Abstract
The recombination activating protein, RAG1, a key component of the V(D)J recombinase, binds multiple Zn2+ ions in its catalytically-required core region. However, the role of zinc in the DNA cleavage activity of RAG1 is not well-resolved. To address this issue, we determined the stoichiometry of Zn2+ ions bound to the catalytically active core region of RAG1 under various conditions. Using metal quantitation methods, we determined that core RAG1 can bind up to four Zn2+ ions. Stripping the full complement of bound Zn2+ ions to produce apo-protein abrogated DNA cleavage activity. Moreover, even partial removal of zinc-binding equivalents resulted in a significant diminishment of DNA cleavage activity, as compared to holo-Zn2+ core RAG1. Mutants of the intact core RAG1 and the isolated core RAG1 domains were studied to identify the location of zinc-binding sites. Significantly, the C-terminal domain in core RAG1 binds at least two Zn2+ ions, with one zinc-binding site containing C902 and C907 as ligands (termed the CC zinc site) and H937 and H942 coordinating a Zn2+ ion in a separate site (HH zinc site). The latter zinc-binding site is essential for DNA cleavage activity, given that the H937A and H942A mutants were defective in both in vitro DNA cleavage assays and cellular recombination assays. Furthermore, as mutation of the active site residue E962 reduces Zn2+ coordination, we propose that the HH zinc site is located in close proximity to the DDE active site. Overall, these results demonstrate that Zn2+ serves an important auxiliary role for RAG1 DNA cleavage activity. Furthermore, we propose that one of the zinc-binding sites is linked to the active site of core RAG1 directly or indirectly by E962.
Keywords: atomic absorption spectroscopy, DNA double strand breaks, Inductively coupled plasma mass spectrometry, site-specific recombination, zinc-binding motif
INTRODUCTION
In the developing immune system, V(D)J recombination yields functional immunoglobulin and T-cell receptor genes via rearrangement of selected gene segments.1,2 Each gene segment, termed variable (V), diversity (D), or joining (J), are marked for possible recombination by a flanking recombination signal sequence (RSS). The RSS consists of conserved heptamer and nonamer sequences separated by 12 or 23 base pairs of spacer length, which yields two classes of RSSs (12-RSS and 23-RSS). Each recombination event joins two gene segments provided that the segments are flanked by RSSs with differing spacer length, a restriction referred to as the 12/23 rule. V(D)J recombination is initiated by the lymphoid-specific recombination activating proteins RAG1 and RAG2, which together create a double-strand break between the RSS heptamer and the adjacent gene segment through a two-step mechanism.3 The RAG proteins first nick the DNA between the gene segment and the 5’ end of the RSS. Second, the resulting 3’OH group forms a covalent bond with the phosphate group between the RSS and the gene segment on the opposite strand via a transesterification reaction. Thus, conventional RAG-mediated DNA cleavage results in two distinct products, which include a gene segment terminated by a covalently sealed hairpin (coding end) and the RSS heptamer terminated by a blunt-ended break (signal end). The production of double-strand breaks in V(D)J recombination requires an ordered assembly of protein and DNA components, with the RAG proteins likely binding to a 12-RSS first followed by recruitment of a 23-RSS to form the paired complex.4,5 Efficient formation of the paired complex requires one of the high mobility group proteins, HMGB1 or HMGB2.6–8 The RAG proteins can introduce a nick on a single 12-RSS. However, nicking at the 23-RSS, along with hairpin formation at both RSSs, occurs in the context of the paired complex.9,10 Processing and joining of the appropriate coding and signal ends is mediated by components of the nonhomologous end-joining machinery.11
The core regions of the murine RAG proteins include residues 384–1008 of 1040 in RAG1 and residues 1–387 of 527 in RAG2.2 The majority of biochemical studies with RAG1 and RAG2 are accomplished with these truncated proteins, which are the minimal catalytically active regions required for double-stranded DNA cleavage activity and also are more soluble than their full-length counterparts.2 Through extensive mutagenesis studies, three essential active site residues (D600, D708 and E962) characteristic of a DDE motif were identified in RAG1.12–14 The DDE motif is found in several transposase and integrase enzymes, and functions by coordinating divalent metal ions (i.e. Mg2+) in the active site.15,16 Previous studies have also shown that RAG1 contains the RSS heptamer17–19 and nonamer binding sites,20,21 and can bind to DNA in the absence of RAG2.18,19,22,23 Core RAG1 is composed of three major regions, which are referred to here as the N-terminal region (residues 384–527), the central domain (residues 528–760) and the C-terminal domain (residues 761–979).24,25 The RSS nonamer and heptamer are recognized by the N-terminal region20,21,26 and the central domain24 of core RAG1, respectively. The latter domain also contains two active-site residues (D600 and D708), as well as a C2H2 zinc finger termed the ZFB (residues 727–750),27 which is the predominant RAG2 binding site.24,28 Although RAG1 contains the active site and RSS binding sites, RAG2 is required for DNA cleavage activity. RAG2 may promote catalytic activity by forming additional contacts with the RSS or by indirectly enhancing recognition of the RSS by RAG1.19,22–24,29–31
RAG-mediated DNA double strand breaks (DSB) at both partner RSSs can be readily generated in in vitro assays in the presence of Mg2+ under conditions that favor formation of the paired complex.2,32 However, only the nicked product is efficiently produced on a single RSS (in the absence of the partner RSS) in Mg2+-containing buffers. In contrast, in the presence of Mn2+, both nicks and hairpins are readily generated in vitro on a single RSS.2,32 Lastly, as is common with other Mg2+-dependent nucleic acid enzymes, the RAG1/RAG2 complex can bind, but not cleave, RSS substrates in the presence of Ca2+.2
Both the full-length and core regions of RAG1 bind multiple Zn2+ ions;18,27,33 however, little is known concerning the role of this metal ion in the V(D)J recombination reaction. Here we demonstrate that the presence of Zn2+ ions bound to core RAG1 is important for the multiple functions of the V(D)J recombinase including nicking and hairpin formation. The core region of RAG1 binds up to four Zn2+ ions with approximately two of these metal ions binding in the C-terminal domain (CTD). Moreover, our results indicate that two conserved His residues, H937 and H942, coordinate a Zn2+ ion in a catalytically required site we refer to as the HH zinc-binding site. By mutagenesis of these Zn2+-coordinating ligands, we show that the HH zinc-binding site is essential for DNA cleavage activity. Other possible ligands to the HH zinc site include the active site residue E962, further indicating a role for the HH zinc site in the DNA cleavage activity of RAG1. Overall, this site represents a novel motif not found in other DDE motif enzymes, and its presence may in part account for key requirements in the catalytic activity of the RAG1-RAG2 V(D)J recombinase.
RESULTS
Core RAG1 binds up to four Zn2+ equivalents
Previous reports have shown that both non-core and core RAG1 bind multiple Zn2+ ions.18,27,33 Only one Zn2+ coordination site in the core region of RAG1 has been previously proposed, namely the C2H2 zinc finger (termed the ZFB site) located in the central domain. However, the overall effect of Zn2+ coordination on the DNA cleavage activity of RAG1 has not been determined, nor has the location of additional zinc-binding sites been identified.
To further characterize the zinc-binding properties of the core region of RAG1, flame atomic absorption spectroscopy (FAAS) was used to quantitate the stoichiometry of Zn2+ ions bound to core RAG1. In this study, murine core RAG1 was fused to maltose-binding protein (mbp), and is referred to as mR1(core). Prior to FAAS measurements, mR1(core) was dialyzed extensively in metal free buffer (20 mM Tris, pH 8.0, 200 mM NaCl, 5 mM β-mercaptoethanol) for ~48 hrs at 4°C. Multiple FAAS measurements showed that wild type (WT) mR1(core) bound 3.9±0.6 Zn2+ (Table 1). The Zn2+ content of mbp* (mbp plus linker, see Materials and Methods) was also determined to be <0.3 Zn2+, indicating that zinc binding was localized to the core RAG1 region of the fusion protein. It is apparent that core RAG1 bound Zn2+ ions with relatively high affinity, since Zn2+ remained bound even after extensive dialysis in metal-free buffers.
Table 1.
Stoichiometry of Zn2+ Ions Bound to mR1(core) and mR1(CTD)
| mR1(core) (fused to MBP) a,b |
Zn2+:mR1(core) (Values from FAAS) c,fg |
mR1(CTD) (fused to MBP)b,d |
Zn2+:mR1(CTD) ICP-MSe,f,g,h |
Zn2+:mR1(CTD) FAASe,f,g,h |
|---|---|---|---|---|
| WT | 3.9 ± 0.6 | WT | 2.3 ± 0.7 | 2.8 ± 0.4 |
| D600,708A | 3.4 ± 0.03 | |||
| E962A | 2.8 ± 0.4 | E962A | 0.6 ± 0.4 | nd |
| H942A | 2.6 ± 0.4 | H942A | 0.7 ±.0.4 | nd |
| H937A | 1.9 ± 0.2 | H937A | 0.6 ± 0.2 | nd |
| C902,907A | 1.7 ± 0.4 | C902,907A | 0.6 ± 0.4 | 1.1 ± 0.4 |
| H937,942A | 1.1 ± 0.5 | 1.2 ± 0.3 | ||
| H795A,C796A | nd | 3.1 ± 0.6 |
The pool of dimeric mR1(core) isolated from size exclusion chromatography (SEC) was used for analysis of Zn2+ content. The stoichiometry is in terms of the number of Zn2+ ions bound per mR1(core) monomeric subunit.
In the nomenclature for each mutant, the first letter is the original amino acid followed by the residue numbers with regard to the full-length murine RAG1 protein, and the last letter is the replacement amino acid in the mutant proteins.
Each sample was dialyzed against 20 mM Tris, pH 8.0, 200 mM NaCl, 5 mM β-mercaptoethanol, for 16 hours at 4°C followed by dialysis against fresh buffer.
The pool of monomeric mR1(CTD) isolated from SEC was used for analysis of Zn2+ content.
Each sample was dialyzed against 20 mM Tris, pH 8.0, 200 mM NaCl, 5 mM β-mercaptoethanol, and 10 mM EDTA for 16 hours at 4°C followed by dialysis against fresh buffer containing 0.1 M instead of 10 mM EDTA for 24 hours at 4°C. Chelex resin (4g per L) was added to each dialysis buffer.
n (number of experiments is at least 3 using multiple protein preparations). Error is reported as standard deviation.
