Abstract
Sequencing of human and other genomes has been at the center of interest in the biomedical field over the past several decades and is now leading toward an era of personalized medicine. During this time, DNA sequencing methods have evolved from the labor intensive slab gel electrophoresis, through automated multicapillary electrophoresis systems using fluorophore labeling with multispectral imaging, to the “next generation” technologies of cyclic array, hybridization based, nanopore and single molecule sequencing. Deciphering the genetic blueprint and follow-up confirmatory sequencing of Homo sapiens and other genomes was only possible by the advent of modern sequencing technologies that was a result of step by step advances with a contribution of academics, medical personnel and instrument companies. While next generation sequencing is moving ahead at break-neck speed, the multicapillary electrophoretic systems played an essential role in the sequencing of the Human Genome, the foundation of the field of genomics. In this prospective, we wish to overview the role of capillary electrophoresis in DNA sequencing based in part of several of our articles in this journal.
1. The early days of slab gel electrophoresis
1.1. Manual read
DNA sequencing has been a main focus of technological development since Nobel laureates Sanger and Gilbert introduced sequencing by chain termination or chemical fragmentation techniques, coupled with gel electrophoresis-based size separation [1, 2]. The key principle of the Sanger method, the use of dideoxynucleotide triphosphates (ddNTPs) as DNA chain terminators, proved to be more efficient, requiring fewer toxic chemicals and lower amounts of radioactivity than that of the Maxam – Gilbert chemical fragmentation method. The classical chain-termination method required a DNA primer (radioactively or fluorescently labeled), a single-stranded template, DNA polymerase enzyme, as well as deoxy- and dideoxy-nucleotides. The template was divided into four aliquots, having all four of the standard deoxynucleotides (dATP, dGTP, dCTP and dTTP) and DNA polymerase. One of the four chain-terminating dideoxynucleotides (ddATP, ddGTP, ddCTP, or ddTTP) was added to each reaction to terminate the DNA strand during the chain elongation reaction. The method resulted in various length DNA fragments which were heat-denatured and size-separated by denaturing polyacrylamide slab gel electrophoresis. After separating each of the four reactions in one of four individual lanes (lanes A, T, G, C), the DNA bands were visualized either by autoradiography or UV light, and the sequence was manually read from the pattern of the four parallel runs.
1.2. Automated read
With further development of the technology, dye labeled primer-based sequencing was introduced, facilitating optical sequence reading [3]. A key advance was introduced by Hood and coworkers [4] in the form of fluorescently labeled ddNTPs that became the basis of the approach used to sequence the Human Genome. Dye-terminator based DNA sequencing allowed the use of four dideoxynucleotide chain terminators, tagged with dyes of different fluorescent emission wavelengths, in a single sequencing reaction. Today, the dye terminating fluorescent tag is still the method of choice in automated long read DNA sequencing. Initial problems with the technique, such as dye effects manifested by unequal peak heights and shapes, were alleviated with the use of modified DNA polymerase enzymes and novel dye chemistries such as energy transfer dyes [5].
2. The advent of capillary gel electrophoresis
Capillary electrophoresis was introduced almost 30 years ago by Jorgensen as a new and automated alternative to slab gel electrophoresis [6]. As early as 1989, Bob Brownlee and coworkers introduced the first commercial instrument with on column UV/VIS detection, automatic injection and computerized data analysis for rapid, high-resolution CE separation [7]. Since that time, others followed the lead in manufacturing commercial capillary electrophoresis units, including several companies developing capillary-based DNA sequencers.
