Abstract
Here we present the use of hydrogen/deuterium exchange (HDX) mass spectrometry to analyze the estrogen receptor β ligand binding domain (ERβLBD) in the absence and presence of a variety of chemical compounds with different binding modes and pharmacological properties. Previously, we reported the use of HDX as a method to predict the tissue selectivity of ERα ligands. HDX profiles of ERαLBD in complex with ligand could differentiate compounds of the same chemotype. In contrast, similar analysis of ERβLBD showed correlation to the compound chemical structures but little correlation with compound tissue selectivity. The different HDX patterns observed for ERβLBD when compared to ERαLBD bound to the same chemical compounds serves as an indication that ERβLBD undergoes a different structural response to the same ligand when compared to ERαLBD. The conformational dynamics revealed by HDX forz ERβLBD together with those for ERαLBD shed light into ER ligand interactions and offer new structural insights. The compound specific perturbations in HDX kinetics observed for each of the two isoforms should aid the development of subtype selective ER ligands.
Estrogen receptors (ERα and ERβ) belong to the nuclear receptor superfamily functioning as ligand-dependent transcription factors and play important roles in a variety of processes including regulation of the reproductive and cardiovascular systems, bone density, cognition, and inflammation (1–4). Although the two isoforms share common domain structure with 94% amino-acid identity in their DNA binding domains (DBDs) and 53% in the ligand binding domains (LBDs) (1), ERα and ERβ have different tissue expression profiles and show distinct biological functions in several disease phenotypes (4–8). Not surprisingly, both ER isoforms bind a wide spectrum of natural and synthetic ligands and recent efforts have focused on the development of isoform selective ER modulators for improved therapies for prostatic diseases, inflammation and arthritis (8–9). Since many of the compounds developed to target ERα also bind to ERβ with comparable affinity and potency, study of the receptor-ligand interaction for each isoform is important to understand how those ligands regulate ER functions from the structural basis.
Previous crystallographic analyses of both ERs revealed a canonical fold for the ER ligand binding domain (10–12). In these X-ray structures, twelve helices are arranged in a three-layered sandwich topology and the receptors hold a hydrophobic cavity for hormone binding. It is believed that agonist or antagonist binding induces a distinct conformation change upon the receptor which initiates activation or suppression of responsive genes as determined by the preferential association/dissociation of coactivator and corepressor proteins. Classic ER binding models were validated by the different ER/ligand complex structures in which ligands reside in the steroid binding site and interact with ER-LBD to modulate the position of helix 12 in distinct positions either favoring or disrupting coactivator binding (10). The cognate steroid binding sites of the two ER isoforms are nearly identical with only two amino acid residues being different (11). Recently, both computational and experimental studies suggested the possibility of other ligand binding sites within the ER LBD although the putative secondary binding sites were distinct from each other in these two studies (13–14). Although X-ray studies of the binding modes of many compounds in complex with two isoforms have greatly helped our understanding of the interactions between ER and ligand, these data are not enough to explain the isoform and tissue selectivity of many ER ligands.
HDX mass spectrometry has been used to evaluate the binding mode of ligands to nuclear receptors (15–19). We have previously described the structure dynamics of ERα in the presence of a variety of natural or synthetic compounds representing agonism, antagonism or selective agonism with tissue specificity (20). Differential HDX analysis of the LBD of ERα in complex with various ligands revealed that alterations in structural dynamics can be related to a compound’s pharmacological properties, and thus provide a novel approach to probe selective estrogen receptor modulators (SERMs). Here we compare the HDX characteristics of the LBD of ERβ with ERα in complex with the same set of ligands. At the same time, we have measured the binding affinity of the same array of compounds to both receptors. Generally, all the ligands are tight binders to both receptors with the binding affinity in the low nanomolar range, with some showing preference for either receptor. In addition, the HDX profiles of both ERα and ERβ in complex with genistein were compared. Genistein is an isoflavonoid phytoestrogen ERβ selective ligand. The binding of genistin to ER β is approximately 10 folder tighter than that to ERα according to previous research (21). Interestingly, distinct HDX patterns were observed for ERβ and ERα upon binding of each ligand in the compound set suggesting a distinct function for ERβ in estrogen action regulation. HDX data for ERβ, together with the published HDX data of ERα, provide unique insight into the ligand-dependent activation of the two ERs.
MATERIALS AND METHODS
Protein and reagents for HDX analysis
The ERβ LBD (amino acids 257–502) was overexpressed as N-terminally His-6-tagged protein in BL21 (DE3) cells by using vector pET19 (Novagen) and the purified protein is provided by PanVera (Madison, WI). All compounds are from Lilly Research Laboratories except raloxifene and genistein which were from Sigma (St. Louis, MO). For HDX analysis, ERβLBD was incubated with compound for 1 hr at a ratio of 1:10. The protein concentration was 12 µM in the deuterium incubation buffer, which is the same condition as the ERα/ligand study we performed earlier. The ERαLBD/genistein experiment followed the conditions that have been reported earlier (20) at the receptor/ligand ratio of 1:10.
