Abstract
Accumulation of stochastic DNA damage throughout organisms’ lifespan is thought to contribute to aging. Conversely, aging appears phenotypically reproducible and regulated through genetic pathways such as the insulin-like growth factor-1 (IGF-1) and growth hormone (GH) receptors, which are central mediators of the somatic growth axis. Here, we report that persistent DNA damage in primary cells elicits similar changes in global gene expression as those occurring in various organs of naturally aged animals. Importantly, we show that, as in aging animals, IGF-1 receptor and GH receptor expression is attenuated resulting in cellular IGF-1 resistance. This cell-autonomous attenuation is specifically induced by persistent lesions leading to RNA polymerase II stalling, in proliferating, quiescent and terminally differentiated cells, is exacerbated and prolonged in cells from progeroid mice and confers resistance to oxidative stress. Our findings suggest that DNA damage accumulation in transcribed genes in most if not all tissues, contributes to the aging-associated shift from growth to somatic maintenance that triggers stress resistance and is thought to promote longevity.
Keywords: Premature aging, longevity, somatotropic axis, IGF-1 receptor, transcription-coupled repair
Aging represents the progressive functional decline exempted from evolutionary selection as it largely occurs after reproduction and beyond the lifespan normally reached in natural habitats 1,2. Accumulation of DNA damage is considered not only a key cause of cancer but also a driving force of aging 3–6. DNA damage invariably accumulates during the lifespan of organisms despite sophisticated DNA repair systems 7–9. In accordance with random damage accumulation, a clonal population of C. elegans shows chance variation in lifespan distribution under identical environmental conditions suggesting that aging may have a strong stochastic component 10,11.
In contrast to models of stochastic damage as an underlying cause of aging, longevity is also regulated genetically 12. Some closely related species, for instance, show great diversity of lifespan, indicating that genetic alterations can determine longevity 13. Single gene mutations in the insulin-like growth factor (IGF-1) pathway extend lifespan in C. elegans and Drosophila 14,15. In mammals, growth hormone (GH) mediated GH receptor (GHR) signaling in various cell types induces secretion of IGF-1, which regulates somatic growth through activation of IGF-1 receptor (IGF-1R) signaling, together comprising the somatotropic axis 16. Mice with reduced IGF-1 signaling either through IGF-1R heterozygocity, Ghr knockout, Klotho over-expression, or reduced GH levels due to pituitary dysfunction (Snell and Ames mice) show extended lifespan 17–20. Thus, there are two principal components of aging: stochastic damage accumulation and genetic pathways that regulate longevity. However, it is unknown how stochastic damage events are linked to genes that regulate longevity.
The effect of DNA damage on organismal aging becomes apparent when DNA damage rapidly accumulates early in life due to defects in genome maintenance leading to segmental progeroid (premature aging-like) conditions 3,6,21–23. Prime examples are nucleotide excision repair (NER) defects 6. NER removes a wide range of helix-distorting lesions, including UV-induced damages, either by global-genome (GG) NER that repairs lesions throughout the genome, or transcription-coupled repair (TCR), which removes damage in actively transcribed genes. Defects in GG-NER lead to skin cancer prone xeroderma pigmentosum (XP), whereas defects in TCR lead to the progeroid conditions Cockayne syndrome (CS), trichothiodystrophy (TTD), and XPF-ERCC1 progeria (XFE), which is additionally linked to interstrand crosslink (ICL) repair21,24. Mouse mutants with engineered mutations in both Csb and Xpa or in Ercc1, mimic the repair deficiencies of CS and XFE patients respectively. Recently, we showed that Csbm/m/Xpa−/− and Ercc1−/− progeroid mice, similarly to wt mice chronically treated with DNA-damaging agents, display downregulation of genes involved in IGF-1/GH signaling 21,25, which normally is associated with lifespan extension 26. It has been hypothesized that somatotropic attenuation becomes progressively installed with advancing age due to gradually accumulating damage, and delays onset of age-related pathology and extends lifespan 21,25. It is unknown how suppression of the somatotropic axis is triggered and whether somatic growth attenuation is a direct response to specific DNA damages or an indirect consequence of damage-driven tissue degeneration. Here, we present evidence that accumulation of transcription-blocking lesions constitute a primary cause for attenuation of the somatotropic axis with age and thus link DNA damage to genes that regulate longevity.
Results
DNA damage impinges on pathways associated with longevity regulation
Inherited defects in NER provide clear-cut links with cancer as well as premature aging 6. We thus examined whether UV-DNA damage induces gene expression responses that normally occur in old age. To this aim primary mouse dermal fibroblasts (MDFs) from selected NER mouse mutants with severe (Csbm/m/Xpa−/−), mild (Csbm/m) or no progeria (Xpa−/−) and wild type (wt) littermates at 15 days of age were exposed at early passage (<6) to very low (0.6 J/m2) and to moderate (4J/m2) UV doses. Sensitivity of NER-deficient cells exposed to 0.6 J/m2 is equivalent to wt cells after 4J/m2 (Supplementary Figure 1). These UV doses give rise to a maximum of 1 lesion in 300kb or 50kb at 0.6 J/m2 and at 4J/m2, respectively 27. Consistent with the presence of randomly distributed lesions in only a small fraction of genes, we did not detect a global transcriptional repression 6 hours subsequent to UV treatment, as an equal number of genes were up-and down-regulated (Supplementary Tables 1–5). A large fraction of UV-induced genes contained relatively long open reading frames, suggesting that the expression changes represent a trans-response rather than a cis effect of transcriptional blockage. To validate the microarray results, we quantitatively confirmed statistically significant gene expression changes greater than ±1.2 fold by quantitative real-time PCR (Supplementary Figure 2A).
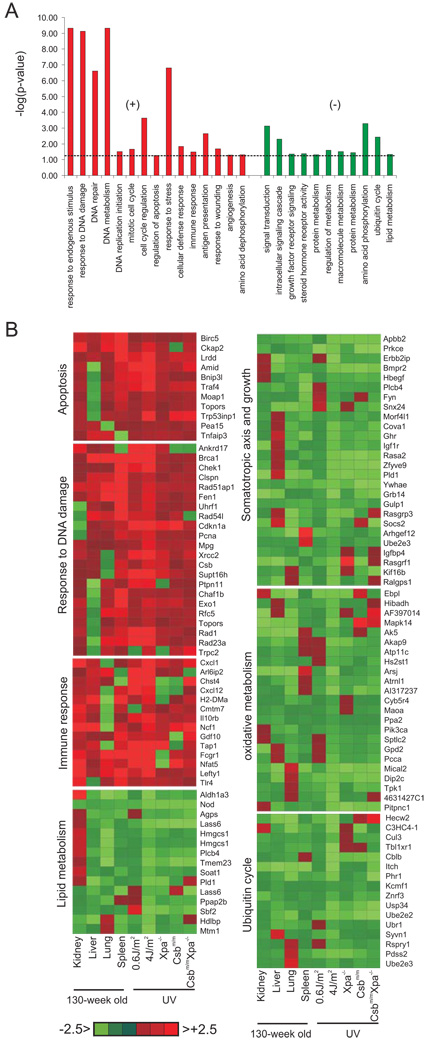
We aligned the significant expression changes of UV-exposed cells to those of the liver, kidney, spleen and lung of naturally aged mice and asked for biological processes containing a significantly disproportionate number of responsive genes relative to the rest of the genome (Methods), thus deriving significantly over-represented biological processes that are affected in response to UV and in aging (Figure 1A). This approach revealed upregulation of genes associated with response to DNA damage, apoptosis, stress, immune and defense as well as a suppression of genes associated with signal transduction, the IGF-1/GH axis and additional growth factor signaling, oxidative metabolism, protein and lipid metabolism, and the ubiquitin cycle (Figure 1B and Supplementary Table 6). We conclude that DNA damage induces a gene expression response that is also seen in natural aging. Attenuation of the expression of genes associated with growth and oxidative metabolism as well as the upregulation of genes involved in genome maintenance, stress and defense responses was previously observed in mice that show extended longevity 28–31. To examine whether genetic programs that extend longevity might be triggered in response to DNA damage, we next asked whether DNA damage might act on central regulators of longevity.
Figure 1. Shared biological processes upon induction of DNA damage in vitro and aging in vivo.
A) List of biological processes that are significantly over-represented upon induction of DNA damage and during aging (left panel: upregulated; right panel: downregulated). B) Expression profiles of genes associated with the significantly over-represented biological processes upon UV treatment in vitro and in aging tissues. For each gene, the average expression value of UV-exposed primary MDFs (4 independent MDF lines were used for each genotype (n=4)) and the indicated tissue from 130-week old mice is compared to that of the corresponding non-treated cells or tissue of 13-week old young mice (average of 3 old vs. 3 young mice (n=3)), respectively. Deeper red or green colors indicate positive and negative fold changes respectively.