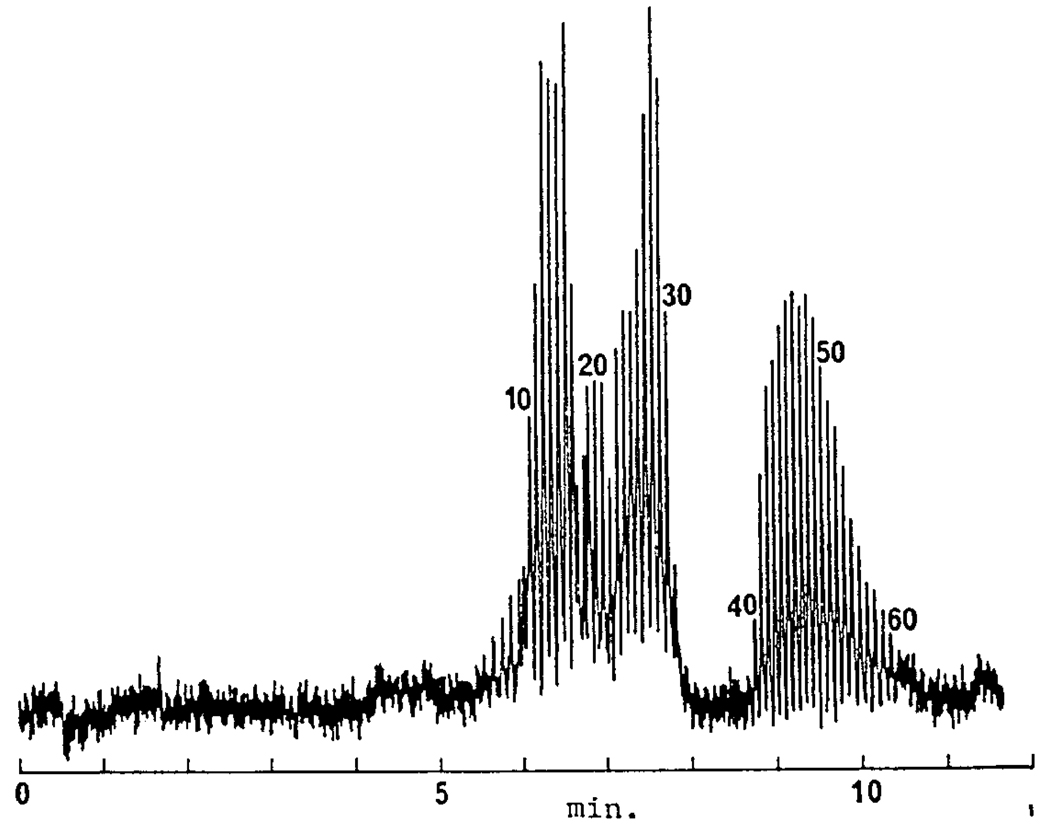
Our laboratory published the first separations of single stranded oligonucleotides by high performance capillary gel electrophoresis in a conference proceeding issue published by Electrophoresis [8]. Picomole amounts of polydeoxyadenylic acids, p(dA)10–30,40–60 were baseline resolved in less than 11 min by capillary gel electrophoresis using 7.5% T / 3.3% C polyacrylamide matrix, as shown in Figure 1.
Figure 1.
Capillary gel electrophoresis separation of polydeoxyadenylic acids, p(dA)10–30,40–60. Capillary: 270 × 0.075 mm i.d.; running buffer: 0.1 M Tris/0.25 M borate/7 M urea, pH 8.3, with crosslinked polyacrylamide gel: 7.5% T and 3.3% C. The applied electric field was 400 V/cm. With permission from [8].
These crosslinked polyacrylamide filled capillary columns were among the highest efficiencies achieved in the early days of DNA analysis with as high as 30,000,000 theoretical plates per meter [9]. Based on these results and others, our group was among the first to recognize the significant potential that existed for sequencing of oligonucleotides with gel filled capillary columns [10]. This fundamental work implied that capillary gel electrophoresis might fulfill the high resolution separation requirement of DNA sequencing by the Sanger approach, and indeed several groups almost immediately started working in the field. The striking increase in separation speed (5-fold) and efficiency capillary gel electrophoresis offered was demonstrated by a comparison with conventional polyacrylamide slab gel electrophoresis for the separation of a fluorescein-labeled C-reaction of M13mp19 DNA [11].
However, fabrication of cross-linked gel filled capillaries was difficult with apparent robustness issues, mainly manifested by bubble formation. Attempts to overcome this problem included optimization of the polymerization temperature, light exposure and pressure assisted polymerization. Swerdlow and Gesteland [12] emphasized that even solving the problem of bubble formation during sieving matrix polymerization, bubbles could also be formed during the electric field mediated separation process. These bubbles usually formed near the injection end of the capillary and in most instances seriously impaired separation performance. As we later showed, the bubbles were due to the osmotic shock, which occurred when the high salt concentration sample plug migrated into the capillary [13]. The low flexibility of the covalently cross-linked polymer could not withstand the resulting extensive Joule heat, and the gel collapsed with concomitant bubble formation, eventually leading to loss of electric contact.
2.1. Linear polymer matrices and replaceable gel technology
The stability problems of cross-linked polyacrylamide gels within microbore columns initiated a rapid development to find novel, more robust sieving matrices. One of the first attempts was the use of high concentration non-crosslinked linear polyacrylamide (LPA, T = 6–12%), demonstrated by our laboratory in 1990 [14]. Based on earlier publications on the use of non cross-linked polymer matrices for size separation of biopolymers in conventional gel electrophoresis [15], other linear polymers such as, cellulose derivatives [16] and polyethylene oxides [17] were evaluated in capillary columns for single and double stranded DNA fragment analysis. It was apparent that non-cross linked polymer matrices (physical gels) were capable to alleviate both temperature and high salt related sample injection problems.
