Abstract
Tannerella forsythia is an anaerobic periodontal pathogen that encounters constant oxidative stress in the human oral cavity due to exposure to air and reactive oxidative species from coexisting dental plaque bacteria as well as leukocytes. In this study, we sought to characterize a T. forsythia ORF with close similarity to bacterial oxidative stress response sensor protein OxyR. To analyse the role of this OxyR homologue, a gene deletion mutant was constructed and characterized. Aerotolerance, survival after hydrogen peroxide challenge and transcription levels of known bacterial antioxidant genes were then determined. Since an association between oxidative stress and biofilm formation has been observed in bacterial systems, we also investigated the role of the OxyR protein in biofilm development by T. forsythia. Our results showed that aerotolerance, sensitivity to peroxide challenge and the expression of oxidative stress response genes were significantly reduced in the mutant as compared with the wild-type strain. Moreover, the results of biofilm analyses showed that, as compared with the wild-type strain, the oxyR mutant showed significantly less autoaggregation and a reduced ability to form mixed biofilms with Fusobacterium nucleatum. In conclusion, a gene annotated in the T. forsythia genome as an oxyR homologue was characterized. Our studies showed that the oxyR homologue in T. forsythia constitutively activates antioxidant genes involved in resistance to peroxides as well as oxygen stress (aerotolerance). In addition, the oxyR deletion attenuates biofilm formation in T. forsythia.
INTRODUCTION
Tannerella forsythia is recognized as one of the microbial pathogens implicated in the development of periodontal disease, a bacterially induced inflammation of tooth-supporting tissue and bone (Grossi et al., 1994, 1995; Tanner et al., 1998; Tran et al., 2001). T. forsythia with Porphyromonas gingivalis and Treponema denticola, known as the red-complex, in dental plaque biofilms, are associated with an increased risk of periodontitis development (Socransky et al., 1998). T. forsythia is an obligate anaerobe that is expected to encounter constant oxidative stress in the oral cavity due to reactive oxidative species produced by other bacteria (mainly oral streptococci) as well as host leukocytes (primarily neutrophils and macrophages). Therefore, to colonize, form biofilms and survive in the oral cavity, the ability of the bacteria to tolerate oxygen and peroxides is critical. Currently, knowledge of the oxidative stress defence mechanisms in T. forsythia is lacking. In this regard, while aerobes such as Escherichia coli contain two independent regulatory systems dependent on the SoxR/SoxS and the OxyR systems for responding to superoxide and peroxide challenges, respectively (Zheng & Storz, 2000), obligate anaerobes such as P. gingivalis express a single OxyR protein to respond to both types of challenge, from peroxide or air (Diaz et al., 2006).
OxyR is a redox-sensitive protein in the LysR family of DNA-binding transcription modulators, and regulates the expression of genes important in defence against oxidative stress in Gram-negative organisms (Zheng & Storz, 2000). The genome of T. forsythia (available at http://www.oralgen.lanl.gov/ under the previously designated organism name T. forsythensis) does not contain genes encoding homologues of the soxR/soxS system. However, putative genes encoding an OxyR homologue and enzymes such as SodF (superoxide dismutase), Dps (a non-specific DNA-binding protein), rubrerythrin and AhpC (alkylhydroperoxide reductase subunit C) involved in antioxidant responses have been identified in the T. forsythia genome. The expression of these antioxidant response genes has been shown to be OxyR-dependent in the Gram-negative anaerobe P. gingivalis (Amano et al., 1990; Diaz et al., 2004; Ohara et al., 2006). Furthermore, a catalase gene homologue is absent in T. forsythia.
Recent studies have also implicated the oxidative stress response sensor OxyR in biofilm development in microbes including E. coli (Reisner et al., 2003), Haemophilus influenzae (Murphy et al., 2005), Pseudomonas aeruginosa (Sauer et al., 2002), Neisseria gonorrhoeae (Seib et al., 2007), Campylobacter jejuni (Sampathkumar et al., 2006), Streptococcus mutans (Wen et al., 2005) and Candida albicans (Murillo et al., 2005). For example, in E. coli, OxyR activation promotes biofilm formation by inducing Ag43 adhesin expression (Danese et al., 2000), and in Serratia marcescens, biofilm formation is severely impaired in oxyR-defective mutants (Shanks et al., 2007). While the mechanisms that relate oxidative stress defences and biofilm formation are poorly understood at present, it is thought that the upregulation of oxidative stress-induced biofilm formation in the host may provide bacteria with a defence strategy. In addition to the role of OxyR in defence against oxidative stress and biofilm formation, this protein has been reported to regulate virulence factor expression in human pathogenic species such as Ps. aeruginosa (Lau et al., 2005), Ser. marcescens (Shanks et al., 2007), E. coli (Johnson et al., 2006), N. gonorrhoeae (Seib et al., 2007) and P. gingivalis (Wu et al., 2008).
In this study we evaluated the role of an OxyR homologue in T. forsythia with respect to its role in defence against oxidative stress and in biofilm formation.
METHODS
Bacterial strains and culture conditions.
T. forsythia strains were grown anaerobically (5 % CO2, 10 % H2, 85 % N2) in BF broth or on BF agar plates (Honma et al., 2007) with or without appropriate antibiotics. P. gingivalis strains were grown anaerobically (85 % N2, 10 % H2, 5 % CO2) at 37 °C in Trypticase soy broth (TSB) or on TSB blood agar plates, supplemented with yeast extract (1 mg ml−1), haemin (5 μg ml−1) and menadione (1 μg ml−1). Fusobacterium nucleatum ATCC 25586 was also maintained anaerobically in TSB or on agar plates (Becton Dickinson). E. coli strains were grown in Luria–Bertani (LB) medium aerobically at 37 °C. E. coli strain DH5α (Invitrogen) was used as a host for cloning and plasmid purification. E. coli strain BL21 (DE3) (EMD Bioscience) was used as a host for expression and purification of the His-tagged OxyR recombinant protein.
Construction of the isogenic T. forsythia oxyR homologue inactivated mutant (TFM104).
T. forsythia gene sequences were retrieved from the Oral Pathogen Sequence Database (Oralgen), Los Alamos National Laboratory, Los Alamos, NM, USA, under the previous name of the organism, T. forsythensis (http://www.oralgen.lanl.gov/), and gene designations correspond to identification (ID) numbers deposited in the database. An oxyR-inactivated insertional mutant was constructed by a previously described allelic replacement strategy (Honma et al., 2007). For this purpose, a DNA fragment containing the ermF gene flanked by upstream and downstream DNA regions of TF0104 (oxyR homologue) was electroporated into T. forsythia ATCC 43037 cells, and transformants were selected on agar-erythromycin plates. The sequences of oligonucleotides used in this study are shown in Table 1. First, a DNA fragment containing TF0104 with flanking sequences was amplified by PCR using primers #1 and #2 from T. forsythia 43037 genomic DNA. This PCR product was then used as template to amplify the upstream (1784 bp) and downstream (1125 bp) fragments of TF0104 with primer sets #1 and #3 and #2 and #5, respectively. The ermF fragment (797 bp) was amplified from pVA2198 (Fletcher et al., 1995) with primers #4 and #6. Primers #3, #4, #5 and #6 contained overlap sequences for ermF and TF0104 to allow generation of fusion fragments by a PCR overlap strategy. A fusion DNA fragment (3708 bp) containing ermF flanked by TF0104 DNA sequences was generated by an overlap PCR strategy using primers #1 and #2 (Horton et al., 1993). Gel-purified PCR fragments of TF10104 flanking regions (1784 and 1125 bp) and ermF (797 bp) served as template for overlap PCR. The fusion product was transformed into T. forsythia 43037 by electroporation, as previously described (Honma et al., 2007). Transformants were plated onto BF agar plates containing 5 μg erythromycin ml−1 and incubated anaerobically at 37 °C for 14 days. Following incubation, 19 erythromycin-resistant colonies were isolated, which were then screened by PCR and Southern blotting. One of the transformants, named TFM104, which was confirmed to have an oxyR deletion, was used for further analyses.
