Abstract
Previous studies evidenced that cystatin B-like gene is specifically expressed and induced in large circulating cœlomic cells following bacterial challenge in the leech Theromyzon tessulatum. In order to understand the role of that cysteine proteinase inhibitor during immune response, we investigated the existence of members of cathepsin family. We cloned a cathepsin L-like gene and studied its tissue distribution. Immunohistochemical studies using anti-cathepsin L and anti-cystatin B antibodies and ultrastructural results demonstrated the presence of three distinct cœlomic cell populations, (1) the chloragocytes which were initially defined as large cœlomocytes, (2) the granular amœbocytes, and (3) small cœlomic cells. Among those cells, while chloragocytes contain cystatin B and cathepsin L, granular amœbocytes do only contain cathepsin L and third cell population contains neither cathepsin nor inhibitor. Finally, results evidenced that cathepsin L immunopositive granular amœbocytes are chemoattracted to the site of injury and phagocyte bacteria.
Keywords: Lophotrochozoa, annelid, leech, innate immunity, cœlomocyte, cathepsin, cystatin, phagocytosis
1. Introduction
Innate immunity in both vertebrates and invertebrates uses many mechanisms like the recognition of pathogen associated molecular patterns (PAMPs), activation of specific receptors depending on the nature of the pathogens (Toll-like receptors), phagocytosis, antimicrobial peptides production, systemic response and cellular recruitment.
In the leech Theromyzon tessulatum, the implication of antimicrobial peptides in immune response has been recently described [1]. It was demonstrated that two peptides respectively named theromacin and theromyzin are rapidly released from various tissues into the cœlomic fluid following bacterial challenge. This challenge strongly enhances the gene expression of both peptides demonstrating that these antimicrobial peptides are inducible.
Simultaneously, in order to evidence other immune inducible genes, a combination of molecular techniques including Differential Display RT-PCR and DNA microarrays has been undertaken in T. tessulatum after a bacterial challenge [2]. The transcriptomic analysis reported up-regulation of several unknown and known genes, of which Threonine Deaminase, Malate Dehydrogenase, Cystatin B, Polyadenylate-binding protein, Ribosomal protein and Alpha Tubulin like genes. In addition, the study of leech cystatin B (Tt-CYSB) revealed that this cysteine proteinases inhibitor is probably involved in the leech immune response [2]. Indeed, we evidenced that leech cystatin B gene (Tt-cysb) is specifically expressed in large circulating cœlomic cells and specifically induced in this cell type following bacterial challenge.
In vertebrates, cystatins are strong inhibitors of cathepsins family [3,4]. Cystatin B has a broad distribution in cells and tissues and targets in vitro lysosomal cathepsins B, H, K, L [5,6]. In addition, cathepsins are involved in processing functions of mammalian Antigen Presenting Cells (APCs) [7-10]. In invertebrates, Drosophila cathepsin L (CP1) is present in small granules of hæmocytes and may be involved in phagocytosis [11].
In this context, we suggest that Tt-CYSB may regulate leech cathepsins and play a role in leech immune response regulation through these circulating cœlomic cells. Through this work, the cœlomic cells were studied to elucidate their implication in the immune response. In order to understand the role of Tt-CYSB, the purpose of this study has been to undertake the characterization of members of the cathepsins family in the leech T. tessulatum. This report presents the cloning of the leech cathepsin L gene (Tt-catl) and the possible implication of cathepsin L protein (Tt-CATL) in the cœlomic cells during phagocytosis.
2. Materials and Methods
2.1. Animals
T. tessulatum leeches were maintained in our laboratory as described elsewhere [12,13].
2.2. Bacterial Challenge
Experimental leeches were injected with a mixture containing Escherichia coli and Micrococcus luteus. After 12 hours of culture, centrifuged heat-killed bacteria were resuspended in PBS, before mixing with PBS (1:1, v/v) to yield a final concentration (109 bacteria per ml). The leeches were inoculated subepidermally by injection of 2.106 bacteria (2 μl). Control animals were injected with a sterile PBS solution (2 μl).
2.3. Cathepsin L cDNA Isolation
Total RNA were extracted from complete animals with TRIzol (Invitrogen) according to the manufacturer's instructions before DNase I treatment. Two different extractions were performed from i) a controle set and ii) a bacteria inoculated set of animals. Total RNA were supplied to Biométhodes S.A. (Genopole Entreprises, Evry, France) in order to construct control and experimental cDNA libraries. After purification of mRNA (Message Maker, Life Technologies), the cDNA synthesis were elaborated from 3 μg of mRNA according to the manufacturer's instructions (Superscript Plasmid System, Life Technologies). Directional cloning of cDNAs in pBM8 vector, screening by differential multiple digestion and enrichment (Biométhodes S.A., Genopole Entreprises) allowed to obtain normalized cDNA libraries from each set of animals. Two microliters of diluted control cDNA library (diluted to 1:200 in sterile water) were then used as template in several Polymerase Chain Reactions (PCR, detailed below) with 1 unit of Taq DNA polymerase (Life Technologies), 0.2 mM dNTP and 300 ng of primers in the reaction buffer (10 mM Tris HCl pH 9, 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X100 and 0.02% gelatin). The amplification steps were: 3 minutes at 94°C; 40 cycles of 30 seconds at 94°C, 1 minute at 55°C, 1 minute at 72°C; and a final step of 7 minutes at 72°C. The alignment of arbitrary known cathepsin L amino-acid sequences from invertebrate species (Fasciola hepatica, Schistosoma mansoni and Drosophila melanogaster) allowed determining similar domains. They were useful to design degenerated primers in order to amplify a putative T. tessulatum cathepsin L gene. The first PCR used cathepsin L fw1 (5′TGCGGGTCGTGYTGGGCNTT3′) and rv1 degenerated primers (5′GTGCCCCAGGARTTYTTNAC3′) in order to amplify an internal fragment of cathepsin L cDNA. Then, a nested PCR used the cathepsin L fw1 and fw2 (5′CTAGTGAAGAAYTCNTGGGG3′) degenerated primers with an oligo (dT) adapter (5′CGAGTCGACATCGATCGTTTTTTTTTTTTTTTTT3′) in order to amplify the 3′ end of cathepsin L cDNA. Finally, a nested PCR used fw-vector1 (5′CGCTTTGC CTGACCCTGCTTGC3′) and fw-vector2 (5′CGCCGTTACAGATCCAAGCTCC3′) with a cathepsin L rv2 degenerated primer (5′AAGGCCCAGCANGANCCRCA3′) in order to amplify the 5′ end of cathepsin L cDNA. All PCR products were cloned in pGEM T-easy vector (Promega) and sequenced using the BigDye Terminator v3.0 polymerisation kit before detection on the ABI Prism 310 Genetic Analyzer (Applied Biosystems). Sequence analysis used the BLAST programs with E=1000 and no filter [14,15]. Cystatin B (GenBank assession number AF542131) cDNA was obtained from a previous study after screening of a cDNA library prepared from total stage 2 T. tessulatum [2].
2.4. Western immunoblotting
Leeches were freezed in liquid nitrogen, then crushed in a mortar and finally resuspended in 800 μl homogenization buffer (0.1 M Tris-HCl, pH 7.5, 20 mM CaCl2, 20 mM MgCl2 and EDTA-free Complete Protease Inhibitor Cocktail (Roche Applied Science)). Samples were centrifuged at 10 000 g for 20 minutes. 100 μl supernatants were mixed 1:1 (v/v) with Laemmli buffer (62.5 mM Tris-HCl, pH 6.8, 1.5% SDS, 10% sucrose, and 0.01% bromophenol blue) and 5% 2-mercaptoethanol. The samples were boiled for 5 minutes prior to loading on SDS-PAGE gel. Stacking gel was 4% Acrylamide:Bis-acrylamide (29:1) in 0.375 M Tris-HCl, pH 6.8 and 0.1% SDS. Resolving gel was 12% Acrylamide:Bis-acrylamide (29:1) in 0.125 M Tris-HCl, pH 8.8 and 0.1% SDS. Gels were polymerised with 10% APS and TEMED. Samples were loaded next to Precision Plus Protein standards (Bio-rad). They were subjected to electrophoresis (100V for 1 hour and 200 V for 3 hours) in 25 mM Tris, pH 8.3, 192 mM glycine, 0.1% SDS buffer and separated proteins were transferred onto nitrocellulose membrane at 0.12 A for 1.5 hour at room temperature in 25 mM Tris, 192 mM glycine, 15% methanol buffer. The nitrocellulose membrane was preincubated with 1% ovalbumin in 10 mM Tris-HCl, pH 8, 150 mM NaCl buffer for 1 hour to block non specific binding. The membrane was then incubated with anti-Tt-CATL antibody (dilution 1:1000) in 1% ovalbumin, 10 mM sodium phosphate, 150 mM NaCl, pH 7.4 buffer overnight at 4°C. After three washes in 10 mM sodium phosphate, 150 mM NaCl, pH 7.4 buffer, the membrane was incubated with goat anti-Rabbit IgG antibody conjugated with horseradish peroxidase (dilution 1:20000) (Jackson Immunoresearch) for 1 hour at room temperature. After final washes, signals were detected with an ECL kit (Pierce) on Biomax light film (Kodak).
