Abstract
The 67LR (67 kDa laminin receptor) enables cells to interact with components of the extracellular matrix. The molecule is derived from the 37LRP (37 kDa laminin receptor precursor); however, the precise molecular mechanism of this conversion is unknown. Recombinant 37LRP, expressed in and purified from Escherichia coli, bound to human laminin in a SPR (surface plasmon resonance) experiment. 67LR isolated from human breast-cancer-derived cells in culture was also shown to bind to laminin by SPR. However, the kinetics of association are qualitatively different. 37LRP, but not 67LR, binds to heparan sulfate. The binding of 37LRP to heparan sulfate did not affect the interaction of 37LRP with laminin. In contrast, heparan sulfate reduces the extent of binding of laminin to 67LR. Taken together, these results show that 37LRP has some of the biological activities of 67LR, even prior to the conversion event. However, the conversion affects the sites of interaction with both laminin and heparan sulfate.
Keywords: heparan sulfate, 37 kDa laminin receptor precursor (37LRP), 67 kDa laminin receptor (67LR), laminin receptor, ribosomal protein SA, surface plasmon resonance (SPR)
Abbreviations: BCIP/NBT, 5-bromo-4-chloroindol-3-yl phosphate/Nitro Blue Tetrazolium; EDC, N-ethyl-N′-(3-dimethylaminopropyl)carbodi-imide; IPTG, isopropyl β-D-thiogalactoside; LB, Luria–Bertani; 37LRP, 37 kDa laminin receptor precursor; 67LR, 67 kDa laminin receptor; NHS, N-hydroxysuccinimide; RU, resonance unit(s); SPR, surface plasmon resonance; TBS, Tris-buffered saline; TBST, TBS, pH 7.4, with 0.1% (v/v) Tween 20
INTRODUCTION
In multicellular organisms, cells must be organized into tissues, organs and other structures. In part, this organization is achieved through interactions between cells and extracellular matrices. Thus it follows that there must be receptors on the surfaces of cells which recognize molecules in the extracellular matrix, bind to that matrix and transduce signals to the interior of the cell. One such receptor is the 67LR (67 kDa laminin receptor) [1]. This molecule has attracted considerable interest since its discovery in 1983 [2–5], not least because of its well documented involvement in the metastasis of cancerous cells [6,7].
The precise molecular nature of this receptor is a mystery. It is now well established that it arises from a smaller polypeptide, the 37LRP (37 kDa laminin precursor). However, the biochemical changes which accompany this alteration in molecular mass are not known and there are a number of conflicting hypotheses. These include homodimerization, heterodimerization with an unidentified partner or fatty acylation [8–10]. Interestingly, 37LRP not only acts as a precursor for 67LR, but also functions as a ribosomal subunit (known as ribosomal protein SA or p40) [11–15] and as a nuclear protein where it interacts with histones [16,17]. The ribosomal functions are conserved across all kingdoms of life, with homologous proteins being found in archea, eubacteria, fungi, plants and animals [1]. However, the laminin receptor functions (and, presumably, the accompanying alterations to the molecule) are unique to multicellular animals. In addition to acting as a laminin receptor, 67LR acts as a receptor for the internalization of some viruses, prions and bacteria [18–21].
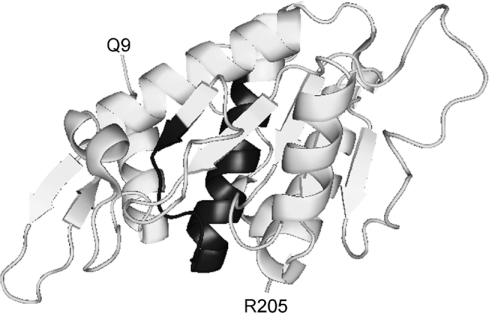
The precise contact sites between 67LR and laminin have not been mapped. However, it is believed that the interaction involves a nonapeptide from the β1 chain of laminin (CDPGYIGSR) and residues 205–229 of human 67LR [22–24]. Additional contacts may be provided by residues 161–180 of 67LR, the so-called peptide G [25]. Residues 205–229 are predicted to adopt a mainly helical secondary structure, presenting two faces: a highly charged polar heparan-sulfate-binding surface and a more hydrophobic surface which interacts with laminin [22]. The functional consequence of this is that laminin and heparan sulfate binding are not mutually exclusive events at this site. The situation is different with peptide G. Here, interaction with laminin and the related sulfated glycoprotein heparin appear to be mutually exclusive [26]. Taken together, this suggests that heparan sulfate (or heparin) binding to 67LR may reduce the affinity of interactions between 67LR and laminin, but not abolish them altogether. Recently, the structure of a fragment of 37LRP was solved [27]. The structure shows a high degree of similarity to ribosomal protein SA from prokaryotes and lower eukaryotes [28–30]. However, this structure (Figure 1) lacks the C-terminal region (which is not present in lower organisms) and thus the bulk of the main putative laminin-binding peptide. Peptide G forms part of a β-strand, a loop and an α-helix, much of which is buried in the interior of the molecule. Thus it has been postulated that considerable conformational changes are required to enable laminin binding [1].