Table 1.
Primers used in this study
| Primer | Use of primer and remarks | Sequence (5′–3′) |
|---|---|---|
| Mutant construction | ||
| #1 | PCR primer for oxyR | ACCAATCAATGTCGAGGAACG |
| #2 | PCR primer for oyxR | GGATACACTTTACTACGGCAACTCA |
| #3 | PCR overlap primer (ermF: lower case type) | gtccctgaaaaatttcatccttcgTGAAGCAACGCTGTATTTTATACTTG |
| #4 | PCR overlap primer (ermF: lower case type) | CAAGTATAAAATACAGCGTTGCTTCAcgaaggatgaaatttttcagggac |
| #5 | PCR overlap primer (ermF: lower case type) | cgggcaatttcttttttgtcatCTCTGTACCATTTATTGATAAAATTGATGC |
| #6 | PCR overlap primer (ermF: lower case type) | GCATCAATTTTATCAATAAATGGTACAGAGatgacaaaaaagaaattgcccg |
| qRT-PCR | ||
| Tf 16S-F | 16S rRNA | GGGTGAGTAACGCGTATGTAACCT |
| Tf 16S-R | ACCCATCCGCAACCAATAAA | |
| dps-F | Tf0105; Dps family DNA-binding protein | CAACCGTCAAGTGTATTATGC |
| dps-R | TCAGATACTCCGAGAAGCG | |
| bcp-F | TF0245; bacterioferritin comigratory protein | GTATTTCTATCCCAAAGACAACAC |
| bcp-R | CAGGGTCGTATCCGTATCG | |
| trx-F | TF2739; thioredoxin | CGGAGTGGAAGTATGAAGG |
| trx-R | AGCGTTGGAAGACTGTTG | |
| ahpC | TF0383; alkyl hydroperoxide reductase subunit C | CGTATCTTTGTGTCGAAATTG |
| ahpc-R | GGAATTTGCGAATTAATAATCGG | |
| trxB-F | TF0375; thioredoxin reductase | CGAAAAAGTGCGTTGTTTGA |
| trxB-R | ACGCCTGCTTTTTCATGTCT | |
| prx-F | TF1518; peroxiredoxin | GGTGGTTGGTGCCCTTATTG |
| prx-R | TCGTTGTTGAGGTCGGAGAG | |
| trxA-F | TF2761; thioredoxin | TCGGAATGGAAATACTTAGGTGAC |
| trxA-R | ACGGGATGGAACGGATGC | |
| trxM-F | TF2732; thioredoxin M | GGCATTAACAGTAACAGAC |
| trxM-R | GTATTCCGAAACCACCTC | |
| fur-F | TF2532; ferric-uptake regulator | ATCCGTCATCAATTCACC |
| fur-R | ATCCCGTAAACATAGAGC | |
| sod-F | TF2927; sodF, Fe-Mn | GCCAGCAGACAATTGAATATCA |
| sod-R | TTCGGCAAGTTTGCCCTTA | |
| oxy-F | TF0104; OxyR homologue | ACCGGCATTTTGCCAAAG |
| oxy-R | ATTGATATACGGACCCTCGC | |
| rub-F | TF0103; rubredoxin | TGTGATTACATTTACGATCCGGTA |
| rub-R | CGACCGGTTCAAAATCATCT | |
| rOxyR | ||
| TF104F | NdeI site underlined | CGCCATATGACGATTCAGCAGTTAGAATATATTATTGC |
| TF104R | BamHI site underlined | GCGCGGATCCCTAGAGGTCTACCTTAATCGTATGACG |
| Mobility shift assay | ||
| Tf-sod-F | PCR; sodF promoter fragment | TCACTTGAAGTCATGCAAATCC |
| Tf-sod-R | AGGGAAGCTTTGGTGTTTCA | |
| Tf-dps-F | PCR; dps promoter fragment | GACAAGCTTCTGCCGCTTT |
| Tf-dps-R | TGTACGGCACTGTTCACTTCTT | |
| Shuttle vector construction | ||
| oxyRCow-Bam | BamHI site underlined | CGCGGATCCGCGTCTCGCGCAAGGTCTTCA |
| oxyRCow-Sal | SalI site underlined | ACGCGTCGACGTCATATGTCAGCTGTTCCTTTATATCAGGG |
Response of the T. forsythia oxyR mutant to oxidative stress.
In order to estimate the sensitivity of T. forsythia strains to hydrogen peroxide (H2O2) and air, the following experiments were performed. H2O2 sensitivity was estimated by a modification of a previously described protocol (Chen et al., 2006). Briefly, T. forsythia strains were grown to mid-exponential phase (OD600 0.5) in BF broth and harvested by centrifugation, washed twice with PBS and adjusted to OD600 1.0. T. forsythia strains were then exposed anaerobically to a twofold diluted series of H2O2 (0 μM to 10 mM) for 20 min at 37 °C. The treated bacterial cells were recovered and inoculated into BF broth. Bacterial cell survivability was expressed as OD600 values of inocula after 48 h incubation.
The aerotolerance assays were carried out as follows. T. forsythia strains were grown to mid-exponential phase (OD600 0.5) in BF broth, harvested by centrifugation, and adjusted to OD600 1.0 after washing twice with PBS. Adjusted T. forsythia strains were diluted 1 to 10 with BF broth, and exposed to air for different time periods with shaking at room temperature. Following aerobic incubation, T. forsythia cells were harvested by centrifugation, resuspended in BF broth, and incubated anaerobically at 37 °C. Aerotolerance of T. forsythia strains was expressed as OD600 values of inocula after 48 h incubation. Both H2O2 survival and the aerotolerance assay were repeated three times.
Estimation of mRNA expression of OxyR-regulated genes.
The relative expression of a select group of genes known to be regulated by the OxyR regulator in bacteria was evaluated by real-time quantitative RT-PCR (qRT-PCR). Briefly, total RNA was prepared from bacterial cells harvested from 2 ml cultures (OD600 0.4) for each wild-type and mutant strain, using the RiboPure Total RNA isolation kit with DNase I treatment (Ambion). In some experiments, catalase (5 μg ml−1; Sigma Aldrich) was added to BF broth 4 h before inoculation to remove any oxidative factors present in broth, as suggested by Diaz et al. (2006). Total RNA samples were isolated from T. forsythia strains, which were exposed to 0.1 mM H2O2 for 30 min. The cDNA was synthesized by utilizing SuperScript III reverse transcriptase (Invitrogen) with random hexamers (Promega) according to the manufacturers' recommendations. Real-time PCR was performed on cDNA samples after adjusting the A260 to 0.1 with gene-specific primers (Table 1) using iQ SYBR green Supermix (Bio-Rad) and an iCycler thermal cycler equipped with the MyIQ real-time PCR detection system as per the manufacturer's recommendations. The 16S rRNA levels were used for normalization and the relative expression levels of each transcript were then calculated using the −ΔΔCt method described for the MyIQ real-time PCR detection system (Bio-Rad).
