Abstract
Clostridial botulinum neurotoxins (BoNTs) inhibit synaptic exocytosis; intoxication requires the di-chain protein to undergo conformational changes in response to pH and redox gradients across the endosomal membrane with consequent formation of a protein-conducting channel by the heavy chain (HC) that translocates the light chain (LC) protease into the cytosol, colocalizing it with the substrate SNARE proteins. We investigate the dynamics of protein-translocation across membranes using a sensitive single molecule assay to track translocation events with millisecond resolution on lipid bilayers and on membrane patches of Neuro 2A cells. Translocation of BoNT/A LC by the HC is observed in real time as changes of channel conductance: the channel is occluded by the light chain during transit, and open after completion of translocation and release of cargo, acting intriguingly similar to the protein-conducting/translocating channels of the endoplasmic reticulum, mitochondria, and chloroplasts. Our findings support the notion of an interdependent, tight interplay between the HC transmembrane chaperone and the LC cargo that prevents LC aggregation and dictates the productive passage of cargo through the channel and completion of translocation. The protein-conducting channel of BoNT, a key element in the process of neurotoxicity, emerges therefore as a target for antidote discovery –a novel paradigm of paramount significance to health science and biodefense.
Introduction
“The dose makes the poison”. This statement, attributed to Paracelsus, the father of toxicology (1493–1541), is embodied by botulinum neurotoxins (BoNTs). On one hand BoNTs are the most poisonous toxins known and the causative agent of botulism; on the other, they are a wonderful drug. Based on their exquisitely powerful neuroparalytic activity, BoNTs have gained tremendous popularity in the past few years as they became the first biological toxin (BoNT serotype A) to receive FDA approval for the treatment of human disease, most notably for dystonias, blepharospasm, cervical torticollis, and strabismus, though it has been elevated to the practical rank of “panacea” for a rapidly growing list of conditions involving muscle spasm, not the least its cosmeceutical blockbuster role. Alarmingly, BoNT is a most feared bioweapon still without an antidote (Arnon et al., 2001).
The targets of the BoNT proteases are their cytosolic SNARE (soluble NSF attachment protein receptor) substrates (Rossetto and Montecucco, 2008; Schiavo et al., 2000; Simpson, 2004). The SNARE proteins form a coil-coil which underlies the assembly of the synaptic fusion core complex required for synaptic vesicle exocytosis. Cleavage of the SNARE components by BoNTs prior to SNARE complex assembly, therefore, abrogates membrane fusion and neurotransmitter release. The mechanism underlying the translocation of the endocytosed BoNT protease from acidic endosomes into the cytosol is now emerging based on the convergence of single molecule studies, high resolution structures and biochemical knowledge. It is clear that BoNT is a modular nanomachine (Chai et al., 2006; Jin et al., 2006; Lacy et al., 1998; Swaminathan and Eswaramoorthy, 2000) in which one of its modules – the heavy chain channel – operates as a specific protein-translocating transmembrane chaperone for another of its component modules – the light chain protease (Fischer et al., 2008; Fischer and Montal, 2006; Fischer and Montal, 2007a; Fischer and Montal, 2007b; Hoch et al., 1985; Koriazova and Montal, 2003; Lacy et al., 1998). The time is ripe to integrate the new in-depth analysis of protein translocation focusing on understanding the intimate relationship between the cargo protease and the protein-conducting channel. The intricate interplay between these two entities is at the root of BoNT action. An in-depth understanding of protein translocation by BoNT (Fischer et al., 2008; Fischer and Montal, 2006; Fischer and Montal, 2007a; Fischer and Montal, 2007b; Koriazova and Montal, 2003) is, therefore, of utmost significance; the progress towards this goal is precisely what we aim to review here.
Protein translocation across membranes is not only a central problem in biology (Rapoport, 2007), but for BoNT, it constitutes a crucial step in the intoxication process and a path for antidote discovery (Rossetto and Montecucco, 2008; Schiavo et al., 2000; Simpson, 2004). The discovery that the heavy chain (HC) of BoNT/A acts as both a protein-conducting channel and a transmembrane chaperone for the light chain (LC) embodied a paradigm shift by providing compelling evidence that the HC is essential to allow translocation of an active LC (Koriazova and Montal, 2003). BoNT exploits its modular design to achieve its potent toxicity which relies on the transmembrane chaperone activity of one of its modules (the HC protein-conducting channel, depicted in orange in Figure 1) to protect the conformation of its LC cargo during transit and ensure its proper refolding at the completion of translocation (Fischer et al., 2008; Fischer and Montal, 2006; Fischer and Montal, 2007a; Fischer and Montal, 2007b; Koriazova and Montal, 2003).
