SUMMARY
Estimating haplotype relative risks in a family-based study is complicated by phase ambiguity and the many parameters needed to quantify relative risks for all possible diplotypes. This problem becomes manageable if a particular haplotype has been implicated previously as relevant to risk. We fit log-linear models to estimate the risks associated with a candidate haplotype relative to the aggregate of other haplotypes. Our approach uses existing haplotype-reconstruction algorithms but requires assumptions about the distribution of haplotypes among triads in the source population. We consider three levels of stringency for those assumptions: Hardy-Weinberg Equilibrium (HWE), random mating, and no assumptions at all. We assessed our method’s performance through simulations encompassing a range of risk haplotype frequencies, missing data patterns, and relative risks for either offspring or maternal genetic effects. The unconstrained model provides robustness to bias from population structure but requires excessively large sample sizes unless there are few haplotypes. Assuming HWE accommodates many more haplotypes but sacrifices robustness. The model assuming random mating is intermediate, both in the number of haplotypes it can handle and in robustness. To illustrate, we reanalyze data from a study of orofacial clefts to investigate a 9-SNP candidate haplotype of the IRF6 gene.
Keywords: log-linear model, candidate haplotype, phase ambiguity, expectation maximization algorithm, Hardy-Weinberg equilibrium, maternal genetic effects, Yin-Yang haplotypes
INTRODUCTION
Family-based association studies are increasingly important in genetic mapping. Analyzed with methods that are robust to population stratification, they can provide good statistical power along with insight into maternally-mediated and parent-of-origin effects. The family relationships also enhance one’s ability to infer phase for haplotype-based analyses. By haplotype, we shall here refer somewhat loosely to any string of closely linked single nucleotide polymorphism (SNP) alleles, which are transmitted in the same gamete from a parent and which may not uniquely specify a resequencing-based haplotype for the given segment of DNA.
Most haplotype-oriented association methods for case-parent triads aim at hypothesis testing; only a few focus on estimation of risk parameters. Approaches that do provide for estimation include: a log-linear model for haplotype analysis (HAPLIN) (Gjessing & Lie, 2006), the projection conditional on parental haplotypes (PCPH) method (Allen & Satten, 2007), a Stochastic Expectation Maximization (SEM) approach (Carayol et al., 2006), a likelihood-based association analysis (Dudbridge, 2008) and case/pseudocontrol analysis (Cordell, 2004; Cordell et al., 2004). By modeling the relative risks and/or haplotype frequencies for all possible haplotypes, HAPLIN and PCPH neither require nor exploit previous specification of a candidate haplotype, and they quickly become computationally intractable because the number of possible haplotypes, hence parameters, grows exponentially with the number of SNPs. We focus on the setting where prior evidence implicated a candidate haplotype, perhaps based on application of a multi-marker method such as TRIMM (Shi et al., 2007). Incorporating such knowledge into relative risk estimation can potentially add efficiency and power.
Reconstructing haplotypes in families from phase-unknown multi-SNP genotypes is a challenge. Several programs for reconstructing haplotypes in a general pedigree are widely used and well tested (Zhang et al., 2005; Marchini et al., 2006; Abecasis et al., 2002; Abecasis & Wigginton, 2005; Zhang & Zhao, 2006). These programs typically handle many loci and perform well in the presence of missing genotypes; some provide a list of possible diplotype (phased genotype) configurations, together with their estimated probabilities, for each pedigree. We set out to design a method for haplotype-relative-risk estimation that effectively takes advantage of existing software for haplotype reconstruction.
With the ever increasing number of genetic studies and the improvement of our understanding of the human genome, a candidate haplotype may be known a priori from previous studies. Our log-linear modeling approach can estimate the relative risk for such a candidate haplotype in a triad-based association study. We assume a candidate haplotype has been tagged using a set of tightly linked SNPs, so that recombination in one generation is negligible. We initially assume that the haplotypes are in HWE in the source population, but later we consider successive relaxations of that assumption. Our method uses triad-specific lists of possible haplotype configurations inferred through a haplotype reconstruction program (Haplore: Zhang et al., 2005). The approach we propose is an extension of the log-linear approach of Weinberg et al.(Weinberg et al., 1998; Wilcox et al., 1998) from a single SNP locus to a candidate haplotype. Gjessing & Lie (2006) proposed a similar approach. Our method has distinct features, however; it relaxes the HWE assumption, it reduces the dimension of the parameter space and achieves high computational efficiency by focusing on a candidate haplotype nominated based on prior knowledge, and it is capable of handling many SNPs.
The assumption that only one haplotype confers risk may sometimes be too restrictive. We show how to extend our approach to allow two candidate risk haplotypes. This extension may prove particularly useful where a pair of candidate haplotypes has complementary alleles at every SNP locus. Such ‘Yin-Yang’ pairs are widespread in the human genome (Zhang et al., 2003). Consider the problem of estimating the relative risk for a single di-allelic SNP; a causative effect of one allele is mathematically and conceptually equivalent to a protective effect of the other allele. This duality also arises in multi-marker analyses. As the number of SNPs increases, however, inference based on our extended model can distinguish between the two complementary interpretations.
We assess the performance of our approach using simulations that encompass a range of risk haplotype frequencies, different rates of missing SNP genotypes and/or of missing parents, and various relative risks involving effects of either offspring or maternal genotypes. We also use simulations to study the robustness of our procedures to various departures from HWE and compare its performance under those circumstances with the performance of competitors Unphased (Dudbridge, 2008) and case/pseudocontrol analysis (Cordell, 2004). As an illustrative example, we applied our method to data on orofacial clefts.
METHODS
Overall modeling strategy
Consider a study of case-parent triads where no two triads have members in common. Suppose we could unambiguously determine the diplotypes for all individuals in that dataset. Estimating the relative risk for a candidate haplotype would become an easy problem. One could group the haplotypes into two classes: the candidate risk haplotype and the aggregate non-risk ‘haplotype’, an amalgam of the remaining haplotypes. After dichotomization, there are effectively two ‘alleles’ yielding 15 distinct triad configurations. A log-linear modeling approach (Weinberg et al., 1998; Wilcox et al., 1998) can then be applied directly.
Nonetheless, in practice, the multi-SNP genotypes observed for any triad may be consistent with several triad diplotype configurations. Assuming HWE in the population, haplotype-phasing programs, such as Haplore (Zhang et al., 2005), assign each triad a list of possible diplotype configurations and estimate each configuration’s relative probability conditional on the observed triad genotypes (even when some may be missing). Although the estimated probabilities are valid for the control population under HWE, they are biased for case families when the relative risks for particular haplotypes differ from 1. Nevertheless, an appropriate likelihood for the case families can be maximized via the Expectation-Maximization (EM) algorithm (Dempster et al., 1977), using the probabilities estimated by Haplore as starting values. The EM algorithm iteratively estimates both the relative risk parameters and the probabilities for each possible diplotype configuration for each case-parent triad, eventually converging to maximum likelihood parameter estimates under the assumed risk model.
For single-SNP data, log-linear models typically include separate stratum parameters for each mating type, thereby conditioning on parental genotypes and protecting inference from bias due to population stratification. This approach can be extended to multiple-linked-SNP data as follows. Let h1, h2, …, hk denote the k possible haplotypes, with corresponding population frequencies p1, p2, …, pk, where , and where subscript i=1 designates the candidate haplotype. For now, we assume that haplotype phase can be unambiguously determined. Adopting convenient shorthand notation that represents both parental diplotypes and also transmissions (Gjessing & Lie, 2006), we let the vector (hi, hj, hr, hs) denote a transmission tetratype, where hi, hj are the two haplotypes of the mother, hr, hs are those of the father, and the second (hj) and fourth (hs) haplotypes are those that were transmitted to the offspring. A generic model for multi-locus SNP genotypes may be represented by:
| (1) |
where D represents the event that the child is affected. We consider three multiplicative and increasingly complex ways to model Pr[(hi,hj,hr,hs)], i.e., the distribution of transmission tetratypes in the population, namely: assume HWE, assume random mating, or use a fully general specification. We also consider several multiplicative specifications of the risk function , as described later. Because we specify both terms in the numerator of (1) multiplicatively and the denominator is a normalizing constant, the probability model specified in (1) describes a log-linear model for the expected counts of transmission tetratypes.
