Abstract
Many bacterial pathogens of plants and animals have evolved a specialized protein-secretion system termed type III to deliver bacterial proteins into host cells. These proteins stimulate or interfere with host cellular functions for the pathogen's benefit. The Salmonella typhimurium pathogenicity island 1 encodes one of these systems that mediates this bacterium's ability to enter nonphagocytic cells. Several components of this type III secretion system are organized in a supramolecular structure termed the needle complex. This structure is made of discrete substructures including a base that spans both membranes and a needle-like projection that extends outward from the bacterial surface. We demonstrate here that the type III secretion export apparatus is required for the assembly of the needle substructure but is dispensable for the assembly of the base. We show that the length of the needle segment is determined by the type III secretion associated protein InvJ. We report that InvG, PrgH, and PrgK constitute the base and that PrgI is the main component of the needle of the type III secretion complex. PrgI homologs are present in type III secretion systems from bacteria pathogenic for animals but are absent from bacteria pathogenic for plants. We hypothesize that the needle component may establish the specificity of type III secretion systems in delivering proteins into either plant or animal cells.
Keywords: bacterial pathogenesis, type III secretion, organelle assembly
A number of bacteria that are pathogenic for plants and animals have evolved a specialized secretion system, termed type III, that is designed to deliver bacterial proteins into host cells (1, 2). These bacterial proteins, in turn, modulate cellular functions for the benefit of the pathogen. Made up of more than 20 protein components, type III secretion systems are among the most complex protein secretion systems known in bacteria. Distinctive features of type III secretion include: (i) the absence of a typical sec-dependent cleavable signal sequence in the secreted proteins; (ii) the requirement of specific cytosolic proteins with chaperone-like activity for the secretion of at least some of the substrate proteins; and (iii) the requirement of an environmental signal, usually derived from contact with the host cell, for their full activation.
Although proteins secreted by type III secretion systems are largely specific for each bacterial species, the structural components of the secretion apparatus itself are highly conserved. These components fall into at least two general categories. One group consists of a number of outer membrane proteins that display typical signal sequences and are thought to be delivered to their destinations in the bacterial envelope by the sec-dependent or general secretion pathway. The other group consists of several integral or peripherally associated membrane proteins that are similar to components of the flagellar export machinery.
The bacterial pathogen Salmonella typhimurium has at least two type III secretion systems. One is encoded within a pathogenicity island at centisome 63 of its chromosome [S. typhimurium pathogenicity island 1 (SPI-1)] and mediates the initial interaction of these bacteria with the intestinal epithelium, allowing them to enter these cells (3). The second type III secretion system is encoded within a pathogenicity island at centisome 31 (SPI-2), is not expressed under standard laboratory growth conditions, and is required for systemic infection (4, 5).
Recent studies demonstrated that a subset of the structural components of the centisome 63 type III secretion system form a supramolecular structure termed “the needle complex” that spans the bacterial envelope (6). This structure is composed of two pairs of rings that are anchored to the inner and outer membranes of the bacterial envelope. The rings, which are ≈40 nm in diameter at their widest section, are joined by a central rod and together form the base of the needle complex. Protruding outward from the base is the needle structure itself, which is a stiff, straight, hollow tube ≈80 nm in length and ≈8 nm wide. Three components of the needle complex have been identified thus far (6): InvG, a member of the secretin family of protein exporters, and the predicted lipoproteins PrgH and PrgK. The particular substructure of the needle complex formed by these proteins presently is unknown. Although the architecture of the needle complex resembles that of the flagellar hook–basal body complex (7, 8), InvG, PrgH, and PrgK exhibit little or no sequence similarity to flagellar proteins.
Currently, nothing is known about the specific composition of the different substructures of the needle complex such as the rings and central rod of the base or the needle segment itself. Furthermore, nothing is known about the assembly of this organelle. In this paper we describe the components of the base of the needle complex and report the identification of a new component that makes up the needle segment of this structure. In addition, we demonstrate that the assembly of the needle portion but not of the base requires the function of the type III secretion export machinery and that the length of the needle is under the strict control of the type III secretion-associated protein InvJ.
Materials and Methods
Bacterial Strains, Culture Conditions, Quantitation of Bacterial Entry and Gene Expression, and Preparation of Culture Supernatant Proteins and Antibodies.