Figure 1. Structure of human 37LRP.
One of the proposed laminin-binding sites, peptide G (residues 161–180) is shown in black. The structure begins at residue 9 (Q9; single-letter amino-acid code) and finishes at residue 205 (R205) (both indicated on the Figure). Thus the structure lacks almost all the C-terminal laminin-binding site, which starts at residue 205. The Figure was produced using the co-ordinates from Protein Data Bank (PDB) file 3BCH [27] and PyMOL (www.pymol.org).
The essential functions of 67LR combined with its key role in a number of different disease processes has resulted in considerable efforts to understand its structure and function. However, these have been hampered by lack of understanding of how 37LRP and 67LR differ. In the present study we expressed 37LRP in the bacterium Escherichia coli, taking advantage of the lack of post-translational modifications in the organism to ensure that we isolated the unmodified precursor, and compared the ability of this protein to interact with laminin of that of 67LR which was shed from human breast-cancer-derived T47D cells in culture.
MATERIALS AND METHODS
Expression and purification of recombinant 37LRP in E. coli
A cDNA IMAGE clone [31] (Clone ID 4820432; Geneservice, Cambridge, U.K.) containing the coding sequence for human 37LRP was used as a PCR template. The amplified product was inserted into pET-46 Ek/LIC (Merck, Nottingham, U.K.) using the ligation-independent cloning protocol according to the manufacturer's instructions. Correctly assembled expression plasmids were identified by restriction digestion and DNA sequencing (MWG Biotech, Ebersford, Germany) of the entire coding sequence.
The expression vector was transformed into E. coli HMS174(DE3) cells (Merck). Single colonies were used to inoculate 5-ml cultures in LB (Luria–Bertani) broth (Miller) supplemented with 100 μg·ml−1 ampicillin. These cultures were grown, with shaking at 37°C overnight, and then diluted into 1 l of LB broth (supplemented with 100 μg·ml−1 ampicillin). This culture was grown, with shaking at 37°C, until the D600 reached between 0.6 and 1.0 (typically 3–4 h), then induced with 1 mM IPTG (isopropyl β-D-thiogalactoside) and grown for a further 2 h. At this point, the cells were harvested by centrifugation at 4200 g, resuspended in approx. 20 ml of cell resuspension buffer [50 mM Hepes-OH, pH 7.5, 150 mM sodium chloride and 10% (v/v) glycerol] and stored, frozen at −80°C, until required.
These cell suspensions were thawed and guanidine hydrochloride was added to a final concentration of 6 M. The cells were disrupted by sonication (three times 30 s pulses at 100 W with 30–60 s gaps between pulses for cooling). Cell debris was then removed by centrifugation (24000 g for 15 min at 4°C) and the supernatant immediately applied to a 1 ml nickel–agarose column (His-Select; Sigma, Poole, Dorset, U.K.), which had been previously equilibrated in buffer A [50 mM Hepes-OH, pH 7.5, 500 mM sodium chloride, 6 M guanidine hydrochloride and 10% (v/v) glycerol]. The supernatant was allowed to flow through by gravity and the column was then washed with 20 ml of buffer A, followed by 20 ml of buffer B (buffer A without guanidine hydrochloride). Elution was achieved by application of three 2 ml aliquots of buffer C (buffer B supplemented with 250 mM imidazole). These aliquots were collected as separate fractions. Protein containing fractions were identified by SDS/PAGE (10% gels) in Tris/glycine buffer (pH 8.3), dialysed against cell resuspension buffer supplemented with 2 mM DTT (dithiothreitol) and frozen in small aliquots at −80°C.
