Abstract
Leukemia inhibitory factor (LIF) promotes the survival of oligodendrocytes both in vitro and in an animal model of multiple sclerosis, but the possible role of LIF signaling in myelination during normal development has not been investigated. We find that LIF-/- mice have a pronounced myelination defect in optic nerve at postnatal day 10. Myelin basic protein (MBP)- and proteolipid protein (PLP)-positive myelin was evident throughout the optic nerve in the wild-type mice, but staining was present only at the chiasmal region in LIF-/- mice of the same age. Further experiments suggest that the myelination defect was a consequence of a delay in maturation of oligodendrocyte precursor cell (OPC) population. The number of Olig2-positive cells was dramatically decreased in optic nerve of LIF-/- mice, and the distribution of Olig2-positive cells was restricted to the chiasmal region of the nerve in a steep gradient toward the retina. Gene expression profiling and cell culture experiments revealed that OPCs from P10 optic nerve of LIF-/- mice remained in a highly proliferative immature stage compared with littermate controls. Interestingly, by postnatal day 14, MBP immunostaining in the LIF-/- optic nerve was comparable to that of LIF+/+ mice. These results suggest that, during normal development of mouse optic nerve, there is a defined developmental time window when LIF is required for correct myelination. Myelination seems to recover by postnatal day 14, so LIF is not necessary for the completion of myelination during postnatal development.
Keywords: myelin, LIF, oligodendrocyte, astrocyte, optic nerve, transcription, OPC, differentiation
Identifying the signals controlling oligodendrocyte differentiation and myelination is an important priority in current research in developmental neurobiology and in clinical research on demyelinating CNS disease. Cytokines have a well-appreciated role in inflammatory responses during nervous system injury and disease, but, in contrast to the case for growth factors and cell adhesion molecules, the possible role of cytokines in regulating oligodendrocyte development and myelination under normal physiological conditions is relatively unexplored. Based on recent research identifying LIF signaling between astrocytes and oligodendrocytes in regulating myelination in response to electrical activity in axons (Ishibashi et al.,[2006]), the present study was undertaken to investigate the possible role of LIF in regulating myelination of early postnatal optic nerve. Using genetic knockout mice, this study is the first analysis of LIF signaling on myelination during early postnatal development in the CNS.
Although cytokines are generally associated with inflammatory disease, there is evidence that nervous system development, including myelination, is regulated by the activation of cytokine signaling at specific stages of development (Bauer et al.,[2007]). Leukemia inhibitory factor (LIF), ciliary neurotrophic factor (CNTF), and interleukin-6 (IL-6) each promote the survival of mature oligodendrocytes in vitro (Barres et al.,[1993]; Kahn and DeVellis,[1994]; Stankoff et al.,[2002]). Cytokines arising from several unrelated genes share a common signaling mechanism via activation of the signaling receptor gp 130 (Ip et al.,[1992]). Supplemental LIF can increase (Stankoff et al.,[2002]; Ishibashi et al.,[2006]) or inhibit (Park et al.,[2001]) myelination in cell culture depending on the concentration. Under states of neural inflammation, up-regulation of LIF, CNTF, and IL-6 expression is observed in patients with multiple sclerosis (Schönrock et al.,[2000]; Vanderlocht et al.,[2006]) and in experimental autoimmune encephalomyelitis (EAE), an animal model of autoimmune demyelination (Butzkueven et al.,[2006]). A more recent study reported that LIF receptor signaling modulates demyelination and remyelination in an in vivo cuprizone-induced demyelination model (Marriott et al.,[2008]).
Some defects in myelination have been reported in an analysis of neuronal and glial phenotype in the brain of adult LIF-/- mice (Bugga et al.,[1998]), but the effects on oligodendrocytes vary with different brain regions, and reduced myelin basic protein (MBP) staining was seen only in female animals. Myelination of optic nerve was not examined. The reasons for the reduced MBP staining in females were not known, and the authors suggest the need for further analysis of LIF in myelination. The objective of the present study was to determine whether there are any effects on myelination of early postnatal optic nerve in LIF-/- mice and, if so, whether the effects are due to an influence of LIF on proliferation, differentiation, or migration of cells in the oligodendrocyte lineage. A second objective is to determine whether the possible effects of LIF signaling during normal development are transient or permanent and to provide a more comprehensive analysis of cellular and molecular biological consequences of LIF gene disruption in early postnatal optic nerve by performing a global analysis of genes that are differentially expressed in normal and LIF-/- mice. The results reveal a delay in oligodendrocyte development and subsequent decreased myelination at postnatal day 10 (P10) in the optic nerve of male and female LIF-/- mice, which largely recovers by P14.
Materials and Methods
Animals
Throughout the study, LIF-/- mice, maintained on a CD1 background, were genotyped using a polymerase chain reaction (PCR)-based method (Stewart et al.,[1992]), and corresponding age-matched wild-type (LIF+/+) control mice were used for in vivo studies. Heterozygotes (LIF+/-) were used for in vitro culture studies.
Immunocytochemistry
LIF-/- as well as LIF+/+ mouse optic nerves were fixed with 4% paraformaldehyde in 0.1 M phosphate buffer (PB), pH 7.4. The optic nerves were cryoprotected with 30% sucrose in PBS, pH 7.4, for 24 hr at 4°C. After embedding in optimal cutting temperature mounting medium (Miles, Elkhart, IN), tissues were cut into 7-10 μm-thick sections. Immunofluorescence staining was performed as described previously (Ishibashi et al.,[2002]). The following primary antibodies were used in overnight incubations at 4°C: anti-NG2 polyclonal (Chemicon, Temecula, CA; 1:1,000), anti-O4 (Chemicon; 1:1,000), antigalactocerebrosidase (GalC; Chemicon; 1:500), anti-GFAP (Dako, Carpinteria, CA; 1:2,000), anti-MBP (Sternberger Monoclonals, Luthersville, MD; 1:1,000), rat anti-PDGFRα monoclonal (BD Pharmingen, San Diego, CA; 1:500), and antproliferating cell nuclear antigen (PCNA; Upstate Biotechnology, Lake Placid, NY; 1:50). Anti-Olig2 polyclonal antibody was kindly provided by Dr. Takebayashi (National Institute of Physiological Sciences, Okazaki, Japan). A rat monoclonal antibody (AA3) against the C-terminal portion of myelin proteolipid protein (PLP; Yamamura et al.,[1991]) was kindly provided by Dr. M. Lees (E.K. Shriver Center, Waltham, MA). For secondary antibodies, Alexa 488 or Alexa 594 conjugated to goat anti-rabbit, goat anti-rat, or goat anti-mouse antibodies (Molecular Probes, Eugene, OR) were applied for 45 min at RT, and images were captured via Axio Imager or confocal microscopy (Zeiss, Oberkochen, Germany). Primary OPC cultures from optic nerves were stained with anti-O4 or anti-GalC antibodies before fixation at 37°C for 30 min. Immunocytochemistry with anti-NG2 or anti-MBP antibodies was carried out using cells fixed with 4% paraformaldehyde in 0.1 M phosphate buffer (PB), pH 7.4.
Cell Culture
Because LIF heterozygous mice (LIF+/-) do not display any defect in myelination, we used LIF+/- mice as a control in these experiments. LIF+/- and LIF-/- mouse optic nerves were dissected as described previously (Mi et al., [1999]). In brief, P10 mixed optic nerve cells were prepared by digesting minced optic nerves in a papain solution (33 U/ml; Worthington, Freehold, NJ) at 37°C for 45-60 min, followed by trituration in DMEM (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (FBS; Hyclone, Logan, UT). The mixed cells were transferred to poly-D-lysine (Sigma, St. Louis, MO)-coated 12 mm glass coverslips in 24-well tissue culture plates containing proliferation medium (10% FBS/DMEM). After 24 hr, the medium was removed and replaced with differentiating medium (DMEM + N1 + 0.5% FBS; Stevens et al.,[2002]). Three days later, cells were fixed with 4% paraformaldehyde in 0.1 M phosphate buffer (PB), pH 7.4, and used for immunofluorescence. There was no LIF or other factors in the proliferation medium or differentiation medium.
Proliferation Assay
After 48 hr in differentiating media, optic nerve cells were pulsed with BrdU (Boehringer Mannheim, Indianapolis, IN) for 6 hr, fixed, and stained according to the manufacturer's instructions. Cells were then counterstained with Hoechst nuclear stain (Molecular Probes, Eugene, OR) at a dilution of 1:2,000 for 10 min. The proliferation rate was calculated as the ratio of BrdU:Hoechst-positive nuclei in each microscope field. All nuclei stained with BrdU and Hoechst were counted in each microscope field. Ten randomly chosen fields were sampled to obtain a mean for each coverslip.
Gene Expression Analysis
Purification and preparation of RNA
Total RNA was extracted from optic nerves using the Trizol reagent method (Invitrogen, Carlsbad, CA; catalog No. 15596-026) from five animals (n = 10 optic nerves) for both LIF+/+ and LIF-/- at P10, from four animals (n = 8 optic nerves) from P14, and from two animals (n = 4 optic nerves) from P35 (adult); all animals used for mRNA profiling were male. The quality of total RNA samples was assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). RNA samples were labeled according to the chip manufacturer's recommended protocols. In brief, for Illumina, 0.5 μg of total RNA from each sample was labeled by using the Illumina TotalPrep RNA Amplification Kit (Ambion, Austin, TX; catalog No. IL1791) in a process of cDNA synthesis and in vitro transcription. Single-stranded RNA (cRNA) was generated and labeled by incorporating biotin-16-UTP (Roche Diagnostics GmbH, Mannheim, Germany; catalog No. 11388908910), and 0.75 μg of biotin-labeled cRNA was hybridized (16 hr) to Illumina's Sentrix MouseRef-8 Expression BeadChips (Illumina, San Diego, CA; catalog No. BD-26-213). The hybridized biotinylated cRNA was detected with streptavidin-Cy3 and quantitated using Illumina's BeadStation 500GX Genetic Analysis Systems scanner.
