Abstract
BACKGROUND
Prostate-infiltrating CD8+ T lymphocytes (CD8+ PIL) are prevalent in men with prostate cancer, however it is unclear whether the presence of such cells reflects a non-specific immune infiltrate or an oligoclonal, antigen-driven adaptive immune response.
METHODS
We investigated the complexity of the T cell receptor (TCR) repertoire in the prostate gland by examining the diversity of CD8+ TCR β chain variable region (Vβ) gene sequences in both the peripheral blood and prostates of cancer patients. Vβ repertoire analysis was performed by family-specific Vβ spectratyping and flow cytometry, as well as direct sequence analysis (5′ RACE and cloning). Programmed death 1 (PD-1 or PDCD1) expression on peripheral blood CD8+ T cells and CD8+ PIL was analyzed by flow cytometry.
RESULTS
CD8+ PIL isolated from cancer patients exhibited restricted TCR Vβ gene usage, and identical clones were identified in multiple sites within the prostate. Furthermore, CD8+ PIL express high levels of the inhibitory receptor PD-1, a cell surface protein associated with an “exhausted” CD8+ T cell phenotype.
CONCLUSIONS
CD8+ PIL appear to have undergone clonal expansion in response to an as yet unidentified antigen; however, due to the high expression of PD-1, these cells are likely incapable of mounting an effective immune response. The results provide an important basis for further efforts aimed at the identification of specific antigens involved in prostatic inflammation, and suggest that PD-1 blockade may be useful in immunotherapy for prostate cancer.
Keywords: Prostate cancer, inflammation, CD8+ T cell, oligoclonal, PD-1
Introduction
Prostate-infiltrating CD8+ T lymphocytes (CD8+ PIL) constitute a major proportion of prostate cancer (PCa) tumor-infiltrating lymphocytes [1,2], yet little is known regarding either the stimulus responsible for this CD8+ T cell infiltrate, or the functional status of these cells. Potential stimuli for the recruitment of inflammatory cells into the prostate gland include chronic infection, autoimmunity or tumor-associated antigens. Both chronic infection and autoimmunity have been shown to result in the accumulation of antigen-driven oligoclonal T cell populations in other organ sites [3,4], and several studies have suggested an association between prostate cancer and prior infection [5-7]. Oligoclonal infiltration of prostate tumors has been observed in animal models of the disease; recent studies in transgenic adenocarcinoma of the mouse prostate (TRAMP) mice revealed a clonal population CD8+ T cells that recognize a peptide derived from histone H4 [8]. This interesting observation suggests that tumorigenesis might alter recognition of widely expressed self-proteins, leading to a specific adaptive immune response in the target organ. Careful studies examining another oligoclonal CD8+ T cell population discovered in TRAMP mice identified a novel prostate-restricted antigen (SPAS-1), highlighting the utility of studying CD8+ T cell responses to identify potential immune target antigens in prostate cancer [9].
In humans, a number of studies have identified circulating CD8+ T cells with specificity for prostate-derived antigens. For example, CD8+ T cells specific for prostate specific antigen (PSA) have been reported in patients with either prostatitis or prostate cancer [10,11]. Another prostate-associated protein recognized by T cells includes prostate stem cell antigen (PSCA), which appears to be recognized by circulating CD8+ T cells from prostate cancer patients [12]. Interestingly, recognition of prostatic acid phosphatase (PAP) has been reported even in healthy donors [13]. Finally, studies have shown humoral immune responses to the androgen receptor (AR) in patients with prostate cancer [14]. These IgG responses suggest CD4+ T cell recognition of AR, with associated “help” for antibody production. The CD4+ T cell response to prostate tumors might be of importance as well; recent studies showed that expression of a “permissive” MHC class II allele can modulate the pattern of tumor rejection in a murine model [15].
