Abstract
Comprehensive identification of polymorphisms among individuals within a species is essential both for studying the genetic basis of phenotypic differences and for elucidating the evolutionary history of the species. Large-scale polymorphism surveys have recently been reported for human1, mouse2, and Arabidopsis thaliana3. Here we report a nucleotide-level survey of genome variation in a diverse collection of 63 S. cerevisiae strains sampled from different ecological niches (beer, bread, vineyards, immunocompromised individuals, various fermentations and nature) and from locations on different continents. We hybridized genomic DNA from each strain to whole-genome tiling microarrays and detected 1.89 million single nucleotide polymorphisms (SNPs), which were grouped into 101,343 distinct segregating sites. We also identified 3,985 deletion events of length >200 bp among the surveyed strains. We analyzed the genome-wide patterns of nucleotide polymorphism and deletion variants, and measured the extent of linkage disequilibrium in S. cerevisiae. These results and the polymorphism resource we have generated lay the foundation for genome-wide association studies in yeast. We also examined the population structure of S. cerevisiae, providing support for multiple domestication events as well as insight into the origins of pathogenic strains.
With their small and compact genomes, the hemiascomycetes (the group of fungi that includes S. cerevisiae) represent a powerful model for comparative genomics and studies of genome evolution4–6. As a result, more than18 hemiascomycetes species are either completely or partially sequenced. The availability of the sequence data has presented an unprecedented opportunity to evaluate DNA sequence variation and genome evolution in a phylum spanning a broad evolutionary range7. This wealth of data on interspecific sequence differences stands in contrast to our limited knowledge of sequence variation within S. cerevisiae. Because of its importance both to human activities and as a model system, we sought to generate a comprehensive view of sequence polymorphism in S. cerevisiae. To determine sequence variation at the nucleotide level, we hybridized genomic DNA from 63 ecologically and geographically diverse strains (Table S1) to a high-density Affymetrix Yeast Tiling Microarray (YTM) and identified positions likely to differ from the reference sequence with the software package SNPscanner8. We detected a total of 1,896,131 SNPs in nonrepetitive regions of the genome (Table S1). Because of variation of up to a few bp in the location of SNPs detected by SNPscanner, we used a grouping procedure (see Materials and Methods) to identify the sites of polymorphic variation across strains. We also removed all singletons (SNPs called in only one strain) to further reduce false positives. This approach detected a total of 1,299,811 individual SNP calls, which were grouped into 101,343 distinct segregating sites. At each of these sites, every strain was classified as having either the same or different nucleotide relative to the reference strain (S288c).
We evaluated the coverage and accuracy of our polymorphism survey by comparing our data to the low-coverage sequence generated by Carter et al.9; 13 strains are shared between the two data sets. The great majority of array-called SNPs with sequence data in the region had corresponding polymorphisms in the sequence data (median of 92% per strain), showing that our data has a low false-positive rate. Array-based polymorphism calls captured most (median of 73% per strain) of the high quality (quality score > 30), independent (> 25 bp from the next closest polymorphism) SNPs present in the sequence data, showing that our data has high coverage. Discrepancies between array-based and sequence-based polymorphism calls likely reflect false positives and false negatives in each type of data, and may also derive from genuine sequence differences between strains with the same name but obtained from different sources by the two studies.