Purification of native 67LR
67LR was obtained from T47D human breast carcinoma cells (European Collection of Cell Cultures, Porton Down, Salisbury, U.K.). This cell line sheds the receptor into the medium on rotational incubation [32]. The cells were cultured in high-glucose DMEM (Dulbecco's modified Eagle's medium), with glutamine and sodium pyruvate (PAA, Pasching, Austria). The cultures were harvested (at 70–75% confluence) by washing the monolayer with calcium-free saline and incubating the monolayer for 15 min with cell dissociation fluid (Sigma). The cells were then centrifuged at 800 g for 10 min. The cell pellet (approx 1.5×107 cells) was resuspended in 500 μl of ice-cold PBS and incubated with rotation at 37°C for 2 h. The crude preparation of shed receptor was obtained by centrifuging the cells at 16000 g at 4°C for 10 min. The supernatant was aliquoted and stored at −80°C. Protein concentration was estimated using the BCA (bicinchoninic acid) protein assay kit (Pierce, Rockford, IL, U.S.A.) in microtitre plates.
Western blotting
37LRP and 67LR were run on 10–20% pre-cast Tricine gels (Invitrogen, Paisley, Renfrewshire, U.K.) under denaturing conditions using TCEP [Tris(2-carboxyethyl)phosphine] (Sigma) as a reducing agent at a final concentration of 5 mM. Following electrophoresis (at 200 V), transfer of the proteins on to a nitrocellulose membrane was carried out using an XCell II MiniBlot apparatus (Novex) at 30 V for 1 h. The membrane was then washed in TBST [TBS (Tris-buffered saline), pH 7.4, with 0.1% (v/v) Tween 20]. The membrane was blocked with 2% (w/v) non-fat dried milk in TBST for 30 min. The membrane was washed and the primary antibody was diluted 1:1000 in 2% (w/v) non-fat dried milk in TBST overnight. The two antibodies used were Pro-20-Ala (a polyclonal antibody raised against a 20 residue peptide from the C-terminus of 67LR) and Ab183 (67LR N-terminal-specific antibody) [33,34]. The membranes were washed with TBST and secondary anti-rabbit alkaline-phosphatase-conjugated antibody (Sigma) was applied to the membrane in a 1:4000 dilution for 1 h. The membrane was washed with TBS and a BCIP/NBT (5-bromo-4-chloroindol-3-yl phosphate/Nitro Blue Tetrazolium) substrate was prepared by dissolving one tablet of Fast BCIP/NBT (Sigma) in 10 ml of de-ionized water and adding it to the membranes for 10 min. Colour development was stopped using de-ionized water.
SPR (surface plasmon resonance)
All experiments were carried out using a BIAcore 3000 biosensor (GE Healthcare, Uppsala, Sweden). Recombinant 37LRP and native 67LR were immobilized using NHS/EDC [N-hydroxysuccinimide/N-ethyl-N′-(3-dimethylaminopropyl)carbodi-imide] coupling, followed by blocking with ethanolamine [35,36]. Two different sets of immobilizations were carried out using 67LR and 37LRP. Both proteins were concentrated in 10 mM acetate buffer (pH 4.0) using YM-3 membrane filters (Microcon). All solutions (made ‘in house’) were filtered before use. Activation was done using 150 μl of 100 mM NHS/390 mM EDC for 30 min at 5 μl·min−1; immobilization was carried out with a 50 μl injection of the desired protein for 10 min at 5 μl·min−1, followed by a 30 min blocking step with 150 μl of 1 M ethanolamine at 5 μl·min−1. The 37LRP was immobilized on lane 2 and the 67LR was immobilized on lane 4 of the same chip. Lanes 1 and 3 served as control lanes, so that any non-specific binding that occurred with the cross-linkers, rather than the protein, could be subtracted from the final RU (resonance units) in the experimental lane. These lanes were subjected to the same NHS/EDC activation followed by blocking with ethanolamine as the experimental lanes. Data were analysed using BIAevaluation software (GE Healthcare), applying the 1:1 Langmuir-binding model, according to the manufacturer's instructions.
Before the start of any binding assay, a regeneration step was carried out using NaOH (5–50 mM, pH 10.0–12.6) to remove any bound proteins and to obtain a stable baseline. The assays were all carried out with a final ligand injection volume of 50 μl and a dissociation time of 300 s, using the KINJECT program (GE Healthcare). Assays were carried out using mouse and human laminin (Sigma), heparan sulfate (Sigma) and the laminin nonapeptide (Peptide Synthesis Service, Queen's University, Belfast, U.K.). All ligands were dissolved in 10 mM Hepes-OH, pH 7.4, 150 mM NaCl, 3 mM EDTA and 0.005% (v/v) Tween 20 (HBS-EP; GE Healthcare). Data were visualized and analysed using BIAevaluation software (GE Healthcare).