As a result of V(D)J recombination, the receptors of the adaptive immune system display tremendous diversity, allowing for the recognition of tens of thousands of different peptides derived from self or foreign proteins. A central tenet of the clonal selection theory is that antigen-driven T cell expansion results in a population of T cells with restricted T cell receptor (TCR) variable region (Vα and Vβ) sequence diversity. Thus, receptor sequences, such as those that comprise the hypervariable complementarity determining region 3 (CDR3) regions of the TCR Vβ chain, can be used to identify clonally expanded T cell populations. Restricted TCR Vβ gene usage as evidenced by detection of clonality and/or oligoclonality of Vβ chains in T cells would be consistent with local T cell activation by one or a few specific antigens. Direct sequence analysis of the Vβ repertoire has been successfully used to identify clonal expansion of T cells in a number of disease states, including rheumatoid arthritis [3], HIV infection [16], and melanoma [17]. To date, two Vβ repertoire studies have been performed on human prostate-derived samples, both of which examined CD4+ PIL. The first study demonstrated the infiltration of oligoclonal CD4+ T cells in the prostates of cancer patients after treatment with androgen ablative therapy [18]. In the second study, an RT-PCR-based semiquantitative TCR repertoire analysis was performed on RNA extracted from prostate tissue cryosections [19]. The authors of this study concluded that the repertoire of prostate tumor-infiltrating lymphocytes was similar to that of healthy prostate tissue, but CD4+ T cells were not separated from CD8+ T cells in this analysis, and TCR repertoire was not compared to a control sample (such as peripheral blood) from the same patient.
In the present study, we assessed the relative clonality of prostate-infiltrating CD8+ T cells by family-specific Vβ spectratyping and flow cytometry, and then confirmed these results by direct sequence analysis (5′ RACE and cloning) of the TCR Vβ CDR3 sequences involved in antigen recognition. To our knowledge, these data represent the first specific analysis of the prostate-infiltrating CD8+ T cell Vβ repertoire in men with prostate cancer. Finally, we assessed these CD8+ T cells for expression of the cell surface marker PD-1, an inhibitory marker on CD8+ T cells associated with a non-functional “exhausted” phenotype [20-22].
Materials and Methods
Patient population and clinical samples
All specimens were acquired under a Johns Hopkins Medicine Institutional Review Board (IRB) approved protocol with written informed consent obtained from each patient. None of the patients were previously treated with immunosuppressive, androgen ablative, or radiation therapy. Patient-matched peripheral blood samples were collected by venupuncture into 8.5 ml whole blood tubes with ACD solution A (BD Biosciences Vacutainer Systems) the evening prior to radical retropubic prostatectomy (RRP) surgery and remained at room temperature with gentle shaking overnight. For TCR Vβ CDR3 sequence analysis, PIL samples were obtained from 7 patients (age 39-62, median 54) undergoing RRP for localized adenocarcinoma of the prostate at the Johns Hopkins Hospital in Baltimore, MD. Within one hour of resection, 15 tissue cores were obtained from both the right and left lobes of each prostate (30 cores total) using a Biopty gun and sterile, single-use Biopty needles (18 gauge × 16 cm, C.R. Bard) and collected into 5 ml RPMI media. Biopsy needles were positioned from apex to base, sampling the posterior aspect (peripheral zone) of the prostate. To digest the tissue and release infiltrating lymphocytes, 100 μl of Liberase Blendzyme 2 solution (28U/ml, Roche Applied Science) and 0.205 mg of DNase I were added and samples were incubated at 37 °C for 1 hour. Digested biopsies were then homogenized by passing through a 1000 μl pipette tip and samples were strained through a 100 μM strainer. The flow-through was spun down and re-suspended in 1 ml Dynal Buffer 1. CD8+ T cells from both peripheral blood and PIL samples were isolated using the Dynal CD8 Positive Isolation Kit (Invitrogen) according to the manufacturer’s protocol. Cells were processed in 1000 μl TRIzol (Invitrogen) and stored in the TRIzol reagent at −80°C until RNA extraction.
TCR Vβ CDR3 spectratyping
The same RNA samples used for 5′ RACE and sequence analysis were reverse transcribed using an oligo d(T) primer. Resulting cDNA was subject to 35 cycles of PCR amplification with a Vβ family-specific forward primer and conserved region reverse primer (CB-R, 5′-CTTCTGATGGCTCAAACAC-3′) labeled on the 5′ end with 6-FAM. PCR products were run on an Applied Biosystems 3700 automated flourescent sequencer coupled with GeneScan software (PE/ABD) for fragment size analysis at the Fragment Analysis Facility at the Johns Hopkins University School of Medicine (Baltimore, MD).