Expression and purification of recombinant OxyR.
The putative OxyR-encoding ORF TF0104 was PCR-amplified from genomic DNA using primers TF104F (engineered NdeI site) and TF104R (engineered BamHI site). The PCR fragment was digested with NdeI and BamHI restriction enzymes and cloned into the pET15b expression vector at the same sites. The recombinant plasmid clone with the correct in-frame insertion sequence, designated pET15-OxyR, allowing expression of OxyR as a fusion protein with a C-terminal polyhistidine tag, was transformed into E. coli BL21 (DE3). A clone transformed with pET15-OxyR was grown in LB medium containing 50 μg ampicillin ml−1 at 37 °C. When the OD600 reached 0.4, IPTG was added to 0.4 mM final concentration and cultures were incubated for an additional 4 h with vigorous shaking. The cells were harvested by centrifugation at 5000 g for 30 min, suspended in PBS and subjected to cell disruption with ultrasonication. The cell lysates were centrifuged at 10 000 g for 30 min to separate the soluble fraction from the insoluble fraction containing bacterial inclusion bodies. The insoluble fraction was found to be enriched in recombinant OxyR protein, as judged by SDS-PAGE analysis. The recombinant protein present in inclusion bodies was then solubilized in 8 M urea and affinity-purified on a PrepEase Ni-TED Column (USB Biochemicals) under denaturing conditions according to the manufacturer's instructions. The eluted recombinant OxyR (rOxyR) protein was dialysed against a series of buffers with stepwise decreases in the urea concentration to allow proper protein folding [10 mM Tris/HCl (pH 7.6) and 1 % (v/v) glycerol, plus urea at 6, 4, 2 and 1 M]. Finally, the protein solution was dialysed against the storage buffer containing 10 mM Tris/HCl (pH 7.6), 1 % (v/v) glycerol, 100 mM KCl, 10 mM MgCl2 and 1 mM EDTA.
Electrophoretic mobility shift assay.
DNA probes containing the promoter regions of the sodF (TF2927) and dps (TF0105) genes were PCR-amplified from genomic DNA with primer sets Tf-sod-F and Tf-sod-R, and Tf-dps-F and Tf-dps-R, respectively; this corresponds to nucleotides −132 to +79 (211 bp fragment) and −201 to +47 (248 bp fragment) relative to the start codons of the sodF and dps genes, respectively. The DNA probes were end-labelled with [γ-32P]ATP (Perkin-Elmer) by T4 polynucleotide kinase (Promega) and purified with a QIAquick PCR purification kit (Qiagen). The standard protein–DNA binding reaction mixture contained 1 μCi (37 kBq; 1 ng) probe DNA and 4 μg purified rOxyR in binding buffer [10 mM Tris/HCl (pH 7.0), 50 mM KCl, 1 mM DTT, 100 μg BSA ml−1, 100 μg poly-(dI-dC) ml−1 and 5 % (v/v) glycerol]. For competitive assays, 50-fold unlabelled DNA fragments were used. After incubation for 20 min at room temperature, protein–DNA complexes were separated from the unbound probe on 5 % native polyacrylamide gels in running buffer [22 mM Tris base, 22 mM boric acid and 0.5 mM EDTA (pH 8.0)] at 220 V for 2 h. The gels were then dried and analysed by autoradiography.
TF0104 complementation in a P. gingivalis oxyR mutant.
Complementation of the TFM0104 mutant with oxyR was not possible due to the lack of a genetic complementation system for T. forsythia. Thus, we attempted heterocomplementation in another obligate oral anaerobe, P. gingivalis. For this purpose, we complemented a P. gingivalis oxyR deletion mutant PgOxyRE (gift from Hua Xie, Meharry Medical College) (Wu et al., 2008) with the T. forsythia TF0104 gene. To construct this complemented mutant, a DNA fragment containing TF0104 with up- and downstream flanking regions was amplified with primers oxyRCow-Bam and oxyRCow-Sal containing restriction sites BamHI and SalI, respectively (Table 1). The PCR fragment was digested with BamHI and SalI and cloned into the Bacteroides/E. coli shuttle vector pT-Cow (Gardner et al., 1996) at the BamHI and SalI sites located within the tetC gene. Purified recombinant plasmid with correct restriction profile and insertion sequences was transferred into a conjugal donor strain E. coli pRK231. This was followed by conjugation of E. coli pRK231 harbouring the recombinant plasmid with PgOxyRE (Gen+, Em+) overnight, anaerobically as described previously (Honma et al., 2001). Transconjugants were selected on BF agar plates supplemented with gentamicin (100 μg ml−1), erythromycin (5 μg ml−1) and tetracycline (1 μg ml−1) to select for the plasmid-resident tetQ gene. Transconjugants appeared after 10 days incubation and were confirmed by PCR and Southern blot analysis. The recombinant insert from plasmid DNA isolated from the complemented strain was sequenced with primers oxy-F and oxy-R and shown to be 100 % identical to wild-type T. forsythia oxyR.
Biofilm formation and aggregation assay.
In order to investigate the role of OxyR in T. forsythia biofilm formation, the effect of oxyR deletion on the development of single and multi-species biofilms was determined by performing biofilm assays as described previously (Honma et al., 2007). Briefly, bacterial cultures of T. forsythia and F. nucleatum were grown in half-strength growth medium to OD600 0.05. Cells were then dispensed (0.5 ml per well) in triplicate wells of 24-well culture plates and incubated anaerobically. For mixed biofilms, each bacterial culture was adjusted to OD600 0.05, dispensed into the wells and incubated as above. After incubation for 2 days, planktonic cells were aspirated and the wells were washed three times with PBS followed by staining of biofilms with crystal violet (0.1 % crystal violet for 15 min). Bound dye was solubilized in 50 % acetic acid and A595 was read. Total biofilm was calculated by normalizing dye binding (A595) to total bacterial growth (biofilm cells+planktonic cells) determined in parallel identical wells by measuring A595.
The aggregation abilities of T. forsythia strains with or without F. nucleatum were evaluated by a modification of the protocol of Levesque et al. (2003). Briefly, bacterial cells were harvested at late-exponential phase, washed with coaggregation buffer [1 mM Tris (pH 7.5), 150 mM NaCl, 0.1 mM CaCl2 and 0.1 mM MgCl2] and adjusted to OD660 1.0. To quantify the time-course of coaggregation, OD660 readings were taken at 0, 15, 30, 60, 90 and 120 min and the percentage of coaggregation calculated using the following equation:
Confocal scanning laser microcopy (CSLM) analysis.
For structural analysis of the biofilms and to obtain a better understanding of the bacterial interactions, CSLM was performed with a Bio-Rad MRC1024 confocal scanning laser (Kr/Ar) microscope attached to a Nikon Diaphot microscope and an APO Plan ×601.4 numerical aperture objective available in the Confocal Microscopy and 3-D Imaging Facility, School of Medicine and Biomedical Sciences, University at Buffalo, as previously described (Honma et al., 2007; Sharma et al., 2005). Briefly, biofilms were formed in two-chamber polystyrene tissue-culture vessels (BD Falcon) as described above and stained with the Bacterial Live-Dead viability kit (Molecular Probes) according to the manufacturer's recommendations. The staining allowed differentiation of live (green) and dead (red) bacteria and identification of F. nucleatum and T. forsythia due to their distinct bacterial morphology (F. nucleatum as long rods vs small fusiform T. forsythia cells).