With the exception of D600,708A mR1(core) and H795,C796A mR1(CTD), the Zn2+ stoichiometry of the remaining mutants significantly differs from the corresponding WT measurement (p<0.05 as determined by Student’s t test).
nd, not determined
These results differ from previous investigations where mR1(core) was reported to bind ~2 Zn2+ ions.18 This discrepancy may be due to the differences in purification protocols, since higher resolution size exclusion chromatography is now used to purify mR1(core), which is more effective in separating mR1(core) oligomers from highly aggregated protein.34
Dialysis of mR1(core) was also performed with the Zn2+ chelators, ethylenediaminetetraacetic acid (EDTA) and diethylenetriaminepentaacetic acid (DTPA). The binding affinity of the Zn2+:DTPA complex is reported to be significantly higher than that for the Zn2+:EDTA complex.35 Likely due to this difference in zinc-binding affinity, each chelator had a differential effect on removing Zn2+ ions from mR1(core). After dialysis in EDTA, the stoichiometry was reduced to 0.95±0.3 Zn2+ equivalents per mR1(core) subunit, whereas dialysis in the DTPA-containing buffer removed essentially all of the coordinated Zn2+ (stoichiometry at 0.3±0.2 equivalents of Zn2+ per mR1(core) monomer). In the case of the 1-Zn2+ form of mR1(core) (dialyzed in EDTA), it is possible that the remaining single Zn2+ equivalent resides in a single site. Alternatively, the 1−Zn2+ form may consist of a mixed population of mR1(core) with varying Zn2+ stoichiometries (from 0 to 4) consisting of Zn2+ ions distributed between the different binding sites.
Zn2+ ions plays an essential role in the DNA cleavage activity of RAG1
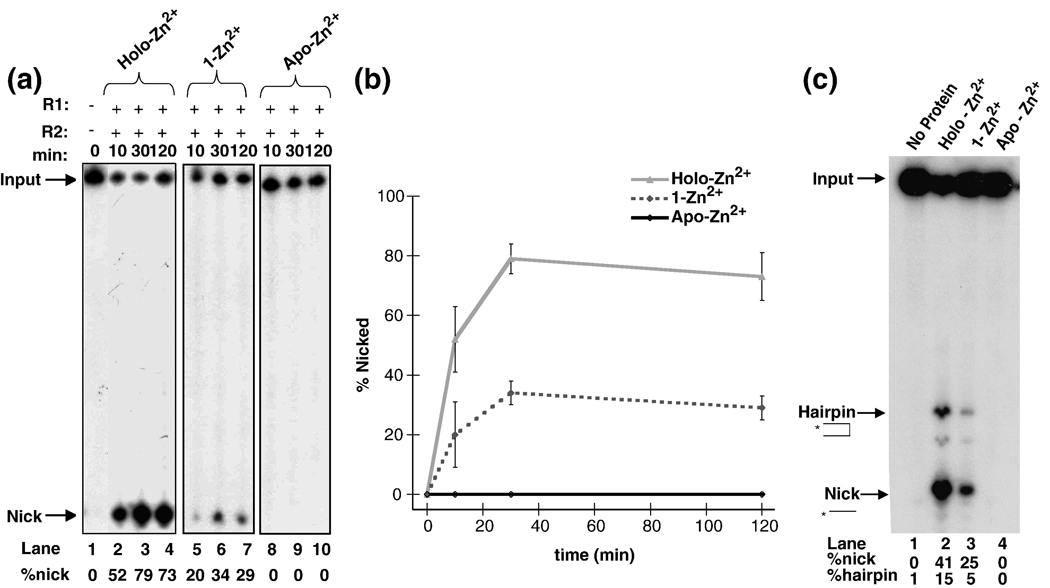
In the presence of RAG2, mR1(core) has been shown to catalyze different DNA cleavage reactions depending on the divalent metal ion present. For example, in Mg2+-containing buffers, nicking readily occurs on a 12-RSS substrate, whereas hairpin formation is most efficient with the addition of a 23-RSS either in trans (as a separate oligonucleotide duplex) or in cis (on the same DNA substrate).1,2 Conversely, in the presence of Mn2+, both nicking and hairpin formation can occur on a single 12-RSS (in the absence of the 23-RSS).2,32 Unlike these metal ions, there is no DNA cleavage activity with Zn2+ as the only divalent metal ion present (data not shown). However, since mR1(core) binds up to four Zn2+ ions, we asked what the effect of Zn2+ bound in these binding sites would have on the different catalytic functions of mR1(core). To address this question, the DNA cleavage activities of the holo-Zn2+, the 1-Zn2+ form (EDTA-treated), and the apo-Zn2+ form (DTPA-treated) of mR1(core) were compared in a single RSS assay containing either Mg2+ or Mn2+ (Figure 1A and 1C, respectively). (Prior to the addition of Mg2+ or Mn2+, the cleavage reaction buffers were rendered metal free by passage over Chelex resin. In addition, since FAAS measurements showed that 1M stock solutions of Mg2+ or Mn2+ contained only a trace amount of Zn2+, no significant amount of exogenous Zn2+ was introduced into the cleavage assay reactions.)
Figure 1.
Zn2+ effects on mR1(core) activity. (A) Timecourse of the nicking reactions of a single 12-RSS substrate by the different Zn2+ forms of mR1(core) in the presence of Mg2+. (B) Quantitation of nicked products from part A. (C) Nicking and hairpin formation of a single 12-RSS substrate by the different Zn2+ forms of mR1(core) in Mn2+ at 2 hours. The percentages of nicked and hairpin products are given below each lane in the autoradiogram. The cleavage reactions in panels A and C consisted of 10 mM Tris pH 8.0, 6% glycerol, 100 mM NaCl, 2 mM dithiothreitol, and 5 mM Mg2+ (in panel A) or Mn2+ (in panel C). mR1(core) and gR2(core) (each at 6 nM) were incubated on ice for 30 min before the addition of 2 nM 32P-labeled 12-RSS. The reactions in the presence of Mg2+ were incubated at 37°C for 10 min, 30 min, and 2 hours. The reactions in the presence of Mn2+ were incubated at 37°C for 2 hours. Products were separated on an 8% denaturing polyacrylamide gel.
Partial removal of Zn2+ from mR1(core), to form the 1-Zn2+ form, resulted in decreased DNA cleavage activities. In the presence of Mg2+, both the holo-Zn2+ and 1-Zn2+ forms formed nicked product, with the majority of nicked product formed within 30 min (Figure 1). The 1-Zn2+ form was significantly less active than the holo-Zn2+ protein over the entire timecourse, with a nearly 4-fold decrease in the amount of nicked product after the 2 hour incubation period (Figure 1B). In the presence of Mn2+, the cleavage activity of the 1-Zn2+ form was also diminished, with the largest effect on hairpin formation. Specifically, there was a > 2.5-fold and a 5-fold decrease in nicking and hairpin formation, respectively, for the 1-Zn2+ form versus the holo-Zn2+ protein after a 2 hour incubation period (Figure 1C). Hairpin formation was similarly diminished in a coupled cleavage assay (with an added 23-RSS) in the presence of Mg2+ (data not shown).
Total removal of Zn2+ ions to form the apo-Zn2+ mR1(core) protein, resulted in a complete absence of DNA cleavage activity in the presence of Mg2+ or Mn2+ (Figure 1). Notably, hairpin formation in the single RSS assay (in the presence of Mn2+) is dependent on the production of nicked products. Therefore, to confirm that the apo-Zn2+ form could not catalyze formation of hairpin products, its activity versus holo-Zn2+ mR1(core) was tested using a pre-nicked 12-RSS substrate in Mn2+-containing buffer. The results show that hairpin products were formed with holo-Zn2+ mR1(core), but not with the apo-Zn2+ form (data not shown), indicating that the presence of at least one equivalent of Zn2+ is required for both nicking and hairpin formation. Significantly, addition of Zn2+ (from 50 µM up to 50 mM) to the 1-Zn2+ form and apo-Zn2+ sample did not increase or rescue mR1(core) activity (data not shown), possibly due to an altered mR1(core) structure that could not be returned to the active state simply by addition of excess Zn2+.
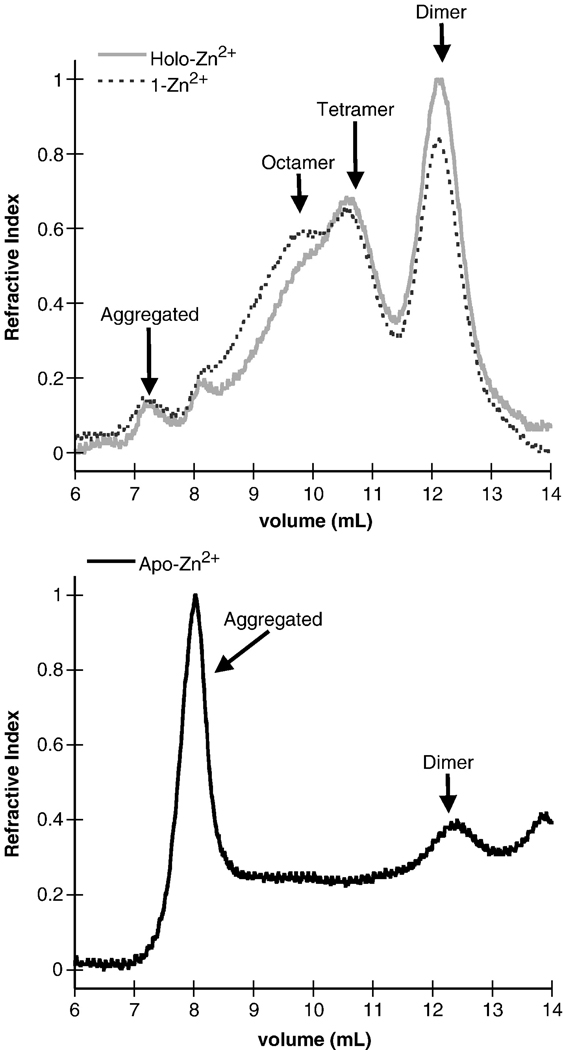
To determine the effect that removal of Zn2+ ions had on the oligomerization properties of mR1(core), the protein self-association properties were compared using size exclusion chromatography of the various Zn2+-bound forms. The oligomerization properties of the 1-Zn2+ form showed a detectable decrease (~15% in terms of peak area) in the amount of dimer with a corresponding increase in the amount of octameric species, as compared to the holo-Zn2+ form (Figure 2). Previous studies have shown that the octameric form of mR1(core) is not active in DNA cleavage assays34 and does not readily equilibrate with the dimer form23, strongly indicating that there are conformational differences between the two oligomeric forms. Overall, the altered oligomerization properties may partially explain the decreased activity of the 1-Zn2+ form of mR1(core).
