Abstract
Xanthorhodopsin (XR), the light-driven proton pump of the halophilic eubacterium Salinibacter ruber, exhibits substantial homology to bacteriorhodopsin (BR) of archaea and proteorhodopsin (PR) of marine bacteria, but unlike them contains a light-harvesting carotenoid antenna, salinixanthin, as well as retinal. We report here the pH dependent properties of XR. The pKa of the retinal Schiff base is as high as in BR, i.e., ≥ 12.4. Deprotonation of the Schiff base and the ensuing alkaline denaturation causes large changes in the absorption bands of the carotenoid antenna, which lose intensity and become broader making the spectrum similar to that of salinixanthin not bound to XR. A small red shift of the retinal chromophore band and increase of its extinction, as well as the pH dependent amplitude of the M intermediate indicate that in detergent-solubilized XR the pKa of the Schiff base counter-ion and proton acceptor is about 6 (compared to 2.6 in BR, and 7.5 in PR). The protonation of the counter-ion is accompanied by a small blue shift of the carotenoid absorption bands. The pigment is stable in the dark upon acidification to pH 2. At pH < 2 a transition to a blue shifted species absorbing around 440 nm occurs, accompanied by loss of resolution of the carotenoid absorption bands. At pH < 3 illumination of XR with continuous light causes accumulation of long-lived photoproducts(s) with an absorption maximum around 400 nm. The photocycle of XR was examined between pH 4 and 10 in solubilized samples. The pH dependence of recovery of the initial state slows at both acid and alkaline pH, with pKa’s of 6.0 and 9.3. The decrease in the rates with pKa 6.0 is apparently caused by protonation of the counter-ion and proton acceptor while that at high pH reflects the pKa of the internal proton donor, Glu94, at the times in the photocycle when this group equilibrates with the bulk.
INTRODUCTION
The cell membrane of Salinibacter ruber, an extremely halophilic bacterium in hypersaline brines (1, 2), contains the carotenoid, salinixanthin, which is responsible for its deep red color (3). Another pigment in these membranes is the retinal protein, xanthorhodopsin (XR), which was shown to be a light-driven proton pump (4) as bacteriorhodopsin (5) and archaerhodopsin (6) of archaea and proteorhodopsin of marine bacteria (7). It exhibits a great deal of homology to these proteins (4), and even more to the rhodopsin of the cyanobacterium Gloeobacter (8).
An unusual feature of XR is that besides the retinal it binds salinixanthin as an additional chromophore, which serves as a light-harvesting antenna. About 40% of the quanta absorbed by the bound salinixanthin are transferred to the retinal chromophore (4). Bound and free carotenoid exhibit different absorption spectra. The spectrum of bound salinixanthin in the complex has higher extinction, is more structured, and contains sharp, well-resolved bands at 521, 488, and 465 nm. As in other rhodopsins, hydrolysis of the retinal Schiff base of XR with hydroxylamine results in disappearance of the absorption band of the retinal chromophore at ca. 565 nm and the appearance of a retinal oxime band at 364 nm. This treatment causes substantial broadening and a slight red shift of the carotenoid bands, and the spectrum of the carotenoid component of the complex becomes almost identical to that of bulk carotenoid. These changes of the carotenoid spectrum are reversed by reconstitution with retinal, indicating close interaction of retinal and carotenoid. The intimate relationship of the two chromophores is further supported by light induced changes of the carotenoid bands during the photocycle (4).
In this paper we describe the fundamental pH dependent properties of the newly discovered retinal protein in the unphotolyzed state and during its photocycle, with the aim to determine the pKa of the Schiff base and its counter-ion. Such information is part of the general knowledge of retinal proteins, but in XR the additional question is how protonation of the counter-ion (proton acceptor) and deprotonation of the donor affects the antenna part of the complex.
Deprotonation of the Schiff base at high pH in BR and other retinal proteins results in a large blue-shift of the retinal chromophore, to 370 nm, with a pKa of 12.5 or even 13 (9, 10). A similar high pKa was reported for PR in E. coli membranes (11), although somewhat lower when solubilized with DM, at 11.3 (12). Protonation of the counter-ion in the unphotolyzed state of both BR and PR, (Asp85 and Asp97, respectively), is accompanied by red-shifts of ca. 38 nm (13, 14) and 32 nm (11, 12, 15). In both retinal proteins this shift correlates with the disappearance of the M intermediate in the photocycle (11, 13, 16). Another feature is that protonation of one or both aspartic acids residues at the Schiff base (Asp85 and Asp212 in BR, and Asp97 and Asp227 in PR) opens the possibility for alternative photochemical reactions leading to long-lived photoproducts (12, 17) which can serve as an indication of the protonation of these residues.
The pH dependence of the rates of photocycle reactions can be used to determine the pKa’s of the groups involved in proton release and uptake. Thus, the recovery of initial state of bacteriorhodopsin at high pH is controlled by the rate of reprotonation of the internal proton donor while slower recovery at low pH reflects the changed photocycle when the counter-ion is protonated in the initial state already.
We describe here the pH dependence of absorption spectrum of XR and some characteristic reactions of the photocycle (M formation and decay, recovery of the initial state). These parameters provide key information on the pKa’s of the groups of XR involved in proton transport (the Schiff base, the counter-ion, and internal proton donor Glu94), and reveal coupling between the binding of carotenoid antenna and protonation state of these groups.
MATERIALS AND METHODS
Growth conditions
Salinibacter ruber M31 was grown in the medium described earlier (1) with minor modifications. The concentrations of salts (per L) were 195 g NaCl, 25 g MgSO4·7H2O, 20.1 g MgCl2·6H2O, 1.25 g CaCl2·2H2O, 5 g KCl, 0.25 g NaHCO3, 0.625 g NaBr, plus 1 g of Yeast Extract, pH 7.0. A 400 ml culture was inoculated with 16 ml of 14 days old starter culture, and grown at 37° C in 2 L flasks loosely covered with foil in an incubator shaker (Series 25, New Brunswick Scientific, NJ, USA) at 180 rpm for 7 days. Fresh medium was then added to each flask to about 1.8 L, sealed to decrease aeration and the culture was grown under illumination for another 7 days while shaking at 250 rpm.
Isolation of cell membranes
The cell membranes were isolated by dialysis and subsequent washing with water (4) at least 6 times (with 60 min centrifugation at 250,000 g). The membranes were solubilized in 0.15 % DM.
Spectroscopic titrations
The titrations were in 100 mM NaCl and a mix of 6 buffers, 5 mM each (BICINE, CAPS, CHES, citric acid, MES, MOPS) that gave an approximately constant buffer capacity between pH 2 and 11. To shift the pH, either 1 to 5 N H2SO4 or NaOH was added, or alternatively, the samples were prepared by mixing with a set of buffers of different pH values. A 1×1 cm quartz cuvette was used. The pH was measured with Thermo Orion Sure-Flow Ross semi-micro glass electrode (98-30) and Orion pH meter, Model 720A. The spectra were recorded on a Shimadzu UV-1601 spectrophotometer. The titrations were performed under dim light. Low light intensity conditions were important for pH below 4 where formation of long-lived photoproducts were observed. At neutral and high pH illumination of the sample with continuous light and subsequent incubation in the dark did not produce any substantial absorption changes analogous to those observed in bacteriorhodopsin during light-dark adaptation cycles and caused by transition between all-trans and 13-cis, 15 syn chromophores (18).
Light-induced transformation to long-lived photoproducts upon continuous illumination
A Cole-Parmer 9741-50 illuminator (150W) in combination with a light guide and band-pass filter (λ>550 nm) was used for illumination. The intensity of actinic light in the range of 550–650 nm was ca. 12 mW/cm2.
Kinetics of photocycle reactions
The laser flash-induced absorption changes in suspensions of solubilized XR were studied on a home-build set up described earlier (19). Excitation was at 532 nm from Continuum Surelite II-10 Nd-YAG laser (Santa Clara, CA). The signal was digitized with analog-to-digital converter (Gage Compuscope 6012/PCI-4M card) with subsequent averaging and reduction of data to 265 points/trace. The samples were thermostated at 20°C. The kinetics were analyzed and fitted with the Fitexp Program (20, 21) as described in (11).
