Abstract
Little is known about how individual olfactory receptor neurons (ORNs) select, from among many odor receptor genes, which genes to express. Abnormal chemosensory jump 6 (Acj6) is a POU domain transcription factor essential for the specification of ORN identity and odor receptor (Or) gene expression in the Drosophila maxillary palp, one of the two adult olfactory organs. However, the mechanism by which Acj6 functions in this process has not been investigated. Here, we systematically examine the role of Acj6 in the maxillary palp and in a major subset of antennal ORNs. We define an Acj6 binding site by a reiterative in vitro selection process. The site is found upstream of Or genes regulated by Acj6, and Acj6 binds to the site in Or promoters. Mutational analysis shows that the site is essential for Or regulation in vivo. Surprisingly, a novel ORN class in acj6 adults is found to arise from ectopic expression of a larval Or gene, which is repressed in wild type via an Acj6 binding site. Thus, Acj6 acts directly in the process of receptor gene choice; it plays a dual role, positive and negative, in the logic of the process, and acts in partitioning the larval and adult receptor repertoires.
Introduction
Animals possess many odor receptor genes and many functionally distinct olfactory receptor neurons (ORNs). Yet in animals ranging from mammals to fruit flies, individual ORNs express only one or a few odor receptor genes (Ache and Young, 2005). How do ORNs select which receptors to express?
Drosophila contains two olfactory organs, the antenna and the maxillary palp. Each contains sensilla that house the dendrites of ORNs. The most numerous antennal sensilla are the basiconic sensilla, which subdivide into large and small subtypes (Shanbhag et al., 1999). Physiological analysis of the antenna and maxillary palp has identified 35 functional ORN classes, housed in 17 sensillum types in stereotyped combinations (Clyne et al., 1997; de Bruyne et al., 1999, 2001; Elmore et al., 2003). The Drosophila larva contains a distinct olfactory organ with 21 ORNs (Stocker, 2008).
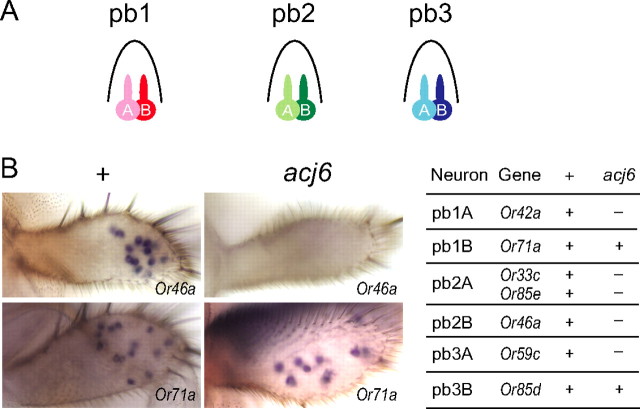
The fly genome contains 60 odorant receptor (Or) genes (Clyne et al., 1999a; Vosshall et al., 1999). Receptor-to-neuron maps were established by physiological and molecular analysis (Hallem et al., 2004; Couto et al., 2005; Fishilevich and Vosshall, 2005; Goldman et al., 2005). The maxillary palp contains three types of sensilla, pb1, pb2, and pb3, each of which contains two ORNs (i.e., pb1A, pb1B, pb2A, pb2B, pb3A, and pb3B) (see Fig. 1A). A single receptor gene has been mapped to each of these six ORN classes, except that two have been mapped to pb2A (Goldman et al., 2005). The larval olfactory organ expresses a subset of Or genes that overlaps partially with the adult Or repertoire.
Figure 1.
Role of acj6 in the maxillary palp. A, The wild-type maxillary palp contains three sensillum types, each housing a pair of ORNs. B, Expression of a subset of Or genes depends on acj6. In situ hybridization of Or genes to wild-type and acj6 maxillary palps is shown.
The first gene shown to be required for proper Or expression was abnormal chemosensory jump 6 (acj6), identified in a behavioral screen (McKenna et al., 1989). In acj66, the maxillary palp does not contain ORNs with the response spectra of pb1A, pb2A, pb2B, or pb3A; in contrast, pb1B and pb3B appear normal (Clyne et al., 1999b). Surprisingly, a novel ORN class, pb2C, was observed. Additional analysis showed that acj6 encodes a POU domain transcription factor (Clyne et al., 1999b) and that some Or genes depend on acj6 for their expression (Clyne et al., 1999a; Goldman et al., 2005). These results did not reveal whether Acj6 acts directly on Or genes or indirectly.
Here, we extend our analysis of Acj6 regulation to include all Or genes of the maxillary palp and all ORNs of the large antennal basiconic sensilla. Through an in vitro selection procedure, we define an Acj6 binding site, find that it lies upstream of Acj6-sensitive Or genes, and determine by mutational analysis that it is required for Acj6-mediated regulation. pb2C is found to derive its response spectrum from the ectopic expression of a larval Or gene, revealing an unexpected role for Acj6 in repression of larval odor receptors in the adult. The negative regulation is mediated by an Acj6 binding site. Thus, Acj6 exerts a dual role in receptor gene choice, positive and negative, adding a degree of freedom to the remarkable process by which each of 60 odor receptors is constrained to a discrete subset of neurons.
Materials and Methods
Drosophila stocks and transformation.
Drosophila stocks were cultured at 25°C. Wild-type flies were Canton-S unless otherwise stated. All DNA constructs were sequenced and then injected into w1118 embryos. More than three transgenic lines were obtained and analyzed for each transgene. Each green fluorescent protein (GFP) reporter line was doubly heterozygous for UAS-mCD8-GFP unless otherwise indicated. In all experiments with acj6, the acj66 allele, a null allele, was used.
Identification of an Acj6 binding site by systematic evolution of ligands by exponential enrichment.
The glutathione S-transferase (GST)-Acj6 expression construct was a gift from Dr. S. Certel (Harvard Medical School, Boston, MA) and contains the coding region (nucleotides 131–1577) of acj6 isoform (1, 4) (Certel et al., 2000). GST::Acj6 fusion protein expression was induced in the Escherichia coli strain BL21 at room temperature, and the fusion protein was purified using glutathione-Sepharose 4B following the manufacturer's instructions (GE Life Sciences).
Systematic evolution of ligands by exponential enrichment (SELEX) was performed using methods previously described (Wey and Schäfer, 1996) with some modifications (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). Double-stranded oligonucleotides were generated by annealing the template random oligonucleotides CAGGTCAGTTCAGCGGATCCTGTCG(N)16 GAGGCGAATTCAGTGCAACTGCAGC, to a 25 bp primer complementary to the 3′-invariant sequence (GCTGCAGTTGCACTGAATTCGCCTC) and then filling in with Klenow DNA polymerase. Unannealed oligonucleotides were removed using MicroSpin G-25 columns (GE Healthcare).
