Abstract
Purpose
The goal of this work was to test the ability of oligonucleotide-based arrays to reproduce the results of focused bacterial artificial chromosome (BAC)-based arrays used clinically in comparative genomic hybridization experiments to detect constitutional copy number changes in genomic DNA.
Methods
Custom oligonucleotide (oligo) arrays were designed using the Agilent Technologies platform to give high-resolution coverage of regions within the genome sequence coordinates of BAC/P1 artificial chromosome (PAC) clones that had already been validated for use in previous versions of clone arrays used in clinical practice. Standard array-comparative genomic hybridization experiments, including a simultaneous blind analysis of a set of clinical samples, were conducted on both array platforms to identify copy number differences between patient samples and normal reference controls.
Results
Initial experiments successfully demonstrated the capacity of oligo arrays to emulate BAC data without the need for dye-reversal comparisons. Empirical data and computational analyses of oligo response and distribution from a pilot array were used to design an optimized array of 44,000 oligos (44K). This custom 44K oligo array consists of probes localized to the genomic positions of >1400 fluorescence in situ hybridization-verified BAC/PAC clones covering more than 140 regions implicated in genetic diseases, as well as all clinically relevant subtelomeric and pericentromeric regions.
Conclusions
Our data demonstrate that oligo-based arrays offer a valid alternative for focused BAC arrays. Furthermore, they have significant advantages, including better design flexibility, avoidance of repetitive sequences, manufacturing processes amenable to good manufacturing practice standards in the future, increased robustness because of an enhanced dynamic range (signal to background), and increased resolution that allows for detection of smaller regions of change.
Keywords: array-CGH, oligonucleotide, focused microarray, CNV, chromosome abnormalities, mosaicism
The advent of array-based copy number analysis using comparative genomic hybridization (CGH) or non-CGH methods, including analysis of single nucleotide polymorphisms, has been a breakthrough in the detection of chromosomal copy number changes in the clinical setting.1 This approach has been shown to be superior to both classical cytogenetic banding methods and fluorescence in situ hybridization (FISH)-based methods because of the greatly improved resolution and highly multiplexed nature of the method.2–4 It is clear that this “molecular cytogenetic” methodology will continue to expand the capabilities for correlations between chromosomal aberrations and clinical phenotypes. This will be invaluable, not only for the diagnostic potential, but also for eventual discovery of the true genotypic basis for specific syndromic features at the molecular level.
Until recently, most clinical applications of array-CGH, other than some cancer studies, have been based on arrays constructed by covalent attachment to glass slides of DNA from whole clones, typically cosmids, P1 artificial chromosomes (PACs), or bacterial artificial chromosomes (BACs), or of polymerase chain reaction products generated from such clones. Although arrays with whole genome coverage have been produced,5–10 the great majority of clinical cases have been analyzed on much more focused arrays, partly because of the problems of production, analysis, and interpretation of the extensive amount of data that can be generated in array experiments.11,12 These arrays have been primarily concentrated in specific genomic regions that have either been shown to correlate with genetic disorders, or which are expected to have that potential (e.g., subtelomeric and pericentromeric regions).13–16
Because of the technical limitations of array production and establishment of rigorous quality control standards for spotted clone-based arrays, it seemed likely that this approach would be only a temporary solution to detect genomic copy number changes and would most likely be supplanted by oligonucleotide arrays.17 The latter have been shown to be very powerful platforms for many types of hybridization-based studies, including analysis of gene expression, DNA methylation, chromatin immunoprecipitation, and single nucleotide polymorphism (reviewed in studies by Koczan and Thiesen, Shaikh, and Zahir and Friedman18–20). Preliminary work has been performed on several of the competing platforms to demonstrate proof-of-principle for applicability to copy number analysis.21–25 However, systematic validations and studies of specific technical aspects have been limited.26 Here, we have undertaken a side-by-side comparison of custom-designed oligonucleotide-focused arrays with our clinical BAC arrays to address these issues.
There has been considerable debate recently about the relative merits of focused versus “whole-genome” array analysis.27–29 The approach described here easily lends itself to future expansion that will blur the distinctions between focused and nonfocused arrays by allowing many options for array design and analysis.
MATERIALS AND METHODS
DNA samples
All patients studied were referred to the BCM cytogenetics laboratory for clinical array-CGH analysis. DNA was extracted from whole blood using the Puregene DNA extraction kit (Gentra, Minneapolis, MN) according to the manufacturer’s instructions.
BAC array designs
Our Version 5.0 BAC microarray (BAC V5) included 853 BAC and PAC clones; it was designed to contain 3–10 BAC/PAC clones for regions corresponding to 75 known genomic disorders as well as all 41 subtelomeric regions and 43 pericentromeric regions.15 The Version 6 BAC microarray (BAC V6) includes 1472 BAC and PAC clones. This version covers approximately 150 genomic disorders with minimum backbone coverage of every chromosome at the 650-band level of cytogenetic resolution (http://www.bcm.edu/cma/table.htm).
Oligonucleotide microarray synthesis and oligo probe selection
Microarrays were synthesized using ink-jet technology with phosporamide chemistry (Agilent Technologies, Inc., Santa Clara, CA).30,31 Probe sequences were chosen from the HD CGH database (eArray, Agilent Technologies), designed in silico, and empirically validated using two-color array-CGH methods.32 The entire human genome was tiled and oligos selected based on melting temperature (Tm), secondary structure, and homology to other sites in the human genome. Oligos specially designed for array-CGH were Tm-matched by trimming, from 60 nucleotide bases (60 mers) to match the target temperature. Although most oligo sequences were 60 bases, some shorter ones (down to 45 mers) were selected to make the oligo selection isothermal. Nonhybridizing nucleotide stilts were used to make all the oligos a uniform 60 bases in length. Oligos were also searched for homology to the human genome (Build 35 hg17) to avoid cross-hybridization, which could lead to confusion for positional mapping of the oligo. Only unique oligos were selected for inclusion in the HD database.
Clinical oligonucleotide array designs
Three oligo array designs corresponding to BAC V5 and V6 arrays were manufactured by Agilent Technologies using standard procedures. First, an oligo array containing 40,937 oligonucleotides of approximately 60 bases mapping within the sequence coordinates of BAC/PAC locations for our BAC V5 array was synthesized (oligo V5). The genome sequence coordinates were determined for BAC clones using the UCSC Genome Browser resources with the May 2004 (hg17) build. Two oligonucleotide arrays were subsequently developed to obtain approximate equivalence in coverage to our BAC V6 array; both of these arrays were based on the March 2006 (hg18) build. To optimize the oligo selection, initially an array containing approximately 100,000 oligos was synthesized with two arrays on each slide. Ten hybridizations were performed with these arrays using male (M) and female (F) reference DNAs (4 × F vs. F, 2 × M vs. M, 4 × M vs. F). An analysis of the intensity distribution from these hybridizations showed consistently low intensities for <5% of the oligos, and these oligos were then eliminated. To further reduce the number of oligos, the range of each BAC interval covered by the oligos was determined as a percentage of the BAC region; a uniform coverage statistic based on splitting each BAC interval into 15K bins and calculating the observed deviation from uniform coverage of each BAC was also computed. Oligos were then eliminated so as to maintain high coverage as a percent of the BAC and high uniformity in the distribution along the BAC. Finally, we enforced the rule that BACs retain a minimum of approximately 10–15 oligos whenever possible. After this preselection process, 42,640 oligonucleotides corresponding to genomic regions covered by the BAC V6 arrays were chosen. This targeted oligo array (oligo V6) was manufactured in a 4 × 44K format, with an average of 28–30 oligos per region previously covered by a single BAC clone.
