Summary
Cytokine signaling via a restricted number of Jak-Stat pathways positively and negatively regulates all cell types involved in the initiation, propagation and resolution of inflammation. Here we focus on Jak-Stat signaling in three major cell types involved in inflammatory responses: T cells, neutrophils and macrophages. We summarize how the Jak-Stat pathways in these cells are negatively regulated by the Suppressor of cytokine signaling (Socs) proteins. We emphasize that common Jak-Stat-Socs signaling modules can have diverse developmental, pro- and anti-inflammatory outcomes depending on the cytokine receptor activated and which genes are accessible at a given time in a cell’s life. Because multiple components of Jak-Stat-Socs pathways are mutated or closely associated with human inflammatory diseases, and cytokine-based therapies are increasingly deployed to treat inflammation, understanding cytokine signaling will continue to advance our ability to manipulate chronic and acute inflammatory diseases.
Introduction
The importance of inflammation as a driver of pathology is no longer confined to autoimmune and infectious diseases. Rather, inflammation is increasingly linked to chronic diseases such as coronary artery disease, obesity and cancer (Lin and Karin, 2007). The role of cytokines in immunoregulation and inflammation is well established and multiple genome-wide association studies have documented that polymorphisms and mutations of cytokine receptors and their signaling components contribute to autoimmune disorders such as diabetes, inflammatory bowel disease, multiple sclerosis, and the spondyloarthropathies. Moreover, anti-cytokine therapies such as anti-tumor necrosis factor-α neutralizing agents are now commonplace in the treatment of chronic inflammatory diseases (Feldmann and Maini, 2003). However, to understand underlying disease mechanisms and to generate new therapies, it is necessary to define how cytokines work to program gene expression, and how their signaling pathways are regulated in different types of immune cells.
For the Type I and Type II cytokine superfamilies, we know a great deal about the mechanisms of signal transduction. Investigation of the signaling pathways emanating from these receptors led to the discovery of the Janus kinase (Jak)-Signal Transducer and Activator of Transcription (Stat) pathway. This field has been reviewed many times, but a number of recent advances have provided important new insights into how the Jak-Stat pathway contributes to inflammation in terms of regulating the differentiation and pro- and anti-inflammatory activity of immune cells that will be the focus of this review. The developmental fates for differentiating T cell subsets such as T helper (Th)17 and Tregs have uncovered new paradigms for inflammatory diseases: Stat family transcription factors, and their correct quantitative and temporal regulation are critical for the development of these T cells subsets. Paradoxically, some factors, like Stat3, have both proinflammatory and anti-inflammatory actions, depending upon the cell- and stimulus-specific context. By focusing on the use of Stat transcription factors and their regulation in the differentiation and function of T cells, granulocytes and macrophages in the context of inflammation, we will attempt to deconvolute the seemingly ubiquitous use of Stat pathways (especially Stat1, Stat3 and Stat5a, Stat5b) for developmental and functional uses, often in the same cell type.
Overview of a Stat signaling module
Type I and II cytokine receptors are a conserved family of ~40 members that includes the receptors for interleukins, interferons and hormones such as growth hormone, leptin and erythropoietin, and colony stimulating factors (CSF) such as granulocyte-CSF and granulocyte-macrophage CSF (Boulay et al., 2003). Unlike other receptors with intrinsic enzyme activity (e.g. kinase or phosphatase), cytokine receptors are associated with a tethered kinase. These cytoplasmic kinases comprise the four members of the Jak family: Jak1, Jak2 and Tyk2 bind to an array of receptors whereas Jak3 binds to only one receptor, the common gamma chain, or γc. Mutations of JAK3 or TYK2 in humans lead to specific primary immunodeficiency syndromes designated severe combined immunodeficiency (SCID) and autosomal recessive hyperimmunoglobulin E syndrome (AR-HIES) (Minegishi et al., 2007; Notarangelo et al., 2001; Watford and O’Shea, 2006). Additionally, the roles of the four Jak proteins have been elucidated through the generation of genetically deficient mice, and specific functions of each Jak member have been assigned (Murray, 2007). Because of their kinase activity, Jak proteins are potential targets for small molecule inhibition. For Jak3, its restricted association with γc has made Jak3 an attractive therapeutic target as an immunosuppressive drug that can primarily target activated T cells (O’Shea et al., 2004b).
Upon cytokine binding to its cognate receptor, the receptor-associated Jaks are activated and in turn phosphorylate tyrosine residues in the receptor cytoplasmic domain. This event provides a docking site for proteins with Src homology 2 domains, one important class of which is the Stat family of transcription factors. With seven members in all (Stat1, Stat2, Stat3, Stat4, Stat5a, Stat5b, and Stat6), these DNA binding proteins provide a rapid membrane to nucleus mechanism for regulation of gene expression (Shuai and Liu, 2003).
Role of Stats in T cell development and differentiation
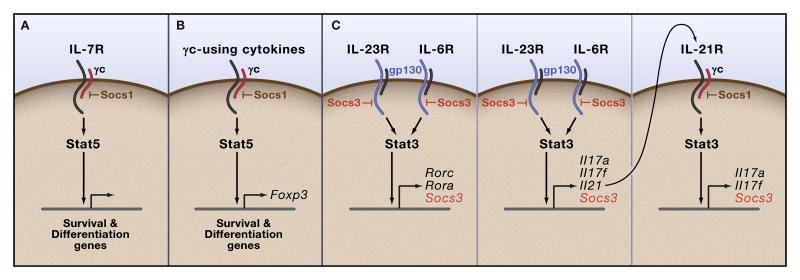
Given the importance of cytokines in T cell development and differentiation and function, it is no surprise that Stat proteins contribute critically to each of these processes (Fig. 1). As an example of the overall importance of cytokine-cytokine receptor-Jak-Stat pathway signaling in thymic T cell development, IL-7 signaling ensures development of appropriate lymphocyte numbers. Mutation of either IL-7R subunit, IL-7Ra or γc (encoded by IL2RG) or its cognate Jak, JAK3, lead to SCID manifested by severely reduced numbers of thymocytes (O’Shea et al., 2004a). IL-7 activates Stat5a and Stat5b and deletion of the locus encoding Stat5a and Stat5b also results in a severe SCID phenotype (Yao et al., 2006). Indeed, Stat5 activity is required for the normal development of all normal lymphoid lineages. However, the absolute role of Stat5 in permitting normal T cell development is only part of Stat5’s contribution to T cell subset development discussed below.
Figure 1. Cytokine signaling in T cell development and function.
(A) Stat5 signaling from cytokines that use γc is essential for T cell development. Mice or humans lacking key components of this pathway (γc, Jak3, Stat5) fail to develop T cells. (B) Stat5 signaling controls the development of FoxP3-positive T regs. (C) Stat3 is crucial to the development and function of Th17 cells. IL-23 and IL-6 enforce Th17 development via the direct or indirect induction of Rorc and Rora expression. Stat3 also regulates the expression of IL-17-encoding genes and Il21, which acts in an autocrine/paracrine way to regulate Th17 cells. Socs3 is an important inhibitor of cytokines that use gp130 (IL-23R and IL-6R) while Socs1 is anticipated to inhibit any cytokines that use γc (IL-7, IL-21 as shown).
Differentiating CD4+ T cells were thought to have two fates – T helper1 (Th1) and Th2 cells. These fates are driven by the cytokine milieu with IL-12 driving Th1 differentiation and IL-4 promoting Th2 differentiation. IL-12 activates Stat4 whereas IL-4 activates Stat6. Stat4- and Stat6-deficient mice have impaired Th1 and Th2 responses respectively (O’Garra and Arai, 2000). The products of Th1 and Th2 T cells, IFN-γ and IL-4 respectively, promote commitment to their respective lineages and inhibit development of the opposing lineage. Surprisingly, a recent genome-wide association study has revealed that polymorphisms in STAT4 confer risk of developing autoimmune diseases including rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE) (Remmers et al., 2007). While RA has typically been view as having elements consistent with Th1-mediated pathology, SLE would not be considered prototypic Th1 disease. In this regard, it is important to note that Type I IFNs also activate Stat4. Depending upon the circumstance, Type I IFN signaling may enhance or inhibit Th1 responses (Nguyen et al., 2002). Although the pathogenesis of SLE is very poorly understood, recent advances have documented that SLE and related autoimmune disorders are characterized by a transcriptional “interferon signature”. Exactly how Stat4 and IFNs contribute to the pathogenesis of SLE is unknown, but this will be an important area to follow.