HDX analysis
HDX experiments were carried out with a LEAP Technologies Twin HTS PAL liquid handling robot interfaced with a Thermo Finnigan LTQ mass spectrometer (Thermo Electron Corporation, San Jose, CA). For a complete experimental description see (17). Briefly, the protein/ligand complex was diluted into a D2O exchange buffer and subjected to HDX for 1, 30, 60, 900 and 4200 sec. HDX experiments were performed at 4°C by dilution of 4 µL of the ER-LBD solution with 16 µL of D2O buffer containing 20 mM Tris-HCl, 100mM KCl, and 1 mM DTT, pD 7.9. After the incubation in D2O at a fixed hydrogen deuterium exchange time aforementioned, the exchange reaction was quenched with 2M urea containing 1% TFA and passed over an in-house packed pepsin column (2mm × 2cm) which was kept at 2°C. The digested ER LBD peptides were then eluted through a 1mm × 1 cm C8 trap cartridge (Eclipse XDB-C8, Agilent, Santa Clara, CA) and desalted. The digestion and desalting takes a total of 2.5 min. Peptides were then eluted across a 2.1mm × 5cm C18 column (Thermo Scientific, Waltham, MA) with a linear gradient of 2%–50% B over 10 min (Solvent A, 0.3% formic acid in water; solvent B, 0.3 % formic acid 80% acetonitrile, 20% water; flow rate 200ul/min) (22). Mass spectrometric analyses were carried out with the capillary temperature at 225°C.
Peptide identification and HDX data processing
Product ion spectra were acquired in a data-dependent MS/MS mode. The precursor ion survey scan was performed and the five most abundant ions were selected for product ion analysis. MS/MS *.raw data was searched against the database containing ER using SEQUEST (Bioworks, Thermo Finnigan, CA). At the same time, the MS/MS *.raw data files were converted to *.dta and *.mgf files and submitted to Mascot (Matrix Science, London, UK) for searching against a database containing ER. All peptide ion assignments were inspected manually.
The weighted average m/z values of each peptide ion isotopic cluster were calculated with the web based software known as “Deuterator” (23). The deuteration level was calculated based on the following equation and the corrections for back-exchange were made based on 70% deuterium recovery and accounting for 80% deuterium content in the on-exchange buffer:
where m/z (P), m/z (N), and m/z (F) are the centroid value of partially deuterated peptide, nondeuterated peptide, and fully deuterated peptide, respectively (24).
ER binding assay
The competition binding assay was performed in a buffer containing 50 mM Hepes, pH 7.5, 1.5 mM EDTA, 150 mM NaCl, 10% glycerol, 1mg/ml ovalbumin and 5 mM DTT, using 0.025 µCi per well 3H-estradiol (NEN #NET517 at 118 Ci/mmol, 1.5 nM E2) (NEN/PerkinElmer, Boston, MA) 10 ng/well ERα or ERβ. Nonspecific binding was determined in the presence of 1 µM of 17-β estradiol (E2). The binding reaction was incubated 4 hrs at room temperature, and then a cold DCC buffer was added to each reaction (The DCC buffer contains per 50 ml of assay buffer, 0.75 g of charcoal (Sigma, St Louis, MO) and 0.25 g of dextran (Pharmacia, Uppsala, Sweden)). Plates were mixed 8 minutes on an orbital shaker at 4 °C, and then centrifuged at 3,000 rpm at 4 °C for 10 minutes. An aliquot of the mix was added to Wallac Optiphase “Hisafe 3” scintillation fluid, incubated for 5 hr, and read in a Wallac Microbeta counter. The Kd for 3H-estradiol was determined by saturation binding for ERα or ERβ receptors. The IC50 values for compounds were converted to Ki using Cheng-Prusoff equation.
Cell Based Functional Assay
Human embryonic kidney HEK 293 cells were cultured in DMEM/Ham F12 (3:1) media without phenol red in the presence of 50 µg/ml gentamicin and 10% heat inactivated FBS (Gibco BRL, Rockville, MD). One day prior to the experiment 3×106 cells were seeded onto DMEM/Ham F12 (3:1) media without phenol red in the presence of 10 % charcoal stripped heat inactivated FBS (Gibco BRL, Rockville, MD), and 50 µg/ml gentamicin in a T225 tissue culture flask. Cells were transfected with 250 µl Fugene (Roche Biologicals, Mannheim, Germany), 100 µg plasmid containing ERE response element upstream of a minimal thymidine kinase promoter driven luciferase gene, and 10 µg of expression vector for hERβ receptor driven viral CMV promoter (pCMVhERβ expression plasmid). After 5 hours, cells were transferred to a 96-well tissue culture dish and the media replaced with DMEM/Ham F12 (3:1) media without phenol red in the presence of 10 % charcoal stripped heat inactivated FBS (Gibco BRL, Rockville, MD) without gentamicin. Cells were treated with compounds at different concentrations from 10 µM to 0.3 nM in 10-point concentration dilutions. Twenty four hours later, cells were harvested and assays for luciferase activity. The activity was measured as relative light units and EC50 for ERβ is calculated after fitting the data to a 4-parameter curve fit. The % stimulation was calculated versus DES maximum response.
RESULTS
HDX analysis of ERβLBD
Exchange kinetics for 26 different regions covering 60 percent of the sequence of the receptor LBD were measured. Figure 1 shows the HDX analysis of the apo ERβLBD with secondary structure alignment. The percent HDX exchange displayed in Figure 1 is the average of five HDX on-exchange experiments for apo ERβLBD (exchange times: 1, 30, 60, 900 and 4200 sec). In the HDX experiment, all the peptides analyzed showed no bimodal deuterium incorporation. In other words, there was no co-existence of deuterated and non-deuterated species, namely, the so-called EX1 exchange regime (25). Each on-exchange time point was measured in triplicate and the standard deviation of the average measurement is shown (17).
Figure 1.
HDX percentage of apo ERβLBD with respect to the secondary structures. HDX percentage from helix 2 to helix 12 was plotted against the secondary structures that have peptides available for HDX analysis. The percentage is the averaged value of five HDX experiments with different exchange times: 1, 30, 60, 900 and 4200 sec. The standard deviation of three independent measurements was showed as error bar.
