Abstract
The establishment of metastasis depends on the ability of cancer cells to acquire a migratory phenotype combined with their capacity to recreate a secondary tumor in a distant tissue. In epithelial cancers, such as those of the breast, the epithelial-mesenchymal transition (EMT) is associated with basal-like breast cancers, generates cells with stem-like properties, and enables cancer cell dissemination and metastasis. However, the molecular mechanism(s) that connects stem cell–like characteristics with EMT has yet to be defined. Using an orthotopic model of human breast cancer metastasis to lung, we identified a poor prognosis gene signature, in which several components of the wnt signaling pathway were overexpressed in early lung metastases. The wnt genes identified in this signature were strongly associated with human basal-like breast cancers. We found that inhibiting wnt signaling through LRP6 reduced the capacity of cancer cells to self-renew and seed tumors in vivo. Furthermore, inhibition of wnt signaling resulted in the reexpression of breast epithelial differentiation markers and repression of EMT transcription factors SLUG and TWIST. Collectively, these results provide a molecular link between self-renewal, EMT, and metastasis in basal-like breast cancers.
Introduction
Breast cancer relapse and subsequent metastatic spread to distant sites, such as lung, liver, brain, and bone, remain the leading cause of morbidity and mortality associated with the disease. Metastasis is thought to be an inefficient process because many thousands of cancer cells are shed into the circulation, yet only a very few have the ability to form distant nodules (1). Because a rare population of cells in human breast cancers have been identified with the capacity to self-renew, produce differentiated daughter cells, and form tumors in immunocompromised mice, these cells are natural candidates to seed metastasis (2, 3). However, little is known about the relationship between the cells that seed and initiate primary tumor growth with those that exhibit metastatic potential and likely seed secondary tumors.
Much recent work has focused on identifying the genetic factors in cancer cells that mediate their migration or propensity to associate with certain tissues in advanced stages of malignancy (4–7). This work has revealed the important role of cell surface molecules, growth factors, extracellular matrix molecules, and proteases as critical components for breast cancer cell tropism and adhesion in certain distant tissues (8). In addition, metastasis has been linked to the expression of embryonic transcription factors (TWIST, SLUG), which promote the migratory and invasive state called the epithelial-to-mesenchymal transition (EMT; refs. 9–11). In an EMT state, epithelial cells lose expression of many markers of differentiation, acquire migratory and mesenchymal features, and exhibit enhanced tumor seeding (12). However, the molecular link between EMT and self-renewal is unclear; whether a shared signaling pathway regulates both processes is unknown.
The wnt signaling pathway mediates a wide variety of processes, including cell proliferation, migration, differentiation, adhesion, and death (13). Furthermore, wnt signals have been reported to promote migration and EMT in breast cancer cells through stabilization of Snail (14). Consistent with this notion, wnt signaling up-regulates the transcription factors Slug and Twist (15, 16), which are both known to be transcriptional repressors of E-cadherin.
The Wnt pathway is composed of two distinct signaling arms: the canonical and noncanonical pathways. In the canonical pathway, the wnt protein binds to the cell surface receptor Frizzled, which uses the coreceptors Lrp5/6 to promote Axin binding to Dishevelled. This leads to the stabilization of β-catenin, which in turn translocates to the nucleus, where it interacts with the DNA-binding proteins from the Tcf/Lef family to activate transcription of an extensive array of genes (17). SFRP1, a negative regulator of wnt signaling, is a member of the family of secreted frizzled-related proteins. These proteins are homologous to the extracellular cysteine-rich domain of the wnt receptor frizzled, thereby preventing wnt ligands from binding (18). DKK1 is another secreted inhibitor that functions by binding directly to the Lrp5/6 coreceptors and inhibiting canonical wnt signal transduction (19). Recent studies report that secreted inhibitors of the wnt pathway, including both SFRP1 and DKK1, are frequently silenced by methylation in many human cancers, including breast cancer (20).
Currently, human tumor models used to study breast cancer metastasis to lung rely on introducing cancer cells directly into the vasculature by injection into the tail vein (4, 21, 22). Whereas these models are useful for identifying and examining factors involved in proliferation of breast cancer cells that have been directly deposited into the lung environment, they do not model all the events necessary for the metastatic process from the primary site.
Given these limitations, we sought to develop a model of breast cancer metastasis that more faithfully recapitulates the biology and progression of the disease when implanted into the orthotopic site. Accordingly, we tested several different human breast cancer cell lines for their ability to spontaneously metastasize after primary tumor growth in the mammary fat pads of NOD/SCID immuno-deficient mice. One cell line, SUM1315, showed the propensity to metastasize to mouse lung and human implanted bone after primary tumor development (23).
In this report, we set out to identify genes that mediate breast cancer metastasis to lung and, in doing so, identified various components of the wnt signaling pathway that are overexpressed in lung micrometastases. We reasoned that wnt signaling might be regulating cancer cell self-renewal and expression of EMT transcription factors, thereby enabling tumor seeding and metastasis. We show herein the effects of inhibiting wnt signaling on cancer cell differentiation states through the expression of the EMT transcription factors Slug and Twist and reveal how this pathway links the EMT state with differentiation, self-renewal, and tumor formation.
Materials and Methods
Cell culture
SUM1315 cells (obtained from Dr. Stephen Ethier, Michigan) were cultured in Ham’s F12 supplemented with 5% fetal bovine serum, insulin (5 µg/mL), and epidermal growth factor (10 µg/mL; see ref. 3 for culturing of other cell lines). Virus was produced as described previously (24).
Real time PCR
RNA was isolated and purified using QIAGEN RNAeasy kit for cells in culture and Trizol reagent (Invitrogen) for whole tissue. RNA was reverse transcribed to cDNA using Bio-Rad iScript cDNA synthesis kit. Quantitative real-time PCR analysis was performed using SyBR Green (Bio-Rad). Custom-designed differentiation arrays were purchased from Superarray (gene list in Supplementary Table S4, catalogue CAPH-0298A). RNA was isolated using the QIAGEN RNAeasy kit. cDNA was prepared using the RT2 First Strand kit, and PCR analysis was performed using Superarray SyBr Green Master Mix. Fold differences were calculated using the software supplied on the Superarray website. Heatmap Builder software was used to create heatmap. Primer sequences used for quantitative real-time PCR are in Supplementary Table S5.
Microarray analysis
Total RNA was collected using Trizol on three fragments of tumor tissue and three fragments of associated lung metastases from four different mice. Total RNA (2 µg) from each pooled sample was hybridized to the Operon human 27K oligonucleotide (70-mer) array chip, which contains 26,791 transcripts representing over 20,000 human genes as previously described (25).