However, it was clear that if capillary electrophoresis were to be used to sequence the Human Genome, a means had to be found for automatic replenishment of the polymer matrix in the separation capillary, i.e., no geneticist was interested in reusing a polymer matrix that had already been employed for the separation of Sanger sequencing fragments. It was also not a viable alternative to change capillaries after each run. In the early 90’s, our group demonstrated that, through the use of polymer solutions of linear polyacrylamide and high pressure, it was possible to automatically replace the polymer after each run [18] and still maintain the high separation efficiency for DNA sequencing [19]. Introduction of replaceable linear polymer matrices in DNA sequencing was a very important step toward the development of automated large scale DNA sequencing by capillary electrophoresis and opened up the horizon to design and implement automated 24/7 instruments capable to sequence the Human Genome within a reasonable time frame.
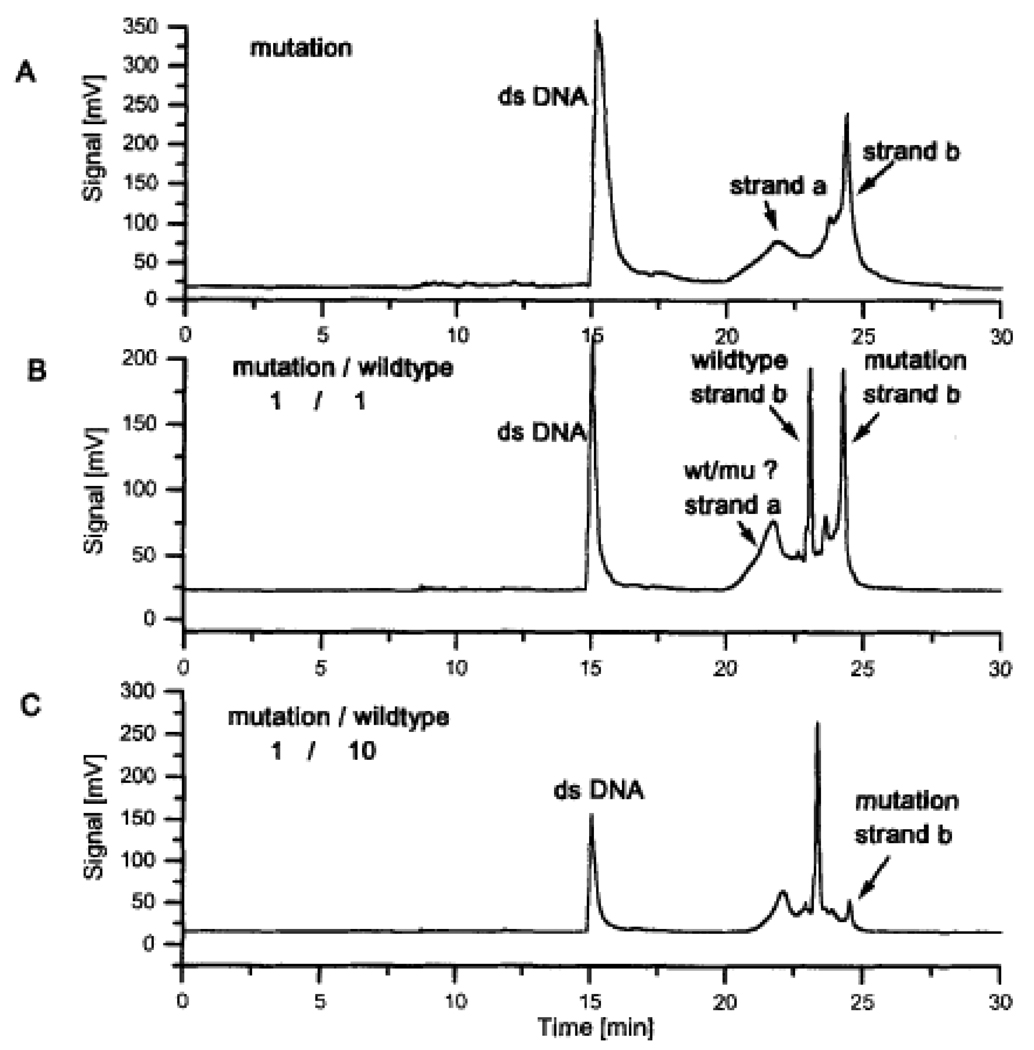
These novel replaceable polymer matrices were not only used in DNA sequencing, but also proved useful in high resolution ssDNA, dsDNA and RNA separations, as well as mutation analysis and large scale purity check of PCR/sequencing primers [20]. An interesting application is shown in Figure 2, published by our group in the mid 1990’s in this journal using replaceable polymer matrices in conjunction with capillary electrophoresis for single strand conformational polymorphism with two-dye laser-induced fluorescence detection [21] demonstrating that the slower migrating strands of the wild type and the mutation had different migration times and could thus be identified in a mixture of the two samples. The method was applied to the detection of different SNPs and offered a rapid and reliable alternative to DNA sequencing. Today, such tests are being done by microarray techniques.