Array-CGH analysis
All array-CGH analyses were performed with gender-matched reference DNA from a single phenotypically normal male or female unless otherwise noted. The procedures for probe labeling and hybridization of our BAC arrays were reported previously.15 The procedures for DNA digestion, labeling, and hybridization for the oligo arrays were performed according to the manufacturer’s instructions, with some modifications. Briefly, 1–2 μg of genomic DNA from experimental and gender-matched reference samples were digested with AluI (10 units) and RsaI (10 units) (Promega, Madison, WI) at 37°C for 2 hours. The labeling reaction was performed using the Bioprime CGH Labeling Module (Invitrogen, Carlsbad, CA) at 37°C for 2 hours in the presence of cyanine 5-dCTP (for the experimental sample) or cyanine 3-dCTP (for the reference sample) (PerkinElmer, Boston, MA). For experiments involving dye-swap labeling, two experiments were performed with reversal of the dye labels incorporated into the control and test samples. Experimental and reference DNAs for each hybridization were purified, pooled, and incubated with human Cot-1 DNA (Invitrogen) and blocking agent (Agilent Technologies). The labeled samples were applied to an array, which was placed in a microarray hybridization chamber (Agilent Technologies), hybridized for more than 20 hours at 65°C in a rotating hybridization oven and washed according to the manufacturer’s protocol (Agilent Technologies).
Imaging and data analysis
The slides were scanned into image files using a GenePix Model 4000B microarray scanner (Molecular Devices, Sunnyvale, CA) or an Agilent Microarray Scanner (PN G2565BA). Microarray image files of oligo arrays were quantified using Agilent Feature Extraction software (v9.0), and text file outputs from the quantitation analysis were imported either into the Agilent CGH Analytics software program or converted to BAC-level emulation data by combining oligo data corresponding to regions encompassed by BAC clones (“emulated BAC clone”) and then using our in-house analysis package for copy number analysis, as described previously.12,15,33
RESULTS
Comparison of BAC arrays with focused oligo arrays
Initially, two basic questions were addressed in this study. First, would oligonucleotide-based arrays reliably recapitulate the findings of both increased and decreased copy number changes detected by focused BAC arrays? Second, was the classical dye-reversal design used for array-CGH with clone-based arrays necessary to obtain statistically valid quantitative data? To address these questions, we initially developed a BAC emulation array by selecting 60-mer oligonucleotides for most of the clones included in our BAC V5 array. Exact coverage equivalence was not possible with the particular clone set used to design this pilot array because approximately 7% of the targets on the clone array lacked sufficient DNA sequence information for precise placement on the human genome sequence assembly. For the remaining 790 clones, genomic sequence coordinates were used to select approximately 41,000 oligonucleotides from the Agilent CGH-HD database, which were printed in a 1 × 44K format. A total of 20 independent hybridizations were performed with these arrays. To perform direct comparisons with BAC data, hybridization ratios from all oligos mapping within the genome sequence coordinates of individual BACs were averaged to give a single regional value.
To test the need for dye-reversal for array-CGH with oligo-based arrays, we performed a hybridization with gender-mismatched normal controls and four hybridizations with gender-matched clinical samples on V5 oligo arrays by dye-swap labeling; representative images are shown in Figure 1. The average log2 ratios showing statistically significant gains or losses for the genomic regions corresponding to the BAC sequences are summarized in Table 1. From these initial data, we concluded that dye-reversal was not necessary because all the predicted changes on each sample were completely consistent between the two experimental designs and, importantly, no new regions were detected by the oligo array that might lead to false-positives. A series of 10 additional clinical samples with a variety of copy number changes were then analyzed with the V5 oligo arrays without dye-swap. In all cases, there was 100% concordance between the original BAC results and the BAC emulation results from the oligo array (data not shown).
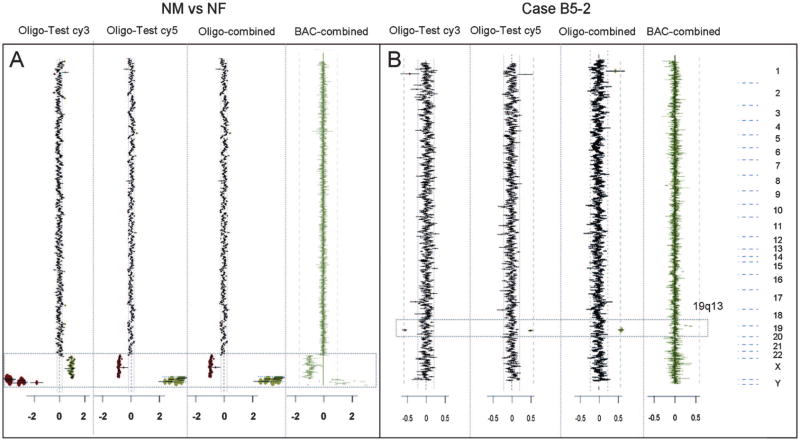
Fig. 1.
Comparison of outcomes of single-label and combined dye reversal data. A, Hybridization of normal male DNA versus normal female DNA. B, Male patient sample (Case B5-2) versus normal male reference showing a gain in copy number of chromosome 19q13. The plots are arranged such that chromosomal data are presented in order from chromosome 1 (top) to chromosome Y (bottom). The log2 ratio scale is shown at the bottom of the plot. The region of interest is indicated by the box. In each panel, Plots 1 and 2 show average log2 ratio values of test DNA labeled with cy3 or cy5 as indicated and reference DNA labeled with the opposite dye. Note that in these plots the direction of deflection from 0 (1:1 ratio) for loss or gain is dependent on the label. In Plot 3 the data sets from Plots 1 and 2 are combined with sign correction so that copy number gains and losses are plotted to the right and left, respectively. Plot 4 shows the cumulative data from a combined dye-reversal experiment on V5 BAC arrays for comparison.
Table 1.
Summary of V5 BAC and oligo array results with and without dye-swap labeling
| No. “BAC clones” detecting aberration |
Average log2 ratio |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case no. | Region | Change | Oligo patient cy5a | Oligo patient cy3b | Oligo combinedc | BAC combinedc | Oligo patient cy5a | Oligo patient cy3b | Oligo combinedc | BAC combinedc |
| M vs. Fd | X | Loss | 45 of 45 | 45 of 45 | 45 of 45 | 58 of 65 | −0.8990 | −0.9065 | −0.9028 | −0.4523 |
| B5-1 | 22q11.2 | Gain | 1 | 1 | 1 | 1 | 0.5960 | 0.5710 | 0.5830 | 0.3770 |
| B5-2 | 19q13 | Gain | 2 | 2 | 2 | 2 | 0.5340 | 0.4830 | 0.5085 | 0.3180 |
| B5-3 | 11:q24.3 | Gain | 1 | 1 | 1 | 1 | 0.5430 | 0.5190 | 0.5310 | 0.4200 |
| B5-5 | N/A | None | 0 | 0 | 0 | 0 | N/A | N/A | N/A | N/A |
BAC emulation oligo array with patient DNA labeled with cy5 and reference DNA labeled with cy3.