All strains were derived from the nonflagellated S. typhimurium strain SJW2941 (9). The invA (10), invC (11), and invJ (12) mutant alleles were introduced into this strain by P22HTint-mediated transduction. A strain of S. typhimurium carrying a nonpolar insertion mutation in prgI was constructed by allele replacement as described previously (13). All S. typhimurium strains were grown under conditions that stimulate the expression of components and substrates of the centisome 63 type III protein-secretion system (14). Salmonella internalization into cultured intestinal epithelial cells was measured by the gentamicin resistance assay as described previously (15). The preparation and analysis of cultured supernatant proteins was also as described previously (16). Antibodies to purified needle filaments and to a synthetic KLH-conjugated PrgI peptide (CKLSETNLTRNAQSN) were prepared by Pocono Rabbit Farm (Canadensis, PA).
Isolation of Base Substructures of the Needle Complex.
Overnight cultures grown 12 h in Luria–Bertani broth containing 0.3 M NaCl were diluted 1:100 into the same medium and grown with aeration at 37°C to an OD600 of approximately 0.8. The cells were pelleted and resuspended in 0.5 M sucrose/0.15 M Tris. After the addition of lysozyme (0.2 mg/ml final) and EDTA (1 mM final), the cells were incubated on ice for 1 h. The bacteria were incubated at 37°C for 15 min and then lysed by the addition of a 3% solution of lauryldimethylamine oxide (LDAO). Lysates were cleared of debris by low-speed centrifugation (10,000 × g for 15 min at 4°C), the pH was adjusted to 10.5, and, after incubation for 1 h at 4°C, the lysates were centrifuged again at 10,000 × g for 15 min. The cleared lysates then were subjected to high-speed centrifugation (250,000 × g for 1 h at 4°C) in a Sorvall microultracentrifuge (model RC-M120GX). Pellets were resuspended in 0.5 M sucrose/0.1 M Tris/0.03% LDAO, pH 10.5, spun briefly (10,000 × g for 10 min) to remove any particulate matter, and centrifuged again at 250,000 × g for 1 h at 4°C. The pellets were resuspended in 0.01 M Tris/0.02 M EDTA/0.03% LDAO, pH 8.0 (TET), centrifuged briefly (10,000 × g for 10 min), and the supernatants were loaded onto 30% (wt/vol) CsCl density gradients. The gradients were centrifuged at 15,000 × g for 15 h at 20°C in a swinging-bucket rotor in a Beckman (model L 80) ultracentrifuge. The different gradient fractions were collected and centrifuged at 250,000 × g for 1 h, and the pellets were resuspended in TET and analyzed by either transmission electron microscopy (TEM) or SDS/PAGE.
Isolation of the Needle Substructures of the Needle Complex.
The S. typhimurium invJ mutant strain SB1077 carrying a plasmid expressing the transcriptional regulator hilA under the paraBAD promoter (17) was grown in 1 liter of Luria–Bertani broth containing 0.3 M NaCl. Cells were harvested at the late log phase of growth and resuspended in 30 ml of TE (10 mM Tris⋅Cl/1 mM EDTA, pH 7.5) containing 1 mM PMSF (phenylmethylsulfonyl fluoride; Sigma). The suspension was vortexed three times for 1.5 min at maximum power and passed five times through a 25-gauge needle resulting in the shearing of the needle filaments. Intact cells were removed by centrifugation (9,000 × g, 20 min at 4°C), and the supernatant was subjected to high-speed centrifugation (85,000 × g, 1 h at 4°C) to collect the macromolecular structures. The pellet was resuspended in TE and applied to a 36% (wt/vol) CsCl density gradient and centrifuged at 55,000 × g for 15 h at 20°C. The fractions were collected and washed twice by ultracentrifugation (100,000 × g, 1 h at 4°C) with TE and finally suspended in 0.05 ml of TE.
Electron Microscopy.
Osmotically shocked cells were prepared as described elsewhere (18). Samples were negatively stained with 2% phosphotungstic acid (pH 7.0) and observed under the electron microscope (EM410; Philips, Eindhoven, the Netherlands). Micrographs were taken at an accelerating voltage of 80 kV. A phenotype was assigned (presence or absence of the needle substructure) to a given strain when at least 20 cells had been scored. A wild-type cell exhibits ≈20 needle complexes in the visible membrane edge of the osmotically shocked bacteria. Therefore, by examining 20 bacterial cells, a minimum of 400 structures were scored for the presence or absence of the needle portion.
Results
The Type III Secretion Export Apparatus Is Required for the Assembly of the Needle Segment of the Needle Complex But Is Dispensable for the Assembly of the Base Substructure.