A likelihood corresponding to (1) can be maximized directly if all haplotypes are known unambiguously, but phase ambiguity is unavoidable in practice. To calculate the likelihood contribution for a triad genotype that is consistent with multiple transmission tetratypes, one sums over all compatible configurations. Denote the set of transmission tetratypes consistent with triad genotype (gM,gF,gC) as Γ(gM,gF,gC), where the subscripts M, F and C mark the genotype for the mother, father and child, respectively. The triad’s contribution to the likelihood is:
| (2) |
The observed-data likelihood to be maximized is the product of all the triad-specific contributions of the form Pr[gM, gF, gC | D]. When some or all SNP genotypes are missing for individuals in a triad, the likelihood in (2) must include an additional sum over all the sets Γ(gM,gF,gC) that are consistent with the observed pattern of missing genotypes for that triad.
Model under HWE
The number of transmission tetratypes quickly becomes intractable as the number of haplotypes increases. Consequently, to reduce the number of parameters in the model, we initially assume, as do most haplotype-phasing programs, that haplotypes are in HWE in the source population. Let h1 be the candidate haplotype with frequency p1 and let R1 and R2 be the relative risks for offspring heterozygous and homozygous for h1 respectively, relative to an offspring with no copies of that haplotype. Under the assumption of population HWE, and modeling offspring genetic effects only, the probability of a particular transmission tetratype for a case-parent triad, conditional on disease in the offspring, is:
| (3) |
Here I1 = 1 if (j=1 and s≠1) or (j≠1 and s=1) and I1 = 0 otherwise; I2 =1 if (j=1 and s=1) and I2 = 0 otherwise. In accord with (2), if all transmission tetratypes are unambiguously known, (3) represents the multiplicative contribution of a single family to the likelihood function. If the pathogenic mechanism is one where the maternal diplotype acts through the mother during gestation, the above formula can be modified so that the arguments of the indicator functions instead flag the maternal diplotype. Maternal relative risk parameters are denoted S1 and S2 and are interpreted in relation to the number of copies of h1 in the mother. One can allow for both maternal and offspring relative risks by extending (3) in the obvious way to include four risk parameters, or more if the maternal and offspring genomes interact. When the frequency of the risk-relevant haplotype is low or the sample size is small, homozygotes may be too rare to estimate R2 and/or S2 reliably. Then one may want to impose a simplified, log-additive risk model, e.g., setting . With this model, only one relative risk parameter, R1 or S1, is estimated, for the offspring or maternal effects, respectively. For testing, this approach remains valid even if the true alternative violates log-additivity, because validity only requires that the model hold under the null hypothesis, which it does. One can use the EM algorithm for data with ambiguous transmission tetratypes, described in Supplement A.
Model under random mating
One can relax the HWE assumption by allowing different parameters for each diplotype frequency to model the transmission tetratype distribution under the assumption of random mating, where the probability of a mating type is the product of parental diplotype frequencies. Let Hij denote the diplotype consisting of haplotypes hi and hj and {Hij ,Hrs}denote an unordered pair of parental diplotypes. When there is no maternal effect, the probability of transmission tetratype (hi, hj, hr, hs) can be written as follows:
| (4) |
This model involves k(k+1)/2 diplotype frequency parameters, more than required for the model under HWE but many fewer than required for the general model described below. The increase in the number of parameters confers added flexibility in modeling parental diplotypes compared to the model based on HWE but the model under random mating is still susceptible to bias from population stratification. The conditional probability in the numerator is calculated from Mendelian assumptions. EM steps analogous to those used under HWE will maximize the likelihood based on (4) (Supplement B).
Fully general model
With adequate sample size, one can consider relaxing the random mating assumption, while still assuming genetic mating symmetry, by including separate mating type parameters for each possible unordered pair of parental diplotypes {Hij ,Hrs}.
| (5) |
This model confers robustness to bias from population stratification but at a cost; the number of parental-diplotype parameters skyrockets as the number of haplotypes increases. One can maximize the appropriate likelihood via EM (Supplement C).
Extension to two candidate haplotypes
We next consider scenarios where two candidate haplotypes are potentially associated with the disease. Let h1, h2 be two candidate haplotypes. Let T1, T2 be the relative risks for offspring heterozygous and homozygous for h2 respectively, relative to offspring with no copies of either h1 or h2, and let R1, R2 also be defined as the relative risks for offspring heterozygous and homozygous for h1, respectively, relative to offspring with no copies of either. Under HWE, the likelihood contribution for transmission tetratype (hi, hj, hr, hs) in a case triad is:
| (6) |
Here I1 = 1 if (j=1 and s≠1) or (j≠1 and s=1) and I1 = 0 otherwise; I2 = 1 if (j=1 and s=1) and I2 = 0 otherwise; I3 = 1 if (j=2 and s≠2) or (j≠2 and s=2) and I3 = 0 otherwise; I4 = 1 if (j=2 and s=2) and I4 = 0 otherwise . Here we modeled the relative risk for offspring with diplotype h1 h2 as the product R1T1; alternatively, one could introduce a distinct risk parameter for that diplotype. The EM steps can be performed as described above, except that two haplotype frequencies and four relative risk parameters are involved, the multinomial has 78 rather than 15 cells, and Supplement A step 4a uses (6) instead of (3). Of course, the distribution of transmission tetratypes in (6) could also be modeled under the random mating assumption or the fully general parameterization with concomitant increased demands on sample size.
Some programs, such as TRIMM (Shi et al., 2007), nominate a candidate haplotype (i.e., a set of risk-tagging SNPs) based on the alleles associated with increased risk for disease. When both members of a Yin-Yang haplotype pair exist in the population, any identified susceptibility associated with one haplotype could alternatively be due to a protective effect of the other, its complement. Model 6 can be used to accommodate such scenarios and identify which is the risk-relevant haplotype by fitting both the Yin and the Yang haplotypes as the two candidates.
Simulations
General methods
We used an approach previously described (Shi et al., 2007) to generate simulated triad data sets under scenarios of offspring or maternal genetic effects. If assuming HWE, we randomly sampled haplotypes for the two parents from the population of haplotypes. If assuming random mating, we randomly sampled diplotypes for the two parents from the population of diplotypes. We then created a random child from the parents on the basis of Mendel’s law, assuming no recombination, and assigned a relative risk of disease to the child based on either the number of inherited copies of the risk haplotype (offspring genetic effects scenario) or the number of maternal copies of the risk haplotype (maternal genetic effects scenario). For simulation efficiency, and without loss of generality, we used the inverse of the larger relative risk as the baseline disease rate. We calculated the risk of the disease and assigned disease status to the child at random based on that risk. Only families with an affected child were retained. If assuming the presence of population structure, we modified this procedure to sample randomly from a mixture of two distinct subpopulations, each in HWE but with different baseline disease risks and risk haplotype frequencies.
We generated 5000 simulated data sets under each of various scenarios. We generated the initial lists of possible transmission tetratypes for each simulated dataset using Haplore. Starting from the Haplore output, we employed the EM algorithm to fit our proposed log-linear models by maximum likelihood. We used a chi-squared likelihood ratio test (LRT) to test for the offspring genetic effects under a log-additive risk model with full adjustment for maternal effects or the maternal genetic effects under a log-additive risk model with full adjustment for offspring effects. The candidate haplotype for our analyses was always the risk haplotype from the simulated scenario.
We evaluated the coverage rate of the profile-based 95% confidence region (CR) for the relative risks (Venzon & Moolgavkar, 1988) without delineating the entire confidence region. For each simulated data set, we first maximized the observed-data log-likelihood over the relative risk(s) and the other parameters (haplotype-frequency, or diplotype-frequency, or mating-type parameters); then, fixing the relative risk(s) at their true value(s), we maximized the log-likelihood over the remaining parameters. The corresponding 95% confidence region contains the true parameter if and only if twice the difference between these maximized log-likelihoods is less than the 95% percentile of a chi-square distribution with degrees of freedom corresponding to the number of risk parameters estimated. The actual confidence limits that we do report are based on the empirical standard errors estimated using the relative risk estimates from 5000 independent simulations.