Figure 2.
Size exclusion chromatography profiles of holo-Zn2+ and 1-Zn2+ (top panel), and apo-Zn2+ (bottom panel) forms of mR1(core). The chromatographic profiles are from an analytical 20 ml Superdex 200 column and monitored by refractive index. Refractive index values were normalized with respect to the total concentration of holo-Zn2+ mR1(core) and are in arbitrary units.
Removal of the majority of Zn2+ ions to form apo-Zn2+ mR1(core) appeared to significantly alter the structure, as the majority of sample eluted in the void volume in size exclusion chromatography (SEC) (Figure 2), likely due to protein unfolding and aggregation. Therefore, the absence of mR1(core) activity when Zn2+ is removed is consistent with an extensively aggregated inactive form of mR1(core).34
Influence of Zn2+ ions on the folding of mR1(core) during protein purification
To assess how excess Zn2+ influences the folding of mR1(core) in vivo, the protein was expressed without addition of excess Zn2+ to the growth media. (Typically 50 µM Zn2+ is added to the growth media to express mR1(core) in bacteria.) Moreover, great care was taken during each step of the purification process to ensure that excess Zn2+ was not present. Specifically, all glassware was acid rinsed and stringent conditions were used to generate metal free buffers for purification, which included treatment of buffers with Chelex resin to remove trace metals including Zn2+.
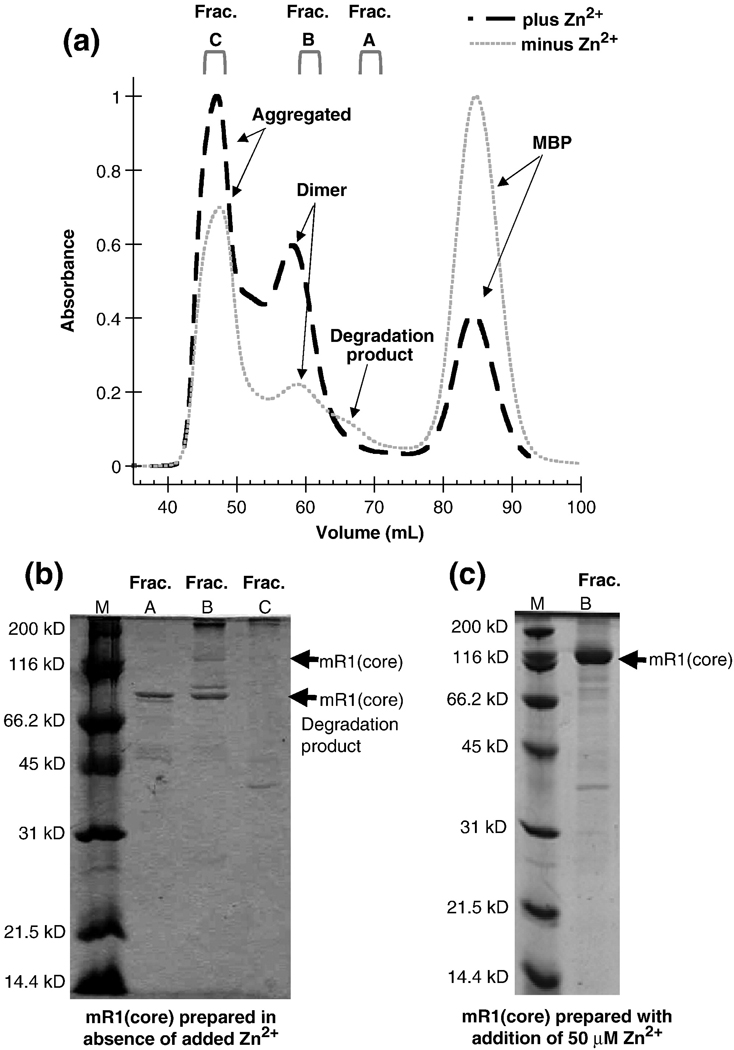
Significantly, the SEC elution profile from the 120 mL Superdex preparative column of the “Zn2+-free” purified form differed from the holo-Zn2+ form (Figure 3A). Using Zn2+-free conditions, SDS-PAGE analysis confirmed that the protein had been proteolyzed and the major degradation product was present in most fractions containing full length mR1(core) (Figure 3B). (The mR1(core) sample used in these experiments contained a PreScission protease site between mbp and core RAG1. The degradation product was susceptible to cleavage with PreScission protease, as evidenced by an ~42 kD band on SDS-PAGE that is consistent with mbp (not shown)). In typical preparations of the enzyme such as in Figure 3C, mR1(core) is not degraded (due to the presence of added Zn2+ to purification buffers as well as the presence of adventitious Zn2+ leached from glassware). Zn2+ analysis of the major degradation product was not feasible due to the difficulty in purifying this species from intact mR1(core). The migration in the gel of the proteolyzed product from the “Zn2+-free” preparation of mR1(core) with respect to the molecular weight markers is consistent with cleavage occurring in the central domain. The central domain contains the two active site Asp residues. In RAG1, it is possible that Zn2+ ions help to stabilize this region, and that insufficient Zn2+ concentrations results in greater accessibility of this region to contaminating proteases during the cell lysis and protein purification procedure.
Figure 3.
(A) Size exclusion chromatography profiles of mR1(core) purified with (—) and without (---) addition of 50 µM Zn2+. The chromatographic profiles are from a preparative 120 ml Superdex 200 column and monitored by UV absorption. The buffer for mR1(core) purified with added Zn2+ contained 50 µM Zn2+ and 0.5 M NaCl, while the buffer for mR1(core) purified in the absence of added Zn2+ contained 0.5 M NaCl and was treated with Chelex to remove any adventitious metals. (B) SDS-PAGE of mR1(core) purified in the absence of added Zn2+. Fractions collected from SEC are A (elution volume 68.2–71.2 mL); B (elution volume 59.2–61.2 mL); and C (elution volume 45.2–48.2 mL). These fractions correspond to lanes A, B, and C on the gel. (C) SDS-PAGE of mR1(core) purified in the presence of Zn2+ shows the characteristic dimer band in Fraction B. Fraction B purified in the presence of added Zn2+ corresponds to the SEC elution volume collected for Fraction B prepared in the absence of added Zn2+.
The activity of three different pooled fractions of mR1(core) purified under Zn2+-free conditions was tested in the single RSS assay in the presence of Mn2+. Compared to holo-Zn2+ mR1(core), fraction B showed similar efficiency in DNA cleavage activity while fractions A and C were inactive (data not shown). Fraction B contained the highest concentration of full-length mR1(core), while fraction A was almost completely degraded to the predominant proteolytic product, and fraction C had a wide range of impurities (Figure 3B).
Location of zinc-binding sites in core RAG1
To deduce the locations of possible zinc-binding sites, Zn2+ analysis was also performed with the isolated central domain and C-terminal domain (both fused to mbp, and referred to as mR1(CD) and mR1(CTD), respectively). The Zn2+ stoichiometry of mR1(CD) (measured by dialyzing the protein into metal free buffer prior to FAAS) was at 1.4 ± 0.2 Zn2+ equivalent per mR1(CD) subunit. This is consistent with previous studies in which at least one Zn2+ ion is bound to the central domain, presumably in the C2H2 ZFB site.27 The Zn2+ stoichiometry of mR1(CTD) was determined by both FAAS and inductively coupled plasma-mass spectrometry (ICP-MS) under metal-free conditions. Both Zn2+ quantitation methods (FAAS and ICP-MS) consistently showed that mR1(CTD) bound 2–3 Zn2+ ions (Table 1). In these measurements, protein samples were dialyzed against metal-free buffer containing EDTA, prior to Zn2+ quantitation. However, no significant change in Zn2+ stoichiometry was found whether protein samples were dialyzed against metal free buffer either containing or lacking EDTA, indicating that the Zn2+ ions were not readily removed from their binding sites in the CTD. Notably, this differs from the results with mR1(core), and may indicate that the zinc-binding sites in the isolated CTD are less accessible to solvent than in the intact core RAG1.
In a previous study, purified mR1(CTD) was shown to elute from size exclusion chromatography (SEC) as both monomeric and dimeric species.24 The monomer fraction from SEC was used for measurements of Zn2+:mR1(CTD) stoichiometry (Table 1). Since Zn2+ can coordinate in the interface of oligomeric complexes36,37, it was conceivable that the stoichiometry of Zn2+ per subunit of mR1(CTD) differed between the monomer and dimer fractions. To address this possibility, it was first necessary to determine the extent of redistribution between monomer and dimer forms during preparation of the samples for Zn2+ quantitation measurements. After preparation of the samples for Zn2+ analysis, which required extensive dialysis, we determined by size exclusion chromatography that the dimer fraction of mR1(CTD) redistributed to a 45:55 mixture of monomer and dimer, whereas the monomer sample did not re-associate to dimer (data not shown). The disparity between dissociation of dimer versus re-association of monomer is not clear, but could reflect an energetic barrier to dimer formation. Nevertheless, we would expect that if Zn2+ ions were coordinated in the dimer interface that the “dimer” fraction (even in a sample with 55% dimer:45% monomer) would show somewhat higher stoichiometries than the monomer fraction. Zn2+ analysis of the two fractions, however, showed similar values as determined by FAAS (dimer fraction at 2.7 ± 0.3 and monomer fraction at 2.8 ± 0.4) confirming that binding of Zn2+ ions to mR1(CTD) was not dependent on dimer formation.
Thus, it appears that the sum of the Zn2+ ions coordinated in mR1(CD) and mR1(CTD) is consistent with the total Zn2+ measured for mR1(core). These results suggest that the zinc-binding sites form in the individual domains in a similar manner as in the intact core RAG1.