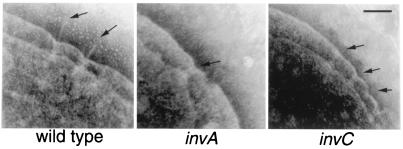
The architecture of the needle complex resembles that of the flagellar hook–basal body complex. The formation of the flagellar hook–basal body follows a sequential, highly coordinated morphological pathway, starting with the assembly of the MS ring and sequentially followed by the assembly of the rod, the outer membrane LP-ring complex, and the extracellular hook (7, 8, 18). Except for the assembly of the MS ring, which is composed of multiple copies of the FliF protein subunit, this assembly pathway cannot proceed in the absence of the flagellar-specific export apparatus, whose components share extensive sequence similarity with those of the type III secretion systems of pathogenic bacteria. Given the architectural similarity between the flagellar hook–basal body and the needle complex, we investigated the role of the type III secretion-associated export apparatus in the assembly of the needle complex. TEM of negatively stained, osmotically shocked cells has been shown previously to be very effective to visualize the needle complex in the bacterial envelope (6). We therefore used this technique to examine S. typhimurium strains carrying a loss-of-function mutation in either invA or invC. Both these genes encode a highly conserved component of the type III export apparatus, and, therefore, these strains are completely defective for type III secretion (10, 11). As shown in Fig. 1, both the S. typhimurium invA and invC mutant strains exhibited apparently intact needle complex base structures in the bacterial envelope but completely lacked the needle portion of this organelle. These results indicate that the assembly of the needle portion of the needle complex requires an intact type III secretion-associated export apparatus. In contrast, the assembly of the base structure itself can take place in the absence of the type III export machinery.
Figure 1.
Role of the type III secretion protein-export apparatus in the assembly of the S. typhimurium needle complex. Electron micrographs of negatively stained, osmotically shocked wild-type S. typhimurium and the isogenic invA and invC mutant derivatives. The positions of needle complexes and bases are indicated by arrows. (Bar = 100 nm.)
InvG, PrgH, and PrgK Are Components of the Base Substructure of the Needle Complex.
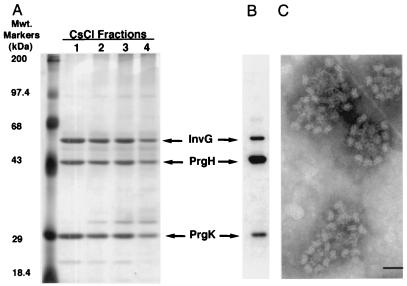
The observation that S. typhimurium strains deficient in the type III secretion export machinery are capable of assembling apparently normal base structures of the needle complex but lack the needle portion provided us with an opportunity to examine the components of the base structure itself. Base substructures from a S. typhimurium invA mutant strain were purified by CsCl gradient centrifugation as described in Materials and Methods and examined both by TEM and by SDS/PAGE. As predicted from the TEM of osmotically shocked bacterial cells, the isolated needle complexes from the invA mutant strain exhibited normal base structures but completely lacked the needle segment (Fig. 2C). Analyses of these purified complexes in SDS-polyacrylamide gels identified three major protein species corresponding to the molecular weights of InvG, PrgH, and PrgK that have been identified previously as components of the needle complex structure (6) (Fig. 2A). Western immunoblot analyses with an antibody specific for these proteins confirmed their identity (Fig. 2B). Therefore, these results demonstrate that InvG, PrgH, and PrgK are the major components of the base structure of the needle complex.
Figure 2.
Identification of the components of the base structure of the needle complex. Type III secretion components were isolated from a S. typhimurium invA mutant strain, purified on a CsCl density gradient, and visualized by silver staining (A) or by Western immunoblotting by using an antibody specific to InvG, PrgH, and PrgK (B). The identity of the components is indicated. (C) Electron micrographs of negatively stained purified complexes from the S. typhimurium invA mutant showing the absence of the needle substructure. (Bar = 50 nm.)
InvJ Controls the Length of the Needle Substructure.