2.5. Bacteria labelling
Live E. coli D31 and M. luteus IFO12708 were used for the antimicrobial assays. Heat killed E. coli or M. luteus were washed in phosphate buffer saline (PBS) and then labelled by incubation for 1 hour at 25°C in 0.1M NaHCO3, pH 9.6, 0.01 mg/ml FITC (sigma). Bacteria were pelleted at 12,500 g for 5 minutes, washed free of unbound fluorochrome with PBS and kept at −20°C until use.
2.6. Phagocytosis assays
Animals were injected individually with 10 μl of a solution containing either 109 FITC-labelled E. coli or 109 FITC-labelled M. luteus. Leeches were killed 30 minutes, 1 hour, 2 hours, 12 hours or 24 hours after injection and treated for confocal microscopy (see below).
2.7. Tissue preparation
For in situ hybridization and confocal microscopy, leeches were fixed overnight in a solution containing 4 % paraformaldehyde, pH 7.4. After dehydration, animals were embedded in paraplast and 7-μm sections were cut, mounted on poly-L-lysine-coated slides, and stored at 4 °C until use.
2.8. In situ hybridization
Probes
Plasmids containing a piece of cathepsin L cDNA (GenBank accession number AF542132) or cystatin B cDNA (GenBank accession number AF542131) were used as templates for the preparation of the probes. Digoxigenin (DIG)-UTP-labelled antisense and sense riboprobes were generated from linearized cDNA plasmids containing cathepsin L cDNA by in vitro transcription using RNA labelling kits, T3 RNA polymerase (Roche). [35S]UTP-labelled antisense and sense riboprobes were generated from linearized cDNA plasmids containing cystatin B cDNA by in vitro transcription using RNA labelling kits, T3 RNA polymerase (Roche) and [35S]UTP (Amersham). Hybridization. DIG-labelled cathepsin L riboprobes (40–100 ng per slide) were diluted in hybridization buffer containing 50% formamide, 10% dextran sulfate, 10 × Denhardt's solution, 0.5 mg·mL-1 tRNA from E. coli, 100 mm dithiothreitol and 0.5 mg·mL-1 salmon sperm DNA. Hybridization was carried out overnight at 55 °C in a humid chamber. Slides were then washed twice (2 ×15 minutes) with 2X SSC, treated with RNase A (20 mg·mL-1 in 2X SSC) for 10 minutes at 37 °C and consecutively rinsed 2 × 10 minutes in 0.1 × NaCl/Cit containing 0.07% 2-mercaptoethanol at 55 °C. The probes labelled with DIG-UTP were revealed using alkaline phosphatase-conjugated antibodies as previously described [16]. For double in situ hybridization, DIG-labelled cathepsin L riboprobes (40–100 ng per slide) and 35S-labelled cystatin B riboprobes (100 ng or 1 × 106 c.p.m. per slide) were mixed in the same hybridization buffer and hybridization was carried out as described before. Hybridization signal for cystatin B was visualized using autoradiography as previously described [17]. Briefly, after revelation of the cathepsin L signal using alkaline phosphatase, samples were coated by dipping in LM1 liquid emulsion (Amersham), immediately dried and finally exposed for a 28-days period. At the end of the exposure period, the autoradiograms were developed in D19b (Kodak), fixed in 30% sodium thiosulfate (10 minutes at room temperature), stained with 1% Toluidine blue and mounted with Xam (Merck).
2.9. Antibodies
The chemically synthesized regions of cathepsin L (X49-X63) and cystatin B (X26-X41) were used for the immunization procedure of rabbits and mice, respectively, according to the manufacturer's instructions (Agro-Bio, La Ferté Saint Aubin, France).
2.10. Confocal microscopy
Double immunofluorescence
Whole animal, seven micrometer-thick paraffin sections were re-hydrated. After 1 hour incubation in TBS (0.1 M Tris, pH 7.5, 0.9% NaCl) containing 1% normal goat serum (NGS), 1% bovine serum albumine (BSA) (TBS/NGS/BSA), sections were incubated overnight at 20°C in TBS containing 1:800 rabbit anti-Tt-CATL antiserum, 1:400 mouse anti-Tt-CYSB antiserum, 1% NGS, 1% BSA, 0.01% triton X-100. After washes, sections were incubated for 2 hours jointly with goat anti-mouse FITC- and goat anti-rabbit Texas-Red tagged antisera (Jackson Immunoresearch) diluted 1:100 in TBS/NGS/BSA. After washes, slides were mounted in glycerol containing 25% TBS and 0.1% p-phenylenediamine. The absence of secondary antibodies cross-reactivity was controlled by omitting one or both of the primary antibodies. Phagocytosis and immunofluorescence. Experimental leeches were injected either with FITC-labelled E. coli or FITC-labelled M. luteus. Thirty minutes, 1 hour, 2 hours, 12 hours or 24 hours after injection, animals were fixed and sections were obtained as previously described. After 1 hour incubation in TBS containing 1% normal goat serum (NGS), 1% BSA (TBS/NGS/BSA), sections were incubated overnight at 20°C in TBS containing 1:1000 rabbit anti-cathepsin L antiserum, 1% NGS, 1% BSA, 0.01% triton X-100. After washes, sections were incubated for 2 hours with goat anti-rabbit Texas-Red tagged antisera (Jackson Immunoresearch) diluted 1:100 in TBS/NGS/BSA. After washes, slides were mounted in glycerol containing 25% TBS and 0.1% p-phenylenediamine and examinated with a confocal microscope. Labelled cells were observed using a Leica laser scanning microscope (TCS NT) equipped with a Leica (DMIRBE) inverted microscope and an argon/krypton laser. FITC signal was detected using a 488 nm band-pass excitation filter and a 575-640 nm pass barrier filter, and Texas-Red signal by exciting samples at 568 nm. Images were acquired sequentially as single transcellular optical sections and averaged over 16 scans per frame.
2.11. Electron microscopy
Ultrastructural study of cœlomocytes
Stage 3 animals were anesthetized with chloretone and cœlomic fluid was collected with a tuberculin syringe. Collected fluid was centrifuged immediately at 800 g for 15 minutes at 4°C. Pellets were fixed in 3% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4, for 2 hours. After postfixation in 0.1 M osmium tetroxide in the same phosphate buffer, pellets were dehydrated with acetone and embedded in Epon in the conventionnal manner (polymerization at 60°C for 48 h). Ultrathin sections (80-90 nm) were cut from the Epon blocks, placed on 200-mesh copper grids, counterstained routinely with uranyl acetate, and lead citrate, and observed in a Jeol CX 100 electron microscope. Immunogold labelling. After cœlomic fluid collection, circulating cell pellets were obtained by 10 minutes centrifugation at 800 g. Cells were fixed for 2 h at 4°C in phosphate buffer saline (PBS) containing 4% paraformaldehyde, 0.2% picric acid, 0.1% glutaraldehyde. Cells were post-fixed in 1% OsO4 for 5 minutes and dehydrated before embedding in LR white (TAAB). Immune staining was performed on 90 nm thick ultrathin sections cut from the embedded pellets and collected on Parlodion-coated nickel grids. Sections were treated as follows: (1) 8 minutes in 10% H2O2; (2) 10 minutes in distilled water; (3) 10 minutes in TBS/NGS/BSA; (4) 48-hours at 4°C in TBS/NGS/BSA containing 1:2000 diluted rabbit anti-cathepsin L antiserum; (5) 3 × 10 minutes in TBS; (6) 1.5 h in TBS/NGS/BSA containing 10 nm colloidal gold-labelled anti-rabbit IgG (Amersham) diluted 1:50; (7) 3× 10 minutes in TBS; (8) 3 minutes in TBS, 1% glutaraldehyde; (9) 2× 5 minutes in distilled water. Sections were then stained 14 minutes with uranyl acetate and examinated with a Jeol JEM 100 CX.