RESULTS
Alkaline titration: pKa of the Schiff base, and pH induced changes in the carotenoid
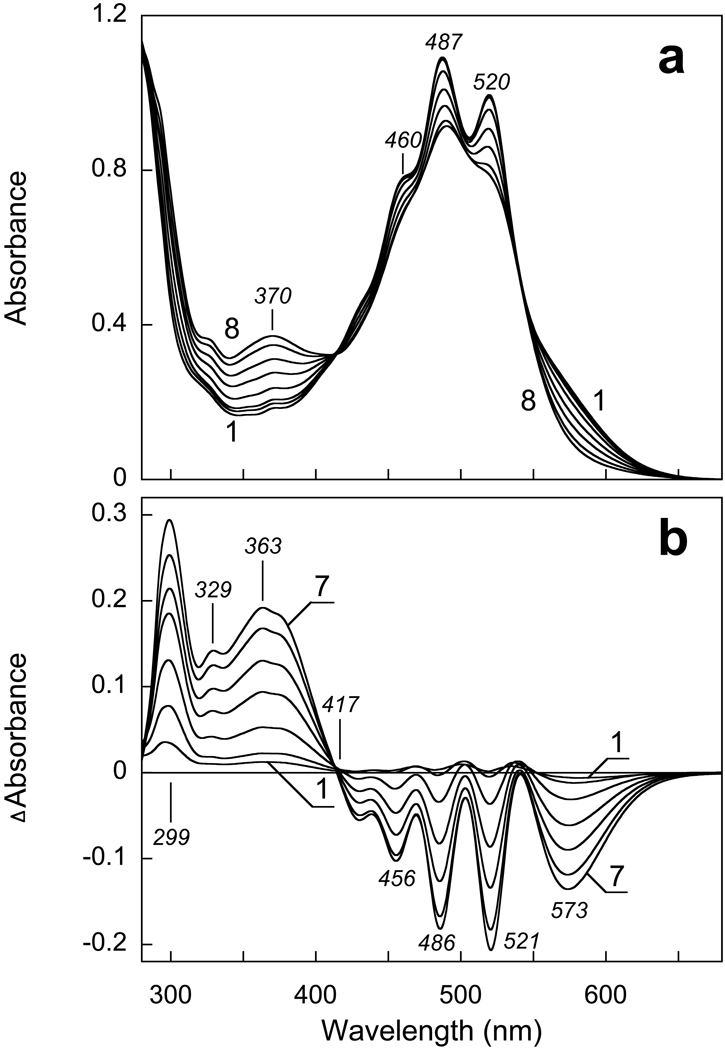
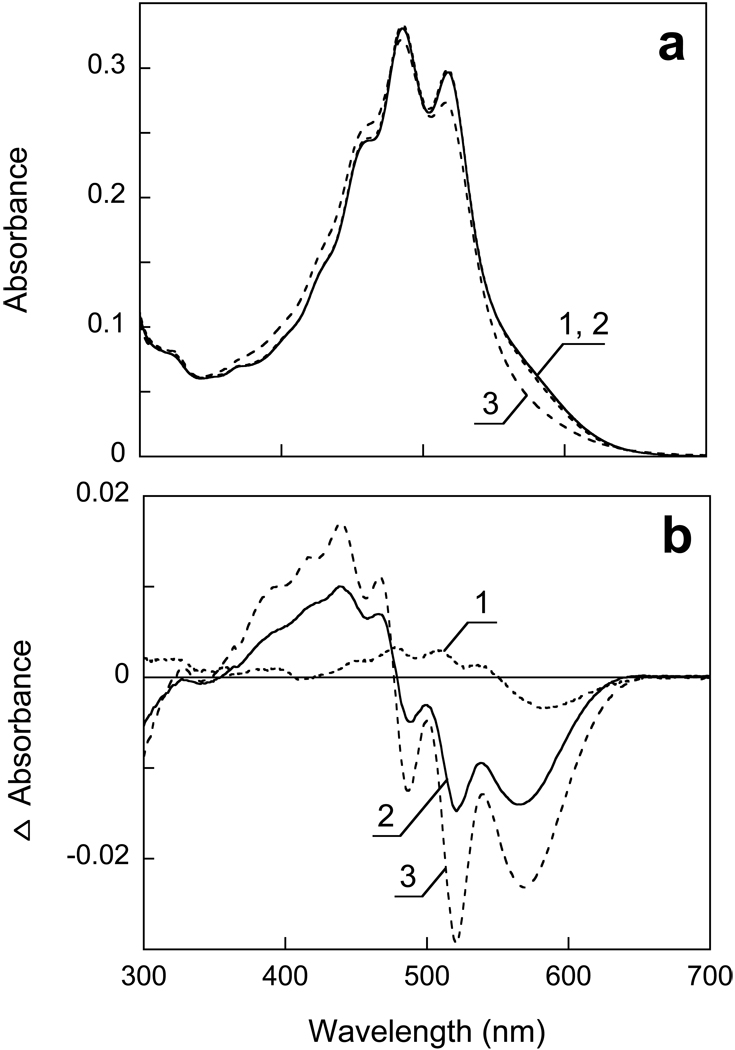
Gradual increase of the pH from 8.0 to 13 causes the absorption changes of solubilized XR shown in Fig. 1a. At pH > 10.5 the band of deprotonated Schiff base begins to appear at 370 nm, along with the disappearance of the retinal chromophore band at ca. 570 nm. These changes are accompanied by dramatic decrease in the resolution and the extinction of carotenoid bands at 520, 487 and 460 nm. In the near UV region a small band appears at 329 nm, which is probably the β-band of the carotenoid. Its increase might indicate a less planar (i.e., less distorted) conformation of the carotenoid chain at high pH. The increase of the absorbance at 299 nm is mainly caused by deprotonation of tyrosines, as reported also in BR (10). All these bands are clearly seen in the difference spectra in Fig. 1b. The band of the deprotonated Schiff base is shifted to 363 nm, apparently as a result of subtraction of the β-band of the retinal chromophore at 370 nm (see curve 1 in Fig. 1a). The negative 573 nm band from the retinal chromophore is somewhat red-shifted from its true maximum in XR [estimated at 560–565 nm (4)], because of overlap with the broadened positive carotenoid band at 540 nm. The negative peaks at 521, 486 and 456 nm originate from changes of the carotenoid spectrum, as after hydroxylamine bleaching of the retinal chromophore (4).
Figure 1.
Absorption changes in suspension of cell membranes solubilized in 0.15% DM and 100 mM NaCl upon increasing the pH from 8 to 13. a) Curves 1 through 8, absolute spectra at pHi 7.9, 9.9, 10.6, 11.2, 11.5, 11.7, 12.1, 12.7, respectively. b) Curves 1 through 7, difference spectra “pHi minus pH 7.9”.