For the initial round of selection, 2 ng of random oligonucleotides was combined with 0.2 μg of GST::Acj6 fusion protein in a 50 μl binding system that contained the following components: 0.5% Nonidet P-40, 20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 4 μg of poly(dI-dC). The binding reaction proceeded at 4°C for 30 min with gentle rocking. Protein–oligonucleotide complexes were precipitated with 10 μl of glutathione-Sepharose beads prewashed and equilibrated in the binding buffer with 0.5% (w/v) skim milk. After three washes with the binding buffer, the beads were resuspended in 100 μl of elution buffer (10 mm Tris·Cl, pH 8.5), and heated at 85°C for 10 min to release bound oligonucleotides, a fraction (10 μl) of which was then amplified by PCR for 20 cycles. The PCR product was column-purified and a fraction (2 ng) was used for the next round of selection. Control binding reactions without GST::Acj6 protein were performed with each selection to ensure that nonspecific binding of oligonucleotides to the Sepharose beads did not yield a visible band after PCR amplification. For the fifth and sixth rounds, oligonucleotides were incubated with GST::Acj6 protein bound to glutathione-Sepharose beads; ∼1 μg of Acj6 protein was used. After six rounds of selection and amplification, PCR products were cloned and 26 clones were randomly selected and sequenced.
Electrophoretic mobility shift assay.
The GST moiety of the GST-Acj6 fusion protein was cleaved with PreScission protease (GE) in a buffer containing 50 mm Tris·Cl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1 mm DTT, and 0.01% NP-40 “substitute” (Sigma-Aldrich) according to the manufacturer's instructions. 5′-end biotin-labeled complementary oligonucleotides and nonlabeled competitor sequences (W. M. Keck facility at Yale) were annealed in a buffer containing 10 mm Tris·Cl, pH 8.0, 1 mm EDTA, and 50 mm NaCl. The annealed double-stranded oligonucleotide probes were purified from a native polyacrylamide gel and run through a Microspin G-50 column (GE). The sequence of the Or85e probe and its cold competitor was 5′-TTTTAAAAACTCATTAATGCATAAAATCAT-3′; the Or85e mutated competitor sequence was 5′-TTTTAAAAACTTCTAGAAAGTTAAAATCAT-3′ (mutated nucleotides are underlined). Binding sites in other Or genes were also tested in competition assays. The Or46a sequence was 5′-ATCGTCGGCTAAATAATTCAGTATTTTTGC-3′; the mutant Or46a sequence was 5′-ATCGTCGGCATCTCTAGACCGTATTTTTGC-3′. The Or45b sequence was 5′-AACTAGTACGCATACTTGATCATATATATA-3′; the mutant Or45b sequence was 5′-AACTAGTAGCTGCCGGATCCCATATATATA-3′; the “switched binding site” sequence Or45bMOr46a was 5′-AACTAGTACTAAATAATTCAGATATATATA-3′. The amounts of protein, probes, and competing sequences are indicated in the figure legend. Protein was diluted in 2.5 mm HEPES, pH 7.6, buffer containing 0.1 m KCl, 1.25 mm MgCl2, 0.01 mm EDTA, 1% glycerol, and 1 mg/ml BSA, when needed. The binding reactions included 20 mm Tris·Cl, 100 mm KCl, 2 mm DTT, 1 μg of poly(dG-dC) (Sigma-Aldrich), 5% glycerol, 0.05% NP-40, 1.25 mm MgCl2, and 1% (w/v) skim milk, and were incubated at room temperature for 15 min. The resulting complexes were then resolved on a 5% native polyacrylamide gel in 0.5× TBE, 2.5% glycerol at 4°C and detected with Pierce chemiluminescent nucleic acid detection module according to the manufacturer's protocol.
Promoter mutagenesis.
Transgenic flies carrying wild-type Or85e-GAL4 (3.1 kb of upstream sequences), Or42a-GAL4 (4.1 kb), and Or46a-GAL4 (1.9 kb) were described previously (Goldman et al., 2005). For Or85eM-GAL4, the Or85e promoter regions upstream and downstream of an Acj6 site (at −85 bp) were cloned separately with primers containing an XbaI site in place of the Acj6 site. The upstream KpnI–XbaI fragment was ligated with the downstream XbaI–NotI fragment and then inserted into pG4PN (Dobritsa et al., 2003), resulting in an 8 nt substitution within the Acj6 binding site (aaaaaCTCATTAATGCAtaaaa was mutated to aaaaaCTTCTAGAAAGTtaaaa). Similar strategies were used to construct Or46aM-GAL4 (tcggcTAAATAATTCAGgtatt was mutated to tcggcATCTCTAGACCGgtatt) and Or42aM310-GAL4 (gaatcATCATTTATTAAtggaa was mutated to gaatcGGATCCTTATAAtggaa). Overlap extension PCR (Higuchi et al., 1988) was used to introduce additional mutations in the Acj6 sites at −249 and −102 bp of Or42a. In Or42aM310-249-GAL4, (gggctTTCATTCTTTAGgtcat) at −249 bp was mutated to (gggctTTCACCGTTTAGgtcat). In Or42aM310-102-GAL4, (cgcctATGAATTATCTAattga) at −102 bp was mutated to (cgcctATGACCGATCTAattga). Or42aM310-249-102-GAL4 bears mutations in all three Acj6 sites.
Wild type Or45b-GAL4 was described by Kreher et al. (2005). Overlap extension PCR was used to mutate the Acj6 site at −392 bp (tagtaCGCATACTTGATcatat) to (tagtaGCTGCCGGATCCcatat) in Or45b (M)-GAL4. In Or45b (MOr46a)-GAL4, this site was replaced by (tagtacTAAATAATTCAGatat), the Acj6 site in the Or46a promoter.
In situ hybridization.
Digoxigenin-labeled RNA probes were hybridized to whole maxillary palps attached to the proboscis, or to antennal sections, and were detected with alkaline phosphatase-conjugated anti-digoxigenin antibody and its substrates BCIP/NBT (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium) or Fast Red tablets (Roche) as previously described (Clyne et al., 1999a; Goldman et al., 2005).
Electrophysiology.
Odor stimuli were presented, and action potentials were recorded as described previously (de Bruyne et al., 1999, 2001; Dobritsa et al., 2003).