3. Results
3.1. Theromyzon tessulatum cathepsin L (Tt-CATL) characterization (NCBI, GenBank Accession Number AF542132)
Molecular approach was used to isolate members of cathepsins family in the leech. From several known cathepsin L (isolated in Fasciola hepatica, Schistosoma mansoni and Drosophila melanogaster), degenerated primers were designated to amplify several regions of cathepsin L cDNA by RT-PCR [11,18,19]. Internal, 5′ end and 3′ end products were necessary to obtain a full length cDNA corresponding to T. tessulatum preprocathepsin L gene, named Tt-catl (Accession number Q8IT42 in the UniProt server of the European Bioinformatics Institute) (Fig. 1A). No enzymological tests were realized yet. Actually, we attributed the name of cathepsin L to this molecule according to the homologies presented with proteins in databases. Indeed, the amino acid sequence was deduced by translation into the right open reading frame and compared in databases with the BLAST-P and BLAST-X programs [14,15]. Tt-CATL sequence contains 351 amino acids (Fig. 1A). The analysis of the sequence by SignalP V1.1 program predicted a putative signal peptide corresponding to region amino acids 1-15 of the protein [20]. The region 16-129 is a proteinase inhibitor I29 domain (Accession number IPR013201) which is found at the N-terminus of some C1 peptidases such as cathepsin L where it acts as a propeptide. The activation process of Tt-CATL might include the removal of this propeptide region. In addition, this proregion contains conserved domains, ERFNIN and GNFD (Fig. 1A), which are specific of cathepsin L-like family [21]. Then, the region 130-348 of Tt-CATL sequence corresponds to the peptidase C1A domain (NCBI Conserved Domain number cd02248.1, papain-like domain) which contains several known catalytic residues (Fig. 1A). They correspond to the cysteine 153 and the histidin 292 which could form a catalytic dyad [22]. Two other residues might play an important role in catalysis, the glutamine 147 preceding the catalytic cysteine, believed to help in the formation of the oxyanion hole and the asparagine 312 which could orient the imidazolium ring of the catalytic histidin 292 [23]. Western blot analysis using anti-Tt-CATL antibody allowed detection of a product (about 24 kDa) which is relevant with the mature form (residues 130-351) of Tt-CATL (Fig. 1B). Finally, Tt-CATL sequence were compared with previously described cathepsins L by the Multalin program [24] (Fig. 1C). We have chosen to show the first molecules matching with Tt-CATL which were isolated from the cnidarian Hydra vulgaris (72%), the flesh fly Sarcophaga peregrina (69%), the fruit fly Drosophila melanogaster (68%), the tick Boophilus microplus (68%), the tick Rhipicephalus haemaphysaloides (65%) and human (63%) [11,25-28].
Figure 1. Molecular characterization of Tt-CATL.
A. Tt-CATL sequence revealed a full length cDNA containing a coding region of 1053 bases (nucleotide positions 48-1100). The amino acid sequence (351 amino acids) which was deduced from this nucleotide sequence is constituted of a putative signal peptide (highlighted in black, 1-15), a propeptide region (highlighted in dark grey, 16-129) and the active peptide (highlighted in light grey, 130-351). ERFNIN and GNFD conserved motifs of cathepsin L like family and catalytic residues are specified in black boxes respectively in propeptide and peptidase C1A regions. Numbers for nucleotides and amino acids are respectively indicated on left and right of the sequence. B. Western blot analysis was performed from whole animal protein extract using anti-Tt-CATL antibody revealed by goat anti-Rabbit IgG antibody conjugated with horseradish peroxidase. A strong signal is observed for a product of about 24 kDa (arrowhead) which is relevant with the active form of Tt-CATL. C. Theromyzon tessulatum cathepsin L (Tt-CATL) has been compared with databases using BLAST-P program. The alignment shows homologies with first identified cathepsins L from other species. Its reveals homologies of 72% with Hydra vulgaris cathepsin L (Accession number AAO65603), 69% with Sarcophaga peregrina cathepsin L (Accession number BAA03970), 68% with Drosophila melanogaster cysteine proteinase 1 (CP1; Accession number AAB18345), 68% with Boophilus microplus cathepsin L (Accession number AAF61565) and 65% with Rhipicephalus haemaphysaloides cathepsin L-like (Accession number AAQ16117). Low consensus regions are highlighted with light grey and medium grey. High consensus regions are represented in black. The high consensus region corresponds to the peptidase domain (139PVEVDWR…ASYP359).
3.2. Distribution of Tt-catl expressing cells by in situ hybridization
Paraffin-embedded sections of animals were hybridized with antisense Tt-catl riboprobes labelled with 35S-UTP. Tt-catl mRNAs were only detected in cœlomic cells (cœlomocytes), circulating through the cœlomic cavity (Figs. 2A, 2B). The lack of signal in controls, i.e. sections hybridized with sense riboprobes, confirms the specificity of the hybridization (Fig. 2C). In order to study the possible interaction between T. tessulatum cathepsin L and a cathepsins inhibitor, the cystatin B (Tt-CYSB) previously described in the leech [2], sections were simultaneously hybridized with antisense Tt-catl riboprobes labelled with Dig-UTP and with antisense Tt-cysb labelled with 35S-UTP. This double in situ hybridization revealed that both probes were hybridized in cœlomocytes (Fig. 2D). Indeed, DIG-UTP labelling from Tt-catl was colocalized with silver staining from Tt-cysb riboprobe in cœlomocytes. Consequently, those circulating cells express both cathepsin L and cystatin B genes.
Figure 2. Tt-catl and Tt-cysb mRNA location by in situ hybridization.
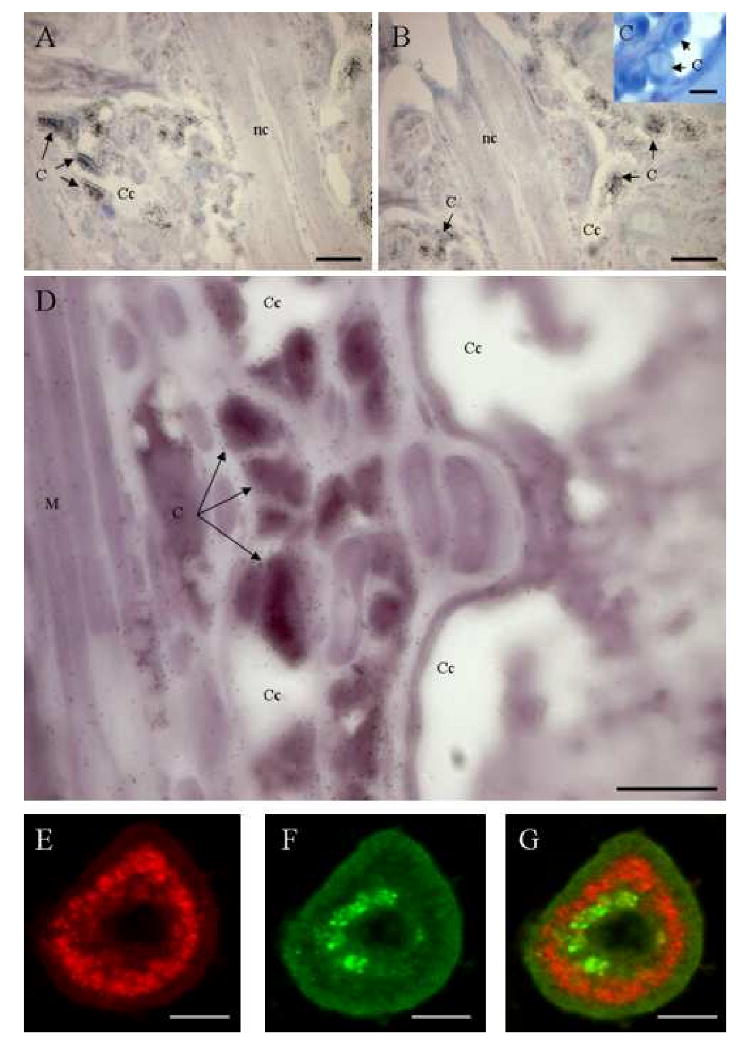
A, B, C. Detection of Tt-catl mRNA in leech tissues by in situ hybridization. Paraffin-embedded sections of animals were hybridized with antisens (A, B) Tt-catl riboprobes were labelled with 35S-UTP. Positive circulating cells are detected in cœlomic cavities and in the ventral sinus. Negative controls consisting of sections hybridized with Tt-catl sense riboprobes (C). D. Double detection of Tt-catl and cystatin b mRNAs (Tt-cysb). Sections were hybridized with antisens Tt-catl riboprobes labelled with Dig-UTP and with antisens Tt-cysb labelled with 35S-UTP. Both probes are co-localized in cœlomocytes. E, F. Confocal microscopic images of Tt-CATL (red) and Tt-CYSB (green) double immune labelling in leech cœlomocytes. G is the merged confocal image of E and F. Merged confocal image suggests that Tt-CATL and Tt-CYSB may be packed in the same cells but in different cell compartments. Abbreviations: C. Cœlomocytes. Cc. Cœlomic cavity. Nc. Nerve cord. M. Muscle. Bars: A, B, C and D: 100 μm. E, F and G: 30 μm.