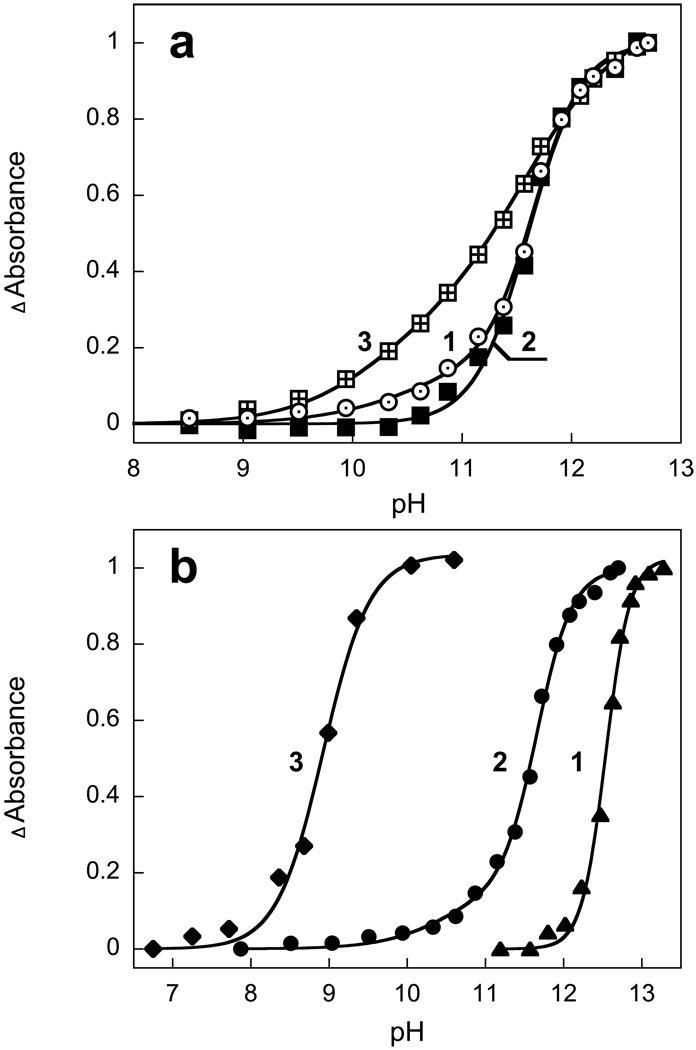
The decrease of absorbance at 570 nm and increase of absorbance at 370 nm occur with a pKa of 11.6 in solubilized XR (Fig. 2a, curves 1 and 2)). This observed pKa does not necessarily correspond to the pKa of the Schiff base, which might be higher, but to the pKa for the structural stability of xanthorhodopsin under alkaline conditions. This is what had been concluded for BR from titration of the infrared spectrum (22). The changes at 521 nm and 486 nm due to perturbation of the carotenoid begin to develop at slightly higher pH than the first sign of deprotonation of the Schiff base (compare curves 1 and 2 in Fig. 2a). This may be because the changes of the carotenoid absorption bands caused by deprotonation of the Schiff base are smaller than those associated with the denaturation of the protein.
Figure 2.
a) pH dependence of absolute values of absorption changes of xanthorhodopsin in 0.15% DM, 100 mM NaCl at 573 nm (curve 1), 521 nm (curve 2) and 299 nm (curve 3). Data are from Fig. 1. The pH dependence of absorption changes at 370 nm (not shown) follows that at 573 nm within experimental error. Curves are normalized to the maximum. b) Effect of detergents on the pKa of the Schiff base deprotonation: curve 1, native cell membrane (pKa 12.4, n=3); curve 2, solubilized in 0.15% DM, 100 mM NaCl (two component fit with A1=0.15, pKa1=10.5, n=1; A2=0.85; pKa2 =11.6, n=1.9); Curve 3, solubilized in 1.5% OG, 100 mM NaCl (pKa 8.9, n=1.5).
Titration of tyrosine residues requires a two component fit (Fig. 2a, curve 3). The pKa of 10 most likely belongs to surface tyrosine residues, and the pKa of 11.6 (n=1.7) is apparently due to buried tyrosines.
Titration of native (non-solubilized) membranes yields a pKa of 12.4 (Fig. 2b, curve 1). Both the retinal chromophore band at ca. 570 nm and the well-structured bands of bound carotenoids are restored after short exposure of membranes to pH 12.1 and subsequent decrease the pH to 8.5. However only a fraction of the pigment (about 30%) undergoes such recovery indicating that in membranes irreversible changes begin to occur above pH 12. The pKa of the deprotonation of the Schiff base and resistance to alkaline denaturation depends on the detergent used. OG destabilizes the protein to a much larger extent than DM, and shifts the pKa of the Schiff base to 8.9 vs 11.3 in DM (Fig. 2b, curve 3).
Acid titration of the absorption spectrum: estimation of the pKa of the counter-ion
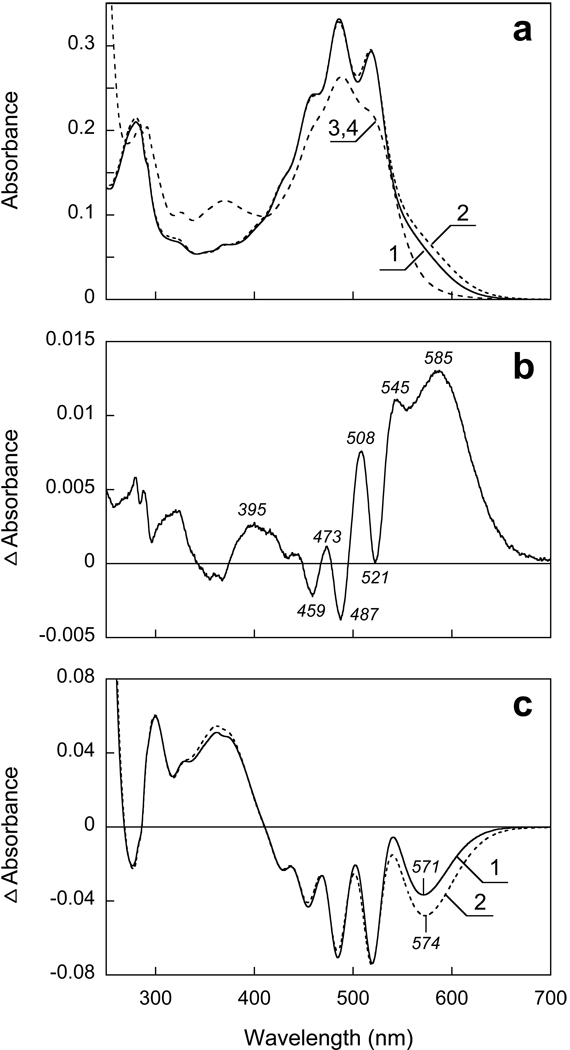
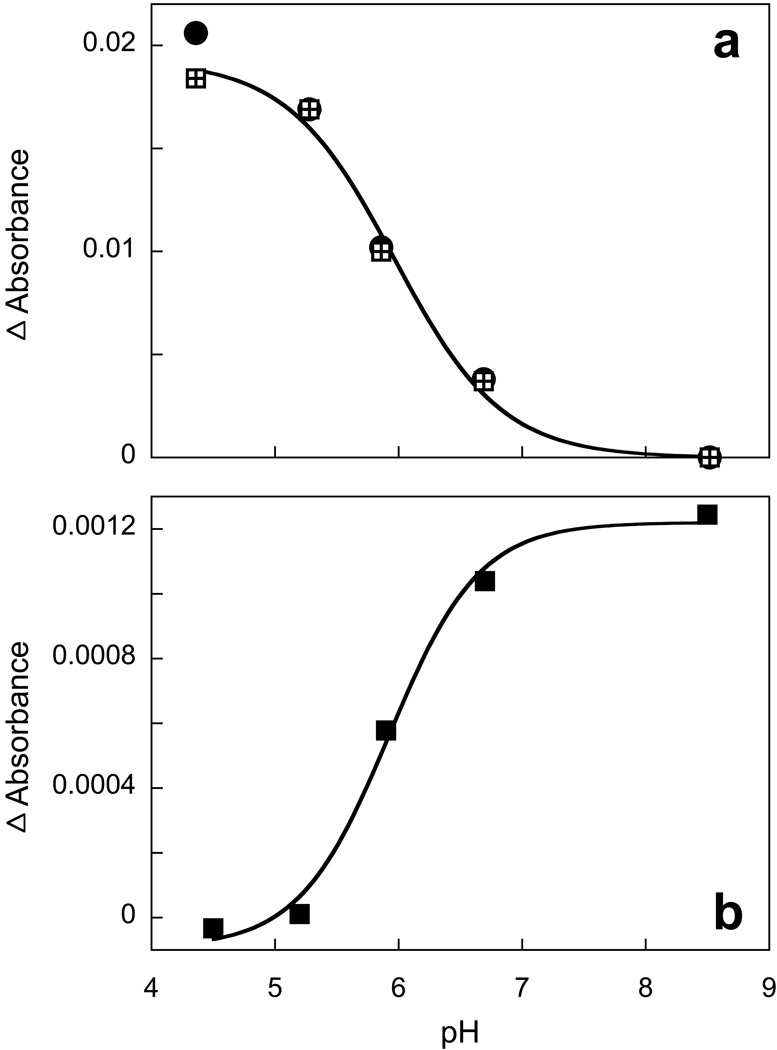
Upon decreasing the pH from 8.5 to 4.5 a small red shift of the retinal chromophore band is seen (Fig. 3a). In the difference spectrum (Fig. 3b) the positive band near 585 nm originates from both an increase of the extinction at low pH and the red shift expected to accompany protonation of the counter-ion. At shorter wavelengths, a set of narrow peaks overlaps the difference spectrum of the retinal chromophore, which originates from blue shift of the carotenoid bands. From the amplitude of the 508 nm peak this blue shift appears to be small, an estimated 0.5 nm (20 cm−1) for the 521 nm band. These changes occur with a pKa of 6.0 (Fig. 4a).