We detected an average of 30,097 SNPs per strain (Table S1). Excluding laboratory strains, most of which are closely related to the reference strain, the frequency of polymorphisms varied between 0.0011 to 0.0041 per bp (0.0028 on average), representing an average density of 2.8 SNPs per kb. Across all strains, we observed 8.35 non-singleton segregating sites per kb (θW/kb = 2.26). The frequency spectrum of the observed polymorphisms is highly skewed toward an excess of low-frequency alleles, even after corrections for the grouping procedure and genotyping errors (Figure S1). This excess of rare alleles resulted in a lowered value for the frequency-weighted measure of nucleotide diversity (π/kb = 1.92). Some of the excess of low-frequency alleles can be attributed to the presence of slightly deleterious variants, which are kept at low frequency by negative selection but have not yet been purged from the population. We expect that deleterious mutations should be more common in coding than in non-coding regions, resulting in a lower overall level of polymorphism in coding regions, and we do observe that coding regions are approximately 17% less polymorphic than noncoding regions (Table 1). The coding regions also show a slightly exaggerated skew in their frequency spectrum (Figure S2). These trends are further emphasized in the set of 1,114 genes known to be essential in the reference strain S288c, which shows both a lower overall level of polymorphism and a greater skew in the frequency spectrum. Noncoding regions are subject to selection on regulatory elements. Short intergenic regions should carry a higher proportion of functional regulatory sequences than longer non-coding regions, and we observe that intergenic regions shorter than 300 bp have significantly lower rates of polymorphism than longer regions (Figure S3). We found a markedly nonrandom distribution of polymorphism levels across the genome. We observed a decrease in SNP density within 25 kb of centromeres (Figure S4A). This observation is consistent with the lack of DNA double-strand breaks (i.e., the presence of meiotic recombination cold spots) near the centromeres10. By contrast, subtelomeric regions, which undergo frequent recombination11, show higher variation at the sequence level in the regions 15–45 kb from telomeres (Figure S4B).
Table 1.
Functional variation in levels of polymorphism
| Number of segregating sites |
θW / kb | π / kb | |
|---|---|---|---|
| Non-coding region | 33465 | 2.69 | 2.33 |
| Coding region | 71420 | 2.23 | 1.89 |
| Paralogs | 14774 | 2.28 | 1.93 |
| Essential genes | 13107 | 2.08 | 1.77 |
In a standard neutral model 4Nμ is the population mutation rate, where N is the effective population size and μ the rate of mutation.
θw is Watterson’s Theta: an estimate of 4Nμ based on the number of segregating sites.
π is an estimate of 4Nμ equal to the mean pairwise difference among individuals.
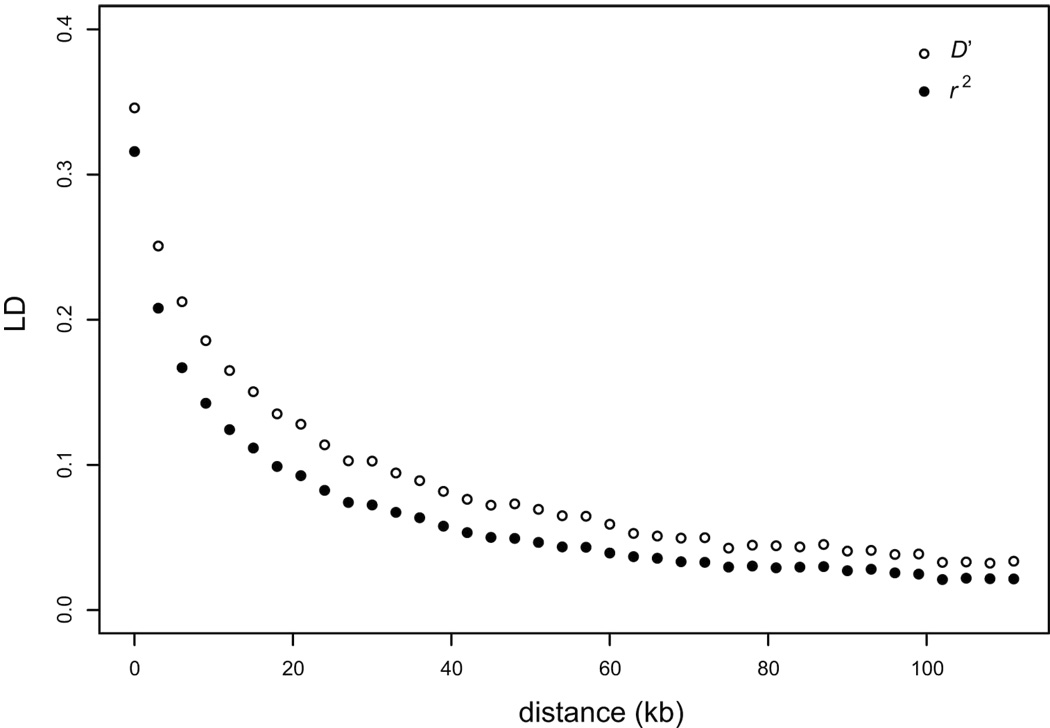
The genomic extent of linkage disequilibrium—nonrandom association of alleles at different polymorphic sites—provides information about recombination and population structure, and is also a critical parameter for population studies of association between genotype and phenotype. Our data provided the first opportunity to measure genome-wide properties of LD across a large collection of diverse strains. We examined pairwise LD for the 101,343 segregating sites and found that LD falls to half of its maximum value at about 11 kb (Figure 1). Because the yeast genome is physically compact (12 Mb), the 101,343 segregating sites reported here (nearly a site every 100 bp, of which close to half have a minor allele frequency >10%) provide a high-density polymorphism resource for S. cerevisiae from which an optimized panel of sites sufficient for whole-genome association studies in yeast can be chosen. To further characterize the architecture of LD, we examined each of the sampling groups that contained at least 10 strains (wine, clinical, distillery and laboratory strains) (Figure S5). In the wine strains, LD falls to half of its maximum value at ~2.5 kb, but is more extensive in clinical (~7 kb), distillery (~9.5 kb), and laboratory (~23.8 kb) strains. Because most of the laboratory strains are recently derived from the same founder strain S288c12, LD is expected to be greater than in the other groups. By contrast, the low level of LD in the wine strains probably reflects a long time since the most recent common ancestor of these strains, and perhaps a higher frequency of outcrossing events.