3.3. Distribution of Tt-CATL protein containing cells by immunohistochemistry
Confocal microscopy was performed using rabbit anti-Tt-CATL antiserum and mouse anti-Tt-CYSB antiserum detected by goat anti-mouse FITC- and goat anti-rabbit Texas-Red tagged antisera. Immunohistochemistry results showed Tt-CATL (red) and Tt-CYSB (green) double immune labelling in leech cœlomocytes (Figs. 2E, 2F). Both proteins were detected into the same cells but the merged confocal image suggests that Tt-CATL and Tt-CYSB may be packed in different cell compartments (Fig. 2G). The absence of secondary antibodies cross-reactivity was controlled by omitting one or both of the primary antibodies (data not shown). At this time, we had no evidence of a possible interaction between cathepsin L and cystatin B (cathepsins inhibitor). In addition, immune labelling revealed Tt-CATL and Tt-CYSB presence in cœlomic cells which were identified as large cœlomocytes (Fig. 2G). In contrast, while Tt-CYSB was detected in large cœlomocytes only, Tt-CATL was detected in the large cœlomocytes and in a smaller size cell population. In order to specify intracellular localization of Tt-CATL in these two cœlomic cell types, we realized an electron microscopic study.
3.4. Typical ultrastructural feature and ultrastructural distribution of Tt-CATL immunoreactivity in circulating cells of T. tessulatum
Tt-CATL was revealed with a 10 nm gold-particle-conjugated secondary antibody.
The ultrastructural analysis allowed us to discriminate three populations of cœlomocytes. Large cœlomocytes (100-150 μm) present large electron-dense granules and electron-lucent vesicles (Fig. 3A). We named these cells chloragocytes according to similar ultrastructural features described from oligochætes [29-31]. These chloragocytes exhibit strong Tt-CATL immunoreactivity in large granules (Figs. 3B, 3C). A second type of cœlomocyte with smaller size (30-70 μm) shows long cytoplasmic pseudopods and large electron-dense granules (Fig. 3D). These cells share ultrastructural features with oligochæte granular amœbocytes (granulocytes) [29-31] and show sparse Tt-CATL immunopositive electron dense granules (Fig. 3E). Finally, a third kind of circulating cells (7-12 μm) was observed in electron microscopy, and is not immunoreactive for Tt-CATL (Fig. 3F). Otherwise, previous studies showed up-regulation of Tt-cysb gene following a bacterial challenge in chloragocytes [2]. That is why, in this context, we wished to elucidate a possible immune function for Tt-CATL in both Tt-CATL-containing cell types.
Figure 3. Typical ultrastructural features (A, D, F) and ultrastructural distribution of Tt-CATL immune reactivity (B, C, E) in circulating cells of T. tessulatum.
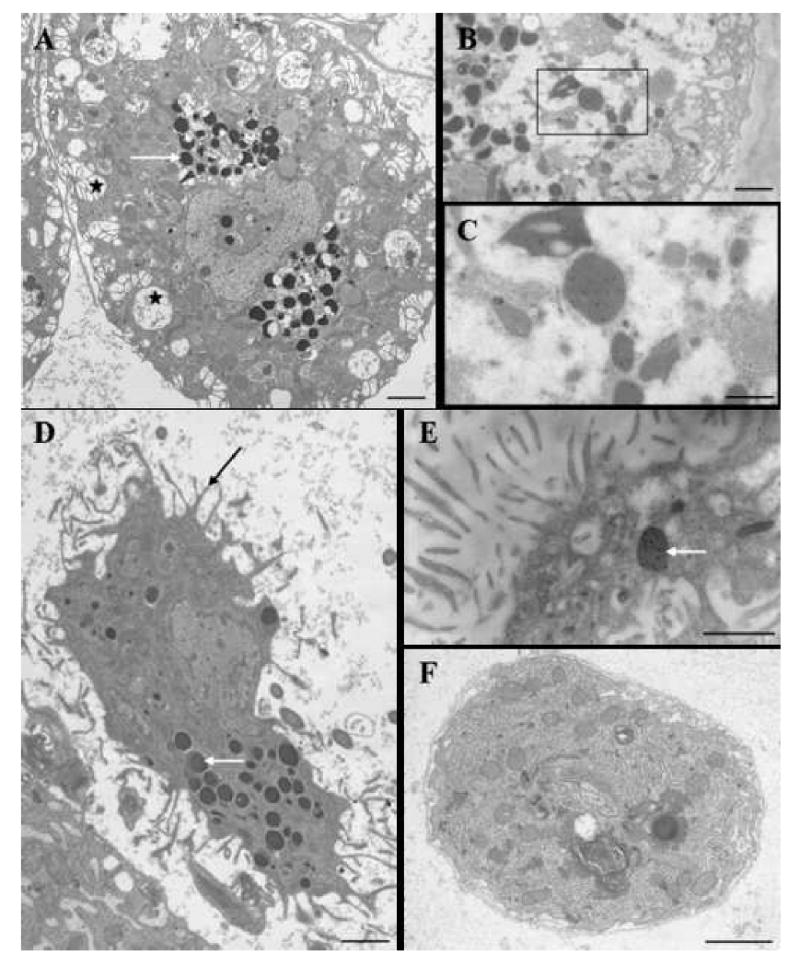
Tt-CATL was revealed with a 10 nm gold-particle-conjugated secondary antibody. A. Large cœlomocytes present large electron-dense granules (white arrow) and electron-lucent vesicles (black stars). B, C. Large cœlomocytes exhibit strong Tt-CATL immunoreactivity in large dense granules. D. Intermediate size cœlomocytes show long cytoplasmic pseudopods (black arrow) and large electron-dense granules (white arrow). E. Large electron dense granules in intermediate size cœlomocyte exhibit a high number of gold particles (white arrow). F. Small circulating cells are rich in endoplasmic reticulum and small granules. They are not immunoreactive for Tt-CATL (data not shown). Bars: A, B: 10 μm; C: 5 μm; D, E: 5 μm; F: 2 μm.
3.5. Dual location of Tt-CATL immunoreactivity and phagocytosed fluorescent bacteria
In order to explore the role of Tt-CATL during the immune response, leeches were injected with FITC-labelled bacteria (M. luteus or E. coli) and submitted to immune detection of Tt-CATL in time course. There was no difference between M. luteus or E. coli. In the text below, the term “bacteria” refers to M. luteus as well as E. coli.