As important as the Th1-Th2 paradigm was in advancing our understanding of T cell biology, CD4+ T cells are now known to have additional fates regulated by Stat3 and Stat5. One subset of CD4+ T cells is termed regulatory T (Treg) cells, which express the transcription factor Foxp3 (Fig 1B). Treg cells are discussed in detail in a review in this issue by Rudensky. Tregs have essential immunosuppressive functions as illustrated by the fact that deletion or mutation of Foxp3 leads to fatal autoimmune disease in mice and humans. CD4+ Treg cells can be generated in the thymus (‘natural’ Tregs) or can be induced in the periphery (iTregs). In both cases, cytokines that use γc are important drivers of Treg development. Deficiency of γc or Jak3 cause a failure to produce Foxp3-positive regulatory T cells (Mayack and Berg, 2006). Accordingly, deficiency of both Stat5a and Stat5b also leads to loss of Tregs and inability to induce Tregs in vitro (Yao et al., 2007) while constitutive activation of Stat5b enforces Foxp3-positive Treg development, bypassing the requirements for upstream cytokine or co-stimulatory signals (Burchill et al., 2008). Stat5 appears to have very direct effects on Tregs in that these transcription factors bind directly to the Foxp3 gene. Thus, even though Stat5 is absolutely required for T cell development, once T cells have developed and exited the thymus, further Stat5-dependent signals are needed to ensure correct subset development and function. One way to think about the requirement for Stat5 to have such diverse functions in T cell development is to consider that cytokine signaling via Stat5, and gene accessibility to Stat5, is partitioned throughout the life of the T cell: while IL-7/Jak3/Stat5 signaling is predominant for thymic development, other Stat5-activating receptors stimulate T cells and activate different combinations of Stat5-dependent genes following maturation and exit from the thymus.
Another recently recognized fate for CD4+ T cells is the Th17 cell (also discussed by the other reviews in this issue) whose development and function is critically dependent on Stat3. Named for their ability to produce the inflammatory cytokine IL-17, Th17 cells recruit and activate neutrophils and other inflammatory cells to sites of tissue inflammation (Korn et al., 2007). Th17 cells can be generated from naïve CD4+ T cells by IL-6 and TGFβ, but also produce another cytokine IL-21, which promotes IL-17 production in an autocrine-paracrine manner (Nurieva et al., 2007; Zhou et al., 2007). Finally, a third cytokine, IL-23, acts on memory cells to expand and maintain Th17 cells. The importance of IL-23 signaling in inflammation is exemplified by recent discoveries that polymorphisms in IL23R are associated with increased risk of inflammatory bowel disease, ankylosing spondylitis and psoriasis (Burton et al., 2007; Cargill et al., 2007; Duerr et al., 2006; Tremelling et al., 2007)
IL-6, IL-21 and IL-23 all activate Stat3 via their cognate receptors (Fig. 1C). Accordingly, selective deletion of Stat3 in T cells abrogates Th17 differentiation in part because the expression of RORγt and RORα, two nuclear hormone receptors essential for Th17 development, is also abrogated (Yang et al., 2008). However, Stat3 also directly regulates the expression of Il21 and Il17 (Chen et al., 2006; Wei et al., 2007). Therefore Th17 fate, peripheral maintenance by IL-21, and effector functions are all regulated by Stat3 signaling from different cytokine receptors. The importance of Stat3 in Th17 development and function is exemplified by the fact that patients with Job’s syndrome, an autosomal dominant disorder due to Stat3 mutations, fail to make Th17 cells (Milner et al., 2008). Parenthetically, it is interesting to note that IL-2 acting through Stat5 inhibits Th17 differentiation (Laurence et al., 2007). Thus, the balance of Treg/Th17 differentiation appears to be regulated by Stat5 and Stat3. Clearly the use of one transcription factor to perform all these functions indicates that Stat3 activity is under tight control throughout the life of a Th17 cell, a task performed in part by Socs3 (discussed below).
An additional complexity of Th17 T cells in inflammation concerns the Stat3-activating cytokine IL-22 (IL-22 signaling is discussed below). Th17 cells preferentially produce IL-22, but its regulation is subtly different from IL-17; whereas IL-6 and TGFβ-1 are important for the differentiation of Th17 cells, IL-6 alone so far appears to be capable of inducing IL-22. The pathways for generating IL-22 are discussed in accompanying reviews in this issue. However, Th17 cells are not the only cell capable of producing IL-22 and the extent to which this cytokine expression is dependent upon which Stat proteins remains to be determined. Perhaps more important are the upstream cytokine signals that drive IL-22 production from Th17 cells at sites of tissue inflammation.
In summary, even though Stat5 is absolutely required for normal T cell development, once T cells have developed and exited the thymus, further Stat5 and Stat3 signals are needed to ensure correct subset development and function. Stats have direct and essential roles in helper T cell development, lineage commitment and function as they bind and presumably regulate genes such as Foxp3, Il17a and Il21. The actions of these Stats may be direct or indirect, but clearly warrant further investigation in defining direct Stat targets in T cells and the mechanisms by which the induce transcriptional programs.
SOCS proteins control inflammatory responses by regulating Stat signaling
Upon cytokine stimulation, a family of cytokine-induced inhibitors termed suppressors of cytokine signaling (Socs proteins) is rapidly induced. The predominant function of Socs proteins is to block the generation of the Stat signal from a cytokine receptor (Alexander and Hilton, 2004; Yoshimura et al., 2007). Importantly, the genes encoding the Socs proteins are direct targets of Stat proteins; the Jak-Stat cascades thereby control their own signaling output by feedback inhibition. Although there are 8 Socs proteins, genetic evidence from mice and cells lacking Socs1 and Socs3 unequivocally shows that these two Socs proteins are necessary to reduce the overall signaling output from their target receptors (Alexander and Hilton, 2004; Yoshimura et al., 2007). The Socs1- and Socs3-mediated modulation in signaling from cytokine receptors therefore has profound effects on the regulation of immunity and inflammation by affecting the activation, development and homeostatic functions of all lineages involved in immune and inflammatory responses.
A major question in understanding the activities of Stat-Socs modules concerns the biochemical mechanism of how Socs proteins block cytokine receptor signaling. Each of the eight Socs proteins have two major domains, an SH2 domain and a Socs box that complexes with elongins B and C, a cullin and Rbx2 to form a E3 ubiquitin ligase (Kile et al., 2002; Zhang et al., 1999). The Socs SH2 domains bind phosphorylated tyrosine residues in their substrates. The best characterized Socs substrates are specific tyrosine residues in the cytoplasmic tails of cytokine receptors. In addition, the Socs SH2 domain has the potential to bind other phospho-tyrosine residues and thereby regulate the activity of a wide range of proteins. The current model of Socs function postulates that the E3 activity of a Socs protein will target the substrate to be ubiquitinated and then directed to the proteosome for degradation. However, genetic studies using mice lacking the Socs box of Socs1 or Socs3, but engineered to retain the SH2 domains of each protein, indicate that the SH2 and Socs box domains don’t always function in concert because the phenotypes of mice lacking the Socs box of Socs1 or Socs3 are dramatically less severe than the corresponding conventional knockouts (Boyle et al., 2007; Zhang et al., 2001). These data suggest that the SH2 domain of Socs1 and Socs3 alone can block cytokine receptor signaling. Thus, the mechanistic relationship between the SH2 and Socs box domains remains unresolved, as does the contribution of E3 ligase activity to Socs function.
A second outstanding question concerns the mechanism by which a Socs protein, tethered to a specific residue of a cytokine receptor, inhibits the generation of activated Stats. An obvious possibility is that a Socs protein directs its receptor substrate to be degraded. At least for gp130, a substrate of Socs3, this does not seem to be the case (Lang et al., 2003). Another possibility is that Socs proteins promote ubiquitination of Stat proteins in the vicinity of the receptor: however this does not agree with the restricted requirement for the Socs box of Socs1 or Socs3 compared to the absolute requirement for the intact proteins and their SH2 domains. A third possibility is that a tethered Socs protein inhibits the activity of tethered JAK proteins through effective concentration-type effects that remain uncharacterized (Kamizono et al., 2001; Stross et al., 2006; Yoshimura et al., 2007). At this stage, the biochemical mechanism(s) of Socs-mediated inhibition of Stat signaling remains unknown.
Stat3-Socs3 regulates homeostatic and emergency granulopoiesis
Neutrophils are made in prodigious numbers every day of our lives to patrol tissue surfaces, especially the lung and skin, for invading microorganisms, which they then target for elimination by a variety of mechanisms including the oxidative burst (Eyles et al., 2006). The role of neutrophils can be seen in the consequences of ablative chemotherapy and in people with severe congenital neutropenias. In chemotherapy, depletion of bone marrow precursors by ablative drugs causes a precipitous drop in numbers of short-lived, mature, circulating neutrophils. Many patients undergoing ablative chemotherapy have infections caused by fungi and bacteria normally innocuous for the immune-competent. A similar situation is found in people with genetic deficiencies in neutrophil number or function. However, G-CSF administration can rescue, in part, the devastating drop in neutrophils numbers by stimulating maturation and exit of neutrophils from the bone marrow (Eyles et al., 2006). G-CSF has become a standard of care in clinical settings where depletion of neutrophil numbers can be anticipated and is therefore a triumph of directed cytokine therapy. G-CSF therapy is also highly effective in treating some cases of congenital neutropenias where bone marrow precursors remain responsive to G-CSF. By contrast to the protective functions of neutrophils, excessive neutrophil numbers are found in a plethora of inflammatory diseases especially those involving tissue surfaces colonized by bacteria and fungi including chronic obstructive pulmonary disease, asthma, cystic fibrosis and different forms of colitis (Eyles et al., 2006). Therefore, neutrophils numbers and function require precise control so that tissue homeostasis can be maintained without causing destructive inflammation. This process is controlled to a large extent by Stat3 and Socs3 (Fig. 2).