Array data processing
Preliminary analysis of the scanned data was performed in Illumina BeadStudio software. The primary Illumina data are returned from the scanner in the form of an “.idat” file which contains single intensity data values/gene following the computation of a trimmed mean average for each probe type represented by a variable number of bead probes/gene on the array. The BeadStudio software returns information on the number and standard deviation of all the bead measurements per probe/gene as well as a detection threshold based on a comparison between the measured intensity calculated for a single probe/gene and the intensities measured for a large number of negative control beads built into the BeadChip arrays (D = percentage above negative/100, 1 = perfect, i.e., the intensity value of a gene is greater than all the intensities for every negative control tested). Any gene consistently below D = .99 for all samples was eliminated from further analysis. This background filter resulted in the removal of approximately 20% of all the genes on the Illumina array (4,938 background genes from among a total of 24,527 RefSeq genes). Further filtering removed 1,688 genes without a meaningful HUGO gene name. Overall differences in gene expression between samples were calculated by a global normalization method based on total intensity counts for each array. A normalization factor was determined by dividing the average total intensity by the true total intensity. Each intensity value was multiplied by its appropriate normalization factor, and differences were calculated as ratios from the normalized values. The data are accessible through GEO Series accession No.: GSE11935 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE11935).
Results
Reduced MBP and PLP Immunoreactivity in the Optic Nerve of P10 LIF-/- Mice
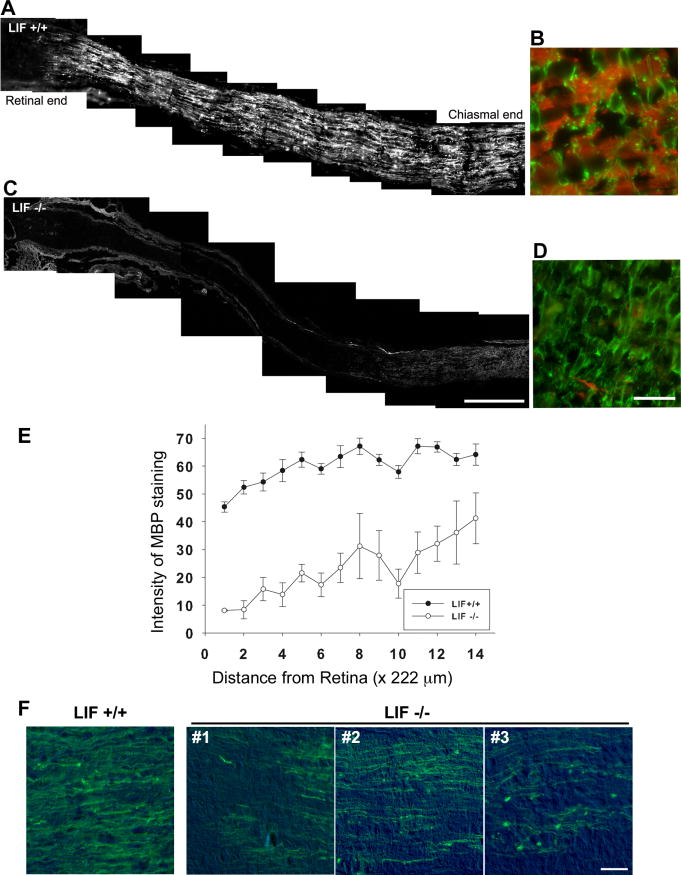
To determine the effect of removal of LIF signaling on myelination in vivo, we examined myelin formation using longitudinal optic nerve sections from LIF knockout (LIF-/-) mice and wild-type (LIF+/+) mice during early postnatal development when myelination is proceeding in mouse optic nerve, P10-P14 (Baba et al.,[1999]). To accomplish this, we used MBP and PLP immunoreactivity as myelin markers in the postnatal optic nerve. At 10 days of age, MBP-positive myelin was present throughout the optic nerve in LIF+/+ mice, as expected (Fig. 1A). However, in LIF-/- mice at the same age, myelination was markedly reduced (Fig. 1C), with MBP-positive myelin seen only at the chiasmal end of the nerve (Fig. 1C). Figure 1B shows MBP staining (red) and GFAP staining (green) from the midpoint of the optic nerve in a LIF+/+ mouse at P10. Figure 1D is from the center of the optic nerve of an LIF-/- animal of the same age (MBP, red; GFAP, green). Little MBP immunoreactivity is apparent in the LIF-/- optic nerve. However, there appears to be no difference in GFAP immunoreactivity between LIF+/+ and LIF-/- optic nerves at P10. Next we quantified the intensity of immunofluoresence of MBP staining in Image J (Fig. 1E). Optic nerves from LIF+/+ mice (n = 7; two male and five female) and LIF-/- mice (n = 7; three male and four female) at P10 were taken, and MBP immunofluorescence was quantified in each field from retina to chiasm. A significant decrease in the intensity of MBP immunoreactivty can be seen in the LIF-/- mice compared with the LIF+/+ mice (Fig. 1E). We also examined optic nerves at P10 for PLP immunoreactivity using sections from the same mice. Figure 1F shows the midpoint of the optic nerves from one LIF+/+ mouse and three examples of LIF-/- mice. PLP immunoreactivity is clearly reduced in the optic nerves of LIF-/- mice. This was confirmed by quantitative analysis of PLP immunofluorescence in Image J. PLP immunoreactivity was significantly reduced in LIF-/- optic nerves (n = 7, LIF+/+ vs. LIF-/-; 41.185 ± 1.819 vs. 14.502 ± 2.196; P < 0.0001). These differences were observed in pups of both sexes.
Figure 1.
MBP and PLP immunoreactivity are markedly reduced in optic nerve of P10 LIF-/- mice. The optic nerves from LIF+/+ mice (A) and LIF-/- mice (C) at 10 days of age were stained with anti-MBP antibody. MBP-positive myelin was observed throughout the entire nerve in LIF+/+ mice (A). In contrast, in optic nerves of LIF-/- mice at the same age, weak MBP immunoreactivity was detected only near the chiasmal region of the nerve (C). B, D: Optic nerve at higher magnification of longitudinal section at the midpoint between retina and chiasma stained with anti-MBP (red) and anti-GFAP (green) antibodies. Note the absence of MBP in LIF-/- mice (D). E: The intensity of MBP staining was quantified in Image J. There was some variability among LIF-/- mice; however, the intensity of MBP immunofluorescence decreased throughout the optic nerves of LIF-/-mice (open circles) compared with LIF+/+ optic nerves (solid circles). F: Optic nerves from LIF+/+ and LIF-/- were assessed for PLP immunoreactivity, revealing less PLP in the optic nerve of LIF-/- mice. Examples from three different nerves from LIF-/- mice are shown (1-3). All four images were taken from the midpoint of each optic nerve. Scale bars = 200 μm in C; 20 μm in D, F.
Decrease in Number of Olig2-Positive Cells and Altered Distribution in a Chiasma-to-Retinal Gradient in LIF-/- Mice During Development
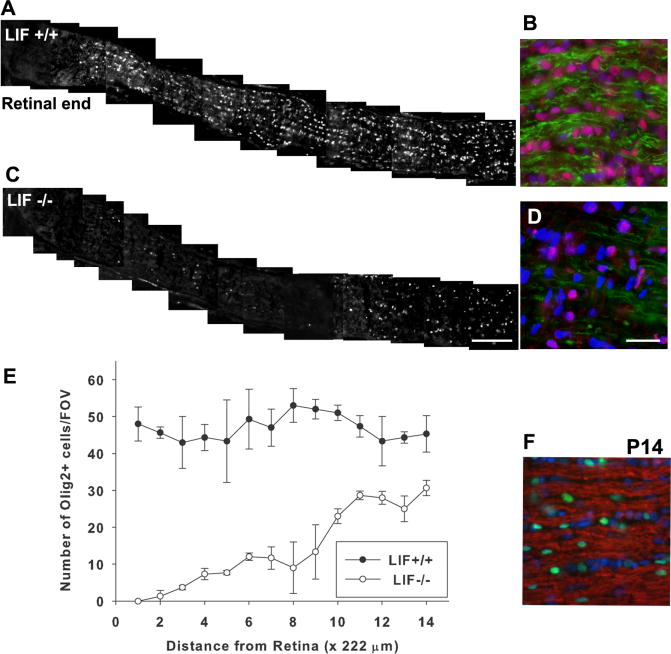
The greatly reduced MBP immunoreactivity observed at P10 in the LIF-/- optic nerve could result from defects in myelin induction or a decrease in the total number of oligodendrocytes and/or OPCs during this stage of development. To determine the OPC population in the optic nerve of LIF+/+ and LIF-/- animals, we carried out immunohistochemistry using the oligodendrocyte progenitor marker Olig2 (Takebayashi et al.,[2000]). The number of Olig2-positive cells was dramatically decreased along the entire length of the optic nerve in LIF-/- mice (Fig. 2C) compared with LIF+/+ mice (Fig. 2A). Olig2-positive cells were concentrated primarily at the chiasmal region in LIF-/- mice (Fig. 2C), although in reduced numbers compared with LIF+/+ optic nerve, where Olig2-positive cells were seen in large numbers along the total length of the optic nerve from the retina to the chiasm. Figure 2B,D shows Olig2 staining in red and MBP staining in green 2.2 mm from the retina of LIF+/+ optic nerve and LIF-/- nerve, respectively. Greatly reduced Olig2 and MBP immunoreactivity is observed in Figure 2D (LIF-/-) compared with Figure 2B (LIF+/+). Results were similar in both sexes.
Figure 2.
