Abstract
Immune cell products such as interferon (IFN)-γ and interleukin (IL)-12 are potent inhibitors of osteoclast formation. We previously characterized the human osteoclast inhibitory peptide-1 (OIP-1/hSca), a Ly-6 gene family member and showed IFN-γ modulation of OIP-1 expression in bone marrow cells. Whether, IL-12 regulates OIP-1 expression in the bone microenvironment is unclear. Real-time PCR analysis revealed that IL-12 treatment significantly enhanced OIP-1 mRNA expression in human bone marrow mononuclear cells. Because IL-12 induces IFN-γ production by T cells, we tested whether IFN-γ participates in IL-12 stimulation of OIP-1 gene expression in these cells. IL-12 treatment in the presence of IFN-γ neutralizing antibody significantly increased OIP-1 mRNA expression, suggesting that IL-12 directly regulates OIP-1 gene expression. Interestingly, real-time PCR analysis demonstrated that IL-12 induces OIP-1 expression (3.2-fold) in CD4+ T cells; however, there was no significant change in CD8+ T cells. Also, IL-12 (10 ng/ml) treatment of Jurkat cells transfected with OIP-1 gene (−1 to −1,988 bp) promoter-luciferase reporter plasmid demonstrated a 5-fold and 2.7-fold increase in OIP-1 gene promoter activity in the presence and absence of antibody against IFN-γ, respectively. We showed that STAT-1,3 inhibitors treatment significantly decreased IL-12 stimulated OIP-1 promoter activity. Chromatin immunoprecipitation (ChIP) assay confirmed STAT-3, but not STAT-1 binding to the OIP-1 gene promoter in response to IL-12 stimulation. These results suggest that IL-12 stimulates the OIP-1 gene expression through STAT-3 activation in CD4+ T cells.
Keywords: OSTEOCLAST INHIBITORY PEPTIDE-1, T CELLS, INTERFERON-γ, STAT, BONE MARROW CELLS
Osteoclast formation and bone resorption activity is regulated by a variety of cell types and its products in the bone microenvironment [Roodman, 1996]. We have previously identified and characterized a novel autocrine/paracrine inhibitor of osteoclast formation termed osteoclast inhibitory peptide-1 (OIP-1/hSca). OIP-1/hSca is also known as retinoic acid induced gene expression (RIG-E) or thymic shared antigen-1 (TSA-1) and is a Ly-6 gene family member expressed on immature thymocytes and thymus epithelial cells. OIP-1/hSca is a glycophosphatidylinositol (GPI)-linked membrane protein (16 kDa) containing a 79 amino acid extracellular peptide and a 32 amino acid carboxy terminal GPI-linked peptide (c-peptide) [Koide et al., 2002]. OIP-1 has 65% homology with mouse Sca-2 at the nucleotide level. High levels of Sca-2 expression in murine bone marrow cells and spleen cells have been reported [Classon and Coverdale, 1996]. Sca-2 is a useful marker in early T cell development and activation, and seems to play a regulatory role in thymocyte differentiation. Sca-2 is expressed in human lymphoid tissues as well as in various nonlymphoid tissues [MacNeil et al., 1993]. Sca-2 has been shown to function as a modulator of the T cell receptor (TCR) signaling pathway. Recently, it has been reported that Sca-2 is physically and functionally associated with CD3 ζ chains of the TCR complex [Kosugi et al., 1998]. Moreover, Ly-6A (Sca-1) knock-out mice had decreased bone mineral density and bone mineral content, implicating an essential role for the LY-6 gene family in normal bone remodeling [Bonyadi et al., 2003].
Immune cell products such as interferon’s (IFNs) that are released in response to inflammatory stimuli have been reported to be important local negative regulators for bone remodeling [Roodman, 1999]. We have shown that IFN-γ significantly enhances OIP-1/hSca mRNA expression in osteoclast precursor cells and that OIP-1/hSca c-peptide specific antibody partially neutralized IFN-γ inhibition of osteoclast formation [Koide et al., 2003]. T cell production of IFN-γ strongly suppresses osteoclastogenesis by interfering with the RANKL-RANK signaling pathway [Takayanagi et al., 2000]. These studies suggested there is cross-talk between TNF and IFN families of cytokines, through which IFN provides a negative link between T cell activation and bone resorption. Furthermore, activated CD8+ T cell subsets have been shown to inhibit osteoclast formation while CD4+ subsets induce osteoclast formation in the presence of M-CSF [Choi et al., 2001]. However, it has been reported that the T cells exerts both stimulatory and inhibitory actions on osteoclastogenesis based on mode of activation and cytokine production by these cells [Grcevic et al., 2001]. IL-12 is primarily produced by macrophages and dendritic cells and has been shown to induce IFN-γ production by T cells [Robinson and O’Garra, 2002]. It has been reported that IL-12 alone and in synergy with IL-18 has been shown to inhibit osteoclast formation in co-cultures of murine osteoblast and spleen cells [Horwood et al., 2001]. Also, the inhibitory effects of these cytokines was shown to be blocked by the addition of an anti-IFN-γ neutralizing antibody suggesting that IFN-γ plays an important role in the IL-12 and IL-18 inhibition of the bone resorbing activity of osteoclasts [Yamada et al., 2002]. In this study, we examine IL-12 transcriptional regulation of OIP-1 expression in T cells and show that IL-12 directly regulates OIP-1 gene expression in CD4+ T cells.