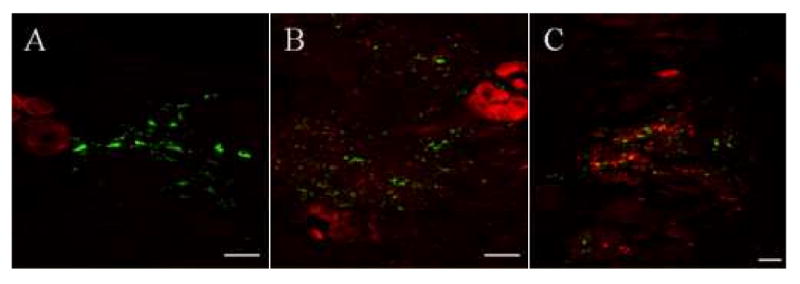
Thirty minutes following the incubation, numerous fluorescent bacteria were present in the cœlomic cavities close to Tt-CATL immunopositive cœlomic cells, i.e. chloragocytes and granular amœbocytes (Figs. 4A, 4B). Some bacteria were seen in contact with Tt-CATL immunoreactive granular amœbocytes (Fig. 4B, white arrow). One hour following the incubation, internalized bacteria were observed into these cells (Fig. 4C). Internalized bacteria may be co-located with Tt-CATL (Fig. 4C, white arrow). Numerous bacteria were also observed in the nephridia which suggests filtration of cœlomic fluid. Nephridian cells which are Tt-CATL immunoreactive (Fig. 4D) are also able to phagocytosis fluorescent bacteria (Fig. 4D, white arrow) as already observed in annelid Arenicola marina [32]. Finally, twenty four hours following injection, bacteria were still observed in close contact with chloragocytes (Fig. 4E) but phagocytosed bacteria were observed in granular amœbocytes only (Fig. 4F). Whatever times of incubation, no fluorescent bacteria were observed into chloragocytes (Figs. 4B, 4E, 4F). Only Tt-CATL immunopositive granular amœbocytes are involved in phagocytosis of injected bacteria. Further studies consisted to observe the movement of Tt-CATL positive granular amœbocytes. Results showed an accumulation of cells at the injection site. Indeed, we initially observed many Tt-CATL positive cœlomocytes in cœlomic cavities near the injection site (Fig. 5A). When we renewed the experiment and analyzed 12 hours following the incubation of fluorescent bacteria, a strong Tt-CATL immunoreactivity was observed at the injection site and concerned granular amœbocytes (Figs. 5B, 5C). As a result, those cells are capable of phagocytosis and seem to answer to chemotactic processes. Otherwise we evidenced that Tt-CATL immunoreactivity highlights the existence of at least two cell populations in cœlomic cavities.
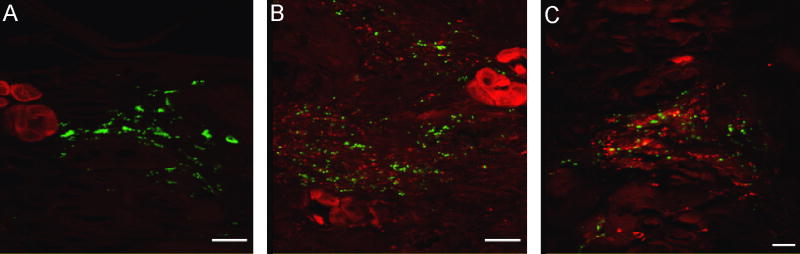
Figure 4. Dual location of Tt-CATL immune reactivity and phagocytosed fluorescent bacteria as observed by confocal microscopy.
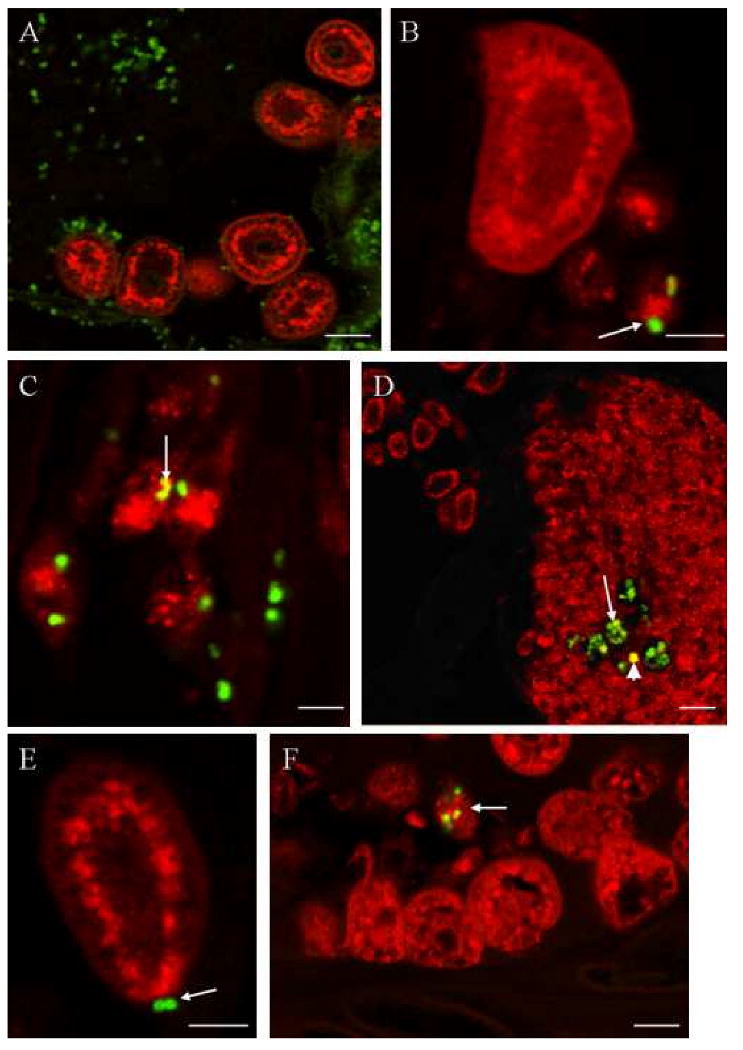
Animals were injected with FITC-labelled bacteria (M. luteus or E. coli) and treated after various incubation times, to immune detection of cathepsin-L using a Texas-Red labelled secondary antibody. A, B. Thirty minutes following the incubation, numerous labelled bacteria are abundant in the cœlomic cavities near Tt-CATL immunopositive cœlomic cells (B, chloragocyte on left and three granular amœbocytes on right). Some bacteria are in close contact with granular amœbocytes (white arrow). C. One hour after injection, internalized bacteria are observed into granular amœbocytes. Internalized bacteria may be co-located with Tt-CATL (white arrow). D. After one hour of incubation, numerous bacteria are also observed in the nephridia which cells are Tt-CATL-immunoreactive (white arrow). Nephridian cells are also able to phagocyte fluorescent bacteria (white arrowhead). E, F. After twenty four hours of incubation, bacteria are still observed near chloragocytes (E, white arrow) but phagocytosed bacteria are observed in granular amœbocytes only (F, white arrow). Bars: A: 50 μm; B: 30 μm; C: 20 μM; D: 100 μm; E: 30 μm and F: 40 μm.
Figure 5. Dual location of Tt-CATL immune reactivity and fluorescent bacteria as observed by confocal microscopy.
Animals were injected with FITC-labelled bacteria (M. luteus or E. coli) and sacrificed after various times. Tt-CATL immune detection was realized using a Texas-Red labelled secondary antibody. An accumulation of Tt-CATL immunopositive granular amœbocytes is observed at the injection site. A. Five minutes after bacterial injection, a few granular amœbocytes only are observed in cœlomic cavities near the injection site. B, C. After 12 hours, strong cathepsin-L immunoreactivity is observed at the injection site. Bars: 100 μm.
4. Discussion
4.1. T. tessulatum cathepsin L (Tt-CATL) characterization
We have previously shown that a cystatin B like gene in the leech Theromyzon tessulatum (Tt-cysb) was upregulated after bacterial challenge in a subpopulation of cœlomocytes [2]. It is the first description of a cystatin B like gene in invertebrates, besides induced at the time of an immune response. Cystatin B is known to inhibit in vitro the papain-like cysteine cathepsins by tight and reversible binding. The inhibitor binds tightly to cathepsins H, L, and S and less tightly to cathepsin B [33]. Because cathepsin L is involved in immunological mechanisms in vertebrates and expressed in Drosophila melanogaster hæmocyte cell line mbn-2 [11], we decided to focus on this cysteine proteinase in leech. We cloned a 1161 bp product (accession number AF542132) with an open reading frame of 1053 bases coding for a protein of 351 amino acids. The amino acids sequence presents active site of the papain like domain, the ERFNIN and GNFD motifs of the cathepsin L family propeptide, and shows significant sequence homology with known cathepsins L. Therefore we named it Theromyzon tessulatum cathepsin L like protein (Tt-CATL). Cysteine proteinases family genes were cloned in many invertebrate organisms, notably arthropod species, but it is the first cathepsin L like gene identified in lophotrochozoans.