The population of cells in the oligodendrocyte lineage is reduced and displays a pronounced chiasm-to-retinal gradient in P10 optic nerve of LIF-/- mice. The optic nerves from LIF+/+ (A) and LIF-/- mice (C) at 10 days of age were stained with anti-Olig2 antibody to identify cells in the oligodendrocyte lineage. Olig2-positive cells were present uniformly along optic nerves in LIF+/+ mice (A). In contrast, Olig2-positive cells were observed only near the chiasmal region in LIF-/- mice (C). B, D: Higher magnification of longitudinal section of optic nerves, taken 2.2 mm from the retina and stained with Olig2 (red) and MBP (green) antibodies and DAPI (blue) to identify cell nuclei. E: To quantify the gradient of Olig2-positive cells along the nerves, Olig2-positive cells were counted in sequential microscope fields in 14 equally spaced increments from the retina to chiasm. These numbers were represented as the mean number of cells per field of view (FOV; 1 FOV = 222 × 166.4 μm). The number of Olig2-positive cells in optic nerves from wild-type mice at P10 (solid circles) was uniform along the entire length of nerve (no correlation between cell number and the distance from retina in LIF+/+ mice). However, there was a significant reduction and a steep gradient in number of Olig2-positive cells in P10 optic nerves of LIF-/- mice (open circles), declining from the chiasm to the retina (y = -3.84 + 2.43x, correlation 0.786, P = 0.001). F: By P14, anti-MBP (red) and Olig2 (green) immunostaining were similar to wild type (not shown) in optic nerves of LIF-/- mice. F shows a longitudinal section of the optic nerve of P14 LIF-/- mice taken at the midpoint between the retina and the chiasm. Scale bars = 200 μm in C (applies to A,C); 20 μm in D (applies to B, D, F).
We then quantified the number of Olig2 cells in the optic nerves of LIF+/+ and LIF-/- animals at P10 (Fig. 2E) to determine whether the decreased myelin protein was the result of fewer cells in the oligodendrocyte lineage. The number of Olig2-positive cells in each field (1 FOV = 222 × 166.4 μm) was almost uniform along the entire length of the optic nerve of LIF+/+ mice (Fig. 2E, solid circles; y = 50.7 - 1.41x y; total number, x; location, no correlation with distance from the retina). In contrast, there were fewer Olig2-positive cells in LIF-/- optic nerve, and a sharp gradient in number of Olig2-positive cells from the chiasm to retina was apparent in LIF-/- mice (Fig. 2E, open circles; y = -3.84 + 2.43x, correlation 0.786, P = 0.001). The decrease in Olig2-positive cells in LIF-/- mice was restored to LIF+/+ mice levels by 14 days of age, and MBP-positive myelin was observed throughout the optic nerve at that stage (Fig. 2F).
Olig2-positive cells migrate out of the brain into the optic nerve during early postnatal development. Our data may suggest that some Olig2-positive cells may be restricted in their migration into the optic nerve in LIF-/- mice. To determine whether optic nerve OPCs from mice lacking LIF exhibit a cell migration defect, we performed an in vitro migration assay using OPCs dissociated from optic nerve of LIF+/+ mice and LIF-/- mice in a Boyden microchemotaxis chamber (Zhang et al.,[2004]) and explant cultures. For the explant culture experiments, the optic nerves were cut into 1-mm-long explants from chiasmal regions and immersed in culture medium and cultured for 5 hr. Neither method revealed any difference in cell migration in the LIF-/- optic nerve compared with LIF+/+ optic nerve (data not shown). However, these data cannot completely rule out the possibility that LIF contributes to OPC migration in vivo. These experiments were carried out in LIF-/- mice of both sexes (five female and six male).
Alteration in Differentiation and Proliferation of OPCs in LIF-/- Optic Nerve Cells In Vitro
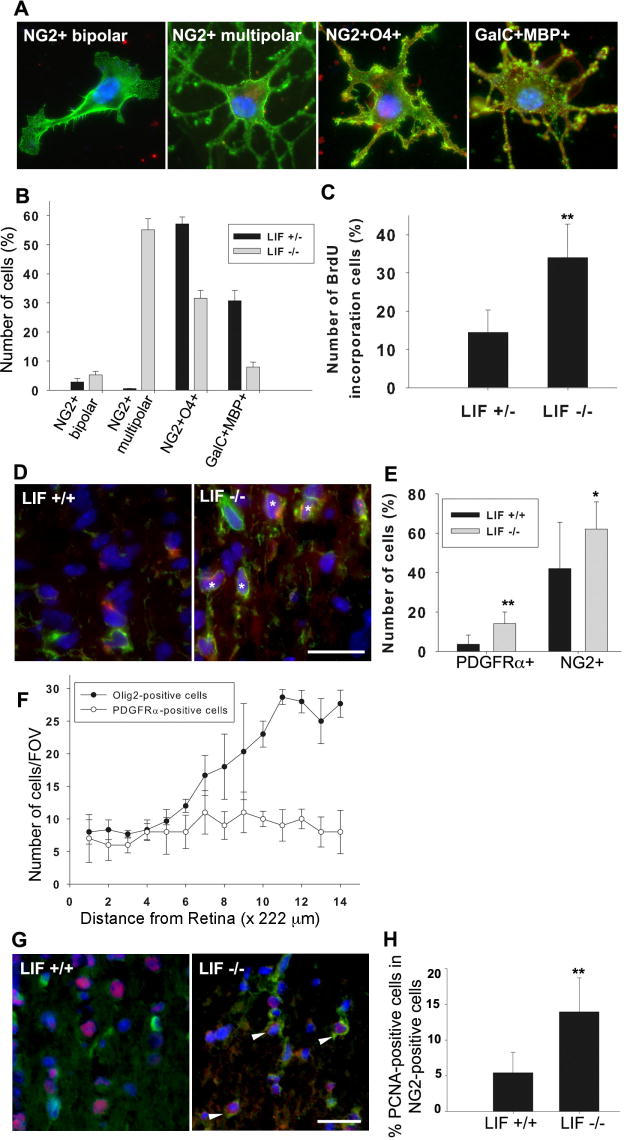
The greatly reduced MBP and PLP immunoreactivity and the reduced Olig2-positive cell population suggest that LIF is involved in differentiation and proliferation of OPCs in the optic nerve during a defined time frame of postnatal development. To examine this possibility, cultured optic nerve cells were characterized with several OPC markers of differentiation and maturation. For these experiments, we used cells from LIF+/- optic nerves as a closely matched control, because optic nerve from these animals displays no defect in MBP immunoreactivity at P10 (data not shown). Cells purified from P10 LIF+/- mice as control and LIF-/- optic nerve were plated into proliferation medium to compare proliferation and differentiation of optic nerve glia in the two conditions. Twenty-four hours after plating, cultures were switched into differentiation medium. Three days later, OPCs from the optic nerve were characterized byimmunohistochemical staining with anti-NG2, -O4, -GalC, and -MBP antibodies. We characterized the OPC population, by staining pattern and morphology, into four broad categories reflecting the sequential differentiation and maturation of oligodendrocytes: first, NG2-positive/unipolar or bipolar shapes (perinatal oligodendrocyte progenitors); second, NG2-positive/multipolar shapes (late oligodendrocyte progenitors); third, NG2-positive/O4-positive cells (immature oligodendrocytes); and, fourth, GalC-positive/MBP-positive cells (premyelinating oligodendrocytes; Fig. 3A). The results showed that OPCs isolated from LIF-/- optic nerves were developmentally immature compared with cells isolated from LIF+/- animals. This is especially apparent in the large increase in number of NG2-positive/multipolar cells isolated from the LIF-/- optic nerve, whereas the number of NG2-positive/O4-positive cells and the number of GalC-positive/MBP-positive cells were decreased compared with LIF+/- cells (Fig. 3B). Cell proliferation in dissociated optic nerve culture was measured by BrdU incorporation into NG2-positive cells. The percentage of BrdU-positive OPCs (NG2-positive cells) in cells isolated from LIF-/- mice was about 2.5 times higher than that in cells isolated from the control mice (Fig. 3C), indicating an increased proliferation rate of OPCs from LIF-/- optic nerve compared with heterozygous mice.
Figure 3.
Proliferation of OPCs was increased and differentiation from the progenitor stage was inhibited in the optic nerve of LIF-/- mice. To determine the effects of endogenous LIF on OPC differentiation, OPCs isolated from optic nerve of P10 mice were grown in cell culture for 5 days and characterized using markers of different stages of oligodendrocyte cell differentiation (A). Cells were classified into four categories based on the staining pattern and morphology, representing the developmental progression of oligodendrocytes from the oligodendrocyte progenitor cell stage: 1) NG2-positive/unipolar or bipolar shapes, 2) NG2-positive/multipolar shapes, 3) NG2-positive/O4-positive cells, and 4) GalC-positive/MBP-positive cells. B: OPCs from LIF-/- mice (gray bars) remained at an immature stage compared with LIF+/- cells (black bars). C: The affects of endogenous LIF on OPC proliferation were measured by BrdU incorporation into cells in culture. Proliferation was about 2.5 times higher in LIF-/- optic nerve cells compared with LIF+/- cells (**P < 0.001). D: The optic nerves from three LIF+/+ and five LIF-/- mice (two male and three female) at 10 days of age were stained with the early OPC markers NG2 (green) and PDGFRα (red) antibodies and a nuclear dye DAPI (blue). Some NG2-positive cells were also PDGFRα-positive in the optic nerves of LIF-/- mice (asterisks). E: The population of PDGFRα-positive cells and NG2-positive cells (averaged throughout the full length of the optic nerve) was increased in the optic nerves of LIF-/- mice compared with LIF+/+ mice *P< 0.01, **P < 0.001, Student's t-test, consistent with in vitro experiments. F: The gradient in Olig2-positive OPCs (solid circles) along P10 optic nerve of LIF-/- mice could not be explained as a defect in migration of immature OPCs into the nerve, insofar as these cells (open circles) were distributed uniformly from the chiasm to the retina. PDGFRα-positive cells as well as Olig2-positive cells in the optic nerves of LIF-/- were quantified in sequential microscopic fields in 14 equally spaced increments from the retina to the chiasm. These numbers are represented as the mean number of cells per field of view (FOV; 1 FOV = 222 × 166.4 μm). G: Consistently with the in vitro proliferation data, OPC proliferation was increased in the absence of endogenous LIF. Optic nerves were immunostained with cell proliferation marker PCNA (red), immature OPC marker NG2 (green), and nuclear marker DAPI (blue). Compared with LIF+/+ optic nerve (left), NG2-positive cells in the LIF-/- optic nerve were more often PCNA positive (arrows), indicative of a more proliferative cell population. H: Quantitative analysis of the total number of proliferating NG2-positive cells (staining for both PCNA and NG2) in LIF-/- nerves was higher than in the LIF+/+ nerves. **P < 0.001, Student's t-test. Scale bars = 20 μm.