Statistical analysis.
Strain comparisons for biofilm formation, aggregation and gene expression in real-time PCR were analysed using Student's t test.
RESULTS
OxyR homologue TF10104 contributes to survivability against air and peroxides
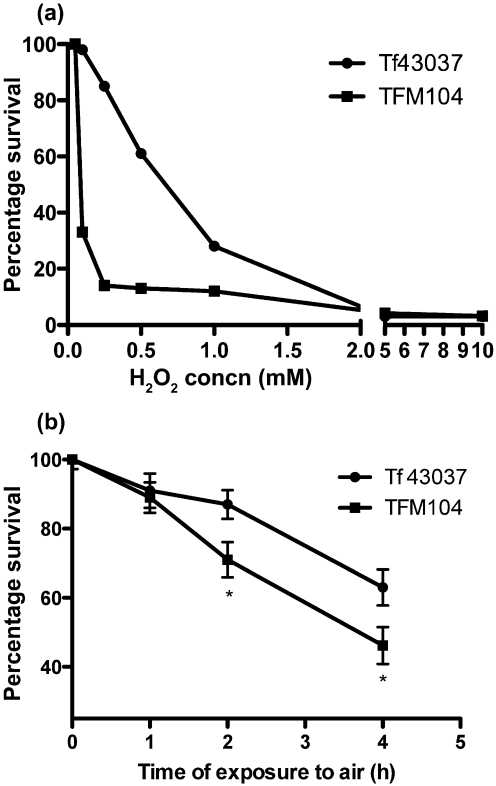
OxyR is a redox-sensitive protein regulator that belongs to the LysR family of DNA-binding transcription modulators. An OxyR homologue has been identified as ORF TF0104 in the T. forsythia genome deposited at the Oralgen database (http://www.oralgen.lanl.gov/ under T. forsythensis). The TF0104-encoded OxyR homologue shows 60 % similarity (42 % identity) with the redox-sensitive transcriptional activator of Bacteroides thetaiotaomicron VPI-5482, 60 % similarity (40 % identity) with the redox-sensitive transcriptional activator OxyR of Bacteroides fragilis NCTC 9343, 58 % similarity (37 % identity) with the redox-sensitive transcriptional activator OxyR of P. gingivalis and 54 % similarity (28 % identity) with the OxyR protein of E. coli. The cysteine residues at positions 199 and 208, present in the prototype E. coli OxyR protein and important for redox activity, are conserved in the TF0104-encoded OxyR homologue. To confirm the identity and role of OxyR-like homologue TF0104 in T. forsythia, a TF0104-defective mutant was constructed and characterized. The chromosomal location of the ermF ORF was confirmed by Southern blotting as well as by sequencing of the chromosomal region at the integration site by DNA sequencing of PCR-generated fragments (data not shown). The double-crossover allelic exchange strategy utilized in this study resulted in replacement of the TF0104 sequence from the translational start to stop codons with that of the ermF gene. Therefore, ermF is expressed from the oxyR promoter and the upstream and downstream regions (putative transcriptional terminators) are kept intact, eliminating the risk of polar effects in the mutant. Furthermore, the oxyR gene in T. forsythia appears to be a monocistronic transcription unit. It is flanked upstream by the dps gene in an opposite transcriptional orientation. The intergenic region between the dps and oxyR genes is 129 nt long and the sequence analysis of this region in the mutant showed that this region was not affected by the ermF insertion (data not shown). Similarly, downstream from oxyR is a putative rubredoxin-encoding gene with a 277 nt long intergenic region, which was also confirmed by sequence analysis to be intact in the mutant. Thus, taken together, in all probability the insertion of the erm cassette did not cause any polar effects on flanking genes. One such mutant with correct integration was named TFM104 and was further characterized. Mutant TFM104 was compared to the parental strain with respect to the functions of OxyR in oxidative stress defence responses. The results of the H2O2 sensitivity assay showed that the maximum H2O2 concentration for growth of the mutant TFM104 was 0.25 mM as compared to 1 mM for T. forsythia wild-type (Fig. 1a). Moreover, while both T. forsythia 43037 and TFM104 showed sensitivity, TFM104 was significantly more sensitive to aerobic stress than 43037 after 2 h incubation (Fig. 1b). These results suggested that TF0104 plays an important role in resistance to oxidative stress.
Fig. 1.
Sensitivity to oxidative stress. (a) Resistance to H2O2 was determined by exposing mid-exponential phase cells to serial dilutions of H2O2 anaerobically for 20 min. Percentage survival for each peroxide concentration was then calculated following normalization with untreated controls under similar conditions (b). To determine aerotolerance, cells harvested at the mid-exponential phase were exposed to air for the indicated times. Percentage survival was then determined for each time point normalized to the initial time 0, as described in the text; *P<0.05.
Expression of oxidative response genes in T. forsythia is OxyR-dependent but does not increase in response to H2O2
The OxyR protein plays an important role in the expression of antioxidant-related genes in the majority of bacteria examined, including the oral Gram-negative obligate anaerobe P. gingivalis (Diaz et al., 2006). According to a DNA microarray analysis, expression of the antioxidant response genes encoding alkyl hydroperoxide reductase C subunit (ahpC), F subunit (ahpF), Dps family protein (dps), superoxide dismutase (sod) and ferritin is dependent upon the expression of OxyR in P. gingivalis (Diaz et al., 2006). In this study we analysed and compared the expression of T. forsythia homologue antioxidant-related genes (Table 2) between T. forsythia 43037 and oxyR-inactivated mutant TFM104 by qRT-PCR. The mRNA expression levels of T. forsythia antioxidant-related genes were downregulated in the oxyR mutant TFM104 as compared to the parental strain.
Table 2.
Genes with decreased levels of expression in the oxyR mutant TFM104 compared with T. forsythia 43037 wild-type (during anaerobic growth)
| Gene* | Description | Fold decrease† | P value |
|---|---|---|---|
| TF0105 dps | DNA-binding stress protein, Dps family | 21.15 | 0.013 |
| TF0245 bcp | Bacterioferritin comigratory protein | 7.28 | 0.00010 |
| TF2739 trx | Thioredoxin | 4.90 | 0.00001 |
| TF0383 ahpC | Alkyl hydroperoxide reductase, subunit C | 4.53 | 0.00056 |
| TF2927 sodF/sodB | Superoxide dismutase, Fe-Mn | 4.46 | 0.00353 |
| TF0103 | Rubredoxin | 2.81 | 0.03356 |
| TF0375 trxB/ahpF | Thioredoxin reductase | 2.75 | 0.00550 |
| TF1518 prx | Peroxiredoxin | 2.53 | 0.00132 |
| TF2761 trxA | Thioredoxin | 2.36 | 0.00781 |
| TF2732 | Thioredoxin M | 2.29 | 0.00187 |
| TF2532 fur | Ferric-uptake regulator | 2.08 | 0.00068 |
*Gene identification corresponds to that of the Oralgen database.
†Only genes exhibiting a greater than twofold decrease in transcription are shown.