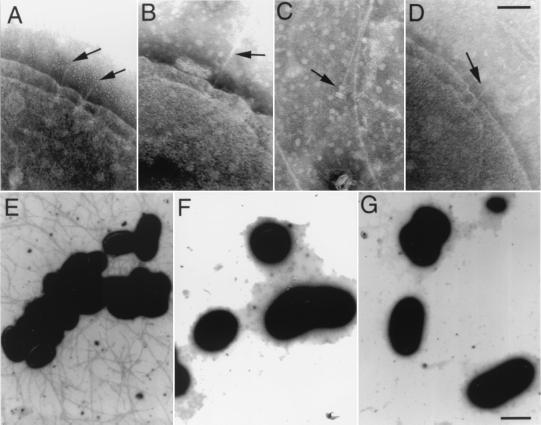
In the process of analyzing the effect of mutations in type III secretion-associated genes on the assembly of the needle complex, we identified a unique phenotype in a S. typhimurium strain carrying a loss-of-function nonpolar mutation in invJ (12). As shown in Fig. 3, invJ mutant cells often exhibited abnormally long needle segments (compare Fig. 3 A with B). When the invJ mutant bacterial cultures were vortexed, long needle structures still attached to the bases occasionally were observed (Fig. 3C). Introduction of a plasmid carrying a wild-type copy of invJ into the invJ mutant strain restored the normal length of the needle structure, indicating that the increased length of the needles observed in this mutant was exclusively a result of the absence of InvJ (Fig. 3D). When a plasmid expressing the type III secretion-associated transcriptional regulatory protein HilA under the control of the strong paraBAD promoter (17) was introduced into the invJ mutant strain (resulting in increased expression of all components of the type III secretion system), very abundant, elongated needle structures were seen upon induction with arabinose (Fig. 3E). The needle structures extended outward from the surface of the bacteria several micrometers, covering the entire surface of the bacteria (Fig. 3E). Under these conditions, profuse clumping of the bacterial culture was observed, which presumably was an indication of bacterial aggregation mediated by these structures. Introduction of a plasmid carrying a wild-type copy of invJ into this strain restored the normal needle length (Fig. 3F) and abrogated bacterial clumping. When a loss-of-function mutation in invA, which encodes an essential component of the export machinery (10), was introduced into the invJ mutant strain, no needle structures or bacterial clumping was observed (Fig. 3G and data not shown). The latter results further support the requirement of an intact type III export apparatus for the export and assembly of the needle structure.
Figure 3.
InvJ controls the length of the needle substructure of the type III secretion complex. Electron micrographs of negatively stained, osmotically shocked wild-type S. typhimurium (A) and its isogenic invJ mutant derivative (B), showing the lengthening of the needle substructure in the absence of the InvJ protein (compare A with B). After vortexing, long needle substructures separated from the bacterial cell but still attached to the type III secretion complex base substructure were seen occasionally (C). Introduction into the invJ mutant strain of a plasmid carrying a wild-type copy of invJ restored the proper needle length (D). Overexpression of hilA, which encodes a transcriptional regulator of the type III secretion system, resulted in the accumulation of large amounts of exceedingly long needle substructures observed on electron micrographs of a negatively stained S. typhimurium invJ mutant (E). Introduction into this strain of a plasmid carrying a wild-type copy of invJ resulted in the disappearance of the unusually elongated needle substructures (F). Likewise, introduction of an invA mutation, which abrogates type III secretion, resulted in the absence of needle filaments (G). Arrows indicate the position of the needle complexes in the bacterial envelope. [Bar = 100 nm (Upper) and 1 μm (Lower).]
The phenotype of the invJ mutant resembles that of the fliK flagellar mutant in S. typhimurium. Absence of FliK results in a phenotype termed “polyhook,” which is characterized by the presence of an abnormal elongation of the hook structure of the hook–basal body complex (19–21). The mechanisms by which FliK controls the length of the hook are not understood. Although InvJ does not have sequence similarity with FliK, its carboxyl-terminal region contains several glutamine residues, a feature also observed in the carboxyl-terminal region of FliK (data not shown). It has been shown previously that the carboxyl terminus of FliK is essential for its hook-length control function (20), therefore suggesting the possibility of a common mechanism of action for these two proteins.
PrgI Is the Main Subunit of the Needle Substructure of the Type III Secretion Complex.