BAC emulation oligo array with patient DNA labeled with cy3 and reference DNA labeled with cy5.
Combined = dye swap results.
Y chromosome changes were not calculated.
Oligo array optimization
Having demonstrated the basic validity of the BAC emulation approach, an optimized oligo version of a higher density BAC array (V6) was developed. As before, BAC endpoint coordinates were determined to direct regional oligonucleotide selection to allow emulation of BAC data from the corresponding clone array. We have found previously that, even with careful selection, the actual performance of some oligos may be inconsistent or suboptimal. Therefore, to use empirical data for better optimization, initially arrays of >100K were tested with a series of normal male and female control hybridizations; it is important to note that these control samples showed no evidence of false-positive changes involving multiple consecutive oligos but did reproducibly give single oligo values that were more than 6 standard deviations from the mean. These data were combined and used to eliminate about 5% of the oligos that gave the most variable signals to eliminate background noise as much as possible. Oligo distributions within each BAC were then plotted and used to select optimal coverage with an average of one oligo per 5 kb of insert sequence. These criteria allowed selection of an approximately 44K oligo array that was essentially equivalent to the V6 BAC array in terms of genomic coverage, but with a potential 3–5-fold increase in resolution within regions previously covered by a single BAC probe because of the oligo probe redundancy at each location.
Oligo array clinical validation
The final 44K oligo V6 array design was validated in two stages. First, the same 14 clinical samples tested on the V5 oligo pilot array, of which 13 had copy number changes of apparent clinical significance based on prior BAC V5 data, plus three additional samples from patients, which had given interesting patterns previously on V5 BAC arrays (B5-4, B5-11, and B5-17), were run for comparison with previous findings; the results are summarized in Table 2. Then 21 patient samples that had been tested previously on V6 BAC arrays were analyzed. Of this group, six had been previously shown to have nonpolymorphic copy number changes on V6 BAC arrays; the results for these patients are also summarized in Table 2. In Stage 2, parallel blind analyses of 62 new patient samples were performed simultaneously on V6 BAC and V6 oligo arrays. Eighteen of the 62 cases (29%) showed one or more chromosomal locations with copy number differences (Table 2). Eleven cases (18%) showed a significant gain or loss of two or more emulated BAC clones that were suggestive of clinically relevant genomic imbalances. The remaining 7 of 18 cases gave changes that could not be dismissed as common polymorphisms and are, therefore, included in the table even though most of them were shown to also be present in a parent.
Table 2.
Summary of V6 oligo array hybridization
| No. clones |
Min. size (Mb) |
Ave. log2 ratio |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case no. | Abnormality | Region | Change | BAC | Oligo | BAC | Oligo | BAC | Oligo | Concordant |
| BAC V5 vs. Oligo V6 | ||||||||||
| B5-1 | dup(22)(q11.2) | 22q11.2 | Gain | 1 | 1 | 0.17 | 0.17 | 0.377 | 0.408 | Yes |
| B5-2 | Familial CNV | 19q13.4 | Gain | 2 | 2 | 0.91 | 0.91 | 0.261 | 0.439 | Yes |
| B5-3 | Familial CNV | 11q24.3 | Gain | 1 | 1 | 0.19 | 0.19 | 0.420 | 0.519 | Yes |
| ins(13;14)a | 13:q21.31 | Loss | 0 | 1b | NA | 0.13 | NA | −0.950 | Addb | |
| 14:q23.1-q23.3 | Gain | 1 | 2b | ~0.1 | 3.07 | 0.324 | 0.570 | Yes | ||
| B5-4 | Trisomy 21 | 21 | Gain | 13 of 16c | 19b | WC | WC | 0.408 | 0.537 | Yes |
| B5-6 | iso(18p) | 18p | Gain | 18 | 18 | 14.14 | 14.14 | 0.639 | 0.966 | Yes |
| B5-7 | der(4)t(4;8) | 4p16 | Loss | 10 | 10 | 3.39 | 3.39 | −0.289 | −0.755 | Yes |
| 8p23 | Gain | 4c | 6b | 6.45 | 6.45 | 0.219 | 0.578 | Yes | ||
| B5-8 | PWS/AS del | 15q11.2-q12 | Loss | 8b | 7 | 3.95 | 3.95 | −0.498 | −0.893 | Yes |
| B5-9 | del(2)(q14q21)d | 2q21.1-q21.3 | Loss | 1 | 3b | 0.17 | 6.97 | −0.571 | −1.029 | Yes |
| B5-10 | WBS del | 7q11.23 | Loss | 9 | 6b | 1.08 | 0.99 | −0.318 | −0.849 | Yes |
| Familial CNV | 7q36.1 | Gain | 1 | 1 | 0.14 | 0.14 | 0.318 | 0.527 | Yes | |
| B5-11 | mar(8) | 8p11.2-q11.2 | Gain | 4 | 4 | 7.46 | 7.46 | 0.424 | 0.734 | Yes |
| B5-12 | STS dup | Xp22.31 | Gain | 3 | 3 | 0.85 | 0.85 | 0.415 | 0.601 | Yes |
| B5-13 | der(6)t(6;10) | 6q27 | Loss | 1 | 4b | 0.13 | 1.58 | −0.455 | −0.861 | Yes |
| 10q24.31-q26.3 | Gain | 16 | 23b | 31.26 | 32.57 | 0.392 | 0.516 | Yes | ||
| B5-14 | WHS del | 4p16.3-p16.2 | Loss | 12 | 12 | 4.84 | 4.84 | −0.604 | −0.774 | Yes |
| B5-15 | der(13)t(1;13) | 1p36.33-p36.32 | Gain | 7 | 4 | 3.22 | 3.22 | 0.340 | 0.432 | Yes |