Antibody staining and flow cytometry
For flow cytometry analyses, peripheral blood and patient-matched PIL samples were obtained as previously described [23]. CD8+ T cells were stained directly ex vivo with the following monoclonal antibodies (mAbs): Vβ Anti-CD8 (APC) and anti-CD3 (PerCPCy5.5), and either anti-Vβ2 (PE) and anti-Vβ13.6 (FITC) or anti-Vβ8 (FITC) and anti-Vβ14 (PE) (all BD Biosciences); PD-1 Anti-CD8 (APC) and anti-PD-1 (PE, a generous gift of Alan Korman, Medarex, Inc.) with an appropriate isotype control. For PD-1 analyses, peripheral blood samples from a cohort of 10 unaffected individuals were included. Flow cytometry was conducted using a FACSCalibur (BD Biosciences) and data were analyzed using WinMDI version 2.8 (TSRI, San Diego, CA) or the FlowJo software package (Tree Star, Inc. San Carlos, CA).
TCR Vβ repertoire analysis
RNA was extracted from positively isolated CD8+ T cells using TRIzol reagent and the manufacturer’s recommended protocol. Reverse transcription of all Vβ RNA transcripts was performed with the 5′/3′ RACE Kit, 2nd Generation (Roche Applied Science) using a conserved constant region primer (CB(E)-R 5′-TTTTGGGTGTGGGAGATCTC-3′). The resultant cDNA was poly A tailed according to the 5′ RACE kit instructions and subject to 40-45 cycles of PCR using the provided forward primer and a nested constant region reverse primer (CB-R, described above). PCR products were agarose gel purified using the QIAquick Gel Extraction Kit (Qiagen) and cloned into the pCR®2.1-TOPO® vector using a TOPO TA Cloning® Kit (K4500-01, Invitrogen). Transformed colonies were picked at random, amplified with M13-F and M13-R primers, and automated sequencing was performed at the Johns Hopkins DNA Analysis Facility (Baltimore, MD) or at Polymorphic DNA Technologies, Inc. (Alameda, CA). At least 50 positive clones were chosen and analyzed per sample if possible. In some cases, cloning did not yield many transformed colonies (< 10). These products were re-cloned at least once in an attempt to increase the number of positive clones. TCR junctional regions were identified using the IMGT V-QUEST alignment software (http://imgt.cines.fr/) [24]. Vβ-family nomenclature is reported according to Arden et al. [25].
Anti-contamination controls
To minimize the possibility of contamination, all RNA extractions were performed under a UV sterilized laminar flow hood. PCR reactions and RNA extractions were performed in separate rooms. After each set of PCR reactions performed, all countertops, thermocyclers, and pipettes were cleaned with DNA AWAY reagent (Molecular BioProducts).
Results
Vβ spectratyping and flow cytometry analysis
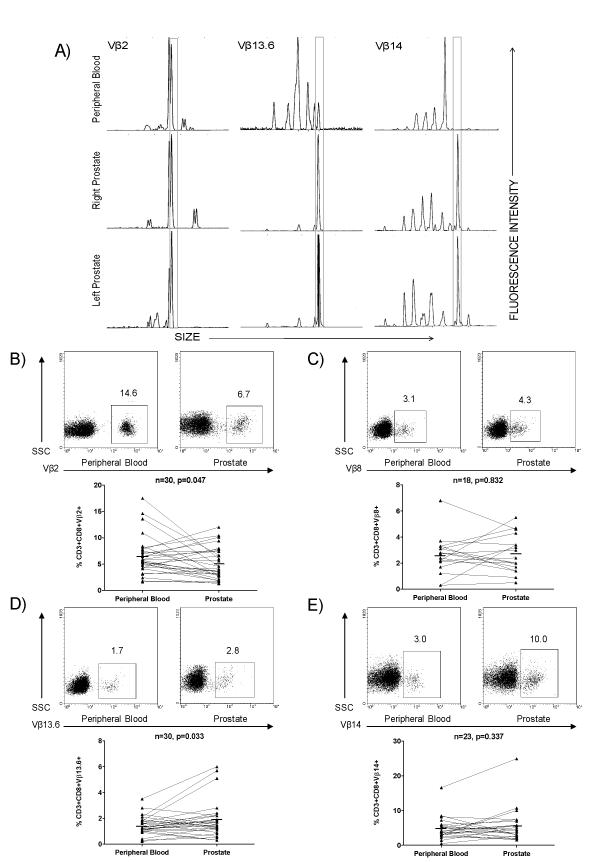
To begin to investigate the repertoire of CD8+ PIL, we performed Vβ spectratyping on CD8+ T cells isolated from the glands of men with prostate cancer using standard spectratyping techniques. This methodology has the advantage of rapidly identifying potential clonality using relatively small samples. Since we aimed to utilize spectratyping as an initial screen to assess the potential of clonality in CD8+ PIL, we chose to query 3 patients and 11 Vβ families. As shown in Fig. 1a, clonality was evident by “skewing” of certain Vβ families in the patient samples analyzed. In all cases, the clonal populations suggested in the prostate gland were not present in matched peripheral blood.