In situ hybridization and immunohistochemistry experiments exclusively showed Tt-catl expression in two types of cœlomic cells. We initially named these cells as large and intermediate cœlomocytes. No signal was detected in other cell types or tissues like nerve chord and muscle close to cœlomic cavities (Figs. 2A, 2B, 2D, 3B). These results differ from observations in arthropods, in which cathepsin L like genes are expressed in many tissues, like digestive and reproductive tracts, eye, muscle or hæmocytes [34]. In order to characterize more finely the Tt-CATL positive cœlomic cells and the cathepsin intracellular distribution, we undertook ultrastructural studies.
4.2. Three distinct populations of cœlomocytes
Concerning large cœlomic cell type (100-150 μm), Tt-CATL immunoreactivity was observed on electron-dense granules (Figs. 5B, C). Moreover, these cells show typical electron-lucent granules (Fig. 5A). Such ultrastructural features have been described for oligochætes chloragocytes (or eleocytes) in cœlomic cavity [35]. Unlike the hirudinoïd species, T. tessulatum, which belongs to glossiphoniid leeches, does not possess botryoïdal tissue into the cœlomic cavity. Because of the absence of botryoïdal tissue, cœlomic cells are freely-floating in the cœlomic fluid or are attached to mesothelia walls as in oligochætes [31]. In addition, botryoïdal cells are considered to be analogous with chloragocytes [36]. Consequently, we name the large cœlomocytes immunopositive for Tt-CATL as chloragocytes.
A second population of cœlomocytes with intermediate size (30-70 μm) was highlighted by ultrastructural studies. They present numerous pseudopods and electron-dense granules that are immunoreactive for Tt-CATL (Figs. 5D, E). Among the cœlomic cells, only those intermediate-size cells are able to phagocyte injected Gram positive and Gram negative bacteria (Fig. 4C). Their ultrastructural features and involvement in phagocytosis are similar to previously described granular amœbocytes in oligochætes [29,30]. Thus, in T. tessulatum, we named this second cœlomic cell population as granular amœbocytes according to their analogies.
Finally, a third cœlomocyte population was revealed by ultrastructural studies. Cells are deprived of cytoplasmic granules as observed in hyaline cells from invertebrates like oligochætes. In contrast, they do not present any phagocytosis ability as described for other invertebrate hyaline cells [16,17,37]. Moreover, the presence of endoplasmic reticulum rich cytoplasm do not correspond to immature cells as described in invertebrates [29-31]. These smaller size cells (7-12 μm) are not immunoreactive for Tt-CATL. Otherwise, they did not present any Tt-CATL mRNA signal by in situ hybridization (data not shown). To date, it seems difficult to determine a biological function and structural analogies with known cells. That is why, we still name them as “small cœlomic cells”.
Consequently, T. tessulatum cœlomic cells are constituted by granular amœbocytes exerting phagocytosis, chloragocytes and small cœlomic cells. This heterogeneous population of cœlomocytes is similar to population described in other annelids. Numerous studies performed in oligochætes as Lumbricus sp. and Eisenia sp. used optical microscopy and electron microscopy. Although monoclonal antibodies were carried out against various invertebrate taxa, the majority of the studies related to insect hæmocytes. Interestingly, a monoclonal antibodies library was realized against Nereis diversicolor cœlomic cells [38,39]. In addition, de Eguileor et al. identified three cœlomic cell populations, i.e. macrophage-like, NK-like and granular cells, using human monoclonal antibodies in the hirudinea Glossiphonia complanata [40]. Recently, Engelmann et al. produced monoclonal antibodies against cœlomic cells in Eisenia fetida earthworm [41]. While anti-EFCC1 antibody (Eisenia fetida cœlomocyte differentiation cluster) is able to recognize antigenic motifs on various tissues, three other antibodies named anti-EFCC2, anti-EFCC3 and anti-EFCC4 allowed to respectively discriminate chloragocytes, hyaline amœbocytes and granular amœbocytes [41,42]. Otherwise, these antibodies do not detect any antigenic motif from mammalian and Drosophila melanogaster cœlomic cells. Because oligochætes and hirudinea belong to annelid group, it should be interesting to test these anti-EFCC antibodies in T. tessulatum. Such studies might confirm our cœlomic cells discrimination and possibly identify other cell types. Indeed, unlike in earthworm, the presence of phagocytic hyaline cells was not evidenced in T. tessulatum.
4.3. Granular amœbocytes are “circulating” phagocyte
Time course analyses for E. coli and M. luteus injected leeches show that only one cœlomic cell population, granular amœbocytes, migrates to the injection site and phagocyte bacteria (Figs. 4 and 5). The molecular mechanism of this recruitment is still unclear. Chemoattractant factors could be released by local cells following the the lesion or the bacteria recognition. Interestingly, recent studies show the presence of chemotactic mechanisms during angiogenic processes following wound healing in leech [43]. Otherwise, experiments using invertebrate hæmocytes and bacteria in Boyden chamber assays revealed that micro-organism contributes to its own recognition. Indeed, bacterial molecules as lipopolysaccharides (LPS) and N-formylmethionine-leucine-phenylalanine (FMLP) molecules act as chemotactic and/or chemokinetic factors on migration of mussel and shrimp hæmocytes [44,45]. Further studies should elucidate molecules involved in migration of leech amœbocytes using their purification and in vitro assays.
4.4. Leech chloragocytes functions
Leech chloragocytes exhibit Tt-cysb and Tt-catl gene expression (Fig. 2) [2]. Confocal microscopy analyses revealed that Tt-CYSB and Tt-CATL proteins are not located in same intracellular compartments (Fig. 2G). In addition, movements of intracellular compartments into the leech chloragocytes were not evidenced following a bacterial challenge. In contrast, while no phagocytosis was detected, analyses allowed observing chloragocytes-bacteria interaction (Fig. 4E) which suggests the presence of recognition molecules. Leech chloragocytes might be involved in encapsulation reactions as previously described in oligochætes [29]. Interestingly, Guillou and collaborators have recently evidenced up-regulation of cystatin B like gene (BgSSHcyst2, accession number # CK988733) in hæmocytes of the snail Biomphalaria glabrata following parasitic challenge with Echinostoma caproni. That molecule is specifically expressed in hæmocytes participating in parasite encapsulation or aggregating at the site of infection [46]. To date, Tt-cysb is up-regulated 24 hours following a bacterial injection in chloragocytes which might be involved in encapsulation mechanisms. Nevertheless, Tt-CYSB belongs to stefin family which is constituted by intracellular proteinase inhibitors [4]. Thus, it is difficult to hypothesize that leech cystatin B might be involved in inhibition of microbial cathepsins following infection. Otherwise, other physiological mechanisms could use cathepsins and their specific inhibitors in chloragocytes. In T. tessulatum, vitellogenesis processes synthesize vitellogenin which is sequestered in the oocyte to give vitellin. This precursor accumulates in the cœlomic fluid before being incorporated in oocytes [47]. Cœlomic cells as chloragocytes may be involved in its production as in nereids [48]. In addition, cathepsin L is involved in processing of yolk proteins in Cænorhabditis elegans and marine teleosts fishes [49-51]. Consequently, chloragocyte cathepsins might participate to yolk proteins processing during the leech vitellogenesis.
4.5. Cathepsin L and cystatin B in leech immunity
While Tt-cysb gene is up-regulated in chloragocytes following microbial challenge [2], Tt-catl expression studies by northern blot and in situ hybridization did not show up or down regulation in any cœlomic cell type (data not shown). These results seem to be in contrast with some reported data in D. melanogaster microarray studies where the proenzyme cathepsin L (Cp1) gene is up-regulated following a bacterial challenge [52]. In addition, other authors showed an enhancement of cathepsin L and B gene expression in sea urchin cœlomocytes in response to LPS [53]. However, Tt-catl might be regulated at the translational or post-translational level and therefore not detected by in situ hybridization, northern-blot or RT-PCR experiences. Loseva and Engstrom showed in D. melanogaster immunocompetent hemocytic cell line mbn-2 that Cp1 proenzyme was processed into the active form in response to LPS treatment [54]. In leech, we detected a putative active form of Tt-CATL (about 24 kDa) by western blot analysis from the whole animal (Fig. 1B). Unfortunately, the inability to collect and purify a large number of cœlomic cells from cavities does not allow the detection of the enzyme precursor and the analysis of Tt-CATL molecular processing at all.