| 13q34 | Loss | 10 | 10 | 9.20 | 9.20 | −0.562 | −0.910 | Yes | ||
| B5-16 | del(5)(p15.32p14.2)d | 5p15.31-p14.3 | Loss | 2 | 4b | 1.18 | 10.89 | −0.525 | −0.966 | Yes |
| B5-17 | mos +13 | 13 | Gain | 20 | 47b,c | WC | WC | 0.074 | 0.114 | Yes |
| BAC V6 vs. Oligo V6 | ||||||||||
| B6-1 | mos 45,X/46,XX | X | Loss | 119 | 111b | WC | WC | −0.056 | −0.092 | Yes |
| B6-2 | Familial CNV | 7p22.2 | Gain | 1 | 1 | 0.18 | 0.18 | 0.388 | 0.540 | Yes |
| B6-6 | del(12)(q13.2q13.3)d | 12q13.3 | Loss | 1 | 1 | 0.13 | 0.13 | −0.477 | −0.952 | Yes |
| B6-9 | Possible CNV | 18q23 | Gain | 1 | 1 | 0.20 | 0.20 | 0.279 | 0.388 | Yes |
| B6-18 | dup(22)(q11.2) | 22q11.2 | Gain | 1 | 1 | 0.17 | 0.17 | 0.452 | 0.429 | Yes |
| B6-21 | dup(8)(p23.3p23.2) | 8p23.3 | Gain | 3 | 3 | 3.47 | 3.47 | 0.355 | 0.607 | Yes |
| Parallel clinical validation | ||||||||||
| C5 | dup(X)(p11.23p11.22) | Xp11.23-p11.22 | Gain | 4 | 4 | 5.22 | 5.22 | 0.133e | 0.533 | Yes? |
| C15 | Familial CNV | 5q23.1 | Gain | 1 | 1 | 0.15 | 0.15 | 0.223 | 0.222 | Yes |
| C17 | Familial CNV | 16p13.3 | Gain | 1 | 1 | 0.16 | 0.16 | 0.273 | 0.271 | Yes |
| C21 | PMD del | Xq22.2 | Loss | 3 | 3 | 0.76 | 0.76 | −0.226 | −0.801 | Yes |
| C28 | STS dup | Xp22.31 | Gain | 3 | 3 | 0.85 | 0.85 | 0.504 | 0.747 | Yes |
| C34 | WBS del | 7q11.23 | Loss | 8 | 6b | 1.08 | 0.99 | −0.364 | −0.594 | Yes |
| C35 | ins(6;X) | 6q25 | Loss | 3 | 3 | 3.69 | 3.69 | −0.428 | −0.923 | Yes |
| Xq28 | Gain | 4 | 4 | 1.13 | 1.13 | 0.404 | 0.808 | Yes | ||
| C37 | Possible CNV | 18:q11.1 | Gain | (1) | 1 | NA | 0.051 | −0.011f | 0.203 | Addg |
| C40 | 47,XXX | X | Gain | 104 of 119c | 110b | WC | WC | 0.194 | 0.508 | Yes |
| C38 | del(9)(q33.3q34.1) | 9q33.3-q34.1 | Gain | 2 | 2 | 1.64 | 1.64 | 0.263 | 0.267 | Yes |
| C42 | Familial CNV | 15q13.3 | Gain | 1 | 1 | 0.22 | 0.22 | 0.320 | 0.485 | Yes |
| C45 | dup(1)(q42.3) | 1q42.3 | Gain | 2 | 2 | 0.83 | 0.83 | 0.322 | 0.512 | Yes |
| C47 | Familial CNV | 7q22.1 | Loss | 1 | 1 | NA | 0.047 | −0.147e | −0.468 | Yesg |
| C48 | PWS/AS BP1-BP2 | 15q11.2 | Loss | 2 | 2 | 0.30 | 0.30 | −0.309 | −0.681 | Yes |
| C50 | del(6)(q25.2q25.3) | 6q25.2-q25.3 | Loss | 4 | 4 | 5.20 | 5.20 | −0.385 | −0.813 | Yes |
| C57 | Familial CNV | 6q11.1c | Loss | (1) | 1 | NA | 0.015 | −0.071f | −0.528 | Addg |
| C60 | mos inv dup(15) | 15q11.2-q13.3 | Gain | 4 of 10c | 9b | 8.88 | 8.88 | 0.706 | 1.336 | Yes |
| C63 | Familial CNV | 13q31.3 | Loss | 1 | 1 | 0.17 | 0.17 | −0.320 | −0.850 | Yes |
FISH analysis showed abnormality.
Change in no. of clones on array.
Not all BACs show gain.
Chromosome analysis showed larger abnormality.
BAC not at threshold.
Not detected by BAC array.
Partial gain or loss on oligo array.
Add, additional findings; WC, whole chromosome.
Representative side-by-side comparisons of the log ratio plots for four hybridizations are shown in Figure 2. In nearly every case there was complete concordance in the detected region of change, with the average log ratio values from the pooled oligo data consistently showing a significantly larger value than was found with the corresponding BAC clone DNA (Table 2). There were a few instances where the oligo array detected additional changes that were not statistically significant on the BAC array. For example, in Cases C5 and C47, the BAC array clone log ratio did not achieve the cutoff value of ±0.2, although in retrospect both had values well above the baseline (Table 2). The BAC array-CGH analysis also failed to detect the single clone copy number change in Cases C37 and C57. Further investigation showed that these copy number differences are caused by gains or losses involving only a portion of the sequence contained within an individual BAC clone region. By using the Web-based software to examine the copy number detected at the level of the oligos instead of at the level of the whole emulated BAC clone, these smaller changes were easily detected (Fig. 3). Furthermore, because of the improved dynamic range observed using the oligo array, these smaller “partial BAC clone changes” are now detected above the normal threshold cutoff value of ±0.2. Importantly, we found that even after careful selection of the oligos used on the final clinical microarray, variability in the hybridization intensities and relative ratios at the level of individual oligos was still observed (Fig. 3 right panel). Therefore, it remains imperative to focus the analysis on binned groups of oligos rather than examining the oligo results independently. It should also be noted that a few regions that we have found previously to be highly polymorphic with BAC arrays were also detected with the appropriate oligo-based emulation arrays. However, we have excluded these from the data discussed because they would not be considered in clinical evaluation.
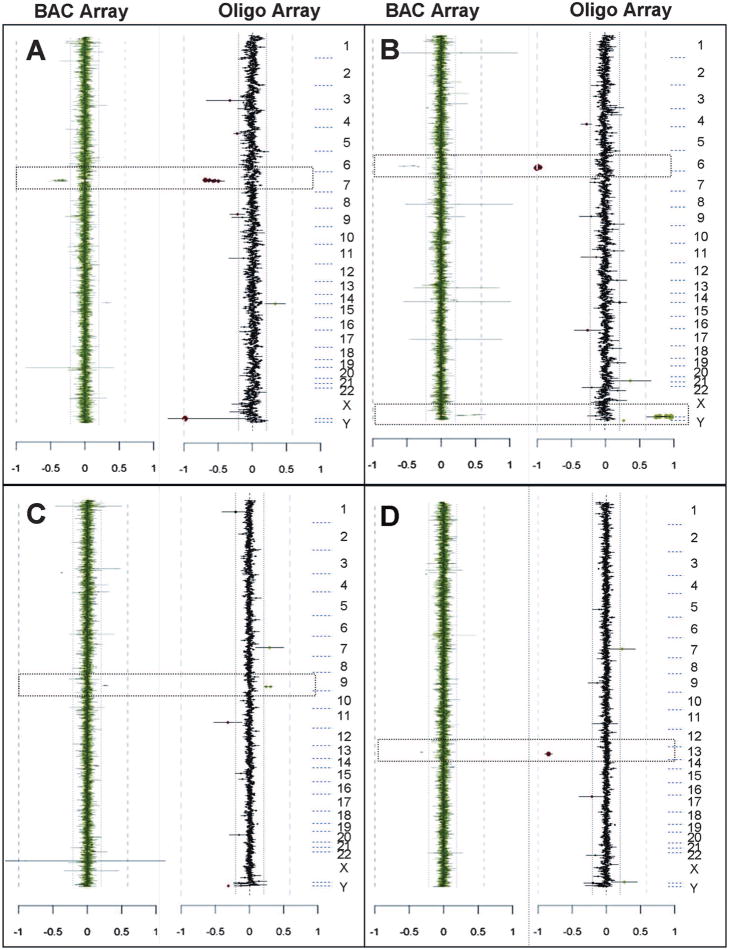
Fig. 2.