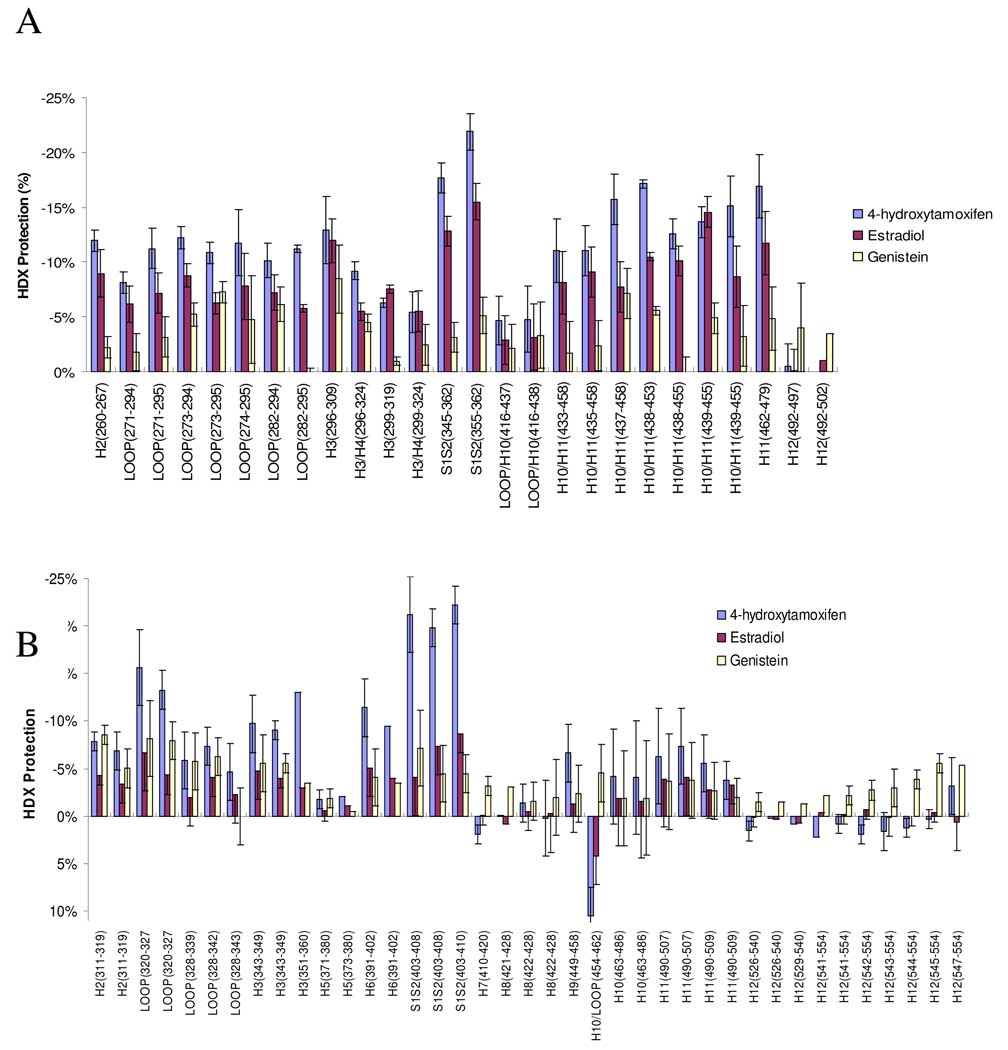
Comprehensive differential HDX analyses were then performed for the 12 ERβLBD-ligand complexes. The chemical structures are presented in Figure 2 and the results are summarized in Table 1. The values in Table 1 represent the average difference in deuterium incorporation percentages for the five on-exchange time points for triplicate experiments (1, 30, 60, 900 and 4200 sec). A negative percentage represents an increase in protection to exchange in that region of the receptor in the presence of ligand, which indicates the region has been protected when bound with ligand. It is clear that different regions of ERβLBD present different degree of protection when binding with the ligand and the same region of ERβLBD present different degree of protection when binding with different ligands. In order to compare ligand induced HDX dynamic change on ERβLBD, a histogram (Figure 3A) was made with respect to the HDX protection (relative HDX exchange in the presence and absence of the ligand) and ER secondary structures. The three compounds plotted are 4-hydroxytamoxifen, estradiol, and genistein. In Figure 3B, the same comparison was made for ERαLBD ± 4-hydroxytamoxifen, and estradiol from our previous data (20), plus the addition of the new data from the analysis of ERαLBD ± genistein. The percent of HDX exchange protection for both receptors are plotted against the secondary structures in Figure 3A and B.
Figure 2.
Chemical structures of 12 ER ligands.
Table 1.
Average difference in deuterium levels of apo ERβLBD in the presence of different ligands with ER binding affinity data and ERβ agonist/antagonist activity.