Figure 1.
TCR Vβ skewing of select Vβ families in peripheral blood and prostate of PCa patients. A) Examples of skewing observed by TCR Vβ CDR3 spectratyping. Skewed CDR3 sizes in CD8+ PIL are observed in both the right and left lobes of the prostate, but not in the peripheral blood. B-E) Positively isolated CD8+ T cells were stained directly ex vivo and analyzed by flow cytometry. Representative FACS plots of CD3+CD8+Vβ2+, CD3+CD8+Vβ8+, CD3+CD8+Vβ13.6+, and CD3+CD8+Vβ14+ T cells in peripheral blood and prostate tissue of PCa patients are shown as well as a summary of the data. Several individual patients demonstrate a relative up-regulation of a particular Vβ family within the prostate gland. P values were calculated by paired Student’s t test, two sided.
We next sought to verify these results at the protein level through the use of flow cytometry. These studies were technically challenging, as needle aspirate samples from prostate gland samples typically yield ~50,000 CD8+ T cells. As we aimed to collect and analyze at least 10,000 CD3+CD8+ events per Vβ stain, we were unable to analyze all Vβ families for which monoclonal antibodies are available. We therefore chose to analyze Vβ families 2, 8, 13.6, and 14, as these were families that appeared to be over-represented in the prostate by spectratyping as well as by sequence analysis (see below). Likewise, all stains were not possible on all samples, with the major limiting factor being the yield of CD8+ T cells from the prostate samples. However, we were able to comprehensively analyze 32 patients by flow cytometry for CD8+ T cell expression of the 4 Vβ families chosen (Fig. 1b-e). The results of these flow cytometry studies are broadly consistent with our spectratyping data, with several individual patients demonstrating a relative up-regulation of a particular Vβ family within the prostate gland. Interestingly, relative expansions were generally patient-restricted, with several patients showing prostate-specific enrichment for each of the Vβ families studied. As shown in Fig. 2, a relative prostatic enrichment of one of these four families was not detected in every patient, but this most likely reflects the small number of Vβ-specific antibodies that we were able to analyze, as well as the HLA diversity of this patient population. Interestingly, these data also showed a relative enrichment of Vβ2 expressing CD8+ T cells in the peripheral blood of a number of patients, the mechanism of which is not immediately obvious.
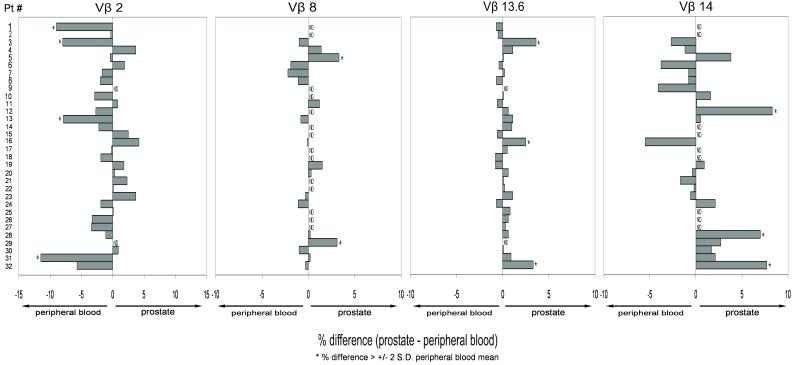
Figure 2.
Variation in clonality by patient and by Vβ family. Flow cytometry data (Fig. 1b-e) are shown as difference between % of prostate CD8+ T cells positive for given Vβ family - % of peripheral blood CD8+ T cells positive for given Vβ family. Positive values indicate higher expression on prostate-infiltrating CD8+ T cells and negative values higher expression on peripheral blood CD8+ T cells. * = Significant difference as defined by % difference > +/−2 standard deviations (S.D.) of expression of Vβ family in peripheral blood of all patients. ND = Not Determined.