MATERIALS AND METHODS
REAGENTS
Cell culture reagents were purchased from Invitrogen (Carlsbad, CA). STAT-1 inhibitor, fludarabine was purchased from Sigma (St. Louis, MO) and the STAT-3 specific peptide inhibitor that contains a C-terminal mts (membrane translocating sequence) which acts as a highly selective, potent blocker of STAT-3 activation (Cat. log # 573096) was obtained from Calbiochem (Gibbstown, NJ). IL-12 recombinant protein and anti-IFN-γ antibody were obtained from R&D systems, Inc. (Minneapolis, MN). Peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Super signal-enhanced chemiluminescence (ECL) reagent was obtained from Amersham Biosciences (Piscat-away, NJ) and nitrocellulose membranes were purchased from Millipore (Bedford, MA). A luciferase reporter assay system was obtained from Promega (Madison, WI).
OIP-1/hSCA GENE PROMOTER ACTIVITY ASSAY
We have previously developed an OIP-1 gene promoter (−1 to −1,988 bp relative to transcription start site)-luciferase reporter plasmid construct (OIP-1 pGL2 Basic#3) as described [Srinivasan et al., 2006]. Jurkat T-cells (2 × 106) plated in a six-well culture plate were transiently transfected with the OIP-1 pGL2 Basic#3 plasmid DNA (2 μg) by using the nucleofector kit V (Amaxa, Gaithersburg, MD) according to the manufacturer’s instructions. The cells were cultured in the presence/absence of IL-12 (10 ng/ml) for 48 h. The cells were washed twice with phosphate buffered saline and incubated at room temperature for 15 min with 0.3 ml cell lysis reagent (Promega). A 20 μl aliquot of each sample was mixed with 100 μl of the luciferase assay reagent and the light emission was measured for 10 s of integrated time using Sirius Luminometer. The transfection efficiency was normalized by co-transfection with pRSV β-gal plasmid (0.2 μg) and measuring the β-galactosidase activity in these cells.
QUANTITATIVE REAL-TIME RT-PCR
OIP-1 mRNA expression levels were determined by quantitative real-time reverse transcription polymerase chain reaction (RT-PCR) as described previously [Srinivasan et al., 2006]. Briefly, total RNA was isolated from human bone marrow mononuclear cells or Jurkat T cells stimulated with and without IL-12 (10 ng/ml) for 48 h, using RNAzol reagent (Tel-Test Inc., Friendswood, TX). A reverse transcription reaction was performed using poly-dT primer and Moloney murine leukemia virus reverse transcriptase (Applied Biosystems) in a 25 μl reaction volume containing total RNA (2 μg), 1× PCR buffer and 2 mM MgCl2, at 42°C for 15 min followed by 95°C for 5 min. The real-time PCR was performed using SYBR Green Supermix in an iCycler (iCycler iQ Single-color Real Time-PCR detection system; Bio-Rad, Hercules, CA). The primer sequences used to amplify glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA were 5′-CCTACCCCCAATGTATCCGTTGTG-3′ (sense) and 5′-GGAGGAATGGGAGTTGCTGTTGAA-3′ (anti-sense); OIP-1 mRNA 5′-TTTATTCACCCAGGAGGAGCTGAAGGTTCC-3′ (sense) and 5′-CCAACATGAGAGTCTTCCTGCCTG-3′ (anti-sense) and for IFN-γ mRNA were 5′-GCGTCATTGAATCACACCTG-3′ (sense), 5′-TGAGCTCATTGAATGCTTGG-3′ (anti-sense). Thermal cycling at 94°C for 3 min, followed by 40 cycles of amplifications at 94°C for 30 s, 66°C for 1 min, 72°C for 1 min and 72°C for 5 min as the final elongation step. Relative levels of OIP-1 mRNA expression were normalized in all the samples analyzed with respect to the levels of GAPDH amplification.
PREPARATION OF HUMAN T CELL SUBSETS
Peripheral blood mononuclear cells (PBMC) obtained following the IRB approved protocol at the Medical University of South Carolina were separated from the heparinized blood by standard Ficoll-Hypaque (Sigma, MO) gradient centrifugation. CD4+ and CD8+ T cell subsets at >95% purity were isolated by negative selection with Dynal CD8 or CD4 magnetic beads (Dynal-Invitrogen, CA). Briefly, the PBMC (2 × 107/ml) were incubated with either CD4 or CD8 beads at the concentration recommended by the manufacturer for 20 min at 4°C. The cell suspensions were then placed on a magnetic plate for 2–3 min. The unbound cells were transferred to another tube and washed three times with PBS. The cells depleted of CD4+ or CD8+ T cells were then suspended in complete medium.
OLIGONUCLEOTIDE PULL-DOWN ASSAY
Jurkat T cells were treated with and without IL-12 (10 ng/ml) and in the presence/absence of excess concentration of anti-IFN-γ neutralizing antibody (50 U/ml) for the indicated time period. Total cell lysates were subjected to nuclear extraction (NE) using a nuclear extraction kit (Pierce, CA). The extracts were incubated for 2 h with biotinylated oligonucleotides from the OIP-1 gene promoter containing STAT binding sequence (−1,629 to −1,639 bp position), sense 5′-GGTGAGCAAGCGTTCCTCTAACAGTGGGAA-3′ and antisense 5′-TTCCCACTGTTAGAGGAACGCTTGCTCACC-3′. The STAT-1 binding site was mutated as underlined (Mut.STAT oligo sense 5′-GGTGAGCAAGCGTTAATTTAACAGTGGGAA 3′). The complexes were isolated by streptavidin conjugated Dynal magnetic beads and analyzed by immunobloting with anti-pSTAT-1 or pSTAT-3 antibodies as described [Srinivasan et al., 2006].