Luciferase assays
Cells were plated in 12-well plates 24 h before transfection. TOPFlash or FOPFlash (150 ng) and Renilla (1 ng) were transfected using FuGene 6 (Roche) according to the manufacturer’s protocol. Wnt3a conditioned media and control conditioned media were added 24 h posttransfection, and luciferase readings were recorded 48 h posttransfection using Promega’s dual-luciferase reporter assay system according to instructions. Luciferase readings were recorded using a Zylux FB12single tube luminometer. Experiments were performed in triplicate on at least three separate days. TOPFlash, FOPFlash, and Wnt1 plasmids were gifts from Dr. Amy Yee (Tufts).
In vivo experiments
Female NOD/SCID mice were injected between 6 and 12wk of age into the fourth inguinal mammary gland. Cells were resuspended in 3:1 media/Matrigel solution and injected at one million cells per gland. Mice were monitored weekly until palpable tumors formed, and tumor growth was measured and recorded using calipers at least once per week thereafter. Tissues were embedded in paraffin and H&E stained at Tufts Medical Center.
Spheres
Cells were plated onto 6-cm nonadherent plates (Corning) at 50,000 cells/mL. Visible spheres were counted on day 6 postplating. SUM1315-shLrp6 spheres, and the respective empty vector control were counted by straining through a 40-µm filter, eluting onto a dish with a grid and counting under a microscope.
Immunoblots
Protein was extracted from cells in culture using radioimmunoprecipitation assay buffer and from tumors using a Brij lysis buffer supplemented with protease inhibitors. Membranes were probed using the following antibodies: sFRP1 (AbCam), Dkk1 (Santa Cruz), active β-catenin (AbCam), glyceraldehyde-3-phosphate dehydrogenase (Chemicon), caspase-3 (Cell Signaling), smooth muscle actin (Vector), β-actin (Abcam), Lrp6 (Cell Signaling), SOX17 (R&D), Twist (Cell Signaling), and Slug (Cell Signaling).
Growth curves
Eight thousand cells were plated in triplicate in 24-well plates. Cells were trypsinized on days recorded in graph and counted using a Coulter Counter. Each well was counted thrice and repeated on three separate days.
Immunohistochemistry
Slides were deparaffinized and rehydrated in graded ethanols from 100% to 70%. Antigen retrieval was performed by boiling slides for 20 min using 10 mmol/L citrate (pH 6.0). Slides were blocked with 1% bovine serum albumin/serum, and primary antibody (Slug, Cell Signaling) was used at 1:50 overnight at 4°C. Secondary antibodies were detected using avidin-biotin complex kit, and signal was developed using Nova Red (Vector Labs.). Slides were dehydrated and coverslipped.
Results
Identification of a wnt signaling signature in an orthotopic model of lung metastases
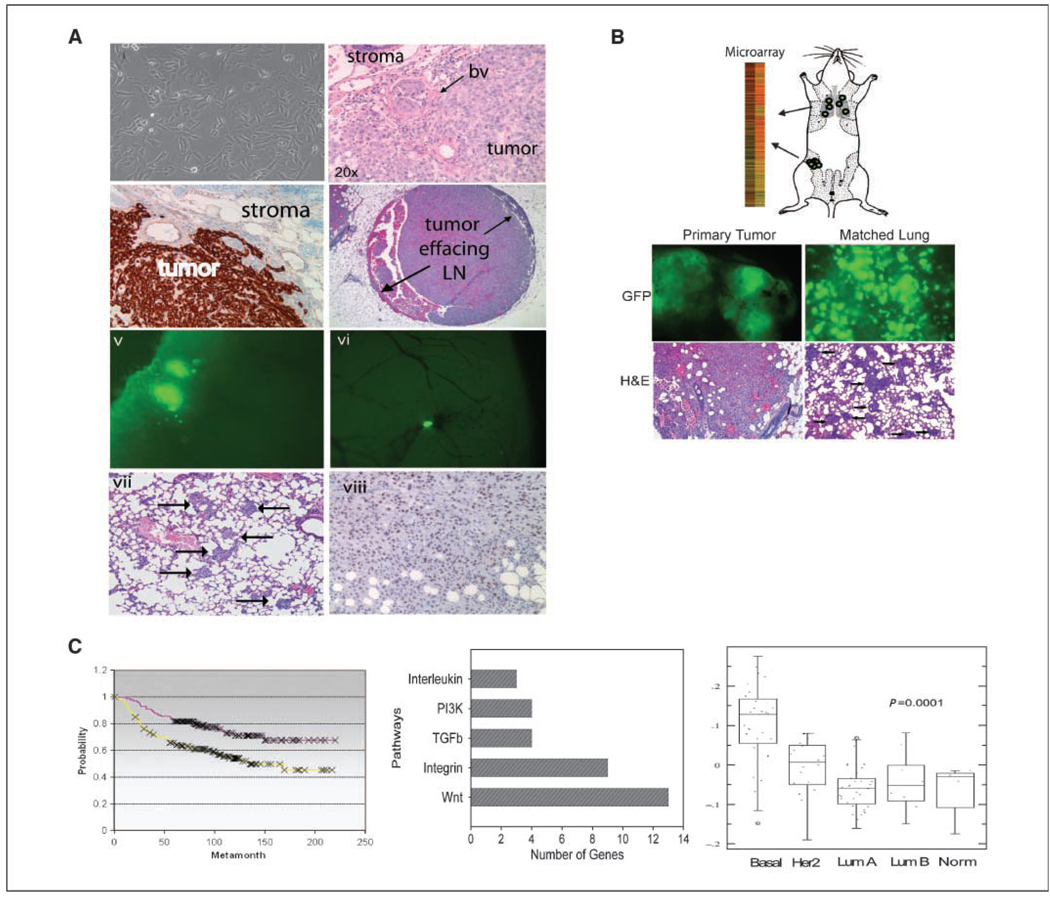
The SUM1315 cell line was derived from a cutaneous metastatic nodule from a patient with advanced invasive breast ductal carcinoma (23) that forms poorly differentiated, highly invasive tumors with 90% (29 of 32) efficiency and exhibits patterns of intravascular and intralymphatic growth within the mammary glands of mice (Fig. 1A). We engineered the SUM1315 cells to express green fluorescent protein (GFP) to visualize distant tissues for the presence of metastatic GFP+ breast cancer cells. Similar to parental SUM1315 cells, SUM1315-GFP cells metastasize to lung in ~30% (6 of 19) of animals and to brain in ~20% (one of five) of animals (Fig. 1A). SUM1315 is an ER/PR/Her2-negative basal-type breast cancer model that exhibits hallmarks of EMT, including loss of E-cadherin and epithelial keratin expression, and acquisition of Vimentin and fibronectin expression. Furthermore, SUM1315 cells express the EMT transcription factors SLUG and TWIST (Fig. 1A; refs. 23, 26).