The mRNA expression levels of the above-mentioned antioxidant genes were also estimated under H2O2-stimulated conditions. The expression levels of these antioxidant genes did not change significantly in response to H2O2 treatment; no significant fold-changes (P>0.05) in the expression levels of the genes were observed when comparison was made between peroxide-treated and untreated wild-type cells under anaerobic growth by qRT-PCR, as described above. The dps gene has been shown to play a role in the protection of E. coli against peroxide stress, and its expression is regulated by OxyR (Altuvia et al., 1994). Dps is also significantly upregulated several fold in E. coli and B. fragilis after H2O2 exposure compared with untreated cultures (Rocha & Smith, 1997). On the other hand, H2O2 does not have any significant effect on the expression of dps in the anaerobe P. gingivalis (Diaz et al., 2006). Since the dps transcript is detected in wild-type cultures grown anaerobically without H2O2 treatment, and it does not increase after treatment with H2O2, it is assumed that OxyR is in a constitutively active state in P. gingivalis. Our findings also suggest that T. forsythia OxyR is at its maximum level of activation during anaerobic growth, as addition of peroxide had no effect on the expression of oxyR-regulated genes. To rule out the possibility that peroxides are present in broth and constitutively keep OxyR in an active state, T. forsythia was grown in BF broth pretreated with catalase, as described by Diaz et al. (2006). Under these conditions, we did not find any changes in the expression of antioxidant genes before and after H2O2 treatment, similar to what was observed with non-catalase-treated broth (data not shown). However, the fact that under anaerobic growth the expression levels of dps, ahpC and sodF were significantly reduced in the oxyR mutant as compared with levels in the wild-type strain suggests that OxyR is necessary for the expression of these antioxidant genes under anaerobic growth.
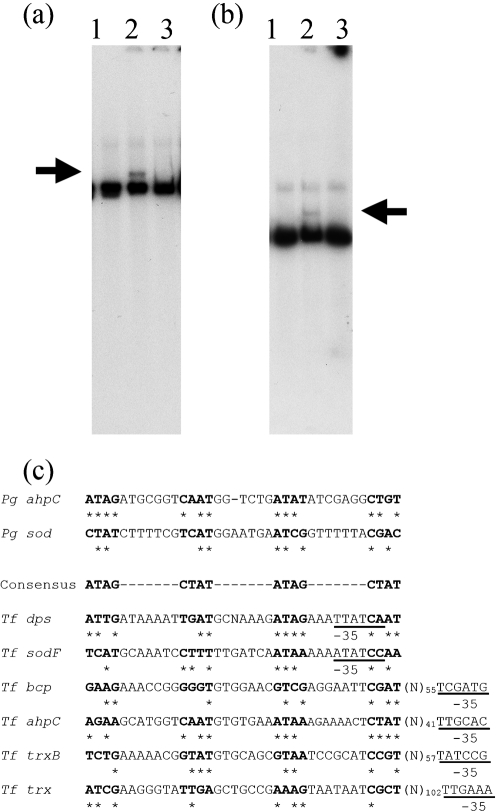
rOxyR binds dps and sodF promoter regions
We next sought to determine whether OxyR binds to the T. forsythia dps and sodF genes, which are regulated by OxyR, utilizing gel mobility shift assays. Our results showed that purified 6His-tagged rOxyR protein encoded by TF0104 at a concentration of at least 10 nM caused a mobility shift of DNA fragments containing these target promoters (Fig. 2a). Weaker than expected shifts due to rOxyR binding to respective promoters were probably due to the following reasons. The rOxyR protein utilized for gel shift assays was obtained following renaturation of urea-solubilized protein by stepwise dialysis; this probably resulted in the recovery of only a fraction of the total protein in a conformationally active state. Furthermore, the promoter regions of antioxidant genes in T. forsythia, including dps and sodF, indicated the presence of putative OxyR-binding sequences when compared with P. gingivalis OxyR-regulated promoters as well as with consensus OxyR-binding sequences found in E. coli (Fig. 2c).
Fig. 2.
Electrophoretic mobility shift analysis of the promoter region DNA of the T. forsythia dps and sod genes with rOxyR. Mobility shift assays were performed with purified rOxyR (1 μg) and 2 ng 32P-labelled dps (a) and sod (b) gene promoter DNA fragments. Lanes: 1, labelled promoter; 2, labelled promoter with rOxyR; 3, same as lane 2 with corresponding unlabelled promoter DNA as competitor. Arrows indicate the complex of probe DNA and rOxyR. (c) Comparison of the putative OxyR-binding sequences within the T. forsythia (Tf) antioxidant genes analysed in the present study with P. gingivalis (Pg) ahpC and sod gene OxyR-binding motifs. The consensus sequence shown is the E. coli OxyR-binding motif. Putative –35 boxes are underlined.
Restoration of OxyR function by heterocomplementation with the putative T. forsythia oxyR
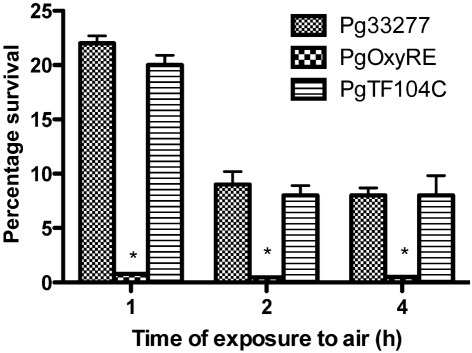
Currently, genetic systems for gene complementation in T. forsythia are unavailable. Therefore, we attempted genetic complementation of a P. gingivalis oxyR deletion mutant (PgOxyRE) with the TF0104 gene carried on a shuttle plasmid. By utilizing this strategy, a heterocomplemented strain designated Pg0270EC was obtained, which was then compared to PgOxyRE in aerotolerance assays. The results (Fig. 3) showed that complementation with TF0104 significantly restored the loss of OxyR function in the P. gingivalis oxyR mutant; the TF0104-complemented strain survived significantly better under oxygen stress than the oxyR deletion mutant or the pT-Cow-complemented strains. These results further demonstrated that TF0104 encodes a functional OxyR protein. Interestingly, several attempts to complement the T. forsythia oxyR mutant TFM104 were unsuccessful. We believe that these results are due to the fastidious and stringent dependence of T. forsythia on anaerobic environments for growth, and also to the fact that a brief exposure to air stress during conjugal mating with E. coli might be lethal to a mutant already defective in the expression of its oxidative stress response sensor.
Fig. 3.
Heterocomplementation with TF0104 restores the OxyR phenotype in the P. gingivalis mutant. Survival of P. gingivalis oxyR mutant (Pg0270E), complemented strain PgTF104C and P. gingivalis strain 33277 was compared following exposure to air for the indicated times. Percentage survival was then determined for each time point normalized to the initial time 0. While survival of Pg0270E was significantly reduced, as expected (P<0.05 compared with the wild-type), survival of the TF104-complemented strain was similar to that of the wild-type strain.