CHROMATIN IMMUNOPRECIPITATION (ChIP) ASSAY
ChIP was performed using the ChIP Assay Kit (Upstate, Temecula, CA). The Jurkat T cells were treated with or without IL-12 (10 ng/ml) and in the presence/absence of excess anti-IFN-γ antibody (50 U/ml). Cells were cross-linked with 1% final concentration of formaldehyde at 37°C for 10 min before harvest. Soluble chromatin was prepared following sonication with a Branson-250 digital sonifier (Branson Ultrasonics, Danbury, CT) to an average DNA length of 200–1,000 bp. Approximately 5 × 105 cell equivalent (1/6th) of the sheared soluble chromatin was precleared with blocked Protein G agarose, and 10% of the precleared chromatin was set aside as input control. Immunoprecipitation was carried out with 5 μg of pSTAT-1 and pSTAT-3 antibodies, overnight at 4°C. Immune complexes were pulled down using Protein G agarose, washed and eluted twice with 250 μl of elution buffer (0.1 M NaHCO3, 1% SDS) and cross-linking reversed in 200 mM NaCl at 65°C overnight with 20 μg RNase A (Sigma). DNA was purified following proteinase K treatment (Invitrogen Life Technologies, Carlsbad, CA) with the Qiagen PCR purification kit (Qiagen, MD). Then, PCR was performed using the OIP-1 gene specific primers for STAT binding region, sense 5′-CCAGAGGCCTGGTGAGCAAG-3′ and antisense 5′-AGACTGC-GATAGACGTCCAT-3′. DNA samples or input DNA fractions were analyzed by 35 cycles of PCR (94°C for 30 s, 55°C for 30 s, and 72°C for 30 s). PCR products were subjected to electrophoresis using 2% agarose gels and visualized by ethidium bromide.
STATISTICAL ANALYSIS
Results are presented as mean ±SE for three independent experiments and were compared by Student’s t-test or one-way ANOVA. Values were considered significantly different for P <0.05.
RESULTS
IL-12 ENHANCES THE OIP-1 mRNA EXPRESSION IN HUMAN BONE MARROW MONOCYTES
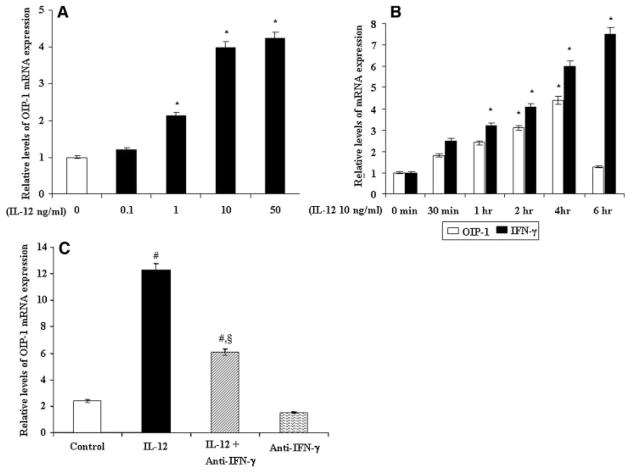
We examined IL-12 stimulation of OIP-1 mRNA expression in human bone marrow derived mononuclear cells by quantitative real-time RT-PCR analysis. As shown in the Figure 1A, IL-12 treatment (0–50 ng/ml) to human bone marrow mononuclear cells demonstrated a significant increase in OIP-1 mRNA expression in a dose-dependent manner. We further examined the IL-12 stimulation of OIP-1 expression in a time (0–6 h) dependent manner. Time course study identified that IL-12 treatment for 4 h period induced high-level expression of OIP-1 in these cells; however this declined by 6 h period of treatment (Fig. 1B). Relative levels of OIP-1 expression were normalized with respect to GAPDH amplification in RT-PCR analysis. We therefore used IL-12 treatment (10 ng/ml) for a 4 h period for subsequent experiments unless indicated.
Fig. 1.
IL-12 stimulation and OIP-1 mRNA expression in human bone marrow cells. A: The human bone marrow mononuclear cells were stimulated with IL-12 (0–50 ng/ml) for 4 h and total RNA isolated was subjected to quantitative real-time PCR analysis for OIP-1 mRNA expression. B: IL-12 stimulation of OIP-1 and IFN-γ expression in human bone marrow cells. Bone marrow mononuclear cells were stimulated with IL-12 (10 ng/ml) for indicated time period (0–6 h) and total RNA isolated was subjected to real-time RT-PCR analysis for OIP-1 and IFN-γ mRNA expression. C: IL-12 stimulation of OIP-1 expression in human bone marrow cells cultured in the presence of anti-IFN-γ neutralizing antibody. Cells stimulated with IL-12 (10 ng/ml) for 4 h with/without IFN-γ antibody (50 U/ml) and total RNA isolated was subjected to quantitative real-time PCR analysis for OIP-1 expression. Relative levels of OIP-1 and IFN-γ gene expression were normalized with respect to the levels of GAPDH amplification. Values expressed as mean ±SE for three independent experiments. *P <0.05 level, #P <0.05 versus control and §P <0.05 versus IL-12 treated group.
