Abstract
Decreased mitochondrial oxidative phosphorylation (OXPHOS) is one of the hallmarks of cancer. To date the identity of nuclear gene(s) responsible for decreased OXPHOS in tumors remains unknown. It is also unclear whether mutations in nuclear gene(s) responsible for decreased OXPHOS affect tumorigenesis. Polymerase γ (POLG) is the only DNA polymerase known to function in human mitochondria. Mutations in POLG are known to cause mtDNA depletion and decreased OXPHOS resulting in mtDNA depletion syndrome (MDS) in humans. We therefore sequenced all coding exons [2-23] and flanking intron/splice junctions of POLG in breast tumors. We found that the POLG gene was mutated in 63% of the breast tumors. We identified a total of 17 mutations across the POLG gene. Mutations were found in all three domains of POLG protein, including T251I (exonuclease domain), P587L (linker region) and E1143G (polymerase domain). We identified two novel mutations that include one silent (A703A) and one missense (R628Q) mutation in the evolutionarily conserved POLG linker region. Additionally, we identified three novel mutations in the intronic region. Our study also revealed that mtDNA was depleted in breast tumors. Consistently, mutant POLG when expressed in breast cancer cells induced depletion of mtDNA, decreased mitochondrial activity, decreased mitochondrial membrane potential, increased levels of reactive oxygen species (ROS), and increased matrigel invasion. Together, our study provides the first comprehensive analysis of the POLG gene mutation in human cancer and suggests a role for POLG in 1) decreased OXPHOS in cancers and 2) in promoting tumorigenicity.
Keywords: Breast Cancer, POLG, MtDNA, Mitochondria, Mutation, Mitochondrial
INTRODUCTION
Decreased mitochondrial oxidative phosphorylation (OXPHOS) is one of the most common and profound phenotypes of cancer cells 1-10. As early as 1930, the German biochemist Otto Warburg described OXPHOS differences in the mitochondria of tumor versus normal cells 1, 2. He proposed that cancer initiates from irreversible injury to OXPHOS 2. He further proposed that decreased OXPHOS led to an increase rate of aerobic glycolysis in most cancers. This phenomenon is described as the Warburg Effect.
In human cells the OXPHOS system is assembled from 13 mtDNA (mitochondrial DNA) genes and over 85 nDNA (nuclear DNA) genes. The entire mitochondrial genome is devoted to the production of 13 protein subunits of OXPHOS complexes (I, III, IV and V) involved in respiration and ATP synthesis. We investigated the underlying reason for decreased OXPHOS in breast cancer and discovered that more than 40% of primary breast tumors lack detectable expression of cytochrome c-oxidase subunit II (OXPHOS complex IV) encoded by mtDNA 11. Other laboratories have measured mtDNA content directly in tumors and report a decrease in mtDNA content in breast, renal, hepatocellular, gastric and prostate tumors 12-17. Depletion of mtDNA is also supported by a decrease in OXPHOS levels in renal tumors 18. It is also noteworthy that drugs used for treating HIV inhibit POLG, which in turn induces mtDNA depletion 19. Tamoxifen, a commonly used drug for the treatment of breast cancer, also depletes mtDNA 20. A recent study also demonstrates that the depletion of mtDNA correlates with tumor progression and prognosis in breast cancer patients 21. To date the identity of nuclear gene(s) responsible for mtDNA depletion and decreased OXPHOS in tumors remains unknown. It is also unclear whether mutations in nuclear gene(s) involved in mtDNA depletion and decreased OXPHOS affect tumorigenesis.
The first mtDNA depletion syndrome (MDS) was described more than 15 years ago 22. MDS results from mutation(s) in nuclear-encoded genes that participate in mtDNA replication, in mitochondrial nucleotide metabolism and in the nucleotide salvage pathway. So far, only six MDS genes have been identified. These nuclear genes include: mtDNA polymerase gamma (POLG), mtDNA helicase twinkle 23, thymidine kinase 2 (TK2) 24,25 deoxyguanosine kinase (DGUOK) 26,27, SUCLA2, the gene-encoding beta-subunit of the adenosine diphosphate-forming succinyl coenzyme A synthetase ligase 28,29, and MPV17, a mitochondrial inner membrane protein 28,30. Of these nuclear genes, POLG is the most frequent target of mutation, and is involved in a variety of mitochondrial diseases. To date, more than 150 mutations in POLG have been identified 31.
POLG is the only DNA polymerase known to date in human mitochondria. POLG is essential for the development of an embryo 32. It contains a large catalytic subunit, POLG (140-kDa), and two smaller identical accessory subunits, POLG2 (55-kDa) 33. POLG belongs to the family of A type DNA polymerases 34 consisting of an exonuclease domain with three exo motifs, I, II and III, and a polymerase domain with three pol motifs, A, B and C, along with an intervening linker region 35. As with any other polymerase, POLG has been involved in DNA polymerase, 3' to 5' exonuclease and the 5'dRP lyase activities of mtDNA replication 36.
The POLG gene maps to 15q25, is 21kb in size and consists of 23 exons. POLG contains CAG trinucleotide repeats that code for polyglutamine in the second exon, which is not present in any of the polymerases or orthologs 37. Since the first identification of POLG mutations in PEO, it has become evident that mutations in POLG are a major cause of many human diseases, ranging from Alpers syndrome to male infertility, Parkinsonism and other mitochondrial diseases 36,38-41. Most disease phenotypes associated with mutations in the POLG are due to mutations and/or depletions in mtDNA.
In this study, we analyzed POLG gene mutations and the associated reduction in mtDNA content in breast tumors. We performed mutational analyses of all coding exons and flanking intron/splice junctions of POLG. This study reports novel somatic mutations in POLG that are frequently found in breast cancer. In addition we provide evidence that mutations in POLG gene promote tumorigenesis.
MATERIALS AND METHODS
Tumor Samples
Tissue samples were collected from the patients with breast tumors undergoing surgery for treatment at the Roswell Park cancer institute and from Cooperative Human Tissue Network (CHTN) with the informed consent.
Cell culture
The breast cell lines MCF7, MDAMB231 and control cell lines MCF12A, MCF12ARho0 were grown in Dulbecco's modified Eagle's media (Cellgro, Herndon, VA, USA) supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA, USA). MCF7 Tet-On Advanced cells (Clontech, Mountain View, CA, USA) were grown in DMEM supplemented with 10% Tet System Approved FBS (Clontech), 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen), 100 μg/ml G418 (Cellgro) and 50 μg/ml uridine (Sigma, St. Louis, MO, USA). Cells were maintained in a 37°C, 5% CO2 environment.
