Abstract
Transfection of small RNAs (si/miRNAs) into cells typically lowers expression of many genes. Unexpectedly, increased expression of genes also occurs. We investigated whether this upregulation results from a saturation effect, i.e. competition for intracellular small RNA processing machinery between the transfected si/miRNAs and the endogenous pool of microRNAs (miRNAs). To test this hypothesis, we analyzed genome-wide transcript responses from more than 150 published transfection experiments in 7 different cell types. We show that endogenous miRNA targets have significantly higher expression levels following transfection, consistent with an impaired effectiveness of endogenous miRNA repression. Further confirmation comes from concentration and temporal dependence. Strikingly, the profile of endogenous miRNAs can largely be inferred by correlating miRNA sites with gene expression changes after transfections. The saturation and competition effects present practical implications for miRNA target prediction, the design of si/shRNA genomic screens and siRNA therapeutics.
Thousands of microRNAs (21–23nt ssRNAs) have been identified in animals over the past seven years [1, 2]. Subsequent research on miRNAs has focused on their biochemical processing and mechanism of action [3], the scope of their regulatory programs and their differential expression profiles in development and disease [4]. Furthermore, various si/miRNA constructs are widely used in functional genomics, miRNA cellular/tissue profiles are measured in medical diagnostics [5] and si/miRNAs (and their inhibitors) are in clinical trials for use as medical therapeutics [6, 7].
However, contrary to expectations, some genes are strongly upregulated in si/miRNA transfections (Supplementary Figure 1). In addition, despite the encouraging success of si/miRNAs in functional genomics and therapeutics, some studies have reported other unusual and unexpected effects suggestive of a compromised endogenous miRNA pathway. For instance, one study claims a non-specific immune response [8], while others implicate saturation of components of the sh/miRNA nuclear export machinery, for example, exportin 5 [9–11]. Some of these latter reports have suggested that saturation-related effects can be avoided by using siRNAs [11] rather than the short-hairpin RNAs (which rely on the nuclear export machinery) and a recent prominent report specifically claims that effective siRNAs used against APOB and F7 do not interfere with endogenous miRNA function [12]. In contrast, in an experiment designed to look at the off-target effects of siRNAs, scrambled siRNAs caused dose dependent upregulation of an observed target gene, SREBF1, in three different cell types [13], and an elegant report on combinatorial delivery of siRNAs in HEK 293 cell lines demonstrated competition for RISC machinery [14].
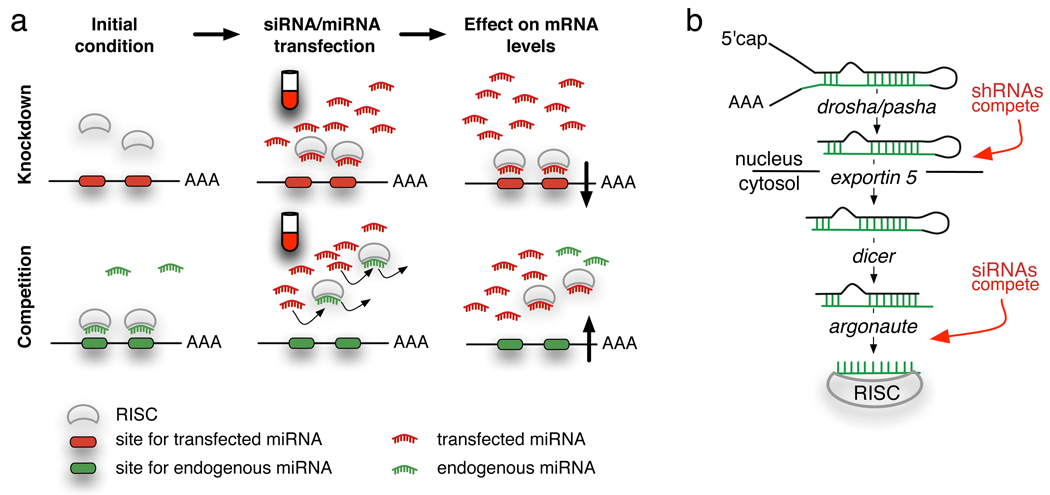
We hypothesize that the unexplained upregulation of genes in si/miRNA transfections are due, at least partly, to a loss of function of the endogenous miRNAs, as modeled in Figure 1 and supported by reports suggesting machinery saturation [14]. In this model, the transfected small RNAs must compete with the endogenous miRNAs for the RISC complex (RNA-induced silencing complex) or other machinery further downstream than exportin 5 in the miRNA pathway, e.g. Argonaute proteins or TRBP [14–17]. Loss of available RISC through competition will relieve the repression of target genes of endogenous miRNAs and upregulate the corresponding mRNAs and proteins.
Figure 1. Competition for small RNA machinery.
A schematic of the hypothesis that there is competition for RISC machinery between transfected si/miRNA and the cell’s endogenous miRNAs. a. Genes with sites (red) for the transfected small RNA (red) are downregulated after transfection; Genes with sites for endogenous miRNAs (green) may be upregulated after small RNA transfections. b. Biogenesis of miRNAs to show where sh- and siRNAs enter the miRNA processing pathway.
To test this hypothesis, we examined more than 150 miRNA and siRNA transfection experiments in 7 different cell lines. We reasoned that if endogenous miRNA activity is altered, we should be able to detect this effect in gene expression profiles taken after si/miRNA perturbations. Finally, if our hypothesis is correct, we may expect to see similar dose response and dynamics of these upregulated genes as for the downregulated target/off-target genes (but in the opposite direction) [18, 19].
Our results show that (i) genes with sites for endogenous miRNAs are significantly upregulated after the transfections, at both the mRNA and protein level, when compared to genes with neither endogenous nor transfected miRNA sites; (ii) genes with sites for the transfected si/miRNAs are more likely to be downregulated if they do not contain sites for endogenous miRNAs; (iii) a regression model predicts these shifts in gene expression from the number and type of miRNA sites in the affected genes; (iv) the transfection dose response of genes with sites for endogenous miRNAs is similar to that of the downregulated genes; and (v) the temporal response of genes with sites for endogenous miRNAs is similar to that of the genes with sites for the transfected si/miRNAs. Our results also highlight specific examples of genes that are consistently upregulated in certain cell types after transfections, including the oncogene HMGA2 and genes involved in cell cycle regulation.