Figure 2.
Comparison of the migration behavior of wild-type and mutated DNA strands of lucl gene. With permission from [21].
The use of replaceable polymers in most instances required elimination of electroosmotic flow, characteristic of fused silica capillaries used with basic buffer systems that is a typical separation condition of DNA sequencing fragments. In these cases a coating of the inner surface of the capillary was necessary to mask the negatively charged fused silica material. Both covalent [22] and non-covalent (dynamic) [23] coatings have been reported to address the problem.
2.2. Multicapillary sequencing
After the first successful attempts in 1988 that proved the potential of capillary electrophoresis in DNA sequencing, it was soon anticipated that single column operation would not satisfy general sequencing needs with higher throughput requirements, such as for the Human Genome Project where 3 billion bases needed to be sequenced. To solve this problem, Mathies et al. [24] developed the approach of multiplexing the separation of DNA sequencing fragments by means of the use of arrays of capillaries, This was a significant and necessary development in capillary based DNA sequencing, leading to a 96-column format as the method of choice for production-scale work. The capillary array systems also required sensitive detection settings applicable to bundles of capillaries. Dovichi and coworkers [25] introduced a high sensitivity post-column laser-induced fluorescence detector with a sheath flow cuvette to minimize the background signal originating from the light scatter of the gel filled capillary. They attained mass detection limits of 10–20 fluorescein-labeled DNA fragment molecules. Later, Yeung’s group [26] developed an axial-beam excitation scheme for capillary electrophoresis using line focusing excitation geometry to simultaneously monitor bundles of capillary tubes. The CCD camera looked at all capillaries at all times, with data rates sufficiently fast for sequencing at >1 base / s / lane. Today, automated multicapillary DNA-sequencing instruments can handle up to hundreds of DNA samples in continuous operation with good sensitivity for long read lengths.
2.3. Electrophoresis microchips
The advent of microfabricated separation devices offered new opportunities in multiplexing the Sanger sequencing scheme [27]. The progress made in this field was quite significant not only in multiplexing but also with system integration, including steps of sample preparation, PCR amplification, template purification, cycle sequencing reaction, injection and separation of the sequencing reaction fragments. As of today, feasibility of most of the individual unit operation parts has been demonstrated opening up the opportunity for a comprehensive handheld DNA analysis device [28].
3. Beyond 1000 bases and above
In the effort of producing longer and longer read lengths, the choice of the appropriate electrophoresis parameters (e.g., separation voltage, temperature, etc.) for the analysis of DNA sequencing fragments played an important role to obtain highly efficient separations. Influence of the separation temperature and sieving matrix composition were considered to be key elements to achieve this goal and studied extensively in the 90’s.
3.1. Effect of temperature
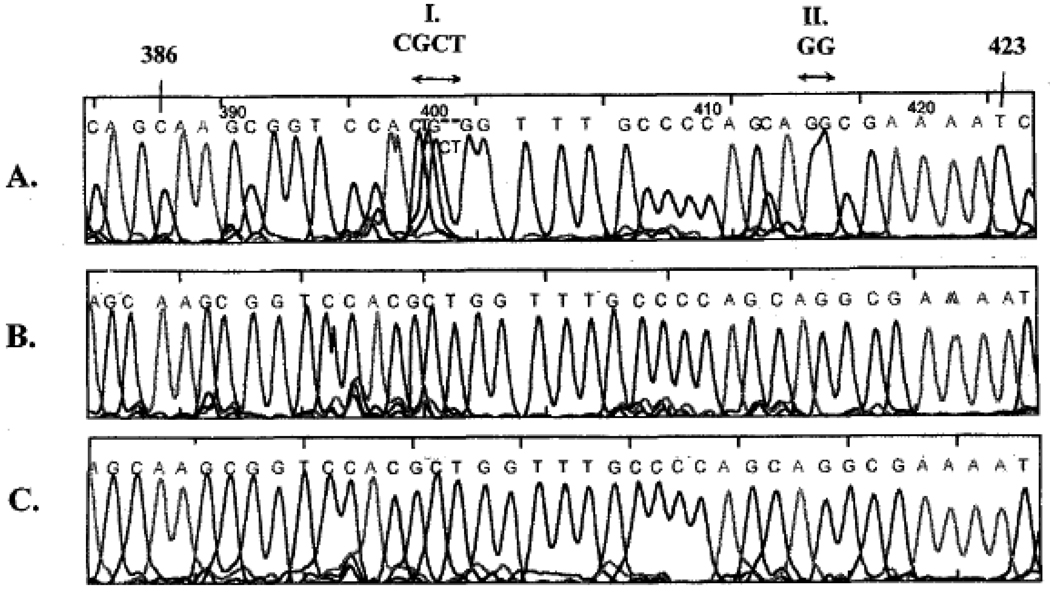
At the beginning, DNA sequencing read lengths with the use of replaceable polymers in capillary columns were only up to around 300 bases. Longer fragments migrated through the gel matrix at a similar rate because of reptation [29] in which, high electric fields DNA molecules become oriented because the field biases the direction of the leading end of the chain as it moves in a snake like motion through the sieving polymer. This phenomenon causes a decrease in size-based electrophoretic mobility differences and ultimately a broad band consisting non-separated fragments at the end of the separation. To increase the read length, reptation must be relaxed by, e.g., increasing the capillary column temperature. Our laboratory reported in this journal that capillary electrophoresis with a replaceable linear polyacrylamide matrix operated at elevated column temperatures of 55 and 60°C extended the separation of DNA sequencing fragments to lengths greater than 800 bases [30]. Figure 3 shows the effect of capillary temperature and denaturants on resolution of two compression regions of the M13mp18 sequence from base 386 to 423. At room temperature, neither of these sites were resolved using the 3% w/v, 30% formamide, 3.5 M urea separation matrix. Separations performed at 55°C, whether with the above buffer in the polymer matrix or 7 M urea only, resolved these regions completely and enabled unambiguous base calling. Through a series of advances, our laboratory reported the first DNA sequencing separation that went beyond 1000 bases [31]. Later we demonstrated that temperatures as high as 60°C could be successfully applied to reach sequencing speeds of more than 1000 bases per hour [32].