Plasmid construction and site directed mutagenesis
The full length POLG cDNA was subcloned into the inducible mammalian expression vector pTRE-Tight-BI-AcGFP1 (Clontech). Site directed mutants were created for the mutations T251I (Exonuclease domain); P587L (Linker domain); T251I and P587L (Double mutant); D1135A and E1143G (Polymerase domain) using the site directed mutagenesis kit (Stratagene/Agilent, Santa Clara, CA, USA). Mutations were confirmed by sequencing the complete ORF of each mutant clones. The primer sequences used for site directed mutagenesis are as follows with the mutated site in upper case:
T251I_F: 5'ccctggaggtccctaTtggtgccagcag 3'
T251I_R: 5'ctgctggcaccaAtagggacctccaggg 3'
P587L_F: 5' tgcatggacccTgggccccagcc 3'
P587L_R: 5' ggctggggcccAgggtccatgca 3'
D1135A_F: 5' gcatcagcatccatgCGgaggttcgctacctgg 3'
D1135A_R: 5' ccaggtagcgaacctcCGcatggatgctgatgc 3'
E1143G_F: 5' cctggtgcgggGggaggaccgct 3'
E1143G_R: 5'agcggtcctccCcccgcaccagg 3'
POLG gene mutational analyses
DNA was isolated from tumors and cell lines with the QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA). All 23 exons and flanking intron/splice junctions of POLG were amplified by PCR with AmpliTaq Gold-polymerase. The primers and PCR conditions are given in the Supplementary Table 1. The PCR products were checked by agarose gel electrophoresis, purified by the QIAEX II Gel Extraction Kit (Qiagen) and sequenced using the BigDye terminator Ready Reaction Kit v.3 on a 3100 Genetic Analyzer Automatic Sequencer (Applied Biosystems, Foster City, CA, USA).
Mitochondrial whole genome sequencing
Complete mtDNA of four representative samples was amplified using the 24 sets of overlapping primers. Direct Sequencing of PCR products were carried out using 100.0 ng of PCR product. The mitochondrial DNA mutations were identified by comparing the sequences with rCRS.
Analysis of mtDNA content
MtDNA content was measured in breast tumor samples and cell lines by SYBR green method (SA biosciences, Frederick, MD, USA) in 7900HT Fast Real time PCR system (Applied Biosystems). Standard curves were obtained using the MCF12A cell line DNA and the reactions were performed in triplicates. Two sets of primers, one amplifying mtDNA tRNA (Leu) gene and other amplifying the nuclear DNA (Beta 2 microglobulin) were used. The ratio of the mtDNA compared to the nuclear DNA was used an index for measuring the mtDNA content 42.
MCF7 Tet-On Advanced cells were transiently transfected with pTRE-Tight-BI-AcGFP1 POLG D1135A vector according to the Fugene HD Transfection Reagent protocol (Roche, Basel, Switzerland). Media containing 1000 ng/ml doxycycline (Clontech) was added 4 h post-transfection. Transiently transfected cells were harvested 2 d after doxycycline treatment and sorted for GFP positive cells on a BD FACAria cell sorter (Becton Dickinson Biosciences, Franklin Lakes, NJ). GFP positive cells were replated with 1000 ng/ml doxycycline. DNA was isolated with the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer's protocol. mtDNA content was analyzed as described above.
GFP Induction
The mean fluorescent intensity of GFP was determined reading the fluorescence of pTRE-Tight-BI-AcGFP1 transfected cells on the FL1 channel of a FACSCalibur (Becton Dickinson Biosciences). Values are represented as mean fluorescence intensity.
Mitochondrial Functional Analyses
MCF7 Tet-On Advanced cells were transiently transfected with pTRE-Tight-BI-AcGFP1 POLG D1135A vector according to the Fugene HD Transfection Reagent protocol. Media containing 1000 ng/ml doxycycline was added 4 h post-transfection. Expression of POLG D1135A was induced by 1000 ng/ml doxycycline for up to 5 days. Cells were analyzed for reactive oxygen species (ROS) production by labeling with 10 μM dihydroethidium (DHE) for 40 min. Mitochondrial membrane potential was assessed by labeling the cells with 100 nM tetramethylrhodamine, ethyl ester, perchlorate (TMRE) for 35 min. Fluorescence of both dyes were analyzed on a FACSCalibur and gated for GFP positive cells.
Mitochondrial respiratory activity was measured by the rate of resazurin reduction as previously described 43,44. MCF7 Tet-On Advanced cells were transiently transfected with the pTRE-Tight-BI-AcGFP1 POLG D1135A vector, treated with 1000 ng/ml doxycycline, and sorted for GFP positive cells as described above. Cells assayed for mitochondrial respiratory activity as measured by the change in resazurin reduction.
Matrigel Invasion Assay
MCF7 Tet-on cells were transfected with mutant POLG plasmid were treated with 1000 ng/ml doxycycline and sorted for GFP as described above. 5 d post-doxycycline treatment cells were analyzed for in vitro matrigel invasion. Cells were plated in serum-free media in an upper Boyden chamber with a Matrigel membrane. Complete media containing 10% FBS was added to the bottom well as a chemoattractant. Cells in the chamber were incubated for 24 h and the membrane was fixed and stained with the Diff-Quick Stain Set.
RESULTS
Mutation in POLG polymerase increase tumorigencity of breast cancer cells
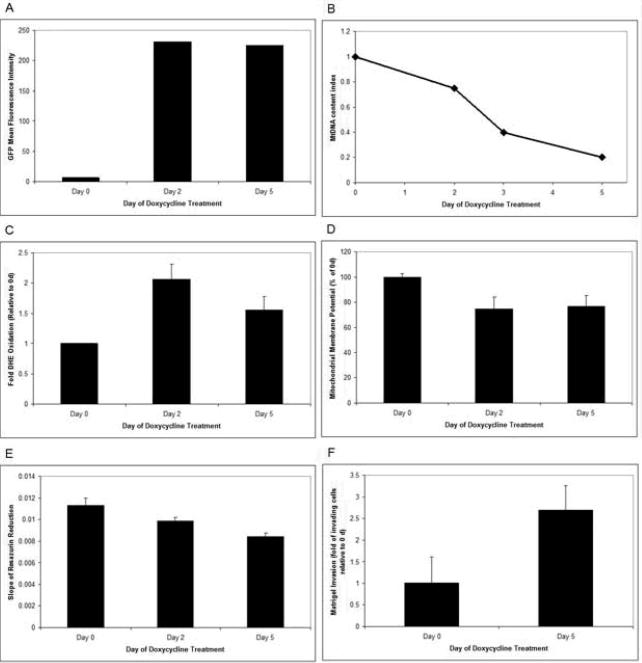
To determine the functional as well as tumorigenic role of POLG, a mutant defective in polymerase domain (D1135A) was cloned under tetracycline inducible promoter and expressed in MCF7 breast cancer cell line. A bicistronic promoter provided the expression of both GFP and POLG simultaneously. GFP expression was used as a guide to identify cells expressing the mutant POLG gene (Figure 1A). The mtDNA content was drastically reduced when expression of D1135A POLG mutant was turned on with addition of doxycycline (Figure 1B).