IL-12 is primarily produced by macrophages and dendritic cells, and has been shown to potently induce the production of IFN-γ by T and NK cells [Horwood et al., 2001]. Real-time RT-PCR analysis of total RNA isolated from human bone marrow mononuclear cells stimulated with IL-12 further confirmed a significant increase in IFN-γ mRNA expression in a time-dependent (0–6 h) manner (Fig. 1B). We have previously reported IFN-γ stimulation of OIP-1 expression in bone marrow cells [Koide et al., 2003]. We further determined if IL-12 stimulation of OIP-1 expression is mediated by IFN-γ. Since OIP-1/hSca and IFN-γ expression is abundant in T lymphocytes, we quantified the levels of OIP-1 mRNA expression in human bone marrow cells in response to IL-12 treatment in the presence and absence of IFN-γ neutralizing antibody. Real-time PCR analysis demonstrated that bone marrow cells treated with IL-12 showed a significant increase (10-fold) in the level of OIP-1 expression. However, IL-12 treatment in the presence of a neutralizing antibody against IFN-γ showed a 4-fold increase in OIP-1 mRNA expression compared to control unstimulated cells (Fig. 1C). These results suggest that IL-12 may have direct effect in stimulating OIP-1 gene expression and confirms our previous results that elevated levels of IFN-γ are in part responsible for IL-12 stimulation of OIP-1 gene expression.
IL-12 SPECIFIC STIMULATION OF OIP-1 EXPRESSION IN CD4+ T CELLS
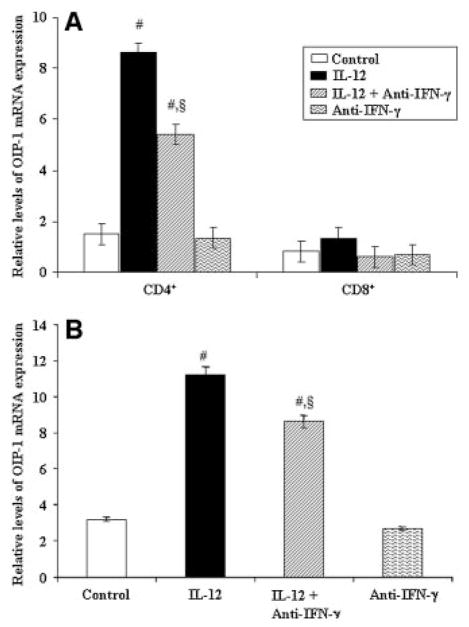
To further delineate the IL-12 stimulation of OIP-1 gene expression in lymphocytes, we isolated the T cell subsets of CD4+ and CD8+ cells from human peripheral blood as described in Materials and Methods Section. Purified CD4+ and CD8+ cells were stimulated with IL-12 in the presence/absence of IFN-γ antibody for 4 h and total RNA isolated was subjected to real-RT-PCR analysis for OIP-1 mRNA expression. As shown in Figure 2A, IL-12 modulation of OIP-1 expression is specific to CD4+ cells and in contrast IL-12 has no significant effect on OIP-1 expression in CD8+ cells. Furthermore, real-time PCR analysis of total RNA isolated from Jurkat T CD4+ cells stimulated with IL-12 in the presence IFN-γ neutralizing antibody revealed significantly increased (2.6-fold) OIP-1 expression, which suggests IL-12 specific modulation of OIP-1 expression in these cells. IFN-γ neutralizing antibody alone, however, has no significant effect on OIP-1 expression in control cultures (Fig. 2B). These results further suggest that IL-12 stimulation of OIP-1 expression is restricted to CD4+ T cells.
Fig. 2.
IL-12 regulation of OIP-1 expression in the T cell subsets cultured in the presence of anti-IFN-γ neutralizing antibody. The peripheral blood mononuclear cells (PBMC) were subjected for CD4+ and CD8+ cells isolation and stimulated with IL-12 (10 ng/ml) for 4 h with/without anti-IFN-γ antibody (50 U/ml) and total RNA was isolated. A: Real-time RT-PCR analysis of OIP-1 mRNA expression in CD4+ and CD8+ T cells. B: Real-time RT-PCR analysis of OIP-1 mRNA expression in Jurkat T cells stimulated with IL-12 (10 ng/ml) for 4 h with/without anti-IFN-γ antibody (50 U/ml). The levels of OIP-1 mRNA expression were normalized with respect to the levels of GAPDH amplification. Values expressed as mean ±SE for three independent experiments. #P <0.05 versus control and §P <0.05 versus IL-12 treated group.
IL-12 REGULATION OF OIP-1 GENE PROMOTER ACTIVITY
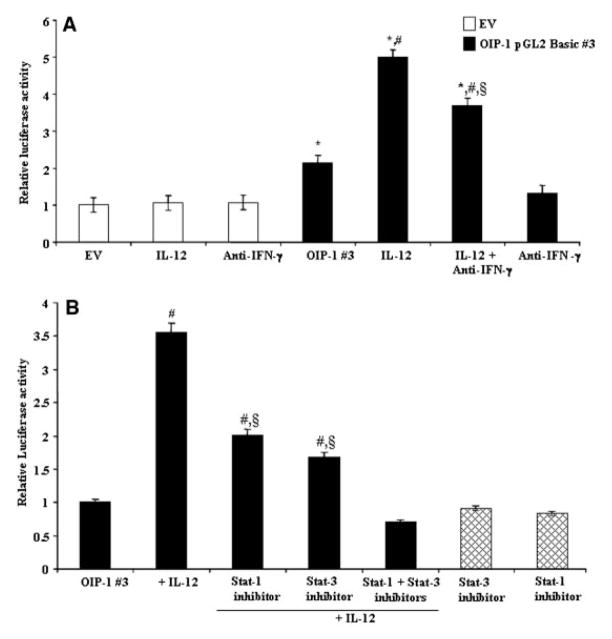
We also examined the transcriptional regulatory mechanism operative in CD4+ T cells with respect to IL-12 modulation of OIP-1 gene expression. We transiently transfected OIP-1 pGL2 Basic#3 luciferase reporter plasmid containing the OIP-1 gene promoter sequence (−1 to −1,988 bp) into Jurkat T cells and measured the luciferase activity levels in response to IL-12 treatment. IL-12 treatment (10 ng/ml) significantly increased (5-fold) in OIP-1 gene promoter activity in these cells (Fig. 3A). Jurkat cells cultured in the presence of excess concentration of neutralizing antibody against IFN-γ showed partial inhibition of IL-12 stimulated OIP-1 gene promoter activity, a finding that further confirms IL-12 specific modulation of OIP-1 gene promoter activity in these cells. The increase in the OIP-1 mRNA expression maximum at 4 h period (Fig. 1B) and stimulation of promoter activity at 48 h period could be explained by experimental conditions of transient transfections using the OIP-1 promoter-luciferase construct.