Figure 3.
The effect of capillary temperature and denaturants on resolution of two compression regions (I and II) in the M13mp18 sequence from base 386 to 423. (A) 25°C, 3.5 M urea, 30% formamide; (B) 50°C, 7 M urea; (C) 50°C, 3.5 M urea, 30 % formamide. The sieving matrix was 3% w/v LPA in 50 mM Tris-TAPS. With permission from [30].
3.2. New sieving matrices
A second way to improve separation performance of DNA sequencing fragments was to increase the chain length of the sieving polymer that resulted in long read lengths (>1000 bases) with high selectivities (Δμ/μaverage > 0.001, where μ is the electrophoretic mobility) [31]. Note that polymer solutions are non-Newtonian fluids, i.e., their viscosities are strongly dependent on the shear forces applied to the liquid. Indeed, the high viscosity of these linear polymers dropped exponentially with the increasing pressure applied to fill the capillaries, practically necessitating only around 1000 psi for sieving polymer replacement. Thus, even very long chain linear polymers are replaceable and can be routinely used for DNA sequencing.
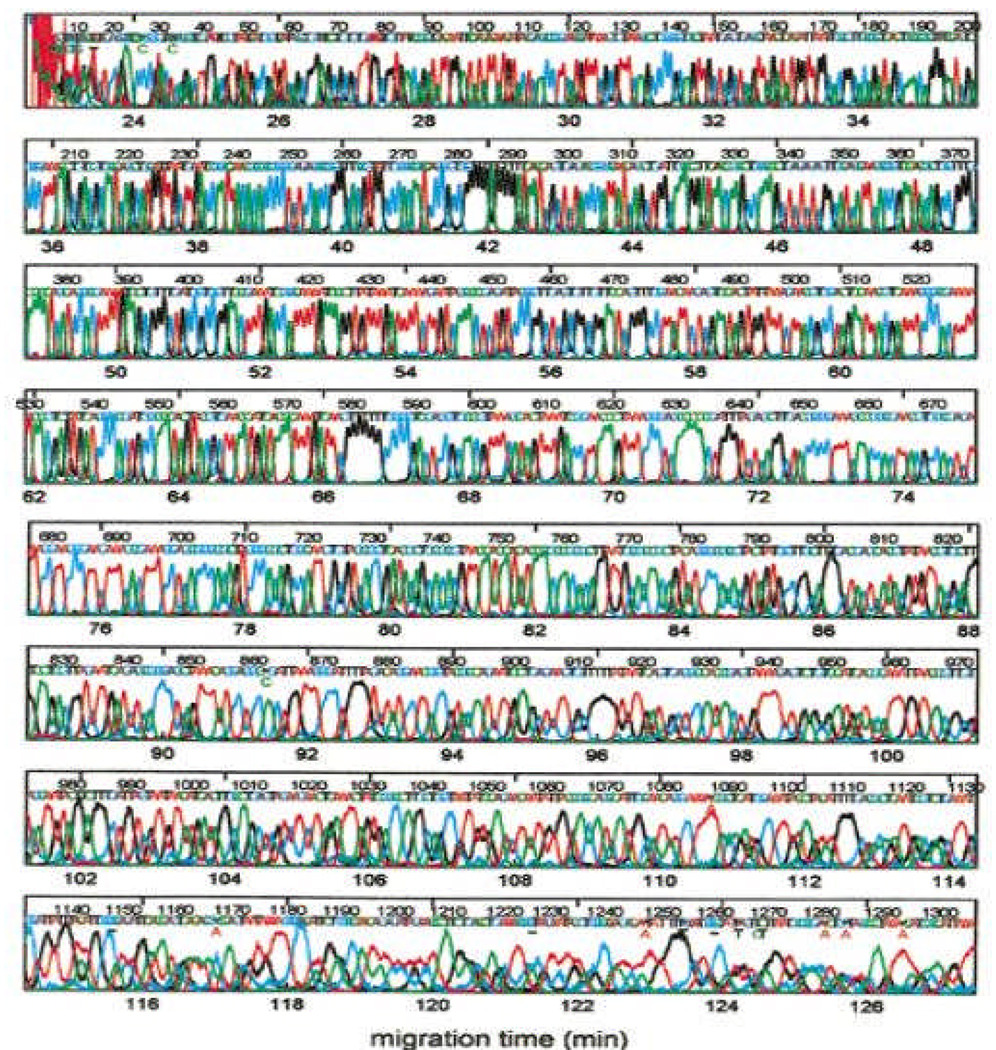
Inverse emulsion polymerization was reported by our laboratory in this journal to be a convenient method to prepare high molecular mass linear polyacrylamide (9 MDa) [33]. The dual liquid phase synthesis system allowed reliable control of the polymer molecular mass, leading to a highly reproducible manufacturing process. By the dawn of the new millennium, our laboratory attained 1300 base read lengths for mp18 templates (Figure 4) by applying 70°C separation temperature with the optimized polymer matrices and electric field strengths. Novel dye chemistries (energy transfer dyes [5]) improved separation performance and advanced base calling algorithms helped decipher the sequence information from highly overlapped peaks [34].
Figure 4.
Sequencing to 1300 bases at 70°C using high molecular mass polymer matrix, optimized electric field and base calling software. With permission from [35].
Besides of the importance of the high-resolution capillary separation for single stranded Sanger DNA sequencing fragments, contributions from other laboratories were essential for the success of the Human Genome Project. The works of Mathies [36], Yeung [37], Dovichi [38] and Kambara [39] were crucial to the development of the automated system. Instrument companies such as Applied BioSystems, Molecular Dynamics, SpectraMedix, Beckman and LiCor also contributed significantly to the development of automated DNA sequencers. On the base calling software side the input of Green [40] should be recognized.
4. Next generation DNA sequencing