Figure 3.
pH dependence of absorption spectrum of xanthorhodopsin in DM between pH 8.5 and 4.5. a) 1 and 2, absolute spectra at pH 8.5 and 4.5; 3, 4 (coincident) after increasing the pH to 12.5 in samples 1 and 2, respectively. b) Difference spectrum “pH 4.5 minus pH 8.5”. c) Curves 1 and 2, difference spectra “pH 4.5 minus 12.5” and “pH 8.5 minus 12.5”, respectively.
Figure 4.
a) pH dependence of absorption changes of the retinal chromophore band (at 590 nm, squares) and carotenoid bands (at 508 nm minus 488 nm, circles). b) pH dependence of the amplitude of the light-induced absorption changes at 410 nm from formation of the M intermediate.
The red shift of the retinal chromophore is also small, and amounts to a few (ca. 3–5) nm, as one can conclude from the absolute spectra in Fig. 3a and the difference spectra in Figs. 3b and 3c. Fig. 3c shows absorption changes produced by raising the pH to 12. 5 in two samples that contain same amount of xanthorhodopsin but at different initial pH (pH 4.5 and 8.5, as shown in Fig. 3a). The larger negative peak from the retinal band for the sample with initial pH of 4.5, and only a minor red-shift of its maximum relative to the other sample, is consistent with the conclusion that the chromophore band experiences only a few nm shift between pH 8.5 and 4.5.
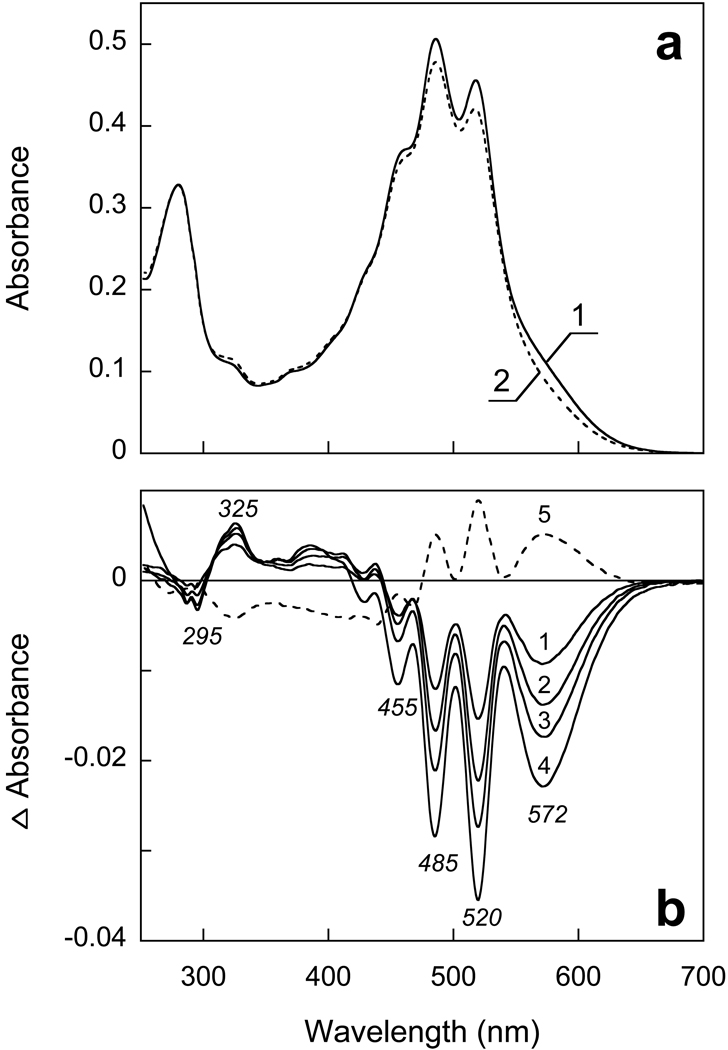
At more acid pH, between 4.5 and 2.5, only minor absorption changes take place, characterized by decrease of absorbance at 580 nm and increase around 500 nm (Fig. 5). There are no substantial changes in the carotenoid bands. Decreasing the pH to below 2.5 causes larger changes, which include decrease of absorbance at 573 nm and increase at 440 nm. This is accompanied by loss of intensity of the carotenoid bands which can be seen both in absolute and difference spectra (Figs. 5a and 5b). This transition occurs with a pKa of about 1.7.
Figure 5.
a) Spectra 1, 2 and 3, absorption spectra of xanthorhodopsin solubilized in 0.15 DM at pH 4.5, 2.5 and 1.0. b) Difference spectra: spectrum 1, pH 2.5-pH 4.5; spectrum 2, pH 2 minus pH 2.5, 3, pH 1 minus pH 2.5.
Light-induced conversion of xanthorhodopsin to long-lived intermediates at low pH
Illumination of XR at λ >550 nm with continuous light for 10 min at pH 4.5 causes transition of a very small fraction of the pigment to a long-lived photoproduct absorbing at shorter wavelengths. At pH 3.3 this becomes a more substantial effect, and a major fraction of the pigment undergoes photoconversion to a product absorbing around 380–400 nm (Fig. 6). The changes in the carotenoid bands indicate loss of specific binding, as one can conclude from a decrease of extinction and resolution of the carotenoid bands similar to what was observed at pH > 11 (in DM). The band at 325 nm indicates decrease in planarity of the carotenoid chromophore (23). The quantum efficiency of the photoreaction and fraction of the photoproduct(s) increases at low pH with a pKa of about 2.6, similarly to what was observed in PR (12), except that in XR the photoproduct absorbs at shorter wavelengths.
Figure 6.
Transformation of xanthorhodopsin to a long-lived photoproduct at pH 3.3 upon illumination at > 580 nm. a) Spectra 1 and 2, initial spectrum of XR in 0.15% DM at pH 3.3 and after illumination for 8 min, respectively. b) Spectra 1 through 4, difference spectra of light-induced conversion to long lived photoproduct after 1, 2, 4 and 8 min of illumination, respectively; 5, difference spectrum of partial thermal recovery after incubation in the dark for 10 min.