Results
Expression of some but not all Or genes in the maxillary palp depends on Acj6
We extended our analysis of Or genes in an acj66 background to include the entire repertoire of maxillary palp receptor genes. Of the seven genes, five depend on acj6, as determined by in situ hybridization: their expression was not detected in the acj66 maxillary palp (Fig. 1B). The genes that depend on Acj6 include the gene that is normally expressed in the pb1A ORN (Or42a), both of the genes that are expressed in pb2A (Or33c and Or85e), the gene expressed in pb2B (Or46a), and the gene expressed in pb3A (Or59c). Or71a and Or85d appear to be expressed normally in acj66. Results for some of these genes have been reported previously (Clyne et al., 1999a; Goldman et al., 2005).
Identification of an Acj6 binding site by in vitro selection
To identify an Acj6 binding site, we used an in vitro selection procedure, SELEX (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). In essence, a pool of random oligonucleotides was allowed to bind to Acj6 protein, and oligonucleotides that bound were eluted, amplified by PCR, and allowed to bind again. Six cycles of enrichment were thus performed for oligonucleotides that bind Acj6, after which oligonucleotides from the enriched population were sequenced. A consensus binding site was defined from the sequenced oligonucleotides, and a positional weight matrix was established (Fig. 2A; supplemental Fig. S2, available at www.jneurosci.org as supplemental material).
Figure 2.
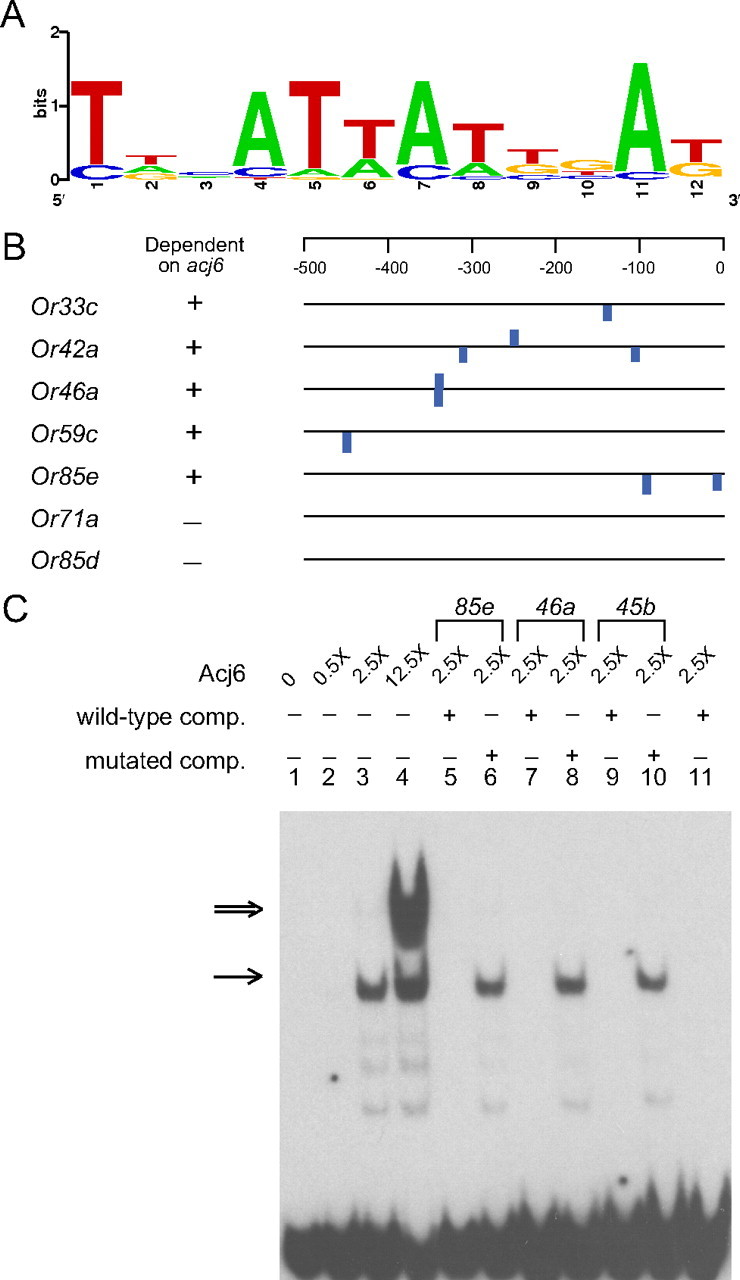
Identification of an Acj6 binding site. A, Acj6 binding site consensus. Alignment of sequences from an in vitro binding selection, SELEX, is represented in a consensus logo, generated with WebLogo 3 (Crooks et al., 2004). The overall height of each stack indicates the sequence conservation at that position (measured in bits), whereas the height of nucleotide symbols within the stack reflects the relative frequency of the nucleotides at that position. B, All five Or genes that depend on Acj6 for expression in the maxillary palp have Acj6 binding sites with scores >6 within 500 bp upstream of their translational start sites. The proximal Acj6 site at Or85e lies at position −17 with respect to the translation start site and seems likely to lie in the 5′-untranslated region of the gene rather than in the promoter region. C, Acj6 binds to sites in Or gene promoters. EMSA was performed with biotin-labeled oligonucleotide probes representing the Acj6 site and its flanking sequences in the Or85e promoter. The amount of Acj6 protein is indicated as a multiple of the molarity of the labeled Or85e oligonucleotide probe used (0.02 pmol). Lanes 5, 7, and 9 contain a 40-fold excess of unlabeled competitor oligonucleotide (comp.) that contains the wild-type Acj6 site in Or85e, Or46a, and Or45b, respectively. Lanes 6, 8, and 10 contain a 40-fold excess of unlabeled competitor oligonucleotide in which the Acj6 sites were mutated. The competitor sequence in lane 11 contains an Or46a site flanked by sequences adjacent to an Acj6 site in the Or45b promoter, as described below. The single arrow indicates bands likely to represent binding of the site to an Acj6 monomer; the double arrow indicates a band likely to represent binding of an Acj6 dimer. Faint bands may represent binding of Acj6 degradation products. Unbound oligonucleotides are at bottom of gel.
Acj6 sites are located upstream of all maxillary palp Or genes that depend on Acj6
We scanned the DNA sequences upstream of each of the seven maxillary palp Or genes for Acj6 binding sites. We focused on the 500 nt upstream of each translation start site, because regions of this length were sufficient to confer faithful expression of a reporter gene in the case of previously analyzed maxillary palp Or genes (Ray et al., 2007).