Figure 2. G-CSFR signaling.
Simplified schematic of G-CSFR signaling to illustrate that Stat3 and Stat5 are regulated by the G-CSFR and that Socs3 is a key downstream target of Stat3. Socs3 is required to feedback inhibit G-CSFR signaling.
The G-CSFR is responsible for transducing the signals from G-CSF via four tyrosine residues located in the cytoplasmic tail of the receptor. G-CSFR signaling via the cytoplasmic tyrosines activates numerous signaling molecules including Stat5, Stat3 and MAP kinases. Deletion of all cytoplasmic tyrosines yields a receptor that does not elicit detectable Stat3 or Stat5 (but can probably activate low levels of Stat activation (McLemore et al., 2001). Mice bearing knockin mutations of the G-CSFR with all tyrosines eliminated have very low (but not entirely absent) circulating neutrophil numbers and severe defects in the emergency mobilization of neutrophils following G-CSF administration (McLemore et al., 2001). A surprising complication of the analysis of Stat3 in neutrophil development and function was observed when Stat3 or Socs3 was conditionally ablated in early hematopoetic development (Croker et al., 2004; Kimura et al., 2004; Lee et al., 2002). In both cases, excessive numbers of late stage neutrophils accumulate in the bone marrow and peripheral blood. A conclusion of these studies was that Stat3 and Socs3 are negative regulators of granulopoiesis (Lee et al., 2002). Indeed, Socs3 binds to one of the tyrosine residues in the G-CSFR (Y729) and restricts the amplitude of Stat3 signaling (Hortner et al., 2002). Thus, loss of Socs3 causes increased G-CSFR signaling leading to increased neutrophil numbers while loss of Stat3 (and failure to induce Socs3 expression) also leads to increased neutrophil numbers. How can we reconcile these data? The logical conclusion is that Socs3 negatively regulates neutrophil numbers by regulating G-CSFR signaling generally and not via specific inhibitory effects on Stat3. In the absence of Socs3 there is likely to be elevated signaling from the G-CSFR, perhaps excessive Stat5 or MAP kinase signaling, because Stat3 is no longer present to induce Socs3 to feedback inhibit the signal from the G-CSFR. Therefore, a more detailed investigation of Stat and MAP kinase activation during neutrophil development is warranted.
The function of the Stat3-Socs3 module in neutrophils is however, more complex than outlined above. Deletion of either Stat3 or Socs3 at a later stage of neutrophil development using the lysMcre deleter strain (where cre activity is predominantly at the committed myeloid progenitor stage) does not lead to the phenotypes described above, where Stat3 or Socs3 loss had been engineered to occur early in hematopoiesis (Panopoulos et al., 2006). Thus, the Stat3-Socs3 module is required to regulate neutrophil numbers at a specific developmental stage. This restriction probably reflects the need for precision in circulating neutrophil numbers as too many neutrophils will drive inflammation. Finally, it is notable that Stat3 has additional Socs3-independent functions that control chemotaxis and neutrophil migration in vivo (Panopoulos et al., 2006; Semerad et al., 2002; Semerad et al., 1999). Therefore, the Stat3-Socs3 signaling module has a restricted but critical role in determining the quantity of neutrophils that mature in the bone marrow and migrate to the peripheral tissues.
The Stat3-Socs3 module in anti-inflammatory signaling
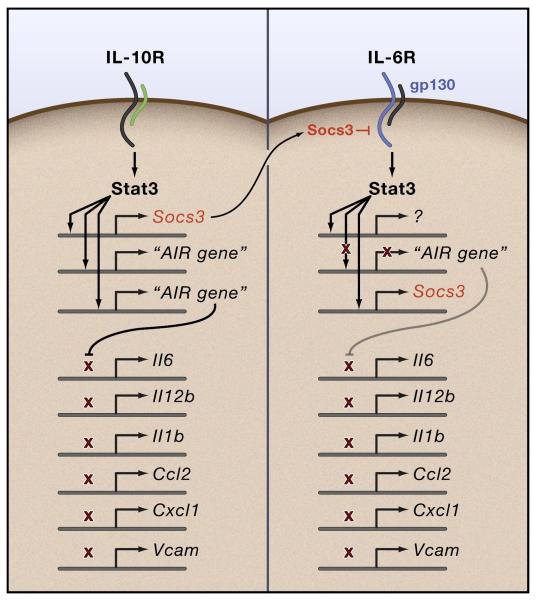
Compared to the effects of the Stat3-Socs3 module on T cell and neutrophil development and function described above, the output of the Stat3-Socs3 pathway in IL-10R signaling is entirely different (Fig. 3). IL-10 is an anti-inflammatory cytokine made by lymphocytes and myeloid lineage cells that is responsible for tempering the output of pro-inflammatory cytokines from activated macrophages (Murray, 2006). The anti-inflammatory functions of IL-10 extend to virtually every type of acute and chronic inflammatory and infectious diseases. Unlike the partial redundancy observed in many cytokine signaling systems, the anti-inflammatory functions of IL-10 cannot be compensated by other factors because deletion of IL-10 in all cells or only in T cells causes excessive inflammation, especially in the gut where IL-10 constitutively blocks inflammation driven by gut flora (Berg et al., 1996; Kuhn et al., 1993; Roers et al., 2004). Socs3 is highly induced by IL-10 but is not required to feedback-inhibit IL-10R signaling or mediate any significant anti-inflammatory effects of IL-10 (Lang et al., 2003; Yasukawa et al., 2003). Instead, Socs3 induction by IL-10 is required to block signaling from other cytokine receptors.
Figure 3. Mechanisms associated with Socs3-mediated suppression of anti-inflammatory signaling by the IL-6R.
The left side depicts IL-10 signaling in a macrophage activated by the TLR pathway (or other similar inflammatory stimuli). Socs3 expression is strongly induced by IL-10, along with the Stat3-dependent genes whose products regulate the anti-inflammatory signaling system (‘anti-inflammatory response’ AIR gene) illustrated as inhibiting the expression at the transcriptional level of classic pro-inflammatory genes. On the right side is shown IL-6 signaling via gp130, which also activates Socs3 and other genes. Unlike the IL-10R, the IL-6R cannot activate the expression of the AIR gene(s) unless Socs3 is absent. Thus, IL-6 and IL-10 (and any other receptors that activate Socs3 expression in macrophages) enforce the inability of the IL-6R to produce the anti-inflammatory response. Note that Socs3 (or any other Socs protein) does not inhibit the IL-10R.
How does IL-10 mediate the anti-inflammatory response? Stat3 is solely responsible for all the effects of IL-10 signaling as shown by both loss of function experiments and gain of function experiments using constitutively activated Stat3, or cytokine receptors unrelated to the IL-10R engineered to activate Stat3 in a way similar to the IL-10R (El Kasmi et al., 2006; Takeda et al., 1999; Williams et al., 2007). Importantly, leukocytes isolated from humans bearing mutations in STAT3 and suffering from Job’s Syndrome, are characterized the overproduction cytokines and chemokines following stimulation with TLR agonists, bacteria and interferons (Holland et al., 2007; Minegishi et al., 2007) (Milner et al. Nature, in press). This phenotype is indicative of a failure of IL-10R signaling. The obligate role of Stat3 in anti-inflammatory signaling suggests a conundrum: if Stat3 is activated by the IL-10R to elicit the anti-inflammatory response then why don’t other receptors that activate Stat3 also activate anti-inflammatory signaling? The answer to this question centers on the highly specific inhibitory effects of Socs3 on gp130, the signaling receptor of the IL-6 family of cytokines. Gp130 has multiple tyrosine residues in its cytoplasmic tail all of which bar one, Y757, serve as docking sites for Stat proteins (especially Stat3). Y757 docks the SH2 domain of Socs3 and is by far the best characterized Socs-cytokine receptor interaction (Hirano and Murakami, 2006; Kamimura et al., 2003). Deletion of Socs3 increases Stat3 signaling from gp130 and, surprisingly, also increases Stat1 signaling leading to an ectopic interferon response (Croker et al., 2003; Lang et al., 2003). Therefore Socs3 controls the quality and quantity of Stat activation (either Stat3 or Stat1) mediated by gp130. Yoshimura and colleagues also showed that when Socs3 was absent, IL-6 via gp130-mediated Stat3 activation, induces an anti-inflammatory response identical to the IL-10R; a finding has since been confirmed using multiple experimental approaches (El Kasmi et al., 2006; Yasukawa et al., 2003). Collectively, these data suggest that Stat3 activation from one receptor, in this case gp130, can generate qualitatively distinct Stat3 signals. Thus, Stat3 signaling from gp130 is convertible between different modes depending on the Socs3 status of the cell. One mode is anti-inflammatory Stat3 signaling like the IL-10R that is actively repressed by Socs3. The other mode is non-anti-inflammatory Stat3 signaling. Since a wide range of stimuli regulates Socs3 expression, repression of anti-inflammatory signaling from gp130 must be advantageous for reasons we do not yet appreciate.