Conserved Cys and His residues in the C-terminal domain of core RAG1 ligate Zn2+ ions
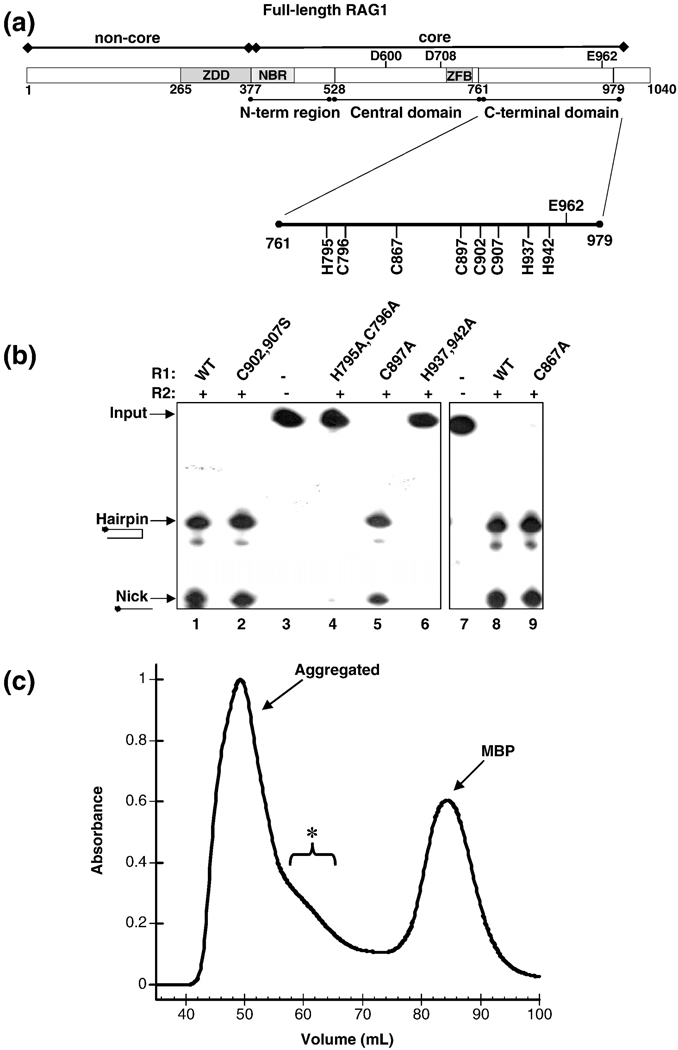
The majority of Zn2+ ions in core RAG1 coordinate to the CTD domain of mR1(core). While the locations of zinc-binding sites in this domain are not known, it does contain multiple, conserved Cys and His residues separated by 2–4 residues (Figure 4A), which are favored ligand spacings for Zn2+ ion coordination. Although the spacing of Cys and His residues in the CTD is not consistent with any of the known zinc-binding motifs, there is a possible C2H2 motif with the spacing: C(902) - X4 – C(907) - X29 – H(937) - X4 – H(942), in which X is any residue and the residue number relative to full length RAG1 sequence is in parentheses. Each of these Cys and His residues are highly conserved among the known RAG1 sequences.
Figure 4.
(A) Domains in core RAG1. The core region is the minimal region required for catalysis. This region contains the nonamer binding region (NBR), a C2H2 zinc finger (ZFB) and the three active site residues (Asp-600, Asp-708 and Glu-962). The location of the central and C-terminal domains are shown below the bar representing core RAG1. (B) The effect of mutating conserved CTD His and Cys residues in mR1(core) on DNA cleavage activity. Analysis of coupled cleavage activity in buffer containing Mg2+. mR1(core) (0.04 µM) and gR2(core) (0.04 µM) proteins were incubated with HMGB1 (0.34 µM) and radiolabeled ds 12-RSS in the presence or absence of unlabeled ds 23-RSS. Lane 1 has no protein in the reaction, only ds 12-RSS. The ds 12-RSS substrate (input) and the nick and hairpin cleavage products are indicated. Also, the mR1(core) protein used in each sample (WT or mutant) is labeled above each lane. H795,C796A mR1(core) and H937,942A mR1(core) showed 0% nicking and hairpin formation. The activity of C902,907S mR1(core), C897A mR1(core), and C867A mR1(core) were approximately equal to the activity level of WT mR1(core). (C) Size exclusion chromatography profile of the C902A,C907A,H942A mR1(core) triple mutant from a preparative 120 ml Superdex 200 column. The chromatography was performed as described in Figure 3A for the WT mR1(core) sample purified in Zn2+-containing buffers. The bracket labeled with an asterisk denotes a shoulder on the aggregated peak that likely contains a mixture of oligomeric forms of the triple mutant.
To determine if these residues are Zn2+ coordinating ligands, double point mutants (C902,907A and H937,942A) and, in some cases, single point mutants (H937A and H942A) were generated in mR1(CTD). Each mutant bound ~1 Zn2+ ion less per subunit than wild type (WT) mR1(CTD) as determined by ICP-MS and/or FAAS (Table 1). In addition, a C902,907S mutant showed a similar reduction in the stoichiometry of bound Zn2+ ions as compared to the C902,907A mutant (data not shown).
In the context of core RAG1, the double point mutant C902,907A, as well as the single point mutants, H937A and H942A, each showed a reduction in Zn2+ stoichiometry relative to WT mR1(core) following dialysis against EDTA-free and Zn2+-free buffers (Table 1). These results provide additional confirmation that the zinc-binding sites that form in the isolated CTD are representative of those in the intact core protein.
Both mR1(core) and mR1(CTD) have distinctive oligomerization properties, and significant structural perturbations would likely be evidenced by variations in these properties. Elution of the mutant forms of mR1(core) and mR1(CTD) were indistinguishable from their WT forms as monitored by SEC (data not shown). Therefore, loss of Zn2+ binding to the mR1(core) and mR1(CTD) mutants is attributed to a direct effect of removing Zn2+ coordinating ligands, and not indirectly from inducing major structural perturbations of the proteins.
Zinc-coordinating residues, H937 and H942, are essential for catalytic activity
The C902,907S and H937,942A mR1(core) mutants were compared to the WT protein in the single RSS cleavage assay to test their ability to nick and generate hairpins on DNA. In this assay, mR1(core), core RAG2 (fused to glutathione S-transferase (gst), and referred to as gR2(core)), and radiolabeled ds 12-RSS were incubated together in the presence of Mn2+. As expected, samples with WT R1(core) showed efficient formation of both nicked and hairpin products (Figure 4B, lanes 1 and 8). The mR1(core) C902,907S mutant, as well as the C902,907A mutant (not shown), showed similar efficiencies in DNA cleavage activities as compared to WT mR1(core) (Figure 4B, lane 2). In contrast, the H937,942A mR1(core) mutant was inactive with no detectable nick or hairpin products formed from the 12-RSS substrate (Figure 4B, lane 6).
DNA cleavage assays with the single point H937A and H942A mR1(core) mutants showed that both His residues are essential for catalytic activity, as no cleavage products were observed for either mutant in either the single RSS (not shown) or coupled cleavage assay (Figure 5A). In addition, neither mutant could form a DNA hairpin from a pre-nicked RSS substrate (data not shown), demonstrating that both nicking and hairpin formation activities were defective. These results also definitively showed that H937 is essential for catalytic activity, since previous H937 mutants with defective activity were in the context of multiple point mutations.38 (It is important to note that the concentrations of mR1(core) and gR2(core) used in the experiments in Figure 4B and Figure 5A were higher than the protein concentrations used in Figure 1 to emphasize the lack of activity of some of the mutants.) These results show that mutation of C902 and C907 has no effect on DNA cleavage activity, while H937 and H942 are essential for DNA cleavage activity. Given the differences in DNA cleavage activities of the H937,942A versus C902,907S mutants, it is unlikely that residues C902 and C907 coordinate the same Zn2+ ion as H937 and H942. We attempted to test this possibility by producing the mR1(core) triple mutant C902A,C907A,H942A. The Zn2+ stoichiometry of the triple mutant was to be compared to the C902,907A and H942A mR1(core) mutants. However, SEC of the purified triple mutant revealed that this protein was predominantly aggregated with no distinct peak in the elution profile for dimeric protein (Figure 4C). As a result, it was not possible to measure the Zn2+ stoichiometry for a dimeric form of the triple mutant. The altered aggregation property of the triple mutant is not consistent with simply removing a third ligand to a single zinc-binding site. Instead, it is possible that the combined effects of removing two separate zinc-binding sites resulted in defects in folding of the triple mutant during its production in E. coli, which yielded the predominantly aggregated protein. From the combined results above, we conclude that the CTD contains two separate zinc-binding sites, with one site containing C902 and C907 as ligands (the CC site) and the second site containing H937 and H942 as ligands (the HH site).
Figure 5.
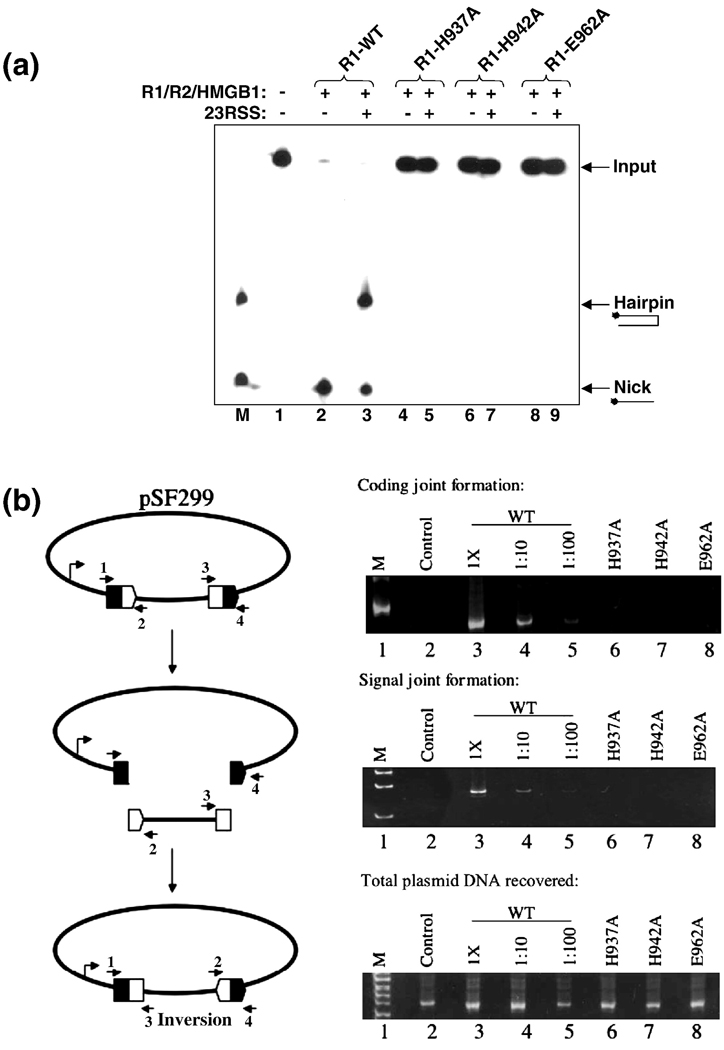
Mutagenesis of the zinc-binding ligands H937 and H942 abolishes DNA cleavage activity of mR1(core). (A) Analysis of coupled cleavage activity, performed as described in Figure 4B. The lane marked M contains oligonucleotides that correspond to the expected nick and hairpin products. Lane 1 contains no proteins in the reaction, only ds 12-RSS. Also, the mR1(core) protein used in each sample (WT or mutant) is labeled above each lane. H937A mR1(core), H942A mR1(core), and E962A mR1 (core) showed 0% nicking and hairpin formation compared to the activity level of WT mR1(core). (B) Cellular V(D)J activity of mR1(core) proteins on an extrachromosomal substrate. The diagram (left panel) is of the pSF299 recombination substrate. Rearrangement of this substrate produces coding joints and signal joints as shown. The 12- and 23-RSS are designated by open and filled pentagons, respectively. The coding flanks are symbolized by open and filled rectangles. The arrows labeled 1–4 represent primers used in the PCR amplifications Right panels: PCR amplifications were used to detect coding joints with primers 1 and 3 (top), signal joints with primers 2 and 4 (middle), and total plasmid DNA recovered from each sample with primers 1 and 4 (bottom). PCR amplifications from transfections using plasmid encoding wild-type (WT) mR1(core) was assayed at 1X, 1:10, and 1:100 dilutions. Transfections using plasmids encoding mR1(core) H937A, H942A and E962A mutants were assayed at 1X concentrations. The lanes labeled “control” represent transfections that lacked the RAG2 expression plasmid.