Representative whole genome plots for V6 emulation arrays. Parallel analysis of clinical cases showing copy number changes detected by both BAC (left) and oligo (right) V6 arrays. The plots are as described in Figure 1. The boxes indicate statistically significant copy number changes detected on the BAC and oligo arrays. On the oligo arrays, the red dots indicate copy number loss and the green dots indicate copy number gain. A, Case C50 shows a loss of chromosome 6q25 detected by four clones. B, Case C35 shows a loss of chromosome 6q25 detected by three clones and a gain of chromosome Xq28 detected by four clones. C, Case C38 shows a gain of chromosome 9q detected by two clones. D, Case C63 shows a loss of chromosome 13q31 detected by one clone.
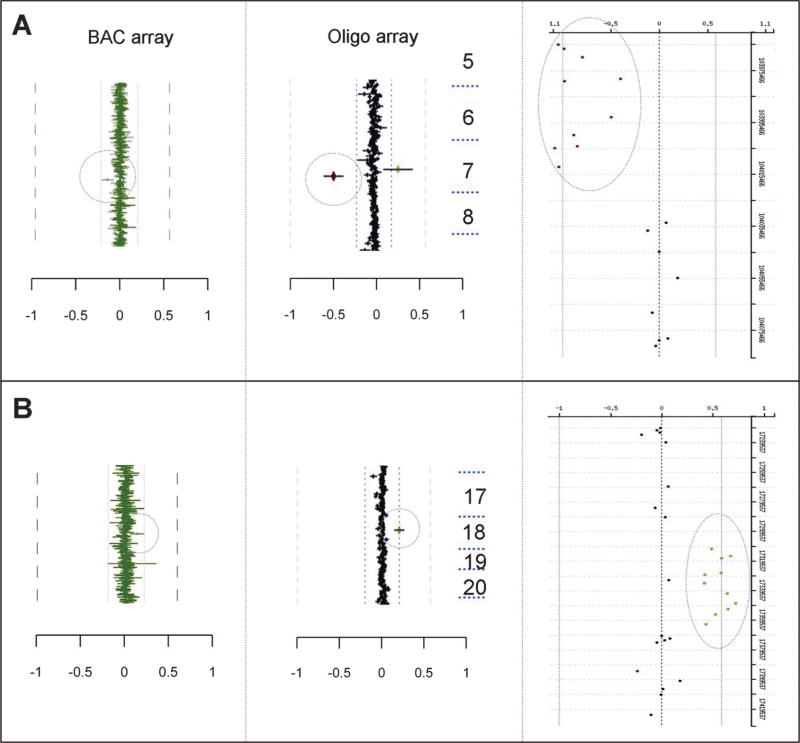
Fig. 3.
Expanded results showing the regions of interest for high-resolution analysis of partial BAC region changes detected by oligo array-CGH analysis (indicated by the circle) at the level of the whole emulated BAC (center panels) and individual oligos (right panels). The BAC array data are shown in the left panels for comparison with the circle indicating the region of interest. A, Case C47. B, Case C37.
Cases with copy number changes on the sex chromosomes were of particular interest to us. In our experience with 7482 clinical samples using gender-matched reference DNA on V5 and V6 BAC arrays, we find that approximately 14% of the abnormal clinical array-CGH cases show abnormalities involving the X or Y chromosomes (unpublished data). In the 62 blinded clinical samples that were tested using the oligo array and gender-matched control samples, we found that five (8%) had genomic imbalances involving the X chromosome ranging from approximately 700 kb to the entire chromosome (Table 2; Cases C5, C21, C28, C35, and C40). Comparative BAC and oligo array results for these are shown in Figure 4. Importantly, the potential for identifying mosaicism involving the sex chromosomes is markedly enhanced by the increased sensitivity of detection as well as by the use of gender-matched controls for all clinical samples (Case B6-1, Fig. 4).
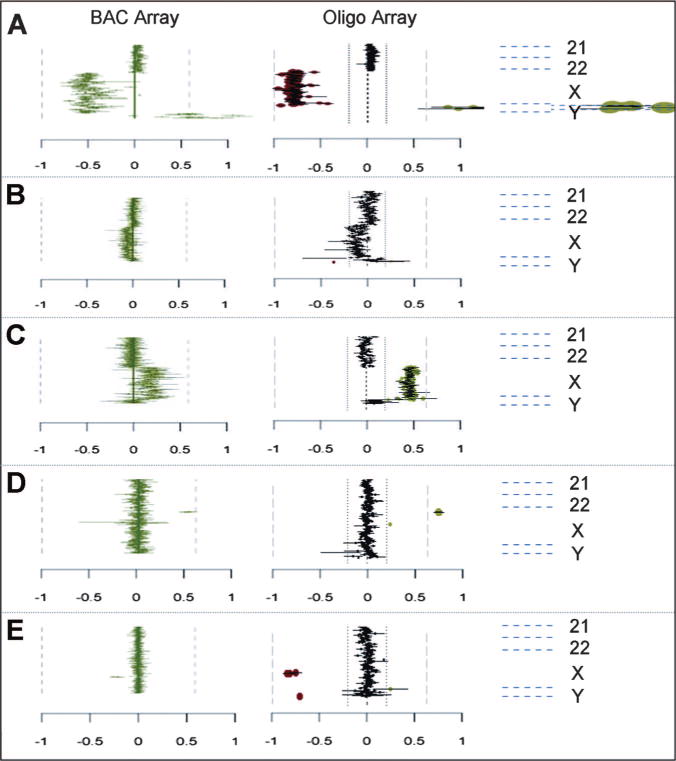
Fig. 4.
Expanded comparisons of X chromosome changes between BAC and oligo arrays. Partial plots from chromosome 21 to Y are shown for each case. A, Normal male hybridized against normal female showing a loss of all clones detecting the X chromosome and a gain of all clones detecting the Y chromosome. B, Case B6-1 shows a mosaic loss detected by clones on the X chromosome (i.e., 45,X/46,XX). C, Case C40 shows a gain detected with all the clones on the X chromosome (i.e., 47,XXX). D, Case C28 shows a gain in the steroid sulfatase (STS) region on chromosome Xp22.31 detected with three clones. E, Case C21 shows a loss in the Pelizaeus-Merzbacher disease (PMD) region on chromosome Xq22.2 detected with three clones.
DISCUSSION
Array-CGH is a powerful new approach to the quantitative determination of genomic copy number changes. As a diagnostic method, it has many advantages over classical cytogenetic or FISH techniques for evaluation of constitutional chromosomal changes leading to phenotypes as general as developmental delay, dysmorphic features, or mental retardation,8,12,22,34–42 as well as for diagnosis of many known specific genetic disorders resulting from deletions and duplications.1 The technology has been developed for clinical use primarily with arrays based on large clones, particularly BAC clones, spotted on glass arrays.13,15,43,44 However, the rapid developments in oligonucleotide-based arrays, including not only technical issues such as probe flexibility and density, but also decreasing costs and improved software capabilities, make these arrays a more attractive approach for the future. To validate a change to this platform for clinical implementation, we have performed comparative studies between our BAC arrays and custom designed oligonucleotide arrays that focus on the same sequences in the genome that are covered by these clones. From a technical viewpoint, the overall similarity between protocols greatly facilitates transition between the two platforms. In addition, this general approach allows the use of prior experience with identification of regions of copy number variation acquired from BAC arrays in interpretation of results. It is also very compatible with the continued application of FISH for independent validation of copy number changes, which we believe is still an important final step.