Figure 1. POLG D1135A mutant depletes mtDNA and promotes tumorigenicity in breast cancer cells.
POLG D1135A cDNA was cloned in the tetracycline inducible plasmid pTRE-Tight-BI-AcGFP1. A bicistronic promoter provided the expression of both GFP and POLG simultaneously. Transfected MCF7 Tet-on Advanced cells were treated with 1000 ng/ml doxycycline for up to 5 day and were sorted by FACS. A) GFP fluorescence was used as a guide to sort cells expressing the mutant POLG gene. Mean fluorescent intensity was determined on the FL1 channel of a FACSCalibur flow cytometer. Data represent geometric mean fluorescence intensity. B). MtDNA index in MCF7 Tet-on Advanced cells expressing POLG D1135A. The ratio of mtDNA to nuclear DNA was used as an index for measuring the mtDNA content C). DHE oxidation of MCF7 Tet-on Advanced cells containing POLG D1135A was measured. Mean fluorescence intensity of each treatment group was normalized to day 0 and expressed as fold DHE oxidation + 1 SD. D). Mitochondrial membrane potential was measured by TMRE fluorescence. Data represents mitochondrial membrane potential as a percent of control (day 0) + 1 SD. E). Mitochondrial respiratory activity was measured by the rate of resazurin reduction. F).Tumorigenicity was measured by Matrigel invasion assay.
These studies demonstrate that mutation(s) in the POLG polymerase domain lead to reduced mtDNA content. We therefore characterized the effect of POLG mutations on mitochondrial function. As figure 1C shows, there is a 2-fold increase in the level of ROS as measured by DHE oxidation 2 d after POLG1 D1135A expression. This change in DHE oxidation decreases by day 5, potentially indicating a shift away from oxidative phosphorylation as a metabolic source. The majority of ROS production in the cell comes from Complex I and Complex III of oxidative phosphorylation. Figure 1D shows that there is a 25% decrease in mitochondrial membrane potential in response to POLG D1135A expression. Mitochondrial respiratory activity was measured by the rate of resazurin reduction as previously described 43, 44. Resazurin is a redox-active dye that acts as an electron acceptor at Complex IV of the ETC and fluoresces upon reduction 44. Expression of POLG D1135A causes a decrease in oxidative phosphorylation when mtDNA is depleted from the POLG D1135A mutation (Figure 1E). Since the mtDNA encodes for 13 subunits of oxidative phosphorylation, loss of mtDNA would be expected to decrease oxidative metabolism. We then measured the in vitro tumorigeneic phenotype of cells expressing mutant POLG by Matrigel Invasion Assay. Figure 1F shows that cells expressing D1135A mutant POLG were more invasive than the vector alone control. We conclude that mutations in the polymerase domain of the POLG gene causes depletion of mtDNA, decreases mitochondrial membrane potential, decreases mitochondrial activity and increases oxidative stress which together promotes tumorigenesis.
POLG mutations identified in primary breast tumors
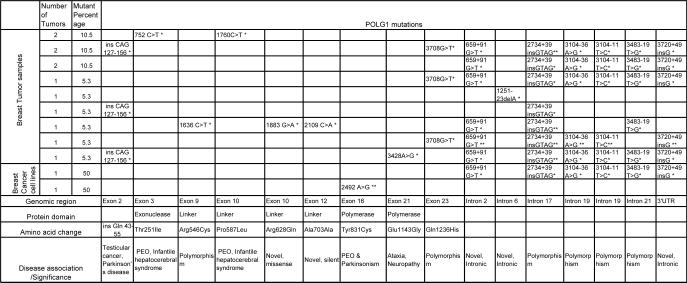
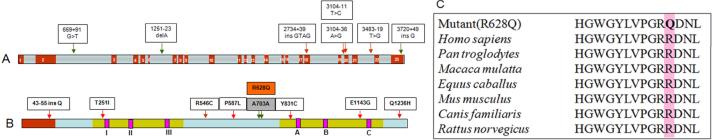
We screened all the coding exons and intron/splice junctions of POLG in 19 breast tumor samples and three cancer cell lines (Supplementary Table 1). The sequence variants found are summarized in Table 1 and depicted in Figures 2A and 2B. We identified novel as well as previously described pathogenic mutations in POLG 33, 45. The electropherograms of key mutations are given in Supplementary Figure 1. In exon 2 of POLG, CAG repeats, was found to be extended in four breast tumor samples. We detected c.752C>T in exon 3 that affects the exonuclease domain of the protein (T251I), which was reported in PEO and infantile hepatocerebral syndrome 33. Four mutations were detected in exons 9 and 10, which encode the linker region, including two novel and two previously reported mutations. The novel variants include c.1883G>A, a missense mutation causing change in the conserved amino acid Arginine to Glutamine at the 628 residue of POLG protein (Figure 2C), and another that is a silent mutation. Mutations were found in exon 16 (c.2492A>G) and exon 21 (c.3428A>G), which encode the POLG polymerase domain. In addition, we identified three novel variants in the intron/splice junctions of POLG. These results suggest that the POLG gene is a frequent target of mutation in breast tumors.
Table 1.
POLG mutations in breast tumors. The table lists POLG mutations identified in primary breast tumor tissues (19) and breast cancer cell lines. The heterozygous mutations are marked by single asterisk and homozygous mutations are marked by double asterisks.
Figure 2. POLG mutations in breast tumors and breast cancer cell lines.
A) Intron/Splice variants in the POLG genome; B) Mutations in the POLG protein with amino acid change. Green and red arrows indicate the novel variants and disease-associated mutations/polymorphisms, respectively. The grey and orange boxes indicate the novel silent and missense mutations, respectively; C) The amino acid conservation at the mutant residue of R628Q, a novel missense mutation observed in the linker region.
MtDNA mutations in primary breast tumors
Mutations in the POLG gene are known to result in the accumulation of mutations in mtDNA 46; therefore, we sequenced the entire mitochondrial genome of four representative tumors samples. Interestingly, in all four samples analyzed, the mutations were concentrated in the control D-loop region (Table 2). These mutations have previously been shown to occur in a variety of tumors 47-51. These results suggest that the identified POLG mutation in breast tumors frequently targets the D-loop region.
Table 2.