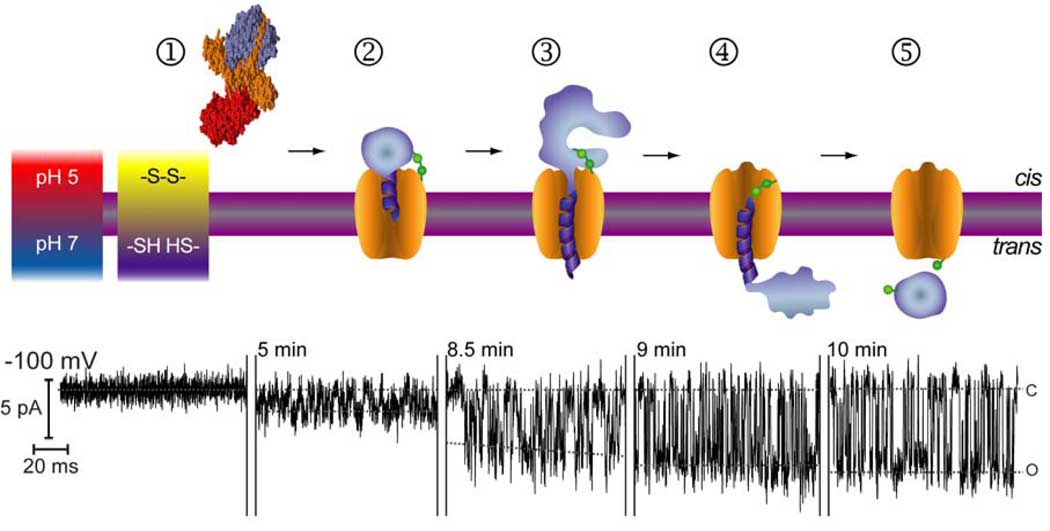
Figure 1.
Sequence of events underlying BoNT LC translocation through the HC channel. ➀ BoNT/A holotoxin prior to insertion in the membrane (grey bar with magenta boundaries); BoNT/A is represented by the crystal structure rendered on YASARA (www.YASARA.org) using Protein Data Bank accession code 3BTA (Lacy et al., 1998). Then, schematic representation of the membrane inserted BoNT/A during an entry event (➁), a series of transfer steps (➂, ➃) and an exit event (➄), under conditions that recapitulate those across endosomes. Segments of a typical record are displayed under the corresponding interpretation for each step. Modified and reproduced with permission from reference (Fischer and Montal, 2007b). Copyright (2007), National Academy of Sciences, U.S.A.
Although the beginning and the end of translocation are known, the translocation process remains largely unknown. The goal is to get insights into the protein-protein interactions between the cargo and the channel that dictate the outcome of translocation: productive passage of cargo or abortive channel occlusion by cargo. An immediate objective is to correlate the occurrence of translocation intermediates with different LC conformers during transit through the HC channel using a single-molecule translocation assay in lipid bilayers and in neuronal cells. The advent of the translocation assay, combined with sensitive assays for the LC catalytic activity, has opened new dimensions for investigation of the conformational transitions of HC and LC, the pivotal role of pH, redox and electrical gradients across endosomes, and the requirements for LC refolding and release after translocation.
Single-molecule translocation assay
Our group’s translocation assay has to date provided us exclusive capability to address questions at single-molecule level of resolution (Fischer et al., 2008; Fischer and Montal, 2006; Fischer and Montal, 2007a; Fischer and Montal, 2007b; Koriazova and Montal, 2003). Translocation of BoNT/A light chain by the heavy chain was observed in real time in excised patches of BoNT/A-sensitive Neuro 2A neuroblastoma cells as a progressive increase of channel conductance: the HC channel is transiently occluded by the LC during transit, then unoccluded after completion of translocation and release of cargo. This process is stringently dependent on conditions that mimic those prevalent across endosomes: a pH gradient - acidic on the inside and neutral on the cytosol; a redox gradient - oxidizing on the inside and reducing on the cytosol; and a transmembrane potential, positive inside.