Upon incubation in the dark the photoproduct partially returns to the initial state at pH 3.3 (Fig. 6). The changes of carotenoid bands are also partially reversed in the dark indicating that loss of the carotenoid binding in the photoproduct is reversed upon returning of the retinal chromophore to the initial state. The formation and partial thermal recovery of long-lived photoproduct(s) apparently involves chromophore isomerization. The exact chromophore configuration was not examined in this study. From previous experiment with bacteriorhodopsin and proteorhodopsin the long lived species might have 9-cis, 11-cis (12, 17, 24) and 13-cis configurations (25).
pH dependence of the yield of the M intermediate of the photocycle
The red-shift of the retinal chromophore absorption band upon decreasing the pH is a typical feature associated with protonation of counter-ion in all retinal-based proton pumps (BR, PR, and archaerhodopsin) and other retinal proteins of the archaeal type including sensory rhodopsins I and II. In XR a red shift occurs only between pH 8 and 4, but the magnitude of the shift is not nearly as large as in BR and PR. The smaller than expected shift raises the question whether it indeed originates from the counter-ion. Another method to titrate the counter-ion is to determine the yield of the M intermediate of the photocycle. As shown in Fig. 7 this method confirms the conclusion from the pH dependent red shift. The amplitude of the light-induced transient absorption change at 410 nm decreases with decreasing pH, with a pKa of 6.0. The kinetics of M formation and decay does not change substantially (the time-constants for rise and decay are ca. 70 µs and 0.6 ms, respectively, throughout), but at pH 5 the amount of M formed decreases from the maximum at pH 8.5 by at least ten-fold. The pKa associated with the accumulation of the M intermediate is 6.0 (Fig 4b). The photocycle kinetics are consistent with the interpretation that the red-shift is caused by a protonation of counter-ion, and that it occurs in all, or at least a major fraction, of XR (> 90%) upon decreasing the pH to 4.4 under the conditions in Fig.7.
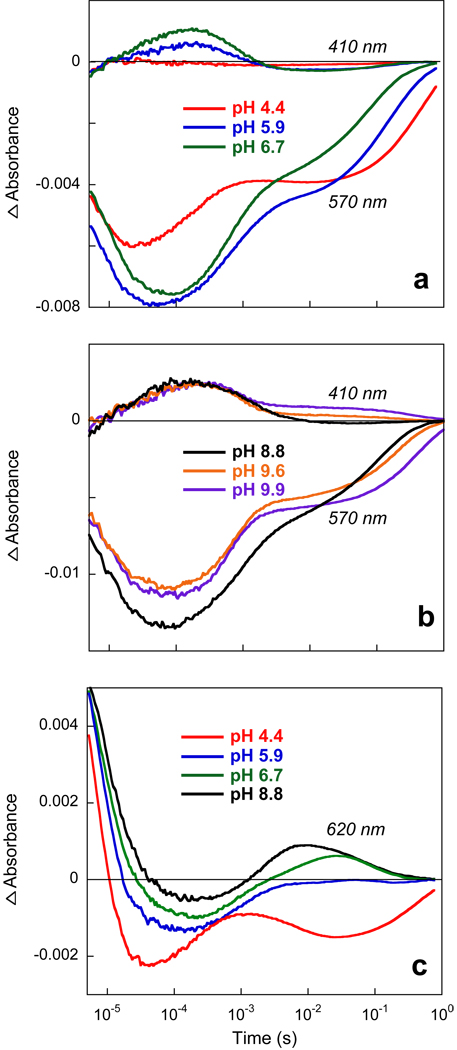
Figure 7.
pH dependence of the kinetics of photocycle of xanthorhodopsin. a) Kinetics of absorption changes at 410 nm and 570 nm at pH 4.4, 5.9 and 6.7. b) As in (a) but at pH 8.8, 9.6, and 9.9. c) Kinetics at 620 nm. All traces were measured using samples with the same concentration of XR.
pH dependence of the photocycle reactions
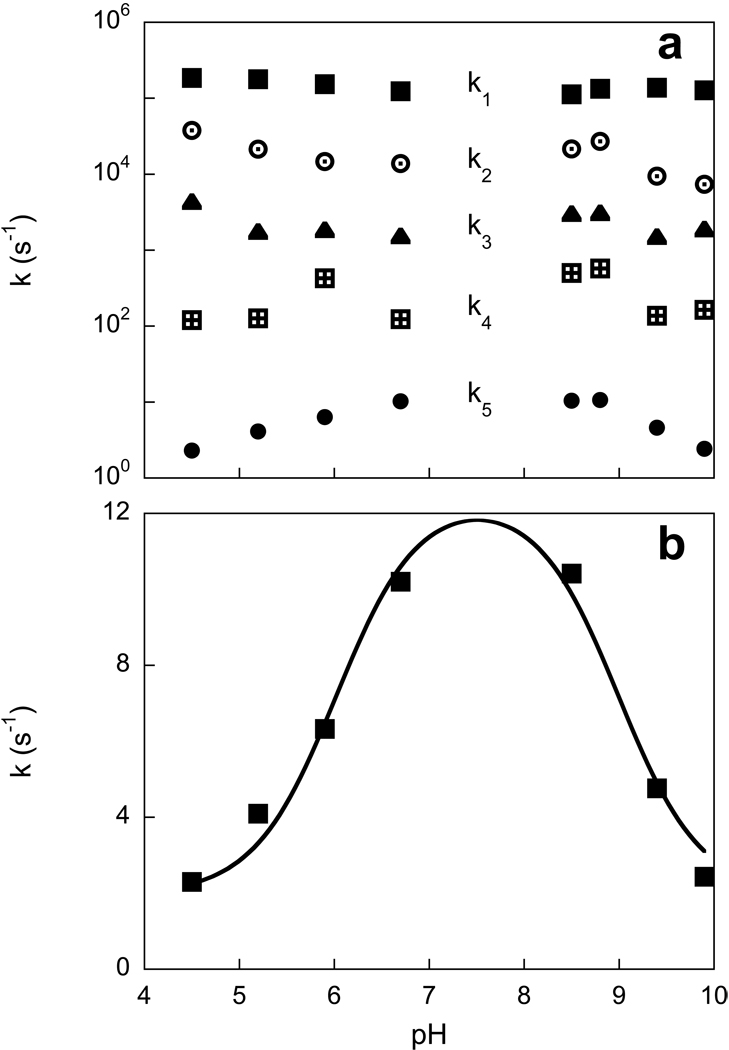
The kinetics of the photocycle reactions of XR can be followed at several characteristic wavelengths (4). Fig. 7 shows absorption changes at 410 and 570 nm and 620 nm at different pH values from 4.4 to 9.9. Absorption changes at 570 nm are mostly from the bleaching and recovery of the initial state, with lesser contributions from all of the intermediates except M. The kinetics at 620 nm is mostly due to decay of K at earlier times, and formation and decay of a red-shifted O-like intermediate at later times. The latter disappears at low pH. The kinetics in the pH range between 4 and 10 reveals pH dependent features additional to the disappearance of the M and O-like intermediates at low pH described above. Most of the rate constants obtained from global fit do not exhibit strong pH dependence, except the slowest time constant, which is found in both the 570 nm and the 410 nm traces and represents the recovery of the initial state (Fig. 8a). This reaction becomes slower at both low and high pH, decreasing from its maximum value at pH between 7 and 8 with pKa’s of 6.0 and 9.3 (Fig. 8b). M formation and the faster component of M decay does not change substantially with pH, whereas the rate constant of a slow component in the decay of the M intermediate, which appears at pH > 8 also slows with pKa 9.3. The origins of these pH dependencies are discussed below.
Figure 8.
pH dependence of the rate constants of the photocycle reactions obtained from the fit of kinetic traces shown in Fig. 7. a) All five rate constants (k1 through k5). b) pH dependence of the slowest rate constant, k5.