Results
Endogenous miRNA targets are upregulated post miRNA transfection
To investigate whether si/miRNA transfections affect gene regulation by endogenous miRNAs, we assembled data from small RNA transfection experiments followed by mRNA profiling and protein mass spectrometry (Methods and Supplementary Figure 2). These data comprise more than 150 experiments from 7 different cell types, involving more than 20 different miRNAs and 40 unique siRNAs (Table 1 and Supplementary Table 1 and Supplementary Table 2). For each cell type, we used available miRNA expression profiles [20–22] to define the 10 most highly expressed endogenous miRNAs, which together make up 70–80% of the measured cellular miRNA content (Supplementary Figure 3 and Methods). Strikingly, a large number of genes are upregulated rather than downregulated in the si/miRNA experiments, see Supplementary Figure 1. We asked whether genes that are predicted targets of the cells’ own (endogenous) miRNAs respond differently to the transfected si/miRNAs as compared to all other genes (Methods). In this analysis, we defined the set D of genes with predicted sites for ‘enDogenous’ miRNAs, the set X of predicted target genes of the ‘eXogenous’ si/miRNA, and a ‘Baseline’ set B of genes with neither endogenous nor exogenous sites. Differences in global expression changes between gene sets following si/miRNA transfection or miRNA inhibition were assessed for statistical significance by a one-sided Kolmogorov-Smirnov (KS) test (Methods).
Table 1.
| Transfected microRNA | Cell type | Reference |
|---|---|---|
| miR-124, miR-1, miR-373, miR-124mut5-6, miR-124mut9-10, chimiR-1-124, chimiR-124-1 | HeLa | Lim, L.P., et al., Microarray analysis shows that some microRNAs down-regulate large numbers of target mRNAs. Nature, 2005. |
| miR-106b, miR-200a/b, miR-141 miR-16, miR-15a/b, miR-103, miR-107, miR-192, miR-215, miR-17-5p, miR-20, let-7c, miR-195 | HeLa, HCT116 HCT116 Dicer−/− | Linsley, P.S., et al., Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol, 2007. |
| miR-7, miR-9, miR-122a, miR-128a, miR-132,miR-133a, miR-142, miR-148, miR-181a | HeLa | Grimson, A., et al., MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell, 2007. |
| miR-34a/b/c | HeLa, A549, TOV21G, HCT116 Dicer−/− | He, L., et al., A microRNA component of the p53 tumour suppressor network. Nature, 2007. |
| miR-1, miR-155, let-7, miR-30 | HeLa | Selbach, M., et al., Widespread changes in protein synthesis induced by microRNAs. Nature, 2008. |
| miR-181a, miR-124, miR-1 | HeLa | Baek, D., et al., The impact of microRNAs on protein output. Nature, 2008. |
| miR-34a | HeLa | Chang, T.C., et al., Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell, 2007. |
| miR-124 | HepG2 | Wang, X. and X. Wang, Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res, 2006. |
| Transfected siRNA | Cell type | Reference |
| igfr-1-16, mapk14-1-8, mapk14-1 (1,2,4,6,12,24,48,72,96) hours, mapk14-1 (.16,.8,4,20,100) nM, makk14-1 (pos 4,5,15) mismatch | HeLa | Jackson A.L., et al., Expression profiling reveals off-target gene regulation by RNAi. Nature Biotechnology, 2003. |
| mphosph1-2692, pik3ca-2692, prkce-1295, vhl-2651, vhl-2652, sos1-1582 | HeLa | Jackson A.L., et al., Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA, 2006. |
| pik3cb-6338, plk-1319, plk-772, pik3cb-6340 | HeLa | Jackson A.L., et al., Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA, 2006. |
| siApoB-Hs1/Hs2/Hs3/Hs4 | Huh7 | Burchard J,et al., MicroRNA-like off-target transcript regulation by siRNAs is species specific. RNA, 2009. |
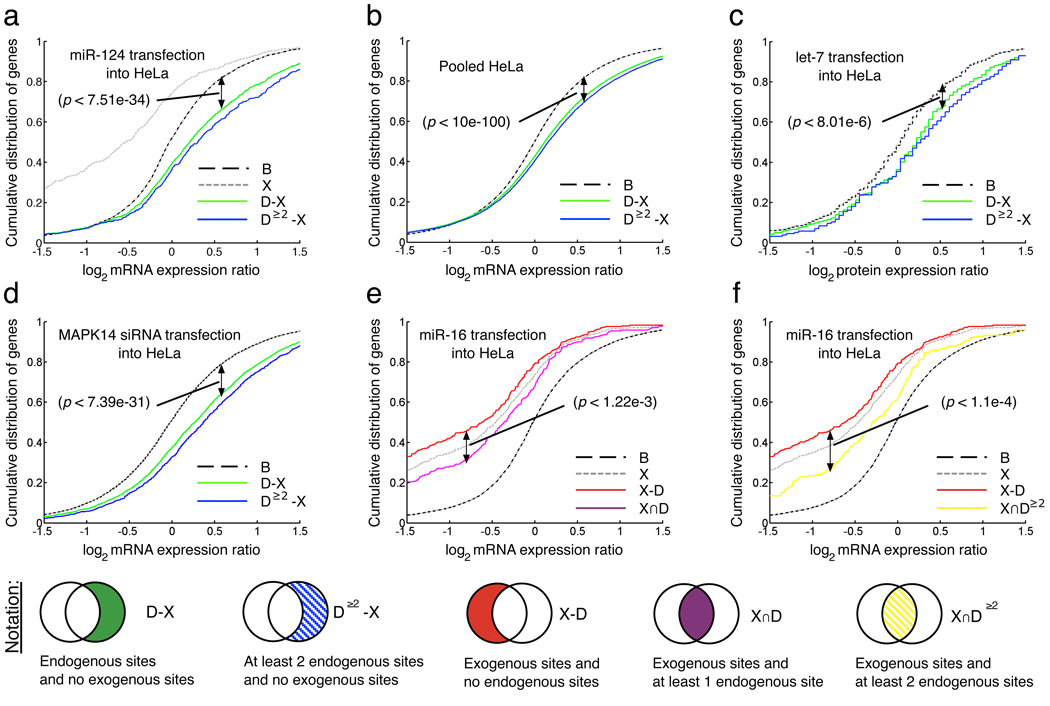
We found that in 90% of the experiments tested, the cumulative distribution of expression changes of the set of genes with endogenous target sites and no exogenous sites (D–X) was significantly up-shifted compared to the baseline set (Supplementary Table 2). For instance, when miR-124 is transfected into HeLa cells [23], genes with sites for HeLa-expressed (endogenous) miRNAs and no miR-124 sites are significantly upregulated compared to the baseline set (p < 7.5e34; Figure 2a, green line and Supplementary Table 2). The size of the effect is even more pronounced when we compare the upregulation of genes with at least 2 endogenous sites and no sites for the transfected miRNA to the baseline set (p < 2.2e-24; Figure 2a, blue line).