Mutation of the active site residue E962 reduces zinc stoichiometry
Since at least two Zn2+ ions can bind to the CTD, other residues in this domain must function as zinc ligands in the CC and HH zinc sites. Because mutation of H937 and H942 abolished the DNA cleavage activity of core RAG1, we sought to identify additional ligands to the HH zinc site. To accomplish this, we systematically mutated residues in the CTD that replaced conserved His and Cys residues with Ala. Our criteria were to find a residue(s) that 1) eliminated DNA cleavage activity when mutated in core RAG1, as well as 2) decreased the number of Zn2+ ions bound to mR1(CTD) and/or mR1(core) when the residue was replaced with Ala. Additional conserved Cys and His in the CTD of RAG1 include residues H795, C796, C867, and C897.
The mR1(core) mutants were tested for their ability to nick and generate hairpins on DNA in the single RSS cleavage assay. Both C867A and C897A showed activity in the cleavage assay (Figure 4B), and thus are not candidates for ligands in the HH site. In contrast, the H795A,C796A mR1(core) mutant demonstrated complete catalytic defects with no evident formation of nicked or hairpin products (Figure 4B, lane 4). Based on these findings, we tested if the H795A,C796A mutant, in the context of the isolated CTD, had lost the equivalent of one Zn2+ ion. However, similar to WT mR1(CTD), the H795A,C796A mutant bound 2–3 Zn2+ ions (Table 1), indicating that the catalytic defect of this mutant is due to another factor besides disruption of a zinc-binding site. Thus, we concluded that the Zn2+ ion coordinated by H937 and H942 was not ligated by any of the other conserved His or Cys residues in the CTD.
Next, we considered the possibility that the Zn2+ ion coordinated by H937 and H942 may include ligands not found in a typical zinc structural site. Whereas, His and Cys residues are the predominant zinc ligands in structural zinc-binding sites, other residues are capable of coordinating Zn2+ ions, in particular Glu and Asp, which often serve as ligands in the active site of zinc-dependent enzymes.36 Extensive mutagenesis studies have been previously performed on core RAG1, with each of the conserved acidic, basic, as well as other selected residues replaced (typically with Ala).12,38,39 Besides the His and Cys residues studied here, E962 is the only residue so far identified in the CTD that when mutated renders the intact core protein inactive and also has the potential to coordinate Zn2+ ions. Thus, E962A mutants were created in both mR1(CTD) and in mR1(core) to test for Zn2+ binding. In summary, the E962A mutant in the isolated mR1(CTD) and in mR1(core) (Table 1) showed a significant decrease in Zn2+ stoichiometry compared to the WT counterparts.
Mutation of E962 is expected to result in inactive protein, as has been previously shown by mutagenesis studies in which all E962 mutants have been defective in both nicking and hairpin formation.13,14 In our hands, as well, the mR1(core) E962A mutant demonstrated no activity in the single RSS (data not shown) or in the coupled cleavage assays (Figure 5A).
The defects in the functions of the R1(core) H937A, H942A, and E962A single point mutants were also demonstrated in a cellular V(D)J recombination assay using an extrachromosomal plasmid substrate. Similar experiments have been previously done with the H942 and H937 residues mutated; however, the latter case was in the context of a double mutant in which both H937 and K938 were replaced with Ala.38 In the present study, the extrachromosomal substrate pSF299 was used (Figure 5B), which can be rearranged by inversion of an insert to form coding and signal joints.40 WT mR1(core), in the presence of R2(core), generated both coding joints and signal joints on the pSF299 substrate, whereas no products were detected using the mR1(core) H937A, H942A, and E962A mutants in place of the WT protein (Figure 5B). Western blot analysis confirmed that the mutant proteins were expressed at WT levels in transiently transfected 293T cells (data not shown).
As E962 is one of the three carboxylate residues that constitutes the DDE active site, we also determined the Zn2+ stoichiometry of a mR1(core) active site mutant, D600,708A. FAAS results demonstrated that the Zn2+ stoichiometry of the D600,708A mutant did not differ significantly from WT mR1(core) (Table 1). Therefore, of the three carboxylate active site residues, only E962 is capable of affecting Zn2+ coordination under the conditions used here. Significantly, this result ruled out the possibility that the decrease in Zn2+ stoichiometry of the E962A mutant (relative to WT) in mR1(core) was not simply due to Zn2+ bound in place of Mg2+ in the active site. However, it should be noted that in this study, the Zn2+ stoichiometric measurements were conducted after extensive dialysis against metal-free buffer. It is possible that if excess zinc was added to the buffer, then Zn2+ ions may be able to coordinate with the Asp active site residues. However, the binding affinity of Zn2+ to an all-oxygen coordination sphere (with the carboxylate active site residues as ligands) may be very low, and thus not detected under the conditions used here.
Therefore, mutation of E962 reduced Zn2+ binding in mR1(CTD), as well as in mR1(core); and also yielded inactive protein. These properties are characteristics of CTD residues uniquely shared by H937 and H942. Given the absence of other candidate zinc ligands with these properties, we propose that E962 can either 1) act directly as a zinc ligand in the HH zinc site along with H937 and H942, or 2) indirectly contribute to Zn2+ coordination by helping to stabilize formation of the HH site, such as through electrostatic stabilization of the bound Zn2+ ions.
The effect on RAG2 and RSS binding upon mutation of zinc ligands in the HH zinc site
To determine if mutation of H937 and H942 disrupted the interaction of mR1(core) with either gR2(core) or the RSS, we performed a panel of binding experiments to test for the ability of H937A and H942A mR1(core) mutants to bind the RSS, interact with gR2(core), and form the paired complex. The assays were also performed with the E962A mR1(core) mutant as a control, since it was previously determined that replacement of E962 does not significantly affect macromolecular interactions relevant to RAG1 function.13,14
Binding of mR1(core) to a single RSS was tested using electrophoretic mobility shift assays (EMSA), in which mR1(core) was incubated with radiolabeled ds 12-RSS in the presence or absence of gR2(core), and the protein-DNA complexes resolved on non-denaturing polyacrylamide gels. WT mR1(core), as well as the H937A, H942A, and E962A mutants, bound to the 12-RSS substrate (Figure 6A, lanes 2, 4, 6, and 8). In addition, with all of the mR1(core) proteins, the highest mobility mR1(core):12-RSS complex (labeled ‘RAG1 oligomer1’ in Figure 6A) was further shifted upon addition of gR2(core) to form a heterotrimeric complex (Figure 6A, lanes 3, 5, 7 and 9). In a previous study, complex formation could not be detected between RAG2, the 12-RSS, and either mR1(core) H942A single point or H937A,K938A double point mutants.38 Our results show that both of the H937A and H942A mutants can form this complex, although it is apparent that there are defects in the formation of the RAG1:RAG2:12-RSS complex for both the H937A and H942A mutants based on the diminished intensities of the supershifted bands (Figure 6A, lanes 3 and 5). The differing DNA binding results for the H937A and H942A mutants versus the E962A mutant is not necessarily inconsistent with E962 acting as a direct zinc ligand in the HH site. Rather, this may be due to the different properties of the His versus Glu sidechains. For example, mutation of carboxylate active site residues in other nucleases has been found to be favorable for DNA interactions.41,42 Similarly with RAG1, the E962A mutation may partially alleviate repulsion of the DNA phosphodiester backbone by the negatively charged DDE active site, and thus potentially counteract any effect that loss of Zn2+ binding may have on DNA interactions.
Figure 6.