Cell-autonomous repression of IGF-1R and GHR in response to UV-induced DNA damage
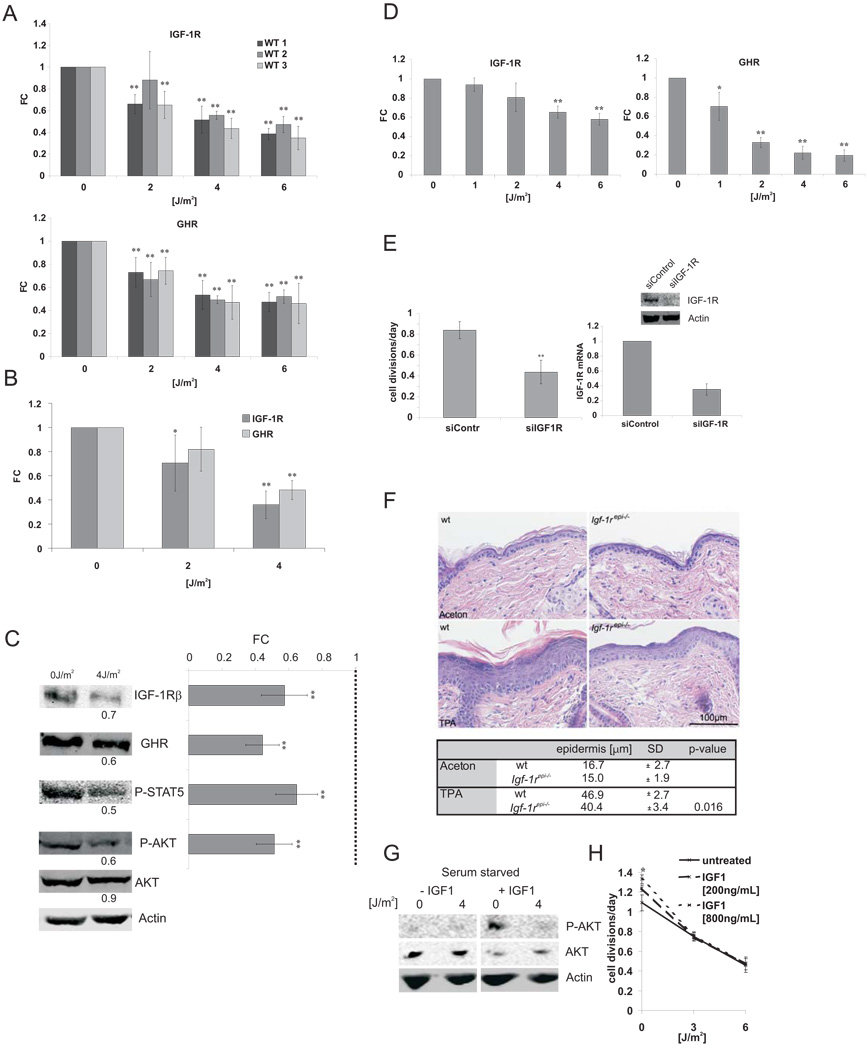
To investigate the causal basis of age-related somatotropic attenuation, we focused on the cellular regulation of IGF-1R and GHR as both genes are central mediators of the somatotropic axis, their attenuation leads to lifespan extension 32, and their expression is down-regulated in both naturally aged and TCR-deficient progeroid mice (Figure 1B) 21,25. We found IGF-1R and GHR expression is suppressed within 6 hours after UV exposure in a dose-dependent manner in primary MDFs and primary chondrocytes from adult wt mice (Figure 2A, B). Accordingly, we observed lower endogenous IGF-1R and GHR protein levels and reduced phosphorylation of their respective targets AKT and STAT5 after UV treatment (Figure 2C). Similarly, we also observed a UV-dependent attenuation of IGF-1R and GHR in human fibroblasts (Figure 2D).
Figure 2. IGF-1R and GHR are repressed upon persistent DNA damage and leads to IGF-1 resistance.
A) Three independent primary mouse dermal fibroblast cell lines (WT1–3; n=3)) at early passage were treated with UV (UV-C, 254 nm) and RNA samples were taken 6 hrs post treatment. IGF-1R and GHR qPCRs were done in duplicate and normalized to gamma-tubulin (gTUB) and beta-2-microglobulin (B2M). B) Primary chondrocytes were treated similarly and IGF-1R and GHR expression levels normalized to B2M, gTUB, and Hprt (average of two independent chondrocyte lines shown) and (C) IGF-1R and GHR protein and phosphorylation levels of STAT5 and AKT were quantified 12 h after UV treatment using fluorescently labeled antibodies and normalized to Actin levels (left panels: representative experiment; right panel: average of three independent experiments, n=3). D) Two independent human fibroblast lines (C5RO and C3RO) were treated with indicated doses of UV. IGF-1R and GHR expression levels were determined 6 hrs post treatment and normalized to Gapdh and B2M. E) IGF-1R down-regulation is sufficient to reduce cell growth. U2OS cells were counted 96 hours after transfection with either non-targeting siRNA oligos or siRNA oligos targeting IGF-1R (daily division rate given, n=4; upper right panel IGF-1R protein levels, lower right panel IGF-1R mRNA levels (qPCR)). F) IGF-1R ablation antagonizes hyperplasia. 8 – 10 weeks old Igf-1repi−/− and control mice were shaved and after 24 hours, treated three times every 48 hours with 10nm TPA/330µl or solvent alone (aceton). Upper panel: H&E section showing epidermal hyperplasia following TPA treatments. Lower panel: Quantification of hyperproliferative response (TPA-treated: n = 3–5, aceton treated: n = 2; two-tailed t-test). UV induces IGF-1 resistance: (G) U2OS cells were serum starved for 2 hours starting at 12 hours after UV treatment and treated with 100ng/mL IGF1 for 10 minutes. H) Medium was supplemented with IGF-1 after UV treatment and cells were counted 48 hours after treatment. (FC = fold change compared to untreated controls; *=p<0.05, **=p<0.01, two-tailed t-test, error bars = SD, n=4)
We next investigated the biological consequences of IGF-1R and GHR attenuation. IGF-1R and GHR inhibition by RNA interference was sufficient to reduce the rate of cell divisions (Figure 2E; data not shown). To validate the significance of IGF-1R for proliferation in vivo 33, we treated wt and epidermal Igf-1r deficient animals (Igf-1repi−/−) 34 with tumor promoting 12-O-tetradecanoylphorbol 13-acetate (TPA) and observed significantly reduced hyperplasia in Igf-1repi−/− mice (Figure 2F). Next, we evaluated the cellular response to IGF-1. Whereas IGF-1 administration induced IGF-1 signaling as assayed by AKT phosphorylation, UV exposure rendered cells refractory to IGF-1 stimulation (Figure 2G). Accordingly, IGF-1 administration stimulated cell growth, whereas UV exposure suppressed the mitogenic effect of IGF-1, indicating that UV damage induces IGF-1 resistance (Figure 2H).
To exclude a potential negative feedback loop, we blocked IGF-1R or GHR signaling with inhibitors of PI3-kinase, MAPK or JAK 35, neither of which had any effect on the UV-induced repression of IGF-1R or GHR suggesting that their repression is not an effect of a potential negative feedback loop of their own signal transduction (Supplementary Figure 2B, C).
UV-induced DNA damage leads to oxidative stress resistance associated with extended longevity
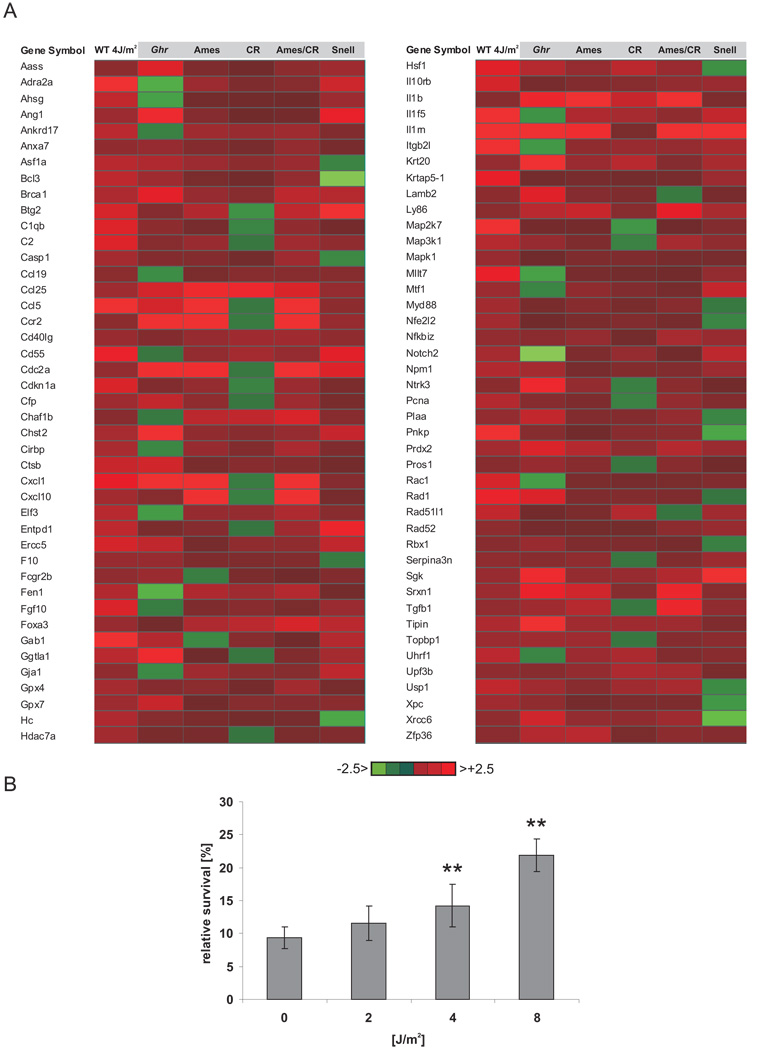
Attenuation of the somatotropic axis has been shown to not only lead to lifespan extension but also enhanced resistance to oxidative stress, in C. elegans, Drosophila, and mice suggesting that increased ability to withstand stress might underlie lifespan extension 11,17,36,37. To investigate whether DNA damage induces stress responses similar to those in long-lived mice, we compared the expression changes of UV-exposed primary MDFs with independent liver microarray datasets of mice that show lifespan extension either due to genetic alteration (Ames and Snell, Ghr−/−) 28–30, calorie restriction (CR) 28 or a combination (Ames-CR) 28. We derived all genes that were upregulated in UV-treated cells and at least 4 out of 5 long-lived mouse models and asked which gene ontology (GO) terms are significantly over-represented, yielding common significantly over-represented biological processes ranked by their relative enrichment score (see Methods) (Supplementary Table 7). We found profound similarities in the expression of genes associated with the response to stress (GO:0006950) between UV-exposed cells and longevity mouse models known to be resistant to oxidative stress (Figure 3A). To examine whether UV-induced stress responses might confer enhanced resistance to oxidative stress, we treated cells with increasing doses of UV and 12 hours later exposed them to hydrogen peroxide. Intriguingly, UV treatment prior to exposure to hydrogen peroxide resulted in a dose-dependent increase in cellular survival upon oxidative stress (Figure 3B). Thus, UV damage leads to the induction of a protective oxidative stress response normally seen in somatotropic mutants with extended longevity.