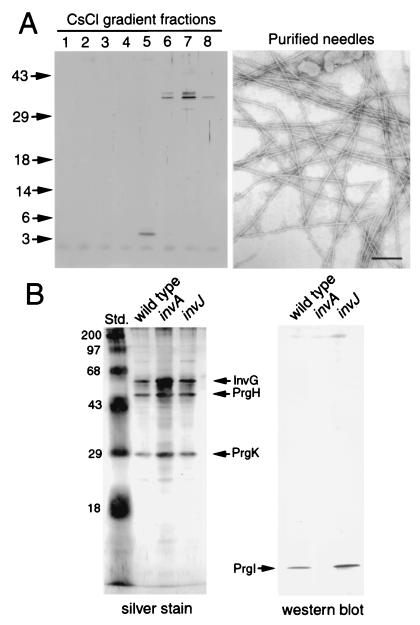
Previous attempts at isolating the component(s) of the needle segment of the type III secretion complex failed largely because of the intrinsic fragility of this structure. The phenotype of the S. typhimurium invJ mutant strain provided us with an opportunity to isolate the needle substructure and determine its composition. A protocol was developed (see Materials and Methods) to obtain highly purified preparations of the needle substructure from this bacterial strain. SDS/PAGE analysis of highly purified fractions of the needle preparations revealed the presence of a single polypeptide species with an apparent molecular mass of ≈5 kDa (Fig. 4). The polypeptide band was excised from the gel and subjected to amino-terminal sequence analysis. The sequence obtained (ATPWSGYL) matched exactly that of the amino terminus (starting at the second residue) of the predicted sequence of PrgI, a protein encoded within a cluster of type III secretion-associated genes in the centisome 63 pathogenicity island (22). PrgI exhibits a predicted propensity to form coiled coils, a feature found often in proteins that assemble into filamentous structures such as flagella. Proteins exhibiting sequence similarity with PrgI are found in type III secretion systems of animal-pathogenic bacteria such as Yersinia spp., Shigella, Escherichia coli, and Pseudomonas aeruginosa (Fig. 5), but are absent from type III secretion systems of plant-pathogenic bacteria.
Figure 4.
Isolation and identification of the main subunit of the needle substructure. (A) Needle structures were isolated from a S. typhimurium invJ mutant strain and purified on a CsCl density gradient, and the different fractions were loaded on a SDS/polyacrylamide gel and visualized by silver staining. An electron micrograph of negatively stained purified needle structures from fraction 5 of the CsCl gradient is shown. (Bar = 100 nm.) (B) Purified needle complexes from wild-type S. typhimurium and the isogenic invA or invJ mutants were separated on a SDS/polyacrylamide gel and visualized by silver staining or transferred to nitrocellulose membranes and probed with an antibody directed against PrgI. The identities of the different proteins are indicated.
Figure 5.
Amino acid sequence alignment of S. typhimurium PrgI and its homologs in type III secretion systems from other animal pathogenic bacteria. Included in the alignment are PrgI from S. typhimurium, MxiH from S. flexneri, YscF from Yersinia enterocolitica, PscF from P. aeruginosa, and EscF from enteropathogenic E. coli.
The predicted molecular mass of PrgI (8,725 Da, residues 2–80) differed from that of the apparent molecular mass of the isolated needle subunit as determined by its migration in 15% SDS-polyacrylamide gels. Although sizing of low-molecular-mass polypeptides in this gel system is unreliable, the apparent difference suggested the possibility of posttranslational processing of the PrgI protein. To further investigate this possibility, we determined the mass of the eluted needle polypeptide subunit by matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) and by electrospray ionization MS (ESI-MS). These techniques can measure the mass of a polypeptide within a margin of error of 0.1% and 0.02%, respectively. MALDI-MS and ESI-MS analyses established that the actual mass of the eluted polypeptide was 8,741.7 ± 1.7 Da, in good agreement with the mass of the predicted PrgI polypeptide. It is unlikely that the small difference of 16 Da between the predicted and observed mass is due to a posttranslational modification because no modification known to occur in bacteria would result in such a small change in mass. Most likely, such a difference is a result of hydroxylation of the isolated polypeptide during the fractionation procedure or the mass spectrometric analysis.
To further confirm the identity of the subunit of the needle structure, we raised antibodies to the purified filament structure and to a peptide derived from the PrgI protein and used them to examine needle complex preparations from wild-type, invA, and invJ S. typhimurium strains for the presence of the PrgI protein. As shown in Fig. 4B, the PrgI protein was detected in needle complex preparations from the wild-type and invJ strains but was not detected in preparations from the secretion-incompetent invA mutant. Because the needle structure, but not the base, is absent from this mutant, these results are consistent with PrgI being a component of the needle portion of the needle complex. Consistent with previous observations (6) (also see Fig. 2), PrgI could not be detected by protein staining of purified needle complex preparations of wild-type and invJ strains (Fig. 2A), underscoring the susceptibility of this substructure to the isolation protocol even when present in the elongated form such as in the invJ mutant. We also constructed a S. typhimurium strain carrying a loss-of-function, nonpolar mutation in prgI. Osmotically shocked cells from the resulting strain then were examined by TEM for the presence of needle structures. As expected, the S. typhimurium prgI mutant strain lacked needle substructures although it did exhibit apparently normal bases (Fig. 6). Introduction of a plasmid encoding a wild-type copy of prgI in the mutant strain restored the production of needles, indicating that the effect of the mutation was solely due to the absence of PrgI. These results further support the notion that the main subunit of the needle structure is PrgI.