Optic nerves were stained with an anti-PCNA antibody and with an anti-NG2 antibody to assess OPC proliferation in vivo (Fig. 3G). The number of NG2-positive cells stained with the cell proliferation marker PCNA was significantly greater in LIF-/- optic nerves (13.95% ± 4.740%) compared with that in LIF+/+ optic nerves (5.43% ± 2.838%; Fig. 3H). Therefore, we conclude that OPCs from LIF-/- optic nerve are developmentally immature and hyperproliferative and that LIF controls the maturation of OPCs in the optic nerve during this stage of postnatal development. The affects of LIF knockout on OPC proliferation and differentiation were not sexually dimorphic (44 pups total, 13 LIF-/- female and 9 LIF-/- male).
We examined the state of differentiation of OPCs in intact optic nerves from three LIF+/+ and five LIF-/- mice (two male and three female) at 10 days of age using the immature OPC markers PDGFRα and NG2. PDGFRα-positive cells were rarely observed in LIF+/+ optic nerves at P10 (Fig. 3D, left), whereas cells coexpressing PDGFRα and NG2 were often observed in LIF-/- optic nerves (Fig. 3D, asterisks in the right panel). The number of PDGFRα-positive cells and NG2-positive cells was significantly increased in LIF-/- optic nerves compared with that in LIF+/+ nerves (Fig. 3E). In this analysis, PDGFRα-positive cells and NG2-positive cells were counted throughout the full length of the optic nerves and averaged. Figure 3F shows no gradient of PDGFRα-positive cells from retinal region to chiasm (open circles), in LIF-/- mice, whereas Olig2-positive cells were concentrated at the chiasmal region (solid circles). This result indicated that PDGFRα-positive immature cells had fully migrated throughout the optic nerve, which is consistent with the in vitro cell migration assays, which failed to detect any difference in migration of OPCs in LIF-/- mice. The results from in vitro experiments are consistent with the in vivo analysis in supporting the conclusion that myelination of P10 optic nerve of LIF-/- is delayed as a result of an LIF-dependent developmental delay maintaining OPCs at an immature and highly proliferative stage.
Gene Expression Analysis of P10 Wild-Type/LIF Knockout Optic Nerves
To characterize gene expression during in vivo postnatal myelination in the optic nerve further, we carried out microarray analysis and functional characterization profiling on LIF-/- and LIF+/+ P10, P14, and adult optic nerves from male mice. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al.,[2002]) and are accessible through GEO Series accession No.: GSE11935 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE11935).
Table I lists the top 50 up-regulated genes in the optic nerve of P10 LIF-/-. Table II lists the corresponding top 50 most down-regulated genes. Further analysis of the gene expression data also shows a decrease in many myelin genes in the LIF-/- optic nerve at P10 and partial recovery by P14, with very little or no difference in the myelin genes in the adult LIF-/- optic nerve (Table III). Many of the genes associated with mature oligodendrocytes and myelination, such as MOG, MAG, CNP, and MAL, were down-regulated in the P10 LIF-/- optic nerve. However, MBP mRNA is only marginally down-regulated in the P10 LIF-/- optic nerve; additionally, PLP mRNA is slightly more abundant in the P10 LIF-/- optic nerve. This apparent contradiction in MBP and PLP immunoreactivity and the level of MBP and PLP mRNA may be explained by a posttranscriptional mechanism regulating the level or localization of these proteins. It is also possible that signaling through LIF regulates the translation of MBP and PLP via an unidentified mechanism. The gene expression analyses are consistent with the earlier data in this study suggesting that LIF regulates oligodendrocyte maturation during postnatal development in the mouse optic nerve. Additionally, partial recovery of myelin gene mRNA to wild-type levels is seen by P14, and very little difference in myelin gene mRNA is seen in the adult LIF-/- optic nerve (Fig. 2F), suggesting that myelination has recovered by P35 (adult).
Table I. Top 50 mRNAs Up-Regulated in P10 LIF Knockout Optic Nerve*.
| Probe Id | Symbol | Gene | P10 LIF-/- | P10 WT | Fold change |
|---|---|---|---|---|---|
| 6020368 | Ucp1 | Ucp1; uncoupling protein 1, mitochondrial | 2,393.25 | 110.06 | 21.75 |
| 4670091 | Sspn | Sspn; sarcospan | 826.41 | 39.58 | 20.88 |
| 6660687 | Cart | Cart; cocaine and amphetamine regulated transcript | 1,494.90 | 123.50 | 12.10 |
| 2450131 | Hbb-b2 | Hbb-b2; hemoglobin, beta adult minor chain | 5,529.12 | 828.72 | 6.67 |
| 1690086 | Sparc | Sparc; secreted acidic cysteine rich glycoprotein | 4,225.86 | 649.88 | 6.50 |
| 6100707 | Eif2s3y | Eif2s3y; eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked | 375.72 | 58.63 | 6.41 |
| 2810403 | Csrp1 | Csrp1; cysteine and glycine-rich protein 1 | 2,696.27 | 488.07 | 5.52 |
| 5550020 | Fcer1g | Fcer1g; Fc receptor, IgE, high affinity I, gamma polypeptide | 247.86 | 46.99 | 5.28 |
| 5340592 | Pdrg1 | Pdrg1; p53 and DNA damage regulated 1 | 303.40 | 67.83 | 4.47 |
| 6350136 | Kera | Kera; keratocan | 200.65 | 48.57 | 4.13 |
| 2450408 | Cxcl10 | Cxcl10; chemokine (C-X-C motif) ligand 10 | 223.50 | 56.72 | 3.94 |
| 520463 | Pttg1 | Pttg1; pituitary tumor-transforming 1. | 339.04 | 86.67 | 3.91 |
| 2850735 | Gabbr1 | Gabbr1; gamma-aminobutyric acid (GABA-B) receptor, 1 | 176.01 | 45.61 | 3.86 |
| 1990451 | Tesk1 | Tesk1; testis specific protein kinase 1 | 484.81 | 131.54 | 3.69 |
| 4280546 | Cntnap2 | Cntnap2; contactin associated protein-like 2 | 445.38 | 122.23 | 3.64 |
| 630487 | Sox2 | Sox2; SRY-box containing gene 2 | 1,080.99 | 299.38 | 3.61 |
| 3940279 | Rere | Rere; arginine glutamic acid dipeptide (RE) repeats | 179.42 | 50.90 | 3.52 |
| 5340577 | Prdx2 | Prdx2; peroxiredoxin 2 | 259.13 | 75.03 | 3.45 |
| 770025 | Ercc5 | Ercc5; excision repair cross-complementing rodent repair deficiency, complementation group 5 | 479.12 | 144.45 | 3.32 |
| 460168 | Rusc2 | Rusc2; RUN and SH3 domain containing 2 | 671.15 | 207.00 | 3.24 |
| 1230707 | Greb1 | Greb1; gene regulated by estrogen in breast cancer protein | 413.82 | 133.34 | 3.10 |
| 5290673 | H2-Aa | H2-Aa; histocompatibility 2, class II antigen A, alpha | 249.47 | 83.39 | 2.99 |
| 3710040 | Tnni1 | Tnni1; troponin I, skeletal, slow 1 | 227.19 | 77.36 | 2.94 |
| 1780538 | Slc15a2 | Slc15a2; solute carrier family 15 (H+/peptide transporter), member 2 | 523.67 | 183.71 | 2.85 |
| 3060037 | Exoc7 | Exoc7; exocyst complex component 7 | 833.61 | 295.78 | 2.82 |
| 3060411 | Tnc | Tnc; tenascin C | 820.91 | 291.34 | 2.82 |
| 430047 | Ccl4 | Ccl4; chemokine (C-C motif) ligand 4 | 250.22 | 89.32 | 2.80 |
| 2120239 | Lgals3 | Lgals3; lectin, galactose binding, soluble 3 | 841.67 | 304.35 | 2.77 |
| 6420138 | Kcnn3 | Kcnn3; potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3. | 574.95 | 208.90 | 2.75 |
| 2340725 | Nell2 | Nell2; nel-like 2 homolog | 175.92 | 63.92 | 2.75 |
| 3060520 | Tnmd | Tnmd; tenomodulin | 190.99 | 69.53 | 2.75 |
| 5900576 | Ndufb10 | Ndufb10; NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 | 1,007.91 | 370.60 | 2.72 |
| 4670592 | Lip1 | Lip1; lysosomal acid lipase 1 | 534.10 | 196.73 | 2.71 |
| 630142 | Psmd8 | Psmd8; proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 | 2,340.46 | 868.09 | 2.70 |
| 6550603 | Sag | Sag; retinal S-antigen | 170.99 | 63.50 | 2.69 |
| 4560600 | Pop4 | Pop4; processing of precursor 4, ribonuclease P/MRP family | 853.32 | 320.02 | 2.67 |
| 1770161 | Phxr4 | Phxr4; per-hexamer repeat gene 4 | 1,012.18 | 384.68 | 2.63 |
| 2510044 | Nup88 | Nup88; nucleoporin 88 | 681.67 | 259.70 | 2.62 |
| 3850717 | Sv2b | Sv2b; synaptic vesicle glycoprotein 2b | 606.89 | 231.23 | 2.62 |
| 4850082 | Gdf10 | Gdf10; growth differentiation factor 10 | 2,362.73 | 907.77 | 2.60 |
| 2230497 | Hs3st3a1 | Hs3st3a1; heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 | 442.44 | 171.86 | 2.57 |
| 3440215 | Fbln2 | Fbln2; fibulin 2 | 927.92 | 360.87 | 2.57 |
| 2940278 | Gpr73 | Gpr73; G protein-coupled receptor 73 | 299.89 | 117.15 | 2.56 |
| 2360280 | Cox8b | Cox8b; cytochrome c oxidase, subunit VIIIb | 614.95 | 242.02 | 2.54 |
| 6020605 | Mgst1 | Mgst1; microsomal glutathione S-transferase 1 | 298.00 | 117.89 | 2.53 |
| 1660301 | Spon1 | Spon1; spondin 1, (f-spondin) extracellular matrix protein | 650.87 | 262.13 | 2.48 |
| 3940736 | Gbx2 | Gbx2; gastrulation brain homeobox 2 | 676.27 | 273.56 | 2.47 |
| 2940138 | Ppnr | Ppnr; per-pentamer repeat gene | 421.21 | 171.44 | 2.46 |
| 4590519 | Cxcr4 | Cxcr4; chemokine (C-X-C motif) receptor 4 | 846.22 | 345.20 | 2.45 |
| 6220398 | Myh7 | Myh7; myosin, heavy polypeptide 7, cardiac muscle, beta | 210.99 | 86.14 | 2.45 |
Gene expression profiling was used to identify the molecular consequences of LIF gene knock out in development of optic nerve. Normalized intensity value for each gene hybridization is listed for P10 LIF knockout and P10 LIF wild-type optic nerve. Fold changes are listed with respect to P10 LIF knockout.