Of the seven Or genes, five contained upstream Acj6 sites, and those were the five genes that depend on Acj6 (Fig. 2B; supplemental Fig. S3, available at www.jneurosci.org as supplemental material). Specifically, all of these five genes contained Acj6 sites with scores >7 [scores are defined in supplemental Fig. S2 (available at www.jneurosci.org as supplemental material)]. If a score of 6 is set as a threshold, then the other two genes have no sites in this region. Moreover, the two genes that did not depend on Acj6 had no sites in a more extensive upstream region extending 2 kb from the translation start site.
Acj6 binds to the predicted sites in Or promoters
We tested whether Acj6 binds to the predicted Acj6 sites in Or promoters using electrophoretic mobility shift assays (EMSAs). We first tested an Acj6 site located 85 bp upstream of the translational start site of Or85e and found that Acj6 protein, expressed in bacteria, indeed bound to it (Fig. 2C, lanes 1–4). The amount of binding depended on the amount of Acj6 added to the binding reaction. At the highest levels of Acj6, two intense bands could be observed, of which the lower and higher molecular weight bands are likely to represent binding of the site to an Acj6 monomer and an Acj6 dimer, respectively. Highly cooperative homodimerization has previously been reported for POU domain proteins (Rhee et al., 1998).
To test whether binding of Acj6 to the Or85e site is sequence specific, we asked whether various sequences could compete with the site for binding to Acj6. We found that an excess of the unlabeled Or85e site, or Acj6 sites from two other genes, Or46a and Or45b (described below), competed effectively; however, these sites did not compete when mutated (Fig. 2C). The simplest interpretation of these results is that Acj6 binds directly to the Acj6 site in a sequence-specific manner.
Acj6 sites are required for Or gene expression
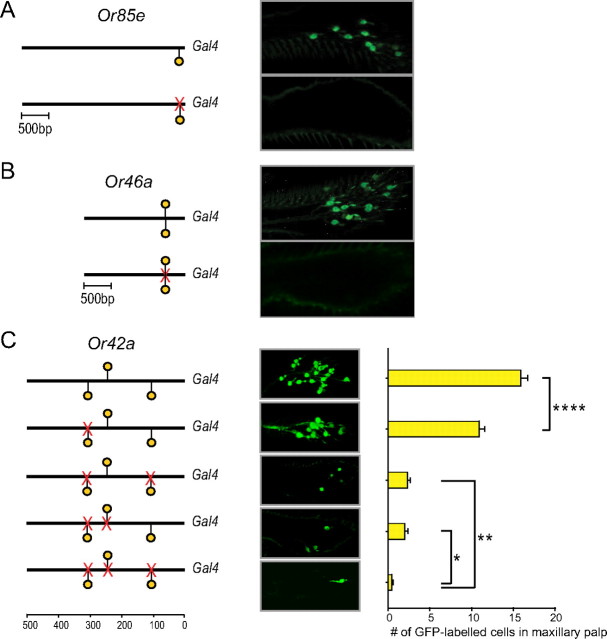
The correlation between the presence of Acj6 sites and Acj6 dependence suggested a role for the sites in Or gene regulation. To test the possibility of a functional role for these sites, we mutated them upstream of three genes, Or85e, Or46a, and Or42a. We examined the effects of these mutations on the expression of Or promoter-GAL4 drivers that have previously been shown to drive faithful expression of a UAS-GFP reporter gene (Goldman et al., 2005).
Upstream of Or85e we mutated the site at −85 bp (Fig. 3A). We note that positions are determined with respect to translational start sites, but the transcriptional start sites as determined by 5′-rapid amplification of cDNA ends are within 50 bp of the predicted translational start sites in all cases examined, including Or85e and Or46a (Ray et al., 2007). We found that mutation of this −85 bp site in the Or85e-GAL4 driver abolished expression, as determined by loss of GFP expression (Fig. 3A) (≥20 maxillary palps examined for each of four independent transgenic mutant lines).
Figure 3.
Acj6 sites are required for expression of Or genes. A, B, GFP expression driven by wild-type Or85e- and Or46a-GAL4 constructs (top panels) and constructs with mutations in predicted Acj6 sites (bottom panels). The proximal Acj6 site in Or85e, which is likely to lie in the 5′-untranslated region of the gene, is not shown. Data were from four independent transgenic lines for the mutant Or85e construct, and three lines for the mutant Or46a construct. C, The effects of mutations in Acj6 sites in Or42a. Data were pooled from three independent transgenic lines for each mutated construct. n = 11 for wild type; 30 < n < 37 for mutated constructs. *p < 0.05; **p < 0.01; ****p < 0.0001 (one-way ANOVA and Holm–Sidak test).
Upstream of Or46a lie two Acj6 sites that overlap. We mutated both simultaneously and saw no remaining expression of the GFP reporter in most maxillary palps (Fig. 3B). A small fraction (∼20%) of maxillary palps showed staining of a few cells (n = 2.3 + 0.7 cells; n = 13 maxillary palps), a much smaller number than is observed when a wild-type promoter is used to drive expression (n = 15.1 + 0.6 cells; n = 8 maxillary palps).
Upstream of Or42a lie three Acj6 sites, at positions −310, −249, and −102. When we mutated the site at −310, there was a small reduction (p < 0.0001, ANOVA) in the number of GFP-labeled cells in the maxillary palp compared with the control (Fig. 3C). When we mutated both the site at −310 and the site at −102, there was a major reduction in labeling. Mutation of the sites at both −310 and −249 also led to a strong reduction in labeling. Mutation of all three sites virtually eliminated labeling.
These results show that Acj6 sites are essential for the normal expression of each of three Or genes tested. The results also show that multiple Acj6 sites are functional upstream of Or42a.
A subset of antennal ORNs depends on Acj6 to acquire their identities
Although the ORNs of the maxillary palp are particularly convenient to study, the great majority of ORNs in the fly's olfactory system are located in the antenna. We wanted to examine the extent to which acj6 is required for the specification of ORN identity and receptor gene expression in the antenna.
We performed an analysis of ORN responses in the antenna of acj66. We systematically measured the responses of all ORNs in the large basiconic sensilla using a panel of diagnostic odorants. There are three types of large basiconic sensilla: ab1 contains four ORNs, ab1A, ab1B, ab1C, and ab1D; ab2 contains two ORNS, ab2A and ab2B; ab3 contains two ORNs, ab3A and ab3B.
In acj66, ab1A and ab1B were severely affected (Fig. 4A). We found no ORNs with the response spectra of these ORN classes. We did, however, observe sensilla containing ORNs with response spectra very similar to those of ab1C and ab1D: one ORN responded only to CO2, as expected of ab1C, and another responded only to methyl salicylate, as expected of ab1D (de Bruyne et al., 2001). In these sensilla, we did not observe the spontaneous action potentials of large amplitudes that are characteristic of ab1A and ab1B, suggesting that these ORNs are either absent or electrically silent.