Figure 2. Genes with predicted target sites for endogenous miRNAs are significantly dysregulated after si/miRNA transfections.
Visual representation of gene sets tested for significance in expression changes against baseline (B). X is the set of genes with (predicted) sites for exogenous si/miRNAs, D–X is the set of genes with (predicted) sites for endogenous miRNA and no (predicted) sites for exogenous si/miRNAs. p-values shown are calculated by one-sided KS test as described in Methods. a. miR-124 transfection results in up-shift in gene expression for D–X with respect to baseline. b. Pooled data from 15 miRNA transfections into HeLa cells; miR-373, miR-124, miR-148b, miR-106b, miR-124, mut9-10, miR-1, miR-181, chimiR-124-1, chimiR-1-124, miR-16, mir-34a, mir-34b, miR-128a, and miR-9. c. Protein expression changes after let-7b transfection. d. mRNA expression changes after MAPK14-siRNA is transfected into HeLa cells showing upregulation of genes with sites for endogenous miRNAs (green line) [19];
e., f., Genes which contain sites for both endogenous miRNA and transfected small RNAs are less downregulated than if they contain only sites for transfected small RNAs. Changes in gene expression after miR-16 transfections, where p-values shown are calculated by one-sided KS test as described in Methods. e. X–D is the set of genes containing exogenous sites and no endogenous sites. X∩D is the set of genes with sites for both endogenous and exogenous miRNAs. f. X–D≥2 is the set of genes containing exogenous sites but less than 2 endogenous sites. X∩D is the set of genes with sites for both endogenous and exogenous miRNAs.
To see if the upregulation of genes with predicted sites for endogenous miRNAs was a general effect, we pooled all the HeLa transfection experiments and repeated the analysis. We found that the ‘competition’ effect is supported by the highly significant upregulation of the set D–X in the pooled HeLa data (p < 10−100; Figure 2b). The same result is also true for pooled data in A549, HCT116, HCT116 Dicer−/− and Tov21G cells, with p < 10−10 for all cell types (Supplementary Table 2). Interestingly, over-expression of one endogenous miRNA also affects the targets of other endogenously expressed miRNAs. For example, in miR-16 and let-7b transfections into HeLa cells, the set D–X was upregulated compared to the baseline set (p < 5.6e-19, p < 6.1e-12 respectively; Supplementary Table 2, Supplementary Figure 4).
As a positive control, we also compared the expression changes for the set of exogenous target genes (X) and the baseline gene set (B), both in individual transfection experiments and in sets of experiments grouped by cell type (Table 1). As expected, we found that the mRNA expression levels of target genes of the transfected small RNAs were significantly downshifted compared to the baseline set (Supplementary Table 2).
We also investigated protein expression levels in HeLa cells using data from mass spectrometry experiments following miRNA transfection [24] and found significant changes in protein expression following the five transfections (Supplementary Table 2). Target genes with sites for endogenous miRNAs and no sites for exogenous miRNAs (D–X) were upregulated in protein expression when compared to the baseline of genes with neither exogenous nor endogenous miRNA sites, (p < 1.3e-9, pooled data). For example, transfection of let-7b into HeLa cells significantly increases protein expression of genes with other endogenous target sites only compared to the baseline gene set (p < 8e-6; Figure 2c, green line).
siRNA transfections display the same effect as miRNA transfections
Next, we investigated 43 independent siRNA transfections in HeLa cells [19, 25, 26] to look for changes in regulation by endogenous miRNAs. We found (Figure 2d) significant upregulation of gene expression after MAPK14-siRNA transfection for targets of endogenous miRNAs. Upregulation of let-7 and miR-15 targets, two miRNAs highly expressed in HeLa cells, was especially significant (Supplementary Table 3). Pooling the data from these siRNA experiments, we see a significant upward shift in expression of genes with endogenous sites only relative to the baseline gene set (p < 10−100; Supplementary Table 2). Five different siRNAs designed to target VHL, PRKCE, MPHOSPH1, SOS1, PIK3CA [26] all showed striking upregulation of highly similar sets of genes including CCND1, DUSP4, DUSP5, ATF3 (Supplementary Table 3). Each one of these upregulated genes contains at least one site for an endogenous miRNA, consistent with upregulation as a consequence of the siRNA transfection, independent of the specific siRNA sequence.
Targets with endogenous sites are less downregulated than expected
Next, we asked whether the response of genes directly targeted by the transfected si/miRNA also showed evidence of the competition effect. Specifically, we partitioned the set of genes with sites for transfected miRNAs (set X) into two subsets: genes with only exogenous sites and no endogenous sites (set X–D); and genes with both exogenous and endogenous sites (X∩D). After transfection of miR-16 into HeLa cells, genes with miR-16 target sites and no endogenous sites (Figure 2e, red line) are significantly more downregulated than target genes with endogenous sites (Figure 2e, magenta line), p < 1.2e-3 (X–D vs. X∩D), with even greater difference when compared to genes with two or more sites for endogenous miRNAs (p < 1.1e-4), Figure 2f, yellow line. Pooling data across a panel of transfection experiments into HeLa cells gave an even more significant result (p < 3.6e-13), Supplementary Table 2.