Fig. 3.
IL-12 Stimulation of OIP-1 gene promoter activity. A: OIP-1 pGL2 Basic#3 promoter-luciferase reporter plasmid was transiently transfected into Jurkat T cells (2 × 106 cells) in a six-well plate using a nucleofector kit V (Amaxa). The cells were stimulated with IL-12 (10 ng/ml) and with/without anti-IFN-γ antibody for 48 h and total cell lysates obtained were assayed for luciferase activity. pGL2 basic empty vector (EV) transfected cells were served as control. B: Effect of STAT-1 and STAT-3 inhibitors on IL-12 stimulated OIP-1 gene promoter activity. OIP-1 pGL2 Basic#3 promoter plasmid was transiently transfected into Jurkat T cells and cultured in the presence/absence of IL-12 (10 ng/ml). STAT-1 specific inhibitor, fludarabine (50 μM) and STAT-3 inhibitor peptide (1 mM) was applied to the cells for 30 min prior to IL-12 stimulation. Total cell lysates prepared after 48 h were assayed for luciferase activity. Transfection efficiency was normalized by measuring β-galactosidase activity co-expressed in these cells. Values expressed as mean ±SE for three independent experiments. *P <0.05 versus EV, #P <0.05 versus OIP-1 #3 and §P <0.05 versus OIP-1 #3 stimulated with IL-12.
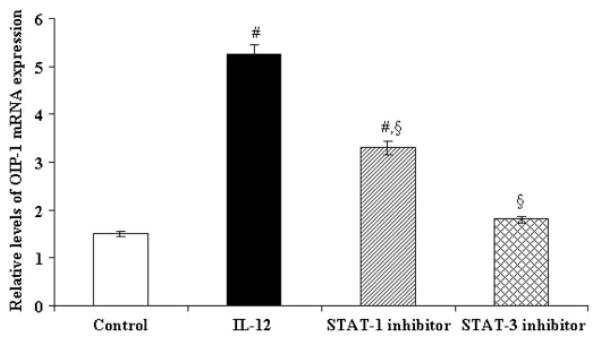
IL-12 has been shown to regulate gene expression through STAT signaling [Bright and Sriram, 1998]. We previously identified the presence of a potential STAT binding motif at −1,629 to −1,639 bp position in the OIP-1 gene promoter region [Srinivasan et al., 2006]. We therefore tested the ability of a STAT-1 specific inhibitor, fludarabine and a STAT-3 specific inhibitor to block the IL-12 stimulation of OIP-1 gene promoter activity. Jurkat T cells transfected with the OIP-1 pGL2 Basic#3 promoter (−1 to −1,988 bp)-luciferase reporter construct were cultured in the presence of STAT-1 inhibitor, fludarabine (50 μM) or a STAT-3 inhibitor for 48 h prior to IL-12 stimulation. Then, total cell lysates obtained were assayed for luciferase activity. As shown in Figure 3B, both STAT-1 and STAT-3 inhibitors significantly decreased IL-12 stimulated OIP-1 gene promoter activity in these cells. Furthermore, cultures treated with both STAT-1 and STAT-3 inhibitors abolished IL-12 stimulated OIP-1 gene promoter activity in Jurkat cells. However, STAT inhibitors in the absence of IL-12 did not significantly effect on OIP-1 promoter activity in control cultures. We further confirmed that STAT-1 specific inhibitor fludarabine and a STAT-3 specific inhibitor peptide suppressed IL-12 stimulated OIP-1 expression in peripheral blood derived CD4+ T cells. Real-time RT-PCR analysis revealed that fludarabine has a modest effect; however, the STAT-3 inhibitor significantly decreased IL-12 induced OIP-1 mRNA expression in isolated CD4+ T cell (Fig. 4). These results implicate STAT participation in IL-12 stimulation of OIP-1 gene expression in CD4+ T cells.
Fig. 4.
Effect of STAT inhibitors on IL-12 stimulation of OIP-1 expression in CD4+ T cells. The CD4+ cells from PBMC were treated with STAT-1 specific inhibitor, fludarabine (50 μM) or STAT-3 inhibitor peptide (1 mM) for 30 min prior to IL-12 (10 ng/ml) treatment. Total RNA isolated was subjected to real time PCR analysis of OIP-1 mRNA expression in these cells. Relative levels of OIP-1 gene expression were normalized with respect to the levels of GAPDH amplification. Values are expressed as the mean ±SE for three independent experiments. #P <0.05 versus control and §P <0.05 versus IL-12 treated group.