Figure 1.
Decay of linkage disequilibrium as a function of distance. Averages of pairwise LD measures r2 (black circles) and D' (open circles) are plotted for each bin of distances between pairs of SNPs. The LD values were corrected for finite-size effects by subtracting the average value computed for a random subset of pairs of SNPs located on different chromosomes.
To examine structural variation, we identified all deletion events >200 bp in the 63 strains (Table S1 and Table S2). We observed 3,985 deletions (an average of 63 per strain). The number of deletions varied from 1 in BY4716 (which is isogenic to the reference but carries an engineered deletion of LYS2) to 106 in YJM320. The deletions ranged in size from 200 bp to 13.8 kb, with nearly half falling between 200 bp and 400 bp (Figure S6). The deletions are unevenly distributed across the genome (Figure S7), with enrichment in subtelomeric regions (45.4 % of events in <10% of the genome; Figure S7B) and a deficit near the centromeres (Figure S7A). These patterns are consistent SNP rates and may similarly be explained by variation in recombination rates. 254 genes contained a whole (119 genes) or partial (135 genes) deletion in at least one strain (Table S3). Most were deleted in one to four strains, but some were deleted in many strains (Figure S8). For example, the gene YAR047C is deleted in 59 of the 63 surveyed strains. This gene is annotated as a dubious ORF (open reading frame) unlikely to encode a protein, based on comparative sequence data of Saccharomyces sensu stricto species4. Our observation within the S. cerevisiae species strongly confirms this hypothesis. Dubious ORFs accounted for 37 of the gene deletions. The set of deleted genes is enriched for those with known functions in transport, and in particular for sugar and hexose transporters (Table S4). Most of these deleted genes are located in the subtelomeric regions. These results provide clear evidence of the importance of variation at subtelomeric regions in adaptation of strains to different carbon sources, as previously suggested12,13.
We looked for deletions in genes known to be essential in the S288c strains14. We observed partial deletions in only 4 of the 1114 essential genes (KRS1, PGS1, SMT3 and ERG20), many fewer than the 49.6 genes that would be expected from the overall deletion frequency (χ2 = 52; P < 0.0001), which shows that the vast majority of the genes defined to be essential in the S288c background are also essential in all other genetic backgrounds of S. cerevisiae. With the exception of KRS1, these deletions were observed in only a few strains (Table S3). Moreover, the deletions observed in the 4 essential genes affect a small fraction of the open reading frame, and the genes may still be functional. We examined more closely the partial deletion in KRS1, which encodes the lysyl-tRNA synthetase. We looked at the spore viability from crosses between the S288c reference strain and several of the strains (K1, CLIB219, K12 and Y9) in which the KRS1 gene is partially deleted (Figure S9). We observed a high spore viability of around 90% in each cross, which shows that the KRS1 gene is still functional in these strains. We also observed a reduced deletion rate in duplicated gene pairs derived from the whole genome duplication event (20 observed vs. 49.4 expected; χ2 = 21.7; P < 0.0001; Table S3).