Figure 3. UV response induces oxidative stress resistance.
A) Common stress responses are induced in response to UV damage in primary cells and in pituitary dwarf, Ghr knockout and calorie restricted mice with extended longevity. B) U2OS cells were treated with indicated UV doses and 12h post radiation treated with 100µM hydrogen peroxide. Relative survival compared to non-hydrogen peroxide treated cells (*=p<0.05, **=p<0.01, two-tailed t-test, error bars = SD, n=4).
DNA damage induces IGF-1R/GHR attenuation in dividing as well as in quiescent and terminally differentiated cells
To investigate how DNA damage leads to somatotropic attenuation, we sought to determine the initial events triggering stress resistance and longevity assurance responses. We first asked whether DNA damage checkpoint activation might be required. ATM deficient (AT2RO) and ATR-defective (Seckel) fibroblasts 38, and simultaneous inactivation/inhibition of both ATM and ATR, as well as p53−/− primary MDFs showed a similar attenuation of IGF-1R and GHR transcriptional levels upon UV exposure as wt cells (Supplementary Figure 3A–D) indicating that canonical DNA damage checkpoint genes are not affecting IGF-1R and GHR transcript levels. In agreement, a p53 deletion neither rescues the progeroid phenotypes of the Csbm/m/Xpa−/− nor the Ercc1−/− mice 25,39.
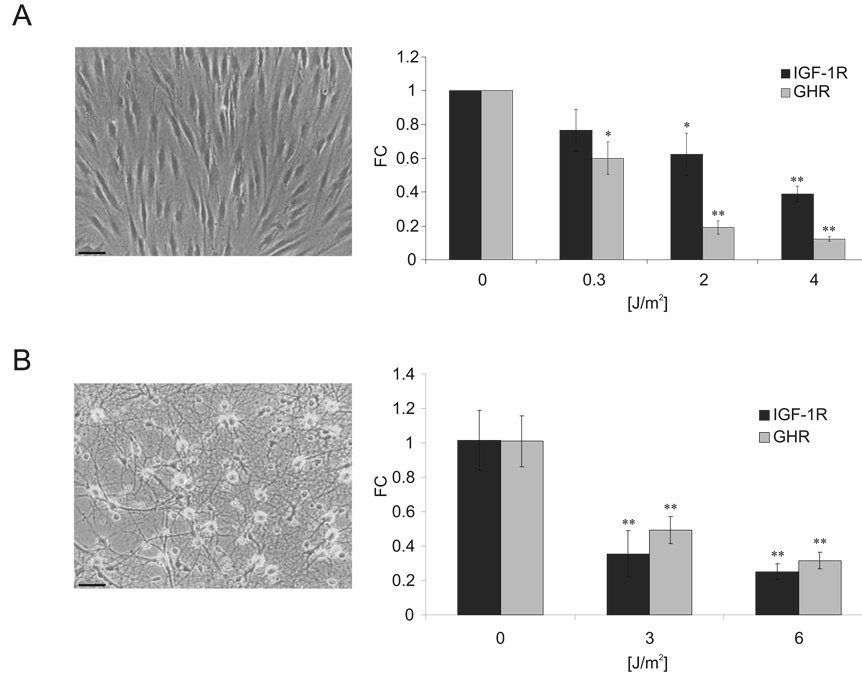
As DNA damage checkpoints are activated in response to UV-induced replication fork stalling 40 but were dispensable for IGF-1R and GHR attenuation, we asked whether this attenuation might be independent of DNA damage-induced perturbations in DNA replication or cell cycle progression and thus examined post-replicative cells. UV-damage led to attenuation of IGF-1R and GHR expression in post-replicative quiescent fibroblasts and terminally differentiated primary neurons (Figure 4A, B). We also find UV-induced repression of other growth-associated genes such as IGF-1, FGF1, EGFR, and PDGF in quiescent cells and neurons, further extending the growth attenuating response triggered by UV damage (Supplementary Figure 4A, B). Thus, UV damage induces a rapid dose-dependent attenuation of IGF-1R/GHR in proliferating as well as quiescent and terminally differentiated cells.
Figure 4. UV irradiation leads to IGF-1R and GHR attenuation in quiescent and terminally differentiated cells.
A) Quiescent C5RO primary human fibroblasts were treated 6 days after transfer to 1% FCS medium and samples taken 6hrs post UV treatment. IGF-1R and GHR expression levels were normalized to Gapdh and gTUB. B) Primary rat neurons were treated with UV and IGF-1R and GHR expression was normalized to Hprt and Gapdh (FC = fold change compared to untreated controls; *=p<0.05, **=p<0.01, two-tailed t-test, error bars = SD; n=4; scale bar 25µm).
Progeroid NER defects lead to exacerbated and prolonged attenuation of IGF-1R and GHR
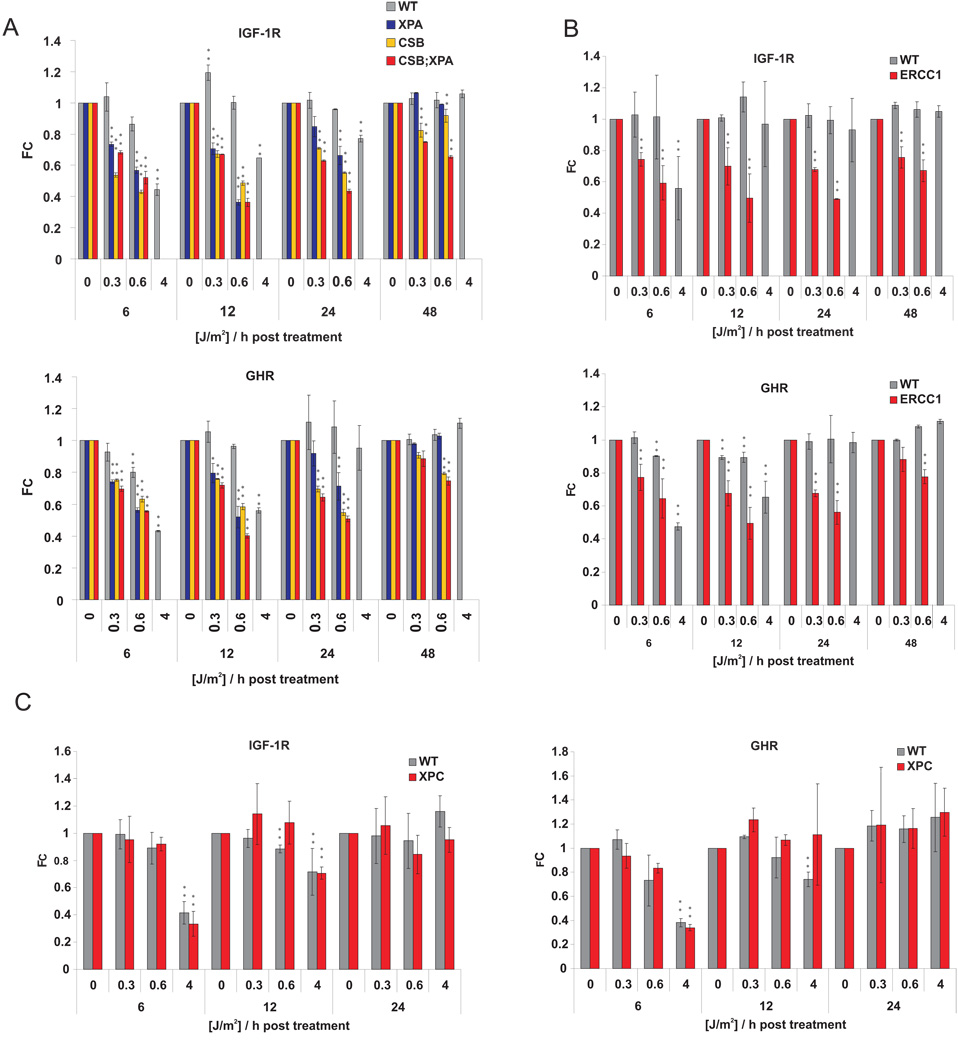
To investigate the genetic components of DNA damage-induced attenuation of IGF-1R/GHR, we investigated this response in primary cells of TCR-deficient progeroid mice. Whereas wt cells showed a transient repression of IGF-1R and GHR (6–12h after 4J/m2), DNA repair-deficient Csbm/mXpa−/−, Csbm/m and Xpa−/− cells showed an exacerbated repression already at very low UV doses (Figure 5A; Supplementary Tables 1–5). Xpa−/− cells fully restored IGF-1R and GHR transcript levels within 48 hours, whereas Csbm/mXpa−/− cells, and Csbm/m cells to a lesser extent, continuously attenuated those genes (Figure 5A). To determine whether prolonged attenuation of IGF-1R and GHR is a general feature of DNA repair-deficient progeroid mutants, we tested primary MDFs from progeroid Ercc1−/− animals. Similarly to Csbm/mXpa−/− cells, Ercc1−/− cells showed an exacerbated and prolonged repression of IGF-1R and GHR in response to low doses of UV as compared to cells derived from wt littermates (Figure 5B). We next evaluated the UV-response in primary MDFs from non-progeroid GG-NER deficient Xpc−/− animals. Xpc−/− cells showed similar sensitivity and recovery of the IGF-1R and GHR repression as wt cells, suggesting that persistent lesions outside of actively transcribed genes do not induce IGF-1R/GHR attenuation (Figure 5C). These observations establish that, in contrast to cancer-prone GG-NER defects, progeroid TCR defects lead to prolonged attenuation of IGF-1R and GHR.