Figure 4.
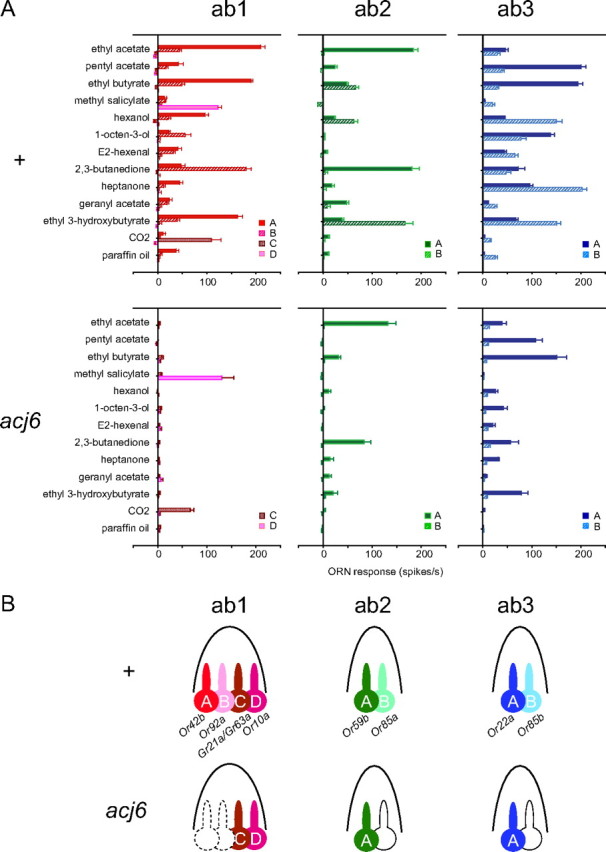
Odor responses of a subset of antennal ORNs are lost in acj66. A, Odor response profiles of large basiconic sensilla of wild-type and acj66 antennae. 6 ≤ n ≤ 12. Error bars indicate SEM. Odorants were applied as vapors of a 10−2 dilution. B, Fate changes of antennal large basiconic neuron classes in acj66. ab1A and ab1B are absent or electrically silent in acj66 and are depicted by dashed lines. Uncolored neurons depicted by solid lines yield spontaneous action potentials but show no response to tested odors.
We observed a second type of sensillum in the acj66 antenna that contained an ORN with a response spectrum very similar to that of ab2A (Fig. 4A). In this sensillum type, we observed a second ORN that exhibited spontaneous action potentials but that did not respond to any tested odorant. Likewise, we observed a third type of sensillum containing an ORN similar to ab3A, paired with an ORN that showed spontaneous action potentials but no odor-evoked activity. We note that, among the four classes of odor-responding ORNs in the acj66 antenna, some yielded responses of smaller magnitude than in the wild-type control, but the overall response profiles of these four ORN classes appeared very similar to those of wild type in each case.
The simplest interpretation of these results is that ab1A, ab1B, ab2B, and ab3B all depend critically on acj6 for the acquisition of their odor sensitivities (Fig. 4B). It is possible that acj6 plays a subtle role in the differentiation of the other ORN classes, but acj6 does not seem to be required by these other ORNs in the process by which they select a receptor gene.
The Or genes of Acj6-dependent antennal ORNs depend on Acj6
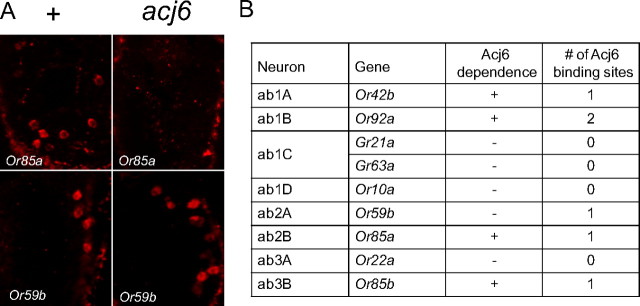
We next tested the expression of Or genes in the acj66 antenna (Fig. 5A) (data not shown). We found by in situ hybridization that the Or genes that are expressed in the ab1A, ab1B, ab2B, and ab3B ORNs, Or42b, Or92a, Or85a, and Or85b, respectively, were not detectable in acj66. As a control, we examined the wild-type antenna and detected expression of all four genes. As an additional control, we tested the expression of Or59b and Or22a, which are expressed in ab2A and ab3A, respectively. Both of these Or genes were found to be expressed in both acj66 and wild type. In summary, the lack of detectable odor responses from four ORN classes of the acj66 antenna correlates with a lack of expression of the receptor genes that these ORNs express in wild type.
Figure 5.
A subset of antennal Or genes require acj6 for expression. A, In situ hybridizations of Or genes to wild-type and acj66 antennae. Cells are labeled in red. B, Effects of acj66 on receptor expression shown by in situ hybridization (Or42b, Or92a, Or59b, Or85a, Or22a, Or85b) or inferred by response properties of antennal neurons (ab1C, ab1D). The number of Acj6 sites within 1 kb upstream of these genes is indicated.
Antennal genes that depend on Acj6 contain Acj6 sites
Having identified four antennal Or genes that depend on Acj6 for their expression, we asked whether they contained Acj6 sites. All four contain Acj6 sites within 1 kb of the predicted ATG translation start site (Fig. 5B); all of the Acj6 sites are within or very near the 500 bp region upstream of this ATG. We also considered the receptor genes of ORNs that are present and show approximately normal response spectra in acj66 (ab1C, ab1D, ab2A, and ab3A). ab1C expresses two gustatory receptor (Gr) genes in wild type, Gr21a and Gr63a (Jones et al., 2007; Kwon et al., 2007), neither of which contains an Acj6 site within 1 kb of the translation start site. ab1D and ab3A express Or10a and Or22a, respectively, which do not contain Acj6 sites. ab2A expresses Or59b, which contains an Acj6 site despite its independence of Acj6. In summary, in large basiconic sensilla, all Acj6-dependent receptor genes contain Acj6 sites and most Acj6-independent receptor genes lack them.
Acj6 represses a larval Or gene in the adult
We previously reported the surprising finding of a novel ORN class in the acj66 maxillary palp (Clyne et al., 1999b). Sensilla containing either pb2A or pb2B were absent in the mutant, but sensilla containing a novel ORN class, pb2C, were detected. The response spectrum of pb2C could not be explained in terms of a linear amplification or summation of the response spectra of other maxillary palp ORN classes, and the simplest interpretation was that either pb2A or pb2B was transformed into pb2C.
