] [PubMed] [Google Scholar]
- Soejima M, Pang H, Koda Y. Genetic variation of FUT2 in a Ghanaian population: identification of four novel mutations and inference of balancing selection. Ann Hematol. 2007;86:199–204. doi: 10.1007/s00277-006-0203-4. [DOI] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature. 1999;400:667–671. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W, 3rd, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- Takahata N, Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Shon AM, Ober C. Evidence of balancing selection at the HLA-G promoter region. Hum Mol Genet. 2005;14:3619–3628. [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Kodama G, Baldi P, Moyzis RK. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc Natl Acad Sci USA. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B. Genetic data analysis II. Sunderland (MA): Sinauer Associates; 1996. [Google Scholar]
- Williamson S, Fledel-Alon A, Bustamante CD. Population genetics of polymorphism and divergence for diploid selection models with arbitrary dominance. Genetics. 2004;168:463–475. doi: 10.1534/genetics.103.024745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SH, Hubisz MJ, Clark AG, Payseur BA, Bustamante CD, Nielsen R. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiuf C, Zhao K, Innan H, Nordborg M. The probability and chromosomal extent of trans-specific polymorphism. Genetics. 2004;168:2363–2372. doi: 10.1534/genetics.104.029488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S, Stone AC, Dunn DM, Mummidi S, Jorde LB, Weiss RK, Ahuja S, Bamshad MJ. Contrasting effects of natural selection on human and chimpanzee CC chemokine receptor 5. Am J Hum Genet. 2004;76:291–301. doi: 10.1086/427927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf LI, Cranston WI, Goodwin BL. Genetics of phenylketonuria. Heterozygosity for phenylketonuria. Nature. 1967;213:882–883. doi: 10.1038/213882a0. [DOI] [PubMed] [Google Scholar]
- Wright S. The distribution of self-sterility alleles in populations. Genetics. 1939;24:538–552. doi: 10.1093/genetics/24.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Nei M. Population dynamics of sex-determining alleles in honey bees and self-incompatibility alleles in plants. Genetics. 1979;91:609–626. doi: 10.1093/genetics/91.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.