Figure 5. Prolonged IGF-1R and GHR repression in cells from NER deficient progeroid mice.
(A), Csbm/m/Xpa−/−, Csbm/m and Xpa−/−, (B) Ercc1−/−, (C) Xpc−/− and wt littermate control primary MDFs at early passage were treated with indicated UV doses and samples taken at indicated time points after treatment. IGF-1R and GHR expression levels were evaluated by qPCR and normalized to gTUB and B2M. (FC = fold change compared to untreated controls; *=p<0.05, **=p<0.01, two-tailed t-test, error bars = SD, n=4).
Persistent but not transient DNA damage leads to IGF-1R/GHR attenuation
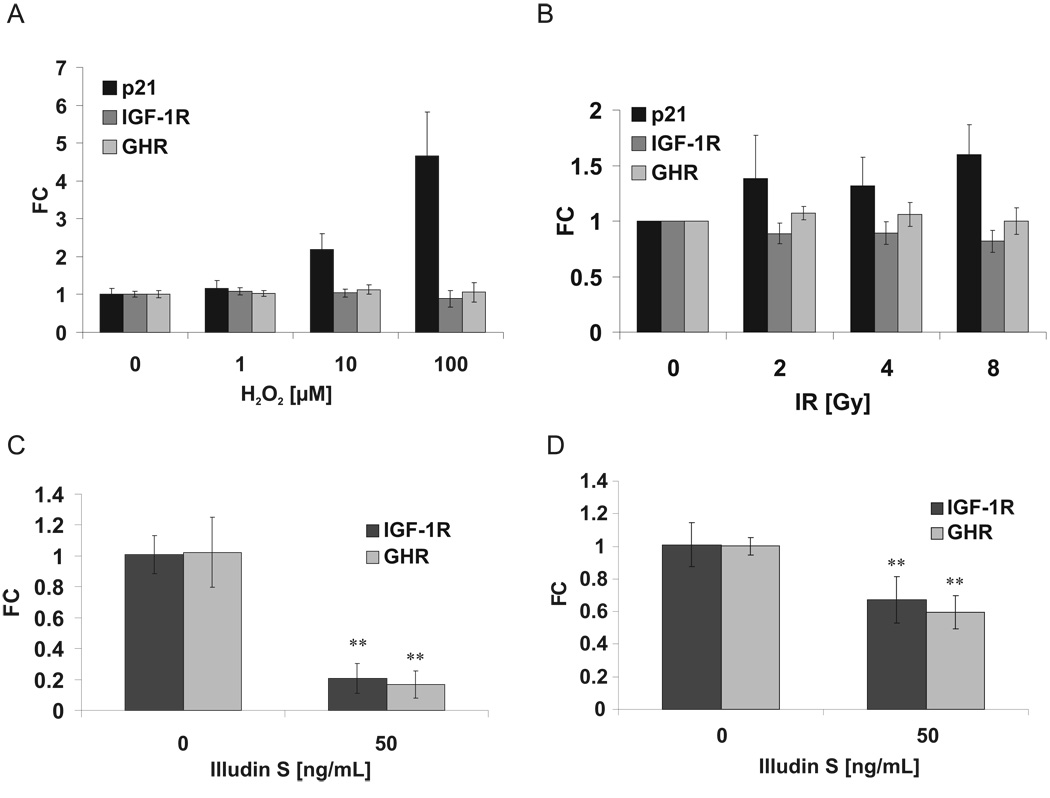
To examine the specificity of UV-induced attenuation of IGF-1R and GHR expression, we investigated the effect of other DNA-damaging agents. H2O2 induces oxidative DNA damage, primarily resulting in base modifications and transient accumulation of single strand breaks; ionizing irradiation (IR) induces, in addition to oxidative damage, single and double strand breaks. Despite induction of p21, indicative of DNA damage checkpoint activation, none of these agents, even over a wide dose range, induced significant IGF-1R/GHR attenuation. (Figure 6A, B). To comprehensively investigate the gene expression response to H2O2 treatment, we employed genome-wide expression analyses and found no attenuation of genes involved in somatotropic and growth signaling (Supplementary Table 8).
Figure 6. Attenuation of IGF1-R and GHR expression in response to illudin S but not to oxidative damage and ionizing irradiation.
Primary MDFs were treated with hydrogen peroxide (A) or ionizing radiation (B) and IGF-1R and GHR expression levels were evaluated after 6 h by qPCR and normalized to gTUB and B2M. p21 expression was included as control for DNA damage response. Illudin S treatment leads to IGF-1R and GHR attenuation in quiescent human fibroblasts (C) and rat neurons (D), normalized to Gapdh and gTUB (C) or Hprt and Gapdh (D) (FC = fold change compared to untreated controls; **=p<0.01, two-tailed t-test, error bars = SD; n=4).
A marked difference between UV-lesions and those induced by H2O2 and IR is the persistence of damage. Most oxidative lesions are rapidly repaired by base excision repair (BER), most single strand DNA breaks are sealed by DNA ligase within seconds, and the vast majority of double strand breaks are joined through non-homologous end-joining within less than 30 minutes 41–43. In contrast, UV-induced cyclobutane pyrimidine dimers (CPDs) persist for days and can even be carried through replication 44–46. Only when CPDs lead to stalling of transcriptional elongation, they are eliminated TCR 47,48. CPDs in actively transcribed genes have a kinetic half-life of 6–8 hours 43,49. To test whether IGF-1R and GHR attenuation constitutes a specific response to persistent transcription-blocking DNA lesions, we used illudin S, which in post-replicative cells specifically induces transcription-blocking lesions 50. Illudin S treatment in quiescent fibroblasts and neurons induced attenuation of IGF-1R and GHR (Figure 6C, D). To further probe for specificity, we also tested rare mitomycin C-induced DNA crosslinks that mostly interfere with replication and mutagenic 4-Nitroquinoline 1-oxide (4-NQO) induced NER and BER lesions, neither of which resulted in IGF-1R/GHR attenuation (Supplementary Figure 4C, D). To exclude that inhibition of transcription per se might elicit this attenuation, we treated cells for 6 hours with transcriptional inhibitors α-amanitin or actinomycin D. Although both treatments strongly reduced RNA-synthesis, we did not observe specific IGF-1R or GHR repression (Supplementary Figure 4E).
Persistent transcription-blocking lesions are the primary instigators of IGF-1R/GHR attenuation and oxidative stress resistance
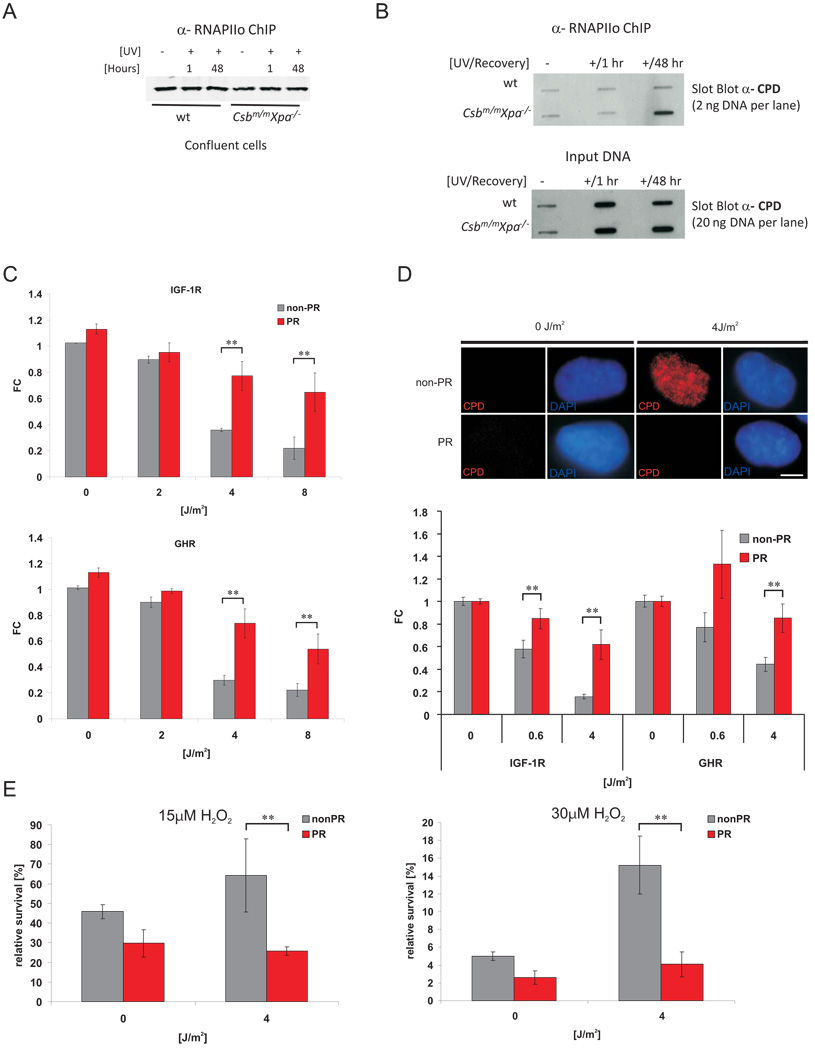
To establish a mechanistic link between persistent transcription-blocking lesions and IGF-1R/GHR attenuation, we investigated whether RNA polymerase II (RNAPII) arrested on persistent CPD lesions represents the critical signal, using the prolonged IGF-1R/GHR attenuation in cells from progeroid CS mouse mutants (Csbm/mXpa−/−; Figure 5A) as a model. We isolated elongating RNAPII complexes (RNAPIIo, Ser 2 phosphorylated) from chromatin of UV-irradiated chrondrocytes of wt and progeroid Csbm/mXpa−/− mice employing in vivo crosslinking and chromatin immunoprecipitation (ChIP) 51 and analyzed the co-precipitated chromatin for CPDs. Equal amounts of RNAPIIo were chromatin-immunoprecipitated from UV-irradiated and mock-treated wt and Csbm/mXpa−/− cells (Figure 7A). Slot blot analysis of the co-precipitated DNA revealed clear enrichment of CPDs in chromatin of Csbm/mXpa−/− but not wt cells 48 hours after UV (Figure 7B), indicating that RNAPIIo was arrested at the site of persistent transcription-blocking lesions in Csbm/mXpa−/− cells. A large number of CPD lesions still remained in both genotypes (Figure 7B, input DNA), further corroborating that persistent lesions outside of active genes are not inducing IGF-1R/GHR attenuation. Thus, progeroid TCR-deficiency leads to persistent RNAPIIo stalling at CPDs and prolongation of UV-induced IGF-1R/GHR attenuation (Figure 5A).