We recently noticed that one receptor, Or45b, which had been characterized during the course of a systematic functional analysis of larval odor receptors (Kreher et al., 2005, 2008), exhibited a response spectrum similar to that observed for pb2C. We therefore compared directly the responses of pb2C to the responses conferred by Or45b in the “empty neuron” system, an in vivo expression system (Dobritsa et al., 2003; Hallem et al., 2004). We found the odor response profiles to be very similar (Fig. 6A), suggesting that Or45b, a larval Or gene, might be ectopically expressed in the maxillary palp of acj66.
Figure 6.
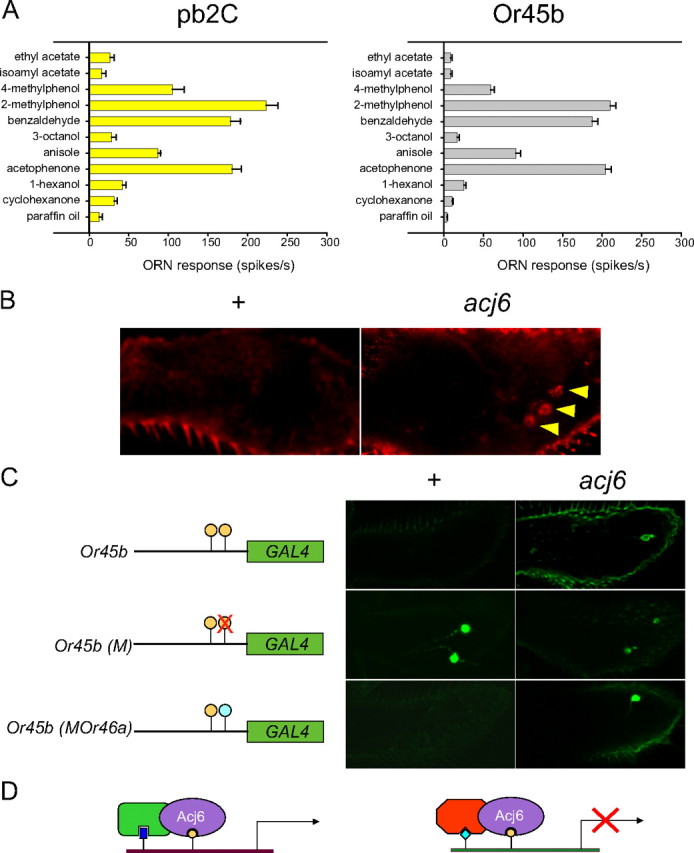
Acj6 represses the larval receptor gene Or45b in the adult maxillary palp. A, Response profiles of pb2C (in acj66) and Or45b (in the empty neuron system). n = 6 for pb2C; n = 19 for Or45b. Error bars indicate SEM. B, In situ hybridization to Or45b mRNA in wild-type and acj6 maxillary palps. The arrowheads indicate labeled cell bodies. C, Effects of altering an Acj6 site on Or45b-GAL4-driven GFP expression. Or45b-GAL4 is not expressed in the wild-type maxillary palp but is ectopically expressed in the acj66 maxillary palp. Mutation of the Acj6 site at −392 caused expression in the wild-type adult, mimicking the effects of the acj66 mutation. Replacement of the Acj6 site at −392 with another Acj6 site, one that directs activation of Or46a, yielded the same expression pattern as that driven by Or45b-GAL4. Two independent lines were examined for each genotype. The Or46a site contains two Acj6 consensus sequences that overlap by 1 bp and are on opposite strands of the DNA. D, Model for dual mode of regulation. Acj6 activates transcription (left) or represses transcription (right) depending on the presence of other proteins that bind in the vicinity.
We performed an in situ hybridization with an Or45b probe to the maxillary palp and found expression in acj66, but not wild type (Fig. 6B). The simplest interpretation of these results is that Acj6 represses Or45b in the adult maxillary palp.
To investigate the mechanism of Acj6-mediated repression, we first examined the upstream DNA sequences of Or45b and found two Acj6 sites, at −499 and −392 (Fig. 6C, top left). The gene also contains two other regulatory sequences that are found upstream of Or genes expressed in the maxillary palp: there are two Dyad-1 elements, CTA(N)9TAA, at −301 and −55, and an Oligo-1 element, CTTATAA, at −193. Dyad-1 is required for Or expression in the maxillary palp, and Oligo-1 both enhances expression of Or genes in the maxillary palp and represses their expression in the antenna (Ray et al., 2007).
The Acj6 site at −392 is in fact capable of binding Acj6: it competed with the positively acting site in Or85e for binding to Acj6 in gel shift analysis (Fig. 2C). To test the hypothesis that Acj6 represses Or45b in the maxillary palp via the Acj6 site, we mutated the site at −392. (Numerous efforts to ablate the site at −499 were unsuccessful.) The loss of the site at −392 resulted in expression in maxillary palps in a wild-type (acj6+) background, consistent with a role for this site in repression of Or45b [Fig. 6C, Or45b (M)]. Thus, loss of either Acj6 or an Acj6 site allowed expression of Or45b in the maxillary palp. We conclude that Acj6 acts negatively on Or45b via the Acj6 site at −392.
Finally, since other Acj6 sites act positively, we wondered whether the negative effect of the Acj6 site at −392 upstream of Or45b was attributable to its context (e.g., on other proteins that are bound in the vicinity). To test this possibility, we replaced the Acj6 site at −392 with the Acj6 site that acts positively at −346 upstream of Or46a [Fig. 6C, Or45b (MOr46a)]. We found that the Acj6 site that promotes expression upstream of Or46a did not promote expression upstream of Or45b in the maxillary palp of wild type. The lack of Or45b expression depended on repression by Acj6: Or45b was expressed in the maxillary palp of acj66 animals carrying the Or46a site upstream of Or45b (Fig. 6C). One possible interpretation of these results is that Acj6 binds to the Or46a site when it is upstream of either Or46a or Or45b, but the mode of regulation that it exerts depends on other proteins that are bound in the vicinity (Fig. 6D).
Discussion
Acj6 and the logic of receptor gene choice
How individual ORNs select individual odor receptors to express is a remarkable problem in molecular neurobiology. Most ORN classes in the fly select one or a small number of receptor genes from a large family of 60 Or genes. The choice dictates the odor response spectrum of the ORN and, consequently, its contribution to the coding of olfactory information.