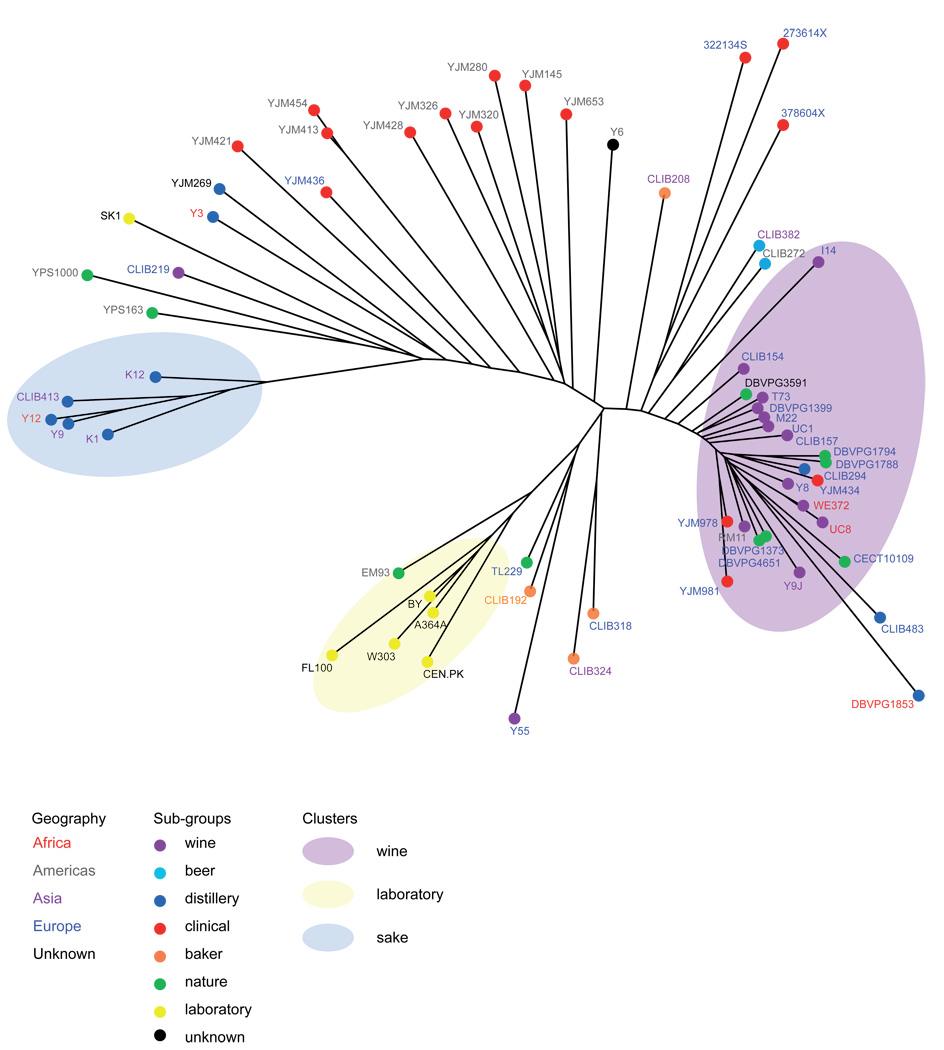
We sought to use the genome-wide genotypes at the 101,343 polymorphic sites across our diverse collection of strains to elucidate the phylogenetic relationships among strains and to evaluate the effects ecological factors and geographic locations on strain diversity. We used standard neighbor joining methods to build a majority-rule consensus tree of the surveyed strains (Figure 2), and also analyzed the data with the model-based clustering algorithm implemented in the program structure15 (Figure 3). Both analyses showed at least 3 distinct subgroups based on the source from which the strains were isolated. The majority of the wine strains (with the exception CLIB219, which was isolated in Russia) are members of a single well defined subpopulation. Because these wine strains were collected from dispersed locations, this observation provides strong evidence of a single domestication event of yeast for winemaking, followed by human-associated migration of wine yeast all over the world. The wine strains show the lowest level of polymorphism among the groups (Table 2), as well as an excess of low-frequency SNPs, consistent with a bottleneck during domestication. This subpopulation also includes a number of strains collected from distilleries, nature (soil, cocoa beans, prickly pear and tuber magnatum) and clinical sources, suggesting that these strains derived from domesticated wine strains, which transited out of this group to other human-associated fermentations as well as back into nature and therefore escaped their man-made environment. The second major population group contains the strains used for sake production and provides strong evidence for a second and independent domestication event, as hypothesized by Fay and Benavides16. The laboratory strains, with the exception of SK1, form a third clear group, a consequence of the fact that most of the commonly used Saccharomyces cerevisiae strains, with the exception of SK1, are derived from the S288c genetic background12. It is worth noting that the EM93 strain, the progenitor of S288c originally isolated from a rotting fig17, is seen to be closely related to the lab strains. A number of strains did not fall into clear groups on the tree and did not cluster into coherent groups in the structure analysis; their genomes appear to be mosaics of contributions from the three genetically distinct subgroups.
Figure 2.
Neighbor-joining tree of 63 Saccharomyces cerevisiae strains. The tree was constructed based on the 101,343 segregating sites identified in the surveyed strains. Branch lengths are proportional to the number of the 101,343 segregating sites that differentiate each pair of strains. Font color of strain name denotes geographic origin and circle color denotes ecological niche as specified in the key.
Figure 3.