Simulations evaluating the performance of our approach
To assess the impact of haplotype frequency and sample size, we obtained haplotypes and their frequencies for a 100-kb region around the replication factor C gene 1 (RFC1) from HapMap-phased genotype data on the sample with European ancestry. This haplotype set is defined by 12 SNPs, which specify 17 different haplotypes (Supplement D). We assigned haplotype 1 log-additive relative risks of , 2 for R1 and R2 respectively. The RFC1 haplotypes with their frequencies formed a convenient and realistic population from which we sampled to perform simulations. We simulated triad datasets over a range of risk haplotype frequencies (p1 = 0.02, 0.05, 0.142, 0.25, or 0.4) and at two sample sizes (400 and 1000 families). For each selected value of p1 , the HapMap frequencies for the non-risk haplotypes were renormalized so that they summed to1- p1 . Because missing data often complicate genetic association studies, we simulated two missing-data scenarios: one where 20% of SNP genotypes are missing randomly, and one where both a random 20% of triads are missing the father and also 20% of the SNP genotypes are missing randomly.
To evaluate the impact of departure from HWE on models that do or do not enforce HWE, we simulated data under Hardy-Weinberg Disequilibrium (HWD). We adopted the definition of the disequilibrium coefficient Dij from Schaid (2004) so that:
as it follows that . We simulated genotype data so that only diplotypes with at least one copy of the risk haplotype exhibit HWD; the remaining diplotypes are in HWE. With haplotype 1 as the risk haplotype, we simulated genotypes where was set to 0.1 and -0.1, respectively, with D1j being proportional to the frequency of the non-risk haplotype j.
Simulations comparing our proposal to competitors
We also carried out simulation studies to compare the performance of our approach and two other published methods: Pseudocontrol, which implements the case/pseudocontrol analysis by Cordell (2004) and Unphased, which implements the likelihood-based association analysis by Dudbridge (2008). Because these two published methods can handle relatively few SNPs, we simulated data sets consisting of three SNPs and four haplotypes. Each data set has 400 case triads. We designated one haplotype as the candidate haplotype with a frequency of 0.4 and kept the frequency of the remaining three haplotypes equal to each other. We simulated scenarios where the parental genotypes are in HWD as defined above, D11= -0.1. To evaluate the methods’ robustness to population stratification, we also simulated data from a population with two subpopulations each in HWE: one with a candidate haplotype frequency of 0.25 and the other 0.75; the ratio of baseline disease risks in the two populations was 3:1. For both kinds of parental populations, HWD and population stratification, we simulated under the null (R1= 1 and R2=1) and under an alternative ( and R2=2) scenario. We simulated scenarios either without missing genotypes or with 20% of genotypes randomly missing. Analyses were based on either a 1-df or 2-df test except for Unphased where the current version did not allow a 2-df test for a candidate haplotype. The program Unphased allows for two options to deal with ambiguous haplotypes and missing genotypes: ‘-certain’ and ‘-missing’. The former option restricts analysis to subjects with unambiguous haplotypes and the latter averages all possible completions of the data. The default setting (with neither input option) includes all individuals with no missing genotypes. We evaluated Unphased under all three settings.
Simulations involving two candidate risk haplotypes
We examined the performance of (6) in simulated scenarios where two non-complementary haplotypes confer risk. The simulations were based on the RFC1 gene described previously. In our simulations, the relative risks for the two haplotypes were R1= 1.2, , and T1= 1.35, , and their respective haplotype frequencies were (0.141, 0.141), (0.021, 0.07) or (0.071, 0.21). We analyzed the data with a log-linear model that enforced HWE, assumed and , and allowed for separate parameters for the two risk haplotypes. As a comparison we also analyzed the same data with a log-linear model that includes parameters only for one of the two risk haplotypes while improperly aggregating the other risk-conferring haplotype with the non-risk haplotypes.
We also examined the performance of (6) in simulated scenarios where the two candidate haplotypes were a Yin-Yang pair. We designated one of the haplotypes of RFC1 as the Yin haplotype and added the complementary Yang haplotype to the pool of RFC1 haplotypes. In our simulations, only one haplotype in the pair was associated with the disease, conferring either a protective () or a causative () effect. The haplotype frequencies for the Yin-Yang pair were either (0.071, 0.21) or (0.141, 0.141) with the first number denoting the frequency of the risk-relevant haplotype. We analyzed the data with a log-linear model that enforced HWE, assumed and for the Yin and the Yang haplotypes, respectively. Whenever a simulation had a likelihood ratio test with p<0.05, we nominated that haplotype from the Yin-Yang pair whose log relative risk most deviated from 0 as the actual risk-relevant haplotype. We studied the behavior of this decision rule by estimating the proportion of simulated studies that nominated the correct haplotype.
RESULTS
Simulations evaluating the performance of our approach
With no data missing, Haplore assigned a unique transmission tetratype to most families (Table 1). With 20% of the SNP genotypes missing sporadically and randomly, Haplore found more than one possible transmission tetratype (i.e., phase ambiguity) for about half of the families, and the average total number of transmission tetratypes per simulated dataset was more than twice the number of families. With 20% of the triads missing the father and also 20% of the genotypes missing randomly, 60% of triads had more than one possible transmission tetratype. The total number of possible transmission tetratypes exceeded ten times the number of families.
Table I.
Performance of the log-linear model that enforced both the random mating assumption and R2=R12 for estimating offspring genetic effects. Genotypes were simulated mimicking RFC1 haplotypes under HWE and R2=R12 with 5,000 simulations per scenario
| Number of triads |
Risk haplotype frequency |
Pattern of missingness1 |
Mean total number of TT |
Mean number of triads with multiple TT |
Empirical coverage of nominal 95% CI |
Geometric mean of estimated R1 (95% CI)2 True |
|---|---|---|---|---|---|---|
| 400 | 0.05 | None | 408.5 | 8.5 | 0.950 | 1.43(1.42,1.43) |
| 20% g | 901.4 | 199.5 | 0.945 | 1.43(1.42,1.44) | ||
| 20% g & 20% f | 4104.1 | 241.2 | 0.946 | 1.43(1.42,1.44) | ||
| 0.142 | None | 411.1 | 11.0 | 0.950 | 1.42(1.41,1.42) | |
| 20% g | 905.9 | 200.8 | 0.951 | 1.42(1.41,1.42) | ||
| 20% g & 20% f | 4141.3 | 242.2 | 0.951 | 1.42(1.41,1.43) | ||
| 0.25 | None | 412.8 | 12.7 | 0.948 | 1.42(1.41,1.42) | |
| 20% g | 904.6 | 200.8 | 0.947 | 1.42(1.41,1.42) | ||
| 20% g & 20% f | 4196.8 | 241.9 | 0.948 | 1.42(1.41,1.42) | ||
| 0.4 | None | 412.4 | 12.2 | 0.949 | 1.41(1.41,1.42) | |
| 20% g | 885.6 | 197.1 | 0.948 | 1.41(1.41,1.42) | ||
| 20% g & 20% f | 4212.6 | 238.4 | 0.947 | 1.41(1.41,1.42) | ||
| 1000 | 0.02 | None | 1018.6 | 18.5 | 0.942 | 1.43(1.42,1.43) |
| 20% g | 2226.7 | 491.9 | 0.945 | 1.43(1.42,1.44) | ||
| 20% g & 20% f | 10057.1 | 597.0 | 0.942 | 1.44(1.42,1.45) | ||
| 0.05 | None | 1021.3 | 21.2 | 0.944 | 1.42(1.41,1.43) | |
| 20% g | 2228.5 | 493.1 | 0.948 | 1.42(1.41,1.43) | ||
| 20% g & 20% f | 10074.4 | 597.8 | 0.948 | 1.42(1.42,1.43) | ||
| 0.142 | None | 1027.9 | 27.7 | 0.949 | 1.42(1.41,1.42) | |
| 20% g | 2240.0 | 497.4 | 0.945 | 1.42(1.41,1.42) | ||
| 20% g & 20% f | 10198.4 | 601.0 | 0.945 | 1.42(1.41,1.42) | ||
| 0.25 | None | 1032.0 | 31.7 | 0.952 | 1.41(1.41,1.42) | |
| 20% g | 2236.9 | 497.8 | 0.954 | 1.42(1.41,1.42) | ||
| 20% g & 20% f | 10333.8 | 601.2 | 0.953 | 1.42(1.41,1.42) | ||
| 0.4 | None | 1031.0 | 30.6 | 0.954 | 1.41(1.41,1.42) | |
| 20% g | 2193.7 | 489.2 | 0.957 | 1.41(1.41,1.42) | ||
| 20% g & 20% f | 10357.3 | 593.2 | 0.958 | 1.41(1.41,1.42) | ||
Abbreviations: TT, transmission tetratype; CI, confidence interval
None: no missing data; 20% g: 20% of SNP genotypes are missing randomly; 20%g & 20% f: 20% of SNP genotypes are missing randomly and 20% of triads have missing father.