The simplest model for receptor gene choice in the fly is that it is governed by a combinatorial code of transcription factors (Ray et al., 2007, 2008) (for review, see Fuss and Ray, 2009). Although there is evidence to support this model, much remains to be learned about the elements of the code and the logic by which it operates. Distinct combinations of six transcription factors could in principle dictate the specification of 26 = 64 distinguishable ORN classes, a number comparable with the number of Or genes and of the same order as the number of ORN classes. Thus for factors A–F, one ORN class might require factors A, B, and D and be specified by the combination (A+B+C−D+E−F−), whereas another class might be specified by the combination (A−B−C+D+E−F+). In this simple model, each factor would be required for the specification of one-half of the 64 ORN classes. Alternatively, a much larger number of such factors could act, each in a small number of classes. In the 14 ORN classes for which data are now available, 6 in the maxillary palp and 8 in the antenna, Acj6 is required for the specification of 8, approximately one-half. Thus, we found evidence that at least one transcription factor acts broadly across the ORN repertoire.
One aspect of the problem of receptor gene choice is that each ORN must express one or a small number of receptor genes; another aspect is that each ORN must not express all the other receptor genes. In principle, each aspect of the problem could be governed by separate sets of factors. We found here that Acj6 plays a role in both positive and negative regulation of Or genes. This duality of Acj6 function imparts economy to the process, an economy that may be critical in a process that controls such a large number of Or genes in a large number of ORN classes. Allowing individual factors to act in two aspects of the problem reduces the number of required factors.
Another example of dual regulation by a single transcription factor was found in the differentiation of photoreceptor subtypes in the Drosophila visual system (Tahayato et al., 2003). Otd, a homeodomain transcription factor, activates the rhodopsin genes rh3 and rh5 but represses rh6 in other photoreceptors, through a common binding site. The modes of regulation are likely to depend on the availability of a coactivator and a corepressor of Otd. An example of dual regulation in a chemosensory system is provided in Caenorhabditis elegans by the UNC-3 Olf/EBF (early B-cell factor) protein (Kim et al., 2005). This protein represses a number of genes, including the odr-10 olfactory receptor gene, in the ASI chemosensory neuron, and it activates other chemoreceptor genes. UNC-3 was shown to repress multiple genes by direct binding to sites in the promoters, but it may activate the expression of ASI-specific genes through an indirect mechanism.
One gene family, two olfactory systems
Drosophila contains two distinct olfactory systems, one in the larva and one in the adult. The two systems differ in morphology and developmental origins, and they operate under different conditions: the larva burrows in semisolid medium, whereas the adult walks and flies through air. Both olfactory systems, however, depend on members of the Or gene family. Some members of the Or family are larval specific, others are adult specific, and some are expressed in both (Couto et al., 2005; Fishilevich et al., 2005; Kreher et al., 2005). By what mechanism are members of the same gene family partitioned into two different systems?
We found that Acj6 acts in the partitioning mechanism. It is required to restrict at least one member, Or45b, to the larval system: when acj6 is mutated, Or45b is ectopically expressed in the adult. Or45b contains in its upstream region two regulatory elements, Dyad-1 and Oligo-1, which are found adjacent to all or most maxillary palp Or genes and are required for their expression in the maxillary palp. We do not know whether the Dyad-1 and Oligo-1 elements play a role in the expression of Or45b in larvae. These observations, together, suggest the possibility that Or45b may have shifted over evolutionary time from a maxillary palp gene to a larval gene. If so, the shift is likely to have occurred before the divergence of Drosophila melanogaster and Drosophila pseudoobscura tens of millions of years ago, since the odor response spectra of the maxillary palp ORNs in these two species are similar (Ray et al., 2008).
The Acj6 site and its context: odor receptors and beyond
Using an in vitro DNA binding assay, we defined a positional weight matrix and a consensus Acj6 binding site. Only one Acj6 site has been experimentally identified and tested previously, in a study of sequences upstream of the gene encoding choline acetyltransferase, Cha (Lee and Salvaterra, 2002). These Cha sequences were scanned for motifs similar to those found for the mammalian POU-IV transcription factors, and of seven identified, one was found to bind Acj6 protein in an EMSA. It does not show close similarity to our site (matrix score, 3.4), and its activity in vivo was not evaluated.
We found a strong correlation between the presence of the Acj6 site that we have identified and Acj6 dependence in vivo. Moreover, we have shown directly, in vivo, that these sites are required for normal expression of flanking Or genes. These results are consistent with a direct role for Acj6 in mediating Or expression, as opposed to an indirect role via another transcription factor.
We have shown that one of the Acj6 sites, the Or45b site, acts in negative regulation. Its score based on the positional weight matrix is intermediate among the sites that act in positive regulation, which is consistent with our finding from a binding-site-swap experiment that repression of Or45b depends not on the specific sequence of an Acj6 site but on its flanking sequences—the Or46a site also mediated repression when placed upstream of Or45b.
In several cases, Acj6 sites lie within 8 bp of conserved sequence elements that are required for appropriate Or expression (Ray et al., 2007, 2008) (supplemental Fig. S4, available at www.jneurosci.org as supplemental material). An Acj6 site also lies very near a putative binding site for Pdm3, another POU domain transcription factor that regulates ORN development and that interacts genetically with Acj6 (Tichy et al., 2008). The proximity of these regulatory elements is likely to reflect the interaction of Acj6 and its cofactors. Identification of the cofactors of Acj6 will further contribute to our understanding of the combinatorial regulation of specific Or expression.
Acj6 is likely to have a number of interesting transcriptional regulatory targets in addition to odor receptors and choline acetyltransferase. It may regulate other molecules that are essential for neuronal signaling: whereas some ORNs in acj66 exhibited spontaneous action potentials but did not respond to odorants, other ORNs showed no electrical activity, consistent with a loss of both odor receptors and other components.
Acj6 is also likely to regulate molecules required for synaptic connectivity. Acj6 is required for axon targeting specificity of a subset of ORN classes, some of which require Acj6 autonomously and some nonautonomously (Komiyama et al., 2004). The second-order neurons of the Drosophila olfactory system, the projection neurons, also require Acj6 for both normal dendritic targeting and axon terminal arborization (Komiyama et al., 2003). Moreover, the role of Acj6 extends beyond the olfactory system: acj6 mutants have defects in retinal axon targeting and synapse selection as well (Certel et al., 2000).
Our definition of an Acj6 binding site may now facilitate the identification of some of its transcriptional targets. There are many sites in the genome with matrix scores >7, and analysis of the adjacent coding regions may be useful in identifying novel components required for synaptic connectivity and other processes.