Figure 1.
A, orthotopic xenograft model of human breast cancer metastasis. i, phase contrast image of SUM1315 cells in culture. ii, H&E-stained section of SUM1315 xenograft (BV, blood vessel). iii, Vimentin-stained section of SUM1315 xenograft. Human tumor cells are Vimentin positive, whereas surrounding mouse stroma is Vimentin negative. iv, H&E-stained section of mouse mammary tissue. The tumor has invaded and replaced most of the normal lymph node tissue. v, GFP image of nodules in lung tissues resulting from a SUM1315 primary tumor. vi, GFP image of metastasis in brain resulting from a SUM1315 primary tumor. GFP-positive cells are tumor cells. vii, H&E-stained section of lungs harvested from mice bearing SUM1315 tumors. Arrows point to metastases. viii, SUM1315 xenograft primary tumor stained with Slug. B, schematic representation of microarray study. GFP images (top) and H&E-stained sections (bottom) of primary tumor and lung metastasis from SUM1315 injections. C, left, lung metastasis gene list (Supplementary Table S1) was compared with van’t Veer poor prognosis data set and is associated with reduced time to metastasis. Middle, pathway analysis of genes identified in lung metastasis signature using the PANTHER database. Wnt gene list (Supplementary Table S3) was compared with published microarray. Right, Wnt gene list was compared with UNC12 data set (Supplementary Fig. S1) and is significantly correlated with basal-like breast cancers.
We used this model system to identify patterns of gene expression associated with metastatic behavior. Accordingly, we performed a microarray study on primary breast tumor tissues from mice bearing SUM1315 xenografts compared with their corresponding lungs harboring visible GFP+ lung micrometastases (Fig. 1B). To minimize selection bias and animal-to-animal variability, three fragments of each tumor tissue and three fragments of associated lung metastases were collected and pooled together from four separate animals, respectively. A list corresponding to genes differentially expressed between lung metastases and primary tumors was generated. In total, 784 genes were differentially expressed in lung metastases compared with primary mammary tumors (Supplementary Table S1).
We next assessed whether this gene signature was associated with clinical outcome or with biological relevance. Using the van’t Veer poor prognosis data set (27), we classified each patient based on tumor outcome and then performed rank order splitting analysis with the genes differentially expressed in lung metastases compared with the primary tumors. Remarkably, this lung metastasis signature was significantly associated with poor prognosis (Supplementary Fig. S1A; P = 0.001), reduced time to metastasis (Supplementary Fig. S1A; P = 0.002), and reduced overall survival (Supplementary Fig. S1; P = 0.001). Because the SUM1315 line is a basal-like breast cancer tumor model, we next examined whether the lung metastasis signature could be used to classify breast cancers according to tumor subtype. However, in contrast to patient outcome, this lung signature was not robustly associated with tumor subtype classification (data not shown).
Functional gene ontology classification of differentially expressed genes revealed that the lung signature was enriched for genes involved in development, metabolism, and cell death (Supplementary Table S2). The gene list was classified by signaling pathways to identify those that might be important in metastasis biology. Remarkably, the most abundantly represented signaling pathway identified in the lung metastatic signature was the Wnt pathway (Fig. 1C), in which 13 wnt genes representing 49 signaling components were differentially expressed. These genes included the cell surface receptors LRP5 and FZD9, as well as the wnt target genes WISP2 and CCND3 (Supplementary Table S3). An additional eight wnt pathway genes were identified in the lung metastasis signature that were differentially expressed 1.4-fold, including AXIN2, DVL2, WNT7A, and engrailed (Supplementary Table S3). Several of the genes that were differentially expressed 1.4-fold was validated by quantitative reverse transcription–PCR (qRT-PCR; Supplementary Fig. S2).
Interestingly, this wnt gene list was significantly correlated with basal-like breast cancers using both the NKI and UNC data sets (refs. 27–29; P = 0.0001 and 0.0006; Supplementary Fig. S1). We also found that several of these wnt genes were associated with poor prognosis, high-grade, metastatic breast cancer (Table 1; ref. 30). Collectively, these data are consistent with and support recent findings that wnt signaling is up-regulated in human basal subtype breast cancers and lung and brain metastasis (31).
Table 1.
Wnt pathway genes expressed in human breast cancer
| Gene | Analysis | P | Study name and reference |
|---|---|---|---|
| NFATc3 | (a) Poorly differentiated | 4.50E–05 | Desmedt (43) |
| (b) Death within 5 y | 1.70E–04 | Pawitan (44) | |
| (c) Relapse/recurrence in 5 y | 5.70E–04 | van de Vijver (45) | |
| CCND3 | (a) Adjuvant hormone | 3.90E–04 | Sotiriou (46) |
| (b) BRCA1 positive | 0.001 | van ’t Veer (27) | |
| (c) Basal-like | 0.005 | Richardson (47) | |
| WISP2 | (a) Positive lymphocytic infiltrate | 1.80E-04 | van ’t Veer (27) |
| (b) Grade 3 | 3.6E-4 and 3.60E-04 | Ivshina (48) | |
| FZD9 | (a) BRCA1 positive | 1.10E-04 | van ’t Veer (27) |
| (b) Grade 3 | 1.60E-04 | van ’t Veer (27) | |
| (c) Poorly differentiated | 9.40E-04 | Desmedt (43) | |
| (d) Positive lymphocytic infiltrate | 0.001 | van ’t Veer (27) | |
| HDAC1 | (a) aggressive lung metastasis | 0.008 | Minn (21) |
| LRP6 | (a) Basal type carcinoma | 3.5 E-5 | Richardson (47) |
| (b) Invasive ductal and invasive lobular carcinoma | 1.6 E-4 | Desmedt (43) | |
| EN1 | (a) Basal-like carcinoma | 1.9E-11 and 5.2E-6 | Farmer (49) |
| (b) Response to chemo | 5.80E-04 | Hess (50) | |
| (c) Poorly differentiated | 7.70E-04 | Desmedt (43) | |
| HOXA7 | (a) Elston Grade 3 | 3E-6 and 5.9E-6 | Ivshina (48) |
| (b) Bone Metastases | 0.004 | Ma (51) | |
| (c) Positive lymph nodes | 0.003 and 0.006 | Miller (52) | |
| CDH23 | (a) BRCA1 tumors | 2.90E-04 | van ’t Veer (27) |
| (b) Lymphocytic infiltrate | 0.003 | van ’t Veer (27) | |
| MYCL1 | (a) Lobular Carcinoma | 0.003 | Ma (51) |
| (b) After chemotherapy | 0.007 | Sørlie (53) | |
| HOXD4 | (a) PR positive | 2.40E-05 | Minn (21) |
| (b) Aggressive Lung Metastasis | 0.006 | Minn (21) |
Wnt signaling through LRP6 is required for tumor formation and metastasis
Whereas mutations in the wnt pathway (e.g., APC) are well known for their causal role in the progression of colon cancer (32), mutations in the wnt pathway have not been identified in breast cancer (33). Despite the lack of genetic mutations, β-catenin is observed in both the cytoplasm and nucleus of many human breast cancer specimens, indicating activation of the pathway (33). Additionally, many negative regulators of the pathway, including SFRP1 and DKK1, are often silenced in breast cancer, a possible contributing factor for the unopposed wnt activity in breast cancer (20). Therefore, we assessed wnt signaling activity of a number of breast cancer cell lines, including SUM1315, SUM149, SUM159, MDA.MB.231, MCF7, and T47D using the TOPFLASH reporter assay. Interestingly, SUM1315 cells, which are capable of completing all the steps necessary for metastasis, exhibited elevated wnt activity in the absence of exogenous wnt stimulation compared with other breast cancer cell lines (Supplementary Fig. S2). Whereas it is unclear whether the SUM1315 cells exhibited the most robust TOPFLASH activity due to further mutations in the wnt pathway or produce elevated levels of wnt ligands compared with the other cell lines, these results support the microarray results above correlating wnt signaling with metastatic behavior.