RESULTS
Soluble recombinant 37LRP can be purified from E. coli
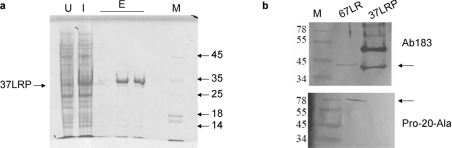
Initial attempts to purify recombinant 37LRP under native conditions using protocols similar to those employed for human S100B expressed using the same vector [37] were not successful. Examination of SDS/PAGE gels showed that an additional protein of approx. 35 kDa was present in E. coli whole-cell extracts following induction with IPTG, but that this protein was not present in the soluble extract following sonication and centrifugation (results not shown). This suggested that, although the protein was expressed, the molecules formed insoluble aggregates in the E. coli cytoplasm. To overcome this problem, the cells were sonicated and applied to the nickel-affinity column in the presence of the strong denaturing agent guanidine hydrochloride. Renaturation was achieved on the column by washing the bound protein with buffer lacking the denaturing agent, and soluble protein could be eluted by the addition of imidazole to the column buffer (Figure 2a). Typical yields of soluble 37LRP were 2 mg per litre of culture. This recombinant protein could be recognized in Western blots by the antibody Ab183, but not the Pro-20-Ala antibody, which only recognized mature 67LR (Figure 2b).
Figure 2. Expression and purification of recombinant 37LRP.
(a) SDS/PAGE (Tris/glycine, pH 8.3, buffer) 10% gel stained with Coomassie Blue showing cell extract prior to induction (U), cell extract 2 h post-induction (I), and samples from three 2 ml fractions following elution with 250 mM imidazole (E). The purified, soluble 37LRP appeared largely in the second and third elution fractions and is indicated by an arrow. (b) 67LR and 37LRP were resolved on 10–20% Tricine-gradient gels and Western blotted on to nitrocellulose. The antibody Pro-20-Ala only recognizes 67LR, whereas the antibody Ab183 recognizes 37LRP. In the 67LR fraction, it recognizes a protein which is slightly larger than 37LRP, which may be a partially processed form of 37LRP or a non-specifically bound contaminant. In addition, a number of unidentified contaminating proteins in the 37LRP fraction are also recognized. Note that different gel and buffer systems were used in the SDS/PAGE and Western blotting, resulting in slight differences in the mobilities of the proteins. Arrows indicate the bands corresponding to 37LRP and 67LR. The sizes (in kDa) of the molecular-mass markers (M) are shown.
Human 37LRP and 67LR both interact with laminin
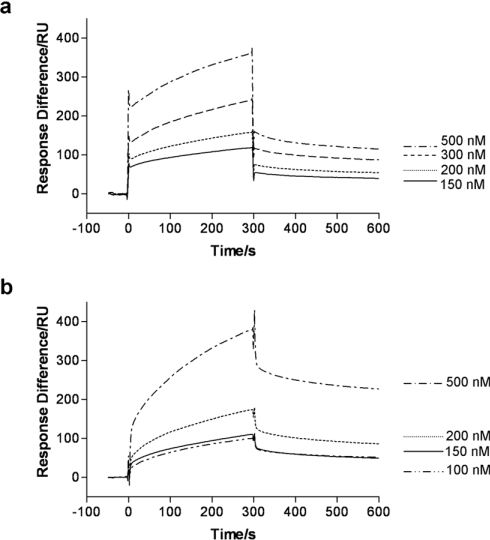
The high level of protein immobilized on the sensor chip combined with the large molecular mass of laminin (approx. 900 kDa [38]) mean that mass transport is likely to be a strong feature in the interaction kinetics, as measured by SPR. Consequently, the results can be regarded, at best, as semi-quantitative. Nevertheless, interactions between human laminin and both 37LRP and 67LR could be detected by SPR measurements (Figure 3). Estimation of the dissociation constants (Kd) for the 37LRP–laminin and 67LR–laminin interactions yielded similar values (410 nM and 320 nM respectively), suggesting similar overall affinities. Qualitatively the two interaction curves look different, with 67LR exhibiting at least two phases of association: an initial rapid phase which gives way to a slower one. In contrast, 37LRP appears to have a single phase of association which is similar to the second, slower phase observed with 67LR. Mouse laminin also interacted with both 37LRP and 67LR, albeit to a lesser extent (results not shown).
Figure 3. An interaction between recombinant human 37LRP and native human 67LR with laminin can be demonstrated by SPR.
In (a) 37LRP was immobilized on to a sensor chip surface, and laminin at various concentrations (as indicated) was passed over the surface. The graphs represent the difference in RU between a lane on the sensor chip in which 37LRP has been immobilized and a control lane in which no protein has been immobilized. In (b) the difference between a lane in which 67LR had been immobilized and the control is shown. In both (a) and (b), the data were analysed (using BIAevaluation software) to derive dissociation constants (Kd) from the ratio of the dissociation and association rate constants. The Kd for the 37LRP–laminin interaction was estimated to be 410 nM and the value for the 67LR–laminin interaction to be 320 nM. Note that these values should be treated with caution, given likely mass-transport complications and the complex nature of the 67LR–laminin interaction.