Although in our experience, the oligo array data are very robust and sensitive, there is additional information that can be uncovered by FISH analysis because it is the only clinical laboratory methodology that provides both copy number information and chromosomal location for gain of genomic material (i.e., insertions and translocations) at the level of an individual cell. This information is important not only for the patient, but also for family counseling and risk assessment.
Clinical validation
To demonstrate proof of principle, in Stage 1, a total of 58 independent hybridizations were performed on oligo arrays that emulated one of our BAC array designs. This included DNA samples from 14 patients that were analyzed by both V5 and V6 oligo arrays, four of which were performed by the classical dye-reversal design. In addition, 24 patient samples were retrospectively analyzed on the V6 oligo array and compared with the known BAC V5 or V6 results. Genomic imbalance was previously identified in 22 of the 38 patients. We found 100% accuracy in detection of all expected copy number changes.
In Stage 2 of the validation process, 62 patient samples were analyzed in parallel on BAC V6 and oligo V6 arrays. This blind analysis of clinical samples had a detection rate of 18% (11 of 62) for clinically relevant copy number changes. The results of the side-by-side analysis showed that the oligo array gave 100% detection rate of all changes identified by BAC arrays. In addition a few additional copy number changes were detected, which can be attributed to the increased sensitivity and resolution of the oligo arrays. We did not detect any false-negatives, demonstrating that the data generated from the BAC-emulated oligo arrays are qualitatively comparable, or superior, to the standard BAC array-CGH analysis.
Advantages of oligo-based analyses
With BAC arrays there is intrinsic signal variability because of the probe complexity resulting from the large size and repetitive DNA content of the clones, as well as issues in array production with large DNA fragments. Therefore, dye-swap experiments are normally used, in which comparing or combining the data helps compensate for some of the experimental variability and, therefore, minimizes the occurrence of false-positive or false-negative results. The demonstration of equivalent data from a single experiment for oligo arrays significantly simplifies the analysis for CGH and reduces the costs.
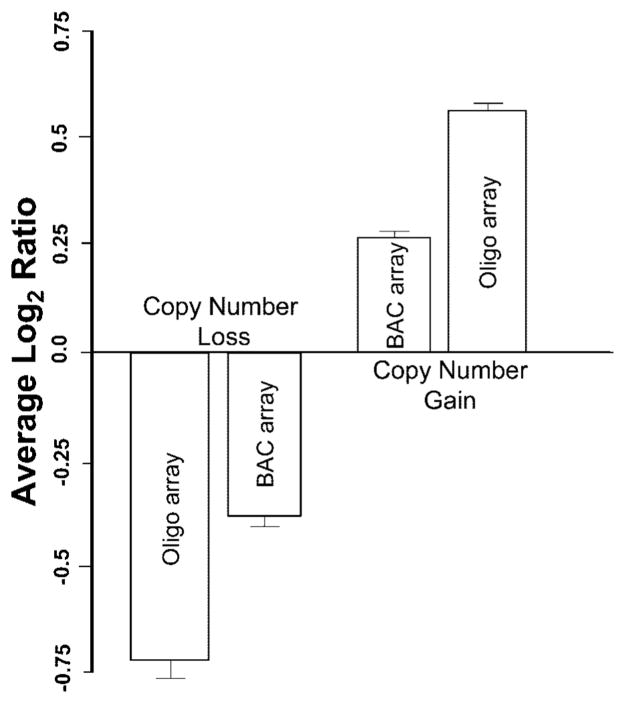
During the course of these experiments it became obvious that there were two other primary experimental advantages of the oligo-based arrays: increased dynamic range and the potential for higher resolution detection of copy number changes. First, an extended dynamic range is extremely important in assessing the validity of experimentally detected changes within regions covered by a single clone. Additionally, this increased dynamic range also facilitates the detection of mosaicism (Fig. 4B). In general, the mean value (log2 ratio) for emulated BAC clone regions showing copy number loss (total =76) was −0.716 for the oligo-based data, compared with a value of −0.379 for the corresponding clones on the BAC arrays (Fig. 5). For gains (total =186), the value was 0.565 for oligo-based data and 0.262 for BAC arrays (Fig. 5). Thus, the copy number changes on the oligo-based arrays were significantly closer to the theoretical log2 ratios for single copy loss or gain (−1 and +0.58, respectively), compared with clone arrays where the lower signals are potentially attributable to the inability to completely block some cross-hybridization from repetitive DNA, even with Cot-1 preassociation. Furthermore, the error and signal-to-noise properties of the binned oligo data were superior to the BAC array results. In more than 90% of instances the oligo data gave T statistics with stronger evidence to detect a copy number change than the corresponding T statistic from the BAC level data (data not shown).
Fig. 5.
A comparison of average log2 ratios between the BAC array data and the oligo array data illustrating the difference in dynamic range for copy number gains and losses. The log2 ratio data from 262 total clone regions across 33 patients assayed on both BAC and oligo array platforms were binned according to the BAC level data and averaged.
A second advantage of the oligo data are the ability to examine changes smaller than the average BAC size (~150 kb). For BAC arrays, confirmation by FISH analysis can sometimes produce ambiguous results. A “diminished” FISH hybridization signal is often interpreted as a possible “partial deletion” of the region detected by the clone used as the probe and a “partial duplication” is extremely challenging to distinguish by FISH analysis. Our BAC-emulation oligo array allows for visualization of the copy number change detected at the level of a BAC clone as well as by each individual oligo, thus verifying that a diminished signal observed by FISH analysis is indeed a partial deletion. This technology further provides the possibility of more accurate mapping of deletion/duplication breakpoints (Fig. 3). However, caution must be taken to avoid “over-calling” copy number changes. In our experience, the performance of individual oligos can vary (see right panels in Fig. 3). At this time, we believe that it is neither practical nor necessary to determine whether copy number changes detected by a single oligo reflect a true loss or gain in the patient or is a technical artifact. Instead, we rely on a large database comprised of all the clinical cases assayed by our laboratory using array-CGH to determine whether the copy number change is significant at the level of the binned BAC-emulation as well as the individual oligos.
Importance of gender-matched controls
Copy number changes involving the sex chromosomes are being detected with increasing frequencies. We estimate that in our experience with 7482 cases analyzed on BAC V5 and V6 arrays, approximately 14% of clinically relevant copy number changes were detected on either the X or Y chromosome (unpublished data). We find that copy number changes involving the sex chromosomes are more difficult to detect using gender mismatched reference DNA (unpublished data). The importance of using gender-matched reference DNA for array-CGH is highlighted in the data shown for the five patient samples with copy number changes involving the X chromosome (Fig. 4) and, in particular, the mosaic case (B6-1) shown in Figure 4B. Furthermore, the marked increase in dynamic range achieved using an oligo platform allows for ease of detecting very subtle changes in copy number of genomic regions on the sex chromosomes that may be missed using gender mismatched controls. This increase in sensitivity becomes more obvious when comparing the average log2 ratio for copy number change detected by BAC and oligo arrays (Table 2; Cases B5-12, B6-1, C28, C35, and C40). For the mosaic 45,X/46,XX case (B6-1), there is a 1.5-fold increase in dynamic range for the detection of the loss of genomic material on the oligo array compared with the BAC array. On average, the increase in dynamic range for copy number changes detected on the X chromosome is 2.55-fold for genomic loss and 2.3-fold for gains.