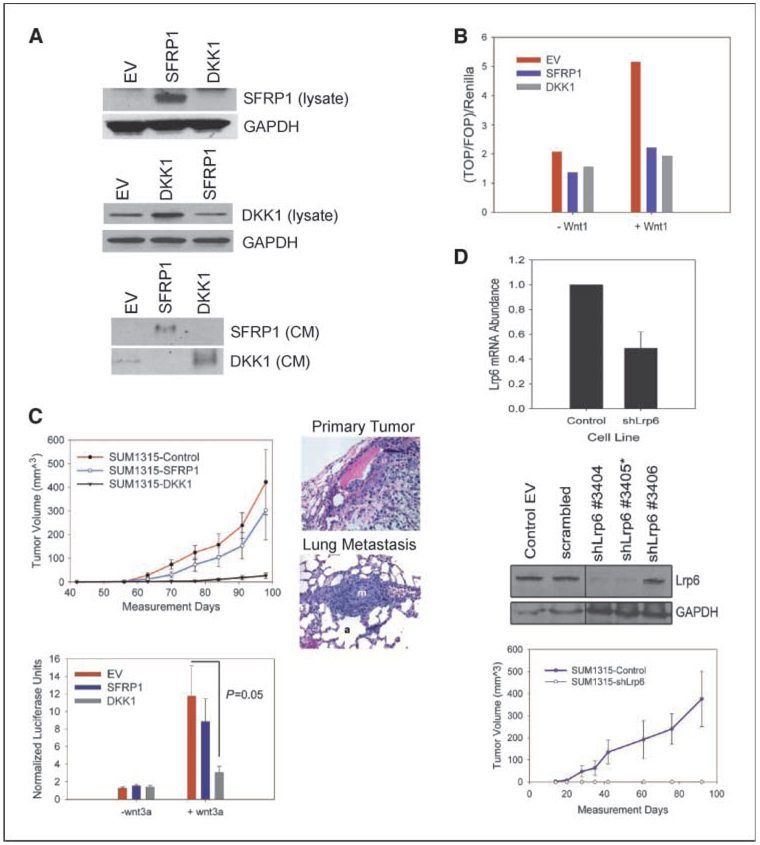
Given these findings, we speculated that inhibition of wnt signaling in SUM1315 cells might affect their ability to form tumors or metastasize. We therefore generated SUM1315 cells that overexpress either SFRP1 or DKK1 (Fig. 2A). SUM1315 cells normally express moderate endogenous DKK1, yet protein expression and secretion into the supernatant were increased upon its ectopic expression in SUM1315-DKK1 cells (Fig. 2A). In contrast, SUM1315 cells do not express endogenous SFRP1 mRNA or protein (Fig. 2A; data not shown), consistent with its methylation (34–36), but SUM1315-SFRP1 cells produce abundant message and protein (Fig. 2A; data not shown). We confirmed that enforced expression of DKK1 and SFRP1 inhibited wnt signaling using the TOPFlash reporter system to assess wnt/β-catenin activity. Exogenous Wnt1 ligand stimulated robust TOPFlash activity in control SUM1315 cells but was blocked in cells overexpressing SFRP1 or DKK1 (Fig. 2B).
Figure 2.
DKK1 inhibits primary tumor growth through inhibition of LRP6. A, Western blot analysis of whole-cell lysate and conditioned media collected from control empty vector (EV), SFRP1, or DKK1-overexpressing cells. B, TOPFlash assay of EV-SUM1315, SFRP1-SUM1315, and DKK1-SUM1315 cells in the absence and presence of Wnt1. C, tumor growth curves of SUM1315 cells overexpressing SFRP1, DKK. Error bars, SE. H&E stained of primary tumor and matched lung from mouse injected with SFRP1-SUM1315 cells. m, metastasis; a, alveolus TOPFlash assay of Control-SUM1315, SFRP1-SUM1315, and DKK1-SUM1315 cells stimulated by wnt3a-conditioned media. D, qRT-PCR analysis of RNA extracted from stable Lrp6 knockdowns. Error bar, SD. Western blot analysis of several Lrp6 stable knockdowns compared with the control EV and scrambled control. Tumor growth curves of SUM1315 expressing shLrp6 3405. Error bars, SE.
We next injected one million SUM1315-SFRP1 and DKK1 cells into the mammary fat pads of NOD/SCID to determine if inhibition of wnt signaling would affect the ability to seed tumors and/or metastasize in mice. SUM1315 cells expressing the empty-vector control or overexpressing SFRP1 formed primary mammary tumors and metastases at similar rates and with identical frequencies (Fig. 2C). In contrast, cells overexpressing DKK1 failed to form palpable tumors or metastases even by 100 days postinjection (Fig. 2C).
Because tumor formation and lung colonization were not affected in cells overexpressing SFRP1, we tested for the possibility that ectopic SFRP1 expression was lost in the cells that formed tumors in vivo. However, we confirmed that tumor tissues growing in mice from SFRP1 expressing SUM1315 cells still retained robust SFRP1 mRNA and protein expression (Supplementary Fig. S3). Therefore, we reasoned that the differences in tumor formation by DKK1 and SFRP1-expressing cells was likely due to the difference in the mechanism by which these inhibitors function rather than due to silencing of the gene.