Table II. Top 50 mRNAs Down-Regulated in P10 LIF Knockout Optic Nerve*.
| Probe Id | Symbol | Gene | P10 LIF-/- | P10 WT | Fold change | |
|---|---|---|---|---|---|---|
| 630403 | Cga | Cga; glycoprotein hormones, alpha subunit | 69.29 | 638.02 | -9.21 | |
| 2760204 | Cntn2 | Cntn2; contactin 2 | 73.08 | 663.95 | -9.09 | |
| 5910341 | Slp | Slp; sex-limited protein | 248.04 | 2,023.39 | -8.16 | |
| 1050193 | Trf | Trf; transferrin | 469.36 | 3,010.32 | -6.41 | |
| 1340092 | Slp | Slp; sex-limited protein | 433.53 | 2,778.56 | -6.41 | |
| 4280487 | Mal | Mal; myelin and lymphocyte protein, T-cell differentiation protein | 692.67 | 3,782.95 | -5.46 | |
| 6770037 | Ccl21c | Ccl21c; chemokine (C-C motif) ligand 21c (leucine) | 68.72 | 371.77 | -5.41 | |
| 4670176 | Ccl21b | Ccl21b; chemokine (C-C motif) ligand 21b (serine) | 49.95 | 248.06 | -4.97 | |
| 4810102 | Fbxw5 | Fbxw5; F-box and WD-40 domain protein 5 | 162.55 | 774.33 | -4.76 | |
| 5340400 | Serpina3n | Serpina3n; serine (or cysteine) proteinase inhibitor, clade A, member 3N | 639.31 | 2,890.42 | -4.52 | |
| 5050338 | Ccl21a | Ccl21a; chemokine (C-C motif) ligand 21a (leucine) | 63.41 | 286.05 | -4.51 | |
| 1940102 | Myh4 | Myh4; myosin, heavy polypeptide 4, skeletal muscle | 35.73 | 161.17 | -4.51 | |
| 3780113 | S100a4 | S100a4; S100 calcium binding protein A4 | 502.06 | 2,253.03 | -4.49 | |
| 4230427 | H2-Ab1 | H2-Ab1; histocompatibility 2, class II antigen A, beta 1 | 140.66 | 618.02 | -4.39 | |
| 3830100 | C4 | C4; complement component 4 (within H-2S) | 116.68 | 493.47 | -4.23 | |
| 360286 | C4 | C4; complement component 4 (within H-2S) | 59.14 | 249.85 | -4.22 | |
| 4920441 | Apod | Apod; apolipoprotein D | 831.62 | 3,398.07 | -4.09 | |
| 3840528 | Tmem10 | Tmem10; transmembrane protein 10 | 1,016.63 | 4,011.12 | -3.95 | |
| 1240441 | Gjb1 | Gjb1; gap junction membrane channel protein beta 1 | 96.68 | 369.01 | -3.82 | |
| 5690441 | Mgl2 | Mgl2; macrophage galactose N-acetyl-galactosamine specific lectin 2 | 54.12 | 196.10 | -3.62 | |
| 5890519 | Prkcn | Prkcn; protein kinase C, nu | 132.98 | 477.49 | -3.59 | |
| 2350079 | H2-Eb1 | H2-Eb1; histocompatibility 2, class II antigen E beta | 41.99 | 145.72 | -3.47 | |
| 5720041 | Map3k4 | Map3k4; mitogen activated protein kinase kinase kinase 4 | 32.70 | 112.92 | -3.45 | |
| 4920053 | Nnat | Nnat; neuronatin (Nnat), transcript variant 2. | 366.05 | 1247.47 | -3.41 | |
| 3870010 | Ccl8 | Ccl8; chemokine (C-C motif) ligand 8 | 35.92 | 122.33 | -3.41 | |
| 4060047 | B3galt5 | B3galt5; UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 | 71.47 | 241.71 | -3.38 | |
| 5420075 | Clu | Clu; clusterin | 1,982.56 | 6,679.09 | -3.37 | |
| 6840110 | Evi2a | Evi2a; ecotropic viral integration site 2a | 171.27 | 561.19 | -3.28 | |
| 2900091 | Stmn4 | Stmn4; stathmin-like 4 | 244.06 | 789.46 | -3.23 | |
| 1740372 | C3 | C3; complement component 3 | 102.36 | 330.49 | -3.23 | |
| 2810551 | Pdlim2 | Pdlim2; PDZ and LIM domain 2 | 472.87 | 1,521.88 | -3.22 | |
| 6400162 | Ii | Ii; Ia-associated invariant chain (Ii). | 132.03 | 414.73 | -3.14 | |
| 4810050 | Rmcs1 | Rmcs1; histocompatibility 2, class II antigen A, beta 1 | 100.66 | 304.99 | -3.03 | |
| 1990056 | Chga | Chga; chromogranin A | 64.74 | 195.78 | -3.02 | |
| 6550463 | Chga | Chga; chromogranin A | 35.92 | 107.84 | -3.00 | |
| 3850100 | Tmod1 | Tmod1; tropomodulin 1 | 56.77 | 170.38 | -3.00 | |
| 540079 | Cdc42ep2 | Cdc42ep2; CDC42 effector protein (Rho GTPase binding) 2 | 514.67 | 1538.92 | -2.99 | |
| 870114 | Ppp1r14a | Ppp1r14a; protein phosphatase 1, regulatory (inhibitor) subunit 14A | 119.52 | 355.47 | -2.97 | |
| 6020722 | Mog | Mog; myelin oligodendrocyte glycoprotein | 1,094.35 | 3,252.24 | -2.97 | |
| 5570450 | Pvalb | Pvalb; parvalbumin | 399.51 | 1185.35 | -2.97 | |
| 6110059 | Serpind1 | Serpind1; serine (or cysteine) proteinase inhibitor, clade D, member 1 | 197.43 | 585.64 | -2.97 | |
| 4220482 | H2-T10 | H2-T10; histocompatibility 2, T region locus 10 | 70.99 | 206.68 | -2.91 | |
| 610364 | Mog | Mog; myelin oligodendrocyte glycoprotein | 933.60 | 2,673.27 | -2.86 | |
| 70451 | Ctps | Ctps; cytidine 5-triphosphate synthase | 94.40 | 264.78 | -2.80 | |
| 6550204 | H2-Q2 | H2-Q2; histocompatibility 2, Q region locus 2 | 223.78 | 627.02 | -2.80 | |
| 4810619 | Cryab | Cryab; crystallin, alpha B | 1,920.86 | 5,376.59 | -2.80 | |
| 2760014 | Admr | Admr; adrenomedullin receptor | 218.19 | 608.39 | -2.79 | |
| 1940600 | Hist1h4a | Hist1h4a; histone 1, H4a | 191.27 | 526.06 | -2.75 | |
| 4850132 | Adssl1 | Adssl1; adenylosuccinate synthase like 1 | 274.87 | 747.45 | -2.72 | |
| 6200541 | Kcnk13 | Kcnk13; potassium channel, subfamily K, member 13 | 184.16 | 500.77 | -2.72 | |
Gene expression profiling identified the most highly down-regulated gene transcripts in developing optic nerve of LIF gene knockout mice. Normalized intensity value for each gene hybridization is listed for P10 LIF knockout and P10 LIF wild-type optic nerve. Fold changes are listed with respect to P10 LIF knockout.