Population structure of 63 Saccharomyces cerevisiae strains. Cluster results from a structure analysis on 101,343 segregating sites identified in the 63 surveyed strains. Structure implements a Bayesian model-based clustering algorithm that attempts to identify genetically distinct subpopulations based on patterns of allele frequencies15. Each strain is represented by a single vertical bar, which is partitioned into K colored segments that represent the strain’s estimated ancestry proportion in each of the K clusters.
Table 2.
Polymorphism and SNP frequency within groups
| Strains | θW / kb | π / kb |
|---|---|---|
| All | 2.26 | 1.92 |
| Wine | 1.38 | 1.25 |
| Clinical | 2.53 | 2.54 |
| Distiller | 2.54 | 2.60 |
| Nature | 2.03 | 1.97 |
In a standard neutral model 4Nμ is the population mutation rate, where N is the effective population size and μ the rate of mutation.
θw is Watterson’s Theta: an estimate of 4Nμ based on the number of segregating sites.
π is an estimate of 4Nμ equal to the mean pairwise difference among individuals.
Although S. cerevisiae is usually considered to be a benign organism, there is a growing recognition that it can be a cause of opportunistic pathogenic fungal infection, typically but not exclusively in immunocompromised individuals18. To investigate the origin of these strains, we examined 16 strains isolated from different clinical sources (e.g. blood, mouth, sputum) in Europe and the Americas (Table S1). The clinical isolates were broadly distributed across the tree, and did not cluster with each other or with any one subgroup of strains in the structure analysis (Figure 2 and Figure 3). Three European clinical strains (YJM434, YJM978 and YJM981) were closely related to wine strains. Three other European strains from the same geographical origin (Newcastle, UK) were closely related to each other, and had some similarity to beer and baker strains. The remaining 10 strains (9 American, 1 European) branched from a similar part of the tree, but did not appear to be closely related to each other or to any other coherent group of strains. Our interpretation of these results is that clinical isolates do not derive from a common ancestor or any one type of strains, but rather represent multiple events in which strains present in the environment opportunistically colonize human tissues. Interestingly, important niches such as being infectious are not associated with dramatic bottlenecks. Our data provide strong evidence that wine strains are capable of such colonization, and suggest that strains from other sources (beer, bakery, lab, nature) can also do this. These results are consistent with clinical reports of patients infected with S. cerevisiae baker’s strains and with the strain S. boulardii, which is used therapeutically to treat diarrhea and is also sold as a probiotic nutritional supplement19. Because the main environmental niches for S. cerevisiae in nature are not known, clinical strains might represent the best approximation of the overall species diversity of S. cerevisiae.