| Structure | AA# | End | Estradiol | 4-hydroxy tamoxifen |
Ralo- xifene |
DES | ICI | Lasodo- xifene |
Bazedo- xifene |
88074 | 156681 | 165176 | 117018 | Genistein | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H2 | 260 | 267 | −9% | −12% | −8% | −11% | −10% | −11% | −11% | −7% | −5% | −6% | −6% | −2% | |
| LOOP | 271 | 294 | −6% | −8% | −7% | −10% | −9% | −11% | −7% | −7% | −7% | −9% | −7% | −2% | |
| LOOP | 271 | 295 | −7% | −11% | −7% | −10% | −10% | −11% | −7% | −8% | −12% | −8% | −7% | −3% | |
| LOOP | 273 | 294 | −9% | −12% | −9% | −12% | −11% | −12% | −6% | −9% | −7% | −15% | −8% | −5% | |
| LOOP | 273 | 295 | −6% | −11% | −7% | −10% | −10% | −9% | −6% | −7% | −7% | −9% | −7% | −7% | |
| LOOP | 274 | 295 | −8% | −12% | −8% | −10% | −11% | −12% | −7% | −8% | −8% | −10% | −8% | −5% | |
| LOOP | 282 | 294 | −7% | −10% | −6% | −8% | −9% | −9% | −4% | −7% | −7% | −9% | −7% | −6% | |
| LOOP | 282 | 295 | −6% | −11% | −7% | −10% | −10% | −9% | −5% | −7% | −7% | −9% | −8% | * | |
| H3 | 296 | 309 | −12% | −13% | −10% | −17% | −14% | −16% | −16% | −15% | −9% | −10% | −7% | −8% | |
| H3/H4 | 296 | 324 | −5% | −9% | −7% | −12% | −10% | −11% | −11% | −8% | −6% | −6% | −5% | −4% | |
| H3 | 299 | 319 | −8% | −6% | −6% | −9% | −7% | −8% | −8% | −5% | −4% | −4% | −4% | −1% | |
| H3/H4 | 299 | 324 | −6% | −5% | −3% | −6% | −5% | −6% | −5% | −5% | −2% | −3% | −3% | −2% | |
| S1S2 | 345 | 362 | −13% | −18% | −14% | −19% | −17% | −19% | −19% | −14% | −14% | −12% | −10% | −3% | |
| S1S2 | 355 | 362 | −16% | −22% | −16% | −23% | −21% | −24% | −21% | −16% | −12% | −14% | −11% | −5% | ≤−20% |
| LOOP/H10 | 416 | 437 | −3% | −5% | −4% | −6% | −4% | −6% | −5% | −5% | −3% | −3% | −3% | −2% | ≤−10% |
| LOOP/H10 | 416 | 438 | −3% | −5% | −3% | −7% | −6% | −7% | −7% | −5% | −3% | −4% | −3% | −3% | <−5% |
| H10/H11 | 433 | 458 | −8% | −11% | −7% | −12% | −10% | −12% | −9% | −9% | −6% | −6% | −5% | −2% | ≤5% |
| H10/H11 | 435 | 458 | −9% | −11% | −8% | −12% | −11% | −14% | −13% | −9% | −7% | −8% | −6% | −2% | |
| H10/H11 | 437 | 458 | −8% | −16% | −8% | −16% | −14% | −19% | −16% | −12% | −10% | −11% | −8% | −7% | |
| H10/H11 | 438 | 453 | −10% | −17% | −10% | −17% | −14% | −19% | −15% | −12% | −8% | −10% | −7% | −6% | |
| H10/H11 | 438 | 455 | −10% | −13% | −7% | −11% | −9% | −12% | −10% | −8% | −6% | −9% | −5% | * | |
| H10/H11# | 439 | 455 | −15% | −14% | −9% | −12% | −13% | −16% | −12% | −9% | −5% | −7% | −6% | −5% | |
| H10/H11# | 439 | 455 | −9% | −15% | −11% | −17% | −15% | −16% | −13% | −10% | −7% | −8% | −7% | −3% | |
| H11 | 462 | 479 | −12% | −17% | −9% | −16% | −11% | −18% | −11% | −12% | −6% | −10% | −6% | −5% | |
| H12 | 492 | 497 | 0% | −1% | −2% | −2% | −2% | −2% | −3% | 1% | −1% | −1% | −2% | −4% | |
| H12 | 492 | 502 | −1% | 2% | 2% | 1% | 1% | 3% | 2% | 3% | 0% | −1% | −2% | −3% | |
| Ki(nM) Rα | 0.16 | 0.20 | 0.37 | 0.09 | 3.00 | 0.34 | 0.65 | 0.67 | 0.44 | 0.21 | 0.32 | 2.60 | |||
| Ki(nM) Rβ | 0.19 | 0.25 | 2.74 | 0.08 | 2.8 | 0.48 | 5.10 | 1.47 | 2.40 | 0.28 | 1.30 | 0.30 | |||
| ERβ Agonist EC50 nM; (% stim) |
0.12 (100) | - | - |
0.14 (97) |
>1000 0 |
>10000 | - |
232 (104) |
>10000 | >10000 | >10000 | - | |||
| ERβ Antag IC50 (% inhib.) |
NA | - | - |
>1000 0 |
12 (109) |
3.62 (103) |
- |
>1000 0 |
202 (112) |
15.6 (114) |
204 (108) |
- | |||
The residue numbers of analyzed peptides relative to the full length ERβ have been tabulated. The percentage numbers listed under each compound name demonstrate the averaged difference in deuteration level of that corresponding peptide in the absence and presence of a ligand (The average calculation is based on five HDX experiments with different exchange times: 1, 30, 60, 900 and 4200 sec). We only consider a change of deuteration level greater than 5% to be significant based on the precision of the platform. A negative percentage represents an increase in protection to exchange in that region of the receptor in the presence of ligand, which indicates the region has been protected when bound with ligand.
The same peptide was detected with different charge states.
Peptide not detected. For details of calculation of Ki IC50 and EC50, see MATERIALS AND METHODS.
Figure 3.
HDX analysis of ER ligands. The percentage numbers on the Y axis demonstrate the averaged difference in deuteration level of that corresponding peptide in the absence and presence of a ligand (The average calculation is based on five HDX experiments with different exchange times: 1, 30, 60, 900 and 4200 sec). A) HDX protection (%) of ERβLBD upon binding with 4-hydroxytamoxifen, E2 and genistein against secondary structures. B) HDX protection (%) of ERαLBD upon binding with 4-hydroxytamoxifen, E2 and genistein against secondary structures. The error bars represent the standard deviation of triplicate experiments.