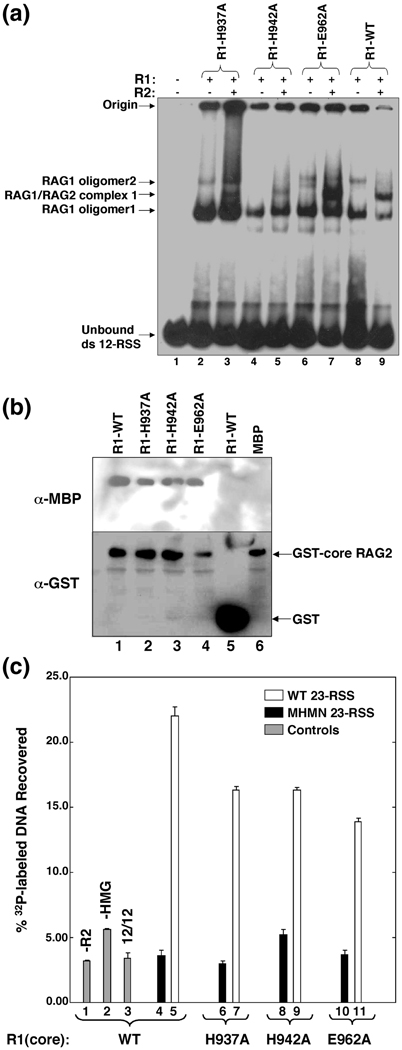
The effect of mutating the zinc-binding ligands H937 and H942 on macromolecular interactions mediated by RAG1. (A) Electrophoretic mobility shift assays of mR1(core) proteins (0.04 µM) and gR2(core) (0.005 µM) combined with 1 nM 32P-labeled 12-RSS. Each lane contained 32P-labeled 12-RSS and mR1(core) and gR2(core) proteins as indicated. The mR1(core) protein used in each lane is labeled above each lane. The shifted bands in lanes containing only mR1(core) correspond to two oligomeric forms of the protein complexed with the radiolabeled 12-RSS and are labeled as RAG1 oligomer1 and oligomer2. The supershifted bands in lanes that contain both mR1(core) and gR2(core) are designated RAG1/RAG2 complex 1. The origin and unbound 32P-labeled 12-RSS are labeled accordingly. (B) Interaction between mR1(core) and gR2(core) proteins. gR2(core) was bound to gluthathione-Sepharose resin in the presence of mR1(core) proteins (WT, H937A, H942A, or E962A) or mbp. After elution from the resin, the proteins were resolved by SDS-PAGE and detected by Western blots. (C) Formation of the paired complex. In each sample (unless labeled otherwise), mR1(core) (WT or mutants) (0.2 µM) and gR2(core) (0.2 µM) proteins, and 0.3 µM HMGB1 were incubated with 10 nM biotinylated WT 12-RSS and 1 nM 32P-labeled RSS. The protein-DNA complexes were captured by streptavidin-coated paramagnetic particles as described in Materials and Methods. The bars in the graph represent the % 32P-labeled RSS captured. The mR1(core) protein used in each sample is labeled beneath the graph. Control samples are in lanes 1–3 (gray bars) and either lacked gR2(core) or HMGB1 (lanes 1 and 2, respectively), or used a 32P-labeled WT 12-RSS in place of the 23-RSS (lane 3). White bars represent samples containing 32P-labeled WT 23-RSS. Black bars represent samples containing 32P-labeled MHMN 23-RSS (where MHMN refers to a 10 RSS with mutated nonamer and heptamer sequences as described in Materials and Methods).Results are from n=4 experiments per sample.
To test the interaction of the mR1(core) mutants with RAG2 in the absence of DNA, we monitored the ability of gR2(core) (bound to glutathione-linked sepharose resin) to associate with the mR1(core) proteins. As expected, WT mR1(core) bound to gR2(core), and furthermore, each of the mR1(core) mutants (H937A, H942A and E962A) successfully bound to gR2(core) (Figure 6B, lanes 1–4). The RAG1:RAG2 interactions were specific given that no apparent complexes were formed between gst and mR1(core) or between mbp* and gR2(core) (Figure 6B, lanes 5 and 6).
An oligonucleotide capture assay was used to test for formation of the paired complex with the H937A, H942A and E962A mR1(core) mutants. In this assay, the paired complex included HMGB1 and both core RAG proteins, as well as biotinylated 12-RSS and radiolabeled 23-RSS. HMGB1 is necessary for paired complex formation in in vitro assays, presumably by enhancing the interaction between the RAG proteins and the 23-RSS.6,7 The 12-RSS used in the assay was biotinylated on the 5’end of the top strand to facilitate capture of the paired complex with streptavidin-coated paramagnetic particles. The extent of paired complex formation was quantified as the percentage of 32P-labeled 23-RSS captured in the paired complex relative to the total amount added to each sample. With samples containing WT mR1(core), 32P-labeled DNA was most efficiently recovered in the presence of gR2(core), HMGB1, and a 12/23 RSS pair (Figure 6C, lane 5). In comparison, a 5 to 7-fold decrease in the levels of 32P-labeled DNA recovered was the result with samples that either lacked a component [gR2(core) or HMGB1] or with the 32P-labeled 23-RSS replaced with either a 12-RSS or a mutated 23-RSS (Figure 6C, lanes 1–4).
The paired complex was formed at comparable levels with samples containing the H937A, H942A, or E962A mR1(core) mutants (Figure 6C, compare lanes 7, 9, and 11). Additionally, any differences in formation of the single RSS complex between the His mutants versus the E962A mutant (Figure 6A), was not apparent in formation of the respective paired complexes. Notably, the paired complexes appeared to form less efficiently with the three mutant proteins than with WT mR1(core) (Figure 6C, compare lanes 7, 9, and 11 to lane 5). Nevertheless, each mutant formed the paired complex with sequence specificity, as the amount of 32P-labeled DNA recovered was substantially decreased in reactions containing a mutated 23-RSS (Figure 6C, lanes 6, 8, 10). Thus, since the H937A and H942A mutants maintain the ability to associate with RAG2 and specifically bind the RSS, there is likely no major structural perturbation of the mutant proteins.
DISCUSSION
We have shown that murine core RAG1 binds up to four Zn2+ ions, with at least two Zn2+ ions bound to the CTD region. Functional assays of WT core RAG1 with varying Zn2+ stoichiometries demonstrated that this metal ion plays an important auxiliary role in the catalytic activity. In the DNA cleavage assays, the apo-Zn2+ form of core RAG1 showed no detectable activity, while the 1-Zn2+ form showed significantly less DNA cleavage activity than the holo-Zn2+ core RAG1. It is important to note that the partial activity of the 1-Zn2+ form cannot necessarily be attributed to the presence of a particular zinc-binding site that remained intact in these samples. For example, the 1-Zn2+ form may have consisted of a mixed population of core RAG1:Zn2+ stoichiometries, with the active fraction containing more than one distinct zinc-binding site. This is the first study that has directly shown that removal of Zn2+ ions from core RAG1 affects the DNA cleavage activity and contributes to its structural integrity.
Only a single zinc-binding site in core RAG1, the C2H2 ZFB in the central domain, has been previously reported.27 In this previous study no attempt was made to remove Zn2+ ions from core RAG1. Rather, formation of the ZFB site was proposed based on sequence analysis and the measurement of Zn2+ content within a RAG1 fragment.27
Here, we used mutagenesis studies combined with Zn2+ quantitation measurements and DNA cleavage assays to identify zinc-binding sites located in the CTD of RAG1. In summary, we show that mutation of H937, H942, and E962 in both the isolated CTD and in the core reduced the stoichiometry of bound Zn2+, as well as eliminated DNA cleavage activity of the core. Significantly, these are the only residues in the CTD capable of binding Zn2+ ions (including conserved Cys, His, and carboxylate residues) that show these combined characteristics.
We postulate that core RAG1 contains a zinc-binding site coordinated by residues H937 and H942 (the HH site), and that E962 may also directly ligate Zn2+ in the HH site. This role for E962 would not be inconsistent with its coordination of Mg2+ in the DDE active site, since carboxylate residues often serve as bridging ligands between two metal ions.36 In this case, E962 could act as a bridging ligand between the D600/D708 Mg2+ binding site and the H937/H942 Zn2+ binding site. Alternatively, E962 may instead be indirectly linked to the HH site by promoting formation of this zinc site without directly ligating to the Zn2+ ion. Direct or indirect linkage of the HH zinc site to the DDE active site could serve to bring residues into the active site that were previously shown to be necessary for hairpin formation, as discussed below.
RAG1 sequences are annotated for >1000 jawed vertebrate species. The intervening sequence between residues 937–962 is one of the most highly conserved regions in RAG1, with a sequence identity of 100% between human and bull shark sequences.43 In addition, the corresponding H937 and H942 residues are conserved in RAG1-like sequences that have been identified in the genomes of several invertebrate species44,45. Notably, the spacings between the RAG1 residues H937, H942, and E962 bears resemblance to the active sites of some zinc-dependent hydrolases37, including thermolysin, in which the Zn2+ ion is coordinated by two His and one Glu separated by the spacing: H-X3-H-X19-E.46
We propose that formation of the HH zinc site serves in a structural role by orientating key residues in the active site of RAG1. Specifically, RAG1 residues Y935 and K938 have been shown to be critical for hairpin formation, but dispensable for nicking.47 These residues are proposed to lie within a modified YREK site, with Y935 and K938 analogous to a Tyr and an Arg residue, respectively, present in the YREK sites of IS4 transposases.47 Crystal structures of Tn5 transposase, one of the IS4 transposases, show both the Tyr and Arg sidechains positioned in the DDE motif active site.48 However, in the IS4 transposases, the Tyr is 7 and the Arg is 4 residues N-terminal to the active site Glu. In contrast, there is a 23 residue insertion between K938 and the active site E962 residue. Notably, in the crystal structures of other DDE motif enzymes, residues that are at similar spacings (>20 residues N-terminal) to the E of the DDE motif are quite distant from the active site.16,48,49 Since in RAG1, H937 is flanked on either side by Y935 and K938, formation of the HH zinc site near E962 would help to position these residues in the active site. Moreover, the HH zinc site may in turn serve to optimally position E962 in the active site, since mutation of each of the zinc ligands resulted in severe defects in both nicking and hairpin formation.
In summary, we have shown the importance of zinc binding to the structure and function of core RAG1. Further, we have shown the presence of previously unknown zinc binding sites, one of which is essential for DNA cleavage activity. Together these results provide new insights into RAG1, and highlight characteristics of this enzyme that are distinct from other DDE motif enzymes. In future studies, a high resolution structure of core RAG1 will be useful in 1) determining the ligands to all of the zinc-binding sites in core RAG1, 2) showing the proximity of the HH zinc site to the DDE active site, and 3) showing if E962 directly coordinates to the Zn2+ ion in the HH zinc site.
MATERIALS AND METHODS
Fusion Protein Cloning, Expression, and Purification
The fusion proteins consisting of the core region, the C-terminal domain (CTD), and the central domain (CD) of RAG1 fused to the C-terminal end of maltose binding protein (mbp) were encoded by pCJM233, pJLA1, and pRS3 respectively.18,24 The fusion proteins are referred to as mR1(core), mR1(CTD), or mR1(CD) depending on the RAG1 fragment that is fused to mbp. The fusion proteins were expressed in Escherichia coli BL21 cells and purified as previously described.24 The protein mbp*, which consists of mbp plus the peptide linker present in the fusion proteins, was expressed and purified as reported.23 mR1(core) and mR1(CTD) mutants were generated from pCJM233 and pJLA1, respectively, using the QuikChangeTM Site-Directed Mutagenesis Kit (Stratagene) and following the manufacturer’s protocols. To confirm that only the residue(s) of interest was modified in each mutant, the entire region of the gene corresponding to the RAG1 coding sequence was examined by DNA sequencing. Some samples of mR1(core) that were purified contained either a Factor X or PreScission Protease site in the linker region between mbp and R1(core). The presence of these sites in the purified protein yielded similar results in activity assays and therefore were used interchangeably.