Table III. Selected Structural Components of Myelin (Fold Changes for P10, P14, and Adult With Respect to LIF-/- Optic Nerve).
| Probe Id | Symbol | Gene | P10 fold change | P14 fold change | Adult fold change |
|---|---|---|---|---|---|
| 4280487 | Mal | Mal; myelin and lymphocyte protein, T-cell differentiation protein | -5.46 | -1.03 | 1.07 |
| 6020722 | Mog | Mog; myelin oligodendrocyte glycoprotein | -2.97 | -1.51 | -1.10 |
| 610364 | Mog | Mog; myelin oligodendrocyte glycoprotein | -2.86 | -1.63 | -1.10 |
| 4590239 | Mal | Mal; myelin and lymphocyte protein, T-cell differentiation protein | -2.28 | -1.24 | -1.16 |
| 2260451 | Cnp1 | Cnp1; cyclic nucleotide phosphodiesterase 1 | -1.85 | -1.18 | 1.05 |
| 6760066 | Omg | Omg; oligodendrocyte myelin glycoprotein | -1.62 | -1.16 | 1.23 |
| 2370037 | Mag | Mag; myelin-associated glycoprotein | -1.34 | 1.03 | -1.02 |
| 7000059 | Mobp | Mobp; myelin-associated oligodendrocytic basic protein | -1.31 | -1.11 | 1.04 |
| 5220369 | Mbp | Mbp; myelin basic protein | -1.10 | -1.01 | 1.01 |
| 460273 | Plp | Plp; proteolipid protein | 1.37 | -1.39 | 1.03 |
We were also interested in transcriptional regulation of oligodendrocyte development in the LIF-/- optic nerve. MBP immunostaining at P10 in the LIF-/- optic nerve indicated a severe defect in myelination and oligodendrocyte development. To characterize the transcriptional network responsible for oligodendrocyte development, we have detailed the expression level and -fold change of many of the transcription factors expressed by cells of the oligodendrocyte lineage (Nicolay et al.,[2007]) at P10, P14, and P35 (adult) in Table IV. The observed up-regulation of transcriptional repressors known to inhibit myelin gene expression, such as Hes5, Idb4, and Tcf4, and a down-regulation of transcription factors known to promote oligodendrocyte development, such as Olig1, Nkx6.2, and Pou3f1 (Table IV), suggest that specific components of the transcriptional network required to promote oligodendrocyte differentiation and myelination are deregulated in the absence of LIF at P10, thus resulting in a severe myelination defect, as seen in Figure 1. At P14 in LIF-/-, the listed transcription factor mRNAs are mostly at wild-type levels. This is consistent with MBP immunostaining at P14, which shows very little difference between LIF-/- optic nerve and LIF+/+ optic nerve at this developmental time point.
Table IV. Selected Transcription Factors Implicated in Oligodendrocyte Development (Fold Changes for P10, P14, and Adult With Respect to LIF-/- Optic Nerve; Nicolay et al.,[2007]; Wegner,[2008]).
| Probe Id | Symbol | Gene | P10 fold change | P14 fold change | Adult fold change | |
|---|---|---|---|---|---|---|
| 4920026 | Hes5 | Hes5; hairy and enhancer of split 5 | 1.69 | -1.03 | 1.30 | |
| 6650100 | Ascl1 | Ascl1; achaete-scute complex homolog-like 1 | 1.60 | 1.03 | 1.03 | |
| 4610347 | Egr1 | Egr1; early growth response 1 | 1.55 | -1.12 | 1.35 | |
| 5270551 | Sox17 | Sox17; SRY-box containing gene 17 | 1.55 | -1.21 | 1.95 | |
| 3830152 | Idb4 | Idb4; inhibitor of DNA binding 4 | 1.55 | -1.35 | -1.11 | |
| 770138 | Tcfe2a | Tcfe2a; transcription factor E2a | 1.45 | -1.04 | -1.11 | |
| 3190128 | Sox5 | Sox5; SRY-box containing gene 5 | 1.44 | 1.11 | 1.07 | |
| 2940458 | Sox6 | Sox6; SRY-box containing gene 6 | 1.37 | 1.01 | -1.00 | |
| 520021 | Tcf4 | Tcf4; transcription factor 4 | 1.37 | -1.08 | -1.23 | |
| 2260091 | Sox4 | Sox4; SRY-box containing gene 4 | 1.18 | 1.73 | -1.37 | |
| 4150731 | Nkx2-2 | Nkx2-2; NK2 transcription factor related, locus 2 | 1.12 | 1.36 | -1.74 | |
| 6770524 | Myt1 | Myt1; myelin transcription factor 1 | 1.07 | 1.16 | -1.01 | |
| 5720681 | Sox9 | Sox9; SRY-box containing gene 9 | 1.06 | 1.14 | -1.45 | |
| 6200538 | Sox10 | Sox10; SRY-box containing gene 10 | -1.10 | -1.12 | -1.30 | |
| 6620484 | Pou3f2 | Pou3f2; POU domain, class 3, transcription factor 2 | -1.15 | 1.17 | 1.07 | |
| 1660594 | Olig1 | Olig1; oligodendrocyte transcription factor 1 | -1.18 | -1.11 | 1.05 | |
| 580026 | Sox8 | Sox8; SRY-box containing gene 8 | -1.21 | 1.15 | -1.01 | |
| 6660402 | Olig2 | Olig2; oligodendrocyte transcription factor 2 | -1.33 | -1.41 | 1.15 | |
| 3710022 | Pou3f1 | Pou3f1; POU domain, class 3, transcription factor 1 | -1.45 | -1.28 | -1.05 | |
| 3120333 | Nkx6-2 | Nkx6-2; NK6 transcription factor related, locus 2 | -1.96 | 1.15 | -1.18 | |
| 2450332 | Nkx6-2 | Nkx6-2; NK6 transcription factor related, locus 2 | -2.24 | 1.10 | -1.24 | |
For OPCs to mature, they must exit the cell cycle and begin a program of differentiation. Our data suggest that LIF promotes this transition at about P10. Our microarray data support the conclusion that, in the LIF-/- optic nerve at P10, the oligodendrocyte population is immature and unable to exit the cell cycle and therefore highly proliferative. Table V lists selected cell cycle components, and the data indicate an increase in several cyclin genes in support of the conclusion that there is a highly proliferative population of cells present in the optic nerve of LIF-/- animals at P10. By P14 in the optic nerve of LIF-/- animals, most cyclins have recovered to wild-type levels, with the exception of cyclin D1, indicating that the cell population is likely to be less proliferative at this time point. In the adult LIF-/- optic nerve, cyclin levels show no change or a decrease from wild type. We have also compared our data set from P10 with that of several published gene expression studies on oligodendrocyte development (Dugas et al., 2006; Cahoy et al.,[2008]; Supp. Info. Tables I and II). Among transcripts regulated in the LIF-/- optic nerve, there are 479 mRNA transcripts that exhibit ≥ 1.5-fold up-regulation; 204 of these transcripts are enriched (4,582 genes ≥ 1.5-fold enrichment) in OPCs over mature oligodendrocytes (Cahoy et al., [2008]); additionally, among the 550 transcripts down-regulated in the LIF-/- optic nerve at P10, ≥ 1.5-fold, 203 are enriched (2,070 genes ≥ 1.5-fold enrichment) in mature oligodendrocytes over OPCs (Cahoy et al.,[2008]). These data strongly suggest that, at P10 in the LIF-/- optic nerve, there are an overrepresentation of highly immature OPCs and an underrepresentation of mature oligodendrocytes.
Table V. Selected Cell Cycle Components (Fold Changes for P10, P14, and Adult With Respect to LIF-/- Optic Nerve).
| Probe Id | Symbol | Gene | P10 fold change | P14 fold change | Adult fold change |
|---|---|---|---|---|---|
| 3190095 | Ccng2 | Ccng2; cyclin G2 | 1.86 | 1.00 | 1.07 |
| 5340167 | Ccnd2 | Ccnd2; cyclin D2 | 1.82 | 1.13 | -2.05 |
| 3120576 | Ccnd1 | Ccnd1; cyclin D1 | 1.62 | 1.64 | -1.51 |
| 4780372 | Ccnb1 | Ccnb1; cyclin B1 | 1.52 | 1.15 | -1.04 |
| 5290075 | Ccna2 | Ccna2; cyclin A2 | 1.47 | 1.13 | -1.06 |
Taken together, the gene expression data strongly suggest that OPCs appear to be held in an immature stage at P10 in the LIF-/- optic nerve, with cells inhibited from exiting the cell cycle and from progressing along a differentiaton/myelination program. This transcriptomic phenotype correlates with immunostaining of myelin components and Olig2 in the optic nerve, and a recovery is seen at P14, with very little difference in myelin gene mRNA at P35 (adult). These results could also reflect a delay in migration of OPCs into the optic nerve of LIF-/- mice; however, we were unable to find evidence of a migration defect in two different in vitro assays or in the distribution of PDGFRα-expressing cells along the P10 optic nerve (Fig. 3F).
Discussion
This study represents the first analysis of the affect of LIF signaling on myelination during early postnatal development in the CNS. We have used an in vivo system with the targeted disruption of the LIF gene to investigate the affect of LIF signaling in myelination, and this revealed that LIF regulates the timing of oligodendrocyte precursor cell (OPC) development and myelination in the optic nerve. We have shown that, in the optic nerve of P10 LIF-/- mice, there are severe defects in myelination, as measured by MBP and PLP immunoreactivity. Microarray analysis of structural components of myelin shows that many myelin gene mRNAs are down-regulated at this developmental time point. We have also shown that the OPC population, as measured by Olig2 immunoreactivity, at P10 in LIF-/- mice is greatly reduced and restricted to the chiasm region of the optic nerve. We have further characterized the OPC population in LIF-/- mice at P10 and have shown this population to be in a highly proliferative and developmentally immature state compared with oligodendrocyte-lineage cells in wild-type optic nerve. This apparent contradiction in reduced oligodendrocyte-lineage (Olig2-positive) cells in the LIF-/- optic nerve and increased proliferation in the OPC (NG2-positive) pool could be explained by yet another potential function of LIF, the conversion of a multipotent Olig2-negative population into an Olig2-positive population in the optic nerve. Further analysis of the microarray data confirms that the cells in the optic nerve at P10 have an overabundance of several cyclin mRNAs and a dysregulation of much of the transcriptional machinery required for oligodendrocyte development. Thus, several lines of evidence strongly implicate LIF in the timing of oligodendrocyte development and myelination in the mouse postnatal optic nerve.