The polymorphism resource we generated, made freely available in the Yeast SNPs Browser database (http://pgbrowse.princeton.edu/cgi-bin/gbrowse/yeast_strains_snps/), enables genome-wide association studies of the phenotypic differences among these and other yeast strains. Phenotypic diversity among yeast isolates is significant, and variation is apparent among the surveyed strains at different levels. The genetic basis of a number of interesting phenotypes can be studied in yeast, including growth at high temperature, sporulation efficiency, telomere length, gene expression, and response to drugs20–24; these studies can now move from linkage in crosses between two strains to the population level. S. cerevisiae provides a powerful model system for studies of complex traits because of the ease with which genetic analyses and phenotyping can be carried out and the ability to engineer and test the effects of individual polymorphisms and their combinations on different genetic backgrounds.
Our analysis also provides insight into the population structure of this yeast species. We show evidence for genetic differentiation of 3 distinct subgroups based on the source from which the strains were isolated: vineyards, sake and related fermentations, and laboratory strains. Thus, population structure at least partly reflects different ecological niches. Surveys of additional strains are needed to fully resolve the roles of ecology vs. geography in the genetic differentiation of this species. Our data strongly support the hypothesis that these three groups represent separate domestication events, and that S. cerevisiae as a whole is not domesticated. Finally, our results suggest that S. cerevisiae strains from a range of environments are capable of opportunistic colonization of human tissues.
Methods Summary
Genomic DNA was extracted from 63 yeast strains (listed in Supplementary Table 1) and hybridized to Affymetrix Yeast Tiling Arrays. We used SNPscanner8 to identify putative SNPs in each of the 63 strains based on the hybridization intensity at each probe. Since there is error in the precise location of SNP calls made by SNPscanner, we employed a grouping procedure (described in Methods) in order to integrate SNP calls across strains and minimize the effects of erroneous positive and negative calls.
We constructed a neighbor-joining tree of the 63 strains from the SNP data using Splitstree25, with branch lengths proportional to the number of segregating sites that differentiate each node. To infer the population ancestry of the strains we used structure15, with ancestral population numbers between 2 and 6. We calculated linkage disequilibrium across the genome using two standard metrics: D’ and r2, both for the whole genome and for each sub-population. We calculated other population genetic summary statistics using code based on the libsequence package26, and performed coalescent simulations of genome evolution using FastCoal27, with corrections for expected error rates and our grouping procedure.
Methods
Yeast strains
Yeast strains were obtained from a number of laboratories: Justin Fay (Washington University, St Louis, USA), Jose Perez-Ortin (University of Valencia, Spain), Gianni Liti and Ed Louis (The University of Nottingham, UK), John McCusker (Duke University, Durham, USA) and Jean-Luc Souciet (Louis-Pasteur University, Strasbourg, France). We also purchased strains from different yeast culture collections: CLIB (Collection de Levures d’Intérêt Biotechnologique), CBS (Centraalbureau voor Schimmelcultures), DBVPG (Dipartimento di Biologia Vegetale e Agroambientale of the University of Perugia) and CECT (Coleccion Espanola de Cultivos Tipo). Strains used in this study are listed in Supplementary Table1.
