Abstract
In cell signaling events, small GTPases are classical on/off switches depending on the nucleotide, GTP or GDP, that is bound; Rac1 and Cdc42 are two of the best studied examples of the Rho-family of proteins which are known to regulate the actin cytoskeleton and are involved with cell motility. While Rac1.GDP assignments are published (Thapar et al., 2003), no NMR assignments have been reported for the active form of Rac1. Rac1, uniformly labeled with 13C and 15N, was loaded with a GTP analogue by nucleotide exchange in presence of excess GMPPNP and alkaline phosphatase. Following gel filtration, heteronuclear NMR experiments were carried out on this active protein at 0.8 mM and 25°C. A mutant C178S, K184Stop was used in physiological buffer (Kremer et al., 2001), with 4 mM DTT and 4 mM MgCl2, at pH 6.8 to improve the quality of spectra. Partially deuterated protein was used in some experiments. Cross peaks for 150 out of 168 possible resonances are observed in 1H–15N HSQC spectra. Backbone assignment was completed for 141 resonances; side-chain carbon assignments for the corresponding residues are nearly complete, e.g. >95% for Cβ. We observe excellent agreement of the secondary structure predicted from the NMR data (based on CSI) with that found in the crystal structure of Rac1.GMPPNP (pdb 1 MH1). Additional materials are given in the online supplements. BMRB deposit number 6970.
Keywords: Rac1, GTPase, NMR assignment, GMPPNP
Biological context
In cell signaling events, small GTPases are classical on/off switches depending on the nucleotide, GTP or GDP, that is bound; Rac1 and Cdc42 are two of the best studied examples of the Rho-family of proteins which are known to regulate the actin cytoskeleton and are involved with cell motility. While Rac1.GDP assignments are published (Thapar et al., 2003), no NMR assignments have been reported for the active form of Rac1. The assignments obtained from the present study will provide the basis for future NMR studies on Rac1 in its active form, and for its interaction with other proteins.
Materials and Methods
A mutant (C178S K184stop) of the Rac1 protein was overexpressed in E.Coli strain BL21 (DE3) in M9 minimum medium containing 15NH4Cl and 13C-glucose as the sole nitrogen and carbon sources. Cells were grown at 32 °C until an OD600 ~ 0.7 and then the protein expression was induced by addition of IPTG to a final concentration of 1mM. The cells were harvested after six hours of expression. The uniformly 13C and 15N labeled sample was purified and then loaded with a GTP analogue by nucleotide exchange in presence of excess GMPPNP and alkaline phosphatase. Following gel filtration, heteronuclear NMR experiments were carried out on this active protein at 0.8mM. A physiological buffer (Kremer et al., 2001) was used with 4mM DTT and 4mM MgCl2 at pH 6.8 to improve the quality of spectra. Partially deuterated protein was used in some experiments.
All NMR experiments were carried out at 25 °C and at 18.8T (1H resonance frequency of 800MHz) on a Bruker spectrometer equipped with cryoprobe technology. Sequence-specific assignments were achieved using 2D 15N-1H HSQC, 3D HNCA, 3D HN(CO)CA, 3D HNCACB, 3D CBCA(CO)NH, 3D C(CO)NH and 3D HNCO experiments. All spectra were processed with nmrPipe and analyzed with NMRView.
In addition to the classical 3-dimensional approach, other experiments were used to complete the assignment, including amino acid type selective experiments based on MUSIC filters (Schubert et al., 1999), and amino acid specific labeling of the Rac1 protein (Lee et al., 1995).