Figure S1 in the supporting information shows the underlying percent deuterium incorporation (%D) vs. Log time plots for three regions of ERβLBD (282–294, 492–502, 355–362) in the presence, and absence, of estradiol and 4-hydroxytamoxifen. The deuterium incorporation vs. log time plots were typical of all peptides measured in this study. ERβLBD showed differential HDX protection in a ligand-dependent and region-specific manner. For example, the β-sheet1/β-sheet2 region (amino acid 355–362) was one region that demonstrated the most statistically significant (p<0.001) differential HDX (i.e., ~ 20% protection to exchange for 4-hydroxytamoxifen) depending on ligand (Figure S1 C and F). Other regions of the binding pocket, such as helix 12 (Figure S1 B and E), in the timeframe tested have no observable protection to exchange following binding of the ligands studied here; an interesting observation which is consistent with our ERαLBD data. Interestingly, the phytoestrogen genistein shows an HDX profile that does not resemble any of the agonists, or SERMs, and provides minimal stabilization to exchange for the ERβLBD.
Comparison of HDX data with atomic resolution structure
The data from HDX analysis (Table 1) was overlaid onto the X-ray structures of ERβLBD/4-hydroxytamoxifen. There are three HDX profiles overlaid, which include ERβLBD/E2 (Figure 4A, HDX data for ERβLBD/E2 is overlaid on the crystal structure for ERβLBD/4-hydroxytamoxifen, PDB: 2FSZ), ERβLBD/4-hydroxytamoxifen (Figure 4B, PDB: 2FSZ), and ERβLBD/genistein (Figure 4C, PDB:1QKM). In the ERβLBD/4-hydroxytamoxifen complex, the β-sheet1/β-sheet2 region (Amino acid 355–362) demonstrated the greatest degree of stabilization, which is consistent with data for the ERαLBD (20). Note that F356 supplies an aromatic π-edge-face interaction with the hydroxyaryl group that is a ubiquitous feature of non-steroidal ER ligands (14). This very important interaction is nearly at a 180° angle from the trialkyl amine substituents that characterize SERM structures. Further studies such as mutagenesis of F356 (e.g. F356A) coupled with HDX analysis of the mutant protein will help to investigate the role of F356. However, these studies are beyond the scope of our current study. Helix 4, which covers the second binding site of 4-hydroxytamoxifen residing in the coactivator binding groove was also stabilized. However, the corresponding peptide spanning this same region in the ERαLBD/4-hydroxytamoxifen complex was not detected with sufficient S/N; therefore a direct comparison is not available. The helix 5 region was detected in the ERαLBD experiments, but showed no protection to exchange.
Figure 4.
HDX profile overlaid onto ERβLBD crystal structures. A) HDX profile of ERβLBD/E2 overlaid onto ERβLBD crystal structure of 4-hydroxytamoxifen since there is no ERβLBD/E2 X-ray data available (PDB:2FSZ), B) HDX profile of ERβLBD/4-hydroxytamoxifen overlaid onto ERβLBD crystal structure of 4-hydroxytamoxifen (PDB:2FSZ), C) HDX profile of ERβLBD/genistein overlaid onto ERβLBD crystal structure of genistein (PDB:1QKM). The color legend shows the deuterium incorporation difference by subtracting deuterium incorporation content of holo ER from apo ER. The regions in the crystal structure that are colored as white belong to peptides that are not detected after pepsin digestion or cannot be measured accurately in the HDX experiments due to co-elution problems. The ligands of 4-hydroxytamoxifen and genistein are shown by space filling.
Statistical analysis of the differential HDX data
In a manner similar to our HDX fingerprint analysis of ERα, a cluster analysis was performed on the twelve ER ligands in the study for which the pharmacological properties are well characterized (20). Cluster analysis was performed for 12 compounds with MultiExperiment Viewer (TM4, V4.0)(26). Different cluster methods including average linkage, complete linkage, and single linkage cluster were used, and all rendered similar results. As shown in Figure 5, two major clusters (A and B) were observed. In addition, a unique HDX profile was observed for genistein. Cluster A includes compounds ICI18278, 4-hydroxytamoxifen, DES, lasofoxifene, and bazedoxifene while cluster B includes 17β-estradiol, raloxifene and the four benzothiophenes; LY88074, LY165176, LY156681 and LY117018. We found that the ligands in cluster A induced a greater overall protection to exchange within the receptor-ligand complex. The β-sheet1/β-sheet2 region experienced the most significant protection to HDX, a pattern consistent across both clusters. Furthermore, ligands in cluster A exhibit a greater protection to exchange in the H10/H11 region than ligands from cluster B. Across both clusters, a rank order of β-sheet1/β-sheet2 region > H10/H11 > the loop region is observed for those areas with >5% protection. Interestingly, genistein represented a unique HDX profile which may suggest a novel mechanism of interaction with the receptor. Genistein is by far the smallest ligand in the set with a water accessible molecular volume of 828 Å3 compared to estradiol at 923 Å3.
Figure 5.
Cluster analysis of twelve ER ligands. The names of the compound are shown on the top of the bar view and peptide regions that have been used for cluster analysis is shown on the right of the bar view. The deuterium incorporation differences of these peptides have been treated as independent variables and each compound has been treated as dependent variables in the cluster analysis. The color represents the differential deuterium level of each peptide in the absence and presence of the compound. With genistin clustered as a single group, we name the other two clusters as cluster A and cluster B. Cluster A includes ICI, 4-hydroxytamoxifin, DES, lasofoxifene and bazodoxifene. Cluster B includes estradiol, raloxifene, LY 88074, LY 165176, LY 156681, and LY 117018.