SNP identification
Yeast strains were grown in yeast extract, peptone, and dextrose (YPD) medium. Total genomic DNA was purified from 30 ml YPD culture using Qiagen Genomic-Tips 100/G and Genomic DNA Buffers as per the manufacturer’s instructions. Genomic DNA was digested with DNaseI, labeled and hybridized to Affymetrix Yeast Tiling Arrays (YTMs) as described in Gresham et al.8
We used SNPscanner8 to identify putative SNPs in each of the 63 strains based on the hybridization intensity of DNA at each probe. SNPs from each strain were independently called against the reference FY3 genome using the following parameters: lod score > 2, number of probes covering a base > 1, and positive region length > 6. These parameters are further described in Gresham et al. 8 and in the SNPscanner documentation (http://genomics-pubs.princeton.edu/SNPscanner/). With these parameters, we previously showed, using the complete genome sequence of strain YJM789, that 90.1% of true SNPs were detected, with only 49 false positive SNP calls over the entire genome (a false-positive rate of 4×10−6 per bp).
Due to the 4 bp resolution of the YTMs and the variance associated with DNA hybridization intensities, the SNP position predicted by SNPscanner may fall at varying positions surrounding the actual site of the SNP. This variance required us to perform a grouping procedure, combining all the calls within 6 bp of each other into a single segregating site. As the average density of putative SNPs is 1 per 6.3 bp, the probability of grouping 2 distinct sites is nontrivial. In order to reduce this probability, we implemented several heuristic filters: First, to reduce false positives, we required that at least one of the called SNPs in each grouping have a lod score >6. Second, we eliminated possible deletion events by removing putative SNPs with large prediction regions (> 100 bp). Finally, we required at least 9 bp between each SNP in a group and the next closest call in the genome. We performed this grouping procedure in a top-down manner, by first grouping the SNPs with the most calls at a given position.
We tested the accuracy of this grouping procedure using a set of known high confidence SNPs from the completely sequenced genomes of the strains S288c, RM11-1a, and YJM145. Specifically, we examined 13,839 SNPs for which YJM145 and RM11-1a had the same allele and differed from the reference sequence. For this set, 12,578 (91%) and 11,518 (83%) SNPs were detected before grouping in YJM145 and RM11-1a, respectively. After grouping, 9,119 SNPs were detected in at least one strain, and 8,086 were correctly called in both strains, from which we infer a false negative rate of 5.7% per strain, given detection in at least one strain. The grouping procedure almost never separated the same site into multiple sites (1 case across the genome), and rarely combined two distinct sites (394 cases; <5% of sites after grouping). These cases are typically SNPs that are located within 4 bp of each other, closer than the theoretical resolution of the YTMs. We also removed all singletons (SNPs called in only one strain) to further reduce false positives.
Tree building and structure analysis
We constructed a neighbor-joining tree of the 63 strains from the SNP data using the software package Splitstree25, with branch lengths proportional to the number of segregating sites that differentiate each node. We ran structure using the linkage model with the population number parameter, K, set from 2 to 6, for 100,000 iterations after a burn-in of 100,000 iterations, the first 50,000 of which were run under the free-recombination model15.
Linkage disequilibrium
We calculated linkage disequilibrium across the genome using two standard metrics: D’ and r2. We computed these statistics for all pairs of sites located within a given distance, both for all the strains and within each predefined sub-population. To correct for finite-size effects and differences in sample size among the sub-populations, we subtracted from each statistic the average value for a random subset of SNP pairs located on different chromosomes (which should not show LD).
Polymorphism and divergence statistics
We calculated population genetic summary statistics of polymorphism using code based on the libsequence package26. To correct for the removal of singleton SNPs in the data set, modified estimators of the population mutation parameters θW and π were used28. An analog of Tajima’s D was calculated as the difference between these modified estimates29. To obtain significance values, we simulated under a modified coalescent model as described below, conditioning on the observed number of segregating sites and the approximate length of the sequence. The significance of an observed statistic was then taken to be the probability of observing a more extreme value in at least 10,000 simulations. Divergence rates were calculated from the multiple species alignments of Kellis et al.4. We used PAML30 to obtain maximum likelihood estimates of the rate of evolution along the S. cerevisiae branch after divergence from S. paradoxus.