The 95% CI is based on the empirical standard error calculated using the estimates from 5000 independent simulations.
The model enforcing HWE and the one enforcing the random mating assumption produced comparable results. We therefore present only results from the model enforcing random mating. The LRT showed an empirical type I error rate that was statistically consistent with the nominal 0.05 level (results not shown). Its power increased as the risk haplotype frequency increased from 0.02 to 0.4 (Fig. 1). Power improved as sample size increased, and missing data had little impact on power (Fig. 1), whether SNPs were missing sporadically or many fathers were also missing. The log-linear model accurately estimated haplotype or diplotype frequencies under all simulated scenarios (data not shown). While relative risk estimates were biased slightly upwards when the candidate haplotype was rare (or with N=200 [data not shown]), they were unbiased when it was relatively common and/or the sample size was larger (Table 1). Empirical coverage for confidence regions was consistent with the nominal 95% level.
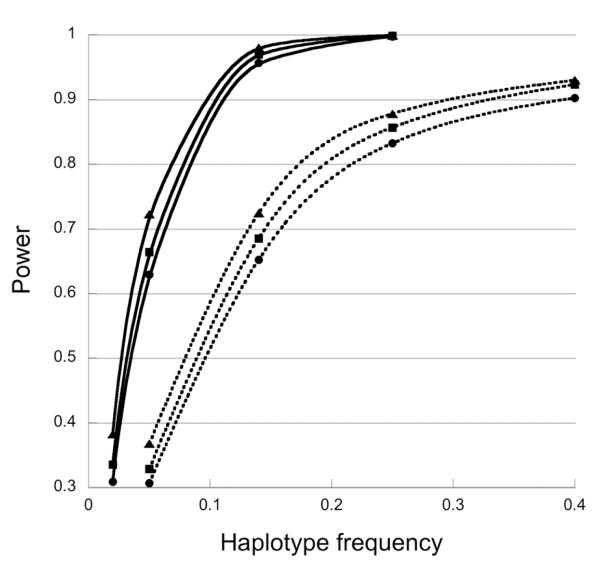
Figure 1.
Power of the one-degree-of-freedom likelihood ratio tests for offspring genetic effects as a function of the risk haplotype frequency. The one-degree-of-freedom likelihood ratio test is based on a log-linear model enforcing random mating and modeling risk with R2= R12 but distinct S1 and S2. Data were simulated under HWE and with , R2=2, S1= S2=1. Solid and dotted lines represent simulations with 400 and 1000 triads per dataset, respectively. Symbols: ▲, no missing data; ■, 20% of SNP genotypes are missing randomly; ●, both 20% of SNP genotypes are missing randomly and 20% of triads are missing the father.
We also performed simulations to assess maternally-mediated genetic effects using scenarios similar to those for offspring genetic effects, but with relative risks depending on the mother’s genotype, not the child’s. Power for maternal effects was almost identical to that for offspring effects (Fig. 2) in the scenarios that we simulated. This equivalence was expected because, generated under HWE, the simulated data naturally follow parental haplotype exchangeability, and testing for maternal effects under a given relative risk alternative should have the same power as testing for offspring effects (Shi et al., 2007).
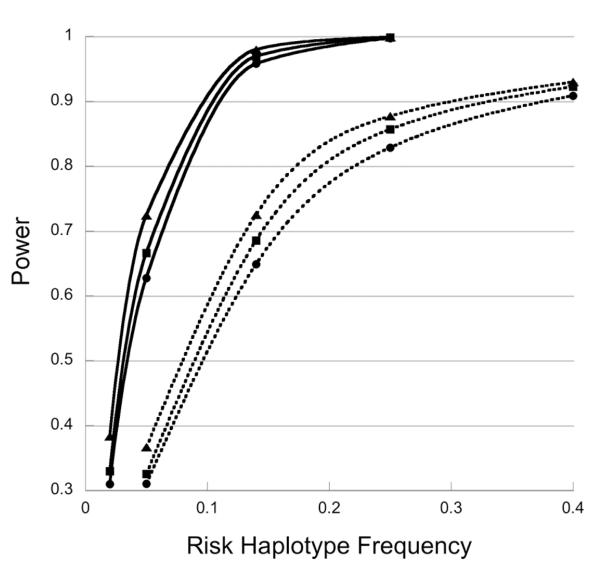
Figure 2.
Power of the one-degree-of-freedom likelihood ratio tests for maternal genetic effects as a function of the risk haplotype frequency. The one-degree-of-freedom likelihood ratio test is based on a log-linear model enforcing random mating and modeling risk with S2= S12 but distinct R1 and R2. Data were simulated under HWE and with , S2=2, R1= R2=1. Solid and dotted lines represent simulations with 400 and 1000 triads per dataset, respectively. Symbols: ▲, no missing data; ■, 20% of SNP genotypes are missing randomly; ●, both 20% of SNP genotypes are missing randomly and 20% of triads are missing the father.
When the simulated data departed from HWE, the model assuming HWE performed badly, as expected. Type I error rates deviated from the nominal 0.05 level and the estimates of the relative risks and genotype frequencies were biased. Confidence-region coverage also deviated markedly from the nominal level. In contrast, the model assuming random mating was robust to deviation from HWE; consistent type I error rate, 95% confidence region coverage and unbiased estimates for the diplotype frequencies and relative risks (Table 2), probably reflecting the fact that data simulated under HWD still honored random mating.
Table II.
Performance of the log-linear model that enforced both the random mating assumption and R2=R12 for estimating offspring genetic effects (N =400). Genotypes were simulated mimicking RFC1 haplotypes under Hardy-Weinberg Disequilibrium and R2=R12 with 5,000 simulations per scenario
| Log-linear model assumption |
HWD coefficient (D11) |
Simulated R1 |
Pattern of missingness1 |
Mean total number of transmission tetratypes |
Mean number of triads with multiple transmission tetratypes |
Power (1df) |
Empirical coverage of nominal 95% confidence interval |
Frequency of carriers of the risk haplotype2 |
Geometric mean of estimated R1 (95% CI)3 |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HWE | -0.1 | 1 | None | 419.4 | 19.1 | 0.071 | 0.929 | 0.36 | 0.48 | 0.16 | 1.00(0.99,1.00) |
| 20% g | 917.4 | 205.0 | 0.073 | 0.927 | 0.36 | 0.48 | 0.16 | 1.00 (1.00,1.00) | |||
| -0.1 | 1.414 | None | 419.3 | 19.0 | 0.988 | 0.876 | 0.39 | 0.47 | 0.14 | 1.51(1.5,1.51) | |
| 20% g | 921.1 | 205.0 | 0.983 | 0.873 | 0.39 | 0.47 | 0.14 | 1.51(1.5,1.51) | |||
| 0.1 | 1 | None | 407.7 | 7.6 | 0.021 | 0.979 | 0.36 | 0.48 | 0.16 | 1.00 (1.00,1.01) | |
| 20% g | 863.1 | 191.9 | 0.021 | 0.979 | 0.36 | 0.48 | 0.16 | 1.00 (1.00,1.00) | |||
| 0.1 | 1.414 | None | 407.1 | 7.0 | 0.618 | 0.916 | 0.33 | 0.49 | 0.18 | 1.29(1.29,1.29) | |
| 20% g | 854.0 | 189.7 | 0.570 | 0.912 | 0.32 | 0.49 | 0.19 | 1.28(1.28,1.29) | |||
| Random Mating |
-0.1 | 1 | None | same as above | 0.049 | 0.951 | 0.26 | 0.68 | 0.06 | 1.00 (1.00,1.00) | |
| 20% g | 0.053 | 0.947 | 0.26 | 0.68 | 0.06 | 1.00 (1.00,1.00) | |||||
| -0.1 | 1.414 | None | 0.982 | 0.946 | 0.26 | 0.68 | 0.06 | 1.41(1.41,1.42) | |||
| 20% g | 0.977 | 0.949 | 0.26 | 0.68 | 0.06 | 1.41(1.41,1.42) | |||||
| 0.1 | 1 | None | 0.050 | 0.950 | 0.46 | 0.28 | 0.26 | 1.00 (1.00,1.01) | |||
| 20% g | 0.048 | 0.952 | 0.46 | 0.28 | 0.26 | 1.00 (1.00,1.01) | |||||
| 0.1 | 1.414 | None | 0.744 | 0.952 | 0.46 | 0.28 | 0.26 | 1.42(1.41,1.42) | |||
| 20% g | 0.702 | 0.953 | 0.46 | 0.28 | 0.26 | 1.42(1.41,1.42) | |||||
None: no missing data; 20% g: 20% of SNP genotypes are missing randomly.