Because of the high importance of de novo sequencing and re-sequencing efforts, new technologies have been emerging to replace the currently used multicapillary format [41], promising orders of magnitude less expensive sequencing costs per genome with the goal of $100,000 in 2009 and $1,000 in 2014 [42]. Attempts in cyclic array sequencing, sequencing by hybridization/ligation, nanopore sequencing and the single molecular approach are in the center of recent developments [43]. Companies behind next generation sequencing technology developments are 454 Life Sciences, Applied Biosystems, Danaher Motion Co, Helicos Biosciences, Illumna, Oxford Nanopore Technologies and Pacific Biosciences.
Cyclic-array sequencing was first implemented in 2005 by Shendure et al. [44] as an alternative sequencing strategy. The process starts with a library preparation by random DNA fragmentation followed by ligation of common adaptor sequences. The clonally clustered amplicons are spatially gathered on a planar substrate or on microbeads, both are amenable to emulsion amplification i.e., template fragments are segregated in tiny aqueous droplets of the emulsion and amplified by PCR in isolation. Alternative cycles of enzyme catalyzed elongation reaction and spectral imaging generate the sequence information, utilizing the sequencing by synthesis phenomenon. As the entire array is interrogated after each cycle, this approach provides a significantly higher degree of multiplexing than that of capillary based sequencing. In the approach of 454 sequencing [45], PCR-based cloning and pyrosequencing [46] technology avoids the requirement to use cloning vectors. Bridge PCR based sequencing [47] first generates clonal libraries while both forward and reversed primers are immobilized onto a solid planar substrate via flexible linkers. Each cycle of extension and denaturation is followed by interrogation of the result of the single base extension. Another approach is sequencing by hybridization in which, the differential hybridization of labeled nucleic acid fragments to an array of oligonucleotide probes can identify variant positions [48]. The platform utilizes clonal sequencing in emulsion PCR. The method of single molecule sequencing is based on the preparation of hundreds of millions of poly(dA) possessing single stranded DNA templates, which are hybridized to tethered poly(dT) primers on a glass substrate [49]. This hybridized primer – template pair readily supports sequencing. Finally, nanopore sequencing is a label free single molecule system utilizing direct electrical identification of DNA bases for massively parallel, high throughput sequencing [50].