HDX Dynamics and Cellular Activity
To investigate the relationship between HDX and the intrinsic cellular function, we evaluated the activity of a subset of ER ligands in order to assess whether ligand-induced protection to exchange resulted in differences in their intrinsic pharmacological properties at ERβ. Ligands were evaluated for their ability to act as agonists or antagonist of estrogen in HEK293 cells co-transfected with human full-length ERβ. As shown at the bottom of Table 1, 17β-estradiol and DES are potent agonist with EC50 values of 0.12 and 0.14 nM, respectively, while ICI is an antagonist with an IC50 of 12 nM. For the benzothiophenes, LY88074 is an agonist (EC50 = 232 nM) with no antagonist activity (antagonist activity defined as EC50 >10 µM) while LY156681, 165176, and LY117108 are ERβ antagonist with no agonist activity (EC50 >10 µM). Thus, compounds with similar HDX profiles, such as DES and ICI, have divergent agonist/antagonist pharmacology in HEK293 cells. Likewise, the benzothiophenes and 17β-estradiol are clustered together based on HDX profile but have divergent functional activity. Taken together, this data indicates that ligand-induced protection to deuterium exchange does not indicate whether a given ligand is an either agonist or antagonist in the environment provided in these HEK cotransfection experiments. This data further supports that the cellular context is a critical component of functional activity. A similar observation was made for ERα in which ligand-dependent HDX protection was more predictive of cellular function in uterine cells (Ishikawa) than breast (MCF-7) cells (20).
DISCUSSION
Comparison of the ligand induced changes to HDX dynamics of ERαLBD and ERβLBD
Under HDX conditions, only including peptide ions with sufficient S/N in all on-exchange time points, the sequence coverage for ERβ was 60% (Table 1) whereas the sequence coverage for ERα was 66% as described in our earlier report (20). This difference in sequence coverage limited a direct comparison of HDX kinetics for all regions of both receptors. Fortunately, most regions of the LBD involved in ligand interaction were observed for both isoforms including helix 2, the LOOP region between helix 2 and 3, helix 3, the β-sheet1/β-sheet2 region, helix 10, helix 11 and helix 12. The loop region between helix 2 and helix 3, the β-sheet1/β-sheet2 region and the helix 10/helix 11 regions in ERβLBD are among the most dynamic regions (Figure 1) and are important for the receptor-ligand interaction based upon the X-ray structures of both receptors (10–11). Surprisingly, the HDX kinetics of helix 12 are not altered upon ligand binding (Table 1) for either of the ER isoforms. This observation is consistent with Yan et al (27) who found that helix 12 in RXRα LBD has the same HDX rate in the agonist- and antagonist-bound receptor as the apo protein. It would be expected that the HDX characteristics of the receptor reflect the context of the experiment, and changes to the dynamics of helix 12 may only be observed when in the presence of coregulatory proteins, presence of the F domain, presence of DNA, or requiring full length receptor. However, it is important to note that in HDX studies on the LBD of PPARγ, ligand-specific changes in helix 12 dynamics were detected in the absence of both coregulatory proteins and DNA (18–19). Regardless, when comparing the HDX behavior of ligands in the present study, the experimental context of the receptor is the same for ERαand ERβ.
Important differences were observed in the differential HDX behavior of ERβLBD as compared to ERαLBD upon ligand interaction. The most acute changes occur with 17β-estradiol which significantly stabilizes the LBD of ERβ but has little effect on ERα LBD. Greater protection to exchange to ERβ than ERα was also observed for the ligands DES, LY88074, and ICI18278. A noteworthy observation was that compounds within the benzothiophene chemical scaffold that includes raloxifene, LY88074, LY165176, LY156681, and LY117018 demonstrate similar HDX profiles upon binding with ERβLBD. This is very different from results obtained with ERαLBD (20) in which subtle structural changes within this same group of benzothiophenes resulted in distinct HDX signatures. From a structure-activity standpoint, the presence of a basic side chain appears to play little role in stabilizing ERβLBD, i.e., the fingerprint for LY88074, which lacks this pharmacophore, is similar to those ligands that contain a basic nitrogen (LY156681, LY165176, LY117018, and raloxifene). This is distinct from the HDX fingerprints for ERαLBD which readily distinguished those ligands including very subtle structural differences in the positioning of this tertiary base (20).
The HDX kinetics of helix 3 was particularly affected. This helix, composed of amino acids 343–360 in ERα and 296–309 in ERβ, contains the aspartic acid that provides the contact point between the tertiary amine of SERMs and helix 3. For ERα, this additional contact point may play a role in stabilizing this helix relative to apo receptor which was reflected in a protection against HDX exchange that ranges from 13 % for 4-hydroxytamoxifen to <5% for 17β-estradiol (20). For ERβ, such a broad range of protection was not observed, i.e., helix 3 is equivalently protected from exchange regardless of the presence or absence of a basic side chain in the ligand. For example, 17β-estradiol, which lacks this pharmacophore, provides 12% stabilization of helix 3 which is similar to SERMS such as 4-hydroxytamoxifen (13% protection) and raloxifene (12% protection). Thus, the nature of the ligand plays a lesser role in ERβ than ERα in the protection of helix 3 to H/D exchange. The reasons for this difference are not clear. When comparing the two ER isoforms, the ligand binding domains share 53% sequence identity (1). However, in the binding cavity there are only two relevant amino acid substitutions, Met421 (ERα) to Ile373 (ERβ) and Leu384 (ERα) to Met336 (ERβ) (11,13) It is intriguing to speculate that the ligand-induced protection that was generally observed for helix 3 in the ERβ-ligand complex was a consequence of enhanced van der Waal interactions between helix 3 and helix 5 as a result of differential interactions between the ligand and the Leu-> Met substitution on helix 5. This phenomenon has been shown to play a critical role in steroid hormone receptor function (28). However more extensive work needs to be done to explore this hypothesis. Regardless, the distinct HDX dynamics we have observed between the two receptor isoforms suggest distinct receptor conformational changes that may lead to unique patterns of coregulator interaction.