Hypothesis
Figure 1 depicts a model consistent with the findings: a sequence of events underlying BoNT LC translocation through the HC channel (Fischer and Montal, 2007b). Step 1 shows BoNT/A prior to insertion into the membrane (grey bar with magenta boundaries) represented by the crystal structure rendered on YASARA (www.YASARA.org) using Protein Data Bank, accession code 3BTA (Lacy et al., 1998): LC is purple, translocation domain (TD) is orange, and receptor binding domain (RBD) is red. Then is shown a schematic representation of BoNT/A inserted in the membrane during an entry event (step 2) with LC (purple) trapped within the HC channel (orange). A series of transfer steps (steps 3 and 4), and an exit event (step 5), are shown. Segments of a typical record are displayed under the corresponding interpretation for each step. The disulfide bridge (green) between LC and HC is intact in the low pH, oxidizing environment of the cis-compartment. The presence of reductant and neutral pH in the trans-compartment promotes release of LC from HC after completion of translocation. We interpret the progressive, stepwise increase in single channel conductance (γ) as the progress of LC translocation, during which the HC channel initially conducts Na+ and partially unfolded LC (illustrated as a helix in Step 2, Figure 1) detected as channel block. After translocation is complete, the channel is unoccluded, attaining a higher, constant γ (Figure 1-Step 5). This is consistent with the γ ≈ 66 pS, a signature of the HC channel (Fischer and Montal, 2006; Koriazova and Montal, 2003), and is characteristic of holotoxin channels after completion of a single translocation event (Figure 1-Step 5) (Fischer and Montal, 2007a; Fischer and Montal, 2007b). The half-time for completion of such an event, estimated from the abrupt transition from low to high conductance, is ~10 s with an initial γ ≈ 13 pS and a final, constant γ ≈ 67 pS characteristic of the unoccluded HC after event completion (Fischer et al., 2008; Fischer and Montal, 2006; Fischer and Montal, 2007a; Fischer and Montal, 2007b; Koriazova and Montal, 2003).
Is translocation affected by a LC or a TD that is tightly bound to a folded protein?
To discern the underlying protein-protein interactions between BoNT/A LC and HC during translocation, we exploited the specificity of several monoclonal antibodies (generated by the Marks Laboratory at the University of California San Francisco) that bind with high affinity to individual modules of BoNT/A or BoNT/E (Table 1). We conjectured that the HC may insert into the membrane and form channels; however, the bound LC-antibody “cargo” might perturb or even arrest translocation. To test this model, Fab fragments were tested on the translocation assay. A Fab specific for BoNT/A LC (ING2, Table 1) induces persistent block of the HC channel. Indeed, Fab binding to the LC allows channel formation and early translocation, however it locks both the channel and the LC in a translocating conformation that is irreversibly incomplete (Fischer et al., 2008; Fischer and Montal, 2007a; Fischer and Montal, 2007b). Next, we asked if mAb 3E6.1 or mAb 4E17.1, selective for BoNT/E TD (Table 1) affect the translocation step. The epitope of 3E6.1 maps, by sequence similarity to BoNT/A, to an extended region parallel to the helical bundle of the TD, whereas the epitope of 4E17.1 maps to the loop at one end of the TD (Fischer et al., 2008). Preincubation of BoNT/E with 3E6.1 or 4E17.1 results in no channel activity during the minimum experimental time of 30 min (n=24 experiments for each condition)(Fischer et al., 2008). Thus, these Fabs bind to the BoNT/E TD with sub nM affinity (Table 1), precluding its insertion into the membrane and selectively disrupting BoNT translocation (Fischer et al., 2008). These Fabs emerge as powerful tools to gain insights into the mechanism of LC translocation (Fischer et al., 2008). As summarized, folding on the cis-side prevents translocation (Fischer and Montal, 2007b), but does refolding on the trans-side promote translocation? This issue is unsolved and needs further study which we are currently exploring by using domain-specific antibodies (Garcia-Rodriguez et al., 2007) and conditions that may promote or accelerate the refolding of the isolated LC in vitro. It will be interesting to visualize structural changes of the HC channel with the LC cargo under conditions that imitate those across endosomes by pursuing the strategy outlined here using a set of serotype-specific (BoNT/A or BoNT/E) and domain-specific (LC or TD) antibodies.
Table 1.
Specificity of mAbs against BoNT/A or BoNT/E
| Antibody | BoNT | Epitope (Residue Number) |
KD (pM) |
|---|---|---|---|
| ING2 | A | LC | 9.6 |
| 4E16.1 | E | LC (around 142) | 3.4 |
| 4E17.1 | E | TD (752–759) | 240 |
| 3E6.1 | E | TD (573–579) | 40 |
Reproduced with permission from reference (Fischer et al., 2008).Copyright, (2008), American Society for Biochemistry and Molecular Biology, U.S.A.