SFRPs exert their inhibitory effects through binding to wnt ligands and preventing their binding to the frizzled receptors. This raised the possibility that ectopic SFRP1 expression might not adequately inhibit wnt ligands produced in vivo by the stromal microenvironment. To test this possibility, we examined wnt activity while stimulating cells with conditioned media collected from mouse stromal fibroblasts expressing Wnt3a. Wnt3a conditioned media induced robust TOPFlash activity in control SUM1315 cells, which was efficiently blocked in cells overexpressing DKK1. However, SFRP1-expressing cells were unable to block wnt3a-stimulated TOPFlash activity (Fig. 2C versus B). Moreover, wnt3a-treated SFRP1-SUM1315 cells exhibited similar gene expression changes as the control SUM1315 cells treated with wnt3a conditioned media (Supplementary Fig. S4). These results imply that the ability of SUM1315-SFRP1 cells to form tumors was due to incomplete inhibition of wnt signaling in vivo.
DKK1 exerts its wnt inhibitory activity by binding directly to the LRP5/6 coreceptor and inhibiting its interaction with frizzled (19). Thus, if DKK1 was inhibiting wnt-signaling through its effects on LRP6, knockdown of the receptor should phenocopy cells overexpressing DKK1. Stable lentiviral integration of hairpins against LRP6 resulted in ~50% reduction in mRNA levels (Fig. 2D) and undetectable protein expression by immunoblotting (Fig. 2D; protein expression of LRP5 was undetectable by Western blot; data not shown). LRP6 short hairpin RNA (shRNA) cells injected into the mammary fat pads of NOD/SCID mice also failed to form tumors indicating that DKK1 overexpression likely inhibits primary tumor formation through its effects on LRP6 (Fig. 2D). Collectively, these data show that wnt signaling through LRP6 is required for tumor formation.
Wnt signaling is necessary for cancer cell self-renewal but not proliferation
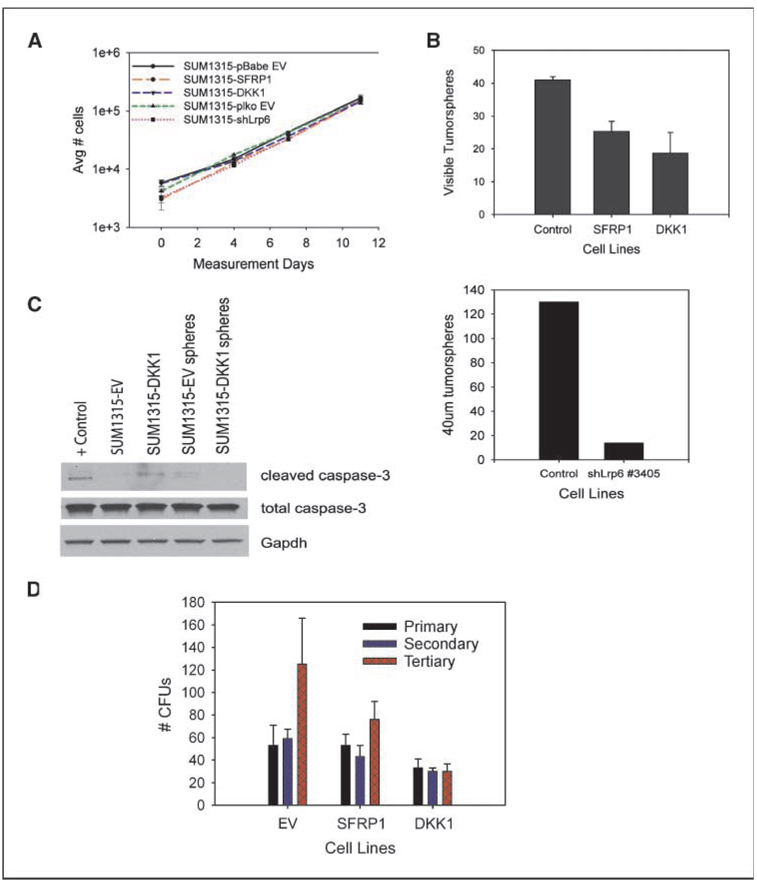
To determine the mechanism by which wnt inhibition blocked tumor formation, we examined the effects of wnt inhibition on cell proliferation in vitro. SUM1315 control cells, as well as cells overexpressing SFRP1, DKK1, or shRNA against LRP6, grew at similar rates in vitro, indicating that wnt is not required for the proliferation of SUM1315 cells in culture (Fig. 3A).
Figure 3.
Wnt signaling is necessary for survival in suspension, but not proliferation. A, growth curve analysis of wnt-inhibited (SFRP1, DKK1, and shLrp6 lines) and control SUM1315 cell lines. B, tumorsphere formation of control-SUM1315, SFRP1-SUM1315, and DKK1-SUM1315 cells. Error bars, SD. C, Western blot for caspase-3. Protein lysates were collected from adherent monolayers (lanes 2 and 3) or cells grown in suspension (lanes 4 and 5). The positive control (lane 1) was SUM1315 cells treated with 5-fluorouracil (1 mmol/L) for 6 d. D, serial CFU assays of SUM1315-EV, SFRP1, and DKK1 cell lines plated at low dilutions. Colonies were defined as having >30 cells.
Sphere formation in nonadherent cultures is reflective of stem-like properties, including the ability to survive suspension-induced cell death, and is a predictor of tumor formation in vivo (3, 12). We and others have previously shown that tumorsphere formation in vitro correlates with tumorigenicity in vivo (3). Interestingly, despite not affecting proliferation, overexpression of SFRP1, DKK1, or knockdown of LRP6 resulted in a reduction in sphere-forming ability (Fig. 3B). In addition, the differences in sphere forming ability were not due to differences in apoptosis, as cleaved caspase-3 expression levels were similar in DKK1-overexpressing cells compared with control cells (Fig. 3C). Given this finding and the fact that the wnt pathway is a critical regulator of self-renewal in other systems, we reasoned that the decrease in sphere formation may be an indication of defects in self-renewal.
Therefore, we used the serial colony-forming unit (CFU) assay to analyze the effect of wnt inhibition on self-renewal. A cell line capable of self-renewal should exhibit an increase in the number of colonies formed after serially passaging in culture. This was indeed observed with the SUM1315 control cell line but also for the SFRP1-SUM1315, which had expanded upon the third serial passage (Fig. 3D). By contrast, the DKK1-SUM1315 line had failed to increase the number of colonies formed after serial passaging, indicating that self-renewal was impeded.