Our findings are consistent with and greatly expand previous evidence suggesting a role for LIF in myelination. A previous study reported that adult female LIF-/- mice (2-5 months old) had decreased immunoreactivity against MBP in several regions in brain, such as cerebellum, corpus callosum, and striatum, but this was not evident in male mice (Bugga et al.,[1998]). Our results, however, show a significant difference in myelination of optic nerve of LIF-/- mice in both sexes at 10 days of age. Neuropoietic cytokines, including LIF, are known to promote the maturation and survival of oligodendrocytes (Barres et al.,[1993]). Also, when EAE is induced in the presence of anti-LIF antibodies, there is enhanced severity of clinical symptoms and increased demyelination, axonal injury, and oligodendrocyte death (Butzkueven et al.,[2006]). Exogenous LIF promotes oligodendrocyte survival and myelin sparing in EAE (Butzkueven et al.,[2002]) and affects CNS myelination in cell culture (Park et al.,[2001]; Stankoff et al.,[2002]; Ishibashi et al.,[2006]). However, the mechanism of LIF signaling during normal oligodendrocyte development in vivo has not been elucidated.
Our data showing a gradient in OPC development in optic nerve of LIF-/- mice are consistent with either a developmental delay or an affect of LIF on OPC migration into optic nerve. In mouse optic nerves, OPCs that are labeled by an antibody against the NG2 proteoglycan (Levine and Stallcup,[1987]) migrate into optic nerve from the brain through the optic chiasm at about P0, then move toward the retina and reach the boundary of future myelinated and unmyelinated regions at P7 (Baba et al.,[1999]). By 10 days of age, MBP-positive myelin sheaths are observed throughout the optic nerves (Fig. 1A), and Olig2-positive OPCs are distributed uniformly in the optic nerve (Fig. 2A). We have shown at P10 in the LIF-/- optic nerve a deficit in MBP and PLP immunoreactivity (Fig. 1C,F) and a reduction in the overall number and an abnormal distribution of Olig2-positive OPCs along the optic nerve (Fig. 2C,E).
The differentiation of OPCs into myelinating oligodendrocytes occurs only once they have reached their final destination (Miller,[1996]). A large percentage of OPCs in the LIF-/- optic nerve remained at an immature developmental stage and NG2-positive/multipolar-shaped (late oligodendrocyte progenitors; Fig. 3B,D,E), and they were observed mainly in the chiasmal region (Fig. 2C). This suggests that OPCs in LIF-/- optic nerve could not reach their final destination at 10 days of age. Some Olig2-positive cells migrated near the retina; however, they did not show any MBP immunoreactivity (Fig. 2E). We did not see any difference in migration of LIF-/- optic nerve glial cells in two in vitro assays of cell migration (data not shown), and, consistent with this, immature PDGFRα-positive OPCs were uniformly distributed throughout the optic nerve of P10 LIF-/- mice. However, additional research may be required to explore possible affects of LIF on OPC migration that could contribute to the established roles of LIF signaling on OPC proliferation, differentiation, and myelination. Taken together, the data presented here strongly suggest that LIF is required for the correct timing of oligodendrocyte development and myelination in the postnatal optic nerve.
Gene expression analysis of P10 optic nerves further characterizes and confirms the in vivo phenotype observed by MBP immunostaining, Olig2 immunostaining, and NG2 immunostaining of optic nerves at P10. LIF appears to play a key role in oligodendrocyte development by controlling, either directly or indirectly, the expression of a network of transcriptional repressors responsible for maintaining an immature stage in OPC development. This could be a consequence of LIF signaling affecting OPC differentiation and myelination or alternatively the consequence of a migration defect enriching the population of optic nerve glia at an immature stage in P10 optic nerve. In either case, the microarray data confirm that the downstream effect of a lack of LIF as a factor required to promote OPC differentiation reduces the number of Olig2-positive cells and increases NG2-positive cells in P10 optic nerve and thus results in the observed defect in myelination as seen by MBP immunoreactivity. Several lines of evidence deriving from the overrepresentation of OPC enriched genes in the optic nerve of P10 LIF-/- animals; deregulation of specific transcription factors, both repressors and activators; and also myelin structural gene expression are all consistent with LIF signaling regulating the transition from a pool of immature OPCs in the optic nerve to a myelinating mature oligodendrocyte phenotype during postnatal development. The apparent recovery of myelination in the optic nerve by P14, as measured by MBP immunoreactivity and many of the myelin gene mRNAs being at wild-type levels, implies that another secreted factor most likely can compensate for a lack of LIF during development at this time point. Other secreted factors that may compensate for the LIF defect include CNTF and IL-6 and others that bind a gp130/LIF receptor complex (for review see Bauer et al.,[2007]). Interestingly, many of the key transcription factors required for repression and/or activation of transcription at myelin gene promoters are at wild-type levels by P14 in the LIF-/- optic nerve (Table IV).
A possible mechanism by which LIF could regulate OPC maturation and myelination is by modulating intracellular signaling pathways regulating many of the transcription factors implicated in oligodendrocyte development and transcription of myelin genes, such as E47, Olig1, Hes5, Nkx6.2, Tcf4, Id4, Mash1, and Krox24 (Table IV). All of these transcription factors interact in transcriptional networks as repressors and/or activators and thereby influence oligodendrocyte maturation and myelination of the CNS. Lack of LIF during postnatal development may perturb notch signaling through a common intermediary, such as phosphorylation of Stat3; notch signaling has previously been implicated in oligodendrocyte development in the optic nerve (Wang et al.,[1998]).
Additionally, the in vivo gene expression data generated during this study will prove to be a useful tool in the identification of novel regulators of optic nerve development and a consequence of a lack of LIF signaling on other aspects of early postnatal and adult optic nerve beyond effects on myelination. This dataset will allow identification of other secreted factors that may compensate for the lack of LIF and rescue the LIF-/- phenotype seen at later stages of development, including into adulthood.
Currently the activity of cytokines as growth factors in regulating nervous system development is an important area of research, which has particular relevance to developmental disorders resulting from inflammatory conditions experienced during the perinatal period. Cytokine production is also characteristic of many demyelinating diseases, and the effects of LIF and other cytokines on oligodendrocyte proliferation and differentiation are relevant to multiple sclerosis and other demyelinating disorders. Interestingly, LIF has been found to regulate the proliferation and formation of glial progenitors in the brain (Bauer et al.,[2006]). This finding may be consistent with our data implicating LIF in the proliferation and differentiation of OPCs in the optic nerve.
Finally, the narrow developmental period when LIF effects on development of optic nerve oligodendrocytes and myelination can be observed is interesting. In general, this is consistent with the transient appearance of other cytokines during nervous system development. We have shown recently that activity-dependent regulation of hippocampal astrocyte development is mediated in part by LIF signaling, stimulated by the activity-dependent release of ATP form neurons (Cohen and Fields,[2008]). Previous research showing that myelination is regulated by impulse activity in axons, in part through a mechanism involving LIF signaling (Ishibashi et al.,[2006]), suggests the possibility that the developmental period when LIF influences OPC proliferation and myelination in optic nerve may be related to regulation of myelination by impulse activity in axons resulting from early visual experience (for review see Fields,[2008]).
Supplementary Material
Acknowledgments
We thank Dr. Hirohide Takebayashi for the polyclonal antibody against Olig2 and Peter Wadeson for assistance with cell culture. We also thank Colin Stewart for the LIF-/- mice, Daniel Abebe for assistance with experimental animals, and Chris Cheadle for help with microarray analysis.