5. Conclusions
In the early days of capillary electrophoresis the hope was to replace some of the slab gel and LC based methods by CE. Genomics provided the killer application for the technique, as both DNA sequencing and DNA analysis (ssDNA, dsDNA, mutation, primers, etc) showed superior performance in capillary gel electrophoresis. Currently used routine Sanger based methods in conjunction with multicapillary gel electrophoresis can directly sequence up to 1000 nucleotides in length in a single reaction. The main obstacle to sequencing DNA fragments above this size limit is insufficient separation power to resolve larger fragments that differ in length by only one nucleotide. While efforts to improvement in sieving matrix composition continue to increase the separation power and speed of electric field mediated separation of DNA sequencing fragments [41], next generation sequencing approaches are on the horizon to address large scale applications, in which the massively parallel approaches have definite cost advantages [42]. For the future of personalized medicine, especially for multifactorial diseases, everyone’s genome will be sequenced using a “sample in / sequence out” method with a likely cost of several hundred dollars per genome.
Recently, capillary electrophoresis has found a new application in the analysis of complex carbohydrates and in the increased associated activities of CE-MS techniques in the biotechnology and biopharmaceutical industry. The high resolving power of capillary electrophoresis and its hyphenation with MS offers a good alternative to currently used separation methods for the analysis of this important class of biopolymers with the hope to decipher the glycome.
Acknowledgment
The author acknowledge the financial support of NIH GM15847. Contribution number 937 of the Barnett Institute.
References
- 1.Gilbert W, Maxam A. Proc Natl Acad Sci U S A. 1973;70:3581–3584. doi: 10.1073/pnas.70.12.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanger F. Proc R Soc Lond B Biol Sci. 1975;191:317–333. doi: 10.1098/rspb.1975.0131. [DOI] [PubMed] [Google Scholar]
- 3.Sanger F, Nicklen S, Coulson AR. Proc. Natl. Acad. Sci. USA. 1997;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith LM, Sanders JZ, Kaiser RJ, Hughes P, et al. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 5.Glazer AN, Mathies RA. Curr Opin Biotechnol. 1997;8:94–102. doi: 10.1016/s0958-1669(97)80163-2. [DOI] [PubMed] [Google Scholar]
- 6.Jorgenson JW, Lukacs KD. Science. 1981;53:266–272. doi: 10.1126/science.6623076. [DOI] [PubMed] [Google Scholar]
- 7.Kasper TJ, Melera M, Gozel P, Brownlee RG. J Chromatogr. 1988;458:303–312. doi: 10.1016/s0021-9673(00)90574-0. [DOI] [PubMed] [Google Scholar]
- 8.Guttman A, Paulus A, Cohen AS, Karger BL, et al. In: Electrophoresis'88. Schafer-Nielsen C, editor. Weinheim, Germany: VCH; 1988. pp. 151–159. [Google Scholar]
- 9.Guttman A, Cohen AS, Heiger DN, Karger BL. Anal. Chem. 1990;62:137–141. [Google Scholar]
- 10.Cohen A, Najarian DR, Paulus A, Guttman A, et al. Proc. Natl. Acad. Sci. USA. 1988;85:9660–9663. doi: 10.1073/pnas.85.24.9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drossman H, Luckey JA, Kostichka AJ, D'Cunha J, Smith LM. Analytical Chemistry. 1990;62:900–903. doi: 10.1021/ac00208a003. [DOI] [PubMed] [Google Scholar]
- 12.Swerdlow H, Gesteland R. Nucleic Acids Res. 1990;18:1415–1419. doi: 10.1093/nar/18.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pariat YF, Berka J, Heiger DN, Schmitt T, et al. J Chromatogr A. 1993;652:57–66. doi: 10.1016/0021-9673(93)80645-O. [DOI] [PubMed] [Google Scholar]
- 14.Heiger DN, Cohen AS, Karger BL. J Chromatogr. 1990;516:33–48. doi: 10.1016/s0021-9673(01)90202-x. [DOI] [PubMed] [Google Scholar]
- 15.Bode HJ. Analytical Biochemistry. 1977;83:364–371. doi: 10.1016/0003-2697(77)90045-8. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez R, Zhu M, Wehr T. American Biotechnology Laboratory. 1992;10:21–22. [PubMed] [Google Scholar]