The concentration of purified RAG1 proteins used in this study was determined by the absorbance at 280 nm using extinction coefficients (calculated from the number of Trp and Tyr residues in each fusion protein) of 95 mM−1 cm−1, 129.5 mM−1cm−1, and 81 mM−1cm−1 for mR1(CTD), mR1(core), and mR1(CD), respectively. Amino acid analyses were performed on each fusion protein to confirm the values used for the extinction coefficients (performed at OUHSC Molecular Biology Proteomics Facility http://wmriokc001.ouhsc.edu/mbrf.htm). For each fusion protein, the concentrations determined by UV spectroscopy and amino acid analysis were within 10%.
The core region of RAG2 (residues 1–387 of the full-length 527 residue murine protein) was produced as a C-terminal fusion protein to glutathione S-transferase (gst), and is referred to as gR2(core). This protein was expressed by transient transfection in 293T cells and purified as previously described.21,50
Zinc Quantitation Methods
The concentration of Zn2+ ions in different protein samples was determined by one of two separate methods including flame atomic absorption spectroscopy (FAAS) and inductively coupled plasma mass spectrometry (ICP-MS). FAAS was performed using a Varian SpectrAA-5 spectrophotometer. Solutions of known Zn2+ concentrations (Sigma) were used to define a standard calibration curve. The concentration of Zn2+ was determined by measurement of absorbance values at 213.9 nm after vaporization/atomization of each sample in an air/acetylene-fueled flame. In ICP-MS, performed at Oxidor Environmental Services (Plano, TX), samples were acid hydrolyzed and atomized/ionized with subsequent peak integration of the mass/charge (m/e) MS peaks assigned to Zn2+ isotopes. In both methods, the Zn2+ concentration of the buffer was also quantitated and subsequently subtracted as background.
Oligonucleotide Substrates for DNA cleavage and Binding Assays
The 59-base sequence of the ‘top’ strand (the strand that contains the CACAGTG and ACAAAAACC heptamer and nonamer sequence, respectively) of the WT 12-RSS is previously reported.29 The 60-base sequence of the WT 23-RSS top strand is: GGCTCGTCTTACACAGTGATGGAAGCTCTATCGGATCTCCGACAAAAACCTGAGCGGAG. The 60-base sequence of the mutant heptamer mutant nonamer (MHMN) 23-RSS oligonucleotide sequence is identical to the WT 12-RSS except the sequence GAGAAGC replaced the WT heptamer sequence (CACAGTG) and the sequence AGGCTCTGA has replaced the WT nonamer sequence (ACAAAAACC). Oligonucleotides were commercially synthesized and PAGE-purified (Integrated DNA Technologies). The double-stranded (ds) 12-RSS and 23-RSS substrates were prepared by annealing the top strands with their respective complementary oligonucleotides. The ds 12-RSS and ds MHMN 23-RSS substrates were labeled with 32P at the 5’-end of the top strand using γ-32P-ATP and T4 polynucleotide kinase.
Preparation of Zn2+ containing R1(core) of differing stoichiometries
First, stock solutions of Tris pH 8.0, and NaCl were rendered metal free by passage over a Chelex-100 column. Samples of mR1(core) containing 3–4 Zn2+ ions (referred to as holo-Zn2+ mR1(core)) were prepared by dialyzing purified protein into 400 mL of metal free buffer (20 mM Tris, pH 8.0, 200 mM NaCl, and 5 mM β-mercaptoethanol) overnight at 4°C and was dialyzed for an additional 16 hours after changing to fresh dialysis buffer. Samples of mR1(core) containing ~1 Zn2+ ion (referred to as the 1-Zn2+ form) were prepared by dialysis into 400 mL of 10 mM EDTA buffer (20 mM Tris pH 8.0, 200 mM NaCl, 5 mM β-mercaptoethanol, and 10 mM EDTA) overnight at 4°C. The protein was then dialyzed into metal free buffer for an additional 16 hours with one change into fresh dialysis buffer. Apo-Zn2+ mR1(core) was prepared by dialysis into 400 mL of DTPA-containing buffer (20 mM Tris, pH 8.0, 200 mM NaCl, 5 mM β-mercaptoethanol, and 5 mM DTPA) overnight at 4°C. DTPA was removed by passage over a Q-sepharose ion exchange column. Typical protein concentrations after dialysis preparation were in the low µM range (2–4 µM) and were subsequently diluted ~300-fold for use in DNA cleavage assays. mR1(core) activity was tested with protein fresh from dialysis.
Size Exclusion Chromatography
SEC was accomplished using a 20 mL Superdex 200 gel filtration column (at room temperature) combined in-line with an Optilab DSP interferometric refractometer (Wyatt Technology, Santa Barbara, CA). The column buffer consisted of 20 mM Tris pH 8.0, 0.2 M NaCl, and 5 mM β-mercaptoethanol. Initial protein concentrations were ~2 µM, respectively. Peak integrations were calculated using Astra 4.72 software.
Single RSS Cleavage Assay
Single RSS assays were performed in 10 mM Tris, pH 8.0, 2 mM dithiothreitol, 6% glycerol, and 100 mM NaCl with the addition of either 5 mM MnCl2 or MgCl2 as appropriate (total volume of each reaction at 10 µl). The RAG proteins, mR1(core) and gR2(core) (see figure legends for concentrations) were incubated together for 30 minutes at 4°C, followed by addition of 1 nM 32P-labeled ds WT 12-RSS. Subsequently, the reaction was incubated for the indicated times at 37°C, an equal volume of formamide gel loading buffer was added to stop the reaction, and the samples were heated at 95°C for 5 minutes. DNA products were resolved on 10% denaturing polyacrylamide gels and visualized by autoradiography.
Coupled Cleavage Assay
Coupled cleavage assays were performed in the same buffer (with 5 mM MgCl2) as the single RSS cleavage assay. mR1(core) and gR2(core), each at 0.04 µM, and 0.3 µM HMGB1 (Sigma) were incubated together at 4°C for 30 minutes, followed by addition of 5nM ds WT 23-RSS and 1 nM 32P-labeled ds WT 12-RSS. The remaining procedure was the same as described above for the single RSS cleavage assay.
Extrachromosomal Plasmid V(D)J Recombination Assays
Plasmids encoding the mR1(core) H937A, H942A, and E962A mutants were derived from a plasmid encoding WT mR1(core) (provided by Patrick Swanson) using QuikChangeTM Site-Directed Mutagenesis Kit (Stratagene) following the manufacturer’s protocols. Mutations were confirmed by DNA sequencing of the entire core RAG1 gene. Fugene 6 transfection reagent (Roche) was used to transiently transfect 293T cells (at 60% confluency in 15 10-cm dishes) with 5µg RAG1 expression vector, 5µg RAG2 expression vector (encoding for gR2(core)) and the 5 µg recombination substrate pSF299 (provided by Sebastian Fugmann). After 48 hours, plasmid DNA was recovered and subjected to PCR amplification (24 cycles for each sample) to detect both coding and signal joints in rearranged pSF299, as previously described.51 PCR products were separated on a 6% polyacrylamide gel and detected with Sybr Green (Invitrogen).
Electrophoretic Mobility Shift Assays
mR1(core), at 0.04 µM, was incubated with 1 nM 32P-labeled ds 12-RSS, in the absence or presence of 0.005 µM gR2(core), and the protein-DNA complexes were resolved on a discontinuous 3.5/8% nondenaturing polyacrylamide gel, as previously described.23 The binding buffer contained 10 mM Tris, pH 8.0, 2 mM dithiothreitol, 6% glycerol, 100 mM NaCl and 5 mM CaCl2.
RAG1 and RAG2 Binding Assay
Glutathione-Sepharose 4B resin (Amersham Pharmacia Biotech) was blocked with 1mg/ml bovine serum albumin in buffer A (20 mM Tris-HCl, pH 8.0, 0.2M NaCl, 10% glycerol, 10µM ZnCl2, and 5mM β-mercaptoethanol) for 45 min at 4°C. The resin was washed three times with buffer A, followed by the addition of the appropriate mbp fusion protein (at 0.1 µM) plus gR2(core) or gst (at 0.15 µM) to the resin. The samples and resin were incubated for 1 h at 4°C. After three washes with buffer A, the bound protein was eluted from the resin with SDS loading buffer. Subsequently, the proteins in each sample were resolved by SDS-PAGE (10% polyacrylamide gel) and electrotransferred to a polyvinylidene difluoride (PVDF) membrane. Western blotting analysis was used to detect both mbp and gst fusion proteins as previously described.24
Isolation of Paired Complex
mR1(core) and gR2(core), each at 0.2 µM, and 0.3 µM HMGB1 were incubated together at 4°C for 15 minutes in buffer B (10 mM Tris, pH 8.0, 100 mM NaCl, 5 mM CaCl2, 2 mM dithiothreitol and 6% glycerol) followed by incubation with 10 nM biotinylated ds WT 12-RSS (biotinylated at the 5’end, Integrated DNA Technologies) and 1 nM 32P-labeled ds 23-RSS (either WT or MHMN) for 30 minutes at 25°C. Streptavidin-coated paramagnetic particles (Promega) were used to capture the protein-DNA complexes via the biotin tag of ds WT 12-RSS.
Following incubation of the particles with the protein-DNA complexes for 20 min. at 25°C, the sample was fractionated under a strong magnetic field via a magnetic separation stand (Promega). Subsequently, the magnetic particles were washed with buffer B and the bound complex eluted with SDS loading buffer. The amount of 32P incorporated in the complex was quantitated by liquid scintillation counting using a TRI-CARB 2300 TR Liquid Scintillation Analyzer (Packard Instrument Company; Meriden, CT). The percentage of 32P-labeled DNA in the captured protein-DNA complexes was calculated by dividing counts of 32P incorporated into the captured complex by the total counts added to each sample.