Future directions
The use of whole-genome oligonucleotide arrays for research studies is a very powerful tool because of the high resolution obtained from such arrays. They have recently been used to screen a variety of patient populations to understand the underlying genetic factors to phenotypes such as developmental delay, dysmorphic features, mental retardation,8,22,34–42 and autism.45–47 We have used them routinely for follow-up of clinical cases to, for example, map deletion or duplication endpoints and examine sequences at translocation breakpoints.48–50 For clinical analysis there are, however, additional practical issues that come into consideration. The human genome has shown much more plasticity than anticipated with regard to copy number variation, which may or may not have clinical relevance.51–53 This creates a significant challenge in data interpretation, in terms of deciding whether observed changes in an individual’s DNA relative to a control is important. Ideally, such changes can be studied for association with inheritance patterns from parents to determine their origin, but this increases both cost and complexity of the analysis. As more knowledge is gained about how to interpret such changes and as robust validation methods are developed for small changes, it is possible that whole genome tiling array analysis will become a routine diagnostic test. The data presented here represent an important intermediate step in that direction, by focusing the analysis within specific regions where clinical interpretation is assisted by precedents from BAC-based diagnostic arrays. This logic may of course be equally applicable to other specific genomic regions, such as genes, depending on the type of analysis and degree of resolution desired; although, as the resolution for genomic imbalances detected by array-CGH increases, FISH analysis may not be an option for validation, and alternative strategies will need to be developed. We believe that the transition to oligo arrays is a very positive step that will greatly improve the quality assurance for production arrays and will offer the possibility of easier upgrades in the content of future arrays as clinical implementation continues to advance to higher-resolution genome analysis.
Acknowledgments
The authors thank the CMA and FISH laboratories at Baylor College of Medicine, Medical Genetics Laboratories for their contributions to this work.
Footnotes
Disclosure: The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis offered in the Clinical Cytogenetics laboratory. C.E.C. is employed by and owns stock in Agilent Technologies, Inc.
References
- 1.Stankiewicz P, Beaudet AL. Use of array CGH in the evaluation of dysmorphology, malformations, developmental delay, and idiopathic mental retardation. Curr Opin Genet Dev. 2007;17:182–192. doi: 10.1016/j.gde.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Pinkel D, Albertson DG. Comparative genomic hybridization. Annu Rev Genomics Hum Genet. 2005;6:331–354. doi: 10.1146/annurev.genom.6.080604.162140. [DOI] [PubMed] [Google Scholar]
- 3.Vermeesch JR, Melotte C, Froyen G, Van Vooren S, et al. Molecular karyotyping: array CGH quality criteria for constitutional genetic diagnosis. J Histochem Cytochem. 2005;53:413–422. doi: 10.1369/jhc.4A6436.2005. [DOI] [PubMed] [Google Scholar]
- 4.Speicher MR, Carter NP. The new cytogenetics: blurring the boundaries with molecular biology. Nat Rev Genet. 2005;6:782–792. doi: 10.1038/nrg1692. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Jiang T, Bejjani B, Rajcan-Separovic E, et al. High-resolution human genome scanning using whole-genome BAC arrays. Cold Spring Harb Symp Quant Biol. 2003;68:323–329. doi: 10.1101/sqb.2003.68.323. [DOI] [PubMed] [Google Scholar]
- 6.Snijders AM, Nowak N, Segraves R, Blackwood S, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- 7.Fiegler H, Carr P, Douglas EJ, Burford DC, et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- 8.Miyake N, Shimokawa O, Harada N, Sosonkina N, et al. BAC array CGH reveals genomic aberrations in idiopathic mental retardation. Am J Med Genet A. 2006;140:205–211. doi: 10.1002/ajmg.a.31098. [DOI] [PubMed] [Google Scholar]
- 9.Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 10.de Vries BB, Pfundt R, Leisink M, Koolen DA, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaffer LG, Bejjani BA. Medical applications of array CGH and the transformation of clinical cytogenetics. Cytogenet Genome Res. 2006;115:303–309. doi: 10.1159/000095928. [DOI] [PubMed] [Google Scholar]
- 12.Lu X, Shaw CA, Patel A, Li J, et al. Clinical implementation of chromosomal microarray analysis: summary of 2513 postnatal cases. PLoS ONE. 2007;2:e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ballif BC, Hornor SA, Sulpizio SG, Lloyd RM, et al. Development of a high-density pericentromeric region BAC clone set for the detection and characterization of small supernumerary marker chromosomes by array CGH. Genet Med. 2007;9:150–162. doi: 10.1097/gim.0b013e3180312087. [DOI] [PubMed] [Google Scholar]
- 14.Bejjani BA, Saleki R, Ballif BC, Rorem EA, et al. Use of targeted array-based CGH for the clinical diagnosis of chromosomal imbalance: is less more? Am J Med Genet A. 2005;134:259–267. doi: 10.1002/ajmg.a.30621. [DOI] [PubMed] [Google Scholar]
- 15.Cheung SW, Shaw CA, Yu W, Li J, et al. Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet Med. 2005;7:422–432. doi: 10.1097/01.gim.0000170992.63691.32. [DOI] [PubMed] [Google Scholar]
- 16.Veltman JA, Schoenmakers EF, Eussen BH, Janssen I, et al. High-throughput analysis of subtelomeric chromosome rearrangements by use of array-based comparative genomic hybridization. Am J Hum Genet. 2002;70:1269–1276. doi: 10.1086/340426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ylstra B, van den Ijssel P, Carvalho B, Brakenhoff RH, et al. BAC to the future! or oligonucleotides: a perspective for micro array comparative genomic hybridization (array CGH) Nucleic Acids Res. 2006;34:445–450. doi: 10.1093/nar/gkj456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koczan D, Thiesen HJ. Survey of microarray technologies suitable to elucidate transcriptional networks as exemplified by studying KRAB zinc finger gene families. Proteomics. 2006;6:4704–4715. doi: 10.1002/pmic.200600010. [DOI] [PubMed] [Google Scholar]
- 19.Shaikh TH. Oligonucleotide arrays for high-resolution analysis of copy number alteration in mental retardation/multiple congenital anomalies. Genet Med. 2007;9:617–625. doi: 10.1097/gim.0b013e318148bb81. [DOI] [PubMed] [Google Scholar]
- 20.Zahir F, Friedman J. The impact of array genomic hybridization on mental retardation research: a review of current technologies and their clinical utility. Clin Genet. 2007;72:271–287. doi: 10.1111/j.1399-0004.2007.00847.x. [DOI] [PubMed] [Google Scholar]
- 21.Bignell GR, Huang J, Greshock J, Watt S, et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome Res. 2004;14:287–295. doi: 10.1101/gr.2012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toruner GA, Streck DL, Schwalb MN, Dermody JJ. An oligonucleotide based array-CGH system for detection of genome wide copy number changes including subtelomeric regions for genetic evaluation of mental retardation. Am J Med Genet A. 2007;143:824–829. doi: 10.1002/ajmg.a.31656. [DOI] [PubMed] [Google Scholar]