ERβ interaction with SERMs, ER Agonists, and ER Antagonists
Cluster analysis of the HDX data for ER ligands including SERMs, agonists, and antagonists is presented in Figure 5. 4-Hydroxytamoxifen, lasofoxifene and bazedoxifene cluster as one sub-group, having larger stabilization effects on ERβLBD than raloxifene. Interestingly, raloxifene together with the other benzothiophene analogs LY88074, LY165176, LY156681, and LY117018 share the same HDX signature demonstrating stabilization within the loop region between helix 2 and helix 3, and the β-sheet1/β-sheet2 region, and the H10/H11 region suggesting that there is a unique interaction between ERβ and the sulfur-containing benzothiophene nucleus. In fact, Phe356 resides in this region of the protein and it may interact with the polarizable sulfur in the B-ring of the benzothiophenes.
Protection to exchange between raloxifene and ERβLBD is weaker than that observed for the ERβLBD/4-hydroxytamoxifen complex, which is consistent with that observed in ERαLBD. A study by Wang et al (14) discovered a second binding site for 4-hydroxytamoxifen in ERβLBD which could explain the magnitude of perturbation in HDX kinetics observed in this study when receptor is bound with 4-hydroxytamoxifen (Figure 4B). This second binding site was located within the coactivator-binding groove of ERβLBD that involves helix 3, helix 4, helix 5 and helix 12. The second binding site could contribute to the stronger interaction in the ERβLBD/4-hydroxytamoxifen complex if there is only one binding site in the ERβLBD/raloxifene complex.
The ERβLBD shows significant change upon binding the agonist ligands estradiol and DES. There is greater stabilization in the ERβLBD/agonist complex versus that of the ERαLBD. It was observed in the HDX analysis of ERαLBD, most regions experience stabilization less than 5% (Table 1 in Reference (20)) while in ERβLBD, most of the regions experience stabilization more than 5% (Table 1). Interestingly, the same observation was obtained for the antagonist ICI 182780 where greater protection is observed when this molecule is bound to ERβ than to ERα.
ER interaction with phytoestrogen genistein
Genistein is a well studied phytoestrogen found in soy products and reported to be protective against breast and prostate cancer (29). It binds both receptors with moderate affinity but has a preference for ERβ (Table 1). It was also discovered that genistein is an ERα agonist but a ERβ partial agonist (30) which may suggest a different regulatory mechanism for coactivator recruitments preference between the two receptors. The ERβLBD/genistein complex has a unique HDX profile among all the compounds that have been analyzed with the least HDX protection. For example, the β-sheet1/β-sheet2 region was stabilized in all ERβLBD/ligand complexes with the exception of the genistein complex. The lack of significant HDX stabilization of ERβLBD upon binding genistein suggests little change in conformational dynamics of the complex and is consistent with the positioning of helix 12 observed in the protein crystal structure for genistein in ERβLBD in which the coactivator recognition site is only partially blocked by helix 12 (11).
CONCLUSION
In this report we have analyzed perturbations in HDX dynamics of ERβLBD upon binding 12 ligands. Our results suggest ERβ has a different degree of plasticity in its LBD cavity compared with ERα. The different HDX patterns observed for ERβLBD when compared to ERαLBD serves as an indication that ERβLBD undergoes a different structural response to the same ligand when compared to ERαLBD. This further suggests distinct receptor conformational changes that may lead to unique patterns of coregulator interaction and, ultimately, tissue selective pharmacological outcomes.
Supplementary Material
Acknowledgements
We are grateful for support from Mark Southern for software to analyze the HDX data.
MJC and PRG acknowledge funding from the National Institutes of Health (U54-MH074404 [PI: H. Rosen] and GM084041). The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
Abbreviations and Textual Footnotes
- HDX
Hydrogen deuterium exchange
- ER
Estrogen receptor
- LBD
Ligand binding domain
- E2
Estradiol
- DES
Diethylstilbestrol.
Footnotes
Supporting Information Available. Deuterium incorporation curves of representative peptides for ERβLBD/ligand complex showing deuterium taken-up versus time (Figure S1) and comparison of the average percentage of HDX protection over 5 hydrogen/deuterium exchange time points of ERαLBD/genistein complex to other ERαLBD/agonist complexes (Table S1) are presented. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pearce ST, Jordan VC. The biological role of estrogen receptors alpha and beta in cancer. Crit Rev Oncol Hematol. 2004;50:3–22. doi: 10.1016/j.critrevonc.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Koehler KF, Helguero LA, Haldosen LA, Warner M, Gustafsson JA. Reflections on the discovery and significance of estrogen receptor beta. Endocr Rev. 2005;26:465–478. doi: 10.1210/er.2004-0027. [DOI] [PubMed] [Google Scholar]
- 3.Zhao L, O'Neill K, Diaz Brinton R. Selective estrogen receptor modulators (SERMs) for the brain: current status and remaining challenges for developing NeuroSERMs. Brain Res Brain Res Rev. 2005;49:472–493. doi: 10.1016/j.brainresrev.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Harris HA, Albert LM, Leathurby Y, Malamas MS, Mewshaw RE, Miller CP, Kharode YP, Marzolf J, Komm BS, Winneker RC, Frail DE, Henderson RA, Zhu Y, Keith JC., Jr Evaluation of an estrogen receptor-beta agonist in animal models of human disease. Endocrinology. 2003;144:4241–4249. doi: 10.1210/en.2003-0550. [DOI] [PubMed] [Google Scholar]