DISCUSSION
pKa of the Schiff base of xanthorhodopsin
Alkaline titration shows that the pKa of the Schiff base of XR in membranes is very high, >12. The stability of the protein might be limited by its alkaline denaturation which occurs in this pH range. This is similar to BR and PR. Solubilization with detergents decreases the limit of stability by about one pH unit in DM and 3.5 units in OG. Deprotonation of the Schiff base and alkaline denaturation of xanthorhodopsin lead to dramatic spectral changes of the carotenoid antenna. These spectral changes are similar to those produced by hydrolysis of the Schiff base with hydroxylamine (4). They indicate that the native retinal-protein controls a specific conformation of the antenna carotenoid.
pKa of the counterion: comparison with bacteriorhodopsin and proteorhodopsin
A large red shift of the absorption maxima of BR (pKa of 2.6) and PR (pKa of 7.5) was correlated with the charge state of the counter-ion (Asp85 in BR and Asp97 in PR) by solid state NMR (26) and the properties of the D85N mutant of BR (27, 28) and the D97N mutant of PR (11). Unlike in these proteins (12, 15), in XR there is only a small red-shift of the retinal chromophore absorption band between pH 1 and 10. This shift, with a pKa 6 (Fig. 4a), is therefore the only candidate for the spectral change from the expected protonation of the counter-ion. Its origin in the counter-ion is confirmed by the fact that the deprotonation of the retinal Schiff base in the photocycle has the same pKa (Figs. 4b). This pKa is 1.5 units lower than in PR, and 3.5 units higher than in BR. The greater similarity of the pKa of XR and PR is consistent with the amino acid sequence similarities of these proteins in the extracellular region (4). Interestingly, protonation of the counter-ion does not affect the spectrum of the carotenoid antenna strongly. The spectrum undergoes only a minor shift (< 1 nm) to shorter wavelengths. This small effect could be caused by either changes in electrostatic interaction with the counter-ion or conformational changes in the protein.
It is not clear why the characteristic red shift from the counter-ion is so small in XR. Sequence comparison among the three retinal proteins (4) reveals no clues. A possible explanation has to do with the binding of carotenoid in XR. In the high-resolution structure of BR with Asp85 deprotonated, the Schiff base is hydrogen-bonded to a water molecule (wat402) which is also bound to Asp85 and Asp212 (29–31). This water is an essential part of the counter-ion. The crystal structure of the acid blue form of BR indicates that protonation of Asp85 is accompanied by loss of this water (32). The nearest water molecule is at a distance of 4.7 Å or 5.4 Å from the Schiff base in the all-trans and 13-cis, 15-syn retinal configurations, respectively (vs. 2.7 Å in the all-trans purple form), excluding the possibility of a hydrogen bond. In addition, the side chain of Arg82 moves downward and several helices undergo substantial movements (32). These changes might contribute to the red shift of the chromophore at acid pH. If the presence of carotenoid antenna were to create a more rigid structure so that water would not be “squeezed” out from the vicinity of the Schiff base, interaction with water (and possibly with Asp223) would be less perturbed by the protonation of counter-ion, and one might expect a smaller shift.
pH dependent yield of long-lived photoproducts at low pH
Illumination of XR at pH < 3 causes transformation to long-lived photoproducts (Fig. 6) absorbing around 400 nm. An analogous transition was observed in PR (12). In the latter pigment the features of D227N mutant indicated that the photoconversion might be facilitated by protonation of Asp227 (12), the residue homologous to Asp212 in BR which is near the C13=C14 bond of the retinal and presumably has some degree of control over chromophore isomerization (33). Based on this analogy we suggest that in XR protonation of the homologous aspartate (Asp223) in the initial state with pKa around 2.6 is responsible for the formation of long-lived photoproducts.
pH dependence of the recovery of the initial state and of other reactions of the xanthorhodopsin photocycle
The pH dependence of slow component of M decay and recovery of initial state of XR can be rationalized in a model similar to that suggested for BR (34, 35). In this model reprotonation of the Schiff base is by the internal proton donor, i.e. it is pH independent during the M to N transition. This corresponds to the faster phase of M decay (36), whereas reprotonation of the internal donor from the bulk (or from an intermediate proton donor) occurs in a pH dependent way during N decay. This pH dependent step is observed both as the slower phase of M decay and the recovery of the initial state. This indicates that, as in BR, M decay involves equilibration with N (M⇔N). The pH dependence reveals the pKa of the proton donor during proton uptake (34, 37). This pKa is 7.5 in BR (37–39). The pH dependence of the rate constant of recovery of XR indicates that this pKa is higher in XR, ca. 9.3. Tentatively, we assign this pKa to Glu94 at the moment of the photocycle when it is reprotonated from the bulk, presumably in a late N state (34, 35, 40). A higher pKa for the internal proton donor means less exposure to the bulk (lesser proton conductivity of the cytoplasmic channel) and correspondingly slower reprotonation, but also the ability to maintain the same turnover rate of the photocycle at a higher pH.
The recovery of the initial state slows also at low pH, with a pKa of ca. 6.0. In BR this occurs with pKa ca. 4.5 and was attributed to the pKa of the proton release complex in the O intermediate (37). The key components of this complex in BR are Arg82 (41, 42), Glu204 (43, 44), and Glu194 (45, 46), that interact with hydrogen-bonded water molecules which share the proton to be released (47, 48). The pKa of the complex is coupled to the pKa of the primary proton acceptor Asp85 in such a way that protonation of Asp85 causes its decrease from ca 9.7 (49, 50) to 5.7 in M (51). The lesser proton affinity is accompanied by proton release in M state at neutral and moderately high pH. Mutation of Arg82, Glu204, or Glu194 disables the complex and eliminates proton release at this time in the photocycle. Instead, proton release occurs at the end of the photocycle, during deprotonation of Asp85. In XR, as in PR, the residue homologous to Glu204 is replaced with a leucine, and the release complex is not functional. In agreement with this, proton release occurs at the end of the photocycle at neutral pH in PR (11).
Unlike in BR, there is no substantial pH dependence of the kinetics of M rise in either PR or XR. This pH dependence in BR (10), abolished in the E204Q (43)and E194C (45) mutants, is attributed to deprotonation of the release complex already in the initial state (42, 46, 49). The absence of an ionizable residue homologous to Glu204 and the absence of the pH dependence of M rise in XR indicates that the proton release mechanism in XR is analogous to that in PR. The slowing of recovery of the initial state at low pH can be caused by switching to a photocycle peculiar to a pigment with an already protonated counter-ion. No M and O-like intermediates accumulate in this cycle, and recovery of initial pigment is slowed by several fold. The observed pKa 5.7 in the pH dependence of the slowest rate constant is close to the pKa of the counter-ion in the initial state (pKa of 6.0), and apparently associated with protonation of Asp83.
CONCLUSIONS
In xanthorhodopsin, as in other retinal-based proton pumps, the retinal Schiff base has a very high pKa. Deprotonation of the Schiff base and alkaline denaturation at pH >12.5 cause large absorption changes in the carotenoid antenna with the same features as those seen upon hydrolysis of the Schiff base by hydroxylamine. The resulting carotenoid spectrum resembles the spectrum of bulk (not bound to XR) salinixanthin. Protonation of the counter-ion (Asp83, from analogy with BR) occurs with a pKa of 6, as indicated by a small red shift of the retinal chromophore band and loss of the M intermediate in the photocycle. It causes a small, 0.5 nm blue shift of the carotenoid bands. The pH dependence of the recovery of the initial state in the photocycle occurs with a pKa of 9.3, which is apparently related to the pKa of the internal proton donor (Glu94). The recovery also slows at low pH, with a pKa of 6.0, which is apparently the pKa of the counter-ion and primary proton acceptor, Asp83. At very low pH transformation to a blue-shifted species occurs thermally and upon illumination. Analogy with a similar transition in PR indicates that it is likely to be associated with protonation of a residue close to the retinal chromophore, Asp223, homologous to Asp212 in BR.
Acknowledgements
The authors thank Prof. Josefa Antón (University of Alicante, Spain) for providing a strain of Salinibacter ruber. This work was supported by grants from NIH (GM29498), DOE (DEFG03-86ER13525) (to J.K.L.) and ARO (W911NF-06-01-0020 to S.P.B. and J.K.L.).