Is translocation coupled to the maintenance of an unfolded conformation of the cargo during passage through the channel?
The translocation domain “belt” is a most intriguing structural feature in the high resolution structures of both BoNT/A (Lacy et al., 1998) and B (Swaminathan and Eswaramoorthy, 2000): It is a ~50-residue-long (amino acids 492–545 in BoNT/A) unstructured loop that wraps around the catalytic domain in the structures solved at pH 7.0 and 6.0, respectively. Given the fact that the belt surrounds the LC, it may facilitate or coordinate the concerted partial unfolding of the LC at acid pH and direct the beginning of its translocation through the HC. A proton gradient has been suggested to drive protein translocation through the anthrax toxin pore which chaperones the unfolded enzymatic moieties (Krantz et al., 2006).
Based on these and other considerations we formulated a hypothesis (Brunger et al., 2007). In essence, we proposed that the belt region of the BoNT HC is a surrogate pseudosubstrate-inhibitor of the LC protease and acts as a chaperone during translocation across the endosomal membrane into the cytosol. The key points are:
The intrinsically unstructured C-terminal domain of SNAP-25 (sn2) adopts partial secondary structure upon binding to the LC in the binary complex crystal structure (Breidenbach and Brunger, 2004) and occupies a similar position as the belt in the holotoxin crystal structures of both BoNT/A (Lacy et al., 1998) and BoNT/B (Swaminathan and Eswaramoorthy, 2000).
In analogy to other “intrinsically unstructured proteins” (IUPs) (Dyson and Wright, 2005), the belt undergoes binding to its LC partner thereby functioning as a chaperone (Koriazova and Montal, 2003).
The belt occupies the exosites, the extensive enzyme surface allocated for substrate binding, yet it does not contain the scissile bond thus potentially inhibiting the LC protease (Chen et al., 2007).
This hypothesis prompts a number of intriguing questions: Is the belt required for channel formation? Is it required for LC translocation? Is the belt the trigger for translocation or a modulator? Are there conformational transitions upon entering the acidic environment of the endosome? Is it practical to probe the pseudo-substrate role of the belt by designing possible substrates based on synthetic peptides which mimic the amino acid sequence of the belt yet incorporate SNAP-25 residues present at the toxin cleavage site. What is the activity of chimeras of BoNT/A and BoNT/E which may differ in the belt sequence? (Foster et al., 2006; Wang et al., 2008). The implication is that the belt region of BoNT/A HC must be subjected to a rigorous structural and functional analysis to evaluate its possible role in the translocation process, in particular with regards to a pH-induced conformational change.
Concluding remarks
From the fundamental viewpoint, BoNT is a modular nanomachine and a marvel of protein design. BoNT represents a fascinating example of molecular partnership: the HC chaperone activity driven by a pH gradient across the endosome which prevents aggregation of the LC in the acidic vesicle interior, maintains the LC unfolded conformation during translocation, and releases the LC after it refolds at the neutral cytosolic pH. In the process, the HC channel is occluded by the LC during transit and open after completion of translocation and release of cargo. In other words, the BoNT HC-LC complex embedded in the membrane is a dynamic structural device that prevents aggregation and achieves translocation of the LC (Fischer and Montal, 2007a; Fischer and Montal, 2007b; Koriazova and Montal, 2003). An analogous mechanism has been invoked for the translocation of the catalytic domains of diphtheria and anthrax toxins (Young and Collier, 2007). While the protease module of BoNT/A (Breidenbach and Brunger, 2004; Lacy et al., 1998; Segelke et al., 2004; Silvaggi et al., 2007), BoNT/B (Hanson and Stevens, 2000), BoNT/C (Jin et al., 2007) BoNT/D (Arndt et al., 2006), BoNT/E (Agarwal et al., 2004), BoNT/F (Agarwal et al., 2005), and BoNT/G (Arndt et al., 2005) and the receptor binding domain of BoNT/B (Chai et al., 2006; Jin et al., 2006) are better understood in terms of the underlying protein structures, no structure is available for the isolated HC of any of the BoNT serotypes. The translocation domain of BoNT/A, which forms channels with properties indistinguishable from those of the HC (unpublished results), was crystallized; however, the crystals were not diffraction quality (Lacy and Stevens, 1997). Therefore, the challenge remains to understand how this protein machine (Alberts, 1998) has evolved to such level of sophistication by exploiting the simplicity of its modular organization and the dynamics emergent from the interplay between its component modules.