Figure 7. Repair of persistent CPD lesions alleviates IGF-1R and GHR repression and oxidative stress resistance.
A) RNAPIIo protein levels in RNAPIIo specific ChIP using chromatin of crosslinked UV- and non-irradiated confluent chondrocytes (as indicated). B) Slot blot analysis of different amounts of RNAPIIo co-ChIPed DNA (top panel) and input crosslinked DNA (lower panel) spotted at the same membrane using a CPD specific antibody (representative experiment shown). CPD and 6-4PP photolyase transgenic MEFs (C) or a CPD photolyase transgenic CS1AN human CS cell line (D) were UV treated and then either photoreactivated (PR) for 1h or kept in the dark (non-PR). IGF-1R and GHR expression levels were determined 6h post treatment and normalized to Gapdh, gTUB and Hprt. Removal of CPDs was assessed with an anti-CPD antibody (D, upper panel; scale bar 10µm) showing presence of CPDs in CSB cells and complete CPD repair after PR (6h post treatment, 1h PR). E) CS1AN cells carrying a CPD-photolyase trangene were treated with UV and either for 1h PR or left in the dark. 12h post UV exposure cells were treated with indicated doses of hydrogen peroxide. Relative survival was determined by comparing cell numbers after 72h to non-H2O2 treated controls (FC = fold change compared to untreated controls; *=p<0.05, **=p<0.01, two-tailed t-test, error bars = SD, n=4).
To assess whether persistent transcription-blocking lesions were the primary instigator, we experimentally removed persistent CPDs. We employed CPD and 6-4PP photolyases (PL), which capture energy of visible light to rapidly split the CPDs and 6-4PPs, respectively (Photo-reactivation (PR)) 52. The UV-induced attenuation of IGF-1R/GHR was largely alleviated in CPD-PL and 6-4PP-PL expressing mouse embryonic fibroblasts upon PR (Figure 7C). To further investigate a link between persistent CPDs in actively transcribed genes and IGF-1R/GHR attenuation, we used TCR-defective Cockayne syndrome (CS) patient fibroblasts that stably expressed a CPD-PL transgene. Specific removal of CPD lesions upon PR alleviated UV-induced IGF-1R/GHR attenuation (Figure 7D). Finally, we examined whether persistent CPDs in actively transcribed genes were the primary instigators of UV-induced oxidative stress resistance. We treated CPD-PL transgenic CS cells with UV and either photoreactived or left the cells in the dark prior to H2O2 treatment. Strikingly, selective CPD repair reversed UV-induced oxidative stress resistance (Figure 7E). We thus conclude that IGF-1R and GHR attenuation and oxidative stress resistance are a consequence of persistent transcription-blocking lesions. Furthermore, the rapid effect of photolyase activation confirms that UV-induced DNA damage as opposed to UV-induced damage to proteins and RNA causes IGF-1R/GHR attenuation and cellular resistance to oxidative stress.
Discussion
We have demonstrated that low doses of UV irradiation lead to changes in gene expression in vitro similar to those in various tissues of naturally aging animals, suggesting that many age-related gene expression changes result from cellular responses to DNA damage accumulation. Gene expression profiles of UV-treated primary cells show induction of tumor suppressive pathways, including apoptosis and cell cycle arrest, and repression of the somatotropic growth axis, which is associated with extended longevity. IGF-1R and GHR, central mediators of the somatotropic axis, are attenuated in response to persistent transcription-blocking lesions such as CPDs and illudin S lesions. In primary cells from TCR-deficient progeroid mice, IGF-1R/GHR suppression is prolonged, suggesting that even low levels of persistent DNA damage in transcribed regions of the genome can result in sustained somatotropic attenuation. However, lesions persisting outside of actively transcribed genes do not result in somatotropic attenuation (Figure 8). As a consequence of this attenuation, cells become resistant to IGF-1. IGF-1 resistance has been observed in natural aging and with the onset of neurodegenerative disorders 53–55 and is supported by the attenuation of somatotropic gene expression in various organs of naturally aged mice (Figure 1). Furthermore, we show that persist ent DNA damage leads to cellular resistance to oxidative stress similar to long-lived mouse mutants. Thus, persistent DNA lesions induce functional consequences of somatotropic attenuation.
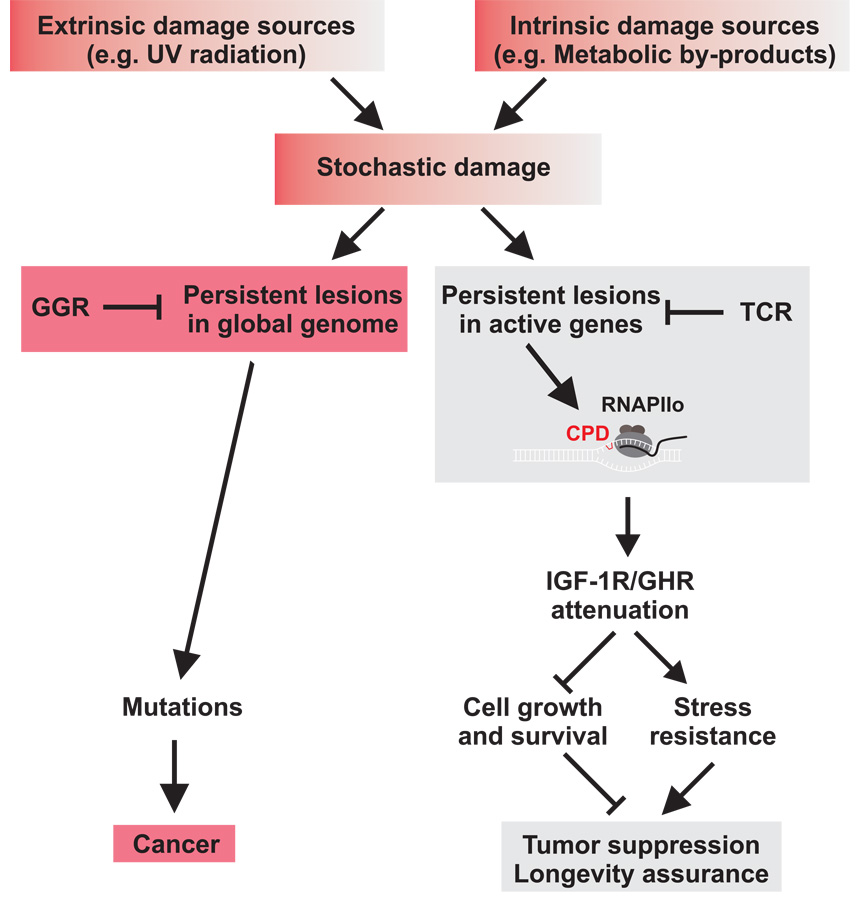
Figure 8. Stochastic DNA damage resulting from extrinsic and intrinsic sources contributes to cancer and aging.
Transcription-coupled sensing of persistent damages acts in differentiated as well as dividing cells and represses the somatotropic axis. Reduced IGF-1R and GHR activity acts tumor suppressive by reducing cellular survival and growth and induces a longevity assuring stress response. Excessive damage, however, leads to loss of cellular reserves and to aging (gray). TCR defects lead to rapid accumulation of damage in active genes resulting in progeroid syndromes. GGR defects lead to failures to repair damages in other regions of the genome, which, in contrast to transcription-coupled somatotropic attenuation, gives rise to mutations and consequently to cancer (red).