A quantitative model resolves the endogenous miRNA profile
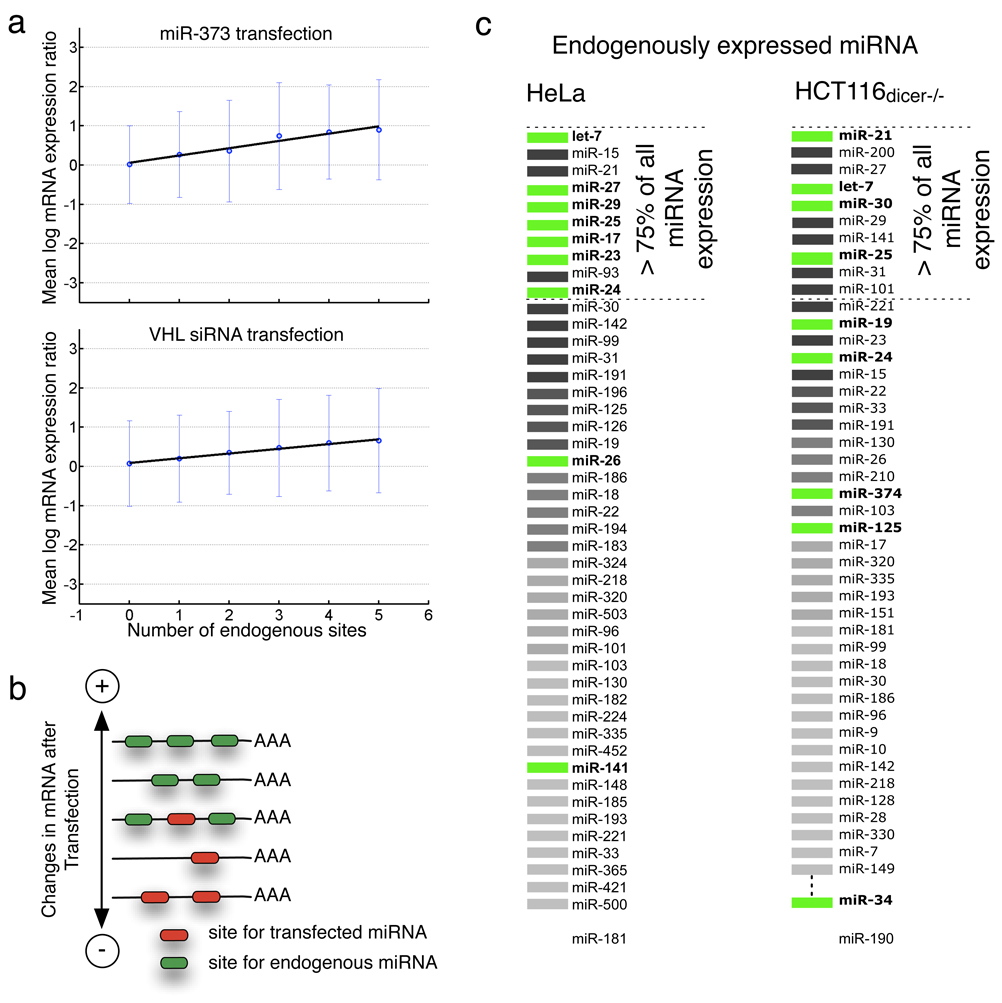
To strengthen our analysis and predict the saturation effect on individual genes, we built a quantitative mathematical model of the change in gene expression after si/miRNA transfection. This model can be used by the siRNA community to predict which genes are likely to be upregulated as well as downregulated (off-target effects) after si/miRNA transfections. Considering each transfection into HeLa cells independently, we first fit a simple linear regression model (Methods) to predict the change in expression of genes from the number of exogenous sites (nX) and the number of endogenous sites (nD) in the 3’ UTR of genes (Figure 3a). In a large majority of experiments, the endogenous count nD was found to be a significant variable for explaining expression changes (84 out of 109 experiments satisfying p < .05 by F statistic, Supplementary Table 2). As expected, the regression coefficient for the endogenous count was always positive when significant, meaning that these sites correlate with upregulation, while the regression coefficient for the exogenous count was always negative. Figure 3b is a cartoon version of the expected effect on expression of a gene that contains different combinations of exogenous and endogenous sites. We then refined the model to assess whether the presence of sites of individual miRNAs could explain upregulation of targets in an experiment, considering all human miRNA families as potential variables. We ranked the importance of each individual miRNA by the number of experiments in which it was included in a forward stepwise regression model (Methods). Among the 10 most frequently included miRNAs, we identified 7/10 of the most highly expressed miRNAs in a HeLa and 4/8 of the most highly expressed in HCT116 Dicer−/− cells, using no prior knowledge of the miRNA profile, Figure 3c. The top ranked miRNAs retrieved by this analysis, let-7 and miR-21 are the most highly expressed miRNA in HeLa and HCT116 Dicer−/− cells respectively, and therefore strongly supports a saturation model. Indeed, taken altogether, these results suggest that the endogenous miRNA profile in a cell can largely be determined simply from expression changes after transfection of small RNAs, plausibly due to competition for cellular resources.
Figure 3. Quantitative model predicting expression change after transfection.
a. Linear regression fitting mean log(expression change) to number of endogenous sites in genes having no exogenous sites, in miR-373 and VHL-siRNA experiments (respectively). One standard deviation from mean depicted with error bars.
b. Cartoon graph of genes with different combinations of sites showing net effect on gene expression.
c. The relative rank of the ten miRNAs that occurred most often with positive regression coefficient in stepwise regression models for >20 HeLa and 16 HCT116 Dicer−/− transfection experiments (see Methods). These miRNAs can be interpreted as a predicted endogenous profile. The true endogenous HeLa and HCT116 Dicer−/− profile is shown, with miRNAs ranked in order of endogenous expression. Correctly predicted endogenous miRNAs are shown in green and other endogenous miRNAs in black-grey; white indicates a miRNA predicted through regression analysis and not expressed endogenously.
The competition effect has a dose response
In a previous study investigating siRNA dose response, a siRNA targeting MAPK14 was transfected into HeLa cells in a range of 5 doses, from 0.16nM-100nM, followed by microarray profiling after 24 hours [19]. We re-analyzed this data and confirmed the original analysis that the off-target effects of the siRNA mimic the dose response of the main target (MAPK14) and are not titrated away at lower transfection concentrations. However, there is also a set of upregulated genes that are consistently regulated in proportion to the dose of the siRNA (Figure 4a): genes with sites for endogenous miRNAs follow a pattern of upregulation that mirrors the downregulation of off-target genes with sites for the transfected siRNA in the 3’ UTR. A 5-fold change in siRNA dose from 4 nM to 20 nM produces a 2-fold change in mean gene expression of the most responsive upregulated genes and the most responsive downregulated genes. The change in expression of both the endogenous target and off-target sets reaches near-maximal dose response at 20 nM; the saturation effect and siRNA off-target effects roughly scale with the dose response of the main target, at least for a significant fraction of genes in these sets, and cannot be titrated away at lower transfection concentrations.