Mitochondrial DNA mutations in breast tumors. Complete mtDNA was sequenced in representative samples (n4), containing POLG mutations T251I (Exonuclease domain) and P587L (Linker domain); ins Gln 43-55 and intronic variants.
| Variant | Mutant percentage (n=4) | Region | Amino Acid Change | Association with Cancer and other diseases |
|---|---|---|---|---|
| A93G | 50 | D-loop | N/A | Colorectal and Gastric tumors |
| T152C | 50 | D-loop | N/A | Ovarian/ Squamous cell carcinoma |
| A263G | 100 | D-loop | N/A | Oral Cancer |
| C309CC | 100 | D-loop | N/A | Multiple tumors |
| C315CC | 100 | D-loop | N/A | Multiple tumors |
| C16169T | 50 | D-loop | N/A | Polymorphism |
| T16172C | 50 | D-loop | N/A | MNGIE/Oral Cancer |
| C16261T | 50 | D-loop | N/A | Oral Cancer |
| T16311C | 50 | D-loop | N/A | Oral Cancer |
| C16320T | 50 | D-loop | N/A | Oral Cancer |
| A750G | 100 | 12S rRNA | N/A | Polymorphism |
| A1438G | 100 | 12S rRNA | N/A | Polymorphism |
| A2706G | 50 | 16S rRNA | N/A | Oral Cancer |
| A4769G | 100 | ND2 | syn | Polymorphism |
| G6755A | 50 | COI | syn | Polymorphism |
| A8860G | 100 | ATPase 6 | T-A | Polymorphism |
| A8869G | 50 | ATPase 6 | M-V | Polymorphism |
| G13759A | 50 | ND5 | A-T | Polymorphism |
| T15214C | 50 | CYT B | syn | Polymorphism |
| A15326G | 100 | CYT B | T-A | Polymorphism |
Reduced mtDNA content in primary breast tumors and cell lines
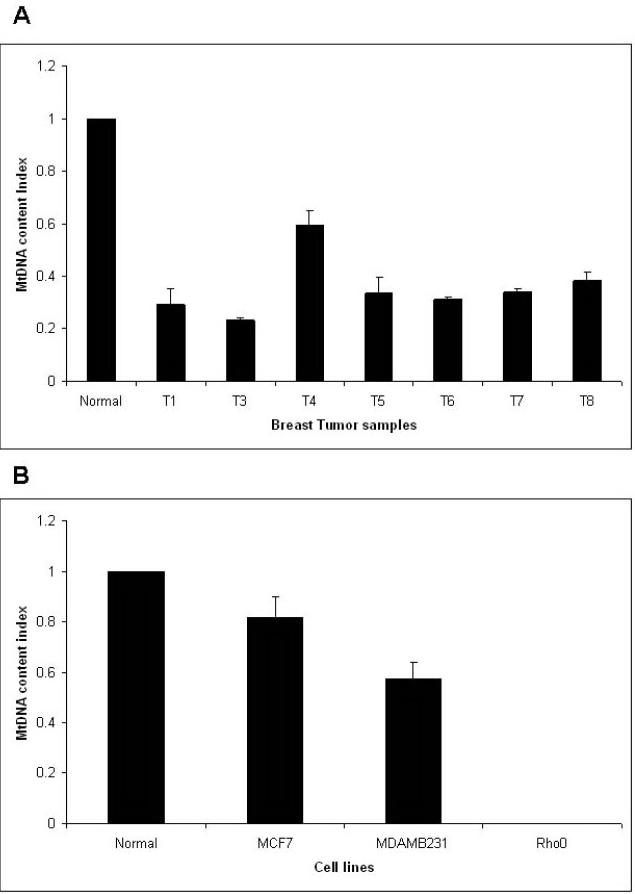
In addition to mutations in mtDNA, a common consequence of POLG mutation in mitochondrial diseases is mtDNA depletion 33. MtDNA depletion is also found in breast tumors and is associated with the prognosis of breast cancer 21. To identify the effect of the POLG mutations described above in breast tumors, we measured the mtDNA content by real-time PCR. The single copy nuclear gene β2microglobulin was used to normalize the mtDNA content. Rho0 cells devoid of mtDNA served as a negative control. Figure 3A shows the mtDNA content index in primary breast tumors. MtDNA content was reduced in samples containing the POLG mutation. Interestingly, a similar observation was made in breast cancer cell lines (Figure 3B). We conclude that POLG mutation leads to decrease in mtDNA content.
Figure 3. Decreased mtDNA content.
A) in breast tumor samples and B) in breast cancer cell lines. The ratio of mtDNA to nuclear DNA was used as an index for measuring the mtDNA content (described in material methods).
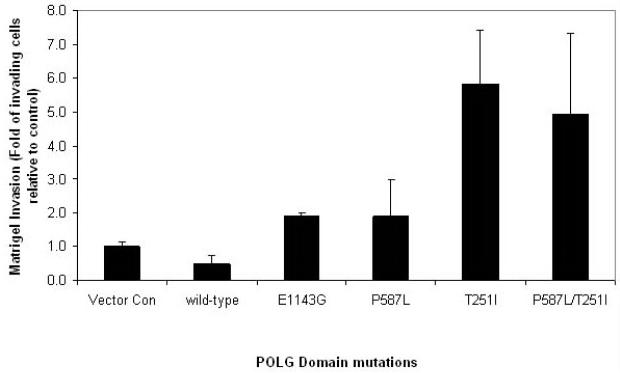
Breast Tumor POLG mutations promote tumorigenesis
The above study demonstrates that the POLG gene is frequently mutated in primary breast tumors (Figure 1). Therefore, by using site-directed mutagenesis we mutagenized the cDNA encoding representative mutations identified in breast tumors in all three functional domains of POLG (E1143G - polymerase domain, P587L - linker domain, and T251I - exonuclease domain), as well as the double mutations P587L and T251I that is often found in cis. Each mutant was tested for in vitro invasion 5 d after doxycycline treatment. Using the Matrigel invasion tumorigenicity assay, we demonstrate that expression of mutant POLG leads to increased invasiveness in vitro (Figure 4). These results suggest that POLG mutations identified in breast tumor indeed promote tumorigenesis by increasing the invasive potential of breast cancer cells.
Figure 4. Breast tumor POLG mutations lead to increased tumorigenicity.
Matrigel invasion of MCF7 Tet-on Advanced cells expressing representative mutations in POLG identified in primary breast tumors. Cell were treated with 1000 ng/ml doxycycline and sorted for GFP fluorescence. Cells were grown in the presence of doxycycline for 5 day and the Matrigel invasion was carried out. Data represents mean percentage of invading cells normalized to negative vector control ± 1 SEM.
DISCUSSION
Although mutation(s) in the POLG gene are shown to result in decreased OXPHOS, decreased mtDNA content and the pathogenesis of human mitochondrial diseases, its role in the pathogenesis of cancer is unclear. Therefore, we screened all coding exons and intron/splice junctions of POLG for mutations in breast tumors. Our analysis identified novel mutations in POLG. We also identified previously described mutations that are known to be involved in the pathogenesis of many mitochondrial diseases. Mutations were found in all three domains of the POLG protein. We identified a mutation in the exonuclease domain (C752T) of the breast tumor that is associated with PEO and infantile hepatocerebral syndrome 52-54.