Figure 6.
Electron micrographs of negatively stained, osmotically shocked S. typhimurium prgI mutant strain (A and B) and the same strain carrying a complementing plasmid encoding wild-type prgI (C). (Bar = 100 nm.)
PrgI Is Essential for Type III Protein Secretion and S. typhimurium Entry into Host Cells.
To investigate the role of PrgI in the stimulation of host cell responses that are dependent on the centisome 63 type III secretion system, we examined the ability of the S. typhimurium prgI mutant strain to enter cultured intestinal epithelial cells. The prgI null mutant was completely defective in its ability to invade cultured intestinal cells. Introduction of a plasmid carrying a wild-type copy of prgI restored the invasion defect, demonstrating that the PrgI protein and the needle structure are required for S. typhimurium signaling to the host cell (invasion values represented as the percentage of the inoculum that resisted gentamicine treatment: for wild-type S. typhimurium strain SJW2941, 2 ± 0.02; for the prgI mutant strain, 0.0002 ± 0.00002; for the prgI mutant carrying a complementing plasmid, 4.2 ± 0.002).
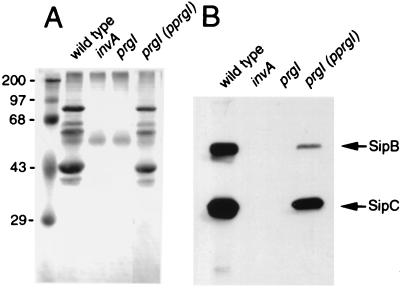
We also examined the ability of the prgI mutant strain to secrete proteins via the type III pathway. Culture supernatants were isolated from wild-type S. typhimurium and its isogenic prgI and invA mutant derivatives. Culture supernatant proteins were either stained with Coomassie blue or probed by Western immunoblot analysis for the presence of SipB and SipC, two type III secreted proteins (23). As shown in Fig. 7, no type III secreted proteins were observed in culture supernatants of the prgI mutant strain. The secretion defect was indistinguishable from that of a strain carrying a null mutation in invA, which encodes a major component of the type III secretion-associated export apparatus (10). Introduction of a plasmid carrying a wild-type copy of prgI restored secretion, indicating that the needle structure composed by PrgI is required for type III protein secretion (Fig. 7).
Figure 7.
S. typhimurium prgI mutants are defective for type III secretion. (A) Total culture supernatant protein profile of wild-type S. typhimurium, the secretion-defective invA mutant, the prgI mutant strain, and the complemented prgI mutant strain. (B) Western immunoblot analysis of culture supernatant proteins from the same strains probed with mAbs directed to the type III secreted proteins SipB and SipC.
Discussion
A central component of the S. typhimurium invasion-associated type III secretion system is a supramolecular structure in the bacterial envelope termed “the needle complex” (6). A similar structure recently has been observed in Shigella flexneri, strongly suggesting that the needle complex is a universal feature of all type III secretion systems (24). This assertion is supported further by the extensive conservation of the type III secretion-associated genes among pathogenic bacteria (1, 2).
The needle complex is a supramolecular cylindrical structure composed of a number of well defined substructures: (i) two inner rings 40 nm in diameter that presumably anchor the structure to the inner membrane; (ii) two outer rings 20 nm in diameter in close contact with the peptidoglycan and outer membranes; (iii) a central rod that links the two pairs of rings to form the base of the structure; and (iv) an external, needle-like extension 80 nm in length and 8 nm in diameter that protrudes outward from the base (6).