What is the relevance of UV-lesions to organismal aging? We specifically observed repression of IGF-1R/GHR in the presence of persistent CPDs and upon exposure to illudin S, which also induces persistent transcription-blocking lesions 50, but not in response to IR or H2O2 both of which induce predominantly transient lesions. In contrast to acute in vitro treatments, oxidative lesions and double-strand breaks might also accumulate over time and persist in aging organisms 7–9. Similarly to persistent CPDs, some oxidative lesions can also lead to stalling of RNAPII when engineered in transcribed DNA 56. A host of metabolic by-products and chemicals can induce many types of DNA lesions some of which also induce helix distortions similar to CPDs. Recently, neurodegeneration in XP patients has been associated with a class of cyclopurines 57, which may be part of the relevant endogenous lesions in progeroid syndromes and in natural aging. UV-induced CPDs represent a unique in vitro system for persistent damage. The remarkable similarities between the cellular response to CPDs and gene expression changes in aging suggest that CPDs mimic various forms of genotoxic insults that accumulate in the course of aging.
What is the significance of attenuating the somatotropic axis in response to distinct DNA damages? Somatotropic attenuation, as in IGF-1R or GHR mutant mice, or in mice with pituitary deficiencies, is associated with extended longevity and reduced tumor growth 17,19,32. Calorie restriction causes down-regulation of the IGF-1 pathway and lifespan extension in worms, flies and mammals, whereas progeroid DNA repair deficiencies lead to somatotropic attenuation but a dramatic shortening of lifespan 21,25. Somatotropic attenuation in the presence of persistent damage, such as in old age or in repair deficiencies, might be an attempt to survive under adverse conditions that result from excessive DNA damage. Beneficial effects of such a response are apparent not only in somatotropic mutants and upon calorie restriction but even in some segmental progerias. For instance, TTD mice are protected against cancer and exhibit delayed aging in a minority of organs despite showing progeroid symptoms in many tissues 58. In contrast to the response to lesions in active genes, non-cytotoxic CPD-type damages in the global genome do not lead to somatotropic attenuation, as demonstrated in Xpc-deficient cells. Accordingly, defects in GG-NER lead to cancer susceptibility but hardly to any premature aging 6. It is noteworthy that transcription-blocking agents such as the illudin S derivative irofulven are currently under clinical trials for treatment of refractory and relapsed tumors 59. We propose that cellular attenuation of somatotropic gene expression might contribute to tumor suppression via IGF-1R attenuation, but also by non-autonomous effects through attenuated GHR mediated IGF-1 production as well as reduced expression of growth factors including IGF-1, FGF1 and PDGF. A number of IGF-1R inhibitors are currently undergoing clinical trials for cancer treatment 60. Taken together, somatotropic attenuation may thus extend organismal survival and hence longevity through a shift from growth to somatic preservation and maintenance and suppression of tumorigenesis. Intriguingly, we have demonstrated that persistent DNA damage induces resistance to oxidative stress, which has been linked to extended longevity in worms, flies, and mammals 11,36,61. The molecular mechanisms through which oxidative stress resistance leads to lifespan extension are yet to be identified 62 as for instance enhanced susceptibility to oxidative stress in various cellular compartments has different effects on lifespan in various organisms 63. IGF-1R and GHR attenuation in vivo has been shown to be sufficient for stress resistance and extended longevity 64. However, in response to persistent DNA damage we observe not only IGF-1R and GHR attenuation but also induction of stress resistance genes that is reminiscent of long-lived and calorie restricted mice. These observations suggest that IGF-1R and GHR attenuation may not be the only mediator of longevity assurance but might rather be part of a larger cellular response program that includes growth suppression and induction of stress resistance. Such beneficial effects of low doses of insult have been described as hormesis 65. Our results suggest that transcription-blocking lesions represent a mechanistic basis for hormetic effects upon persistent DNA damage.
How and why does persistent damage in active genes lead to somatotropic attenuation? TCR acts not only in dividing but also in differentiated cells, which only need to maintain the integrity of expressed genes 66. RNAPII transcription has been suggested to function as dosimeter of DNA damage recognition in post-replicative cells 67. We identified stalled RNAPIIo as a primary signal leading to somatotropic attenuation. According to the disposable soma theory of aging, organisms need to optimally allocate their resources between maintenance of their soma and other biological functions, in order to maximize their Darwinian fitness 68. Transcription-coupled damage sensing might thus constitute a mechanism through which organisms orchestrate the reallocation of their resources between growth and somatic preservation amid increasing DNA damage loads with age. DNA damage responses that originate from transcription-coupled damage sensing not only in proliferating but also in terminally differentiated cells are still unknown and likely act through novel DNA damage response pathways. Here, we reveal that persistent transcription-blocking lesions act as instigators of somatotropic attenuation and enhanced oxidative stress resistance. Unlike checkpoint mechanisms that induce cell cycle arrest and apoptosis in response to acute DNA damage, transcription-coupled damage sensing of already low doses of damage might be more suitable for long-term adjustments such as somatic growth attenuation in aging. Thus, transcription-coupled damage sensing links stochastic damage events to genes that determine longevity.
Methods
Animals, Cell culture and treatments
All mice were inbred, isogenic C57/Bl6 genetic background except for Ercc1−/− (uniform F1 mixed C57/Bl6 and FVB background) and housed at the Animal Resource Center (Erasmus University Medical Center) and RIVM (Bilthoven). Both institutions operate in compliance with the “Animal Welfare Act” of the Dutch government, using the “Guide for the Care and Use of Laboratory Animals” as its standard. As required by Dutch law, formal permission to generate and use genetically modified animals was obtained from the responsible local and national authorities. All animal studies were approved by an independent Animal Ethical Committee (Dutch equivalent of the IACUC). Primary MDFs and chondrocytes were derived from the tail and the ear of the mice, respectively, cut and incubated with 1.6mg/ml collagenase II overnight, filtered, and cultured at 3% oxygen in DMEM/Hams F10 medium, 20% fetal calf serum (FCS) and antibiotics. Human fibroblasts were cultured in Hams F10 medium, 15% FCS and antibiotics. For UV-treatments 250’000 cells were seeded in a 6 cm culture dish. For quiescent cells, confluent cells were shifted to Hams F10 medium containing 1% FCS and antibiotics for 6 days. Photoreactivation was performed as described in 45. CPD lesions were visualized with anti-CPD antibodies as described 44. For H2O2 experiments tert-Butyl hydroperoxide solution (Sigma) was used, for IR a Caesium-137 source, Illudin S treatments as previously described 50. Primary hippocampal cultures were prepared from embryonic day 18 rat brains as described 69.
Protein and protein phosphorylation levels were quantified using Li-cor odyssey fluorescence technology with antibodies against IGF-1R (Santa Cruz sc-713), GHR (Sigma G8919), P-STAT5 (Upstate mouse monoclonal 8-5-2), P-AKT (Cell Signalling 9271), AKT (Cell Signalling 9272), Actin (Chemicon MAB1501R), RNAPIIo (Abcam H5). IGF1 was purchased from Sigma. ATM was inhibited using KU-55933 kindly provided by Graeme Smith of Kudos Pharmaceuticals.
siRNA transfection were performed using Lullaby reagents (Oz Biosciences) with siRNA oligos (Dharmacon) targeting CHK1: UCG UGA GCG UUU GUU GAA C, ATR: GCC GCA UGA GCA CAC CGU C and IGF-1R: GGA AGC ACC CUU UAA GAA U, GGA CUC AGU ACG CCG UUU A, AAA UAC GGA UCA CAA GUU G, AGU GAG AUC UUG UAC AUU C and non-targeting UGG UUU ACA UGU CGA CUA A, UGG UUU ACA UGU UGU GUG A, UGG UUU ACA UGU UUU CUG A, UGG UUU ACA UGU UUU CCU A.
In vivo Crosslinking and Chromatin Immunoprecipitation
For ChIP experiments chondrocytes were grown to confluence for approximately 10 days (15% FCS, 3% Oxygen). Cells were mock- or UV-treated (8 J/m2) and incubated at 37°C for 48 hours prior to in vivo crosslinking (11 min at 4°C). In vivo crosslinking, lysis of cells (2 ml/per 25 ×106 cells), resuspension of crude nuclei in 1 X RIPA buffer (1 ml/per 25 ×106 cells), sonication (9 × 25”, Bioruptor, maximum output), chromatin purification, immunoprecipitation and reversal of crosslinks prior to protein and DNA analysis were performed as previously described 51. For each ChIP reaction 100µg of crosslinked chromatin from mock or UV-irradiated cells was immunoprecipitated with 2µg RNAPIIo (H5) antibody. Immunocomplexes were collected by adsorption to protein A Sepharose beads (Fast Flow, Amersham Pharmaceuticals) precleared by o/n incubation with 1 × RIPA, 0.1 mg/ml sonicated salmon sperm DNA (Invitrogen) and 1 mg/ml BSA (BioLabs).
Slot Blot analysis
For DNA analysis of CPDs, RNAPIIo-ChIPed DNA was treated and urified as previously described 51. DNA concentration of input crosslinked DNA was estimated using Nanodrop ND-1000 Spectrophotometer (NanoDrop Technologies, Inc.). DNA content of the ChIP samples and dilutions of input DNA samples were verified using Agilent 2100 Bioanalyzer and 6000 Pico kit (detection limit 0.25–5 ng/µl DNA). 2ng of each of the ChIPed DNA and 20ng of relevant input DNA were spotted onto Hybond N+ membrane (Amersham Biosciences) using PR 648 Slot Blot Manifold (Schleicher & Shuell). Readout of ChIPed DNA was compared to values obtained with input DNA. Analysis was performed as described 70 using mouse monoclonal CPD antibody (TDM-2) for immunodetection (ECL-Plus, Amersham Biosciences).