Several mutations in the POLG linker region that lead to neuromuscular diseases, including Alpers's disease and Parkinson's disease have been described 35,55. However, we identified two novel linker region mutations in breast tumors. These include: (a) a missense mutation in the evolutionarily conserved (R628Q) linker region and (b) a silent linker region mutation (A703A). Previous functional analysis of the linker region mutants shows decreased enzyme activity, DNA binding and processivity of the polymerase 56. The mutants in the linker region of the fruit fly enzyme also affect its enzyme activity, processivity and DNA-binding affinity 57. The codon usage analysis for human POLG suggest that 56/103 Alanines use the GCC codon, but only 13/103 alanines use the GCA codon. This is important in the context of identified c.2109C>A (A703A) substitution in the Linker region. It is conceivable that base substitution causes ribosome stalling because Alanyl-tRNAs don't recognize the GCA codon so well which may slow the synthesis of protein. In some proteins, this type of substitution results in improper folding of protein leading to reductions in activity.
Breast tumors also harbored mutations in the polymerase domain (Y831C and E1143G) of POLG. Previous studies suggest that these mutations inhibit mtDNA polymerase activity and, hence, may lead to mtDNA depletion 58. Targeting POLG polymerase mutations in mice hearts also provides in vivo evidence for the depletion of mtDNA 59.
One of the common features associated with mitochondrial diseases is the co-occurrence of mutations in POLG. The mutation T251I is found to occur in cis with P587L in many mitochondrial diseases 34. Likewise, T251I was found in cis with P587L in two breast tumors. However, the E1143G mutation, frequently found in conjunction with W748S in ataxia 60, was uniquely present in breast tumors. POLG contains trinuleotide repeats (CAG) in the coding region 37. CAG trinucleotide repeat sequences are highly unstable, leading to the expansion or contraction of the repeat sequence, and are known to be involved in the pathogenesis of many human diseases 61. Our study revealed that the expansion of CAG repeats in more than 20% of breast tumors analyzed.
We also identified novel intron/splice junction variants in conjunction with CAG repeats. Mutations in the intron/splice junctions of other genes are known to induce exon skipping, activation of the cryptic splice sites or alteration of the balance of the alternative spliced isoforms 62. Variants in the splice junctions, particularly the GTAG insertion into intron 17, are predicted to alter splicing and POLG activity, as is also observed in PEO patients 63,64. The CAG in 43-55Q was found to co-occur with seven variants in the intron/splice junction in two breast cancer cases. Interestingly, all breast tumors with CAG repeat expansion contained at least one splice site variant c.2734+39 insGTAG. POLG repeat expansion is reported to be associated with testicular cancer 65. The POLG CAG repeats variation is also a predisposing genetic factor in idiopathic sporadic Parkinson's disease 55. The expansion of CAG located in number of genes has been shown to cause many dominantly inherited neurodegenerative diseases, described as polyglutamine diseases 66. The CAG repeats variation in other genes, such as androgen and estrogen receptors, plays an important role in breast and other cancers 67-69. The contraction of CAG repeats in POLG affects its expression 58. However, it is unknown at this time whether the expansion of CAG repeats in the POLG gene described in this paper affects its expression. An expanded CAG tract seems to affect the function of the host protein through protein-protein interaction 66. It is conceivable that CAG expansion in POLG affects its function and may contribute to tumorigenesis. However, further studies are required to identify the exact role of POLG CAG expansion in cancer.
Mutations in POLG are known to deplete mtDNA in multiple tissues of mitochondrial disease patients 70. Interestingly, our analysis also revealed 1) decreased mtDNA content in primary breast tumors and 2) when mutant POLG was expressed in breast cancer cells it led to depletion of mtDNA. Furthermore we identified mutations that were predominantly present in the D-loop control region of mtDNA. An increased incidence of novel mtDNA point mutations has been demonstrated in patients with POLG mutations 71,72. The highest incidence of the mtDNA D-loop mutations could be due to the mutations affecting exonuclease and the polymerase domains of POLG. These findings suggest that reduced mtDNA content in breast tumors may arise due to 1) inefficient enzyme activity associated with POLG mutations and/or 2) mutations in the D-loop region affecting the binding of nuclear factors involved in mtDNA replication. Irrespective of POLG-induced depletion, our studies 11,73 and those of others 74,75 suggest that mtDNA depletion leads to tumorigenicity. Indeed, we recently demonstrated that depletion of mtDNA in breast epithelial cells lead to neoplastic transformation, and that this process is mediated by p53 9. These studies led us to ask whether POLG mutations, particularly the one in the polymerase domain that causes mtDNA depletion play a role in tumorigenesis. Studies presented in this paper demonstrate that D1135A polymerase domain mutant when expressed in MCF7 cells functions as dominant negative and promote tumorigenesis in vitro. We also show that expression of mutant protein results in decreased mtDNA content, decreased OXPHOS, decreased mitochondrial membrane potential and increased oxidative stress which together contribute to increased tumorigenic phenotype. We also asked whether other POLG mutations play a role in tumorigenesis. The data presented in this paper show that with the exception of linker domain mutation (P587L), all other mutants (Polymerase domain E1143G; and exonuclease domain T251I) show increased tumorigenicity in breast cancer cells. Since mutations P587L and T251I are often found in cis in many mitochondrial diseases we also determined the effect of double mutant on Matrigel invasion. Our results show lack of synergistic effects on tumorigenicity in double mutants. The single T251I mutant was as invasive as the double P587L/T251I mutant. These studies suggest that P587L is not a significant player towards increased invasive property of MCF7 cells.
Apart from depletion, breast tumors contained mutations in mtDNA. Mutations in POLG are known to cause mutations in mtDNA. The mtDNA mutator mice that harbor the mutation in the exonuclease domain (that abolishes the POLG proof reading activity) show a marked reduction in lifespan due to the increased rate of mtDNA mutation 46, 76. To date, there is no published report that describes the incidence of tumor development in these mice. It is possible that mtDNA mutations observed in these mice do not initiate tumorigenesis, i.e., transform normal cells, but rather are involved in the promoting tumoringenesis (as described in this paper) once cells are transformed. This argument is substantiated by our report which demonstrates that mtDNA mutations in normal cells do not confer tumorigenicity. In contrast, mutant mtDNA from breast tumors when transferred to transformed cells show metastasis 77. In summary, our studies described in this paper provide the first comprehensive analyses of POLG gene mutations in human cancer that suggest a role for POLG in human tumorigenesis.
Supplementary Material
ACKNOWLEDGMENTS
The research in our laboratory was supported by National Institute of Health grant [RO1 121904 to KKS]. It was also supported, in part, by the National Cancer Institute Support Grant to Roswell Park Cancer Institute [CA 16056]. We thank Ms Paula Jones for help in writing this manuscript.