Abbreviations
- BR
bacteriorhodopsin
- PR
proteorhodopsin
- XR
xanthorhodopsin
- DM
n-Dodecyl-α-D-Maltopyranoside
- OG
n-Octyl-β-D-Glucopyranoside
- BICINE
N,N-bis[2-Hydroxyethyl]glycine
- CAPS
3-Cyclohexylamino-1-propanesulfonic acid
- CHES
2-[N-Cyclohexylamino]ethanesulfonic acid
- MES
2-[N-Morpholino]ethanesulfonic acid
- MOPS
3-[N-Morpholino]propanesulfonic acid
The numbering of amino acid residues in the XR sequence is used based on alignment with BR: Asp83, Glu94, Asp223 are the residues of XR homologous to Asp85, Asp96, and Asp212
References
- 1.Antón J, Rosselló-Mora R, Rodríguez-Valera F, Amann R. Extremely halophilic Bacteria in crystallizer ponds from solar salterns. Appl. Environ. Microbiol. 2000;66:3052–3057. doi: 10.1128/aem.66.7.3052-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antón J, Oren A, Benlloch S, Rodríguez-Valera F, Amann R, Rosselló-Mora R. Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int. J. Syst. Evol. Microbiol. 2002;52:485–491. doi: 10.1099/00207713-52-2-485. [DOI] [PubMed] [Google Scholar]
- 3.Lutnaes BF, Oren A, Liaaen-Jensen S. New C-40-carotenoid acyl glycoside as principal carotenoid in Salinibacter ruber, an extremely halophilic eubacterium. J. Nat. Prod. 2002;65:1340–1343. doi: 10.1021/np020125c. [DOI] [PubMed] [Google Scholar]
- 4.Balashov SP, Imasheva ES, Boichenko VA, Antón J, Wang JM, Lanyi JK. Xanthorhodopsin: A proton pump with a light-harvesting carotenoid antenna. Science. 2005;309:2061–2064. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oesterhelt D, Stoeckenius W. Functions of a new photoreceptor membrane. Proc. Natl. Acad. Sci. U.S.A. 1973;70:2853–2857. doi: 10.1073/pnas.70.10.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukohata Y, Ihara K, Uegaki K, Miyashita Y, Sugiyama Y. Australian Halobacteria and their retinal-protein ion pumps. Photochem. Photobiol. 1991;54:1039–1045. doi: 10.1111/j.1751-1097.1991.tb02127.x. [DOI] [PubMed] [Google Scholar]
- 7.Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 8.Mongodin EF, Nelson KE, Daugherty S, DeBoy RT, Wister J, Khouri H, Weidman J, Walsh DA, Papke RT, Sanchez Perez G, Sharma AK, Nesbø CL, MacLeod D, Bapteste E, Doolittle WF, Charlebois RL, Legault B, Rodriguez-Valera F. The genome of Salinibacter ruber: Convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18147–18152. doi: 10.1073/pnas.0509073102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Druckmann S, Ottolenghi M, Pande A, Pande J, Callender RH. Acid-base equilibrium of the Schiff base in bacteriorhodopsin. Biochemistry. 1982;21:4953–4959. doi: 10.1021/bi00263a019. [DOI] [PubMed] [Google Scholar]
- 10.Balashov SP, Govindjee R, Ebrey TG. Red shift of the purple membrane absorption band and the deprotonation of tyrosine residues at high pH. Origin of the parallel photocycles of trans-bacteriorhodopsin. Biophys. J. 1991;60:475–490. doi: 10.1016/S0006-3495(91)82074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dioumaev AK, Brown LS, Shih J, Spudich EN, Spudich JL, Lanyi JK. Proton transfers in the photochemical reaction cycle of proteorhodopsin. Biochemistry. 2002;41:5348–5358. doi: 10.1021/bi025563x. [DOI] [PubMed] [Google Scholar]
- 12.Imasheva ES, Balashov SP, Wang JM, Dioumaev AK, Lanyi JK. Selectivity of retinal photoisomerization in proteorhodopsin is controlled by aspartic acid 227. Biochemistry. 2004;43:1648–1655. doi: 10.1021/bi0355894. [DOI] [PubMed] [Google Scholar]
- 13.Mowery PC, Lozier RH, Chae Q, Tseng Y-W, Taylor M, Stoeckenius W. Effect of acid pH on the absorption spectra and photoreactions of bacteriorhodopsin. Biochemistry. 1979;18:4100–4107. doi: 10.1021/bi00586a007. [DOI] [PubMed] [Google Scholar]
- 14.Fischer U, Oesterhelt D. Chromophore equilibria in bacteriorhodopsin. Biophys. J. 1979;28:211–230. doi: 10.1016/S0006-3495(79)85172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedrich T, Geibel S, Kalmbach R, Chizhov I, Ataka K, Heberle J, Engelhard M, Bamberg E. Proteorhodopsin is a light-driven proton pump with variable vectoriality. J. Mol. Biol. 2002;321:821–838. doi: 10.1016/s0022-2836(02)00696-4. [DOI] [PubMed] [Google Scholar]
- 16.Lakatos M, Lanyi JK, Szakács J, Váró G. The photochemical reaction cycle of proteorhodopsin at low pH. Biophys. J. 2003;84:3252–3256. doi: 10.1016/S0006-3495(03)70049-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeda A, Iwasa T, Yoshizawa T. Formation of 9-cis and 11-cis-retinal pigments from bacteriorhodopsin by irradiating purple membrane in acid. Biochemistry. 1980;19:3825–3831. doi: 10.1021/bi00557a027. [DOI] [PubMed] [Google Scholar]
- 18.Harbison GS, Smith SO, Pardoen JA, Winkel C, Lugtenburg J, Herzfeld J, Mathies R, Griffin RG. Dark-adapted bacteriorhodopsin contains 13-cis, 15-syn and all-trans, 15-anti retinal Schiff bases. Proc. Natl. Acad. Sci. U.S.A. 1984;81:1706–1709. doi: 10.1073/pnas.81.6.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown LS, Dioumaev AK, Needleman R, Lanyi JK. Connectivity of the retinal Schiff base to Asp85 and Asp96 during the bacteriorhodopsin photocycle: The local-access model. Biophys. J. 1998;75:1455–1465. doi: 10.1016/S0006-3495(98)74064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharonov AY, Tkachenko NV, Savransky VV, Dioumaev AK. Time-resolved ultraviolet-absorption changes in the photocycle of bacteriorhodopsin. Photochem. Photobiol. 1991;54:889–895. [Google Scholar]
- 21.Dioumaev AK. Evaluation of intrinsic chemical kinetics and transient product spectra from time-resolved spectroscopic data. Biophys. Chem. 1997;67:1–25. doi: 10.1016/s0301-4622(96)02268-5. [DOI] [PubMed] [Google Scholar]
- 22.Száraz S, Oesterhelt D, Ormos P. pH-induced structural changes in bacteriorhodopsin studied by Fourier transform infrared spectroscopy. Biophys. J. 1994;67:1706–1712. doi: 10.1016/S0006-3495(94)80644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koyama Y, Fujii R. Cis-trans Carotenoids in Photosynthesis: Configurations, Excited-State Properties and PhysiolgicalFunctions. In: Frank HA, Young AJ, Britton G, Cogdell RJ, editors. The Photochemistry of Carotenoids. Dordrecht / Boston / London: Kluwer Academic Publishers; 1999. pp. 161–188. [Google Scholar]
- 24.Pande C, Callender RH, Chang CH, Ebrey TG. Resonance Raman study of the pink membrane photochemically prepared from the deionized blue membrane of H. halobium. Biophys. J. 1986;50:545–549. doi: 10.1016/S0006-3495(86)83493-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imasheva ES, Shimono K, Balashov SP, Wang JM, Zadok U, Sheves M, Kamo N, Lanyi JK. Formation of a long-lived photoproduct with a deprotonated Schiff base in proteorhodopsin, and its enhancement by mutation of Asp227. Biochemistry. 2005;44:10828–10838. doi: 10.1021/bi050438h. [DOI] [PubMed] [Google Scholar]