The architecture of the needle complex resembles that of the flagellar hook–basal body, consistent with a distant evolutionary relationship between these two structures. The actual components of the needle complex identified so far, however, share little or no amino acid sequence similarity with those of the flagellar structure. Nevertheless, other components of the type III secretion system not associated with the needle complex, such as a group of integral membrane proteins and an ATPase, are highly conserved between these two systems. In flagella, this group of conserved proteins is essential for flagellar assembly and constitute what is known as the “export apparatus” (8), whereas in type III systems these proteins are required for the secretion of effector proteins (1, 2). We report here that two of these conserved proteins, InvA and InvC, also are required for the full assembly of the needle complex. InvA is a highly conserved polytopic integral membrane protein with sequence similarity to the flagellar FlhA protein (10). InvC is an ATPase with amino acid sequence similarity to the flagellar FliI protein and other members of the F0F1-related family of ATPases (11). We found that null mutations in either invA or invC resulted in complete absence of the needle segment of the needle complex. However, these mutations did not affect the assembly of the base substructure. Purified base complexes from these strains exhibited a morphology indistinguishable from that of the base of a wild-type needle complex. These results indicate that the assembly of the base substructure of the needle complex does not require the type III secretion export machinery and that the export of the components of the bases most likely occurs through the general “sec-dependent“ secretory pathway. Consistent with this hypothesis, all components of the base structure identified so far (see below) exhibit a sec-dependent signal sequence. This is in sharp contrast to the flagellar system in which the absence of the flagellar-export machinery prevents the assembly of the hook–basal body complex (18). In fact, in the absence of the export machinery, the MS ring is the only membrane-embedded structure that is assembled (18). This structure is composed of a single protein, FliF, which also exhibits a sec-dependent secretion signal sequence.
Previous studies have identified protein components of the needle complex, but these components were not assigned to any particular substructure of the needle complex (6). The availability of S. typhimurium mutants that lacked the needle portion of the type III secretion complex allowed us to identify the components of the base structure. Highly purified preparations of the base structure exhibited only three major protein species, InvG, PrgH, and PrgK, which have been identified previously as components of the needle complex. Although our studies did not allow us to assign each protein to a particular base substructure, we hypothesize that InvG forms the outer rings and PrgH and PrgK form the inner rings and central rod. This hypothesis is based on the sequence similarities exhibited by these proteins. InvG is a member of the secretin family of outer membrane proteins (13). Members of this protein family have been shown to be involved not only in type III but also in type II protein secretion as well as in bacteriophage assembly (25). Both PrgH and PrgK exhibit consensus lipidation sites and typical sec-dependent secretion signals (22). Interestingly, the PrgH signal sequence is not cleaved upon export, suggesting the possibility that this protein remains associated with the inner membrane and, thus, forms part of the inner rings of the needle complex (6). PrgK exhibits regions of sequence similarity with FliF, the component of the flagellar MS ring and the proximal part of the rod substructure (26).
We made the observation that introduction of a loss-of-function mutation in invJ results in the production of abnormally long needle substructures of the type III secretion complex. This is particularly pronounced if the expression of components of the type III secretion system is increased by the overexpression of the transcriptional regulator HilA. In this case, very long needle structures could be seen covering the entire bacterial surface, resulting in bacterial clumping. This phenotype allowed us to obtain highly purified needle preparations and to identify the S. typhimurium protein PrgI as the main subunit of this filamentous structure. PrgI is an 80-aa protein encoded within the prg cluster of genes in the centisome 63 pathogenicity island (22). Although PrgI does not share sequence similarity with any flagellar proteins, its predicted secondary structure is characterized by a high α-helical content and propensity to form coiled coils, properties consistent with its role as the main subunit of a filamentous structure. Proteins exhibiting sequence similarity with PrgI are found in type III secretion systems of animal pathogenic bacteria such as Yersinia spp., Shigella, E. coli, and P. aeruginosa. Intriguingly, homologous proteins are not found in type III secretion systems from plant pathogenic bacteria. This suggests that the structure of the needle portion of the needle complex may exhibit different characteristics in plant pathogenic bacteria, perhaps reflecting an adaptation to deliver proteins to host cells that exhibit significantly different properties than animal cells. The structure of the needle of the type III secretion complex may represent the major difference between type III secretion systems from plant or animal pathogenic bacteria.
The absence of InvJ results in bacterial cells exhibiting very long needles, indicating that this protein is involved in controlling the length of this structure. How does InvJ control the needle length? Certain structural features of InvJ, such as the presence of several glutamines at its carboxyl terminus, also are present in FliK, a protein involved in determining the length of the flagellar hook, suggesting that these two proteins may exert their functions in a similar manner. However, the mechanism by which FliK controls hook length is not understood. It is possible that InvJ may exert its function by directing the type III secretion machinery to switch from a state in which it secretes mainly PrgI to a state that is competent for the export of other type III secreted proteins. Therefore, in the absence of the InvJ “switch,” the type III secretion machine would continue secreting PrgI, resulting in longer needles. Consistent with this model, mutations in InvJ not only result in elongated needles but also in the complete absence of type III secretion (27). Furthermore, with the exception of the Shigella spp. type III secretion-associated protein Spa32 (28), InvJ exhibits little similarity to any type III secretion-associated protein from other bacteria. Most type III secreted proteins are specific for a particular pathogen with little similarity across bacterial species (1, 2). Therefore, the lack of similarity of InvJ to proteins from other type III systems that may exert a similar function may be consistent with a requirement of a functional interaction between InvJ (or its functional homologs) and the secreted effector proteins as part of its “switch” function. In this context, the similarity of InvJ with Spa32 is consistent with the similarity of several of the S. typhimurium and Shigella spp. secreted proteins (16, 23).