Coalescent simulations of genome evolution were performed using FastCoal27. Output from each coalescent simulation was run through a series of steps to mirror the sources of error inherent in the SNPscanner data. First, called SNPs were randomly removed with a probability of 5%. The addition of randomly missed calls creates a characteristic dearth of high frequency SNPs in the data set; simulations under a 5% false negative rate fit very closely with the observed pattern of polymorphism at high frequency. To correct for incorrectly grouped SNPs, we performed the previously described grouping procedure on all simulated data.
Supplementary Material
Acknowledgments
We are grateful to all the researchers and institutions, and especially Justin Fay, for sharing yeast strains. We thank K. Dolinski and J. Matese for technical support and D. Gresham for comments on the manuscript. The authors gratefully acknowledge helpful discussions with members of the Kruglyak and Botstein laboratories. This work was supported by NIH grant R37 MH059520 and a James S. McDonnell Foundation Centennial Fellowship (L.K.) and NIH grant GM071508 to the Lewis-Sigler Institute.
Footnotes
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions
The authors declare no competing financial interests.
References
- 1.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frazer KA, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- 3.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 4.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 5.Cliften P, et al. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 6.Dujon B, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 7.Dujon B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 2006;22:375–387. doi: 10.1016/j.tig.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Gresham D, et al. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science. 2006;311:1932–1936. doi: 10.1126/science.1123726. [DOI] [PubMed] [Google Scholar]
- 9.Carter DM, et al. Population genomics of domestic and wild yeasts. doi: 10.1038/nature07743. submitted to Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerton JL, et al. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pryde FE, Gorham HC, Louis EJ. Chromosome ends: all the same under their caps. Curr Opin. Genet. Dev. 1997;7:822–828. doi: 10.1016/s0959-437x(97)80046-9. [DOI] [PubMed] [Google Scholar]
- 12.Schacherer J, et al. Genome-wide analysis of nucleotide-level variation in commonly used Saccharomyces cerevisiae strains. PLoS ONE. 2007;2:e322. doi: 10.1371/journal.pone.0000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winzeler EA, et al. Genetic diversity in yeast assessed with whole-genome oligonucleotide arrays. Genetics. 2003;163:79–89. doi: 10.1093/genetics/163.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 15.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fay JC, Benavides JA. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 2005;1:66–71. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mortimer RK, Johnston JR. Genealogy of principal strains of the yeast genetic stock center. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enache-Angoulvant A, Hennequin C. Invasive Saccharomyces infection: a comprehensive review. Clin. Infect. Dis. 2005;41:1559–1568. doi: 10.1086/497832. [DOI] [PubMed] [Google Scholar]
- 19.de Llanos R, Querol A, Peman J, Gobernado M, Fernandez-Espinar MT. Food and probiotic strains from the Saccharomyces cerevisiae species as a possible origin of human systemic infections. Int. J. Food. Microbiol. 2006;110:286–290. doi: 10.1016/j.ijfoodmicro.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 20.Steinmetz LM, et al. Dissecting the architecture of a quantitative trait locus in yeast. Nature. 2002;416:326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- 21.Deutschbauer AM, Davis RW. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 2005;37:1333–1340. doi: 10.1038/ng1674. [DOI] [PubMed] [Google Scholar]
- 22.Gatbonton T, et al. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2006;2:e35. doi: 10.1371/journal.pgen.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 24.Perlstein EO, Ruderfer DM, Roberts DC, Schreiber SL, Kruglyak L. Genetic basis of individual differences in the response to small-molecule drugs in yeast. Nat. Genet. 2007;39:496–502. doi: 10.1038/ng1991. [DOI] [PubMed] [Google Scholar]
- 25.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 26.Thornton K. Libsequence: a C++ class library for evolutionary genetic analysis. Bioinformatics. 2003;19:2325–2327. doi: 10.1093/bioinformatics/btg316. [DOI] [PubMed] [Google Scholar]
- 27.Marjoram P, Wall JD. Fast "coalescent" simulation. B.M.C. Genet. 2006;7:16. doi: 10.1186/1471-2156-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu YX. Estimating effective population size or mutation rate using the frequencies of mutations of various classes in a sample of DNA sequences. Genetics. 1994;138:1375–1386. doi: 10.1093/genetics/138.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.