References
- 1.Warburg O. Metabolism of Tumors. Arnold Constable; London, UK: 1930. [Google Scholar]
- 2.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 3.Pedersen PL. Bioenergetics of cancer cells. J Bioenerg Biomembr. 1997;29:301–302. doi: 10.1023/a:1022494613613. [DOI] [PubMed] [Google Scholar]
- 4.Pedersen PL. Warburg, me and Hexokinase 2: Multiple discoveries of key molecular events underlying one of cancers' most common phenotypes, the “Warburg Effect”. J Bioenerg Biomembr. 2007;39:349–355. doi: 10.1007/s10863-007-9094-x. [DOI] [PubMed] [Google Scholar]
- 5.Singh KK. Mitochondrial DNA mutations in Aging, Disease, and Cancer. Springer; New York, USA: 1998. [Google Scholar]
- 6.Modica-Napolitano J, Singh KK. Mitochondria as Targets for Detection and Treatment of Cancer. Expert Rev. Mol. Med. 2002;4:1–19. doi: 10.1017/S1462399402004453. [DOI] [PubMed] [Google Scholar]
- 7.Modica-Napolitano J, Singh KK. Cancer: The first mitochondrial disease. Mitochondrion. 2004;4:755–762. doi: 10.1016/j.mito.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 8.Modica-Napolitano JS, Kulawiec M, Singh KK. Mitochondria and human cancer. Curr. Mol. Med. 2007;7:121–131. doi: 10.2174/156652407779940495. [DOI] [PubMed] [Google Scholar]
- 9.Kulawiec M, Safina A, Desouki MM, Still I, Matsui SI, Bakin A, et al. Tumorigenic transformation of human breast epithelial cells induced by mitochondrial DNA depletion. Cancer Biol. Ther. 2008;7:1732–1743. doi: 10.4161/cbt.7.11.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh KK, Costell L, editors. Mitochondria and Cancer. Springer; New York, USA: 2008. [Google Scholar]
- 11.Desouki MM, Kulawiec M, Bansal S, Das GM, Singh KK. Cross talk between mitochondria and superoxide generating NADPH oxidase in breast and ovarian tumors. Cancer Biol. Ther. 2005;4:1367–1373. doi: 10.4161/cbt.4.12.2233. [DOI] [PubMed] [Google Scholar]
- 12.Lee HC, Yin PH, Lin JC, Wu CC, Chen CY, Wu CW, et al. Mitochondrial genome instability and mtDNA depletion in human cancers. Ann N Y Acad. Sci. 2005;1042:109–122. doi: 10.1196/annals.1338.011. [DOI] [PubMed] [Google Scholar]
- 13.Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM, et al. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes & Cancer. 2006;45:629–638. doi: 10.1002/gcc.20326. [DOI] [PubMed] [Google Scholar]
- 14.Selvanayagam P, Rajaraman S. Detection of mitochondrial genome depletion by a novel cDNA in renal cell carcinoma. Lab. Invest. 1996;74:592–599. [PubMed] [Google Scholar]
- 15.Yin PH, Lee HC, Chau GY, Wu YT, Li SH, Lui WY, et al. Alteration of the copy number and deletion of mitochondrial DNA in human hepatocellular carcinoma. Br. J. Cancer. 2004;90:2390–2396. doi: 10.1038/sj.bjc.6601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu CW, Yin PH, Hung WY, Li AF, Li SH, Chi CW, et al. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes Chromosomes & Cancer. 2005;44:19–28. doi: 10.1002/gcc.20213. [DOI] [PubMed] [Google Scholar]
- 17.Moro L, Arbini AA, Yao JL, di Sant'agnese PA, Marra E, Greco M. Mitochondrial DNA depletion in prostate epithelial cells promotes anoikis resistance and invasion through activation of PI3K/Akt2. Cell Death Differ. 2009;16:571–583. doi: 10.1038/cdd.2008.178. [DOI] [PubMed] [Google Scholar]
- 18.Simonnet H, Alazard N, Pfeiffer L, Gallou C, Beroud C, Demont J, et al. Low mitochondrial respiratory chain content correlates with tumor aggressiveness in renal cell carcinoma. Carcinogenesis. 2002;23:759–768. doi: 10.1093/carcin/23.5.759. [DOI] [PubMed] [Google Scholar]
- 19.Kakuda TN. Pharmacology of nucleoside and nucleotide reverse transcriptase inhibitor-induced mitochondrial toxicity. Clin. Ther. 2000;22:685–708. doi: 10.1016/S0149-2918(00)90004-3. [DOI] [PubMed] [Google Scholar]
- 20.Larosche I, Letteron P, Fromenty B, Vadrot N, Abbey-Toby A, Feldmann G, et al. Tamoxifen inhibits topoisomerases, depletes mitochondrial DNA, and triggers steatosis in mouse liver. J Pharmacol. Exp. Ther. 2007;321:526–535. doi: 10.1124/jpet.106.114546. [DOI] [PubMed] [Google Scholar]
- 21.Yu M, Zhou Y, Shi Y, Ning L, Yang Y, Wei X, et al. Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life. 2007;59:450–457. doi: 10.1080/15216540701509955. [DOI] [PubMed] [Google Scholar]
- 22.Moraes CT, Shanske S, Tritschler HJ, Aprille JR, Andreetta F, Bonilla E, et al. mtDNA depletion with variable tissue expression: a novel genetic abnormality in mitochondrial diseases. Am. J. Hum. Genet. 1991;48:492–501. [PMC free article] [PubMed] [Google Scholar]
- 23.Sarzi E, Goffart S, Serre V, Chretien D, Slama A, Munnich A, et al. Twinkle helicase (PEO1) gene mutation causes mitochondrial DNA depletion. Ann. Neurol. 2007;62:579–587. doi: 10.1002/ana.21207. [DOI] [PubMed] [Google Scholar]
- 24.Saada A, Shaag A, Mandel A, Nevo Y, Eriksson S, Elpeleg O. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat. Genet. 2001;29:342–344. doi: 10.1038/ng751. [DOI] [PubMed] [Google Scholar]
- 25.Mancuso M, Salviati L, Sacconi S, Otaegui D, Camano P, Marina A, et al. Mitochondrial DNA depletion: mutations in thymidine kinase gene with myopathy and SMA. Neurology. 2002;59:1197–1202. doi: 10.1212/01.wnl.0000028689.93049.9a. [DOI] [PubMed] [Google Scholar]
- 26.Mandel H, Szargel R, Labay V, Elpeleg O, Saada A, Shalata A, et al. The deoxyguanosine kinase gene is mutated in individuals with depleted hepatocerebral mitochondrial DNA. Nat. Genet. 2001;29:337–341. doi: 10.1038/ng746. [DOI] [PubMed] [Google Scholar]
- 27.Salviati L, Sacconi S, Mancuso M, Otaegui D, Camao P, Marina A, et al. Mitochondrial DNA depletion and dGK gene mutations. Ann. Neurol. 2002;52:311–317. doi: 10.1002/ana.10284. [DOI] [PubMed] [Google Scholar]