37LRP interacts with heparan sulfate, but 67LR does not
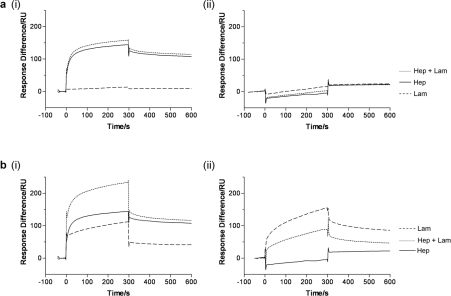
When heparan sulfate (0.2 mg·ml−1) was passed over immobilized 37LRP, an interaction was clearly detected (Figure 4a, panel i). Binding to the laminin nonapeptide could not be detected by SPR (but it should be noted that the response change to this much smaller molecule would be reduced compared with those observed with laminin). This peptide caused little or no change to the binding of heparan sulfate. Interestingly, no interaction between 67LR and heparan sulfate could be detected, nor could any interaction be detected with the laminin nonapeptide or a mixture of heparan and nonapeptide (Figure 4a, panel ii). The binding of laminin and heparan sulfate to 37LRP were not mutually exclusive; the responses observed with mixtures of these molecules were essentially the sum of responses seen with the individual molecules at the same concentrations (Figure 4b, panel i). This is not the case with 67LR, where heparan sulfate reduces the binding of laminin (Figure 4b, panel ii).
Figure 4. Interactions of 37LRP and 67LR with heparan sulfate.
(a, panel i) An interaction between recombinant human 37LRP (immobilized) and heparan sulfate (Hep, continuous line, soluble phase, 0.2 mg·ml−1) can be demonstrated by SPR. In contrast, no interaction could be detected between 37LRP and the laminin nonapeptide (Lam, dashed line, soluble phase, 100 μM), nor does this peptide greatly affect the heparan–37LRP interaction (Hep+Lam, dotted line). (a, panel ii) No interaction could be detected between 67LR (immobilized) and heparan sulfate (soluble phase, 0.2 mg·ml−1), laminin nonapeptide (soluble phase, 100 μM) or a mixture of the two molecules. (b, panel i) Heparan and laminin interactions with 37LRP are not mutually exclusive. The results observed (Hep+Lam, dotted line) with a mixture of heparan sulfate (soluble phase, 0.2 mg·ml−1) and mouse laminin (soluble phase, 250 nM) are essentially the sum of the responses observed with heparan sulfate (Hep, continuous line) and mouse laminin (Lam, dashed line) individually at the same concentrations. Similar results were observed with human laminin (results not shown). (b, panel ii) Heparan sulfate (soluble phase, 0.2 mg·ml−1) reduces the extent of interaction between mouse laminin (soluble phase, 200 nM) and 67LR (immobilized).
DISCUSSION
It has been demonstrated previously that intact 37LRP is insoluble upon expression in bacteria [39]. Two solutions have been pursued to address this problem. In one, a complex purification procedure involving the isolation of exclusion bodies and their solubilization in urea resulted in purified soluble 37LRP [39]. In the second approach, used to obtain the structure of 37LRP, a truncated form of the protein was expressed, which proved to be soluble. On purification, this protein could be crystallized [27]. In the present study, we demonstrated a simpler and faster method for obtaining purified soluble full-length 37LRP. Our method couples total denaturation of the E. coli proteins, extraction of the His-tagged 37LRP by affinity chromatography and renaturation of protein on the column prior to elution. The protein can be purified from bacterial cells in less than 1 day and yields material with biological activity, as shown by its interactions with an antibody in Western blotting and with laminin and heparan sulfate in SPR experiments.
This interaction between 37LRP and these ligands is interesting, as it infers that the processing of 37LRP to 67LR is not required for the protein to interact with them. It has been speculated that the C-terminal region of 37LRP/67LR (which is not present in the prokaryotic and lower eukaryotic ribosomal proteins) is at least partly responsible for interaction with these ligands [1]. Our observations are consistent with that hypothesis. Although we were able to detect interactions between laminin and both 37LRP and 67LR, the qualitative differences between the binding curves suggests differences in the molecular mechanisms of interaction. These differences may arise due to intrinsic differences between the two molecules. Indeed, if 67LR results from dimerization [9], there would be two laminin-binding sites, possibly with different affinities. Alternatively, it is possible that heterogeneity of the 67LR preparation (which, potentially, includes some unmodified 37LRP and intermediate stages between the two forms) gives rise to the multiphasic nature of the association kinetics. Thus it seems likely that the modifications resulting in maturation of 37LRP to 67LR directly, or indirectly, affect the sites of interaction with laminin.