Acknowledgments
The research summarized here was conducted in collaboration with Audrey Fischer, Lilia Koriazova, and Myrta Oblatt-Montal. The original publications have been accordingly cited. This work was supported by the Pacific Southwest Regional Center of Excellence NIH AI065359.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The author declares that there are no conflicts of interest.
References
- Agarwal R, Binz T, Swaminathan S. Structural analysis of botulinum neurotoxin serotype F light chain: implications on substrate binding and inhibitor design. Biochemistry. 2005;44:11758–11765. doi: 10.1021/bi0510072. [DOI] [PubMed] [Google Scholar]
- Agarwal R, Eswaramoorthy S, Kumaran D, Binz T, Swaminathan S. Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212-->Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway. Biochemistry. 2004;43:6637–6644. doi: 10.1021/bi036278w. [DOI] [PubMed] [Google Scholar]
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- Arndt JW, Chai Q, Christian T, Stevens RC. Structure of botulinum neurotoxin type D light chain at 1.65 A resolution: repercussions for VAMP-2 substrate specificity. Biochemistry. 2006;45:3255–3262. doi: 10.1021/bi052518r. [DOI] [PubMed] [Google Scholar]
- Arndt JW, Yu W, Bi F, Stevens RC. Crystal structure of botulinum neurotoxin type G light chain: serotype divergence in substrate recognition. Biochemistry. 2005;44:9574–9580. doi: 10.1021/bi0505924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon SS, Schechter R, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Hauer J, Layton M, Lillibridge S, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Swerdlow DL, Tonat K. Botulinum toxin as a biological weapon: medical and public health management. JAMA. 2001;285:1059–1070. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- Breidenbach MA, Brunger AT. Substrate recognition strategy for botulinum neurotoxin serotype A. Nature. 2004;432:925–929. doi: 10.1038/nature03123. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Breidenbach MA, Jin R, Fischer A, Santos JS, Montal M. Botulinum neurotoxin heavy chain belt as an intramolecular chaperone for the light chain. PLoS Pathog. 2007;3:1191–1194. doi: 10.1371/journal.ppat.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Q, Arndt JW, Dong M, Tepp WH, Johnson EA, Chapman ER, Stevens RC. Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature. 2006;444:1096–1100. doi: 10.1038/nature05411. [DOI] [PubMed] [Google Scholar]
- Chen S, Kim JJ, Barbieri JT. Mechanism of substrate recognition by botulinum neurotoxin serotype a. J Biol Chem. 2007;282:9621–9627. doi: 10.1074/jbc.M611211200. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Fischer A, Garcia-Rodriguez C, Geren I, Lou J, Marks JD, Nakagawa T, Montal M. Molecular architecture of botulinum neurotoxin e revealed by single particle electron microscopy. J Biol Chem. 2008;283:3997–4003. doi: 10.1074/jbc.M707917200. [DOI] [PubMed] [Google Scholar]
- Fischer A, Montal M. Characterization of Clostridial botulinum neurotoxin channels in neuroblastoma cells. Neurotox Res. 2006;9:93–100. doi: 10.1007/BF03033926. [DOI] [PubMed] [Google Scholar]
- Fischer A, Montal M. Crucial role of the disulfide bridge between botulinum neurotoxin light and heavy chains in protease translocation across membranes. J Biol Chem. 2007a;282:29604–29611. doi: 10.1074/jbc.M703619200. [DOI] [PubMed] [Google Scholar]
- Fischer A, Montal M. Single molecule detection of intermediates during botulinum neurotoxin translocation across membranes. Proc Natl Acad Sci U S A. 2007b;104:10447–10452. doi: 10.1073/pnas.0700046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KA, Adams EJ, Durose L, Cruttwell CJ, Marks E, Shone CC, Chaddock JA, Cox CL, Heaton C, Sutton JM, Wayne J, Alexander FC, Rogers DF. Re-engineering the target specificity of Clostridial neurotoxins - a route to novel therapeutics. Neurotox Res. 2006;9:101–107. doi: 10.1007/BF03354881. [DOI] [PubMed] [Google Scholar]