T. forsythia oxyR contributes to biofilm formation with F. nucleatum
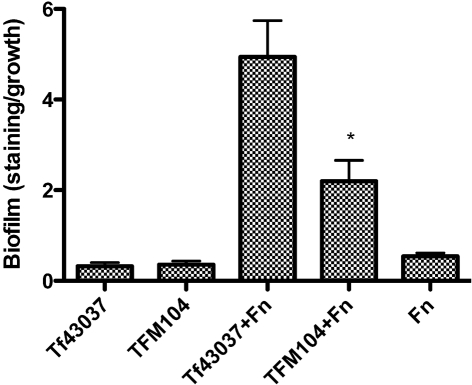
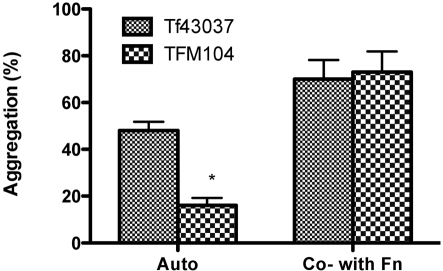
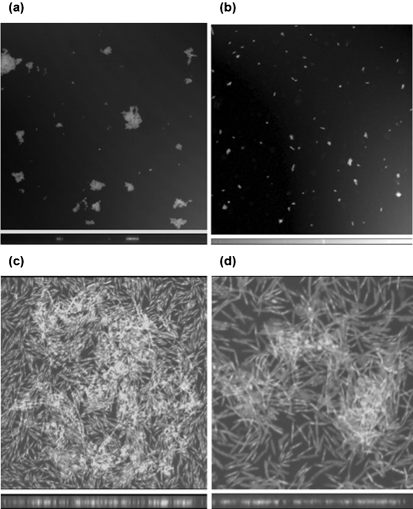
We have previously shown that T. forsythia 43037 forms synergistic biofilms with F. nucleatum (Honma et al., 2007; Sharma et al., 2005). In this study, we tested the ability of TFM104 to form synergistic biofilms with F. nucleatum. Our results showed that TFM104 and F. nucleatum form weaker mixed biofilms than that of T. forsythia 43037 and F. nucleatum (Fig. 4). To determine whether surface adhesin expression was affected in TFM104, resulting in reduced biofilm-forming ability, aggregation assays were performed. As per the aggregation assay results, the autoaggregation activity of TFM104 was found to be lower than that of the wild-type T. forsythia strain (Fig. 5). T. forsythia biofilm morphology was further analysed by CSLM (Fig. 6). In agreement with the results of autoaggregation assays, CSLM analysis showed that wild-type T. forsythia cells formed small autoaggregated microcolonies, whereas the oxyR mutant TFM104 cells remained uniformly dispersed with minimum to no microcolony formation. When viewed in the x-z plane, microcolonies of the wild-type strain extended away from the attached surface and were taller than microcolonies of the mutant strain TFM104 (Fig. 6a, b). Moreover, the mixed bioflms of wild-type T. forsythia and F. nucleatum were thicker than those of the TFM104 mutant and F. nucleatum, 2.88 and 1.64 μm, respectively, under x-z digital view (Fig. 6c, d). Within the mixed biofilms, F. nucleatum cells were observed as long rods and T. forsythia was observed as fusiform. More than 99 % of total bacterial cells were live (stained green). A qualitative estimation also indicated that a higher proportion of the wild-type T. forsythia cells were present in mixed biofilms than mutant TFM104 cells in the respective mixed biofilms.
Fig. 4.
Analysis of biofilm activity of T. forsythia strains. Monospecies or mixed-species biofilms were formed in polystyrene wells and biofilm formation was calculated as crystal violet staining (A595) normalized to total bacterial growth (OD600). Tf43037, monospecies wild-type T. forsythia biofilm; TFM104, monospecies mutant TFM104 biofilm; Tf43037+Fn, mixed T. forsythia and F. nucleatum biofilm; TFM104+Fn, mixed F. nucleatum and TFM104 biofilm; Fn, monospecies F. nucleatum biofilm. Data points represent mean±sd of triplicate samples. Data are representative of three independent experiments. *P<0.01 compared to mixed T. forsythia wild-type and F. nucleatum biofilm.
Fig. 5.
Aggregation activity of T. forsythia strains. For autoaggregation activity, T. forsythia strains were harvested at mid-exponential phase, washed in aggregation buffer and adjusted to OD660 1.0, and incubated at 37 °C in glass cuvettes. Aggregation activity was calculated based on absorbance of the cells after 30 min incubation by utilizing the equation given in Methods. For coaggregation activity, T. forsythia cells were mixed with F. nucleatum cells prewashed in aggregation buffer and adjusted to OD660 1.0. Data points represent mean±sd of triplicate samples. Data are representative of three independent experiments. *P<0.05.
Fig. 6.
CSLM images of autoaggregation and mixed biofilms. (a, b) Autoaggregation. Monospecies cultures were incubated for 3 h in two-well culture dishes and surface-attached bacterial cells were stained with live-dead bacterial stain as described in the text [(a) T. forsythia wild-type; (b) mutant TFM104]. (c, d) Mixed biofilms. T. forsythia wild-type and F. nucleatum (c) or TFM104 mutant and F. nucleatum (d) were coincubated in two-well polystyrene tissue-culture vessels for 36 h and stained with a bacterial live-dead staining kit as described in the text. x-z reconstruction (biofilm thickness) for each condition is shown below the x-y image: 2.88 μm (c) and 1.68 μm (d).
DISCUSSION
This report describes the identification and functional characterization of an oxyR homologue encoded by the ORF TF0104 in T. forsythia. The OxyR-like proteins are redox sensors that belong to the LysR family of DNA-binding transcription modulators and control the expression of a number of antioxidant genes in bacteria. In aerobic organisms such as E. coli, the OxyR protein is activated in response to peroxide stress through formation of disulfide bonds and regulates antioxidant genes involved in defence against peroxide stress (Zheng & Storz, 2000). While OxyR is mainly responsible for tolerance to peroxide stress in aerobic E. coli and the facultative anaerobe B. fragilis, it appears to play an essential role against peroxide as well as oxygen (air) stress in obligate anaerobes such as P. gingivalis (Diaz et al., 2006). The aerotolerance of the facultative anaerobe B. fragilis depends on alternative mechanisms that are not OxyR-dependent (Sund et al., 2006; Tang et al., 1999). To date, except for the role of OxyR in P. gingivalis, this protein has not been well studied in Gram-negative obligate anaerobes. Thus, the role of the OxyR regulator in T. forsythia was examined by characterizing a deletion mutant of the oxyR homologue. Our results showed that, in comparison with the wild-type T. forsythia strain 43037, an oxyR deletion mutant TFM104 was defective in its ability to resist H2O2 and aerobic stress. The mutant showed significantly reduced expression of antioxidant gene-encoding enzymes involved in the detoxification of H2O2 in comparison with the parental strain under anaerobic growth conditions. Since T. forsythia lacks catalase activity (Hudspeth et al., 1999), bacterioferritin comigratory protein (BCP; an alkyl hydroperoxide reductase C family protein) (Jeong et al., 2000) and AhpC may compensate for the absence of catalase activity in peroxide detoxification. Superoxide dismutase activity has been shown to be important in the obligate anaerobe P. gingivalis for protection against aerobic exposure (Ohara et al., 2006). In addition, the Dps protein with ferritin-like activity has the capacity to attenuate the production of reactive oxygen species (Chiancone et al., 2004). Dps assists in controlling iron pools that can lead to hydroxyl radical production via the Fenton reaction. Taken together, we predicted that OxyR-dependent expression of antioxidant homologue genes should be critical for protection against oxidative stress in T. forsythia. Interestingly, the levels of oxidative stress-induced genes did not increase in response to peroxide stress, indicating a constitutively activated stage of OxyR in this bacterium. This is analogous to the situation in another Gram-negative obligate anaerobe, P. gingivalis (Diaz et al., 2006). It is hypothesized that a constitutively activated state of OxyR can benefit obligate anaerobes living in the oral cavity by protecting them against constant challenge from atmospheric oxygen and antibacterial oxidative responses of host leukocytes. The mechanisms underlying constitutive activation of OxyR in P. gingivalis and T. forsythia have yet to be determined. However, it is likely that OxyR proteins in anaerobes have evolved due to amino acid substitutions that induce conformational changes favouring disulfide bond formation and locking OxyR in an active state. This notion is supported by the fact that Bacteroides mutants have been isolated with constitutively active OxyR conformational states. Single amino acid substitutions in the OxyR protein in these strains have been shown to cause constitutive activation (Rocha et al., 2000).