That both heparan sulfate and laminin can interact with 37LRP simultaneously suggest that the two molecules bind at different sites and that there is limited (or no) allosteric communication between these sites. Since both laminin and heparan sulfate are extracellular molecules, we assume that 37LRP does not interact with these ligands in vivo. In these experiments, we were unable to detect interactions between 67LR and heparan sulfate, suggesting that processing of 37LRP abolishes this activity. It should be noted that immobilized heparin is often used to purify nucleic-acid-binding proteins. Therefore, it is possible that the interaction between 37LRP and heparan sulfate may reflect the welldocumented interactions of the molecule with nucleic acids [40]. Furthermore, heparan sulfate appears to inhibit the 67LR–laminin interaction. It has been shown that the related polysaccharide, heparin, also inhibits 67LR–laminin interactions, and it was suggested that this is caused by the polysaccharide competing for the 67LR-binding site on laminin [26]. A similar mechanism may be occurring here. If it is, then 37LRP may interact, at least in part, at a separate site on laminin, which is not affected by heparan sulfate competition.
Overall, the results of the present study demonstrate that 37LRP has at least some of the biological activities of 67LR, even prior to processing. However, the processing events modify these biological activities. Thus they also further underline the need to understand this processing event at the molecular level.
ACKNOWLEDGEMENTS
We thank Emma Chambers who completed some of preliminary work on the construction of the 37LRP expression vector as part of her final year BSc project.
FUNDING
This research received no specific grants from any funding agency in the public, commercial or not-for-profit sectors.
References
- 1.Nelson J., McFerran N. V., Pivato G., Chambers E., Doherty C., Steele D., Timson D. J. The 67 kDa laminin receptor: structure, function and role in disease. Biosci. Rep. 2008;28:33–48. doi: 10.1042/BSR20070004. [DOI] [PubMed] [Google Scholar]
- 2.Rao N. C., Barsky S. H., Terranova V. P., Liotta L. A. Isolation of a tumor cell laminin receptor. Biochem. Biophys. Res. Commun. 1983;111:804–808. doi: 10.1016/0006-291x(83)91370-0. [DOI] [PubMed] [Google Scholar]
- 3.Lesot H., Kuhl U., Mark K. V. Isolation of a laminin-binding protein from muscle cell membranes. EMBO J. 1983;2:861–865. doi: 10.1002/j.1460-2075.1983.tb01514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terranova V. P., Rao C. N., Kalebic T., Margulies I. M., Liotta L. A. Laminin receptor on human breast carcinoma cells. Proc. Natl. Acad. Sci. U.S.A. 1983;80:444–448. doi: 10.1073/pnas.80.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malinoff H. L., Wicha M. S. Isolation of a cell surface receptor protein for laminin from murine fibrosarcoma cells. J. Cell Biol. 1983;96:1475–1479. doi: 10.1083/jcb.96.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wewer U. M., Taraboletti G., Sobel M. E., Albrechtsen R., Liotta L. A. Role of laminin receptor in tumor cell migration. Cancer Res. 1987;47:5691–5698. [PubMed] [Google Scholar]
- 7.Wewer U. M., Liotta L. A., Jaye M., Ricca G. A., Drohan W. N., Claysmith A. P., Rao C. N., Wirth P., Coligan J. E., Albrechtsen R. Altered levels of laminin receptor mRNA in various human carcinoma cells that have different abilities to bind laminin. Proc. Natl. Acad. Sci. U.S.A. 1986;83:7137–7141. doi: 10.1073/pnas.83.19.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buto S., Tagliabue E., Ardini E., Magnifico A., Ghirelli C., van den Brule F., Castronovo V., Colnaghi M. I., Sobel M. E., Menard S. Formation of the 67-kDa laminin receptor by acylation of the precursor. J. Cell. Biochem. 1998;69:244–251. doi: 10.1002/(sici)1097-4644(19980601)69:3<244::aid-jcb2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 9.Landowski T. H., Dratz E. A., Starkey J. R. Studies of the structure of the metastasis-associated 67 kDa laminin binding protein: fatty acid acylation and evidence supporting dimerization of the 32 kDa gene product to form the mature protein. Biochemistry. 1995;34:11276–11287. doi: 10.1021/bi00035a037. [DOI] [PubMed] [Google Scholar]