These findings affect how we interpret signals the drive pro- and anti-inflammatory signaling from cells receptive to multiple cytokines for two reasons. First, the anti-inflammatory signal generated from the IL-10R is not unique to the IL-10R but is actively suppressed from other receptors by Socs3. Second, Stat3 activation is not generic and the read-out of tyrosine phosphorylation as an activation marker is insufficient to tell us about the downstream consequences of Stat3 activation from one receptor versus another (Murray, 2007). Thus in macrophages, Stat3 tyrosine phosphorylation is activated by signaling through both the IL-10R and IL-6R but activates overlapping but distinct gene expression profiles (Socs3 is an example of a common gene). ChIP-sequencing techniques will have the final say on this issue because it should be possible to ask what genes bind Stat3 at a given time after IL-6 or IL-10 stimulation.
Is IL-22 pro- or anti-inflammatory?
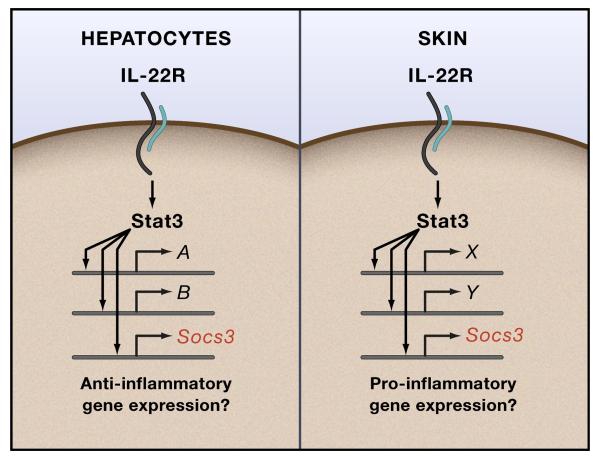
IL-22 is grouped with IL-10 because both the IL-10R and IL-22R share the IL-10Rβ chain (along with IL-26 and IL-28 that also use the IL-10Rβ chain) and like the IL-10R, the IL-22R activates Stat3 (Donnelly et al., 2004). However, the IL-22R is not expressed on hematopoietic lineage cells but rather expressed on cells of the skin epithelia, pancreas and hepatocytes (Donnelly et al., 2004). The source of IL-22 is predominantly T cells, leading to the idea that IL-22 is a pro-inflammatory cytokine made by T cells to drive tissue inflammation. How correct is this assumption? New information has now linked IL-22 more closely to Th17 T cells suggesting the potential for IL-22 to drive tissue inflammation and to function in host defense (Liang et al., 2006) (and discussed in this issue by Ouyang and Kolls). However, other experiments suggest the opposite: that IL-22 may be an anti-inflammatory cytokine and has the potential to behave as an IL-10-like cytokine for non-hematopoietic tissues (Fig. 4). The Flavell and Reynaud groups have recently described two independent IL-22-deficient mouse strains. The former used a mouse model of hepatitis to demonstrate that IL-22 is an essential anti-inflammatory mediator in the liver because IL-22-deficient mice had greatly increased liver damage and inflammation after concanavalin treatment (Zenewicz et al., 2007). Additional studies in liver damage models also support the notion that IL-22 plays an anti-inflammatory, protective role in the liver (Pan et al., 2004; Radaeva et al., 2004). In an elegant and technically challenging model of ulcerative colitis, IL-22 administration via pressurized local microinjection of IL-22-expressing vectors was shown to have a robust anti-inflammatory effect in the intestine mediated in part via Stat3 activation in colonic epithelial cells (Sugimoto et al., 2008). These results implicate IL-22 as a cytokine that protects against inflammatory damage and can therefore be considered anti-inflammatory. By contrast, however, IL-22-deficient mice develop EAE indistinguishable from controls suggesting IL-22 plays neither a protective nor disease-exacerbating role in this key model of Th17 T cell function (Kreymborg et al., 2007). These experiments do not readily square with skin inflammation models where a pro-inflammatory role of IL-22 has been described by multiple laboratories (Boniface et al., 2005; Boniface et al., 2007; Ma et al., 2008; Wolk et al., 2004; Zheng et al., 2007), and recent studies that show protective roles of IL-22 in mucosal defense against pathogens (Aujla et al., 2008; Zheng et al., 2008) where IL-22 induces the expression of anti-microbial proteins, including S1008A, a zinc and manganese chelating protein that deprives bacteria of essential cations (Corbin et al., 2008). Clearly, more detailed experiments in tissue inflammation models need to be performed with IL-22-deficient mice along with mice yet to be reported that can track IL-22 producing cells and IL22Ra knockout mice.
Figure 4. IL-22R signaling.
Hypothetical schematic of tissue-specific effects of Stat3 activation by the IL-22R. In hepatocytes IL-22 activates an anti-inflammatory gene expression program while the opposite occurs in skin. Socs3 is expected to be a common target gene in each tissue.
Reconstitution experiments performed with the IL-22Rα argue however that IL-22 generates anti-inflammatory signals via Stat3 (Fig. 3). When the IL-22Rα chain is expressed in primary macrophages it can use the endogenous IL-10Rβ chain to form a mature signaling complex. Upon stimulation with IL-22 in the presence of a strong inflammatory signal from LPS, the IL-22R activates a Stat3-dependent signaling cascade indistinguishable from the IL-10R itself (El Kasmi et al., 2006). Furthermore, like the IL-10R, Socs proteins do not regulate the IL-22R in this system. Therefore, the IL-22 is functionally equivalent to the IL-10R in macrophages. The key question is whether an anti-inflammatory Stat3 pathway can be elicited by the IL-22R in naturally IL-22-responsive cells such as hepatocytes and keratinocytes, and whether any of the Socs proteins regulate IL-22R signaling in these cells. The in vivo data described above indicate that hepatocytes and keratinocytes respond very differently to IL-22. Therefore, ChIP-seq experiments for IL-22-activated Stat3 in these two cell types would be an ideal experiment. It should be pointed out that anti-inflammatory functions of Stat3 have been recognized in non-myeloid cells for some time. For example, deletion of Stat3 in epithelial lineage cells renders mice sensitive to the pro-inflammatory effects of LPS while established non-hematopoietic tumor lines use Stat3 as a means to suppress the production of inflammatory markers as a means to escape host immune recognition (Kano et al., 2003; Kortylewski et al., 2005; Wang et al., 2004). Additionally, Stat3 inactivation in keratinocytes causes fulminant skin inflammation demonstrating the protective effects of Stat3 function in skin (Sano et al., 1999). An important question that stems from these findings is whether the Stat3-dependent anti-inflammatory gene expression patterns generated in myeloid lineage cells by the IL-10R are functionally identical to those generated by the IL-22R in non-hematopoietic cells. This problem should be resolved when the downstream Stat3-dependent mediators of the anti-inflammatory signal are identified and functionally linked to inhibition of inflammatory mediator production.
The Stat1-Socs1 module and inflammation
The Stat1-Socs1 module primarily regulates interferon signaling (Alexander and Hilton, 2004). Disruption of this pathway has profound effects on immune and inflammatory responses in addition to controlling cross talk with Stat3-Socs3 signaling from other cytokine receptors. The regulation of Stat1 activation by the interferon response, and its downstream effects has been comprehensively reviewed (Platanias, 2005; Shuai and Liu, 2003; van Boxel-Dezaire et al., 2006). We will focus instead on Socs1 because of its unique role in controlling inflammation. Socs1, unlike the broad expression of Socs3, is activated predominantly by interferon signaling, although other cytokines such as IL-4 also activate Socs1 expression but in a cell-type dependent way. The effects of interferon activation of Socs1 were first shown to have a critical effect in blocking IFN-γ signaling: mice lacking Socs1 die a few days after birth from a massive systemic inflammatory response that can be largely rescued by deleting Ifng. Socs1−/−; Ifng−/− mice outlived Socs1−/− mice by months-years but nevertheless still die prematurely compared to wild-type controls (Alexander et al., 1999). (Socs1+/− also die prematurely but of a different inflammatory syndrome) (Metcalf et al., 2000). Subsequent extensive genetic analysis using Socs1−/− mice or Socs1 conditionally-deficient mice crossed to deficiencies in immune and inflammatory pathways, has revealed that while the IFN-γR is the primary target of Socs1, other cytokine receptors also respond to the negative effects of Socs1 including the γc, IL-12R, IFN-αβR and IL-4Rα (Alexander and Hilton, 2004; Yoshimura et al., 2007). Therefore, Socs1 has a dominant effect on the IFN-γR but additional inhibitory effects have been revealed by compound mutations that regulate other cytokine receptors.