Figure 4. siRNA transfections result in upregulation of genes with sites for endogenous miRNAs, have a dose-response and dynamics similar to targeted effect but in opposite direction.
b. A bar graph of the dose of siRNA transfection ranging 0.16nM–100nM versus mean log(expression change) in XNC(grey) and 90th percentile of DNC ≥2 – XNC (green). The graph shows that the putative saturation effect on the endogenous target genes has a dose response similar to the siRNA targeted genes but in the opposite direction. c. A bar graph of mean log(expression change) of XNC(grey) and 90th percentile of DNC ≥2 – XNC (green) versus time. d. A bar graph of log(expression change) versus putative endogenously regulated genes SCML2, TNRC6, YOD1, CX3CL1, AKAP12, and PGM2L1, and a set of MAPK14-siRNA (‘off-targets’) MAPK14 (light-green), MARK2, SLC35F3, HMGB3, FZD7, RPA2, IER5L, at a series of time points over 3 days (genes displayed in this order, from left to right).
Evidence for a transitory saturation effect
We also examined the dynamics of the gene expression changes over 96 hours after siRNA transfections to measure the time dependence of the response of genes with sites for endogenous miRNAs. If genes under endogenous miRNA regulation are de-repressed, we expect to see the response to have a similar time progression as that of the intended siRNA target genes and its off-targets. We compared the mRNA changes of the putative off-target genes of the siRNA to the MAPK14 mRNA itself. Although the off-target genes of the siRNA (genes with non-conserved seed matches, XNC) follow a temporal downregulation pattern similar to MAPK14 in the first 48 hours, the expression level of the XNC set of genes returns to near its original expression level by 92 hours. Indeed, the intended target MAPK14 has a gradually increasing downregulatory effect, with a half maximal effect seen at ~12 hours and a sustained effect from 24–96 hours (Figure 4c; light-green bar).
We investigated the dynamics of a set of genes with at least 2 non-conserved endogenous sites (90th percentile for expression change, pooling all time points, ~ 1000 genes), compared to a set of siRNA targeted genes (Methods). The genes in the endogenous set have maximal upregulation at 24–48 hours with similar dynamics across the 92 hours, consistent with being ‘on-targets’ of endogenous miRNAs competing for components of the RISC (Figure 4b). To examine particular examples, we compared the expression patterns of a set of the 6 most downregulated ‘off-target genes’ with 6 of the most upregulated genes in set DNC-XNC and found strikingly similar temporal effects (Figure 4c). The upregulated genes, SCML2, TNRC6, YOD1, CX3CL1, AKAP12, and PGM2L1, have maximal upregulation at 24–48 hours with similar dynamics across the 92 hours and contain at least 4 sites for highly expressed endogenous miRNAs. These genes are consistent with being ‘on-targets’ of endogenous miRNAs competing for components of the RISC (Figure 4c). Notably, TNRC6 is associated with AGO2 in P-bodies.
We also investigated a recent set of experiments that were designed to examine the off-target effects of a therapeutic siRNA targeting APOB [18]. Our results showed a highly significant saturation effect with all 4 siRNAs designed to target the human APOB (p < 1e-8 at 6 hours, Supplementary Table 2). We noticed that this siRNA effect reached its maximum effect at 6 hours, in line with the faster response time of the experiment as noted by the authors. The upregulated genes with sites for endogenous miRNAs also reached their maximum effect rapidly. Taken together these investigations of dynamics of small RNA gene regulation after transfection show that the upregulatory effect mirrors the expected downregulatory effect and supports the proposed competition model.
Cell cycle genes are upregulated after si/miRNA transfections
Dysregulation of endogenous miRNAs is known to contribute to tumorigenesis [27], and the experiments we analyzed were conducted in immortalized cell lines (e.g., HeLa cells). We were therefore not surprised to find a significant number of cell cycle, oncogene, and tumor suppressor genes (Supplementary Figure 5a) consistently upregulated across transfection experiments (Supplementary Table 1). For instance, known miRNA targets, including the oncogene HMGA2 [28], CCND1 [29, 30] and DUSP2 are upregulated after many different independent HeLa transfection experiments, including siRNA transfections. We also find that cell cycle genes are significantly enriched in endogenous miRNA target sites compared to other genes expressed in HeLa (Supplementary Figure 5b). Together, this suggests that cell cycle and oncogenes are particularly susceptible to the proposed saturation effect.
miRNA inhibition may cause upregulation of other endogenous targets
Finally, we examined mRNA expression changes after miRNA inhibition. miR-16 and miR-106b ‘antagomirs’ [31], 2’-O-methyl inhibitors, produced a significant upregulation of genes which contained only endogenous sites, p < 5e-16 (D – X) and p < 2e-30 (D≥2–X). There is a set of genes significantly de-repressed in both experiments, including, for example, SSR3, PLSCR4 and PTRF, (Supplementary Table 3), although they contain no predicted sites for transfected inhibitors. They do, however, contain sites for endogenously expressed miRNAs, and so they serve as examples for the general effect that we observed statistically. Inhibition of miR-122 with LNA molecules [32] also produced significant upregulation of genes with sites for other endogenous miRNAs when compared with a saline transfection (p < 2.5e-6). Elmen et al. observed dose-dependent accumulation of a shifted heteroduplex band, implying that the LNA-antimiR binds stably to the miRNA [32]. This finding is consistent with hypothesis that the heteroduplex of miR-122∷antimiR prevents the availability of free RISC machinery, (Supplementary Figure 6) but clearly more experiments are needed to distinguish between the possible models and assess the size of the inhibition effect on the function of endogenous miRNAs.
Discussion
We have shown that the expression of genes predicted to be under endogenous miRNA regulation is affected by small RNA transfection; that the effect is observable both at the mRNA and protein levels; and that it occurs following transfection of siRNAs designed to inhibit particular genes, as well as miRNA mimics and miRNA inhibitors introduced to test the biological effects of miRNAs. In a quantitative approach, we built a regression model that can to a large extent recover the endogenous miRNA profile simply from the changes in gene expression following small RNA transfections. The purpose of this approach is not to infer the miRNA profiles per se but to provide independent strong evidence of the indirect perturbation of miRNA function. Finally, we used a series of published data to show that the dynamics and dose response of the genes affected by the proposed competition effect follow the same patterns as that of the genes directly targeted by the transfection.