- 28.Alberio S, Mineri R, Tiranti V, Zeviani M. Depletion of mtDNA: syndromes and genes. Mitochondrion. 2007;7:6–12. doi: 10.1016/j.mito.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Elpeleg O, Miller CH, Hershkovitz E, Bitner-Glindzicz M, Bondi-Rubinstein G, Rahman S, et al. Deficiency of the ADP-Forming Succinyl-CoA Synthase Activity Is Associated with Encephalomyopathy and Mitochondrial DNA Depletion. Am. J. Hum. Genet. 2005;76:1081–1086. doi: 10.1086/430843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spinazzola A, Viscomi C, Fernandez-Vizarra E, Carrara F, D'Adamo P, Calvo S, et al. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat. Genet. 2006;38:570–575. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- 31.Chan SS, Copeland WC. DNA polymerase gamma and mitochondrial disease: Understanding the consequence of POLG mutations. Biochim. Biophys. Acta. 2008;1787:312–319. doi: 10.1016/j.bbabio.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hance N, Ekstrand MI, Trifunovic A. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum. Mol. Genet. 2005;14:1775–1783. doi: 10.1093/hmg/ddi184. [DOI] [PubMed] [Google Scholar]
- 33.Copeland WC. Inherited mitochondrial diseases of DNA replication. Ann. Rev. Med. 2008;59:131–146. doi: 10.1146/annurev.med.59.053006.104646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graziewicz MA, Longley MJ, Bienstock R, Zeviani M, Copeland WC. Structure-function defects of human mitochondrial DNA polymerase in autosomal dominant progressive external ophthalmoplegia. Nat. Struct. Mol. Biol. 2004;11:770–776. doi: 10.1038/nsmb805. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen KV, Ostergaard E, Ravn SH, Balslev T, Danielsen ER, Vardag A, et al. POLG mutations in Alpers syndrome. Neurology. 2005;65:1493–1495. doi: 10.1212/01.wnl.0000182814.55361.70. [DOI] [PubMed] [Google Scholar]
- 36.Hudson G, Chinnery PF. Mitochondrial DNA polymerase-gamma and human disease. Hum. Mol. Genet. 2006;15(Spec No 2):R244–R252. doi: 10.1093/hmg/ddl233. [DOI] [PubMed] [Google Scholar]
- 37.Ropp PA, Copeland WC. Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase gamma. Genomics. 1996;36:449–458. doi: 10.1006/geno.1996.0490. [DOI] [PubMed] [Google Scholar]
- 38.Van GG, Dermaut B, Lofgren A, Martin JJ, Van BC. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat. Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- 39.Naviaux RK, Nguyen KV. POLG mutations associated with Alpers' syndrome and mitochondrial DNA depletion. Ann. Neurol. 2004;55:706–712. doi: 10.1002/ana.20079. [DOI] [PubMed] [Google Scholar]
- 40.Longley M,J, Graziewicz MA, Bienstock RJ, Copeland WC. Consequences of mutations in human DNA polymerase gamma. Gene. 2005;354:125–131. doi: 10.1016/j.gene.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 41.Davidzon G, Greene P, Mancuso M, Klos KJ, Ahlskog JE, Hirano M, et al. Early-onset familial parkinsonism due to POLG mutations. Ann. Neurol. 2006;59:859–862. doi: 10.1002/ana.20831. [DOI] [PubMed] [Google Scholar]
- 42.Mambo E, Chatterjee A, Xing M, Tallini G, Haugen BR, Yeung SC, et al. Tumor-specific changes in mtDNA content in human cancer. Int. J. Cancer. 2005;116:920–924. doi: 10.1002/ijc.21110. [DOI] [PubMed] [Google Scholar]
- 43.Abu-Amero KK, Bosley TM. Detection of mitochondrial respiratory dysfunction in circulating lymphocytes using resazurin. Arch. Pathol. Lab. Med. 2005;129:1295–1298. doi: 10.5858/2005-129-1295-DOMRDI. [DOI] [PubMed] [Google Scholar]
- 44.Perrot S, Dutertre-Catella H, Martin C, Rat P, Warnet JM. Resazurin metabolism assay is a new sensitive alternative test in isolated pig cornea. Toxicol. Sci. 2003;72:122. doi: 10.1093/toxsci/kfg014. [DOI] [PubMed] [Google Scholar]
- 45.Horvath R, Hudson G, Ferrari G, Futterer N, Ahola S, Lamantea E, et al. Phenotypic spectrum associated with mutations of the mitochondrial polymerase gamma gene. Brain. 2006;129:1674–1684. doi: 10.1093/brain/awl088. [DOI] [PubMed] [Google Scholar]
- 46.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 47.Alonso A, Martin P, Albarran C, Aquilera B, Garcia O, Guzman A, et al. Detection of somatic mutations in the mitochondrial DNA control region of colorectal and gastric tumors by heteroduplex and single-strand conformation analysis. Electrophoresis. 1997;18:682–685. doi: 10.1002/elps.1150180504. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki M, Toyooka S, Miyajima K, Iizasa T, Fujisawa T, Bekele NB, et al. Alterations in the mitochondrial displacement loop in lung cancers. Clin. Cancer Res. 2003;9:5636–5641. [PubMed] [Google Scholar]
- 49.Brandon MC, Lott MT, Nguyen KC, Spolim S, Navathe SB, Baldi P, et al. MITOMAP: a human mitochondrial genome database--2004 update. Nucleic Acids Res. 2005;33:D611–D613. doi: 10.1093/nar/gki079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumimoto H, Yamane Y, Nishimoto Y, Fukami H, Shinoda M, Hatooka S, et al. Frequent somatic mutations of mitochondrial DNA in esophageal squamous cell carcinoma. Int. J. Cancer. 2004;108:228–231. doi: 10.1002/ijc.11564. [DOI] [PubMed] [Google Scholar]
- 51.Prior SL, Griffiths AP, Baxter JM, Baxter PW, Hodder SC, Silvester KC, et al. Mitochondrial DNA mutations in oral squamous cell carcinoma. Carcinogenesis. 2006;27:945–950. doi: 10.1093/carcin/bgi326. [DOI] [PubMed] [Google Scholar]
- 52.Lamantea E, Tiranti V, Bordoni A, Toscano A, Bono F, Servidei S, et al. Mutations of mitochondrial DNA polymerase gammaA are a frequent cause of autosomal dominant or recessive progressive external ophthalmoplegia. Ann, Neurol. 2002;52:211–219. doi: 10.1002/ana.10278. [DOI] [PubMed] [Google Scholar]
- 53.Van GG, Schwartz M, Lofgren A, Dermaut B, Van BC, Vissing J. Novel POLG mutations in progressive external ophthalmoplegia mimicking mitochondrial neurogastrointestinal encephalomyopathy. Eur J. Hum. Genet. 2003a;11:547–549. doi: 10.1038/sj.ejhg.5201002. [DOI] [PubMed] [Google Scholar]