It has been shown that sphere formation, basal-like breast cancers, and EMT (epithelial dedifferentiation) correlate with a CD44hi/CD24lo antigen phenotype (3, 12, 37, 38). Because the SUM1315 cell line already exhibits a high CD44hi/CD24lo profile (3, 38), we wanted to determine whether this phenotype was altered in response to overexpression of DKK1. We indeed found that expression of DKK1 increased the proportion CD44lo/CD24lo by 50% (Supplementary Fig. S5). Taken together, these data imply that wnt signaling is necessary for cancer cell self-renewal and regulation of cell differentiation phenotypes independent of proliferation and apoptosis.
Wnt signaling maintains the dedifferentiated EMT state
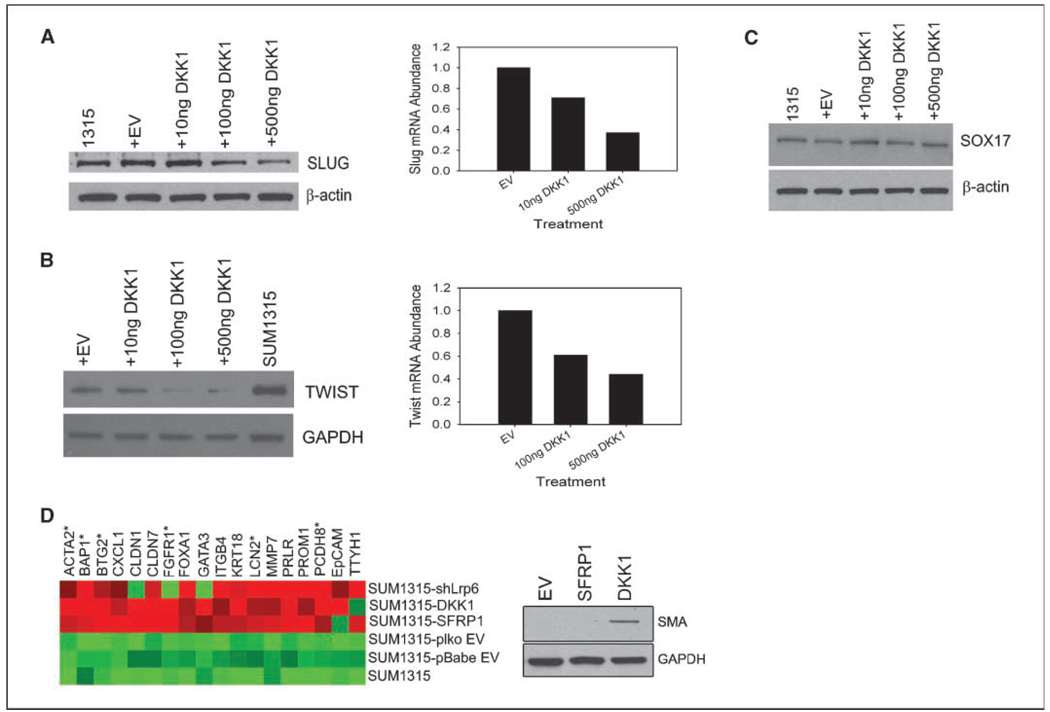
As previously mentioned, the SUM1315 line is a mesenchymal/basal cell line that expresses high levels of the EMT transcription factors Slug and Twist (Fig. 1A; ref. 26). SLUG is a reported wnt target gene (39, 40), whereas Twist has been shown to be up-regulated in response to Wnt1 stimulation (16). Based on the recent findings that ectopic expression of EMT transcription factors, including TWIST, could enhance tumorigenicity (12), we wished to determine whether the effects of wnt inhibition on tumor seeding might be through the expression of EMT genes. Accordingly, SUM1315 cells treated with DKK1 exhibited a dose-dependent reduction in mRNA expression and protein levels of both Slug and Twist (Fig. 4A and B). In contrast, expression levels of Sox17, a transcription factor not targeted by wnt signaling, was unaffected by treatment with DKK1 (Fig. 4C), indicating that the reduction in expression of Slug and Twist were specifically due to inhibition of wnt signaling.
Figure 4.
Wnt signaling represses differentiation. Western blot and qRT-PCR analysis for Slug (A), Twist (B), and SOX17 (C) in SUM1315 cells transfected with DKK1. D, differential gene expression of differentiation genes from wnt-inhibited cell lines. Heatmap Builder Software was used to analyze the data. Starred (*) genes represent those of myoepithelial differentiation, whereas unstarred genes represent luminal differentiation. Western blot analysis of α-smooth muscle actin expression in Control-SUM1315, SFRP1-SUM1315, and DKK1-SUM1315 cells to confirm the qRT-PCR results from left.
Because the EMT transcription factors SLUG and TWIST were reduced upon DKK1 treatment, we reasoned that such cells would display features of epithelial differentiation and lineage commitment. We, therefore, examined the differentiation state of the wnt-inhibited lines using a custom qRT-PCR array targeting 86 genes known to be associated with either myoepithelial, luminal epithelial, or mammary epithelial stem cell differentiation (Supplementary Table S4). Remarkably, all the wnt-inhibited cell lines exhibited an increased mRNA expression of mammary epithelial markers, including those of committed luminal (KRT18, EPCAM, GATA3, PRLR) and myoepithelial cells (TTHY1, ACTA2; Fig. 4D). Expression of α-smooth muscle actin (ACTA2), a marker of mature differentiated myoepithelial cells, was further confirmed by immunoblotting (Fig. 4D). These results indicated that wnt signaling was necessary for the maintenance of a dedifferentiated epithelial phenotype consistent with EMT.