Expected frequencies of diplotypes with 0, 1, 2 copies of the risk haplotype are 0.26, 0.68, and 0.06, respectively, when D11 =-0.1 and 0.46, 0.28, and 0.26, respectively, when D11 = 0.1.
The 95% CI is based on the empirical standard error calculated using the estimates from 5000 independent simulation.
The fully general model that relaxes the random mating assumption to accommodate population stratification failed to produce unbiased results at the sample sizes used in our RFC1 simulations, presumably due to the large number of mating type parameters (>10,000) involved (data not shown). When we simulated scenarios using four or fewer haplotypes, the general model performed well. For a scenario with six haplotypes, the relative risk estimates were slightly biased, but increasing the sample size from 1000 triads to 2500 eliminated the bias (data not shown), suggesting that the bias was due to the limited sample size relative to the number of mating type parameters.
Simulations comparing our approach to competitors
Consider first the scenario where a population exhibits HWD, a setting where procedures enforcing HWE should perform poorly. Under the null hypothesis (R1=1), among the methods compared, only the log-modeling approach enforcing HWE demonstrated empirical type-I error rates that were inflated compared to the nominal 0.05 (Table 3). It was also the only method that exhibited bias in the point and interval estimates (Table 3). All the other procedures studied performed well under the null. Under the alternative hypothesis (R1=1.414) and when no data were missing, the power of all the valid 2-df tests was the same; the power of the 1-df tests was also the same. When 20% of genotypes were missing at random, however, the power of the valid 2-df versions of our approach was markedly superior to the power of the 2-df Pseudocontrol test; the power of the 1-df fully general version of our approach was similar to the power of Unphased 1-df tests with the “-missing” option but both were markedly superior to the power of Unphased 1-df tests with the “-certain” option or no options (Table 3). Whether data were missing or not, estimates of R1 showed slight upward bias (Table 3) for most procedures; this bias disappeared, however, with a sample size of 1000 triads (data not shown). Any bias was most pronounced with 20% missing data for the Pseudocontrol procedure.
Table III.
Comparisons of several competing modeling options. Genotypes were generated with four haplotypes under HWD or under population stratification (N=400) with 1000 simulations per scenario. Details of the scenarios are described in the Methods.
| Missingness | No missing genotypes | 20% missing genotypes | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Scenarios | Relative Risk (R1) |
df | Method | Power | Geometric mean and 95% CI | Power | Geometric mean and 95% CI | ||
| R1 | R2 | R1 | R2 | ||||||
| One population HWD D= -0.1 |
1 | 2 | Log-linear (HWE)1 | 0.132 | 0.94(0.93,0.95) | 1.02(1.00,1.04) | 0.129 | 0.95(0.94,0.96) | 1.03(1.01,1.05) |
| 2 | Log-linear (RM)2 | 0.061 | 1.00(0.99,1.01) | 1.00(0.99,1.01) | 0.056 | 1.00(0.99,1.01) | 1.00(0.98,1.01) | ||
| 2 | Log-linear (full)3 | 0.055 | 1.00(0.99,1.01) | 1.00(0.99,1.01) | 0.044 | 1.00(0.99,1.01) | 1.00(0.99,1.01) | ||
| 2 | Pseudocontrol | 0.052 | 1.00(0.99,1.01) | 1.00(0.99,1.01 ) | 0.051 | 1.03(1.00,1.06) | 1.00(0.97,1.04) | ||
| 1 | Log-linear (full) | 0.054 | 1.00(0.99,1.01) | na | 0.049 | 1.00(0.99,1.01) | na | ||
| 1 | Unphased | 0.05 | 1.00(0.99,1.01) | na | 0.056 | 1.01(0.99,1.03) | na | ||
| 1 | Unphased (certain)4 | 0.054 | 1.00(0.99,1.01) | na | 0.061 | 1.01(0.99,1.03) | na | ||
| 1 | Unphased (missing)5 | na6 | na | na | 0.050 | 1.00(1.00,1.01) | na | ||
| One population HWD D= -0.1 |
1.414 | 2 | Log-linear (HWE) | 0.983 | 1.57(1.55,1.58) | 2.77(2.73,2.82) | 0.958 | 1.63(1.61,1.65) | 2.96(2.91,3.02) |
| 2 | Log-linear (RM) | 0.944 | 1.42(1.41,1.43) | 1.99(1.97,2.01) | 0.912 | 1.42(1.41,1.43) | 1.99(1.96,2.01) | ||
| 2 | Log-linear (full) | 0.945 | 1.42(1.41,1.43) | 1.99(1.97,2.01) | 0.916 | 1.42(1.41,1.44) | 2.00(1.97,2.02) | ||
| 2 | Pseudocontrol | 0.947 | 1.42(1.41,1.44) | 1.99(1.97,2.01) | 0.237 | 1.48(1.44,1.53) | 2.03(1.97,2.11) | ||
| 1 | Log-linear (full) | 0.973 | 1.41(1.40,1.42) | na | 0.953 | 1.41(1.40,1.42) | na | ||
| 1 | Unphased | 0.974 | 1.41(1.40,1.42) | na | 0.336 | 1.43(1.41,1.46) | na | ||
| 1 | Unphased (certain) | 0.976 | 1.43(1.42,1.44) | na | 0.303 | 1.43(1.40,1.45) | na | ||
| 1 | Unphased (missing) | na | na | na | 0.956 | 1.46(1.45,1.47) | na | ||
| Population stratification |
1 | 2 | Log-linear (HWE) | 0.934 | 0.59(0.59,0.6) | 0.82(0.81,0.83) | 0.924 | 0.58(0.57,0.58) | 0.81(0.80,0.82) |
| 2 | Log-linear (RM) | 0.942 | 0.58(0.58,0.59) | 0.81(0.80,0.83) | 0.933 | 0.57(0.56,0.57) | 0.80(0.79,0.82) | ||
| 2 | Log-linear (full) | 0.041 | 1.01(1.00,1.02) | 1.01(0.99,1.02) | 0.045 | 1.01(0.99,1.03) | 1.01(0.99,1.03) | ||
| 2 | Pseudocontrol | 0.052 | 1.01(0.99,1.02) | 1.01(0.99,1.02) | 0.059 | 1.27(1.11,1.46) | 1.30(1.13,1.50) | ||
| 1 | Log-linear (full) | 0.044 | 1.00(0.99,1.01) | na | 0.041 | 1.00(0.99,1.01) | na | ||
| 1 | Unphased | 0.045 | 1.00(0.99,1.01) | na | 0.048 | 1.02(1.00,1.04) | na | ||
| 1 | Unphased (certain) | 0.039 | 1.00(0.99,1.01) | na | 0.054 | 1.02(1.00,1.04) | na | ||
| 1 | Unphased (missing) | na | na | na | 0.046 | 1.00(0.99,1.01) | na | ||
| Population stratification |
1,414 | 2 | Log-linear (HWE) | 0.973 | 0.76(0.75,0.77) | 1.29(1.28,1.31) | 0.969 | 0.73(0.72,0.74) | 1.26(1.24,1.27) |
| 2 | Log-linear (RM) | 0.983 | 0.78(0.77,0.79) | 1.44(1.41,1.46) | 0.981 | 0.75(0.74,0.76) | 1.41(1.39,1.43) | ||
| 2 | Log-linear (full) | 0.732 | 1.45(1.42,1.47) | 2.04(2.00,2.08) | 0.643 | 1.46(1.43,1.49) | 2.06(2.02,2.10) | ||
| 2 | Pseudocontrol | 0.719 | 1.45(1.42,1.47) | 2.04(2.00,2.08) | 0.163 | 3.38(2.60,4.40) | 4.86(3.74,6.32) | ||