- 54.Barthelemy C, de Baulny HO, Lombes A. D-loop mutations in mitochondrial DNA: link with mitochondrial DNA depletion? Hum. Genet. 2002;110:479–487. doi: 10.1007/s00439-002-0708-4. [DOI] [PubMed] [Google Scholar]
- 55.Luoma PT, Eerola J, Ahola S, Hakonen AH, Hellstrom O, Kivisto KT, et al. Mitochondrial DNA polymerase gamma variants in idiopathic sporadic Parkinson disease. Neurology. 2007;69:1152–1159. doi: 10.1212/01.wnl.0000276955.23735.eb. [DOI] [PubMed] [Google Scholar]
- 56.Luoma PT, Luo N, Loscher WN, Farr CL, Horvath R, Wanschitz J, et al. Functional defects due to spacer-region mutations of human mitochondrial DNA polymerase in a family with an ataxia-myopathy syndrome. Hum. Mol. Genet. 2005;14:1907–1920. doi: 10.1093/hmg/ddi196. [DOI] [PubMed] [Google Scholar]
- 57.Luo N, Kaguni LS. Mutations in the spacer region of Drosophila mitochondrial DNA polymerase affect DNA binding, processivity, and the balance between Pol and Exo function. J. Biol. Chem. 2005;280:2491–2497. doi: 10.1074/jbc.M411447200. [DOI] [PubMed] [Google Scholar]
- 58.Spelbrink JN, Toivonen JM, Hakkaart GA, Kurkela JM, Cooper HM, Lehtinen SK, et al. In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J. Biol. Chem. 2000;275:24818–24828. doi: 10.1074/jbc.M000559200. [DOI] [PubMed] [Google Scholar]
- 59.Lewis W, Day BJ, Kohler JJ, Hosseini SH, Chan SS, Green EC, et al. Decreased mtDNA, oxidative stress, cardiomyopathy, and death from transgenic cardiac targeted human mutant polymerase gamma. Lab. Invest. 2007;87:326–335. doi: 10.1038/labinvest.3700523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hakonen AH, Heiskanen S, Juvonen V, Lappalainen I, Luoma PT, Rantamaki M, et al. Mitochondrial DNA polymerase W748S mutation: a common cause of autosomal recessive ataxia with ancient European origin. Am. J. Hum. Genet. 2005;77:430–441. doi: 10.1086/444548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 62.Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 63.Van GG, Martin JJ, Dermaut B, Lofgren A, Wibail A, Ververken D, et al. Recessive POLG mutations presenting with sensory and ataxic neuropathy in compound heterozygote patients with progressive external ophthalmoplegia. Neuromuscul. Disord. 2003b;13:133–142. doi: 10.1016/s0960-8966(02)00216-x. [DOI] [PubMed] [Google Scholar]
- 64.Tiangyou W, Hudson G, Ghezzi D, Ferrari G, Zeviani M, Burn DJ, et al. POLG in idiopathic Parkinson disease. Neurology. 2006;67:1698–1700. doi: 10.1212/01.wnl.0000238963.07425.d5. [DOI] [PubMed] [Google Scholar]
- 65.Nowak R, Zub R, Skoneczna I, Sikora K, Ligaj M. CAG repeat polymorphism in the DNA polymerase gamma gene in a Polish population: an association with testicular cancer risk. Ann. Oncol. 2005;16:1211–1212. doi: 10.1093/annonc/mdi205. [DOI] [PubMed] [Google Scholar]
- 66.Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, et al. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsezou A, Tzetis M, Gennatas C, Giannatou E, Pampanos A, Malamis G, et al. Association of repeat polymorphisms in the estrogen receptors alpha, beta (ESR1, ESR2) and androgen receptor (AR) genes with the occurrence of breast cancer. Breast. 2008;17:159–166. doi: 10.1016/j.breast.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 68.Li AJ, Scoles DR, Armstrong KU, Karlan BY. Androgen receptor cytosine-adenine-guanine repeat polymorphisms modulate EGFR signaling in epithelial ovarian carcinomas. Gynecol. Oncol. 2008;109:220–225. doi: 10.1016/j.ygyno.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 69.Lange EM, Sarma AV, Ray A, Wang Y, Ho LA, Anderson SA, et al. The androgen receptor CAG and GGN repeat polymorphisms and prostate cancer susceptibility in African-American men: results from the Flint Men's Health Study. J. Hum. Genet. 2008;53:220–226. doi: 10.1007/s10038-007-0240-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spinazzola A, Zeviani M. Disorders of nuclear-mitochondrial intergenomic signaling. Gene. 2005;354:162–168. doi: 10.1016/j.gene.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 71.Del Bo R, Bordoni A, Sciacco M, Di Fonzo A, Galbiati S, Crimi M, et al. Remarkable infidelity of polymerase gammaA associated with mutations in POLG1 exonuclease domain. Neurology. 2003;61:903–8. doi: 10.1212/01.wnl.0000092303.13864.be. [DOI] [PubMed] [Google Scholar]
- 72.Wanrooij S, Luoma P, van Goethem G, van Broeckhoven C, Suomalainen A, Spelbrink JN. Twinkle and POLG defects enhance age-dependent accumulation of mutations in the control region of mtDNA. Nucleic Acids Res. 2004;32:3053–64. doi: 10.1093/nar/gkh634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singh KK, Kulawiec M, Still I, Desouki MM, Geradts J, Matsui S. Inter-genomic cross talk between mitochondria and the nucleus plays an important role in tumorigenesis. Gene. 2005;354:140–146. doi: 10.1016/j.gene.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 74.Guha M, Srinivasan S, Biswas G, Avadhani NG. Activation of a novel calcineurin-mediated insulin-like growth factor-1 receptor pathway, altered metabolism, and tumor cell invasion in cells subjected to mitochondrial respiratory stress. J. Biol. Chem. 2007;282:14536–14546. doi: 10.1074/jbc.M611693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Biswas G, Srinivasan S. Anandatheerthavarada,H.K. and Avadhani,N.G. Dioxin-mediated tumor progression through activation of mitochondriato-nucleus stress signaling. Proc. Natl. Acad. Sci. U S A. 2008;105:186–191. doi: 10.1073/pnas.0706183104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, et al. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat. Genet. 2007;39:540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- 77.Kulawiec M, Owens KM, Singh KK. Cancer cell mitochondria confer apoptosis resistance and promote metastasis. Cancer Biol. Ther. 2009 Jul 15; doi: 10.4161/cbt.8.14.8751. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.