In annelids, a cystatin B-like gene has recently been detected in E. fetida midgut EST library whereas no data concerning cathepsin L existence is available in this worm [55]. Pattern expression of earthworm cystatin B gene seems to be different from leech one. The specificity of cystatin B for cathepsins family is still unclear too. Thus, recent data show the presence of cystatin B like molecules in annelids and in mollusks but no functional study was reported to date [55,56]. Further studies might elucidate the distribution of cathepsin L-like and cystatin B-like molecules in oligochætes in order to determine the possible similarities with leech and their implication in immune response.
Because cystatin B and cathepsin L are not associated in cœlomocytes during immune response, further experiments will relate to the characterization of other T. tessulatum cathepsin B and Z which were partially described (Genbank accession number # AF542133 and AF542134). Moreover, Tettamanti and collaborators evidenced the existence of cathepsin B in H. medicinalis by immunochemistry studies [57]. Finally, recent data were obtained thanks to “The Hirudo medicinalis transcriptome project” [58] and highlighted the presence of cathepsin B, C and L like mRNA in leech nervous system.
Acknowledgments
Supported by grants from Centre National de la Recherche Scientifique (CNRS), Ministère de L'Education Nationale, de L'Enseignement Supérieur et de la Recherche, National Institute of Health (NIH)-Fogarty. The authors are grateful to Mrs Céline Boidin-Wichlacz for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tasiemski A, Vandenbulcke F, Mitta G, Lemoine J, Lefebvre C, Sautiere PE, et al. Molecular characterization of two novel antibacterial peptides inducible upon bacterial challenge in an annelid, the leech Theromyzon tessulatum. J Biol Chem. 2004;279:30973–82. doi: 10.1074/jbc.M312156200. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre C, Cocquerelle C, Vandenbulcke F, Hot D, Huot L, Lemoine Y, et al. Transcriptomic analysis in the leech Theromyzon tessulatum: involvement of cystatin B in innate immunity. Biochem J. 2004;380:617–25. doi: 10.1042/BJ20040478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anastasi A, Brown MA, Kembhavi AA, Nicklin MJ, Sayers CA, Sunter DC, et al. Cystatin, a protein inhibitor of cysteine proteinases. Improved purification from egg white, characterization, and detection in chicken serum. Biochem J. 1983;211:129–38. doi: 10.1042/bj2110129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett AJ. The cystatins: a diverse superfamily of cysteine peptidase inhibitors. Biomed Biochim Acta. 1986;45:1363–74. [PubMed] [Google Scholar]
- 5.Cimerman N, Mesko Brguljan P, Krasovec M, Suskovic S, Kos J. Serum concentration and circadian profiles of cathepsins B, H and L, and their inhibitors, stefins A and B, in asthma. Clin Chim Acta. 2001;310:113–22. doi: 10.1016/s0009-8981(01)00530-7. [DOI] [PubMed] [Google Scholar]
- 6.Kos J, Lah TT. Cysteine proteinases and their endogenous inhibitors: target proteins for prognosis, diagnosis and therapy in cancer (review) Oncol Rep. 1998;5:1349–61. doi: 10.3892/or.5.6.1349. [DOI] [PubMed] [Google Scholar]
- 7.Barrera C, Ye G, Espejo R, Gunasena S, Almanza R, Leary J, et al. Expression of cathepsins B, L, S, and D by gastric epithelial cells implicates them as antigen presenting cells in local immune responses. Hum Immunol. 2001;62:1081–91. doi: 10.1016/s0198-8859(01)00281-6. [DOI] [PubMed] [Google Scholar]
- 8.Honey K, Rudensky AY. Lysosomal cysteine proteases regulate antigen presentation. Nat Rev Immunol. 2003;3:472–82. doi: 10.1038/nri1110. [DOI] [PubMed] [Google Scholar]
- 9.Muno D, Kominami E, Mizuochi T. Generation of both MHC class I- and class II-restricted antigenic peptides from exogenously added ovalbumin in murine phagosomes. FEBS Lett. 2000;478:178–82. doi: 10.1016/s0014-5793(00)01849-4. [DOI] [PubMed] [Google Scholar]
- 10.Riese RJ, Chapman HA. Cathepsins and compartmentalization in antigen presentation. Curr Opin Immunol. 2000;12:107–13. doi: 10.1016/s0952-7915(99)00058-8. [DOI] [PubMed] [Google Scholar]
- 11.Tryselius Y, Hultmark D. Cysteine proteinase 1 (CP1), a cathepsin L-like enzyme expressed in the Drosophila melanogaster haemocyte cell line mbn-2. Insect Mol Biol. 1997;6:173–81. doi: 10.1111/j.1365-2583.1997.tb00085.x. [DOI] [PubMed] [Google Scholar]
- 12.Malecha J. Osmoregulation in Hirudinea Rhynchobdellida Theromyzon tessulatum (O.F.M.). Experimental localization of the secretory zone of a regulation factor of water balance. Gen Comp Endocrinol. 1983;49:344–51. doi: 10.1016/0016-6480(83)90198-3. [DOI] [PubMed] [Google Scholar]
- 13.Malecha J, Tramu G, Cardon C, Verger-Bocquet M. Evidence of apparent vasopressin and oxytocin peptides in the brain of the leech Rhynchobdelle Theromyzon tessulatum (O.F.M.) Gen Comp Endocrinol. 1986;64:13–20. doi: 10.1016/0016-6480(86)90022-5. [DOI] [PubMed] [Google Scholar]
- 14.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karlin S, Altschul SF. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Sci U S A. 1990;87:2264–8. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitta G, Vandenbulcke F, Noel T, Romestand B, Beauvillain JC, Salzet M, et al. Differential distribution and defence involvement of antimicrobial peptides in mussel. J Cell Sci. 2000;113(Pt 15):2759–69. doi: 10.1242/jcs.113.15.2759. [DOI] [PubMed] [Google Scholar]
- 17.Munoz M, Vandenbulcke F, Saulnier D, Bachere E. Expression and distribution of penaeidin antimicrobial peptides are regulated by haemocyte reactions in microbial challenged shrimp. Eur J Biochem. 2002;269:2678–89. doi: 10.1046/j.1432-1033.2002.02934.x. [DOI] [PubMed] [Google Scholar]
- 18.Heussler VT, Dobbelaere DA. Cloning of a protease gene family of Fasciola hepatica by the polymerase chain reaction. Mol Biochem Parasitol. 1994;64:11–23. doi: 10.1016/0166-6851(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 19.Michel A, Ghoneim H, Resto M, Klinkert MQ, Kunz W. Sequence, characterization and localization of a cysteine proteinase cathepsin L in Schistosoma mansoni. Mol Biochem Parasitol. 1995;73:7–18. doi: 10.1016/0166-6851(95)00092-f. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst. 1997;8:581–99. doi: 10.1142/s0129065797000537. [DOI] [PubMed] [Google Scholar]
- 21.Turk B, Turk D, Turk V. Lysosomal cysteine proteases: more than scavengers. Biochim Biophys Acta. 2000;1477:98–111. doi: 10.1016/s0167-4838(99)00263-0. [DOI] [PubMed] [Google Scholar]
- 22.Lewis SD, Johnson FA, Shafer JA. Effect of cysteine-25 on the ionization of histidine-159 in papain as determined by proton nuclear magnetic resonance spectroscopy. Evidence for a his-159--Cys-25 ion pair and its possible role in catalysis. Biochemistry. 1981;20:48–51. doi: 10.1021/bi00504a009. [DOI] [PubMed] [Google Scholar]
- 23.Rawlings ND, Barrett AJ. Evolutionary families of peptidases. Biochem J. 1993;290(Pt 1):205–18. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–90. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen LZ, Zhou JL, Zhou YZ, Gong HY, Li PY. Molecular cloning of two Rhipicephalus haemaphysaloides haemaphysaloides cathepsin L-like cysteine proteinase gene. Sheng Wu Gong Cheng Xue Bao. 2004;20:203–8. [PubMed] [Google Scholar]
- 26.Homma K, Kurata S, Natori S. Purification, characterization, and cDNA cloning of procathepsin L from the culture medium of NIH-Sape-4, an embryonic cell line of Sarcophaga peregrina (flesh fly), and its involvement in the differentiation of imaginal discs. J Biol Chem. 1994;269:15258–64. [PubMed] [Google Scholar]