Reexpression of EMT transcription factors in vivo bypasses wnt inhibition
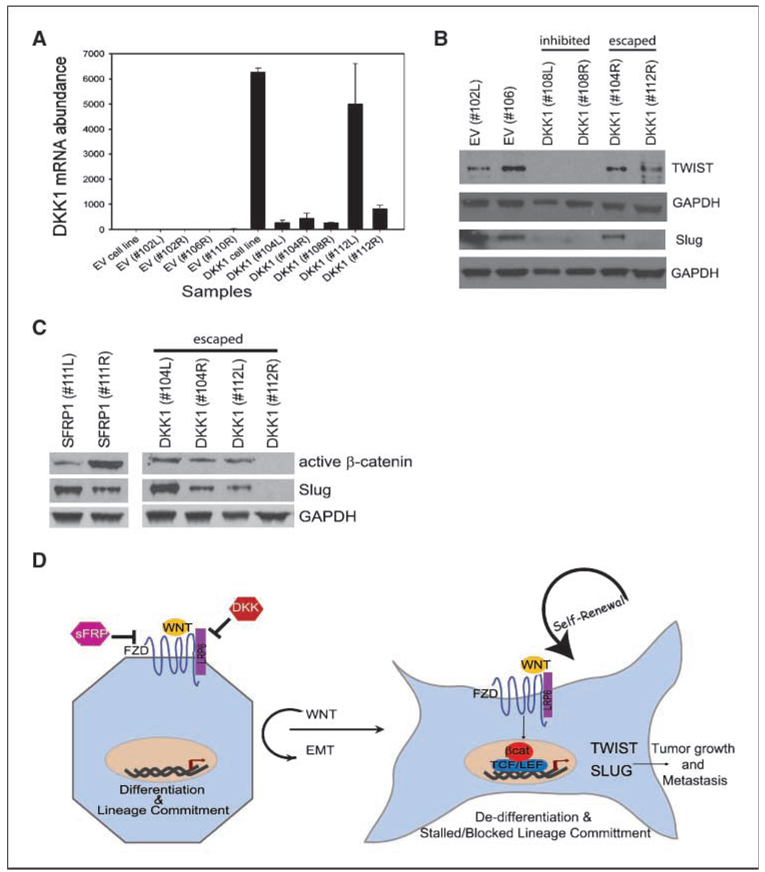
The EMT state generates cells with many of the properties of self-renewing stem cells, including suppression of epithelial differentiation and the ability to seed tumors in vivo (12, 24). Because SLUG and TWIST were reduced upon DKK1 treatment, we, therefore, wanted to determine whether the lack of tumor seeding in SUM1315-DKK1 cells was a consequence of repressed SLUG or TWIST expression. To this end, one million SUM1315-DKK1 cells were injected orthotopically into NOD/SCID female mice. Whereas tumors did not form in any animal by 100 days postinjection, tumorigenic clones eventually escaped and tumors developed in 50% of the mice (“escaped”); the other 50% of the mice still had not formed palpable tumor masses (“inhibited”) within this time. We speculated that if the expression of Slug and Twist facilitates tumor growth, then tumor cells, which had eventually escaped wnt inhibition, would have reexpressed such transcription factors. To test this hypothesis, tumors and injection sites were harvested and examined for expression of DKK1, TWIST, and SLUG. Whereas all “escaped” and “inhibited” DKK1 tumors maintained robust mRNA and protein expression of DKK1 (Fig. 5A; data not shown), we observed reexpression of Slug and Twist only in the escaped DKK1-SUM1315 tumors compared with the inhibited DKK1-SUM1315 tumors (Fig. 5B and C). Furthermore, the escaped DKK1-SUM1315 tumors exhibited high levels of active β-catenin, an indication that the wnt–β-catenin pathway had been reactivated despite DKK1 expression (Fig. 5C). Taken together, these data suggest that inhibition of wnt signaling blocks tumor formation by promoting epithelial differentiation and repressing transcriptional repressors.
Figure 5.
Reexpression of Slug and Twist in vivo bypasses wnt inhibition. A, qRT-PCR analysis of DKK1 expression in tumor and injection sites from mammary glands of mice injected with control SUM1315-EV or DKK1-expressing cells. B, Western blot analysis of Twist and Slug expression in tumors and injection sites isolated from mice injected with SUM1315-Control EV or DKK1 expressing cells. C, Western blot analysis of DKK1 and SFRP1 tumors for active β-catenin and Slug. D, schematic model of wnt in breast epithelial differentiation, EMT, and metastasis. In the absence of wnt signaling (left; or in the presence of wnt inhibitors), cells undergo lineage commitment and differentiation. In the presence of wnt signals, β-catenin partners with TCF/LEF to activate target genes, such as SLUG and TWIST, which promote an EMT, repress differentiation, and enhance tumor seeding and metastasis.
Discussion
Several studies have documented EMT in enabling both tumor seeding at low dilutions and lung metastasis (12, 41). Our results provide evidence of a unifying signaling pathway that mediates both of these processes. Wnt signaling through LRP6 in a model of basal-like breast cancer repressed cancer cell differentiation and induced the expression of the EMT transcription factors Slug and Twist. Ectopic expression of DKK1 affected wnt signaling by inactivation of the LRP6 receptor, leading to a reduction in tumor formation and survival in suspension culture. DKK1 also reduced expression of Slug and Twist that was attendant with reexpression of epithelial characteristics. Because Slug and Twist are regulators of EMT, our results imply that wnt signals integrate cell fate decisions with differentiation and cell behavior (Fig. 5D).
In addition to identifying a molecular link between EMT, differentiation, and metastasis, these data also validate the use of the SUM1315 model of human breast cancer metastasis from the orthotopic site and the identification of genes mediating breast cancer metastasis to the lungs. Interestingly, very few of the genes identified using this model system overlapped with the lung metastasis signature previously reported (21). However, this is likely due to the significant differences in model systems and experimental designs. Previous studies used serial passaging of MDA.MB.23l cells after tail vein injection to generate subpopulations of MDA.MB.23l cells with increased ability for lung colonization. In addition, transcriptional profiling compared the in vitro cultured lung-selected clones to the in vitro cultured parental MDA.MB.231 cell line. By contrast, we used a heterogeneous population of cells and performed microarray analysis on the fresh primary tumor tissues compared with fresh lung metastases. Because it has been shown that profiling of MDA.MB.231 cells grown in culture compared with the same cells injected into the mammary fat pad of mice results in profound differences (42), the transcriptome profiling of the MDA.MB.231 subpopulations likely represent a small set of the potential genes differentially expressed in lung metastasis and poor prognosis tumors.
The inhibition of wnt signaling in SUM1315 cells affected tumor growth, thereby precluding the examination of lung metastasis in wnt-inhibited cells. However, previous studies have shown that reduction in Slug and/or Twist expression reduces lung metastasis in vivo (24, 41). Therefore, it would seem likely that cells that cannot seed a primary tumor will likely be unable to seed a secondary tumor as well. Taken together with the data presented here, this implies that wnt signaling can regulate cell-cell adhesion, and cell behaviors that are characterized by EMT through its effects on Slug and Twist but can also connect EMT with cell fate and differentiation. Hence, it will be of great importance to determine whether pharmacologic or antibody-based therapies targeting the wnt pathway will affect tumor recurrence and/or metastasis.
Supplementary Material
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Acknowledgments
Grant support: Susan G. Komen Foundation grant (T.A. DiMeo), Raymond and Beverly Sackler Foundation grant, NIH/NCI RO1CA12555 (C. Kuperwasser), and Breast Cancer Research Foundation grant (T.A. DiMeo and C. Kuperwasser). C. Kuperwasser is a Raymond and Beverly Sackler Foundation scholar.