IL-12 STIMULATION OF STAT-3 BINDING TO OIP-1 PROMOTER SEQUENCE
We also examined the status of STAT binding to the OIP-1 gene promoter element in response to IL-12 treatment of Jurkat T cells by oligonucleotide pull-down assay as previously described [Ikeda et al., 2004]. Total cell lysates prepared from the Jurkat T cells stimulated with IL-12 in the presence/absence of IFN-γ neutralizing antibody were immunoblot analyzed for pSTAT-3 expression. IL-12 stimulation of Jurkat cells induced expression of pSTAT-3, which was modestly decreased (0.92 ± 0.03) in cells cultured in the presence of IFN-γ neutralizing antibody relative to IL-12 treatment alone. However, IFN-γ antibody treatment alone did not significantly effect on pSTAT-3 and STAT-3 expression in these cells (Fig. 5A). We then examined STAT binding to the OIP-1 promoter element in response to IL-12 stimulation by oligonucleotide pull-down assay as described. The nuclear extracts (NE) obtained from Jurkat cells showed both STAT-1 and STAT-3 binding with biotinylated oligonucleotides containing the intact STAT binding sequence present in the OIP-1 gene promoter. NE obtained from cultures stimulated with IL-12 in the presence of excess anti-IFN-γ antibody demonstrated a partially inhibited STAT-3, but completely inhibited STAT-1 binding to these oligomers. In contrast, mutant oligo (Mut.STAT oligo) which lacks the STAT recognition site did not demonstrate specific affinity binding to pSTAT-3 and pSTAT-1 (Fig. 5B).
Fig. 5.
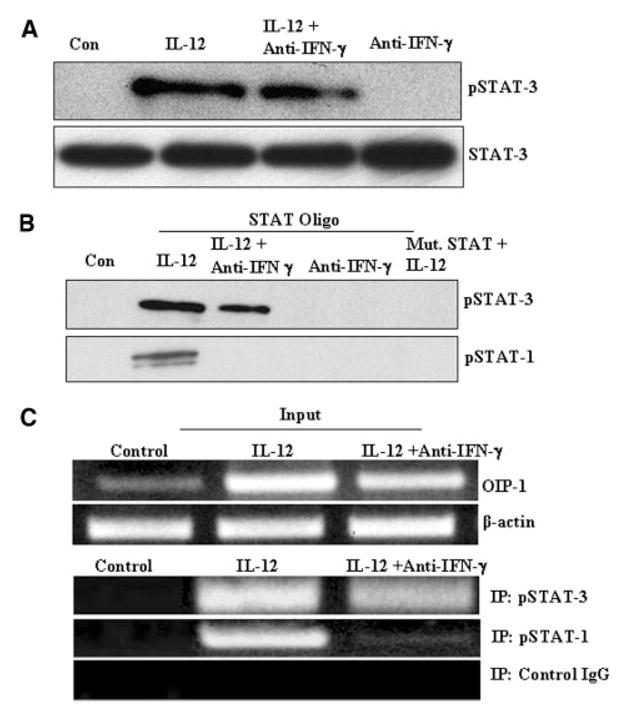
Oligonucleotide pull-down and ChIP assay for STAT-3 binding to OIP-1 gene promoter sequence. A: Jurkat T cells stimulated with IL-12 (10 ng/ml) for 30 min in the presence/absence of anti-IFN-γ antibody. pSTAT-3 and STAT-3 expression in the total cell lysates were confirmed by Western blot analysis. Data represent three independent experiments (P <0.05). B: The nuclear extracts (NE) prepared from Jurkat cells were incubated for 2 h with biotinylated oligonucleotides containing the intact STAT binding sequence present in OIP-1 gene promoter and STAT Mut-oligo; analyzed by immunoblotting with specific anti-p-STAT-3, anti-p-STAT-1 antibody. (C) ChIP assay for STAT binding to OIP-1 promoter region. The input sample was an aliquot of nuclear extract (NE) from Jurkat T cells stimulated with IL-12 and in the presence/absence of anti-IFN-γ antibody. The input samples from Jurkat T cells were cross-linked, lysed, sonicated, and subjected to immunoprecipitation using polyclonal antibodies against pSTAT-1 and pSTAT-3. Rabbit IgG was used as a negative control antibody. The precipitated DNA fragments, input DNA, were subjected to PCR analysis using specific primers for the STAT binding region in the OIP-1 gene promoter. Data represent three independent experiments.
We further confirmed STAT-3 binding to the OIP-1 gene promoter element using the ChIP assay. Jurkat T cells stimulated with IL-12 in the presence/absence of anti-IFN antibody subsequently were subjected to chromatin immune precipitation with anti-pSTAT-1 and anti-pSTAT-3 antibodies as described. PCR analysis of chromatin immune complexes was then performed using OIP-1 gene specific primers for the STAT binding region. As shown in Figure 5C, treatment with IFN-γ antibody partially inhibited pSTAT-3 binding and completely inhibited pSTAT-1 binding to the OIP-1 gene promoter element. These results suggest that STAT-3 signaling play an important role in IL-12 modulation of OIP-1 gene expression in CD4+ T cells.