| 1 | Log-linear (full) | 0.813 | 1.42(1.40,1.43) | na | 0.758 | 1.42(1.41,1.43) | na | ||
| 1 | Unphased | 0.810 | 1.42(1.41,1.43) | na | 0.184 | 1.43(1.40,1.46) | na | ||
| 1 | Unphased (certain) | 0.814 | 1.41(1.40,1.42) | na | 0.191 | 1.45(1.42,1.48) | na | ||
| 1 | Unphased (missing) | na | na | na | 0.751 | 1.40(1.39,1.41) | na | ||
Log-linear (HWE): log-linear model enforcing HWE
Log-linear (RM): log-linear model enforcing random mating
Log-linear (full): fully general log-linear model
Unphased (certain): Unphased with “-certain” option
Unphased (missing): Unphased with “-missing” option
Unphased with “-missing” option has the same performance as Unphased (with no input options) when there are no missing genotypes
In the population stratification scenario, procedures that enforce HWE or our random mating assumption should perform poorly. Under the null hypothesis, our procedures enforcing either of these two assumptions exhibited inflated Type-I error rates and substantial bias (Table 3). All the other procedures performed well under the null whether data were missing or not. Under the alternative hypothesis, with no missing data, the power of the valid 2-df tests was similar; the power of the 1-df tests was also similar. When 20% of genotypes were missing at random, however, the power of the fully general 2-df version of our approach again was markedly superior to the power of the 2-df Pseudocontrol test. The 1-df version of our fully general test had similar performance to Unphased with ‘-missing’ option. With no missing data, estimates of R1 showed no bias under the null and slight upward bias under the alternative for the valid 2-df tests (Table 3). With 20% missing data, estimates of R1 showed slight upward bias under both the null and the alternative, except for the Pseudocontrol procedure where the bias was substantial (Table 3). Again, for all the valid procedures, bias declined with larger sample sizes (data not shown).
Simulations involving two candidate risk haplotypes
Simulations showed that our method performed well when there are two non-complementary risk-conferring haplotypes. The two-candidate-haplotype model with different risk parameters for each haplotype produced unbiased relative risk estimates for both haplotypes with either sample size. Power for a two-degree-of-freedom test where the model included both candidate haplotypes was generally higher than or close to that for a one-degree-of-freedom test where the model included risk parameters for only one of the candidate haplotypes (Table 4).
Table IV.
Performance of the two-candidate-haplotype log-linear model enforcing HWE. Genotypes were simulated mimicking RFC1 haplotypes under HWE with two haplotypes conferring risk: haplotype 1, R1 =1.2 and R2=R12; haplotype 2, T1= 1.35 and T2=T12. Each scenario reflects 5000 simulations.
| N | Pattern of Missing data 1 |
Simulated haplotype 1 frequency |
Simulated haplotype 2 frequency |
Power of test including only one haplotype as candidate2 |
Power of test including both haplotypes as candidates |
Empirical coverage of nominal 95% confidence region |
Geometric mean of estimated R1, T1 (both haplotypes included as candidate haplotypes in the model, 95% CI)3 |
||
|---|---|---|---|---|---|---|---|---|---|
| haplotype 1 | haplotype 2 | haplotype 1 | haplotype 2 | ||||||
| 400 | None | 0.141 | 0.141 | 0.152 | 0.500 | 0.537 | 0.956 | 1.20(1.20,1.21) | 1.35(1.35,1.36) |
| 20% g | 0.127 | 0.492 | 0.514 | 0.956 | 1.20(1.20,1.21) | 1.35(1.35,1.36) | |||
| None | 0.0707 | 0.212 | 0.093 | 0.671 | 0.632 | 0.953 | 1.21(1.20,1.21) | 1.35(1.35,1.35) | |
| 20% g | 0.082 | 0.663 | 0.620 | 0.951 | 1.21(1.20,1.21) | 1.35(1.35,1.35) | |||
| None | 0.212 | 0.0707 | 0.235 | 0.283 | 0.420 | 0.947 | 1.20(1.19,1.20) | 1.35(1.34,1.36) | |
| 20% g | 0.212 | 0.276 | 0.399 | 0.952 | 1.20(1.20,1.20) | 1.35(1.34,1.36) | |||
| 1000 | None | 0.141 | 0.141 | 0.292 | 0.868 | 0.915 | 0.948 | 1.20(1.20,1.20) | 1.35(1.35,1.35) |
| 20% g | 0.243 | 0.865 | 0.904 | 0.950 | 1.20(1.20,1.20) | 1.35(1.35,1.35) | |||
| None | 0.0707 | 0.212 | 0.138 | 0.965 | 0.960 | 0.950 | 1.20(1.20,1.21) | 1.35(1.35,1.35) | |
| 20% g | 0.105 | 0.962 | 0.955 | 0.949 | 1.20(1.20,1.21) | 1.35(1.35,1.35) | |||
| None | 0.212 | 0.0707 | 0.498 | 0.598 | 0.833 | 0.942 | 1.20(1.20,1.20) | 1.35(1.35,1.36) | |
| 20% g | 0.446 | 0.588 | 0.803 | 0.943 | 1.20(1.20,1.20) | 1.35(1.35,1.36) | |||
None: no missing; 20% g: 20% of SNP genotypes are missing randomly.
Only one risk haplotype is included as the candidate haplotype and the other is aggregated with the non-risk haplotypes.
The 95% CI is based on the empirical standard error calculated using the estimates from 5000 independent simulations.
For Yin-Yang haplotypes, although data were simulated so that only one of the pair determined risk, we fit a model with different risk parameters for each haplotype, assuming HWE and log-additivity in both relative risks. Our procedure that nominated the haplotype whose log relative risk deviated most from 0 as the one more likely to be risk-relevant, identified the correct haplotype in more than 95% of simulations with a significant LRT (p<0.05) when the sample size was 400 triads (Table 5) and in more than 99% of such simulations when the sample size was 1000 triads (data not shown). Thus we could separate Yin from Yang. As expected, the power of the two-degree-of-freedom LRT involving separate risk parameters for the Yin and the Yang haplotypes was lower than the power of the one-degree-of-freedom LRT where the risk haplotype was regarded as known a priori and was the only candidate in the model. Overall, scenarios with a causative effect had higher power than scenarios with a similar-sized (on the log scale) protective effect. Again, relative risk estimates were unbiased when the haplotype was relatively common, and confidence region coverage was consistent with the nominal 95% level (Table 5).
Table V.