- 23.Hehir-Kwa JY, Egmont-Petersen M, Janssen IM, Smeets D, et al. Genome-wide copy number profiling on high-density bacterial artificial chromosomes, single-nucleotide polymorphisms, and oligonucleotide microarrays: a platform comparison based on statistical power analysis. DNA Res. 2007;14:1–11. doi: 10.1093/dnares/dsm002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carvalho B, Ouwerkerk E, Meijer GA, Ylstra B. High resolution microarray comparative genomic hybridisation analysis using spotted oligonucleotides. J Clin Pathol. 2004;57:644–646. doi: 10.1136/jcp.2003.013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Ijssel P, Tijssen M, Chin SF, Eijk P, et al. Human and mouse oligonucleotide-based array CGH. Nucleic Acids Res. 2005;33:e192. doi: 10.1093/nar/gni191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wicker N, Carles A, Mills IG, Wolf M, et al. A new look towards BAC-based array CGH through a comprehensive comparison with oligo-based array CGH. BMC Genomics. 2007;8:84. doi: 10.1186/1471-2164-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bejjani BA, Shaffer LG. Application of array-based comparative genomic hybridization to clinical diagnostics. J Mol Diagn. 2006;8:528–533. doi: 10.2353/jmoldx.2006.060029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veltman JA, de Vries BB. Diagnostic genome profiling: unbiased whole genome or targeted analysis? J Mol Diagn. 2006;8:534–537. doi: 10.2353/jmoldx.2006.060131. discussion 537–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aradhya S, Cherry AM. Array-based comparative genomic hybridization: clinical contexts for targeted and whole-genome designs. Genet Med. 2007;9:553–559. doi: 10.1097/gim.0b013e318149e354. [DOI] [PubMed] [Google Scholar]
- 30.Cleary MA, Kilian K, Wang Y, Bradshaw J, et al. Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nat Methods. 2004;1:241–248. doi: 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- 31.Hughes TR, Mao M, Jones AR, Burchard J, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 32.Barrett MT, Scheffer A, Ben-Dor A, Sampas N, et al. Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA. Proc Natl Acad Sci U S A. 2004;101:17765–17770. doi: 10.1073/pnas.0407979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw CJ, Shaw CA, Yu W, Stankiewicz P, et al. Comparative genomic hybridisation using a proximal 17p BAC/PAC array detects rearrangements responsible for four genomic disorders. J Med Genet. 2004;41:113–119. doi: 10.1136/jmg.2003.012831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bar-Shira A, Rosner G, Rosner S, Goldstein M, et al. Array-based comparative genome hybridization in clinical genetics. Pediatr Res. 2006;60:353–358. doi: 10.1203/01.pdr.0000233012.00447.68. [DOI] [PubMed] [Google Scholar]
- 35.Schoumans J, Ruivenkamp C, Holmberg E, Kyllerman M, et al. Detection of chromosomal imbalances in children with idiopathic mental retardation by array based comparative genomic hybridisation (array-CGH) J Med Genet. 2005;42:699–705. doi: 10.1136/jmg.2004.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw-Smith C, Redon R, Rickman L, Rio M, et al. Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet. 2004;41:241–248. doi: 10.1136/jmg.2003.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyson C, Harvard C, Locker R, Friedman JM, et al. Submicroscopic deletions and duplications in individuals with intellectual disability detected by array-CGH. Am J Med Genet A. 2005;139:173–185. doi: 10.1002/ajmg.a.31015. [DOI] [PubMed] [Google Scholar]
- 38.Fan YS, Jayakar P, Zhu H, Barbouth D, et al. Detection of pathogenic gene copy number variations in patients with mental retardation by genomewide oligonucleotide array comparative genomic hybridization. Hum Mutat. 2007;28:1124–1132. doi: 10.1002/humu.20581. [DOI] [PubMed] [Google Scholar]
- 39.Friedman JM, Baross A, Delaney AD, Ally A, et al. Oligonucleotide microarray analysis of genomic imbalance in children with mental retardation. Am J Hum Genet. 2006;79:500–513. doi: 10.1086/507471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krepischi-Santos AC, Vianna-Morgante AM, Jehee FS, Passos-Bueno MR, et al. Whole-genome array-CGH screening in undiagnosed syndromic patients: old syndromes revisited and new alterations. Cytogenet Genome Res. 2006;115:254–261. doi: 10.1159/000095922. [DOI] [PubMed] [Google Scholar]
- 41.Menten B, Maas N, Thienpont B, Buysse K, et al. Emerging patterns of cryptic chromosomal imbalance in patients with idiopathic mental retardation and multiple congenital anomalies: a new series of 140 patients and review of published reports. J Med Genet. 2006;43:625–633. doi: 10.1136/jmg.2005.039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagenstaller J, Spranger S, Lorenz-Depiereux B, Kazmierczak B, et al. Copy-number variations measured by single-nucleotide-polymorphism oligonucleotide arrays in patients with mental retardation. Am J Hum Genet. 2007;81:768–779. doi: 10.1086/521274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sahoo T, Cheung SW, Ward P, Darilek S, et al. Prenatal diagnosis of chromosomal abnormalities using array-based comparative genomic hybridization. Genet Med. 2006;8:719–727. doi: 10.1097/01.gim.0000245576.47154.63. [DOI] [PubMed] [Google Scholar]
- 44.Shaffer LG, Kashork CD, Saleki R, Rorem E, et al. Targeted genomic microarray analysis for identification of chromosome abnormalities in 1500 consecutive clinical cases. J Pediatr. 2006;149:98–102. doi: 10.1016/j.jpeds.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 45.Ullmann R, Turner G, Kirchhoff M, Chen W, et al. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 2007;28:674–682. doi: 10.1002/humu.20546. [DOI] [PubMed] [Google Scholar]
- 46.Jacquemont ML, Sanlaville D, Redon R, Raoul O, et al. Array-based comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J Med Genet. 2006;43:843–849. doi: 10.1136/jmg.2006.043166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sebat J, Lakshmi B, Malhotra D, Troge J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berg JS, Brunetti-Pierri N, Peters SU, Kang SH, et al. Speech delay and autism spectrum behaviors are frequently associated with duplication of the 7q11.23 Williams-Beuren syndrome region. Genet Med. 2007;9:427–441. doi: 10.1097/gim.0b013e3180986192. [DOI] [PubMed] [Google Scholar]
- 49.Probst FJ, Roeder ER, Enciso VB, Ou Z, et al. Chromosomal microarray analysis (CMA) detects a large X chromosome deletion including FMR1, FMR2, and IDS in a female patient with mental retardation. Am J Med Genet A. 2007;143:1358–1365. doi: 10.1002/ajmg.a.31781. [DOI] [PubMed] [Google Scholar]
- 50.Kang SH, Scheffer A, Ou Z, Li J, et al. Identification of proximal 1p36 deletions using array-CGH: a possible new syndrome. Clin Genet. 2007;72:329–338. doi: 10.1111/j.1399-0004.2007.00876.x. [DOI] [PubMed] [Google Scholar]
- 51.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 52.Redon R, Ishikawa S, Fitch KR, Feuk L, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sebat J, Lakshmi B, Troge J, Alexander J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]