DISCUSSION
We have previously reported that IFN-γ enhances OIP-1 expression in bone marrow cells and in osteoclast precursors [Koide et al., 2003]. Also, it is known that immune regulatory cytokines such as IL-12 and IL-4 are known to be potent inhibitors of osteoclastogenesis [Abu-Amer, 2001; Horwood et al., 2001]. IL-12 has been shown to inhibit osteoclast formation and bone resorption activity alone and/or in synergy with IL-18 [Horwood et al., 2001; Kitaura et al., 2002]. Also, IL-12 and IL-18 were shown to synergistically elevate IFN-γ production [Yamada et al., 2002]. The present study delineates IL-12 specific regulation of OIP-1 expression in T cells. We show that IL-12 stimulates OIP-1 expression in both bone marrow derived monocytes and Jurkat T cells. Partial inhibition of IL-12 stimulated OIP-1 expression in the presence of IFN-γ neutralizing antibody is consistent with that IL-12 increasing IFN-γ production by T cells. The modest decrease in OIP-1 expression observed in bone marrow cells treated with IFN-γ antibody alone suggest an autocrine/paracine effect of basal levels of IFN-γ present in these cultures. It has also been shown that IFN-γ produced from non-T cell population of bone marrow contributes to IL-12 inhibition of osteoclastogenesis [Nagata et al., 2003]. IL-12 has been shown to modulate IFN-γ production in both CD4 and CD8+ T cells [Gerosa et al., 1996]. However, our results suggest that IL-12 specific stimulation of OIP-1 expression is restricted to CD4+ T cells. Therefore, CD4+ T cells may be involved in IL-12 inhibition of osteoclatogenesis. In contrast, CD8+ cells have been shown to regulate osteoclast differentiation by a mechanism not mediated by IL-4 and TGF-β [John et al., 1996]. Although T cells have been shown to mediate IL-4 inhibition of osteoclast formation, IL-4 does not affect TSA-1 (thymic sheared antigen-1) and osteoclast inhibitory lectin (OCIL) expression [Mirosavljevic et al., 2003]. IL-12 stimulation of OIP-1 gene promoter activity in Jurkat T cells implicated an IL-12 specific transcriptional regulatory mechanism in these cells. Modest decrease in OIP-1 promoter activity in cells cultured in presence of anti-IFN antibody indicates low level basal IFN production. However, IL-12 significantly stimulated OIP-1 gene promoter activity in the presence of excess concentrations of anti-IFN neutralizing antibody, which suggests IL-12 specific regulation of OIP-1 expression.
STAT signaling molecules have been implicated in IL-12 and IFN-γ signaling to modulate gene expression [Watford et al., 2003]. We previously identified the potential STAT binding element in the OIP-1 gene promoter region and further demonstrated STAT-1 binding in response to IFN-γ treatment of osteoclast progenitor cells. This study further demonstrates that a combination treatment of STAT-1 and STAT-3 inhibitors treatment to Jurkat T cells completely abolished IL-12 stimulated OIP-1 gene promoter activity and suggests a functional role for STAT signaling in IL-12 modulation of OIP-1 expression in CD4+ T cells. Also, our results show that IL-12 upregulated the level of pSTAT-1 and pSTAT-3 expression in Jurkat T cells, implicating STAT signaling in IL-12 modulation of OIP-1 expression. Both the oligonucleotide pull-down assay and the ChIP assay revealed that IFN-γ neutralizing antibody completely abolished binding of pSTAT-1 to the OIP-1 gene promoter element in IL-12 stimulated cells. This confirms our previous findings that STAT-1 is primarily involved in IFN-γ regulation of OIP-1 expression. However, pSTAT-3 binding to the OIP-1 promoter element in response to IL-12 stimulation in the presence of anti-IFN-γ antibody indicates the STAT-3 specific signaling in IL-12 modulated OIP-1 gene expression. Similarly, IL-12 has been shown to upregulate T-bet (Th1-specific T box transcription factor) expression independently of IFN-γ in human CD4+ cells [Ylikoski et al., 2005]. The partial inhibition of OIP-1 promoter activity and abolished STAT-1 binding to the OIP-1 promoter in response to IL-12 stimulation in the presence of anti-IFN-γ antibody implicates a role for STAT-1 in IL-12 induced IFN-γ expression. These results suggest that IL-12 stimulates the OIP-1 gene expression through STAT-3 activation and binding to OIP-1 promoter element in CD4+ T cells.
Acknowledgments
NIH; Grant number: DE 12603; Grant sponsor: DOD; Grant number: DAMD 17-03-1-0763.
Footnotes
The authors declare no conflict of interest associated with this work.
References
- Abu-Amer Y. IL-4 abrogates osteoclastogenesis through STAT6-dependent inhibition of NF-kappaB. J Clin Invest. 2001;107:1375–1385. doi: 10.1172/JCI10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonyadi M, Waldman SD, Liu D, Aubin JE, Grynpas MD, Stanford WL. Mesenchymal progenitor self-renewal deficiency leads to age-dependent osteoporosis in Sca-1/Ly-6A null mice. Proc Natl Acad Sci USA. 2003;100:5840–5845. doi: 10.1073/pnas.1036475100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright JJ, Sriram S. TGF-beta inhibits IL-12-induced activation of Jak-STAT pathway in T lymphocytes. J Immunol. 1998;161:1772–1777. [PubMed] [Google Scholar]