The most plausible model for these observations is saturation of the RISC complex (or other necessary small RNA processing or transport machinery) and competition between the transfected small RNA and endogenous miRNA for binding (Figure 1). Other models cannot be ruled out by this analysis and may be consistent with the observed effect. While the precise mechanism of this competition effect remains to be established, the statistical significance of the observed shifts in transcript levels is clear, and the results of this analysis strongly support the thesis that small RNA transfections unexpectedly and unintentionally (from the point of view of the investigators) disturb gene regulation by endogenous miRNA.
Our results have potentially important practical consequences for the use of siRNAs, as well as shRNAs, in functional genomics experiments. While it is already known that siRNAs can produce unwanted off-target effects, i.e. unintended downregulation of mRNAs via a partial sequence match between the siRNA and target, the effects observed here are distinct and involve the de-repression of miRNA-regulated genes.
Our findings also have consequences for the development of miRNA target prediction methods, in two ways. First, as measuring mRNA expression changes after si/miRNA perturbations is a standard way to validate miRNA target prediction methods [23, 25, 33], one should take the saturation effect into consideration. Despite concerted efforts, bioinformatic si/miRNA target prediction methods still significantly over-predict the number of targets by at least 7 fold [24, 34–36]. Elegant work showing the dynamic (condition and cell-type dependent) regulation of UTR lengths [37] may explain some of these false positives, since shortening of UTRs may lead to loss of target sites, but is unlikely to explain all. The proposed competition effect may offer an explanation for false positive target prediction in cases where UTRs have target sites for both the transfected and endogenous miRNAs (Figure 3b). Second, as miRNAs may compete with each other, for target sites in mRNAs, the very idea of a ‘target’ mRNA should be re-assessed.
Further, our results have consequences for the development of small RNA therapeutics, considered to hold substantial promise [38]. miRNA inhibitors, e.g., anti-miR-122, have been used to target cholesterol synthesis [39] as well as HCV (hepatitis C virus) [39, 40] and HSV2 (herpes simplex virus) [41]. Therapeutic siRNAs have also been designed for potential treatment of cancer, including in melanoma, against VEGF-A/-C [42], and through anti-miR-21 in glioma [38, 43, 44]. Our work illustrates the potentially broad consequences of the perturbation of the cell's miRNA activity profile after introduction of si/miRNA inhibitors and suggests that these effects be considered quantitatively during development of small RNA therapies. Experiments that quantify the relative concentrations of protein machinery and small RNAs in a particular cellular context, as well as a fuller exploration of the kinetics of the various binding events involved in small RNA biogenesis and function, are clearly required. Our quantitative model implies a procedure for calibrating and potentially avoiding unwanted effects of the designed small RNA therapeutics.
Our work tests the hypothesis that transfections of small RNAs can perturb endogenous miRNA function, subject to some limitations. In particular, this report does not attempt to resolve details of the mechanism behind the competition effect. The calculations of the effect, though carefully evaluated in statistical terms, are subject to the inaccuracies of miRNA target prediction, which entails both false positives and false negatives at the level of particular target genes. We therefore argue in terms of overall distributions, rather than attempting to quantify the involvement of individual target sites in transfection-mediated expression changes. In future work, a number of quantitative criteria will determine the extent of the competition between exogenous and endogenous miRNAs and their effects on gene targeting. Quantitative detail will depend on knowing the concentration of the RISC complex and of other components of the small RNA machinery in the cell, the concentration of the transfected and endogenous miRNAs, the concentrations of the target mRNAs, and the number of actual targets in the cell for a specific small RNA, as well as kinetic parameters such as the on and off rates of small RNAs in the RNA-protein complexes. Models that posit different concentration-dependent and kinetic scenarios could help focus the range of experiments needed to quantify these effects.
Finally, our results may have an important biological correlate, as plausibly the competition effect may have a role in normal biological or disease-related cellular processes, e.g., in affecting miRNA-dependent regulatory programs. For example, during both differentiation and disease processes such as cancer, miRNA profiles can change dramatically both in the identity of the dominant miRNAs and in total cellular miRNA concentration. Such changes, via competition for limited resources, may orchestrate observable changes in cellular regulatory programs with potential physiological consequences.
In summary, the proposed and statistically supported competition effect for small RNAs may point to new biological mechanisms and likely has important practical consequences for the use of small RNAs in functional genomics experiments, for the development of miRNA target and siRNA off-target prediction methods, and for the development of small RNA therapeutics.
Methods
mRNA and protein experimental datasets
We collected data from four types of experiments: (i) transfection of a miRNA followed by mRNA profiling using microarrays [4, 23, 24, 29, 35, 45]; (ii) transfection of an siRNA followed by mRNA profiling [18, 19, 26] ; (iii) inhibition of miRNA followed by mRNA profiling [32]; and (iv) transfection of miRNA followed by protein profiling using mass spectrometry [24]. These four types of data sets of 150 experiments encompass 7 different cell types, 20 different miRNAs, and 40 different siRNAs (Supplementary Table 2). The synthetic transfected miRNAs are all commercially available siRNA/miRNA mimics (Dharmacon, Inc.). Sequences of mimics can be found in the respective references. When possible, we used normalized microarray expression data as provided with the original publications. In all other cases, we used the "affy" package in the "R" software package to perform RMA normalization of microarray probe-level data. For statistical analysis over multiple mRNA microarray profiling experiments, each experiment was independently centered using the mean log(expression change) of genes lacking conserved endogenous or exogenous sites and normalized to have unit variance in log(expression change) across all genes. This normalization results in a modified Z-transformation of the data, where genes with no exogenous or endogenous sites have mean 0. For the transfection experiments followed by mass spectrometry, we used normalized protein expression levels as provided by the authors of the original publication, Supplementary Figure 2.