In this study we also showed that the deletion of oxyR results in attenuation of mixed biofilm formation of T. forsythia with F. nucleatum. We have previously shown that T. forsythia forms weak biofilms alone but forms synergistic biofilms in mixed cultures with F. nucleatum (Sharma et al., 2005). Here we showed that the OxyR mutant TFM104 was significantly attenuated in its ability to form mixed biofilms with F. nucleatum. In this regard, links between biofilm formation and oxidative stress defences have been observed in several microbes, including E. coli (Reisner et al., 2003), H. influenzae (Murphy et al., 2005), Ps. aeruginosa (Sauer et al., 2002), C. jejuni (Sampathkumar et al., 2006), Strep. mutans (Blumer et al., 2005), Can. albicans (Murillo et al., 2005), Ser. marcescens (Shanks et al., 2007) and P. gingivalis (Wu et al., 2008). To our knowledge, F. nucleatum is not known to increase oxidative stress, which could explain the reduction in synergistic biofilm formation by the oxyR mutant TFM104 with F. nucleatum in mixed cultures. In fact, the growth of F. nucleatum has been shown to maintain low redox potentials [E(h)=−274 mV] with no detectable oxygen in the medium and to support the growth of anaerobic P. gingivalis (Diaz et al., 2000). An alternative explanation for the reduced biofilm phenotype of TFM104 compared with the parental strain might be differences in the expression of surface components, which could affect biofilm formation. For example, in E. coli, OxyR regulates the expression of a surface adhesin, Ag43 (Danese et al., 2000). Ag43 promotes biofilm formation via its strong cell-to-cell aggregation characteristics (autoaggregation). Interestingly, it is the reduced form of OxyR which then acts as a repressor for derepression of the Ag43 encoding gene upon exposure to oxidative stress, leading to Ag43 expression (Schembri et al., 2003). It is believed that Ag43-mediated cell aggregation and biofilm formation confer protection against H2O2 killing (Schembri et al., 2003). In Ser. marcescens, OxyR regulates biofilm formation through induction of fimbriae production (Shanks et al., 2007). In P. gingivalis, OxyR acts as a repressor of the surface adhesin FimA involved in biofilm formation, and therefore an oxyR deletion mutant forms increased biofilms compared with its parental strain (Wu et al., 2008). Our results presented here with respect to T. forsythia show that in comparison with the parental strain, an oxyR mutant, TFM104, is significantly reduced in its ability to autoaggregate and form mixed biofilms with F. nucleatum. The coaggregation ability of the mutant with F. nucleatum did not differ significantly from that of the parental strain, indicating that the adhesins involved in autoaggregation and coaggregation may be different. With regard to the effect of autoaggregation in biofilm development, in a previous study we also observed that the ability of T. forsythia to form autoaggregated microcolonies is important in biofilm development (Honma et al., 2007). We showed that the surface exopolysaccharide-deficient mutants with greater surface hydrophobicity than the wild-type strain make aggregated microcolonies and subsequently better biofilms than the wild-type strain. While the underlying mechanisms linking oxidative stress defences and biofilm formation are not well understood, the OxyR regulation of biofilm formation in T. forsythia is expected to promote bacterial adaptation in the oxidative environment of the oral cavity. Taken together, our findings provide evidence for a role for the OxyR pathways in antioxidant responses and suggest that OxyR also affects biofilm formation in T. forsythia.
In summary, TF0104 encodes an OxyR homologue in T. forsythia that acts as a positive regulator of the expression of oxidative defence responses and also affects biofilm formation. Thus, T. forsythia OxyR likely plays roles in defence against oxidative stress mediated by neutrophils and other bacteria, as well as in biofilm development, critical to the ability of the bacterium to persist in the oral cavity.
Acknowledgments
We thank Howard K. Kuramitsu for critical reading of this manuscript. This study was supported by a grant from the National Institute of Dental and Craniofacial Research (NIDCR) (DE014749).
Abbreviations
CSLM, confocal scanning laser microcopy
qRT-PCR, real-time quantitative RT-PCR
rOxyR, recombinant OxyR
References
- Altuvia, S., Almiron, M., Huisman, G., Kolter, R. & Storz, G. (1994). The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol Microbiol 13, 265–272. [DOI] [PubMed] [Google Scholar]
- Amano, A., Shizukuishi, S., Tamagawa, H., Iwakura, K., Tsunasawa, S. & Tsunemitsu, A. (1990). Characterization of superoxide dismutases purified from either anaerobically maintained or aerated Bacteroides gingivalis. J Bacteriol 172, 1457–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer, C., Kleefeld, A., Lehnen, D., Heintz, M., Dobrindt, U., Nagy, G., Michaelis, K., Emödy, L., Polen, T. & other authors (2005). Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology 151, 3287–3298. [DOI] [PubMed] [Google Scholar]
- Chen, W., Honma, K., Sharma, A. & Kuramitsu, H. K. (2006). A universal stress protein of Porphyromonas gingivalis is involved in stress responses and biofilm formation. FEMS Microbiol Lett 264, 15–21. [DOI] [PubMed] [Google Scholar]
- Chiancone, E., Ceci, P., Ilari, A., Ribacchi, F. & Stefanini, S. (2004). Iron and proteins for iron storage and detoxification. Biometals 17, 197–202. [DOI] [PubMed] [Google Scholar]
- Danese, P. N., Pratt, L. A., Dove, S. L. & Kolter, R. (2000). The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol Microbiol 37, 424–432. [DOI] [PubMed] [Google Scholar]
- Diaz, P. I., Zilm, P. S. & Rogers, A. H. (2000). The response to oxidative stress of Fusobacterium nucleatum grown in continuous culture. FEMS Microbiol Lett 187, 31–34. [DOI] [PubMed] [Google Scholar]
- Diaz, P. I., Zilm, P. S., Wasinger, V., Corthals, G. L. & Rogers, A. H. (2004). Studies on NADH oxidase and alkyl hydroperoxide reductase produced by Porphyromonas gingivalis. Oral Microbiol Immunol 19, 137–143. [DOI] [PubMed] [Google Scholar]
- Diaz, P. I., Slakeski, N., Reynolds, E. C., Morona, R., Rogers, A. H. & Kolenbrander, P. E. (2006). Role of oxyR in the oral anaerobe Porphyromonas gingivalis. J Bacteriol 188, 2454–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, H. M., Schenkein, H. A., Morgan, R. M., Bailey, K. A., Berry, C. R. & Macrina, F. L. (1995). Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun 63, 1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, R. G., Russell, J. B., Wilson, D. B., Wang, G. R. & Shoemaker, N. B. (1996). Use of a modified Bacteroides-Prevotella shuttle vector to transfer a reconstructed beta-1,4-d-endoglucanase gene into Bacteroides uniformis and Prevotella ruminicola B(1)4. Appl Environ Microbiol 62, 196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi, S. G., Zambon, J. J., Ho, A. W., Koch, G., Dunford, R. G., Machtei, E. E., Norderyd, O. M. & Genco, R. J. (1994). Assessment of risk for periodontal disease. I. Risk indicators for attachment loss. J Periodontol 65, 260–267. [DOI] [PubMed] [Google Scholar]