RNA samples and qPCR
RNA was extracted with Trizol (Invitrogen). DNA was generated through RT-PCR using Superscript II (Invitrogen). All qPCR reactions were done in duplicate using Biorad MyIQ using SYBR GreenI (Sigma) and Platinum Taq polymerase (Invitrogen). Generation of specific PCR products was confirmed by melting curve analysis, gel electrophoresis and sequencing. Primer pairs were tested with a logarithmic dilution cDNA to generate a linear standard curve (crossing point (CP) plotted versus log of template concentration), which was used to calculate the primer pair efficiency (E = 10(−1/slope)). For data analysis, the second derivative maximum method was applied, and induction of target cDNA was calculated according to Pfaffl 71: (Etarget ΔCP(cDNAuntreated-cDNAtreated)target)/ (EcontrolΔCP(cDNAuntreated-cDNAtreated)control). Primers used for qPCR: Mouse sequences: Growth Hormone Receptor (GHR) 5’- aaggtgaaggtcggagtcaa-3 ’ , 5 ’-accatgtagttgaggtcaatg-3’; Insulin-like Growth Factor-1 Receptor (IGF-1R) acgacaacacaacctgcgt, aacgaagccatccgagtca; Hypoxanthine-Guanine Phosphoribosyl Transferase (Hprt) cgaagtgttggatacaggcc, ggcaacatcaacaggactcc; beta-2-microglobulin ( B 2 M ) c c c t g g tctttctggtgctt, atttcaatgtgaggcgggtg; Glyceraldehyde-3-Phosphate Dehydrogenase (Gapdh) accacagtccatgccatcac, tccaccaccctgttgctgta; Gamma-Tubulin (gTUB) cagaccaaccactgctacat, agggaatgaagttggccagt; Human sequences: GHR tatggcgagttcagtgaggt, atagcatcactgttagcccaa; IGF-1R gcaccatcttcaagggcaat, aggacaaggagaccaaggca; B2M ctcttgtactacactgaattca, cctccatgatgctgcttaca; Gapdh aaggtgaaggtcggagtcaa, accatgtagttgaggtcaatg; Hprt ttggtcaggcagtataatcca, atagtcaagggcatatcctac; gTUB acatgaacaatgacctcatcg, atcctctgcaggctcttgtg; Rat sequences: IGF-1R acgacaacacaacctgcgt, gacgaagccatctgagtca; GHR aaggtgaaggtcggagtcaa, tccattccggggtccattca; Hprt cgaagtgttggatacaggcc, ggccacatcaacaggactct; Gapdh accacagtccatgccatcac, ttcagctctgggatgacctt;
Microarray hybridizations
Total RNA was isolated with Trizol (Invitrogen) from liver, kidney, spleen and lung of naturally aged wt mice (3 animals per age group) at 13 and 130 weeks of age as well as from the irradiated cells (4 cell lines per group). Synthesis of double stranded cDNA and biotin labeled cRNA was performed according to instructions of manufacturer (Affymetrix). Fragmented cRNA preparations were hybridized to full mouse genome oligonucleotide arrays (Affymetrix, mouse expression 430 V2.0 arrays), using Affymetrix hybridization Oven 640, washed, and subsequently scanned on GeneChip Scanner 3000. Initial data extraction and normalization within each array was performed using GCOS software (Affymetrix). Data intensities were log transformed and normalized within and between arrays with quantile normalization method using the R open statistical package. All microarray experiments complied with the “minimum information about microarray experiments; (MIAME)” and are available through the public repository ArrayExpress at accession codes: E-MEXP-1968 (UV-treated cells), EMEXP-1504 (wt mouse liver, spleen, kidney, lung) and E-MEXP-1975 (H2O2 treated cells).
Data analysis
Two-tail, pair wise analysis or a two-way analysis of variance was employed using Spotfire Decision Site software package 7.2 v10.0 (Spotfire Inc.) to extract the statistically significant data from each group of mice indicated in this study. The criteria for significance were set at p≤0.05 and a ≥ ±1.2-fold change.
GO Classification and Overrepresentation of Biological Themes
All significant gene entries were subjected to GO classification (http://www.geneontology.org). Significant over-representation of GO-classified biological processes was assessed by comparing the number of pertinent genes in a given biological process to total number of relevant genes printed on the array for that particular biological process (Fisher exact test, p≤0.01 False discovery rate (FDR) ≤0.1) using the publicly accessible software Ease and/or DAVID (http://david.abcc.ncifcrf.gov/summary.jsp). Due to the redundant nature of GO annotations, we employed Kappa statistics to measure the degree of the common genes between two annotations, and heuristic clustering to classify the groups of similar annotations according to kappa values (http://david.abcc.ncifcrf.gov/summary.jsp).
TPA induced hyperproliferation
8 – 10 weeks old Igf-1repi−/− and control mice (TPA-treated: n = 3–5, aceton treated: n = 2) were shaved and after 24 hours, treated three times every 48 hours with 10nm TPA/330µl or solvent alone (aceton). 48 hours after the last treatment skin was isolated and either fixed in 5% formalin/PBS at 4°C overnight and embedded in paraffin for histological sections or taken for cryosections. 40 high power fields per back skin were scored and quantified for epidermal thickness. TPA stock: 5mg/ml in ethylacetat (LC labs, USA), TPA solution: 10 nm (5 µg) TPA in aceton: 330µl for each mouse 215µl TPA (5mg/ml) + 58 ml aceton = 10 nm/330µl.
Supplementary Material
Acknowledgements
We thank Christel Kockx, Zeliha Ozgur, and Anja Raams for technical assistance, Casper Hoogenraad for neuron cultures, Anton Gartner and Francis Barr for comments on the manuscript. This research was supported by the Netherlands Organization for Scientific Research (NWO) and Netherlands Genomics Initiative (NGI) (NGI/NWO 05040202), SenterNovem IOP-Genomics (IGE03009), NIH (1PO1 AG17242), NIEHS (1UO1 ES011044), EC (QRTL-1999-02002), and the Dutch Cancer Society (EUR 99-2004). G.G. acknowledges support from the Cancer Genomics Centre and B.S. from EMBO long-term, Marie Curie Intra-European fellowships and Veni grant of the NWO. JHJH is CSO of DNage/Pharming. The authors declare competing financial interest.
References
- 1.Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 2.Kirkwood TB, Cremer T. Cytogerontology since 1881: a reappraisal of August Weismann and a review of modern progress. Hum Genet. 1982;60:101–121. doi: 10.1007/BF00569695. [DOI] [PubMed] [Google Scholar]
- 3.Hasty P, Campisi J, Hoeijmakers J, van Steeg H, Vijg J. Aging and genome maintenance: lessons from the mouse? Science. 2003;299:1355–1359. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- 4.Lombard DB, et al. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 5.Campisi J. Aging, tumor suppression and cancer: high wire-act. Mech.Ageing Dev. 2005;126:51–58. doi: 10.1016/j.mad.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 6.Schumacher B, Garinis GA, Hoeijmakers JH. Age to survive: DNA damage and aging. Trends Genet. 2008;24:77–85. doi: 10.1016/j.tig.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Sedelnikova OA, et al. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat.Cell Biol. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- 8.Dolle ME, et al. Rapid accumulation of genome rearrangements in liver but not in brain of old mice. Nat Genet. 1997;17:431–434. doi: 10.1038/ng1297-431. [DOI] [PubMed] [Google Scholar]
- 9.Sedelnikova OA, et al. Delayed kinetics of DNA double-strand break processing in normal and pathological aging. Aging Cell. 2008;7:89–100. doi: 10.1111/j.1474-9726.2007.00354.x. [DOI] [PubMed] [Google Scholar]
- 10.Herndon LA, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 11.Rea SL, Wu D, Cypser JR, Vaupel JW, Johnson TE. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat Genet. 2005;37:894–898. doi: 10.1038/ng1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Gems D. Longevity and ageing in parasitic and free-living nematodes. Biogerontology. 2000;1:289–307. doi: 10.1023/a:1026546719091. [DOI] [PubMed] [Google Scholar]
- 14.Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 15.Giannakou ME, Partridge L. Role of insulin-like signalling in Drosophila lifespan. Trends Biochem Sci. 2007;32:180–188. doi: 10.1016/j.tibs.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Carter CS, Ramsey MM, Sonntag WE. A critical analysis of the role of growth hormone and IGF-1 in aging and lifespan. Trends Genet. 2002;18:295–301. doi: 10.1016/S0168-9525(02)02696-3. [DOI] [PubMed] [Google Scholar]
- 17.Holzenberger M, et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 18.Kurosu H, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonkowski MS, et al. Long-lived growth hormone receptor knockout mice show a delay in age-related changes of body composition and bone characteristics. J Gerontol A Biol Sci Med Sci. 2006;61:562–567. doi: 10.1093/gerona/61.6.562. [DOI] [PubMed] [Google Scholar]
- 20.Bartke A, Brown-Borg H. Life extension in the dwarf mouse. Curr Top Dev Biol. 2004;63:189–225. doi: 10.1016/S0070-2153(04)63006-7. [DOI] [PubMed] [Google Scholar]