Even though the majority of pro-inflammatory effects of excessive IFN-γ signaling caused by loss of Socs1 can be rescued by removal of IFN-γ or its signaling components (IFN-γR, Stat1), Socs1−/−; Ifng−/− mice remain extremely sensitive to systemic challenge with LPS (Kinjyo et al., 2002; Nakagawa et al., 2002). This finding led to the idea that Socs1 additionally regulates components of the TLR cascade, including IRAK1 and the p65 subunit of NF-κB (Kinjyo et al., 2002; Nakagawa et al., 2002; Ryo et al., 2003). These findings have been challenged because analysis of TLR signaling in macrophages isolated from Socs1−/−; Ifng−/− mice or macrophages engineered to constitutively express Socs1, Socs2 or Socs3 showed no direct effects of Socs1 on the main TLR signaling pathways, including tolerance to LPS (Baetz et al., 2004; Gingras et al., 2004). The same studies also concluded that instead of direct effects of Socs1 on TLR signaling components, Socs1 instead caused indirect effects by regulating the signaling output of the IFN-αβR: because TLR signaling induces autocrine-paracrine IFN-α–β production this might underlie the cause of LPS sensitivity in Socs1−/−; Ifng−/− mice.
Subsequent work using sensitive assays for the effects of Socs1 on the IFN-αβR has definitively shown that Socs1 is an irreplaceable regulator of IFN-αβR activity (Fenner et al., 2006). At this stage however, the potential for Socs proteins to regulate one or more non-cytokine receptor signaling components in TLR signaling remains open. Mansell et al. have demonstrated that Socs1 can bind to, and regulate the degradation of Mal (also known as TIRAP), an adapter molecule specifically associated with TLR2 and TLR4 signaling (Mansell et al., 2006). Previous studies had shown that the tyrosine kinase Btk phosphorylates Mal, providing binding sites for Socs1 (Gray et al., 2006). However, enforced expression of Socs1 has no effect on LTA signaling via TLR2, LPS signaling via TLR4 and, as noted above, loss of Socs1 has no obvious effects on LPS signaling (Baetz et al., 2004; Gingras et al., 2004). Collectively, these studies indicate that more work is required to establish specific targets of Socs proteins during inflammatory response, and in this circumstance, TLR signaling.
Socs1 and Socs3 have precise roles in regulating T cell development and function
So far we have emphasized that Socs1 and Socs3 have precise functions in regulating a subset of cytokine receptors. Consistent with this idea, Socs1 and Socs3 also have very defined roles in controlling T cell development and function. To test the T cell-specific functions of Socs1 and Socs3, conditionally deficient mice have been employed; these mice sidestep the complexities associated with conventional genetic deficiency of these key Socs proteins. Consistent with preference of Socs3 for gp130, loss of Socs3 in T cells has no effect on Th1 or Th2 development (or overall T cell development in the thymus) but is instead required for the IL-6 and IL-23-mediated effects on Th17 T cells, both of which signal via gp130 (Chen et al., 2006; Wong et al., 2006). Indeed, loss of Socs3 enhances IL-17 production from Th17 cells by increasing the amount of Stat3 recruited to Il17a and Il17f (Fig. 1C). Once again, the essential effects of Socs3 are highly restricted.
Although deficiency in Socs1 causes a profound and lethal inflammatory syndrome mediated largely by IFN-γ-producing T cells, loss of Socs1 only in T cells does not recapitulate any of the inflammatory pathology seen in the conventional Socs1 deficient mice (Chong et al., 2003). Instead, Socs1 regulates T cell numbers and especially CD8+ cells by controlling responsiveness to cytokines that signal through γc and Jak3 such as IL-7 and IL-15 (Chong et al., 2003; Ramanathan et al., 2006). Therefore, the central role of Socs1 activity in inhibiting inflammation is partitioned between cell types and receptors. On one hand, Socs1 regulates T cell development by inhibiting γc signaling. On the other hand, once these cells enter the periphery and begin secreting IFN-γ it is the IFN-γ-responsive cells that must be regulated by Socs1 to constrain the lethal IFN-γ-mediated inflammation.
Do Socs proteins have substrates other than cytokine receptors?
The widespread use of microarrays for interrogating gene expression patterns in any number of biological systems has revealed increased Socs expression is a very common phenomenon. The apparent ubiquity of Socs expression raises the issue of the number, specificity and relevance of the Socs client proteins. As we have discussed above, Socs proteins have confirmed roles in cytokine receptor signaling and more controversial functions in regulating TLR signaling. A major question in this area concerns the physiological targets of the Socs proteins in diverse signaling scenarios. Potentially any phosphorylated tyrosine residue with sufficient affinity for a Socs SH2 domain could be targeted for binding and potentially led to the protein destruction machinery. However, as we have noted above, the number of definitive Socs targets is so far largely limited to cytoplasmic tails of a subset of cytokine receptors. Are cytokine receptors the only targets of Socs proteins? The answer to this question is harder to address than the determination of the essential targets of, for example Socs1 and Socs3, because loss of function studies cannot readily expose the full range of targets in a natural (i.e. non-deficient) setting. Therefore, Socs overexpression or experimentally regulated expression has the potential to illuminate additional target proteins that might not be observed as ‘essential’ in genetic studies. However, promiscuous overexpression of Socs proteins has often led to misleading data because the increased expression of SH2 domains in the cytoplasm allows binding to a huge range of tyrosine phosphorylated proteins. It is also important to consider the tight temporal regulation of Socs expression by cytokines, hormones, TLR agonists and other factors that signal transient increases in Socs expression not mirrored by constitutive overexpression. Nevertheless, a number of potential Socs targets have been identified that might regulate inflammatory cascades beyond cytokine receptor regulation.
An example of a non-cytokine receptor substrate of Socs3 has been described in recent studies on chemokine receptor signaling (Le et al., 2007). The chemokine CXCL12 activates CXCR4 and induces phosphorylation of FAK, a ubiquitous tyrosine kinase, in addition to other pathways. Le et al. have shown that Socs3 is crucial to regulate the amounts of CXCL12-activated phospho-FAK as Socs3-deficient B cells have increased amount of phospho-FAK. The consequence of increased phospho-FAK in absence of Socs3 is accumulation of immature B cells in the bone marrow mediated by increased CXCR4 signaling. Link and colleagues have also postulated that signals from the G-CSFR negatively regulate CXCL12 amounts in the bone marrow allowing escape of neutrophils into the circulation (Semerad et al., 2002). Therefore, the assignment of FAK as a substrate of Socs3 potentially explains previous data that suggested that Socs3 directly regulates CXCR4 to inhibit its activity (Soriano et al., 2002). Further work is need to tie together how Stat3 and Socs3 control mature immune cell numbers leaving the bone marrow for the tissues.
The non-cytokine receptor targets of the Socs proteins need to be evaluated to the same standard as the Socs binding residues definitively identified in cytokine receptors: mutations must be generated to mutate the target tyrosine to create a protein that can no longer be inhibited by the suspect Socs protein. Together with the conventional and conditional loss-of-function alleles of Socs1 and Socs3, an arsenal of experimental approaches can be used to link Socs expression with a downstream effect.