- Grossi, S. G., Genco, R. J., Machtei, E. E., Ho, A. W., Koch, G., Dunford, R., Zambon, J. J. & Hausmann, E. (1995). Assessment of risk for periodontal disease. II. Risk indicators for alveolar bone loss. J Periodontol 66, 23–29. [DOI] [PubMed] [Google Scholar]
- Honma, K., Kuramitsu, H. K., Genco, R. J. & Sharma, A. (2001). Development of a gene inactivation system for Bacteroides forsythus: construction and characterization of a BspA mutant. Infect Immun 69, 4686–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma, K., Inagaki, S., Okuda, K., Kuramitsu, H. K. & Sharma, A. (2007). Role of a Tannerella forsythia exopolysaccharide synthesis operon in biofilm development. Microb Pathog 42, 156–166. [DOI] [PubMed] [Google Scholar]
- Horton, R. M., Ho, S. N., Pullen, J. K., Hunt, H. D., Cai, Z. & Pease, L. R. (1993). Gene splicing by overlap extension. Methods Enzymol 217, 270–279. [DOI] [PubMed] [Google Scholar]
- Hudspeth, M. K., Hunt Gerardo, S., Maiden, M. F., Citron, D. M. & Goldstein, E. J. (1999). Characterization of Bacteroides forsythus strains from cat and dog bite wounds in humans and comparison with monkey and human oral strains. J Clin Microbiol 37, 2003–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, W., Cha, M.-K. & Kim, I.-H. (2000). Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/alkyl hydroperoxide peroxidase C (AhpC) family. J Biol Chem 275, 2924–2930. [DOI] [PubMed] [Google Scholar]
- Johnson, J. R., Clabots, C. & Rosen, H. (2006). Effect of inactivation of the global oxidative stress regulator oxyR on the colonization ability of Escherichia coli O1 : K1 : H7 in a mouse model of ascending urinary tract infection. Infect Immun 74, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, G. W., Britigan, B. E. & Hassett, D. J. (2005). Pseudomonas aeruginosa OxyR is required for full virulence in rodent and insect models of infection and for resistance to human neutrophils. Infect Immun 73, 2550–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque, C., Lamothe, J. & Frenette, M. (2003). Coaggregation of Streptococcus salivarius with periodontopathogens: evidence for involvement of fimbriae in the interaction with Prevotella intermedia. Oral Microbiol Immunol 18, 333–337. [DOI] [PubMed] [Google Scholar]
- Murillo, L. A., Newport, G., Lan, C. Y., Habelitz, S., Dungan, J. & Agabian, N. M. (2005). Genome-wide transcription profiling of the early phase of biofilm formation by Candida albicans. Eukaryot Cell 4, 1562–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, T. F., Kirkham, C., Sethi, S. & Lesse, A. J. (2005). Expression of a peroxiredoxin-glutaredoxin by Haemophilus influenzae in biofilms and during human respiratory tract infection. FEMS Immunol Med Microbiol 44, 81–89. [DOI] [PubMed] [Google Scholar]
- Ohara, N., Kikuchi, Y., Shoji, M., Naito, M. & Nakayama, K. (2006). Superoxide dismutase-encoding gene of the obligate anaerobe Porphyromonas gingivalis is regulated by the redox-sensing transcription activator OxyR. Microbiology 152, 955–966. [DOI] [PubMed] [Google Scholar]
- Reisner, A., Haagensen, J. A., Schembri, M. A., Zechner, E. L. & Molin, S. (2003). Development and maturation of Escherichia coli K-12 biofilms. Mol Microbiol 48, 933–946. [DOI] [PubMed] [Google Scholar]
- Rocha, E. R. & Smith, C. J. (1997). Regulation of Bacteriodes fragilis katB mRNA by oxidative stress and carbon limitation. J Bacteriol 179, 7033–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha, E. R., Owens, G., Jr & Smith, C. J. (2000). The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J Bacteriol 182, 5059–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar, B., Napper, S., Carrillo, C. D., Willson, P., Taboada, E., Nash, J. H., Potter, A. A., Babiuk, L. A. & Allan, B. J. (2006). Transcriptional and translational expression patterns associated with immobilized growth of Campylobacter jejuni. Microbiology 152, 567–577. [DOI] [PubMed] [Google Scholar]
- Sauer, K., Camper, A. K., Ehrlich, G. D., Costerton, J. W. & Davies, D. G. (2002). Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184, 1140–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schembri, M. A., Hjerrild, L., Gjermansen, M. & Klemm, P. (2003). Differential expression of the Escherichia coli autoaggregation factor antigen 43. J Bacteriol 185, 2236–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seib, K. L., Wu, H. J., Srikhanta, Y. N., Edwards, J. L., Falsetta, M. L., Hamilton, A. J., Maguire, T. L., Grimmond, S. M., Apicella, M. A. & other authors (2007). Characterization of the OxyR regulon of Neisseria gonorrhoeae. Mol Microbiol 63, 54–68. [DOI] [PubMed] [Google Scholar]
- Shanks, R. M., Stella, N. A., Kalivoda, E. J., Doe, M. R., O'Dee, D. M., Lathrop, K. L., Guo, F. L. & Nau, G. J. (2007). A Serratia marcescens OxyR homolog mediates surface attachment and biofilm formation. J Bacteriol 189, 7262–7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, A., Inagaki, S., Sigurdson, W. & Kuramitsu, H. K. (2005). Synergy between Tannerella forsythia and Fusobacterium nucleatum in biofilm formation. Oral Microbiol Immunol 20, 39–42. [DOI] [PubMed] [Google Scholar]
- Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C. & Kent, R. L., Jr (1998). Microbial complexes in subgingival plaque. J Clin Periodontol 25, 134–144. [DOI] [PubMed] [Google Scholar]
- Sund, C. J., Greg Wells, W. & Jeffrey Smith, C. (2006). The Bacteroides fragilis P20 scavengase homolog is important in the oxidative stress response but is not controlled by OxyR. FEMS Microbiol Lett 261, 211–217. [DOI] [PubMed] [Google Scholar]
- Tang, Y. P., Dallas, M. M. & Malamy, M. H. (1999). Characterization of the BatI (Bacteroides aerotolerance) operon in Bacteroides fragilis: isolation of a B. fragilis mutant with reduced aerotolerance and impaired growth in in vivo model systems. Mol Microbiol 32, 139–149. [DOI] [PubMed] [Google Scholar]
- Tanner, A., Maiden, M. F., Macuch, P. J., Murray, L. L. & Kent, R. L., Jr (1998). Microbiota of health, gingivitis, and initial periodontitis. J Clin Periodontol 25, 85–98. [DOI] [PubMed] [Google Scholar]
- Tran, S. D., Rudney, J. D., Sparks, B. S. & Hodges, J. S. (2001). Persistent presence of Bacteroides forsythus as a risk factor for attachment loss in a population with low prevalence and severity of adult periodontitis. J Periodontol 72, 1–10. [DOI] [PubMed] [Google Scholar]
- Wen, Z. T., Suntharaligham, P., Cvitkovitch, D. G. & Burne, R. A. (2005). Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect Immun 73, 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Lin, X. & Xie, H. (2008). OxyR is involved in coordinate regulation of expression of fimA and sod genes in Porphyromonas gingivalis. FEMS Microbiol Lett 282, 188–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, M. & Storz, G. (2000). Redox sensing by prokaryotic transcription factors. Biochem Pharmacol 59, 1–6. [DOI] [PubMed] [Google Scholar]