- Garcia-Rodriguez C, Levy R, Arndt JW, Forsyth CM, Razai A, Lou J, Geren I, Stevens RC, Marks JD. Molecular evolution of antibody cross-reactivity for two subtypes of type A botulinum neurotoxin. Nat Biotechnol. 2007;25:107–116. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]
- Hanson MA, Stevens RC. Cocrystal structure of synaptobrevin-II bound to botulinum neurotoxin type B at 2.0 A resolution. Nat Struct Biol. 2000;7:687–692. doi: 10.1038/77997. [DOI] [PubMed] [Google Scholar]
- Hoch DH, Romero-Mira M, Ehrlich BE, Finkelstein A, DasGupta BR, Simpson LL. Channels formed by botulinum, tetanus, and diphtheria toxins in planar lipid bilayers: relevance to translocation of proteins across membranes. Proc Natl Acad Sci U S A. 1985;82:1692–1696. doi: 10.1073/pnas.82.6.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Rummel A, Binz T, Brunger AT. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature. 2006;444:1092–1095. doi: 10.1038/nature05387. [DOI] [PubMed] [Google Scholar]
- Jin R, Sikorra S, Stegmann CM, Pich A, Binz T, Brunger AT. Structural and biochemical studies of botulinum neurotoxin serotype C1 light chain protease: implications for dual substrate specificity. Biochemistry. 2007;46:10685–10693. doi: 10.1021/bi701162d. [DOI] [PubMed] [Google Scholar]
- Koriazova LK, Montal M. Translocation of botulinum neurotoxin light chain protease through the heavy chain channel. Nat Struct Biol. 2003;10:13–18. doi: 10.1038/nsb879. [DOI] [PubMed] [Google Scholar]
- Krantz BA, Finkelstein A, Collier RJ. Protein Translocation through the Anthrax Toxin Transmembrane Pore is Driven by a Proton Gradient. J Mol Biol. 2006;355:968–979. doi: 10.1016/j.jmb.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Lacy DB, Stevens RC. Recombinant expression and purification of the botulinum neurotoxin type A translocation domain. Protein Expr Purif. 1997;11:195–200. doi: 10.1006/prep.1997.0772. [DOI] [PubMed] [Google Scholar]
- Lacy DB, Tepp W, Cohen AC, DasGupta BR, Stevens RC. Crystal structure of botulinum neurotoxin type A and implications for toxicity. Nat Struct Biol. 1998;5:898–902. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Rossetto O, Montecucco C. Presynaptic neurotoxins with enzymatic activities. Handb Exp Pharmacol. 2008:129–170. doi: 10.1007/978-3-540-74805-2_6. [DOI] [PubMed] [Google Scholar]
- Schiavo G, Matteoli M, Montecucco C. Neurotoxins affecting neuroexocytosis. Physiol Rev. 2000;80:717–766. doi: 10.1152/physrev.2000.80.2.717. [DOI] [PubMed] [Google Scholar]
- Segelke B, Knapp M, Kadkhodayan S, Balhorn R, Rupp B. Crystal structure of Clostridium botulinum neurotoxin protease in a product-bound state: Evidence for noncanonical zinc protease activity. Proc Natl Acad Sci U S A. 2004;101:6888–6893. doi: 10.1073/pnas.0400584101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvaggi NR, Boldt GE, Hixon MS, Kennedy JP, Tzipori S, Janda KD, Allen KN. Structures of Clostridium botulinum Neurotoxin Serotype A Light Chain complexed with small-molecule inhibitors highlight active-site flexibility. Chem Biol. 2007;14:533–542. doi: 10.1016/j.chembiol.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Simpson LL. Identification of the major steps in botulinum toxin action. Annu Rev Pharmacol Toxicol. 2004;44:167–193. doi: 10.1146/annurev.pharmtox.44.101802.121554. [DOI] [PubMed] [Google Scholar]
- Swaminathan S, Eswaramoorthy S. Structural analysis of the catalytic and binding sites of Clostridium botulinum neurotoxin B. Nat Struct Biol. 2000;7:693–699. doi: 10.1038/78005. [DOI] [PubMed] [Google Scholar]
- Wang J, Meng J, Lawrence GW, Zurawski TH, Sasse A, Bodeker MO, Gilmore MA, Fernandez-Salas E, Francis J, Steward LE, Aoki KR, Dolly JO. Novel chimeras of botulinum neurotoxin /A and /E unveil contributions from the binding, translocation and protease domains to their functional characteristics. J Biol Chem. 2008 doi: 10.1074/jbc.M710442200. [DOI] [PubMed] [Google Scholar]
- Young JA, Collier RJ. Anthrax toxin: receptor binding, internalization, pore formation, and translocation. Annu Rev Biochem. 2007;76:243–265. doi: 10.1146/annurev.biochem.75.103004.142728. [DOI] [PubMed] [Google Scholar]