- 21.Niedernhofer LJ, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 22.Ramirez CL, Cadinanos J, Varela I, Freije JM, Lopez-Otin C. Human progeroid syndromes, aging and cancer: new genetic and epigenetic insights into old questions. Cell Mol Life Sci. 2007;64:155–170. doi: 10.1007/s00018-006-6349-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garinis GA, van der Horst GT, Vijg J, Hoeijmakers JH. DNA damage and ageing: new-age ideas for an age-old problem. Nat Cell Biol. 2008;10:1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehmann AR. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003;85:1101–1111. doi: 10.1016/j.biochi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 25.van der Pluijm I, et al. Impaired genome maintenance suppresses the growth hormone--insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 2006;5:e2. doi: 10.1371/journal.pbio.0050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masternak MM, et al. Effects of caloric restriction on insulin pathway gene expression in the skeletal muscle and liver of normal and long-lived GHR-KO mice. Exp Gerontol. 2005;40:679–684. doi: 10.1016/j.exger.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 27.van Hoffen A, Venema J, Meschini R, van Zeeland AA, Mullenders LH. Transcription-coupled repair removes both cyclobutane pyrimidine dimers and 6-4 photoproducts with equal efficiency and in a sequential way from transcribed DNA in xeroderma pigmentosum group C fibroblasts. Embo J. 1995;14:360–367. doi: 10.1002/j.1460-2075.1995.tb07010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuchiya T, et al. Additive regulation of hepatic gene expression by dwarfism and caloric restriction. Physiol Genomics. 2004;17:307–315. doi: 10.1152/physiolgenomics.00039.2004. [DOI] [PubMed] [Google Scholar]
- 29.Rowland JE, et al. In vivo analysis of growth hormone receptor signaling domains and their associated transcripts. Mol Cell Biol. 2005;25:66–77. doi: 10.1128/MCB.25.1.66-77.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boylston WH, et al. Altered cholesterologenic and lipogenic transcriptional profile in livers of aging Snell dwarf (Pit1dw/dwJ) mice. Aging Cell. 2004;3:283–296. doi: 10.1111/j.1474-9728.2004.00115.x. [DOI] [PubMed] [Google Scholar]
- 31.Schumacher B, et al. Delayed and accelerated aging share common longevity assurance mechanisms. PLoS Genet. 2008;4:e1000161. doi: 10.1371/journal.pgen.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang H, et al. Genetic mouse models of extended lifespan. Exp.Gerontol. 2003;38:1353–1364. doi: 10.1016/j.exger.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 33.Verma AK, Wheeler DL, Aziz MH, Manoharan H. Protein kinase Cepsilon and development of squamous cell carcinoma, the nonmelanoma human skin cancer. Mol Carcinog. 2006;45:381–388. doi: 10.1002/mc.20230. [DOI] [PubMed] [Google Scholar]
- 34.Stachelscheid H, et al. Epidermal insulin/IGF-1 signalling control interfollicular morphogenesis and proliferative potential through Rac activation. Embo J. 2008;27:2091–2101. doi: 10.1038/emboj.2008.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piwien-Pilipuk G, Huo JS, Schwartz J. Growth hormone signal transduction. J Pediatr Endocrinol Metab. 2002;15:771–786. doi: 10.1515/jpem.2002.15.6.771. [DOI] [PubMed] [Google Scholar]
- 36.Partridge L, Gems D. Mechanisms of ageing: public or private? Nat Rev Genet. 2002;3:165–175. doi: 10.1038/nrg753. [DOI] [PubMed] [Google Scholar]
- 37.Salmon AB, et al. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am J Physiol Endocrinol Metab. 2005;289:E23–E29. doi: 10.1152/ajpendo.00575.2004. [DOI] [PubMed] [Google Scholar]
- 38.O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 39.Nunez F, Chipchase MD, Clarke AR, Melton DW. Nucleotide excision repair gene (ERCC1) deficiency causes G(2) arrest in hepatocytes and a reduction in liver binucleation: the role of p53 and p21. Faseb J. 2000;14:1073–1082. doi: 10.1096/fasebj.14.9.1073. [DOI] [PubMed] [Google Scholar]
- 40.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 41.Dizdaroglu M. Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat Res. 2005;591:45–59. doi: 10.1016/j.mrfmmm.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 42.Dianov GL, Parsons JL. Co-ordination of DNA single strand break repair. DNA Repair (Amst) 2007;6:454–460. doi: 10.1016/j.dnarep.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 43.DiBiase SJ, et al. DNA-dependent protein kinase stimulates an independently active, nonhomologous, end-joining apparatus. Cancer Res. 2000;60:1245–1253. [PubMed] [Google Scholar]
- 44.Essers J, et al. Dynamics of relative chromosome position during the cell cycle. Mol Biol Cell. 2005;16:769–775. doi: 10.1091/mbc.E04-10-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garinis GA, et al. Transcriptome analysis reveals cyclobutane pyrimidine dimers as a major source of UV-induced DNA breaks. Embo J. 2005;24:3952–3962. doi: 10.1038/sj.emboj.7600849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You YH, et al. Cyclobutane pyrimidine dimers are responsible for the vast majority of mutations induced by UVB irradiation in mammalian cells. J Biol Chem. 2001;276:44688–44694. doi: 10.1074/jbc.M107696200. [DOI] [PubMed] [Google Scholar]
- 47.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 48.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 49.Riou L, et al. Differential repair of the two major UV-induced photolesions in trichothiodystrophy fibroblasts. Cancer Res. 2004;64:889–894. doi: 10.1158/0008-5472.can-03-2070. [DOI] [PubMed] [Google Scholar]
- 50.Jaspers NG, et al. Anti-tumour compounds illudin S and Irofulven induce DNA lesions ignored by global repair and exclusively processed by transcription- and replication-coupled repair pathways. DNA Repair (Amst) 2002;1:1027–1038. doi: 10.1016/s1568-7864(02)00166-0. [DOI] [PubMed] [Google Scholar]
- 51.Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 52.Jans J, et al. Powerful skin cancer protection by a CPD-photolyase transgene. Curr Biol. 2005;15:105–115. doi: 10.1016/j.cub.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 53.Dardevet D, Sornet C, Attaix D, Baracos VE, Grizard J. Insulin-like growth factor-1 and insulin resistance in skeletal muscles of adult and old rats. Endocrinology. 1994;134:1475–1484. doi: 10.1210/endo.134.3.8119189. [DOI] [PubMed] [Google Scholar]
- 54.Rivera EJ, et al. Insulin and insulin-like growth factor expression and function deteriorate with progression of Alzheimer's disease: link to brain reductions in acetylcholine. J Alzheimers Dis. 2005;8:247–268. doi: 10.3233/jad-2005-8304. [DOI] [PubMed] [Google Scholar]
- 55.Li M, Li C, Parkhouse WS. Age-related differences in the des IGF-I-mediated activation of Akt-1 and p70 S6K in mouse skeletal muscle. Mech Ageing Dev. 2003;124:771–778. doi: 10.1016/s0047-6374(03)00124-6. [DOI] [PubMed] [Google Scholar]
- 56.Charlet-Berguerand N, et al. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. Embo J. 2006;25:5481–5491. doi: 10.1038/sj.emboj.7601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brooks PJ. The case for 8,5'-cyclopurine-2'-deoxynucleosides as endogenous DNA lesions that cause neurodegeneration in xeroderma pigmentosum. Neuroscience. 2007;145:1407–1417. doi: 10.1016/j.neuroscience.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wijnhoven SW, et al. Accelerated aging pathology in ad libitum fed Xpd(TTD) mice is accompanied by features suggestive of caloric restriction. DNA Repair (Amst) 2005;4:1314–1324. doi: 10.1016/j.dnarep.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 59.Paci A, et al. Pharmacokinetics, metabolism, and routes of excretion of intravenous irofulven in patients with advanced solid tumors. Drug Metab Dispos. 2006;34:1918–1926. doi: 10.1124/dmd.106.010512. [DOI] [PubMed] [Google Scholar]
- 60.Hartog H, Wesseling J, Boezen HM, van der Graaf WT. The insulin-like growth factor 1 receptor in cancer: old focus, new future. Eur J Cancer. 2007;43:1895–1904. doi: 10.1016/j.ejca.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 61.Vermeulen CJ, Loeschcke V. Longevity and the stress response in Drosophila. Exp Gerontol. 2007;42:153–159. doi: 10.1016/j.exger.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 62.Daitoku H, Fukamizu A. FOXO transcription factors in the regulatory networks of longevity. J Biochem. 2007;141:769–774. doi: 10.1093/jb/mvm104. [DOI] [PubMed] [Google Scholar]
- 63.Doonan R, et al. Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev. 2008;22:3236–3241. doi: 10.1101/gad.504808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murakami S. Stress resistance in long-lived mouse models. Exp Gerontol. 2006;41:1014–1019. doi: 10.1016/j.exger.2006.06.061. [DOI] [PubMed] [Google Scholar]
- 65.Gems D, Partridge L. Stress-response hormesis and aging: "that which does not kill us makes us stronger". Cell Metab. 2008;7:200–203. doi: 10.1016/j.cmet.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 66.Nouspikel T, Hanawalt PC. DNA repair in terminally differentiated cells. DNA Repair. 2002;1:59–75. doi: 10.1016/s1568-7864(01)00005-2. [DOI] [PubMed] [Google Scholar]
- 67.Ljungman M, Lane DP. Transcription - guarding the genome by sensing DNA damage. Nat.Rev.Cancer. 2004;4:727–737. doi: 10.1038/nrc1435. [DOI] [PubMed] [Google Scholar]
- 68.Kirkwood T. Repair and its evolution: survival versus reproduction. Oxford: Blackwell; 1981. pp. 165–189. [Google Scholar]
- 69.Teuling E, et al. Motor neuron disease-associated mutant vesicle-associated membrane protein-associated protein (VAP) B recruits wild-type VAPs into endoplasmic reticulum-derived tubular aggregates. J Neurosci. 2007;27:9801–9815. doi: 10.1523/JNEUROSCI.2661-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ford JM, Hanawalt PC. Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 71.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.