Extent of assignment and data deposition
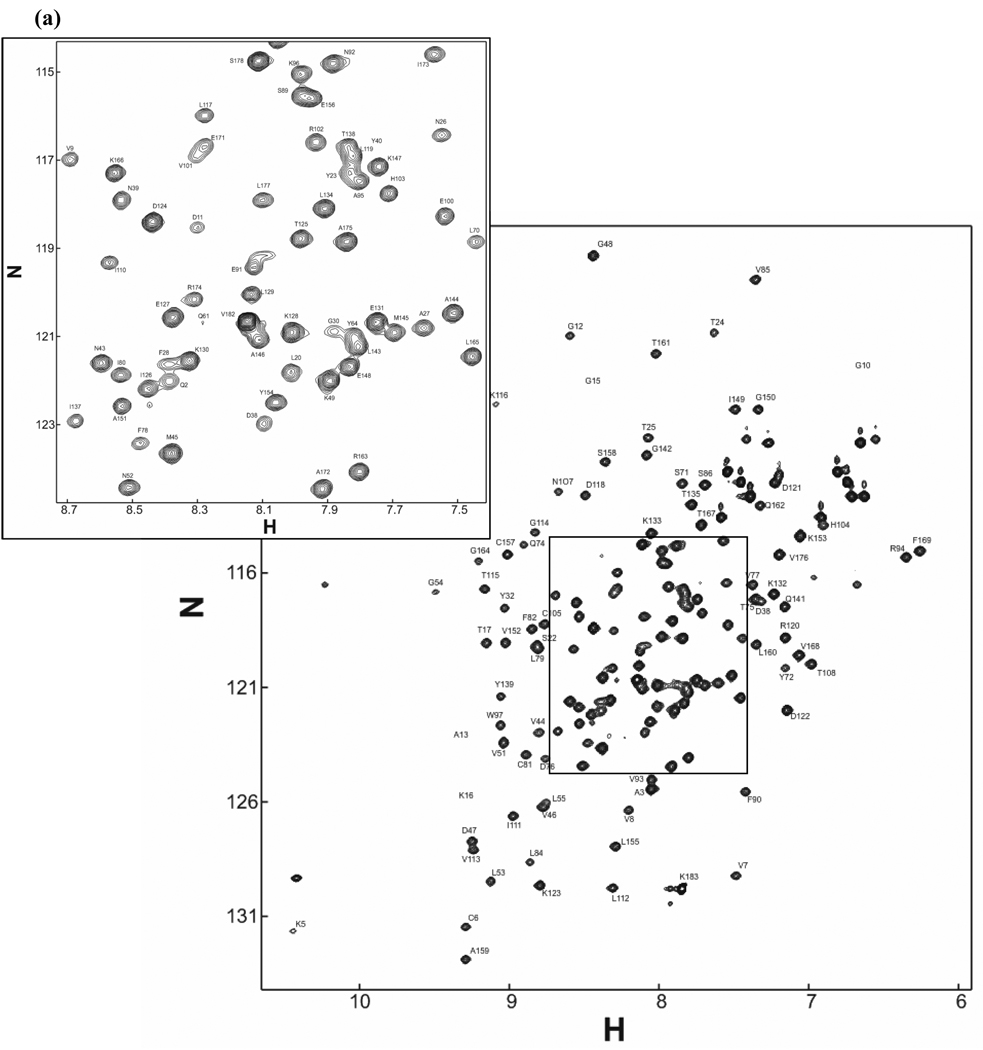
The 1H-15N HSQC spectrum of the 15N-labeled Rac1.GMPPNP is shown in Figure 1a. Cross peaks for 150 out of 168 possible resonances are observed. Backbone assignment was completed for 141 resonances; side-chain carbon assignment for the corresponding residues is complete to > 95% for Cβ. In both forms of Rac1 (GDP- and GTP-bound), it is noticeable that signals from the switch regions are missing or have weak intensities (due to exchange phenomena). The same observation has been made for another GTPase homologous to Rac1, namely Cdc42 (Feltham et al., 1997).
Figure 1.
(a) 1H–15N HSQC spectrum of uniformly 15N-labeled and partially deuterated Rac1 C178S K184Stop loaded with GMPPNP, collected at 800MHz (1H resonance frequency) at 25°C and pH 6.8. (b) Difference between experimental Cα chemical shift and random-coil chemical shift value in ppm (Wishart et al., 1994). Assignment in the switch regions (green lines) is incomplete due to conformational exchange observed in small GTPases (e.g. Cdc42).
As shown in Figure 1b, we observe excellent agreement of the secondary structure predicted from the NMR data (based on CSI: Wishart et al, 1994) with that found in the crystal structure of Rac1.GMPPNP (pdb 1MH1). The chemical shifts have been deposited in the BioMagResBank database (accession number 6970).
Acknowledgements
We thank Dr. Yufeng Tong for help in the initial phase of the assignment and for discussion. This work is partially funded by a Basil O’Connor Starter Grant from the March of Dimes Foundation for Birth Defects.
Footnotes
Supplementary material is available in electronic format at http://www.dx.doi.org/10.1007/s10858-006-9029-6.
References
- Feltham JL, Dötsch V, Raza S, Manor D, Cerione RA, Sutcliffe MJ, Wagner G, Oswald RE. Biochemistry. 1997;36:8755–8766. doi: 10.1021/bi970694x. [DOI] [PubMed] [Google Scholar]
- Kremer W, Kalbitzer HR. Methods Enzymo. 2001;339:3–19. doi: 10.1016/s0076-6879(01)39306-0. [DOI] [PubMed] [Google Scholar]
- Lee KM, Androphy EJ, Baleja JD. J.Biomol.NMR. 1995;5:93–96. doi: 10.1007/BF00227474. [DOI] [PubMed] [Google Scholar]
- Schubert M, Smalla M, Schmieder P, Oschkinat H. J.Magn.Reson. 1999;141:34–43. doi: 10.1006/jmre.1999.1881. [DOI] [PubMed] [Google Scholar]
- Thapar R, Moore CD, Campbell SL. J.Biomol.NMR. 2003;27:87–88. doi: 10.1023/a:1024774230562. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]