Funded by:
NICHD
Japan Society for the Promotion of Science; Grant Number: 19800043
References
- Baba H, Akita H, Ishibashi T, Inoue Y, Nakahira K, Ikenaka K. Completion of myelin compaction, but not the attachment of oligodendroglial processes triggers K+ channel clustering. J Neurosci Res. 1999;58:752–764. [PubMed] [Google Scholar]
- Barres BA, Schmid R, Sendnter M, Raff MC. Multiple extracellular signals are required for long-term oligodendrocyte survival. Development. 1993;118:283–295. doi: 10.1242/dev.118.1.283. [DOI] [PubMed] [Google Scholar]
- Bauer S, Patterson PH. Leukemia inhibitory factor promotes neural stem cell self-renewal in the adult brain. J Neurosci. 2006;26:12089–12099. doi: 10.1523/JNEUROSCI.3047-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Kerr BJ, Patterson PH. The neuropoietic cytokine family in development, plasticity, disease and injury. Nat Rev Neurosci. 2007;8:221–232. doi: 10.1038/nrn2054. [DOI] [PubMed] [Google Scholar]
- Bugga L, Gadient RA, Kwan K, Stewart CL, Patterson PH. Analysis of neuronal and glial phenotypes in brains of mice deficient in leukemia inhibitory factor. J Neurobiol. 1998;36:509–524. doi: 10.1002/(sici)1097-4695(19980915)36:4<509::aid-neu5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Butzkueven H, Zhang JG, Soilu-Hanninen M, Hochrein H, Chionh F, Shipham KA, Emery B, Turnley AM, Petratos S, Ernst M, Bartlett PF, Kilpatrick TJ. LIF receptor signaling limits immune-mediated demyelination by enhancing oligodendrocyte survival. Nat Med. 2002;8:613–619. doi: 10.1038/nm0602-613. [DOI] [PubMed] [Google Scholar]
- Butzkueven H, Emery B, Cipriani T, Marriott MP, Kilpatrick TJ. Endogenous leukemia inhibitory factor production limits autoimmune demyelination and oligodendrocyte loss. Glia. 2006;53:696–703. doi: 10.1002/glia.20321. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagnoni CW, Kampf K, Mason B, Handley VW, Campagnoni AT. Isolation and characterization of a cDNA encoding the zebra finch myelin proteolipid protein. Neurochem Res. 1994;19:1061–1065. doi: 10.1007/BF00968717. [DOI] [PubMed] [Google Scholar]
- Cohen JE, Fields RD. Activity-dependent neuron-glial signaling by ATP and leukemia-inhibitory factor promotes hippocampal glial cell development. Neuron Glia Biol. 2008;4:43–55. doi: 10.1017/S1740925X09000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B, Raff M. A cell-intrinsic timer that operates during oligodendrocyte development. Bioessays. 2000;22:64–71. doi: 10.1002/(SICI)1521-1878(200001)22:1<64::AID-BIES11>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruttiger M, Calver AR, Richardson WD. Platelet-derived growth factor is constitutively secreted from neuronal cell bodies but not from axons. Curr Biol. 2000;10:1283–1286. doi: 10.1016/s0960-9822(00)00757-0. [DOI] [PubMed] [Google Scholar]
- Fields RD. White matter in learning, cognition, and psychiatric disorder. Trends Neurosci. 2008;31:361–370. doi: 10.1016/j.tins.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Macklin W, Gerson J, Miller RH. Intrinsic and extrinsic inhibition of oligodendrocyte development by rat retina. Dev Biol. 2006;290:277–286. doi: 10.1016/j.ydbio.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Ip NY, Nye SH, Boulton TG, Davis S, Taga T, Li Y, Birren SJ, Yasukawa K, Kishimoto T, Anderson DJ, Stahl N, Yancopoulos GD. CNTF and LIF act on neuronal cells via shared signaling pathways that involve the IL-6 signal transducing receptor component gp130. Cell. 1992;69:1121–1132. doi: 10.1016/0092-8674(92)90634-o. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Dupree JL, Ikenaka K, Hirahara Y, Honke K, Peles E, Popko B, Suzuki K, Nishino H, Baba H. A myelin galactolipid, sulfatide, is essential for maintenance of ion channels on myelinated axon but not essential for initial cluster formation. J Neurosci. 2002;22:6507–6514. doi: 10.1523/JNEUROSCI.22-15-06507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T, Dakin KA, Stevens B, Lee PR, Kozlov SV, Stewart CL, Fields RD. Astrocytes promote myelination in response to electrical impulses. Neuron. 2006;49:823–832. doi: 10.1016/j.neuron.2006.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarjour AA, Manitt C, Moore SW, Thompson KM, Yuh SJ, Kennedy TE. Netrin-1 is a chemorepellent for oligodendrocyte precursor cells in the embryonic spinal cord. J Neurosci. 2003;23:3735–3744. doi: 10.1523/JNEUROSCI.23-09-03735.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn MA, DeVellis J. Regulation of an oligodendrocyte progenitor cell line by the interleukin-6 family of cytokines. Glia. 1994;12:87–98. doi: 10.1002/glia.440120202. [DOI] [PubMed] [Google Scholar]
- Kerr BJ, Patterson PH. Leukemia inhibitory factor promotes oligodendrocyte survival after spinal cord injury. Glia. 2005;51:73–79. doi: 10.1002/glia.20177. [DOI] [PubMed] [Google Scholar]
- Levine JM, Stallcup WB. Plasticity of developing cerebellar cells in vitro studied with antibodies against the NG2 antigen. J Neurosci. 1987;7:2721–2731. doi: 10.1523/JNEUROSCI.07-09-02721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriott MP, Emery B, Cate HS, Binder MD, Kemper D, Wu Q, Kolbe S, Gordon IR, Wang H, Egan G, Murray S, Butzkueven H, Kilpatrick TJ. Leukemia inhibitory factor signaling modulates both central nervous system demyelination and myelin repair. Glia. 2008;56:686–698. doi: 10.1002/glia.20646. [DOI] [PubMed] [Google Scholar]
- Mi H, Barres BA. Purification and characterization of astrocyte precursor cells in the developing rat optic nerve. J Neurosci. 1999;19:1049–1061. doi: 10.1523/JNEUROSCI.19-03-01049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikoshiba K, Okano H, Tamura T, Ikenaka K. Structure and function of myelin protein genes. Annu Rev Neurosci. 1991;14:201–217. doi: 10.1146/annurev.ne.14.030191.001221. [DOI] [PubMed] [Google Scholar]
- Miller RH. Oligodendrocyte origins. Trends Neurosci. 1996;19:92–96. doi: 10.1016/s0166-2236(96)80036-1. [DOI] [PubMed] [Google Scholar]
- Miller RH, Payne J, Milner L, Zhang H, Orentas DM. Spinal cord oligodendrocytes develop from a limited number of migratory highly proliferative precursors. J Neurosci Res. 1997;50:157–168. doi: 10.1002/(SICI)1097-4547(19971015)50:2<157::AID-JNR5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Taga T. Astrocyte differentiation mediated by LIF in cooperation with BMP2. FEBS. 1999;457:43–46. doi: 10.1016/s0014-5793(99)00997-7. [DOI] [PubMed] [Google Scholar]
- Nicolay DJ, Doucette JR, Nazarali AJ. Transcriptional control of oligodendrogenesis. Glia. 2007;55:1287–1299. doi: 10.1002/glia.20540. [DOI] [PubMed] [Google Scholar]
- Park SK, Solomon D, Vartanian T. Growth factor control of CNS myelination. Dev Neurosci. 2001;23:327–337. doi: 10.1159/000048716. [DOI] [PubMed] [Google Scholar]
- Raff MC, Durand B, Gao FB. Cell number control and timing in animal development: the oligodendrocyte cell lineage. Int J Dev Biol. 1998;42:263–267. [PubMed] [Google Scholar]
- Schönrock LM, Gawlowski G, Brück W. Interleukin-6 expression in human multiple sclerosis lesions. Neurosci Lett. 2000;294:45–48. doi: 10.1016/s0304-3940(00)01543-3. [DOI] [PubMed] [Google Scholar]
- Sherry DM, Mitchell R, Li H, Graham DR, Ash JD. Leukemia inhibitory factor inhibits neuronal development and disrupts synaptic organization in the mouse retina. J Neurosci Res. 2005;82:316–332. doi: 10.1002/jnr.20619. [DOI] [PubMed] [Google Scholar]
- Spassky N, de Castro F, Le Bras B, Heydon K, Queraud-LeSaux F, Bloch-Gallego E, Chedotal A, Zalc B, Thomas JL. Directional guidance of oligodendroglial migration by class 3 semaphorins and netrin-1. J Neurosci. 2002;22:5992–6004. doi: 10.1523/JNEUROSCI.22-14-05992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankoff B, Aigrot MS, Noel F, Wattilliaux A, Zalc B, Lubetzki C. Ciliary neurotrophic factor (CNTF) enhances myelin formation: a novel role for CNTF and CNTF-related molecules. J Neurosci. 2002;22:9221–9227. doi: 10.1523/JNEUROSCI.22-21-09221.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B, Fields RD. Response of Schwann cells to action potentials in development. Science. 2000;287:2267–2271. doi: 10.1126/science.287.5461.2267. [DOI] [PubMed] [Google Scholar]
- Stevens B, Porta S, Haak LL, Gallo V, Fields RD. Adenosine: a neuron-glial transmitter promoting myelination in the CNS in response to action potentials. Neuron. 2002;36:855–868. doi: 10.1016/s0896-6273(02)01067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CL, Kaspar P, Brunet LJ, Bhatt H, Gadi I, Köntgen F, Abbondanzo SJ. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359:76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- Sugimoto Y, Taniguchi M, Yagi T, Akagi Y, Nojyo Y, Tamamaki N. Guidance of glial precursor cell migration by secreted cues in the developing optic nerve. Development. 2001;128:3321–3330. doi: 10.1242/dev.128.17.3321. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Yoshida S, Sugimori M, Kosako H, Kominami R, Nakafuku M, Nabeshima Y. Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech Dev. 2000;99:143–148. doi: 10.1016/s0925-4773(00)00466-4. [DOI] [PubMed] [Google Scholar]
- Vanderlocht J, Hellings N, Hendriks JJ, Vandenabeele F, Moreels M, Buntinx M, Hoekstra D, Antel JP, Stinissen P. Leukemia inhibitory factor is produced by myelin-reactive T cells from multiple sclerosis patients and protects against tumor necrosis factor-alpha-induced oligodendrocyte apoptosis. J Neurosci Res. 2006;83:763–774. doi: 10.1002/jnr.20781. [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres BA. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- Wegner M. A matter of identity: transcriptional control in oligodendrocytes. J Mol Neuroscience. 2008;35:3–12. doi: 10.1007/s12031-007-9008-8. [DOI] [PubMed] [Google Scholar]
- Yamamura T, Konola JT, Wekerle H, Lees MB. Monoclonal antibodies against myelin proteolipid protein: identification and characterization of two major determinants. J Neurochem. 1991;57:1671–1680. doi: 10.1111/j.1471-4159.1991.tb06367.x. [DOI] [PubMed] [Google Scholar]
- Zalc B, Fields RD. Do action potentials regulate myelination? Neuroscientist. 2000;6:5–13. doi: 10.1177/107385840000600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Vutskits L, Calaora V, Durbec P, Kiss JZ. A role for the polysialic acid-neural cell adhesion molecule in PDGF-induced chemotaxis of oligodendrocyte precursor cells. J Cell Sci. 2004;117:93–103. doi: 10.1242/jcs.00827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.