ACKNOWLEDGEMENTS
The authors wish to thank Matthew Junker for assistance with the atomic absorption measurements as well as for helpful comments and suggestions. We also thank SWOSU for use of their instrument laboratory. This work was supported by National Institutes of Health grant AI-054467 to KKR. LMG was supported by National Institutes of Health award F32 AI069784, and MMP was supported by NIH Training Grant T32 AI07633.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 2.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 3.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 4.Curry JD, Geier JK, Schlissel MS. Single-strand recombination signal sequence nicks in vivo: evidence for a capture model of synapsis. Nat Immunol. 2005;6:1272–1279. doi: 10.1038/ni1270. [DOI] [PubMed] [Google Scholar]
- 5.Jones JM, Gellert M. Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. Embo J. 2002;21:4162–4171. doi: 10.1093/emboj/cdf394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawchuk DJ, Weis-Garcia F, Malik S, Besmer E, Bustin M, Nussenzweig MC, Cortes P. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J Exp Med. 1997;185:2025–2032. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Gent DC, Hiom K, Paull TT, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. Embo J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.West RB, Lieber MR. The RAG-HMG1 complex enforces the 12/23 rule of V(D)J recombination specifically at the double-hairpin formation step. Mol Cell Biol. 1998;18:6408–6415. doi: 10.1128/mcb.18.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eastman QM, Schatz DG. Nicking is asynchronous and stimulated by synapsis in 12/23 rule-regulated V(D)J cleavage. Nucleic Acids Res. 1997;25:4370–4378. doi: 10.1093/nar/25.21.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu K, Lieber MR. The nicking step in V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol Cell Biol. 2000;20:7914–7921. doi: 10.1128/mcb.20.21.7914-7921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 12.Fugmann SD, Villey IJ, Ptaszek LM, Schatz DG. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- 13.Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haren L, Ton-Hoang B, Chandler M. Integrating DNA: transposases and retroviral integrases. Annu Rev Microbiol. 1999;53:245–281. doi: 10.1146/annurev.micro.53.1.245. [DOI] [PubMed] [Google Scholar]
- 16.Rice PA, Baker TA. Comparative architecture of transposase and integrase complexes. Nat Struct Biol. 2001;8:302–307. doi: 10.1038/86166. [DOI] [PubMed] [Google Scholar]
- 17.Nagawa F, Ishiguro K, Tsuboi A, Yoshida T, Ishikawa A, Takemori T, Otsuka AJ, Sakano H. Footprint analysis of the RAG protein recombination signal sequence complex for V(D)J type recombination. Mol Cell Biol. 1998;18:655–663. doi: 10.1128/mcb.18.1.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodgers KK, Villey IJ, Ptaszek L, Corbett E, Schatz DG, Coleman JE. A dimer of the lymphoid protein RAG1 recognizes the recombination signal sequence and the complex stably incorporates the high mobility group protein HMG2. Nucleic Acids Res. 1999;27:2938–2946. doi: 10.1093/nar/27.14.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akamatsu Y, Oettinger MA. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol Cell Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Difilippantonio MJ, McMahan CJ, Eastman QM, Spanopoulou E, Schatz DG. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 21.Spanopoulou E, Zaitseva F, Wang FH, Santagata S, Baltimore D, Panayotou G. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 22.Ciubotaru M, Ptaszek LM, Baker GA, Baker SN, Bright FV, Schatz DG. RAG1-DNA binding in V(D)J recombination. Specificity and DNA-induced conformational changes revealed by fluorescence and CD spectroscopy. J Biol Chem. 2003;278:5584–5596. doi: 10.1074/jbc.M209758200. [DOI] [PubMed] [Google Scholar]
- 23.Godderz LJ, Rahman NS, Risinger GM, Arbuckle JL, Rodgers KK. Self-association and conformational properties of RAG1: implications for formation of the V(D)J recombinase. Nucleic Acids Res. 2003;31:2014–2023. doi: 10.1093/nar/gkg281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arbuckle JL, Fauss LA, Simpson R, Ptaszek LM, Rodgers KK. Identification of two topologically independent domains in RAG1 and their role in macromolecular interactions relevant to V(D)J recombination. J Biol Chem. 2001;276:37093–37101. doi: 10.1074/jbc.M105988200. [DOI] [PubMed] [Google Scholar]
- 25.De P, Rodgers KK. Putting the pieces together: identification and characterization of structural domains in the V(D)J recombination protein RAG1. Immunol Rev. 2004;200:70–82. doi: 10.1111/j.0105-2896.2004.00154.x. [DOI] [PubMed] [Google Scholar]
- 26.Yin FF, Bailey S, Innis CA, Ciubotaru M, Kamtekar S, Steitz TA, Schatz DG. Structure of the RAG1 nonamer binding domain with DNA reveals a dimmer that mediates DNA synapsis. Nat. Struct. Mol. Biol. 2009 doi: 10.1038/nsmb.1593. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodgers KK, Bu Z, Fleming KG, Schatz DG, Engelman DM, Coleman JE. A zinc-binding domain involved in the dimerization of RAG1. J Mol Biol. 1996;260:70–84. doi: 10.1006/jmbi.1996.0382. [DOI] [PubMed] [Google Scholar]
- 28.Aidinis V, Dias DC, Gomez CA, Bhattacharyya D, Spanopoulou E, Santagata S. Definition of minimal domains of interaction within the recombination-activating genes 1 and 2 recombinase complex. J Immunol. 2000;164:5826–5832. doi: 10.4049/jimmunol.164.11.5826. [DOI] [PubMed] [Google Scholar]
- 29.Peak MM, Arbuckle JL, Rodgers KK. The central domain of core RAG1 preferentially recognizes single-stranded recombination signal sequence heptamer. J Biol Chem. 2003;278:18235–18240. doi: 10.1074/jbc.M302041200. [DOI] [PubMed] [Google Scholar]
- 30.Aidinis V, Bonaldi T, Beltrame M, Santagata S, Bianchi ME, Spanopoulou E. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol Cell Biol. 1999;19:6532–6542. doi: 10.1128/mcb.19.10.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao S, Gwyn LM, De P, Rodgers KK. A Non-Sequence-Specific DNA Binding Mode of RAG1 Is Inhibited by RAG2. J Mol Biol. 2009;387:744–758. doi: 10.1016/j.jmb.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santagata S, Aidinis V, Spanopoulou E. The effect of Me2+ cofactors at the initial stages of V(D)J recombination. J Biol Chem. 1998;273:16325–16331. doi: 10.1074/jbc.273.26.16325. [DOI] [PubMed] [Google Scholar]
- 33.Bellon SF, Rodgers KK, Schatz DG, Coleman JE, Steitz TA. Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nat Struct Biol. 1997;4:586–591. doi: 10.1038/nsb0797-586. [DOI] [PubMed] [Google Scholar]
- 34.De P, Zhao S, Gwyn LM, Godderz LJ, Peak MM, Rodgers KK. Thermal dependency of RAG1 self-association properties. BMC Biochem. 2008;9:5. doi: 10.1186/1471-2091-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaberek S, Martell AE. Organic Sequestering Agents. New York: John Wiley & Sons, Inc; 1959. [Google Scholar]
- 36.Auld DS. Zinc coordination sphere in biochemical zinc sites. Biometals. 2001;14:271–313. doi: 10.1023/a:1012976615056. [DOI] [PubMed] [Google Scholar]
- 37.Maret W. Exploring the zinc proteome. J Anal At Spectrom. 2004;19:15–19. [Google Scholar]
- 38.Huye LE, Purugganan MM, Jiang MM, Roth DB. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Mol Cell Biol. 2002;22:3460–3473. doi: 10.1128/MCB.22.10.3460-3473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ko JE, Kim CW, Kim DR. Amino acid residues in RAG1 responsible for the interaction with RAG2 during the V(D)J recombination process. J Biol Chem. 2004;279:7715–7720. doi: 10.1074/jbc.M311471200. [DOI] [PubMed] [Google Scholar]
- 40.Fugmann SD, Schatz DG. Identification of basic residues in RAG2 critical for DNA binding by the RAG1-RAG2 complex. Mol Cell. 2001;8:899–910. doi: 10.1016/s1097-2765(01)00352-5. [DOI] [PubMed] [Google Scholar]
- 41.Lagunavicius A, Siksnys V. Site-directed mutagenesis of putative active site residues of MunI restriction endonuclease: replacement of catalytically essential carboxylate residues triggers DNA binding specificity. Biochemistry. 1997;36:11086–11092. doi: 10.1021/bi963125i. [DOI] [PubMed] [Google Scholar]
- 42.Feng M, Patel D, Dervan JJ, Ceska T, Suck D, Haq I, Sayers JR. Roles of divalent metal ions in flap endonuclease-substrate interactions. Nat Struct Mol Biol. 2004;11:450–456. doi: 10.1038/nsmb754. [DOI] [PubMed] [Google Scholar]
- 43.Bernstein RM, Schluter SF, Bernstein H, Marchalonis JJ. Primordial emergence of the recombination activating gene 1 (RAG1): sequence of the complete shark gene indicates homology to microbial integrases. Proc Natl Acad Sci U S A. 1996;93:9454–9459. doi: 10.1073/pnas.93.18.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fugmann SD, Messier C, Novack LA, Cameron RA, Rast JP. An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci U S A. 2006;103:3728–3733. doi: 10.1073/pnas.0509720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holmes MA, Matthews BW. Structure of thermolysin refined at 1.6 A resolution. J Mol Biol. 1982;160:623–639. doi: 10.1016/0022-2836(82)90319-9. [DOI] [PubMed] [Google Scholar]
- 47.Lu CP, Sandoval H, Brandt VL, Rice PA, Roth DB. Amino acid residues in Rag1 crucial for DNA hairpin formation. Nat Struct Mol Biol. 2006;13:1010–1015. doi: 10.1038/nsmb1154. [DOI] [PubMed] [Google Scholar]
- 48.Steiniger-White M, Rayment I, Reznikoff WS. Structure/function insights into Tn5 transposition. Curr Opin Struct Biol. 2004;14:50–57. doi: 10.1016/j.sbi.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 49.Hickman AB, Perez ZN, Zhou L, Musingarimi P, Ghirlando R, Hinshaw JE, Craig NL, Dyda F. Molecular architecture of a eukaryotic DNA transposase. Nat Struct Mol Biol. 2005;12:715–721. doi: 10.1038/nsmb970. [DOI] [PubMed] [Google Scholar]
- 50.Rahman NS, Godderz LJ, Stray SJ, Capra JD, Rodgers KK. DNA cleavage of a cryptic recombination signal sequence by RAG1 and RAG2. Implications for partial V(H) gene replacement. J Biol Chem. 2006;281:12370–12380. doi: 10.1074/jbc.M507906200. [DOI] [PubMed] [Google Scholar]
- 51.Jung D, Bassing CH, Fugmann SD, Cheng HL, Schatz DG, Alt FW. Extrachromosomal recombination substrates recapitulate beyond 12/23 restricted VDJ recombination in nonlymphoid cells. Immunity. 2003;18:65–74. doi: 10.1016/s1074-7613(02)00507-1. [DOI] [PubMed] [Google Scholar]