Footnotes
This work was supported by grants from the National Institutes of Health and a McKnight Investigator Award (J.R.C.). We thank Sarah Certel for GST-Acj6 fusion protein and its expression constructs, Scott Kreher for UAS-Or45b transgenic flies, W. van der Goes van Naters for help with electrophysiology, Chih-Ying Su for critical reading of this manuscript, and Anandasankar Ray for discussion.
References
- Ache BW, Young JM. Olfaction: diverse species, conserved principles. Neuron. 2005;48:417–430. doi: 10.1016/j.neuron.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Certel SJ, Clyne PJ, Carlson JR, Johnson WA. Regulation of central neuron synaptic targeting by the Drosophila POU protein, Acj6. Development. 2000;127:2395–2405. doi: 10.1242/dev.127.11.2395. [DOI] [PubMed] [Google Scholar]
- Clyne P, Grant A, O'Connell R, Carlson JR. Odorant response of individual sensilla on the Drosophila antenna. Invert Neurosci. 1997;3:127–135. doi: 10.1007/BF02480367. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999a;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Certel SJ, de Bruyne M, Zaslavsky L, Johnson WA, Carlson JR. The odor specificities of a subset of olfactory receptor neurons are governed by Acj6, a POU-domain transcription factor. Neuron. 1999b;22:339–347. doi: 10.1016/s0896-6273(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15:1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyne M, Clyne PJ, Carlson JR. Odor coding in a model olfactory organ: the Drosophila maxillary palp. J Neurosci. 1999;19:4520–4532. doi: 10.1523/JNEUROSCI.19-11-04520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyne M, Foster K, Carlson JR. Odor coding in the Drosophila antenna. Neuron. 2001;30:537–552. doi: 10.1016/s0896-6273(01)00289-6. [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- Elmore T, Ignell R, Carlson JR, Smith DP. Targeted mutation of a Drosophila odor receptor defines receptor requirement in a novel class of sensillum. J Neurosci. 2003;23:9906–9912. doi: 10.1523/JNEUROSCI.23-30-09906.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- Fishilevich E, Domingos AI, Asahina K, Naef F, Vosshall LB, Louis M. Chemotaxis behavior mediated by single larval olfactory neurons in Drosophila. Curr Biol. 2005;15:2086–2096. doi: 10.1016/j.cub.2005.11.016. [DOI] [PubMed] [Google Scholar]
- Fuss SH, Ray A. Mechanisms of odorant receptor gene choice in Drosophila and vertebrates. Mol Cell Neurosci. 2009;41:101–112. doi: 10.1016/j.mcn.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Goldman AL, Van der Goes van Naters W, Lessing D, Warr CG, Carlson JR. Coexpression of two functional odor receptors in one neuron. Neuron. 2005;45:661–666. doi: 10.1016/j.neuron.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Ho MG, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- Kim K, Colosimo ME, Yeung H, Sengupta P. The UNC-3 Olf/EBF protein represses alternate neuronal programs to specify chemosensory neuron identity. Dev Biol. 2005;286:136–148. doi: 10.1016/j.ydbio.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Komiyama T, Johnson WA, Luo L, Jefferis GS. From lineage to wiring specificity. POU domain transcription factors control precise connections of Drosophila olfactory projection neurons. Cell. 2003;112:157–167. doi: 10.1016/s0092-8674(03)00030-8. [DOI] [PubMed] [Google Scholar]
- Komiyama T, Carlson JR, Luo LQ. Olfactory receptor neuron axon targeting: intrinsic transcriptional control and hierarchical interactions. Nat Neurosci. 2004;7:819–825. doi: 10.1038/nn1284. [DOI] [PubMed] [Google Scholar]
- Kreher SA, Kwon JY, Carlson JR. The molecular basis of odor coding in the Drosophila larva. Neuron. 2005;46:445–456. doi: 10.1016/j.neuron.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Kreher SA, Mathew D, Kim J, Carlson JR. Translation of sensory input into behavioral output via an olfactory system. Neuron. 2008;59:110–124. doi: 10.1016/j.neuron.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon JY, Dahanukar A, Weiss LA, Carlson JR. The molecular basis of CO2 reception in Drosophila. Proc Natl Acad Sci U S A. 2007;104:3574–3578. doi: 10.1073/pnas.0700079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Salvaterra PM. Abnormal chemosensory jump 6 is a positive transcriptional regulator of the cholinergic gene locus in Drosophila olfactory neurons. J Neurosci. 2002;22:5291–5299. doi: 10.1523/JNEUROSCI.22-13-05291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna M, Monte P, Helfand SL, Woodard C, Carlson J. A simple chemosensory response in Drosophila and the isolation of acj mutants in which it is affected. Proc Natl Acad Sci U S A. 1989;86:8118–8122. doi: 10.1073/pnas.86.20.8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, van Naters WG, Shiraiwa T, Carlson JR. Mechanisms of odor receptor gene choice in Drosophila. Neuron. 2007;53:353–369. doi: 10.1016/j.neuron.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, van der Goes van Naters W, Carlson JR. A regulatory code for neuron-specific odor receptor expression. PLoS Biol. 2008;6:e125. doi: 10.1371/journal.pbio.0060125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee JM, Gruber CA, Brodie TB, Trieu M, Turner EE. Highly cooperative homodimerization is a conserved property of neural POU proteins. J Biol Chem. 1998;273:34196–34205. doi: 10.1074/jbc.273.51.34196. [DOI] [PubMed] [Google Scholar]
- Shanbhag SR, Muller B, Steinbrech RA. Atlas of olfactory organs of Drosophila melanogaster. 1. Types, external organization, innervations and distribution of olfactory sensilla. Int J Insect Morphol Embryol. 1999;28:377–397. [Google Scholar]
- Stocker RF. Design of the larval chemosensory system. Adv Exp Med Biol. 2008;628:69–81. doi: 10.1007/978-0-387-78261-4_5. [DOI] [PubMed] [Google Scholar]
- Tahayato A, Sonneville R, Pichaud F, Wernet MF, Papatsenko D, Beaufils P, Cook T, Desplan C. Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Dev Cell. 2003;5:391–402. doi: 10.1016/s1534-5807(03)00239-9. [DOI] [PubMed] [Google Scholar]
- Tichy AL, Ray A, Carlson JR. A new Drosophila POU gene, pdm3, acts in odor receptor expression and axon targeting of olfactory neurons. J Neurosci. 2008;28:7121–7129. doi: 10.1523/JNEUROSCI.2063-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Wey E, Schäfer BW. Identification of novel DNA binding sites recognized by the transcription factor mPOU (POU6F1) Biochem Biophys Res Commun. 1996;220:274–279. doi: 10.1006/bbrc.1996.0395. [DOI] [PubMed] [Google Scholar]