In conclusion, our studies have identified the components of the different substructures of the needle complex and, in particular, a protein, PrgI, that makes up the needle portion of this supramolecular structure. In addition, we have identified a function for InvJ, which is involved in controlling the proper length of the needle structure. These findings have profound functional and evolutionary implications for the understanding of type III secretion in plant and animal pathogenic bacteria.
Acknowledgments
We thank members of the Galán laboratory for critical review of this manuscript. T.K. was supported by a fellowship from the Human Frontier Science program. This work was supported by Public Health Service Grant AI30492 from the National Institutes of Health to J.E.G.
Abbreviation
- TEM
transmission electron microscopy
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170128997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170128997
References
- 1.Galán J E, Collmer A. Science. 1999;284:322–328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 2.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galán J E. Curr Opin Microbiol. 1999;2:46–50. doi: 10.1016/s1369-5274(99)80008-3. [DOI] [PubMed] [Google Scholar]
- 4.Ochman H, Soncini F C, Solomon F, Groisman E A. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shea J E, Hensel M, Gleeson C, Holden D W. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán J E, Aizawa S-I. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 7.Aizawa S I. Mol Microbiol. 1996;19:1–5. doi: 10.1046/j.1365-2958.1996.344874.x. [DOI] [PubMed] [Google Scholar]
- 8.Macnab R M. J Bacteriol. 1999;181:7149–7153. doi: 10.1128/jb.181.23.7149-7153.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaguchi S, Fujita H, Ishihara A, Aizawa S, Macnab R M. J Bacteriol. 1986;166:187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galán J E, Ginocchio C, Costeas P. J Bacteriol. 1992;17:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichelberg K, Ginocchio C, Galán J E. J Bacteriol. 1994;176:4501–4510. doi: 10.1128/jb.176.15.4501-4510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collazo C M, Zierler M K, Galán J E. Mol Microbiol. 1995;15:25–38. doi: 10.1111/j.1365-2958.1995.tb02218.x. [DOI] [PubMed] [Google Scholar]
- 13.Kaniga K, Bossio J C, Galán J E. Mol Microbiol. 1994;13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen L M, Kaniga K, Galán J E. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 15.Galán J E, Curtiss R., III Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaniga K, Trollinger D, Galán J E. J Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eichelberg K, Galán J E. Infect Immunol. 1999;67:4099–4105. doi: 10.1128/iai.67.8.4099-4105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S-I. J Mol Biol. 1992;226:433–446. doi: 10.1016/0022-2836(92)90958-m. [DOI] [PubMed] [Google Scholar]
- 19.Hirano T, Yamaguchi S, Oosawa K, Aizawa S. J Bacteriol. 1994;176:5439–5449. doi: 10.1128/jb.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawagishi I, Homma M, Williams A W, Macnab R M. J Bacteriol. 1996;178:2954–2959. doi: 10.1128/jb.178.10.2954-2959.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muramoto K, Makishima S, Aizawa S I, Macnab R M. J Mol Biol. 1998;277:871–882. doi: 10.1006/jmbi.1998.1659. [DOI] [PubMed] [Google Scholar]
- 22.Pegues D A, Hantman M J, Behlau I, Miller S I. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- 23.Kaniga K, Tucker S C, Trollinger D, Galán J E. J Bacteriol. 1995;177:3965–3971. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blocker A, Gounon P, Larquet E, Niebuhr K, Cabiaux V, Parsot C, Sansonetti P. J Cell Biol. 1999;147:683–693. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russel M. J Mol Biol. 1998;279:485–499. doi: 10.1006/jmbi.1998.1791. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki H, Yonekura K, Murata K, Hirai T, Oosawa K, Namba K. J Struct Biol. 1998;124:104–114. doi: 10.1006/jsbi.1998.4048. [DOI] [PubMed] [Google Scholar]
- 27.Collazo C, Galán J E. Infect Immunol. 1996;64:3524–3531. doi: 10.1128/iai.64.9.3524-3531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkatesan M M, Buysse J M, Oaks E V. J Bacteriol. 1992;174:1990–2001. doi: 10.1128/jb.174.6.1990-2001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]