- 17.Chang H-T, Yeung ES. Journal of Chromatography, B: Biomedical Applications. 1995;669:113–123. doi: 10.1016/0378-4347(95)00044-j. [DOI] [PubMed] [Google Scholar]
- 18.Guttman A. 5,332,481. U.S. Patent. 1994
- 19.Ruiz-Martinez MC, Berka J, Belenkii A, Foret F, et al. Anal. Chem. 1993;65:2851–2858. doi: 10.1021/ac00068a023. [DOI] [PubMed] [Google Scholar]
- 20.Karger BL, Foret F, Berka J. Methods Enzymol. 1996;271:293–319. doi: 10.1016/s0076-6879(96)71015-7. [DOI] [PubMed] [Google Scholar]
- 21.Hebenbrock K, Williams PM, Karger BL. Electrophoresis. 1995;16:1429–1436. doi: 10.1002/elps.11501601236. [DOI] [PubMed] [Google Scholar]
- 22.Hjerten S. J. Chromatogr. 1985;347:191–197. [Google Scholar]
- 23.Madabhushi RS. Electrophoresis. 1998;19:224–230. doi: 10.1002/elps.1150190215. [DOI] [PubMed] [Google Scholar]
- 24.Huang XC, Quesada MA, Mathies RA. Anal. Chem. 1992;64:2149–2154. doi: 10.1021/ac00042a021. [DOI] [PubMed] [Google Scholar]
- 25.Swerdlow H, Wu SL, Harke H, Dovichi NJ. J Chromatogr. 1990;516:61–67. doi: 10.1016/s0021-9673(01)90204-3. [DOI] [PubMed] [Google Scholar]
- 26.Taylor JA, Yeung ES. Anal. Chem. 1992:1741–1749. [Google Scholar]
- 27.Blazej RG, Kumaresan P, Mathies RA. Proc Natl Acad Sci U S A. 2006;103:7240–7245. doi: 10.1073/pnas.0602476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, et al. Proc Natl Acad Sci U S A. 2006;103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lupmkin OJ, Dejardin P, Zimm BH. Biopolymers. 1985;24:1573–1593. doi: 10.1002/bip.360240812. [DOI] [PubMed] [Google Scholar]
- 30.Kleparnik K, Foret F, Berka J, Goetzinger W, et al. Electrophoresis. 1996;17:1860–1866. doi: 10.1002/elps.1150171210. [DOI] [PubMed] [Google Scholar]
- 31.Carrilho E, Ruiz-Martinez MC, Berka J, Smirnov I, et al. Anal Chem. 1996;68:3305–3313. doi: 10.1021/ac960411r. [DOI] [PubMed] [Google Scholar]
- 32.Salas-Solano O, Carrilho E, Kotler L, Miller AW, et al. Anal. Chem. 1998;70:3996–4003. doi: 10.1021/ac980457f. [DOI] [PubMed] [Google Scholar]
- 33.Goetzinger W, Kotler L, Carrilho E, Ruiz-Martinez MC, et al. Electrophoresis. 1998;19:242–248. doi: 10.1002/elps.1150190217. [DOI] [PubMed] [Google Scholar]
- 34.Zhou H, Miller AW, Sosic Z, Buchholz B, et al. Anal Chem. 2000;72:1045–1052. doi: 10.1021/ac991117c. [DOI] [PubMed] [Google Scholar]
- 35.Karger BL, Guttman A. Genomic/Proteomic Technology. 2003;3:12–14–16. [Google Scholar]
- 36.Kheterpal I, Mathies RA. Anal. Chem. 1999;71:A31–A37. doi: 10.1021/ac990099w. [DOI] [PubMed] [Google Scholar]
- 37.Li Q, Yeung ES. Appl. Spetrosc. 1995;49:1528–1533. [Google Scholar]
- 38.Dovichi NJ, Zhang J. Angew. Chem. Int. Ed. 2000;39:4463–4468. doi: 10.1002/1521-3773(20001215)39:24<4463::aid-anie4463>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 39.Kamahori M, Kambara H. Methods Mol Biol. 2001;163:271–287. doi: 10.1385/1-59259-116-7:271. [DOI] [PubMed] [Google Scholar]
- 40.Gordon D, Desmarais C, Green P. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hert DG, Fredlake CP, Barron AE. Electrophoresis. 2008;29:4618–4626. doi: 10.1002/elps.200800456. [DOI] [PubMed] [Google Scholar]
- 42.Mukhopadhyay R. Anal Chem. 2009;81:1736–1740. doi: 10.1021/ac802712u. [DOI] [PubMed] [Google Scholar]
- 43.Shendure J, Ji H. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 44.Shendure J, Porreca GJ, Reppas NB, Lin X, et al. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 45.Margulies M, Egholm M, Altman WE, Attiya S, et al. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ronaghi M, Pettersson B, Uhlen M, Nyren P. Biotechniques. 1998;25:876–878. doi: 10.2144/98255rr02. 880–872, 884. [DOI] [PubMed] [Google Scholar]
- 47.Turcatti G, Romieu A, Fedurco M, Tairi AP. Nucleic Acids Res. 2008;36:e25. doi: 10.1093/nar/gkn021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drmanac S, Kita D, Labat I, Hauser B, et al. Nat Biotechnol. 1998;16:54–58. doi: 10.1038/nbt0198-54. [DOI] [PubMed] [Google Scholar]
- 49.Harris TD, Buzby PR, Babcock H, Beer E, et al. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 50.Branton D, Deamer DW, Marziali A, Bayley H, et al. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]