Performance of the two-candidate-haplotype log-linear model enforcing HWE and R2=R12 and T2=T12 for Yin-Yang haplotypes. Genotypes were simulated mimicking RFC1 haplotypes under HWE with “Yin” as the risk-relevant haplotype, conferring either risk or protection. Each scenario reflects 5000 simulations with 400 triads in each simulation.
| Ratio of frequencies (Yin:Yang) |
Pattern of Missing data 1 |
Simulated R1 (Yin haplotype) |
Power (1df Yin) |
Power (2df Yin+Yang) |
Empirical coverage of nominal 95% confidence region (2 risk parameters) |
Geometric mean of Estimated R1 of risk haplotype (95% CI)2 |
Error Rate3 |
|
|---|---|---|---|---|---|---|---|---|
| Yin | Yang | |||||||
| 1:1 (0.141:0.141) |
None | 1.414 | 0.730 | 0.631 | 0.954 | 1.42(1.41,1.43) | 1.00(1.00,1.01) | 0.040 |
| 0.707 | 0.617 | 0.506 | 0.950 | 0.71(0.70,0.71) | 1.00(0.99,1.00) | 0.043 | ||
| 20% g | 1.414 | 0.686 | 0.585 | 0.956 | 1.42(1.42,1.43) | 1.00(1.00,1.01) | 0.039 | |
| 0.707 | 0.574 | 0.467 | 0.948 | 0.71(0.70,0.71) | 1.00(0.99,1.00) | 0.042 | ||
| 1:3 (0.071:0.212) |
None | 1.414 | 0.479 | 0.373 | 0.949 | 1.42(1.41,1.43) | 1.00(1.00,1.01) | 0.036 |
| 0.707 | 0.385 | 0.297 | 0.949 | 0.70(0.70,0.71) | 1.00(1.00,1.00) | 0.033 | ||
| 20% g | 1.414 | 0.430 | 0.337 | 0.950 | 1.42(1.41,1.43) | 1.00(1.00,1.01) | 0.039 | |
| 0.707 | 0.344 | 0.261 | 0.950 | 0.70(0.70,0.71) | 1.00(1.00,1.00) | 0.037 | ||
None: no missing; 20% g: 20% of SNP genotypes are missing randomly.
The 95% CI is based on the empirical standard error calculated using the estimates from 5000 independent simulations.
Error rate incurred by designating the haplotype whose log relative risk deviated most from 0 as the risk-relevant one.
Example
We applied our approach to data from an orofacial cleft study (Zucchero et al., 2004) using the 9-SNP risk haplotype identified by Zucchero et al. as the candidate haplotype. The data, from 296 Filipino case-parents triads, include genotypes of 36 SNPs in a 300-kb region around interferon regulatory factor 6 (IRF6). Based on Haplore, this dataset included 56 haplotypes; we therefore applied models enforcing HWE or random mating but not the fully general model. The model enforcing HWE and a log-additive risk structure produced an estimated risk haplotype frequency of 39.4%, a highly significant LRT (, p<0.00002), and an R1 estimate of 1.83 [95% CI = (1.38, 2.42)]. The model enforcing random mating produced similar results: estimated genotype frequencies were (0.44, 0.40, 0.16) for diplotypes with zero, one, and two copies of the risk haplotype, respectively; the LRT was highly significant (, p<0.00001); and the R1 estimate was 2.03 [95% CI = (1.48, 2.77)]. Thus a fetus inheriting one copy of the risk haplotype would experience a doubling of risk of orofacial cleft.
DISCUSSION
Statistical methods for estimating relative risks associated with haplotypes face two difficulties. The first is phase ambiguity, a difficulty that is exacerbated as the number of SNPs defining haplotypes increases and as the proportion of missing genotype data grows. The second difficulty is that the highly parameterized models needed to estimate relative risks for one or two copies of each haplotype offer little precision for estimation of those individual parameters. The more SNPs that are used to define haplotypes, the more haplotypes are possible, and the more parameters are needed to capture the full haplotype relative risk structure. Thus, methods for estimating haplotype relative risks are typically limited to haplotype groupings defined by a relatively small number of SNPs.
We have described a log-linear modeling approach for estimating relative risks associated with a candidate multi-SNP haplotype using case-parents data. The proposed method can detect and quantify either offspring or maternal genetic effects in genotype data consisting of multiple SNP markers. It achieves computational efficiency in dealing with phase ambiguity by using an available haplotype reconstruction program to generate an initial list of transmission tetratypes compatible with the observed triad genotypes. We used Haplore for this purpose because of its ability to handle family-based genotypes, computational efficiency, and good performance (Zhang & Zhao, 2006), although we expect the use of other similar software would achieve substantially the same results. Haplore estimates the number of haplotypes present, so that, with a large number of SNPs, our method works with a reduced set of haplotypes compared to methods that allow for every possible haplotype. Consequently, our method can handle more SNPs. Our approach effectively reduces the number of risk parameters in the model by comparing a candidate haplotype nominated based on prior knowledge to an aggregate of the remaining haplotypes. Our simulation studies indicated that, for data that meet its assumptions, this method is powerful and can provide unbiased estimates for both maternal and offspring relative risk parameters. Confidence-interval coverage was also consistent with the nominal level. We described models that required successively less restrictive assumptions and showed via simulations that the model assuming random mating appeared to include a rich enough parameterization to be robust to HWD. One can also extend the proposed models to allow multiple candidate haplotypes. Extended to two haplotypes under HWE, our approach was able to accurately select the risk-relevant member of a Yin-Yang haplotype pair. Software implementing the proposed methods is available from our website. Likelihood based methods such as ours can fail when the nuisance parameters become too numerous, so caution must be exercised in applying them with a large number of SNPs/haplotypes.
Our approach is related to one proposed by Carayol et al. (2006) in that both methods reduce the number of parameters in the risk model by recoding the full set of haplotypes into a smaller set of interest, such as a single candidate haplotype. Our method differs from theirs in several important ways, however. The SEM algorithm that Carayol et al. employ generates the initial probabilities of parental paired-diplotypes by a simple count of diplotypes in parents. As suggested by their simulations, the reliability of this calculation goes down as phase ambiguity increases with the number of SNPs or the number of missing genotypes. Our approach instead builds on previous research for dealing with these sources of phase ambiguity by using available phase construction software to generate an initial list of possible transmission tetratypes and their corresponding probabilities. Our EM implementation iteratively updates those initial estimates of transmission tetratype probabilities through updated risk estimates. One product of this approach is an unbiased estimate of the candidate haplotype’s/diplotypes’ prevalence in the source population in the absence of bias from population structure. Our proposed models allow researchers to enforce different assumptions based on the population under investigation. These two methods also differ in the way confidence intervals are estimated: Carayol et al. compute Wald-type intervals by taking advantage of the SEM algorithm’s repeated sampling process to estimate the variances of the estimated risk parameters whereas we use profile-likelihood-based intervals, which provided more consistent coverage in our simulations.
Despite some important differences in implementation between these two approaches, the risk models are similar. One might expect then that both would perform similarly when applied to data that met similar assumptions. A comparison of our simulation results to those of Carayol et al. (2006) as reported in their Tables IV and V suggests that differences in implementation have an impact and that our approach offers better performance. For samples of 1000 triads, Carayol et al. observed some discrepancies between average estimated relative risks and their true values as well as below nominal confidence-interval coverage, particularly in scenarios with greater phase ambiguity either from more haplotypes or from missing genotypes, although such discrepancies disappeared with larger sample sizes. By contrast, we saw little bias in relative risk estimates and essentially nominal coverage for sample sizes as low as 400 triads even in the face of substantial missing data. These comparisons, however, are based on different underlying haplotype populations between the two studies as well as on slightly different risk models. We also tested our method mimicking the same simulation scenarios reported in their Tables IV, V, and VIII and again our method performed better in terms of bias and coverage (data available from the authors’ website) for scenarios either with or without HWE.