- 5.Couse JF, Lindzey J, Grandien K, Gustafsson JA, Korach KS. Tissue distribution and quantitative analysis of estrogen receptor-alpha (ERalpha) and estrogen receptor-beta (ERbeta) messenger ribonucleic acid in the wild-type and ERalpha-knockout mouse. Endocrinology. 1997;138:4613–4621. doi: 10.1210/endo.138.11.5496. [DOI] [PubMed] [Google Scholar]
- 6.Neubauer BL, McNulty AM, Chedid M, Chen K, Goode RL, Johnson MA, Jones CD, Krishnan V, Lynch R, Osborne HE, Graff JR. The selective estrogen receptor modulator trioxifene ( LY133314) inhibits metastasis and extends survival in the PAIII rat prostatic carcinoma model. Cancer Res. 2003;63:6056–6062. [PubMed] [Google Scholar]
- 7.Ali SH, O'Donnell AL, Balu D, Pohl MB, Seyler MJ, Mohamed S, Mousa S, Dandona P. Estrogen receptor-alpha in the inhibition of cancer growth and angiogenesis. Cancer Res. 2000;60:7094–7098. [PubMed] [Google Scholar]
- 8.Harris HA. Estrogen receptor-beta: recent lessons from in vivo studies. Mol Endocrinol. 2007;21:1–13. doi: 10.1210/me.2005-0459. [DOI] [PubMed] [Google Scholar]
- 9.Imamov O, Shim GJ, Warner M, Gustafsson JA. Estrogen receptor beta in health and disease. Biol Reprod. 2005;73:866–871. doi: 10.1095/biolreprod.105.043497. [DOI] [PubMed] [Google Scholar]
- 10.Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 11.Pike AC, Brzozowski AM, Hubbard RE, Bonn T, Thorsell AG, Engstrom O, Ljunggren J, Gustafsson JA, Carlquist M. Structure of the ligand-binding domain of oestrogen receptor beta in the presence of a partial agonist and a full antagonist. Embo J. 1999;18:4608–4618. doi: 10.1093/emboj/18.17.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 13.van Hoorn WP. Identification of a second binding site in the estrogen receptor. J Med Chem. 2002;45:584–589. doi: 10.1021/jm0109661. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Chirgadze NY, Briggs SL, Khan S, Jensen EV, Burris TP. A second binding site for hydroxytamoxifen within the coactivator-binding groove of estrogen receptor beta. Proc Natl Acad Sci U S A. 2006;103:9908–9911. doi: 10.1073/pnas.0510596103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan X, Broderick D, Leid ME, Schimerlik MI, Deinzer ML. Dynamics and ligand-induced solvent accessibility changes in human retinoid X receptor homodimer determined by hydrogen deuterium exchange and mass spectrometry. Biochemistry. 2004;43:909–917. doi: 10.1021/bi030183c. [DOI] [PubMed] [Google Scholar]
- 16.Yan X, Deinzer ML, Schimerlik MI, Broderick D, Leid ME, Dawson MI. Investigation of ligand interactions with human RXRalpha by hydrogen/deuterium exchange and mass spectrometry. J Am Soc Mass Spectrom. 2006;17:1510–1517. doi: 10.1016/j.jasms.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 17.Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2006;78:1005–1014. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- 18.Hamuro Y, Coales SJ, Morrow JA, Molnar KS, Tuske SJ, Southern MR, Griffin PR. Hydrogen/deuterium-exchange (H/D-Ex) of PPARgamma LBD in the presence of various modulators. Protein Sci. 2006;15:1883–1892. doi: 10.1110/ps.062103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruning JB, Chalmers JM, Prasad D, Busby SA, Kamenecka KM, He Y, Nettles KM, Griffin pR. Partial Agonists Activate PPARg Using a Helix 12 Independent Mechanism. Structure. 2007 doi: 10.1016/j.str.2007.07.014. In press. [DOI] [PubMed] [Google Scholar]
- 20.Dai SY, Chalmers MJ, Bruning J, Bramlett KS, Osborne HE, Montrose-Rafizadeh C, Barr RJ, Wang Y, Wang M, Burris PT, Dodge JA, Griffin PR. Prediction of the Tissue-Specificity of Selective Estrogen Receptor Modulators using a Single Biochemical Method. Proc Natl Acad Sci U S A. 2007;105:7171–7176. doi: 10.1073/pnas.0710802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuiper GG, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, Gustafsson JA. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 22.Chalmers MJ, Busby SA, Pascal BD, Southern MR, Griffin PR. A two-stage differential hydrogen deuterium exchange method for the rapid characterization of protein/ligand interactions. J Biomol Tech. 2007;18:194–204. [PMC free article] [PubMed] [Google Scholar]
- 23.Pascal BD, Chalmers MJ, Busby SA, Mader CC, Southern MR, Tsinoremas NF, Griffin PR. The Deuterator: software for the determination of backbone amide deuterium levels from H/D exchange MS data. BMC bioinformatics. 2007;8:156. doi: 10.1186/1471-2105-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. Journal of the American Society for Mass Spectrometry. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 27.Yan X, Perez E, Leid M, Schimerlik MI, de Lera AR, Deinzer ML. Deuterium exchange and mass spectrometry reveal the interaction differences of two synthetic modulators of RXRalpha LBD. Protein Sci. 2007;16:2491–2501. doi: 10.1110/ps.073019707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Simisky J, Tsai FT, Geller DS. A critical role of helix 3-helix 5 interaction in steroid hormone receptor function. Proc Natl Acad Sci U S A. 2005;102:2707–2712. doi: 10.1073/pnas.0409663102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adlercreutz H, Mazur W. Phyto-oestrogens and Western diseases. Ann Med. 1997;29:95–120. doi: 10.3109/07853899709113696. [DOI] [PubMed] [Google Scholar]
- 30.Barkhem T, Carlsson B, Nilsson Y, Enmark E, Gustafsson J, Nilsson S. Differential response of estrogen receptor alpha and estrogen receptor beta to partial estrogen agonists/antagonists. Mol Pharmacol. 1998;54:105–112. doi: 10.1124/mol.54.1.105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.