- 10.Rao C. N., Castronovo V., Schmitt M. C., Wewer U. M., Claysmith A. P., Liotta L. A., Sobel M. E. Evidence for a precursor of the high-affinity metastasis-associated murine laminin receptor. Biochemistry. 1989;28:7476–7486. doi: 10.1021/bi00444a047. [DOI] [PubMed] [Google Scholar]
- 11.Ardini E., Pesole G., Tagliabue E., Magnifico A., Castronovo V., Sobel M. E., Colnaghi M. I., Menard S. The 67-kDa laminin receptor originated from a ribosomal protein that acquired a dual function during evolution. Mol. Biol. Evol. 1998;15:1017–1025. doi: 10.1093/oxfordjournals.molbev.a026000. [DOI] [PubMed] [Google Scholar]
- 12.Montero M., Marcilla A., Sentandreu R., Valentin E. A Candida albicans 37 kDa polypeptide with homology to the laminin receptor is a component of the translational machinery. Microbiology. 1998;144:839–847. doi: 10.1099/00221287-144-4-839. [DOI] [PubMed] [Google Scholar]
- 13.Rosenthal E. T., Wordeman L. A protein similar to the 67 kDa laminin binding protein and p40 is probably a component of the translational machinery in Urechis caupo oocytes and embryos. J. Cell Sci. 1995;108:245–256. doi: 10.1242/jcs.108.1.245. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Hernandez M., Davies E., Staswick P. E. Arabidopsis p40 homologue. A novel acidic protein associated with the 40 S subunit of ribosomes. J. Biol. Chem. 1994;269:20744–20749. [PubMed] [Google Scholar]
- 15.Davis S. C., Tzagoloff A., Ellis S. R. Characterization of a yeast mitochondrial ribosomal protein structurally related to the mammalian 68-kDa high affinity laminin receptor. J. Biol. Chem. 1992;267:5508–5514. [PubMed] [Google Scholar]
- 16.Kinoshita K., Kaneda Y., Sato M., Saeki Y., Wataya-Kaneda M., Hoffmann A. LBP–p40 binds DNA tightly through associations with histones H2A, H2B, and H4. Biochem. Biophys. Res. Commun. 1998;253:277–282. doi: 10.1006/bbrc.1998.9699. [DOI] [PubMed] [Google Scholar]
- 17.Sato M., Kinoshita K., Kaneda Y., Saeki Y., Iwamatsu A., Tanaka K. Analysis of nuclear localization of laminin binding protein precursor p40 (LBP/p40) Biochem. Biophys. Res. Commun. 1996;229:896–901. doi: 10.1006/bbrc.1996.1899. [DOI] [PubMed] [Google Scholar]
- 18.Kim K. J., Chung J. W., Kim K. S. 67-kDa laminin receptor promotes internalization of cytotoxic necrotizing factor 1-expressing Escherichia coli K1 into human brain microvascular endothelial cells. J. Biol. Chem. 2005;280:1360–1368. doi: 10.1074/jbc.M410176200. [DOI] [PubMed] [Google Scholar]
- 19.Hundt C., Peyrin J. M., Haik S., Gauczynski S., Leucht C., Rieger R., Riley M. L., Deslys J. P., Dormont D., Lasmezas C. I., Weiss S. Identification of interaction domains of the prion protein with its 37-kDa/67-kDa laminin receptor. EMBO J. 2001;20:5876–5886. doi: 10.1093/emboj/20.21.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gauczynski S., Peyrin J. M., Haik S., Leucht C., Hundt C., Rieger R., Krasemann S., Deslys J. P., Dormont D., Lasmezas C. I., Weiss S. The 37-kDa/67-kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein. EMBO J. 2001;20:5863–5875. doi: 10.1093/emboj/20.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K. S., Kuhn R. J., Strauss E. G., Ou S., Strauss J. H. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J. Virol. 1992;66:4992–5001. doi: 10.1128/jvi.66.8.4992-5001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kazmin D. A., Hoyt T. R., Taubner L., Teintze M., Starkey J. R. Phage display mapping for peptide 11 sensitive sequences binding to laminin-1. J. Mol. Biol. 2000;298:431–445. doi: 10.1006/jmbi.2000.3680. [DOI] [PubMed] [Google Scholar]