Target prediction
We conducted four different types of miRNA target site searches using miRNA sequences grouped into families, and 3’ UTR alignment of 5 species. miRNAs were grouped into families as defined by identical nucleotides in positions 2–8. We searched for target sites for miRNA families in 3’UTRs using four different types of seed matches: (i) 6-mers (position 2–7 and 3–8), (ii) 7-mers (position 2–8), (iii) 7-mer positions 2–7 m1A (the first nucleotide an A in the mRNA) and (iv) 8-mers (position 1–8). 7-mer positions 2–8 were selected for analysis since this choice gave the most significant p-values for downregulation of targets with sites for the transfected si/miRNA based as compared to baseline genes based on a one-sided KS statistic (set X versus set B, as described below).
For target matches, we considered both non-conserved and conserved targets in human 3’UTRs. 3' UTR sequences for human (hg18), mouse (mm8), rat (rn4), dog (canFam2), and chicken (galGal2) were derived from RefSeq and the UCSC genome browser (http://hgdownload.cse.ucsc.edu/goldenPath/hg18/multiz17way/). We used multiple genome alignments across the 5 species as derived by multiZ. The RefSeq annotation with the longest UTR mapped to a single gene was always used. To establish a conservation filter, we required that the 7-mer target site in human be present in at least three of the other four species, i.e. exact matching in a 7 nucleotide window of the alignment in at least 3 other species, to be flagged as conserved. Restricting to conserved sites led to more significant p-values for downregulation of targets with exogenous sites as compared to baseline genes (one-sided KS statistic, set X versus set B, as defined below). We chose these stringent requirements so that our prediction method would be conservative and err on the side of under-prediction rather than over-prediction. However, we acknowledge that there are indeed functional siRNA and miRNA target sites that have mismatches, G:U wobbles in the 5’ end and are not conserved (see for example work of the Hobert and Slack groups [46, 47]).
Endogenous miRNA expression
We used endogenous miRNA profiles from the Landgraf et al [20] compendium for HeLa, A549, HepG2 and TOV21G, which provide relative miRNA expression levels from cloning and sequencing small RNA libraries. We used miRNA profiles from the Cummins et al [48] cloning and sequencing data for HCT116 and HCT116 Dicer−/−. For consistency across cell types, we took the top 10 miRNAs with highest expression levels (clone counts), which corresponds to at least 75% of the miRNA content in each cell type, to be the set of endogenous miRNAs in our statistical analysis.
KS statistics
To compare the expression changes for two gene sets, we compared their distributions of Z-transformed log(expression change) using a one-sided Kolmogorov-Smirnov (KS) statistic, which assesses whether the distribution of expression changes for one set is significantly shifted downwards (downregulated) compared to the distribution for the other set. We chose the KS statistic to apply a uniform treatment of data despite the heterogeneity of the transfection experiments, which involve different cell types, different numbers of target genes with sites for the transfected si/miRNA, and different apparent transfection efficiencies. The KS statistic has the advantages that (i) it is non-parametric and hence does not rely on distributional assumptions about expression changes; (ii) it does not rely on arbitrary thresholds; and (iii) it measures significant shifts between the entire distributions rather than just comparing the tails. The KS statistic computes the maximum difference in value of the empirical cumulative distribution functions (cdfs):
, where is the empirical cdf for gene set j = 1, 2, based on nj (Z-transformed) log(expression change) values. We used the Matlab function kstest2 to calculate the KS test statistic and asymptotic p-value. Full KS test results are provided in Supplementary Table 2.
Notation
We use the following notation to describe sets of genes based on the number of sites for exogenous and endogenous miRNAs in their 3’UTRs:
Non-conserved sites: sets with subscript NC denote non-conserved sites have been used; subscript NC ≥2 denotes 2 or more non-conserved sites
Endogenous sites: sites for endogenous miRNAs, i.e., miRNAs expressed in the cell
Exogenous sites: sites for exogenous si/miRNAs, i.e., small RNAs introduced into the cell
X (“eXogenous”): set of genes containing at least one site for the exogenous (transfected) si/miRNA
D (“enDogenous”): set of genes containing at least one site for a miRNA endogenously expression in the cell type
B (“Baseline”): set of genes containing neither exogenous nor endogenous sites
D–X: set of genes containing at least one endogenous site and no exogenous sites
X∩D: set of genes containing at least one exogenous site and at least one endogenous site
X–D: set of genes containing at least one exogenous site and no endogenous sites
D≥2: set of genes containing 2 or more endogenous sites
X∩D≥2: set of genes containing at least one exogenous site and at least 2 endogenous sites
Regression analysis to model expression
We performed multiple linear regression to fit a linear model expressing the Z-transformed log(expression change), denoted as y, in terms of the number of a gene’s exogenous and endogenous target sites, denoted as nX and nD, respectively:
We use the Matlab regress function to fit the model and assess the significance of the fit as measured by the R2 statistic.We used the F statistic, also computed by the regress function, to assess whether the linear model with 2 independent variables, nX and nD, significantly improves the fit over the simpler model: y = cX nX + b, given the number of sites for exogenous si/miRNAs a priori. All p-values from the F statistic across experiments are reported in Supplementary Table 2.
Forward stepwise regression analysis
As an extension to the linear model with 2 independent variables, we performed forward stepwise regression to fit the number of target sites for each of the (162) miRNA families to the Z-transformed log(expression change) data. Starting again with the simpler model, y = cX nX + b, we incrementally added the number of target sites for the miRNA seed family with highest F statistic to the model. The procedure was continued until the p-value from the F-statistic for the best remaining seed family failed to satisfy a significance threshold of p < .05. The final model can be viewed as a linear combination of the number exogenous target sites and the additive contribution of other miRNAs represented by their number of target sites ni:
Since we did not enforce a stringent significance criterion for including miRNA sites in the model, we do not expect every miRNA added to the model to be correct; however, miRNAs added consistently across different transfections experiments are likely to be significant. We repeated the forward stepwise regression for multiple experiments in HeLa and HCT116 Dicer−/− cells and computed the frequency of the most statistically significant additive factors with positive regression coefficient in the model for each cell type; we reported the 10 most frequent of these miRNAs. All p-values from the F statistic across experiments are reported in Supplementary Table 4.