We thank Annette Shepard-Barry at Tufts Medical Center in Histology-Special Procedures Laboratory for Histologic Staining, Ina Klebba for assistance with maintenance of the animal colony, and Nicolas Kuperwasser for critical reading of the manuscript.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–72. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hajj M, Wicha MS, ito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 5.Kang Y, He W, Tulley S, et al. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc Natl Acad Sci U S A. 2005;102:13909–13914. doi: 10.1073/pnas.0506517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weigelt B, Glas AM, Wessels LF, Witteveen AT, Peterse JL, van’t Veer LJ. Gene expression profiles of primary breast tumors maintained in distant metastases. Proc Natl Acad Sci U S A. 2003;100:15901–15905. doi: 10.1073/pnas.2634067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woelfle U, Cloos J, Sauter G, et al. Molecular signature associated with bone marrow micrometastasis in human breast cancer. Cancer Res. 2003;63:5679–5684. [PubMed] [Google Scholar]
- 8.Lu X, Kang Y. Organotropism of breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2007;12:153–162. doi: 10.1007/s10911-007-9047-3. [DOI] [PubMed] [Google Scholar]
- 9.Kang Y, Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 10.Bates RC, Mercurio AM. The epithelial-mesenchymal transition (EMT) and colorectal cancer progression. Cancer Biol Ther. 2005;4:365–370. doi: 10.4161/cbt.4.4.1655. [DOI] [PubMed] [Google Scholar]
- 11.Vincent-Salomon A, Thiery JP. Host microenvironment in breast cancer development: epithelial-mesenchymal transition in breast cancer development. Breast Cancer Res. 2003;5:101–106. doi: 10.1186/bcr578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dihlmann S, von Knebel DM. Wnt/β-catenin-pathway as a molecular target for future anti-cancer therapeutics. Int J Cancer. 2005;113:515–524. doi: 10.1002/ijc.20609. [DOI] [PubMed] [Google Scholar]
- 14.Yook JI, Li XY, Ota I, et al. A Wnt-Axin2-3β cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol. 2006;8:1398–1406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 15.Conacci-Sorrell M, Simcha I, Ben-Yedidia T, Blechman J, Savagner P, Ben-Ze’ev A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of β-catenin signaling, Slug, and MAPK. J Cell Biol. 2003;163:847–857. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howe LR, Watanabe O, Leonard J, Brown AM. Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Res. 2003;63:1906–1913. [PubMed] [Google Scholar]
- 17.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Bhat RA, Stauffer B, Komm BS, Bodine PV. Structurefunction analysis of secreted frizzled-related protein-1 for its Wnt antagonist function. J Cell Biochem. 2007;102:1519–1528. doi: 10.1002/jcb.21372. [DOI] [PubMed] [Google Scholar]
- 19.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki H, Toyota M, Carraway H, et al. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer. 2008;98:1147–1156. doi: 10.1038/sj.bjc.6604259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta GP, Perk J, Acharyya S, et al. ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc Natl Acad Sci U S A. 2007;104:19506–19511. doi: 10.1073/pnas.0709185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuperwasser C, Dessain S, Bierbaum BE, et al. A mouse model of human breast cancer metastasis to human bone. Cancer Res. 2005;65:6130–6138. doi: 10.1158/0008-5472.CAN-04-1408. [DOI] [PubMed] [Google Scholar]
- 24.Gupta PB, Kuperwasser C, Brunet JP, et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulka M, Fukuishi N, Rottem M, Mekori YA, Metcalfe DD. Mast cells, which interact with Escherichia coli, up-regulate genes associated with innate immunity and become less responsive to Fc(ε)RI-mediated activation. J Leukoc Biol. 2006;79:339–350. doi: 10.1189/jlb.1004600. [DOI] [PubMed] [Google Scholar]
- 26.Blick T, Widodo E, Hugo H, et al. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin Exp Metastasis. 2008;25:629–642. doi: 10.1007/s10585-008-9170-6. [DOI] [PubMed] [Google Scholar]
- 27.van ’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 28.Herschkowitz JI, Simin K, Weigman VJ, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray plat-forms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smid M, Wang Y, Zhang Y, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68:3108–3114. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 32.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7537. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 33.Lin SY, Xia W, Wang JC, et al. β-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci U S A. 2000;97:4262–4266. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo PK, Mehrotra J, D’Costa A, et al. Epigenetic suppression of secreted frizzled related protein 1 (SFRP1) expression in human breast cancer. Cancer Biol Ther. 2006;5:281–286. doi: 10.4161/cbt.5.3.2384. [DOI] [PubMed] [Google Scholar]
- 35.Klopocki E, Kristiansen G, Wild PJ, et al. Loss of SFRP1 is associated with breast cancer progression and poor prognosis in early stage tumors. Int J Oncol. 2004;25:641–649. [PubMed] [Google Scholar]
- 36.Dahl E, Wiesmann F, Woenckhaus M, et al. Frequent loss of SFRP1 expression in multiple human solid tumours: association with aberrant promoter methylation in renal cell carcinoma. Oncogene. 2007;26:5680–5691. doi: 10.1038/sj.onc.1210345. [DOI] [PubMed] [Google Scholar]
- 37.Shipitsin M, Campbell LL, Argani P, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Sheridan C, Kishimoto H, Fuchs RK, et al. CD44+/ Breast Cancer Res. 2006;8:R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallin J, Thuret R, Giacomello E, Faraldo MM, Thiery JP, Broders F. Cloning and characterization of three Xenopus slug promoters reveal direct regulation by Lef/β-catenin signaling. J Biol Chem. 2001;276:30350–30358. doi: 10.1074/jbc.M103167200. [DOI] [PubMed] [Google Scholar]
- 40.Taneyhill LA, Bronner-Fraser M. Dynamic alterations in gene expression after Wnt-mediated induction of avian neural crest. Mol Biol Cell. 2005;16:5283–5293. doi: 10.1091/mbc.E05-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Jessani N, Humphrey M, McDonald WH, et al. Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc Natl Acad Sci U S A. 2004;101:13756–13761. doi: 10.1073/pnas.0404727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;11:3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 44.Pawitan Y, Bjohle J, Amler L, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–R964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van de Vijver MJ, He YD, van ’ Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 46.Sotiriou C, Neo SY, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;10018:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;2:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 48.Ivshina AV, George J, Senko O, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 49.Farmer P, Bonnefoi H, Becette V, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 50.Hess KR, Anderson K, Symanns WF, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 51.Ma XJ, Wang Z, Ryan PD, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 52.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).