- 23.Starkey J. R., Uthayakumar S., Berglund D. L. Cell surface and substrate distribution of the 67-kDa laminin-binding protein determined by using a ligand photoaffinity probe. Cytometry. 1999;35:37–47. doi: 10.1002/(sici)1097-0320(19990101)35:1<37::aid-cyto6>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 24.Landowski T. H., Uthayakumar S., Starkey J. R. Control pathways of the 67 kDa laminin binding protein: surface expression and activity of a new ligand binding domain. Clin. Exp. Metastasis. 1995;13:357–372. doi: 10.1007/BF00121912. [DOI] [PubMed] [Google Scholar]
- 25.Castronovo V., Taraboletti G., Sobel M. E. Functional domains of the 67-kDa laminin receptor precursor. J. Biol. Chem. 1991;266:20440–20446. [PubMed] [Google Scholar]
- 26.Guo N. H., Krutzsch H. C., Vogel T., Roberts D. D. Interactions of a laminin-binding peptide from a 33-kDa protein related to the 67-kDa laminin receptor with laminin and melanoma cells are heparin-dependent. J. Biol. Chem. 1992;267:17743–17747. [PubMed] [Google Scholar]
- 27.Jamieson K. V., Wu J., Hubbard S. R., Meruelo D. Crystal structure of the human laminin receptor precursor. J. Biol. Chem. 2008;283:3002–3005. doi: 10.1074/jbc.C700206200. [DOI] [PubMed] [Google Scholar]
- 28.Badger J., Sauder J. M., Adams J. M., Antonysamy S., Bain K., Bergseid M. G., Buchanan S. G., Buchanan M. D., Batiyenko Y., Christopher J. A., et al. Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins. 2005;60:787–796. doi: 10.1002/prot.20541. [DOI] [PubMed] [Google Scholar]
- 29.Spahn C. M., Gomez-Lorenzo M. G., Grassucci R. A., Jorgensen R., Andersen G. R., Beckmann R., Penczek P. A., Ballesta J. P., Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yusupov M. M., Yusupova G. Z., Baucom A., Lieberman K., Earnest T. N., Cate J. H., Noller H. F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 31.Lennon G., Auffray C., Polymeropoulos M., Soares M. B. The I.M.A.G.E. consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 32.Keydar I., Chen L., Karby S., Weiss F. R., Delarea J., Radu M., Chaitcik S., Brenner H. J. Establishment and characterization of a cell line of human breast carcinoma origin. Eur. J. Cancer. 1979;15:659–670. doi: 10.1016/0014-2964(79)90139-7. [DOI] [PubMed] [Google Scholar]
- 33.Nelson J., Scott W. N., Allen W. E., Wilson D. J., Harriott P., McFerran N. V., Walker B. Murine epidermal growth factor peptide (33–42) binds to a YIGSR-specific laminin receptor on both tumor and endothelial cells. J. Biol. Chem. 1996;271:26179–26186. doi: 10.1074/jbc.271.42.26179. [DOI] [PubMed] [Google Scholar]
- 34.Donaldson E. A., McKenna D. J., McMullen C. B., Scott W. N., Stitt A. W., Nelson J. The expression of membraneassociated 67-kDa laminin receptor (67LR) is modulated in vitro by cell-contact inhibition. Mol. Cell Biol. Res. Commun. 2000;3:53–59. doi: 10.1006/mcbr.2000.0191. [DOI] [PubMed] [Google Scholar]
- 35.Fagerstam L. G., Frostell A., Karlsson R., Kullman M., Larsson A., Malmqvist M., Butt H. Detection of antigen–antibody interactions by surface plasmon resonance. application to epitope mapping. J. Mol. Recognit. 1990;3:208–214. doi: 10.1002/jmr.300030507. [DOI] [PubMed] [Google Scholar]
- 36.O'shannessy D. J., Brigham-Burke M., Peck K. Immobilization chemistries suitable for use in the BIAcore surface plasmon resonance detector. Anal. Biochem. 1992;205:132–136. doi: 10.1016/0003-2697(92)90589-y. [DOI] [PubMed] [Google Scholar]
- 37.Pathmanathan S., Elliott S. F., McSwiggen S., Greer B., Harriott P., Irvine G. B., Timson D. J. IQ motif selectivity in human IQGAP1: binding of myosin essential light chain and S100B. Mol. Cell. Biochem. 2008;318:43–51. doi: 10.1007/s11010-008-9855-9. [DOI] [PubMed] [Google Scholar]
- 38.Tzu J., Marinkovich M. P. Bridging structure with function: structural, regulatory, and developmental role of laminins. Int. J. Biochem. Cell Biol. 2008;40:199–214. doi: 10.1016/j.biocel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siianova E. I. Expression of human LSB 32/67 KD gene in E. coli and analysis of its interactions with laminin. Biull. Eksp. Biol. Med. 1992;113:70–72. [PubMed] [Google Scholar]
- 40.Kjellen L., Lindahl U. Proteoglycans: structures and interactions. Annu. Rev. Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]