Besides our proposed approach and that of Carayol et al. discussed earlier, few other methods exist for the estimation of haplotype relative risks from case-parents data with possibly missing genotypes. HAPLIN (Gjessing & Lie, 2006) employs a log-linear modeling approach to estimate offspring or maternal relative risk parameters for one or two copies of each haplotype. This software has a flexible parameterization of relative risks, tolerates phase ambiguity, and maximizes the log-linear likelihood via the EM algorithm under an assumption of HWE in the source population, an assumption that substantially reduces the number of nuisance mating-type parameters. HAPLIN is effective for haplotypes defined by a few SNPs but its current implementation does not function well when the number of SNPs exceeds four or five. The PCPH approach of Allen and Satten (Allen & Satten, 2007) employs a novel estimating-equation strategy that can estimate the full complement of offspring relative risk parameters while avoiding explicit assumptions (such as HWE) about the distribution of parental haplotypes. This approach requires working with large matrices that expand with the number of SNPs under consideration, a feature that again raises difficulties when handling relatively many SNPs. The case/pseudocontrol approach of Cordell et al. (2004) relies on generating pseudocontrols based on parental genotypes and retains only families where haplotype phase is resolvable. Unless the number of families is exceedingly large, the sacrifice of families with phase ambiguity will quickly become a major problem as the number of SNPs increases. This feature was starkly illustrated in our simulation study involving only three SNPs. A likelihood-based approach of Dudbridge (Dudbridge, 2008) as implemented in the software Unphased is also a competitor. It performed validly under population stratification in our simulations involving three SNPs and its power was similar to that our full model using 400 triads with 20% missing data. Unphased, however, is limited by the number of SNPs that it can handle. With the ever-increasing availability of multi-SNP data, the ability to handle a relatively large number of SNPs is becoming more important for association studies. Our approach appears to outperform other haplotype relative risk estimation methods when the analysis includes many SNPs.
Our approach’s ability to handle larger numbers of SNPs does not come without trade-offs. First, we focus attention on one or two candidate haplotypes and aggregate the remaining haplotypes into a non-risk referent grouping. This tactic makes sense when prior information has identified a strong candidate or two and supports little effect on risk by other haplotypes. Information needed to make such a judgment may become increasingly common as haplotype-based analyses become more widespread. If the wrong haplotype is chosen or if more distinct haplotypes confer risk, our models will be mis-specified. On the other hand, if knowledge of the risk-relevance of haplotypes is lacking, one could apply our method in a haplotype-by-haplotype manner. As shown by our simulations (Table 3), even if a risk-increasing haplotype is mistakenly included in the non-risk group, the power of one-candidate haplotype approach may not suffer much or may perform even better when the risk and frequency of the designated haplotype are relatively high. This one-candidate-haplotype-at-a-time approach requires, however, adjustment for multiple testing. Second, the fully general model provides robustness to bias from population stratification but it is limited by the number of haplotypes that can be handled, unless the sample is very large. On the other hand, bias from population structure may not be ubiquitous and severe (Wacholder et al., 2002) so that methods that assume HWE or random mating may be adequate in many populations to handle data involving a large number of haplotypes.
Our simulations have shown that randomly missing genotypes/fathers only had small impact on the power of the test and the accuracy of parameter estimation. We did not simulate scenarios where mothers are missing because, when testing either offspring or maternal effects under mating symmetry, the power should be the same whether mothers or fathers are missing. Thus our missing-father scenarios should appropriately reflect the more general missing-parent scenarios encountered in practice.
In conclusion, the proposed log-linear modeling approach can efficiently analyze multi-SNP genotype data when previous studies have identified one or two candidate risk haplotypes. The proposed method can provide unbiased relative risk estimates and can provide robustness to departure from HWE and to bias from population stratification.
Supplementary Material
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01-ES04007-12). We thank Drs. Jeffrey Murray and Theresa Zucchero for sharing the genotype data of their orofacial cleft study and Drs. Rolv Terje Lie and Dmitri Zaykin for helpful comments on the manuscript.
Footnotes
Electronic Resources Authors’ website: http://www.niehs.nih.gov/research/atniehs/labs/bb/staff/weinberg/index.cfm (for the Triad Multi-Marker relative risk Estimation (TRIMMEST) program) Program Haplore website: http://bioinformatics.med.yale.edu/group/software.html
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Wigginton JE. Handling marker-marker linkage disequilibrium: pedigree analysis with clustered markers. Am J Hum Genet. 2005;77:754–67. doi: 10.1086/497345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen AS, Satten GA. Inference on haplotype/disease association using parent-affected-child data: the projection conditional on parental haplotypes method. Genet Epidemiol. 2007;31:211–223. doi: 10.1002/gepi.20203. [DOI] [PubMed] [Google Scholar]
- Carayol J, Philippi A, Tores F. Estimating haplotype relative risks in complex disease from unphased SNPs data in families using a likelihood adjusted for ascertainment. Genet Epidemiol. 2006;30:666–76. doi: 10.1002/gepi.20178. [DOI] [PubMed] [Google Scholar]
- Cordell HJ. Properties of case/pseudocontrol analysis for genetic association studies: Effects of recombination, ascertainment, and multiple affected offspring. Genet Epidemiol. 2004;26:186–205. doi: 10.1002/gepi.10306. [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Barratt BJ, Clayton DG. Case/pseudocontrol analysis in genetic association studies: A unified framework for detection of genotype and haplotype associations, gene-gene and gene-environment interactions, and parent-of-origin effects. Genet Epidemiol. 2004;26:167–85. doi: 10.1002/gepi.10307. [DOI] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum Likelihood from Incomplete Data via the EM Algorithm. Journal of the Royal Statistical Society. Series B (Methodological) 1977;39:1–38. [Google Scholar]
- Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008;66:87–98. doi: 10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjessing HK, Lie RT. Case-parent triads: estimating single- and double-dose effects of fetal and maternal disease gene haplotypes. Ann Hum Genet. 2006;70:382–96. doi: 10.1111/j.1529-8817.2005.00218.x. [DOI] [PubMed] [Google Scholar]
- Marchini J, Cutler D, Patterson N, Stephens M, Eskin E, Halperin E, Lin S, Qin ZS, Munro HM, Abecasis GR, Donnelly P. A comparison of phasing algorithms for trios and unrelated individuals. Am J Hum Genet. 2006;78:437–50. doi: 10.1086/500808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ. Linkage disequilibrium testing when linkage phase is unknown. Genetics. 2004;166:505–12. doi: 10.1534/genetics.166.1.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Umbach DM, Weinberg CR. Identification of Risk-Related Haplotypes with the Use of Multiple SNPs from Nuclear Families. Am J Hum Genet. 2007;81:53–66. doi: 10.1086/518670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venzon DJ, Moolgavkar SH. A Method for Computing Profile-Likelihood-Based Confidence Intervals. Applied Statistics. 1988;37:87–94. [Google Scholar]
- Wacholder S, Rothman N, Caporaso N. Counterpoint: bias from population stratification is not a major threat to the validity of conclusions from epidemiological studies of common polymorphisms and cancer. Cancer Epidemiol Biomarkers Prev. 2002;11:513–20. [PubMed] [Google Scholar]
- Weinberg CR, Wilcox AJ, Lie RT. A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am J Hum Genet. 1998;62:969–78. doi: 10.1086/301802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox AJ, Weinberg CR, Lie RT. Distinguishing the effects of maternal and offspring genes through studies of “case-parent triads”. Am J Epidemiol. 1998;148:893–901. doi: 10.1093/oxfordjournals.aje.a009715. [DOI] [PubMed] [Google Scholar]
- Zhang J, Rowe WL, Clark AG, Buetow KH. Genomewide distribution of high-frequency, completely mismatching SNP haplotype pairs observed to be common across human populations. Am J Hum Genet. 2003;73:1073–81. doi: 10.1086/379154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Sun F, Zhao H. HAPLORE: a program for haplotype reconstruction in general pedigrees without recombination. Bioinformatics. 2005;21:90–103. doi: 10.1093/bioinformatics/bth388. [DOI] [PubMed] [Google Scholar]
- Zhang K, Zhao H. A comparison of several methods for haplotype frequency estimation and haplotype reconstruction for tightly linked markers from general pedigrees. Genet Epidemiol. 2006;30:423–37. doi: 10.1002/gepi.20154. [DOI] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, Caprau D, Christensen K, Suzuki Y, Machida J, Natsume N, Yoshiura K, Vieira AR, Orioli IM, Castilla EE, Moreno L, Arcos-Burgos M, Lidral AC, Field LL, Liu YE, Ray A, Goldstein TH, Schultz RE, Shi M, Johnson MK, Kondo S, Schutte BC, Marazita ML, Murray JC. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–80. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.