Cross-talk between the Stat1-Socs1 and Stat3-Socs3 modules
Many examples of cytokines stimulating the production of other cytokines have been described, and in principle many of these pathways can be, and are controlled by, feedback inhibition by Socs proteins. The production of IL-10 in the myeloid lineage and T cell lineages, however, offers new insights into a complex regulatory hierarchy. As noted above IL-10, via Stat3, is essential for inhibiting inflammatory responses, especially those driven by TLR signaling. Many groups have therefore focused the when and the how of IL-10 production in inflammation. Mice lacking IL-10 in T cells recapitulate many of the chronic effects of complete IL-10 deficiency including severe inflammatory bowel disease (Roers et al., 2004). However, T cell-specific IL-10-deficient mice have identical responses to control mice in LPS challenge experiments while mice bearing a complete IL-10 deficiency are extremely sensitive to LPS. These data suggest that cells other than T cells must make IL-10 that (partially) protects against excessive acute inflammation. These non-T cells are myeloid-derived cells and especially macrophages and dendritic cells that are capable of prodigious IL-10 production. What are the signals that control IL-10 production in myeloid cells? Addressing this question is complicated by the diversity of stimuli and downstream signaling molecules that contribute to myeloid IL-10 production including TLR agonists via TLRs and the p38 MAP kinase pathways, and zymosan via the dectin and ERK pathways to list a few. Despite this complexity, it is now clear that TLR-induced interferons enforce further IL-10 secretion via a feedforward loop. Cheng and colleagues have demonstrated that TLR-mediated activation of type I interferon production is essential for IL-10 synthesis: in the absence of the type I IFN-αβR, TRIF or IRF-3, sustained IL-10 production by LPS-activated macrophages fails (Chang et al., 2007). However, since IL-10 regulates its own production by Stat3 (Cheng et al., 2003; Staples et al., 2007), the feed-forward loop via Stat3 also fails. Therefore, myeloid lineage cells have developed a complex feed-forward loop to generate IL-10. The question is how these pathways are controlled in vivo. Aspects of this complex regulatory pathway including the role of IL-27 in driving IL-10 production via Stat1 and Stat3 are covered in an accompanying review by Li and Flavell. It is worth returning to the findings noted above for the T cell-specific IL-10-deficient mice because these mice emphasize that the T cell-specific production of IL-10 is the main arbiter of the anti-inflammatory response in chronic infection, and that for tissues such as the gut, continuous exposure to IL-10 is essential for ‘homeostatic’ inflammation in the intestines (Denning et al., 2007). Therefore, the multiple IL-10 reporter mice recently reported will prove decisive in delineating who makes IL-10 and when during inflammation (Kamanaka et al., 2006; Maynard et al., 2007).
Conclusions
Despite the many tools we have to dissect the function of each JAK, Stat and Socs protein in each cell type involved in inflammatory responses, we still know little about how cytokine signaling is integrated in cells and tissues, especially when a cell receives inputs from more than one cytokine: for example, defining how a cell responds in vivo to signals from a pro- and anti-inflammatory cytokine at the same time remains an unmet goal. We have stressed that approaches such as ChIP-seq will be essential to interpreting cytokine signaling from the point of ongoing gene expression changes that will be converted to functional changes in cellular behavior. Despite the current limitations in understanding the Jak-Stat-Socs pathways, the clinical application of exogenous cytokine therapy or blocking individual cytokines correlated with pro-inflammatory pathology is advancing rapidly.
Acknowledgements
We apologize to the many researchers whose primary citations could not be included because of space restriction. We thank Stephanie Watowich and members of our laboratories for discussion. PJM is supported by grants from the NIH, Sandler Program for Asthma Research, NIH CORE grant P30 CA21765 and the American Lebanese Syrian Associated Charities.
References
- Alexander WS, Hilton DJ. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu Rev Immunol. 2004;22:503–529. doi: 10.1146/annurev.immunol.22.091003.090312. [DOI] [PubMed] [Google Scholar]
- Alexander WS, Starr R, Fenner JE, Scott CL, Handman E, Sprigg NS, Corbin JE, Cornish AL, Darwiche R, Owczarek CM, et al. SOCS1 is a critical inhibitor of interferon gamma signaling and prevents the potentially fatal neonatal actions of this cytokine. Cell. 1999;98:597–608. doi: 10.1016/s0092-8674(00)80047-1. [DOI] [PubMed] [Google Scholar]
- Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, McAllister F, Edeal J, Gaus K, et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetz A, Frey M, Heeg K, Dalpke AH. Suppressor of cytokine signaling (SOCS) proteins indirectly regulate toll-like receptor signaling in innate immune cells. J Biol Chem. 2004;279:54708–54715. doi: 10.1074/jbc.M410992200. [DOI] [PubMed] [Google Scholar]
- Berg DJ, Davidson N, Kuhn R, Muller W, Menon S, Holland G, Thompson-Snipes L, Leach MW, Rennick D. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J Clin Invest. 1996;98:1010–1020. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniface K, Bernard FX, Garcia M, Gurney AL, Lecron JC, Morel F. IL-22 inhibits epidermal differentiation and induces proinflammatory gene expression and migration of human keratinocytes. J Immunol. 2005;174:3695–3702. doi: 10.4049/jimmunol.174.6.3695. [DOI] [PubMed] [Google Scholar]
- Boniface K, Guignouard E, Pedretti N, Garcia M, Delwail A, Bernard FX, Nau F, Guillet G, Dagregorio G, Yssel H, et al. A role for T cell-derived interleukin 22 in psoriatic skin inflammation. Clin Exp Immunol. 2007;150:407–415. doi: 10.1111/j.1365-2249.2007.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay JL, O’Shea JJ, Paul WE. Molecular phylogeny within type I cytokines and their cognate receptors. Immunity. 2003;19:159–163. doi: 10.1016/s1074-7613(03)00211-5. [DOI] [PubMed] [Google Scholar]
- Boyle K, Egan P, Rakar S, Willson TA, Wicks IP, Metcalf D, Hilton DJ, Nicola NA, Alexander WS, Roberts AW, Robb L. The SOCS box of suppressor of cytokine signaling-3 contributes to the control of G-CSF responsiveness in vivo. Blood. 2007;110:1466–1474. doi: 10.1182/blood-2007-03-079178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchill MA, Yang J, Vang KB, Moon JJ, Chu HH, Lio CW, Vegoe AL, Hsieh CS, Jenkins MK, Farrar MA. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28:112–121. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, Kwiatkowski DP, McCarthy MI, Ouwehand WH, Samani NJ, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill M, Schrodi SJ, Chang M, Garcia VE, Brandon R, Callis KP, Matsunami N, Ardlie KG, Civello D, Catanese JJ, et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–290. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EY, Guo B, Doyle SE, Cheng G. Cutting edge: involvement of the type I IFN production and signaling pathway in lipopolysaccharide-induced IL-10 production. J Immunol. 2007;178:6705–6709. doi: 10.4049/jimmunol.178.11.6705. [DOI] [PubMed] [Google Scholar]
- Chen Z, Laurence A, Kanno Y, Pacher-Zavisin M, Zhu BM, Tato C, Yoshimura A, Hennighausen L, O’Shea JJ. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc Natl Acad Sci U S A. 2006;103:8137–8142. doi: 10.1073/pnas.0600666103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Wang HW, Cuenca A, Huang M, Ghansah T, Brayer J, Kerr WG, Takeda K, Akira S, Schoenberger SP, et al. A critical role for Stat3 signaling in immune tolerance. Immunity. 2003;19:425–436. doi: 10.1016/s1074-7613(03)00232-2. [DOI] [PubMed] [Google Scholar]
- Chong MM, Cornish AL, Darwiche R, Stanley EG, Purton JF, Godfrey DI, Hilton DJ, Starr R, Alexander WS, Kay TW. Suppressor of cytokine signaling-1 is a critical regulator of interleukin-7-dependent CD8+ T cell differentiation. Immunity. 2003;18:475–487. doi: 10.1016/s1074-7613(03)00078-5. [DOI] [PubMed] [Google Scholar]
- Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, et al. Metal Chelation and Inhibition of Bacterial Growth in Tissue Abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- Croker BA, Krebs DL, Zhang JG, Wormald S, Willson TA, Stanley EG, Robb L, Greenhalgh CJ, Forster I, Clausen BE, et al. SOCS3 negatively regulates IL-6 signaling in vivo. Nat Immunol. 2003;4:540–545. doi: 10.1038/ni931. [DOI] [PubMed] [Google Scholar]
- Croker BA, Metcalf D, Robb L, Wei W, Mifsud S, DiRago L, Cluse LA, Sutherland KD, Hartley L, Williams E, et al. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity. 2004;20:153–165. doi: 10.1016/s1074-7613(04)00022-6. [DOI] [PubMed] [Google Scholar]
- Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol. 2007;8:1086–1094. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- Donnelly RP, Sheikh F, Kotenko SV, Dickensheets H. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J Leukoc Biol. 2004;76:314–321. doi: 10.1189/jlb.0204117. [DOI] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kasmi KC, Holst J, Coffre M, Mielke L, de Pauw A, Lhocine N, Smith AM, Rutschman R, Kaushal D, Shen Y, et al. General nature of the STAT3-activated anti-inflammatory response. J Immunol. 2006;177:7880–7888. doi: 10.4049/jimmunol.177.11.7880. [DOI] [PubMed] [Google Scholar]
- Eyles JL, Roberts AW, Metcalf D, Wicks IP. Granulocyte colony-stimulating factor and neutrophils--forgotten mediators of inflammatory disease. Nat Clin Pract Rheumatol. 2006;2:500–510. doi: 10.1038/ncprheum0291. [DOI] [PubMed] [Google Scholar]
- Feldmann M, Maini RN. Lasker Clinical Medical Research Award. TNF defined as a therapeutic target for rheumatoid arthritis and other autoimmune diseases. Nat Med. 2003;9:1245–1250. doi: 10.1038/nm939. [DOI] [PubMed] [Google Scholar]
- Fenner JE, Starr R, Cornish AL, Zhang JG, Metcalf D, Schreiber RD, Sheehan K, Hilton DJ, Alexander WS, Hertzog PJ. Suppressor of cytokine signaling 1 regulates the immune response to infection by a unique inhibition of type I interferon activity. Nat Immunol. 2006;7:33–39. doi: 10.1038/ni1287. [DOI] [PubMed] [Google Scholar]
- Gingras S, Parganas E, de Pauw A, Ihle JN, Murray PJ. Re-examination of the role of suppressor of cytokine signaling 1 (SOCS1) in the regulation of toll-like receptor signaling. J Biol Chem. 2004;279:54702–54707. doi: 10.1074/jbc.M411043200. [DOI] [PubMed] [Google Scholar]
- Gray P, Dunne A, Brikos C, Jefferies CA, Doyle SL, O’Neill LA. MyD88 adapter-like (Mal) is phosphorylated by Bruton’s tyrosine kinase during TLR2 and TLR4 signal transduction. J Biol Chem. 2006;281:10489–10495. doi: 10.1074/jbc.M508892200. [DOI] [PubMed] [Google Scholar]
- Hirano T, Murakami M. Grasp a pTyr-peptide by its SOCS. Dev Cell. 2006;10:542–544. doi: 10.1016/j.devcel.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, Freeman AF, Demidowich A, Davis J, Turner ML, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357:1608–1619. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- Hortner M, Nielsch U, Mayr LM, Johnston JA, Heinrich PC, Haan S. Suppressor of cytokine signaling-3 is recruited to the activated granulocyte-colony stimulating factor receptor and modulates its signal transduction. J Immunol. 2002;169:1219–1227. doi: 10.4049/jimmunol.169.3.1219. [DOI] [PubMed] [Google Scholar]
- Kamanaka M, Kim ST, Wan YY, Sutterwala FS, Lara-Tejero M, Galan JE, Harhaj E, Flavell RA. Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity. 2006;25:941–952. doi: 10.1016/j.immuni.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Kamimura D, Ishihara K, Hirano T. IL-6 signal transduction and its physiological roles: the signal orchestration model. Rev Physiol Biochem Pharmacol. 2003;149:1–38. doi: 10.1007/s10254-003-0012-2. [DOI] [PubMed] [Google Scholar]
- Kamizono S, Hanada T, Yasukawa H, Minoguchi S, Kato R, Minoguchi M, Hattori K, Hatakeyama S, Yada M, Morita S, et al. The SOCS box of SOCS-1 accelerates ubiquitin-dependent proteolysis of TEL-JAK2. J Biol Chem. 2001;276:12530–12538. doi: 10.1074/jbc.M010074200. [DOI] [PubMed] [Google Scholar]
- Kano A, Wolfgang MJ, Gao Q, Jacoby J, Chai GX, Hansen W, Iwamoto Y, Pober JS, Flavell RA, Fu XY. Endothelial cells require STAT3 for protection against endotoxin-induced inflammation. J Exp Med. 2003;198:1517–1525. doi: 10.1084/jem.20030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile BT, Schulman BA, Alexander WS, Nicola NA, Martin HM, Hilton DJ. The SOCS box: a tale of destruction and degradation. Trends Biochem Sci. 2002;27:235–241. doi: 10.1016/s0968-0004(02)02085-6. [DOI] [PubMed] [Google Scholar]
- Kimura A, Kinjyo I, Matsumura Y, Mori H, Mashima R, Harada M, Chien KR, Yasukawa H, Yoshimura A. SOCS3 is a physiological negative regulator for granulopoiesis and granulocyte colony-stimulating factor receptor signaling. J Biol Chem. 2004;279:6905–6910. doi: 10.1074/jbc.C300496200. [DOI] [PubMed] [Google Scholar]
- Kinjyo I, Hanada T, Inagaki-Ohara K, Mori H, Aki D, Ohishi M, Yoshida H, Kubo M, Yoshimura A. SOCS1/JAB is a negative regulator of LPS-induced macrophage activation. Immunity. 2002;17:583–591. doi: 10.1016/s1074-7613(02)00446-6. [DOI] [PubMed] [Google Scholar]
- Korn T, Oukka M, Bettelli E. Th17 cells: Effector T cells with inflammatory properties. Semin Immunol. 2007 doi: 10.1016/j.smim.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, Niu G, Kay H, Mule J, Kerr WG, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11:1314–1321. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- Kreymborg K, Etzensperger R, Dumoutier L, Haak S, Rebollo A, Buch T, Heppner FL, Renauld JC, Becher B. IL-22 is expressed by Th17 cells in an IL-23-dependent fashion, but not required for the development of autoimmune encephalomyelitis. J Immunol. 2007;179:8098–8104. doi: 10.4049/jimmunol.179.12.8098. [DOI] [PubMed] [Google Scholar]
- Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- Lang R, Pauleau AL, Parganas E, Takahashi Y, Mages J, Ihle JN, Rutschman R, Murray PJ. SOCS3 regulates the plasticity of gp130 signaling. Nat Immunol. 2003;4:546–550. doi: 10.1038/ni932. [DOI] [PubMed] [Google Scholar]
- Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Le Y, Zhu BM, Harley B, Park SY, Kobayashi T, Manis JP, Luo HR, Yoshimura A, Hennighausen L, Silberstein LE. SOCS3 protein developmentally regulates the chemokine receptor CXCR4-FAK signaling pathway during B lymphopoiesis. Immunity. 2007;27:811–823. doi: 10.1016/j.immuni.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Lee CK, Raz R, Gimeno R, Gertner R, Wistinghausen B, Takeshita K, DePinho RA, Levy DE. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity. 2002;17:63–72. doi: 10.1016/s1074-7613(02)00336-9. [DOI] [PubMed] [Google Scholar]
- Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WW, Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest. 2007;117:1175–1183. doi: 10.1172/JCI31537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HL, Liang S, Li J, Napierata L, Brown T, Benoit S, Senices M, Gill D, Dunussi-Joannopoulos K, Collins M, et al. IL-22 is required for Th17 cell-mediated pathology in a mouse model of psoriasis-like skin inflammation. J Clin Invest. 2008 doi: 10.1172/JCI33263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansell A, Smith R, Doyle SL, Gray P, Fenner JE, Crack PJ, Nicholson SE, Hilton DJ, O’Neill LA, Hertzog PJ. Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat Immunol. 2006;7:148–155. doi: 10.1038/ni1299. [DOI] [PubMed] [Google Scholar]
- Mayack SR, Berg LJ. Cutting edge: an alternative pathway of CD4+ T cell differentiation is induced following activation in the absence of gamma-chain-dependent cytokine signals. J Immunol. 2006;176:2059–2063. doi: 10.4049/jimmunol.176.4.2059. [DOI] [PubMed] [Google Scholar]
- Maynard CL, Harrington LE, Janowski KM, Oliver JR, Zindl CL, Rudensky AY, Weaver CT. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3− precursor cells in the absence of interleukin 10. Nat Immunol. 2007;8:931–941. doi: 10.1038/ni1504. [DOI] [PubMed] [Google Scholar]
- McLemore ML, Grewal S, Liu F, Archambault A, Poursine-Laurent J, Haug J, Link DC. STAT-3 activation is required for normal G-CSF-dependent proliferation and granulocytic differentiation. Immunity. 2001;14:193–204. doi: 10.1016/s1074-7613(01)00101-7. [DOI] [PubMed] [Google Scholar]
- Metcalf D, Di Rago L, Mifsud S, Hartley L, Alexander WS. The development of fatal myocarditis and polymyositis in mice heterozygous for IFN-gamma and lacking the SOCS-1 gene. Proc Natl Acad Sci U S A. 2000;97:9174–9179. doi: 10.1073/pnas.160255197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, Kanno Y, Spalding C, Elloumi HZ, Paulson ML, et al. Impaired Th17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008 doi: 10.1038/nature06764. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, Kawamura N, Ariga T, Pasic S, Stojkovic O, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- Murray PJ. Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response. Curr Opin Pharmacol. 2006;6:379–386. doi: 10.1016/j.coph.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Murray PJ. The JAK-STAT signaling pathway: input and output integration. J Immunol. 2007;178:2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- Nakagawa R, Naka T, Tsutsui H, Fujimoto M, Kimura A, Abe T, Seki E, Sato S, Takeuchi O, Takeda K, et al. SOCS-1 participates in negative regulation of LPS responses. Immunity. 2002;17:677–687. doi: 10.1016/s1074-7613(02)00449-1. [DOI] [PubMed] [Google Scholar]
- Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, Gadina M, O’Shea JJ, Biron CA. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science. 2002;297:2063–2066. doi: 10.1126/science.1074900. [DOI] [PubMed] [Google Scholar]
- Notarangelo LD, Mella P, Jones A, de Saint Basile G, Savoldi G, Cranston T, Vihinen M, Schumacher RF. Mutations in severe combined immune deficiency (SCID) due to JAK3 deficiency. Hum Mutat. 2001;18:255–263. doi: 10.1002/humu.1188. [DOI] [PubMed] [Google Scholar]
- Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, Dong C. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- O’Garra A, Arai N. The molecular basis of T helper 1 and T helper 2 cell differentiation. Trends Cell Biol. 2000;10:542–550. doi: 10.1016/s0962-8924(00)01856-0. [DOI] [PubMed] [Google Scholar]
- O’Shea JJ, Husa M, Li D, Hofmann SR, Watford W, Roberts JL, Buckley RH, Changelian P, Candotti F. Jak3 and the pathogenesis of severe combined immunodeficiency. Mol Immunol. 2004a;41:727–737. doi: 10.1016/j.molimm.2004.04.014. [DOI] [PubMed] [Google Scholar]
- O’Shea JJ, Pesu M, Borie DC, Changelian PS. A new modality for immunosuppression: targeting the JAK/STAT pathway. Nat Rev Drug Discov. 2004b;3:555–564. doi: 10.1038/nrd1441. [DOI] [PubMed] [Google Scholar]
- Pan H, Hong F, Radaeva S, Gao B. Hydrodynamic gene delivery of interleukin-22 protects the mouse liver from concanavalin A-, carbon tetrachloride-, and Fas ligand-induced injury via activation of STAT3. Cell Mol Immunol. 2004;1:43–49. [PubMed] [Google Scholar]
- Panopoulos AD, Zhang L, Snow JW, Jones DM, Smith AM, El Kasmi KC, Liu F, Goldsmith MA, Link DC, Murray PJ, Watowich SS. STAT3 governs distinct pathways in emergency granulopoiesis and mature neutrophils. Blood. 2006;108:3682–3690. doi: 10.1182/blood-2006-02-003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Radaeva S, Sun R, Pan HN, Hong F, Gao B. Interleukin 22 (IL-22) plays a protective role in T cell-mediated murine hepatitis: IL-22 is a survival factor for hepatocytes via STAT3 activation. Hepatology. 2004;39:1332–1342. doi: 10.1002/hep.20184. [DOI] [PubMed] [Google Scholar]
- Ramanathan S, Gagnon J, Leblanc C, Rottapel R, Ilangumaran S. Suppressor of cytokine signaling 1 stringently regulates distinct functions of IL-7 and IL-15 in vivo during T lymphocyte development and homeostasis. J Immunol. 2006;176:4029–4041. doi: 10.4049/jimmunol.176.7.4029. [DOI] [PubMed] [Google Scholar]
- Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, de Bakker PI, Le JM, Lee HS, Batliwalla F, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–986. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roers A, Siewe L, Strittmatter E, Deckert M, Schluter D, Stenzel W, Gruber AD, Krieg T, Rajewsky K, Muller W. T cell-specific inactivation of the interleukin 10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. J Exp Med. 2004;200:1289–1297. doi: 10.1084/jem.20041789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, Rottapel R, Yamaoka S, Lu KP. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell. 2003;12:1413–1426. doi: 10.1016/s1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- Sano S, Itami S, Takeda K, Tarutani M, Yamaguchi Y, Miura H, Yoshikawa K, Akira S, Takeda J. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. Embo J. 1999;18:4657–4668. doi: 10.1093/emboj/18.17.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semerad CL, Liu F, Gregory AD, Stumpf K, Link DC. G-CSF is an essential regulator of neutrophil trafficking from the bone marrow to the blood. Immunity. 2002;17:413–423. doi: 10.1016/s1074-7613(02)00424-7. [DOI] [PubMed] [Google Scholar]
- Semerad CL, Poursine-Laurent J, Liu F, Link DC. A role for G-CSF receptor signaling in the regulation of hematopoietic cell function but not lineage commitment or differentiation. Immunity. 1999;11:153–161. doi: 10.1016/s1074-7613(00)80090-4. [DOI] [PubMed] [Google Scholar]
- Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- Soriano SF, Hernanz-Falcon P, Rodriguez-Frade JM, De Ana AM, Garzon R, Carvalho-Pinto C, Vila-Coro AJ, Zaballos A, Balomenos D, Martinez AC, Mellado M. Functional inactivation of CXC chemokine receptor 4-mediated responses through SOCS3 up-regulation. J Exp Med. 2002;196:311–321. doi: 10.1084/jem.20012041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples KJ, Smallie T, Williams LM, Foey A, Burke B, Foxwell BM, Ziegler-Heitbrock L. IL-10 induces IL-10 in primary human monocyte-derived macrophages via the transcription factor Stat3. J Immunol. 2007;178:4779–4785. doi: 10.4049/jimmunol.178.8.4779. [DOI] [PubMed] [Google Scholar]
- Stross C, Radtke S, Clahsen T, Gerlach C, Volkmer-Engert R, Schaper F, Heinrich PC, Hermanns HM. Oncostatin M receptor-mediated signal transduction is negatively regulated by SOCS3 through a receptor tyrosine-independent mechanism. J Biol Chem. 2006;281:8458–8468. doi: 10.1074/jbc.M511212200. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, Blumberg RS, Xavier RJ, Mizoguchi A. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118:534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Forster I, Akira S. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- Tremelling M, Cummings F, Fisher SA, Mansfield J, Gwilliam R, Keniry A, Nimmo ER, Drummond H, Onnie CM, Prescott NJ, et al. IL23R variation determines susceptibility but not disease phenotype in inflammatory bowel disease. Gastroenterology. 2007;132:1657–1664. doi: 10.1053/j.gastro.2007.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boxel-Dezaire AH, Rani MR, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S, Bhattacharya R, Gabrilovich D, Heller R, Coppola D, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10:48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- Watford WT, O’Shea JJ. Human tyk2 kinase deficiency: another primary immunodeficiency syndrome. Immunity. 2006;25:695–697. doi: 10.1016/j.immuni.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Wei L, Laurence A, Elias KM, O’Shea JJ. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J Biol Chem. 2007;282:34605–34610. doi: 10.1074/jbc.M705100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LM, Sarma U, Willets K, Smallie T, Brennan F, Foxwell BM. Expression of constitutively active STAT3 can replicate the cytokine-suppressive activity of interleukin-10 in human primary macrophages. J Biol Chem. 2007;282:6965–6975. doi: 10.1074/jbc.M609101200. [DOI] [PubMed] [Google Scholar]
- Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R. IL-22 increases the innate immunity of tissues. Immunity. 2004;21:241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Wong PK, Egan PJ, Croker BA, O’Donnell K, Sims NA, Drake S, Kiu H, McManus EJ, Alexander WS, Roberts AW, Wicks IP. SOCS-3 negatively regulates innate and adaptive immune mechanisms in acute IL-1-dependent inflammatory arthritis. J Clin Invest. 2006;116:1571–1581. doi: 10.1172/JCI25660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, et al. T Helper 17 Lineage Differentiation Is Programmed by Orphan Nuclear Receptors RORalpha and RORgamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang Q, Bhandoola A, et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci U S A. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, Laurence A, Robinson GW, Shevach EM, Moriggl R, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa H, Ohishi M, Mori H, Murakami M, Chinen T, Aki D, Hanada T, Takeda K, Akira S, Hoshijima M, et al. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat Immunol. 2003;4:551–556. doi: 10.1038/ni938. [DOI] [PubMed] [Google Scholar]
- Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Karow M, Flavell RA. Interleukin-22 but not interleukin-17 provides protection to hepatocytes during acute liver inflammation. Immunity. 2007;27:647–659. doi: 10.1016/j.immuni.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JG, Farley A, Nicholson SE, Willson TA, Zugaro LM, Simpson RJ, Moritz RL, Cary D, Richardson R, Hausmann G, et al. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc Natl Acad Sci U S A. 1999;96:2071–2076. doi: 10.1073/pnas.96.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JG, Metcalf D, Rakar S, Asimakis M, Greenhalgh CJ, Willson TA, Starr R, Nicholson SE, Carter W, Alexander WS, et al. The SOCS box of suppressor of cytokine signaling-1 is important for inhibition of cytokine action in vivo. Proc Natl Acad Sci U S A. 2001;98:13261–13265. doi: 10.1073/pnas.231486498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Danilenko DM, Valdez P, Kasman I, Eastham-Anderson J, Wu J, Ouyang W. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- Zhou L, Ivanov, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]