Cell cycle and cancer genes
A list of expertly annotated genes for which mutations (both germline and somatic) have been causally implicated in cancer was obtained from the Cancer Genome Project (Cancer Gene Census catalogue version 2008.12.16, http://www.sanger.ac.uk/genetics/CGP/Census) [49]. A list of genes that have consistently showed a periodic expression pattern during the cell cycle in several mRNA microarray studies was obtained from the Cyclebase data base [50]. From these lists, we could match 312 and 651 genes to the mRNA data sets collected in this work, respectively. The gene sets were designated “oncogenes” and “cell cycle genes”, respectively. To investigate if oncogenes or cell cycle genes were enriched for miRNA targets in Hela cells compared to all genes we used Fisher’s exact tests.
Supplementary Material
Acknowledgements
We thank Nick Stroustrup, Yifat Merbl, Grégoire Altan-Bonnet, Aaron Arvey, Nick Gauthier and Teresia Dahl for useful discussions.
References
- 1.Ruvkun G. The perfect storm of tiny RNAs. Nat Med. 2008;14(10):1041–1045. doi: 10.1038/nm1008-1041. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Tam W. The emergent role of microRNAs in molecular diagnostics of cancer. J Mol Diagn. 2008;10(5):411–414. doi: 10.2353/jmoldx.2008.080067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossi J, Zamore P, Kay MA. Wandering eye for RNAi. Nat Med. 2008;14(6):611. doi: 10.1038/nm0608-611. [DOI] [PubMed] [Google Scholar]
- 7.Leachman SA, et al. Therapeutic siRNAs for dominant genetic skin disorders including pachyonychia congenita. J Dermatol Sci. 2008;51(3):151–157. doi: 10.1016/j.jdermsci.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robbins M, et al. Misinterpreting the therapeutic effects of siRNA caused by immune stimulation. Hum Gene Ther. 2008 doi: 10.1089/hum.2008.131. [DOI] [PubMed] [Google Scholar]
- 9.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 10.Stewart CK, Li J, Golovan SP. Adverse effects induced by short hairpin RNA expression in porcine fetal fibroblasts. Biochem Biophys Res Commun. 2008;370(1):113–117. doi: 10.1016/j.bbrc.2008.03.041. [DOI] [PubMed] [Google Scholar]
- 11.McBride JL, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105(15):5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.John M, et al. Effective RNAi-mediated gene silencing without interruption of the endogenous microRNA pathway. Nature. 2007;449(7163):745–747. doi: 10.1038/nature06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vankoningsloo S, et al. Gene expression silencing with 'specific' small interfering RNA goes beyond specificity - a study of key parameters to take into account in the onset of small interfering RNA off-target effects. Febs J. 2008;275(11):2738–2753. doi: 10.1111/j.1742-4658.2008.06415.x. [DOI] [PubMed] [Google Scholar]
- 14.Castanotto D, et al. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35(15):5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruber J, et al. RNAi of FACE1 protease results in growth inhibition of human cells expressing lamin A: implications for Hutchinson-Gilford progeria syndrome. J Cell Sci. 2005;118(Pt 4):689–696. doi: 10.1242/jcs.01652. [DOI] [PubMed] [Google Scholar]
- 16.Wargelius A, Ellingsen S, Fjose A. Double-stranded RNA induces specific developmental defects in zebrafish embryos. Biochem Biophys Res Commun. 1999;263(1):156–161. doi: 10.1006/bbrc.1999.1343. [DOI] [PubMed] [Google Scholar]
- 17.Oates AC, Bruce AE, Ho RK. Too much interference: injection of double-stranded RNA has nonspecific effects in the zebrafish embryo. Dev Biol. 2000;224(1):20–28. doi: 10.1006/dbio.2000.9761. [DOI] [PubMed] [Google Scholar]
- 18.Burchard J, et al. MicroRNA-like off-target transcript regulation by siRNAs is species specific. Rna. 2009;15(2):308–315. doi: 10.1261/rna.1326809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson AL, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21(6):635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 20.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cummins JM, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103(10):3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 24.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008 doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 25.Jackson AL, et al. Widespread siRNA "off-target" transcript silencing mediated by seed region sequence complementarity. Rna. 2006;12(7):1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson AL, et al. Position-specific chemical modification of siRNAs reduces "off-target" transcript silencing. Rna. 2006;12(7):1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136(4):586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315(5818):1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linsley PS, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27(6):2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonci D, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 31.Krutzfeldt J, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 32.Elmen J, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36(4):1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson AL, Linsley PS. Noise amidst the silence: off-target effects of siRNAs? Trends Genet. 2004;20(11):521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008 doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.John B, et al. Human MicroRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandberg R, et al. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320(5883):1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stenvang J, Lindow M, Kauppinen S. Targeting of microRNAs for therapeutics. Biochem Soc Trans. 2008;36(Pt 6):1197–1200. doi: 10.1042/BST0361197. [DOI] [PubMed] [Google Scholar]
- 39.Fabani MM, Gait MJ. miR-122 targeting with LNA/2'-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. Rna. 2008;14(2):336–346. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupberger J, Brino L, Baumert TF. RNAi: a powerful tool to unravel hepatitis C virus-host interactions within the infectious life cycle. J Hepatol. 2008;48(3):523–525. doi: 10.1016/j.jhep.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 41.Palliser D, et al. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature. 2006;439(7072):89–94. doi: 10.1038/nature04263. [DOI] [PubMed] [Google Scholar]
- 42.Shibata MA, et al. Combination therapy with short interfering RNA vectors against VEGF-C and VEGF-A suppresses lymph node and lung metastasis in a mouse immunocompetent mammary cancer model. Cancer Gene Ther. 2008;15(12):776–786. doi: 10.1038/cgt.2008.43. [DOI] [PubMed] [Google Scholar]
- 43.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 44.Corsten MF, et al. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67(19):8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 45.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA::target interaction. Chem Biol. 2004;11(12):1619–1623. doi: 10.1016/j.chembiol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 47.Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3' UTR. Rna. 2008;14(7):1297–1317. doi: 10.1261/rna.1082708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cummins JM, Velculescu VE. Implications of micro-RNA profiling for cancer diagnosis. Oncogene. 2006;25(46):6220–6227. doi: 10.1038/sj.onc.1209914. [DOI] [PubMed] [Google Scholar]
- 49.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gauthier NP, et al. Cyclebase.org--a comprehensive multi-organism online database of cell-cycle experiments. Nucleic Acids Res. 2008;36(Database issue):D854–D859. doi: 10.1093/nar/gkm729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.