- 26.Metz G, Siebert F, Engelhard M. Asp85 is the only internal aspartic acid that gets protonated in the M intermediate and the purple-to-blue transition of bacteriorhodopsin. A solid-state 13C CP-MAS NMR investigation. FEBS Lett. 1992;303:237–241. doi: 10.1016/0014-5793(92)80528-o. [DOI] [PubMed] [Google Scholar]
- 27.Subramaniam S, Marti T, Khorana HG. Protonation state of Asp (Glu)-85 regulates the purple-to-blue transition in bacteriorhodopsin mutants Arg-82 -> Ala and Asp-85 -> Glu: The blue form is inactive in proton translocation. Proc. Natl. Acad. Sci. U.S.A. 1990;87:1013–1017. doi: 10.1073/pnas.87.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner GJ, Miercke LJW, Thorgeirsson TE, Kliger DS, Betlach MC, Stroud R. Bacteriorhodopsin D85N: three spectroscopic species in equilibrium. Biochemistry. 1993;32:1332–1337. doi: 10.1021/bi00056a019. [DOI] [PubMed] [Google Scholar]
- 29.Luecke H, Richter H-T, Lanyi JK. Proton transfer pathways in bacteriorhodopsin at 2.3 Angstrom resolution. Science. 1998;280:1934–1937. doi: 10.1126/science.280.5371.1934. [DOI] [PubMed] [Google Scholar]
- 30.Luecke H, Schobert B, Richter H-T, Cartailler J-P, Lanyi JK. Structure of bacteriorhodopsin at 1.55 Å resolution. J. Mol. Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 31.Belrhali H, Nollert P, Royant A, Menzel C, Rosenbusch JP, Landau EM, Pebay-Peyroula E. Protein, lipid and water organization in bacteriorhodopsin crystals: a molecular view of the purple membrane at 1.9 Å resolution. Structure. 1999;7:909–917. doi: 10.1016/s0969-2126(99)80118-x. [DOI] [PubMed] [Google Scholar]
- 32.Okumura H, Murakami M, Kouyama T. Crystal structure of acid blue and alkaline purple forms of bacteriorhodopsin. J. Mol. Biol. 2005;351:481–495. doi: 10.1016/j.jmb.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 33.Song L, Yang D, El-Sayed M, Lanyi JK. Retinal isomer composition in some bacteriorhodopsin mutants under light and dark adaptation conditions. J. Phys. Chem. 1995;99:10052–10055. [Google Scholar]
- 34.Balashov SP. Protonation reactions and their coupling in bacteriorhodopsin. Biochim. Biophys. Acta. 2000;1460:75–94. doi: 10.1016/s0005-2728(00)00131-6. [DOI] [PubMed] [Google Scholar]
- 35.Dioumaev AK, Brown LS, Needleman R, Lanyi JK. Coupling of the reisomerization of the retinal, proton uptake, and reprotonation of Asp-96 in the N photointermediate of bacteriorhodopsin. Biochemistry. 2001;40:11308–11317. doi: 10.1021/bi011027d. [DOI] [PubMed] [Google Scholar]
- 36.Otto H, Marti T, Holtz M, Mogi T, Lindau M, Khorana HG, Heyn MP. Aspartic acid-96 is the internal proton donor in the reprotonaion of the Schiff base of bacteriorhodopsin. Proc. Natl. Acad. Sci. U.S.A. 1989;86:9228–9232. doi: 10.1073/pnas.86.23.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balashov SP, Lu M, Imasheva ES, Govindjee R, Ebrey TG, Chen Y, Crouch RK, Menick DR. The proton release group of bacteriorhodopsin controls the rate of the final step of its photocycle at low pH. Biochemistry. 1999;38:2026–2039. doi: 10.1021/bi981926a. [DOI] [PubMed] [Google Scholar]
- 38.Ames JB, Mathies RA. The role of back-reactions and proton uptake during the N -> O transition in bacteriorhodopsin's photocycle: A kinetic resonance Raman study. Biochemistry. 1990;29:7181–7190. doi: 10.1021/bi00483a005. [DOI] [PubMed] [Google Scholar]
- 39.Zscherp C, Schlesinger R, Tittor J, Oesterhelt D, Heberle J. In situ determination of transient pKa changes of internal amino acids of bacteriorhodopsin by using time-resolved attenuated total reflection Fourier-transform infrared spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 1999;96:5498–5503. doi: 10.1073/pnas.96.10.5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borucki B, Otto H, Heyn MP. Time-resolved linear dichroism and linear birefringence of bacteriorhodopsin at alkaline pH: Identification of two N substates with different orientations of the transition dipole moment. J. Phys. Chem. B. 2004;108:2076–2086. [Google Scholar]
- 41.Balashov SP, Govindjee R, Kono M, Imasheva E, Lukashev E, Ebrey TG, Crouch RK, Menick DR, Feng Y. Effect of the arginine-82 to alanine mutation in bacteriorhodopsin on dark adaptation, proton release, and the photochemical cycle. Biochemistry. 1993;32:10331–10343. doi: 10.1021/bi00090a008. [DOI] [PubMed] [Google Scholar]
- 42.Balashov SP, Govindjee R, Imasheva ES, Misra S, Ebrey TG, Feng Y, Crouch RK, Menick DR. The two pKa's of aspartate-85 and control of thermal isomerization and proton release in the arginine-82 to lysine mutant of bacteriorhodopsin. Biochemistry. 1995;34:8820–8834. doi: 10.1021/bi00027a034. [DOI] [PubMed] [Google Scholar]
- 43.Brown LS, Sasaki J, Kandori H, Maeda A, Needleman R, Lanyi JK. Glutamic acid 204 is the terminal proton release group at the extracellular surface of bacteriorhodopsin. J. Biol. Chem. 1995;270:27122–27126. doi: 10.1074/jbc.270.45.27122. [DOI] [PubMed] [Google Scholar]
- 44.Govindjee R, Misra S, Balashov SP, Ebrey TG, Crouch RK, Menick DR. Arginine-82 regulates the pKa of the group responsible for the light-driven proton release in bacteriorhodopsin. Biophys. J. 1996;71:1011–1023. doi: 10.1016/S0006-3495(96)79302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balashov SP, Imasheva ES, Ebrey TG, Chen N, Menick DR, Crouch RK. Glutamate-194 to cysteine mutation inhibits fast light-induced proton release in bacteriorhodopsin. Biochemistry. 1997;36:8671–8676. doi: 10.1021/bi970744y. [DOI] [PubMed] [Google Scholar]
- 46.Dioumaev A, Richter H-T, Brown LS, Tanio M, Tuzi S, Saito H, Kimura Y, Needleman R, Lanyi JK. Existence of a proton transfer chain in bacteriorhodopsin: Participation of Glu-194 in the release of protons to the extracellular surface. Biochemistry. 1998;37:2496–2506. doi: 10.1021/bi971842m. [DOI] [PubMed] [Google Scholar]
- 47.Rammelsberg R, Huhn G, Lübben M, Gerwert K. Bactetriorhodopsin’s intramolecular proton-release pathway consists of a hydrogen-bonded network. Biochemistry. 1998;37:5001–5009. doi: 10.1021/bi971701k. [DOI] [PubMed] [Google Scholar]
- 48.Garczarek F, Brown LS, Lanyi JK, Gerwert K. Proton binding within a membrane protein by a protonated water cluster. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3633–3638. doi: 10.1073/pnas.0500421102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kono M, Misra S, Ebrey TG. pH dependence of light-induced proton release by bacteriorhodopsin. FEBS Lett. 1993;331:31–34. doi: 10.1016/0014-5793(93)80291-2. [DOI] [PubMed] [Google Scholar]
- 50.Balashov SP, Imasheva ES, Govindjee R, Ebrey TG. Titration of aspartate-85 in bacteriorhodopsin: What it says about chromophore isomerization and proton release. Biophys. J. 1996;70:473–481. doi: 10.1016/S0006-3495(96)79591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zimányi L, Váró G, Chang M, Ni B, Needleman R, Lanyi JK. Pathways of proton release in the bacteriorhodopsin photocycle. Biochemistry. 1992;31:8535–8543. doi: 10.1021/bi00151a022. [DOI] [PubMed] [Google Scholar]