- Choi Y, Woo KM, Ko SH, Lee YJ, Park SJ, Kim HM, Kwon BS. Osteoclastogenesis is enhanced by activated B cells but suppressed by activated CD8(+) T cells. Eur J Immunol. 2001;31:2179–2188. doi: 10.1002/1521-4141(200107)31:7<2179::aid-immu2179>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Classon BJ, Coverdale L. Genomic organization and expression of mouse thymic shared antigen-1 (TSA-1): Evidence for a processed pseudogene. Immunogenetics. 1996;44:222–226. [PubMed] [Google Scholar]
- Gerosa F, Paganin C, Peritt D, Paiola F, Scupoli MT, Aste-Amezaga M, Frank I, Trinchieri G. Interleukin-12 primes human CD4 and CD8 T cell clones for high production of both interferon-gamma and interleukin-10. J Exp Med. 1996;183:2559–2569. doi: 10.1084/jem.183.6.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grcevic D, Katavic V, Lukic IK, Kovacic N, Lorenzo JA, Marusic A. Cellular and molecular interactions between immune system and bone. Croat Med J. 2001;42:384–392. [PubMed] [Google Scholar]
- Horwood NJ, Elliott J, Martin TJ, Gillespie MT. IL-12 alone and in synergy with IL-18 inhibits osteoclast formation in vitro. J Immunol. 2001;166:4915–4921. doi: 10.4049/jimmunol.166.8.4915. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Nishimura R, Matsubara T, Tanaka S, Inoue J, Reddy SV, Hata K, Yamashita K, Hiraga T, Watanabe T, Kukita T, Yoshioka K, Rao A, Yoneda T. Critical roles of c-Jun signaling in regulation of NFAT family and RANKL-regulated osteoclast differentiation. J Clin Invest. 2004;114:475–484. doi: 10.1172/JCI19657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John V, Hock JM, Short LL, Glasebrook AL, Galvin RJ. A role for CD8+ T lymphocytes in osteoclast differentiation in vitro. Endocrinology. 1996;137:2457–2463. doi: 10.1210/endo.137.6.8641199. [DOI] [PubMed] [Google Scholar]
- Kitaura H, Nagata N, Fujimura Y, Hotokezaka H, Yoshida N, Nakayama K. Effect of IL-12 on TNF-alpha-mediated osteoclast formation in bone marrow cells: Apoptosis mediated by Fas/Fas ligand interaction. J Immunol. 2002;169:4732–4738. doi: 10.4049/jimmunol.169.9.4732. [DOI] [PubMed] [Google Scholar]
- Koide M, Kurihara N, Maeda H, Reddy SV. Identification of the functional domain of osteoclast inhibitory peptide-1/hSca. J Bone Miner Res. 2002;17:111–118. doi: 10.1359/jbmr.2002.17.1.111. [DOI] [PubMed] [Google Scholar]
- Koide M, Maeda H, Roccisana JL, Kawanabe N, Reddy SV. Cytokine regulation and the signaling mechanism of osteoclast inhibitory peptide-1 (OIP-1/hSca) to inhibit osteoclast formation. J Bone Miner Res. 2003;18:458–465. doi: 10.1359/jbmr.2003.18.3.458. [DOI] [PubMed] [Google Scholar]
- Kosugi A, Saitoh S, Noda S, Miyake K, Yamashita Y, Kimoto M, Ogata M, Hamaoka T. Physical and functional association between thymic shared antigen-1/stem cell antigen-2 and the T cell receptor complex. J Biol Chem. 1998;273:12301–12306. doi: 10.1074/jbc.273.20.12301. [DOI] [PubMed] [Google Scholar]
- MacNeil I, Kennedy J, Godfrey DI, Jenkins NA, Masciantonio M, Mineo C, Gilbert DJ, Copeland NG, Boyd RL, Zlotnik A. Isolation of a cDNA encoding thymic shared antigen-1. A new member of the Ly6 family with a possible role in T cell development. J Immunol. 1993;151:6913–6923. [PubMed] [Google Scholar]
- Mirosavljevic D, Quinn JM, Elliott J, Horwood NJ, Martin TJ, Gillespie MT. T-cells mediate an inhibitory effect of interleukin-4 on osteoclastogenesis. J Bone Miner Res. 2003;18:984–993. doi: 10.1359/jbmr.2003.18.6.984. [DOI] [PubMed] [Google Scholar]
- Nagata N, Kitaura H, Yoshida N, Nakayama K. Inhibition of RANKL-induced osteoclast formation in mouse bone marrow cells by IL-12: Involvement of IFN-gamma possibly induced from non-T cell population. Bone. 2003;33:721–732. doi: 10.1016/s8756-3282(03)00213-8. [DOI] [PubMed] [Google Scholar]
- Robinson DS, O’Garra A. Further checkpoints in Th1 development. Immunity. 2002;16:755–758. doi: 10.1016/s1074-7613(02)00331-x. [DOI] [PubMed] [Google Scholar]
- Roodman GD. Advances in bone biology: The osteoclast. Endocr Rev. 1996;17:308–332. doi: 10.1210/edrv-17-4-308. [DOI] [PubMed] [Google Scholar]
- Roodman GD. Cell biology of the osteoclast. Exp Hematol. 1999;27:1229–1241. doi: 10.1016/s0301-472x(99)00061-2. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Ito M, Kajiya H, Key LL, Jr, Johnson-Pais TL, Reddy SV. Functional characterization of human osteoclast inhibitory peptide-1 (OIP-1/hSca) gene promoter. Gene. 2006;371:16–24. doi: 10.1016/j.gene.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayanagi H, Ogasawara K, Hida S, Chiba T, Murata S, Sato K, Takaoka A, Yokochi T, Oda H, Tanaka K, Nakamura K, Taniguchi T. T-cell-mediated regulation of osteoclastogenesis by signalling cross-talk between RANKL and IFN-gamma. Nature. 2000;408:600–605. doi: 10.1038/35046102. [DOI] [PubMed] [Google Scholar]
- Watford WT, Moriguchi M, Morinobu A, O’Shea JJ. The biology of IL-12: Coordinating innate and adaptive immune responses. Cytokine Growth Factor Rev. 2003;14:361–368. doi: 10.1016/s1359-6101(03)00043-1. [DOI] [PubMed] [Google Scholar]
- Yamada N, Niwa S, Tsujimura T, Iwasaki T, Sugihara A, Futani H, Hayashi S, Okamura H, Akedo H, Terada N. Interleukin-18 and interleukin-12 synergistically inhibit osteoclastic bone-resorbing activity. Bone. 2002;30:901–908. doi: 10.1016/s8756-3282(02)00722-6. [DOI] [PubMed] [Google Scholar]
- Ylikoski E, Lund R, Kylaniemi M, Filen S, Kilpelainen M, Savolainen J, Lahesmaa R. IL-12 up-regulates T-bet independently of IFN-gamma in human CD4+ T cells. Eur J Immunol. 2005;35:3297–3306. doi: 10.1002/eji.200526101. [DOI] [PubMed] [Google Scholar]