- 27.Joseph LJ, Chang LC, Stamenkovich D, Sukhatme VP. Complete nucleotide and deduced amino acid sequences of human and murine preprocathepsin L. An abundant transcript induced by transformation of fibroblasts. J Clin Invest. 1988;81:1621–9. doi: 10.1172/JCI113497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renard G, Garcia JF, Cardoso FC, Richter MF, Sakanari JA, Ozaki LS, et al. Cloning and functional expression of a Boophilus microplus cathepsin L-like enzyme. Insect Biochem Mol Biol. 2000;30:1017–26. doi: 10.1016/s0965-1748(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 29.Cooper EL, Stein EA. Oligochaetes. In: Ratcliffe RA, Rowley AF, editors. Invertebrate blood cells. Academic Press; London: 1981. pp. 75–140. [Google Scholar]
- 30.Cooper EL. Earthworm immunity. Prog Mol Subcell Biol. 1996;15:10–45. doi: 10.1007/978-3-642-79735-4_2. [DOI] [PubMed] [Google Scholar]
- 31.Hartenstein V. Blood cells and blood cell development in the animal kingdom. Annu Rev Cell Dev Biol. 2006;22:677–712. doi: 10.1146/annurev.cellbio.22.010605.093317. [DOI] [PubMed] [Google Scholar]
- 32.Fitzgerald SW, Ratcliffe NA. In vivo cellular reactions and clearance of bacteria from the coelomic fluid of the marine annelid, Arenicola marina L. (polychaeta) Journal of Experimental Zoology. 1989;249:293–307. [Google Scholar]
- 33.Turk B, Turk V, Turk D. Structural and functional aspects of papain-like cysteine proteinases and their protein inhibitors. Biol Chem. 1997;378:141–50. [PubMed] [Google Scholar]
- 34.Hu KJ, Leung PC. Shrimp cathepsin L encoded by an intronless gene has predominant expression in hepatopancreas, and occurs in the nucleus of oocyte. Comp Biochem Physiol B Biochem Mol Biol. 2004;137:21–33. doi: 10.1016/j.cbpc.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Engelmann P, Molnar L, Palinkas L, Cooper EL, Nemeth P. Earthworm leukocyte populations specifically harbor lysosomal enzymes that may respond to bacterial challenge. Cell Tissue Res. 2004;316:391–401. doi: 10.1007/s00441-004-0874-x. [DOI] [PubMed] [Google Scholar]
- 36.Emden v M. Bau und Funktion des Bothryoidgewebes von Herpobdella atomaria carena. Z wiss Zool. 1929;134:1–83. [Google Scholar]
- 37.Mitta G, Vandenbulcke F, Hubert F, Salzet M, Roch P. Involvement of mytilins in mussel antimicrobial defense. J Biol Chem. 2000;275:12954–62. doi: 10.1074/jbc.275.17.12954. [DOI] [PubMed] [Google Scholar]
- 38.Porchet-Hennere E. Co-operation between different coelomocyte populations during the encapsulation response on Nereis diversicolor demonstrated using monoclonal antibodies. J Invert Pathol. 1990;56:353–361. [Google Scholar]
- 39.Porchet-Hennere E, Vernet G. Cellular immunity in an annelid (Nereis diversicolor, Polychaeta): production of melanin by a subpopulation of granulocytes. Cell Tissue Res. 1992;269:167–74. doi: 10.1007/BF00384737. [DOI] [PubMed] [Google Scholar]
- 40.de Eguileor M, Grimaldi A, Tettamanti G, Valvassori R, Cooper EL, Lanzavecchia G. Lipopolysaccharide-dependent induction of leech leukocytes that cross-react with vertebrate cellular differentiation markers. Tissue Cell. 2000;32:437–45. doi: 10.1054/tice.2000.0132. [DOI] [PubMed] [Google Scholar]
- 41.Engelmann P, Palinkas L, Cooper EL, Nemeth P. Monoclonal antibodies identify four distinct annelid leukocyte markers. Dev Comp Immunol. 2005;29:599–614. doi: 10.1016/j.dci.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 42.Engelmann P, Cooper EL, Nemeth P. Anticipating innate immunity without a Toll. Mol Immunol. 2005;42:931–42. doi: 10.1016/j.molimm.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 43.Grimaldi A, Tettamanti G, Perletti G, Valvassori R, de Eguileor M. Hematopoietic cell formation in leech wound healing. Curr Pharm Des. 2006;12:3033–41. doi: 10.2174/138161206777947443. [DOI] [PubMed] [Google Scholar]
- 44.Schneeweiss H, Renwrantz L. Analysis of the attraction of haemocytes from Mytilus edulis by molecules of bacterial origin. Dev Comp Immunol. 1993;17:377–87. doi: 10.1016/0145-305x(93)90029-p. [DOI] [PubMed] [Google Scholar]
- 45.Yip EC, Wong YH, Wong JT. Bacterial formyl peptide mediated chemotaxis and extracellular acidification in shrimp haemocytes. Dev Comp Immunol. 2001;25:269–77. doi: 10.1016/s0145-305x(00)00060-4. [DOI] [PubMed] [Google Scholar]
- 46.Guillou F, Mitta G, Galinier R, Coustau C. Identification and expression of gene transcripts generated during an anti-parasitic response in Biomphalaria glabrata. Dev Comp Immunol. 2007;31:657–71. doi: 10.1016/j.dci.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Baert JL, Britel M, Slomianny MC, Delbart C, Fournet B, Sautiere P, et al. Yolk protein in leech. Identification, purification and characterization of vitellin and vitellogenin. Eur J Biochem. 1991;201:191–8. doi: 10.1111/j.1432-1033.1991.tb16273.x. [DOI] [PubMed] [Google Scholar]
- 48.Bonnier P, Porchet-Hennere E, Baert JL. Identification of the eleocytes as the vitellogenin producing cells in nereids. Biol Cell. 1991;73:179–81. doi: 10.1016/0248-4900(91)90101-r. [DOI] [PubMed] [Google Scholar]
- 49.Britton C, Murray L. Cathepsin L protease (CPL-1) is essential for yolk processing during embryogenesis in Caenorhabditis elegans. J Cell Sci. 2004;117:5133–43. doi: 10.1242/jcs.01387. [DOI] [PubMed] [Google Scholar]
- 50.Carnevali O, Cionna C, Tosti L, Cerda J, Gioacchini G. Changes in cathepsin gene expression and relative enzymatic activity during gilthead sea bream oogenesis. Mol Reprod Dev. 2007 doi: 10.1002/mrd.20768. [DOI] [PubMed] [Google Scholar]
- 51.Carnevali O, Cionna C, Tosti L, Lubzens E, Maradonna F. Role of cathepsins in ovarian follicle growth and maturation. Gen Comp Endocrinol. 2006;146:195–203. doi: 10.1016/j.ygcen.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 52.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci U S A. 2001;98:12590–5. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nair SV, Del Valle H, Gross PS, Terwilliger DP, Smith LC. Macroarray analysis of coelomocyte gene expression in response to LPS in the sea urchin. Identification of unexpected immune diversity in an invertebrate. Physiol Genomics. 2005;22:33–47. doi: 10.1152/physiolgenomics.00052.2005. [DOI] [PubMed] [Google Scholar]
- 54.Loseva O, Engstrom Y. Analysis of signal-dependent changes in the proteome of Drosophila blood cells during an immune response. Mol Cell Proteomics. 2004;3:796–808. doi: 10.1074/mcp.M400028-MCP200. [DOI] [PubMed] [Google Scholar]
- 55.Lee MS, Cho SJ, Tak ES, Lee JA, Cho HJ, Park BJ, et al. Transcriptome analysis in the midgut of the earthworm (Eisenia andrei) using expressed sequence tags. Biochem Biophys Res Commun. 2005;328:1196–204. doi: 10.1016/j.bbrc.2005.01.079. [DOI] [PubMed] [Google Scholar]
- 56.Kang YS, Kim YM, Park KI, Kim Cho S, Choi KS, Cho M. Analysis of EST and lectin expressions in hemocytes of Manila clams (Ruditapes philippinarum) (Bivalvia: Mollusca) infected with Perkinsus olseni. Dev Comp Immunol. 2006;30:1119–31. doi: 10.1016/j.dci.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 57.Tettamanti G, Grimaldi A, Rinaldi L, Arnaboldi F, Congiu T, Valvassori R, et al. The multifunctional role of fibroblasts during wound healing in Hirudo medicinalis (Annelida, Hirudinea) Biol Cell. 2004;96:443–55. doi: 10.1016/j.biolcel.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 58.The Hirudo medicinalis transcriptome project. http://www.genoscope.cns.fr/externe/English/Projets/Projet_PE/organisme_PE.html.