Analysis of prostate TCR Vβ CDR3 gene sequences
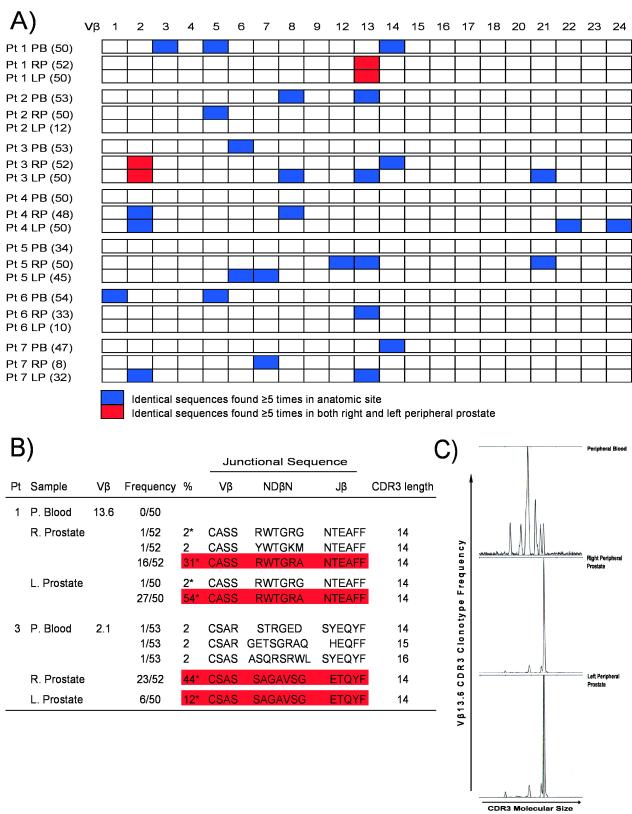
As previously discussed, direct sequence analysis of CDR3 regions has been successfully used to identify clonal expansion of T cells associated with several disease states such as rheumatoid arthritis [3], HIV infection [16], and melanoma [17]. In the present study, we utilized a similar approach which amplifies all Vβ sequences present in positively isolated CD8+ T cells from both peripheral blood and prostate using 5′ RACE with a TCR Cβ constant region primer. To decrease the chances for repeated clones based on contamination, we utilized anti-contamination measures as outlined in the Materials and Methods. As show in Fig. 3, we found significant heterogeneity in Vβ family usage between different patients as well as between peripheral blood and prostate CD8+ T cells within an individual patient. Surprisingly, a large fraction of the TCR clonotypes recovered from the prostate were identical in an individual patient, consistent with clonal expansion (Fig. 3a). For these studies, we set the criteria for a Vβ expansion as observing ≥ 5 identical sequences in one sample. These criteria are quite stringent - since the number of possible TCR β chain gene sequences in a given individual is high (on the order of 1010) [26], the a priori probability of finding two identical sequences by chance alone is extemely low. An average of 1.3 expansions were identified in the peripheral blood of the patients studied as opposed to an average of 3 expansions in the prostate. Thus, our findings are highly suggestive of localized antigen-driven CD8+ T cell expansion in the prostate of the PCa patients studied. The observation of a small degree of CDR3 clonality in the peripheral blood of several of these patients is not surprising since clonal expansions are known to occur in the peripheral blood of older individuals [27] and the median age of the patient population studied was 54 years old. In support of this notion, the youngest patient studied was 39 years old (Patient 4) and this patient showed no evidence clonality in the peripheral blood sample, but had 5 clonally expanded T cell populations in the prostate gland. Furthermore, the Vβ expansions observed in the prostate were almost always from different families than those observed in the peripheral blood, again indicating prostate-specific expansion in these patients, and arguing strongly against peripheral blood contamination of our prostate samples. Strikingly, in two patients (Patients 1 and 3), a sizeable proportion of the sequences analyzed (up to 54%) were identical in both the right and left peripheral prostate (Fig. 3b). These results were consistent with the spectratyping analysis with Vβ family-specific primers (Fig. 3c). These studies confirmed our spectratyping and flow cytometry results – oligoclonal spikes representing specific Vβ family expansions were identified in both the right and the left peripheral prostate in 5 of the 7 patients studied (Patients 1,2,3,4 and 6).
Figure 3.
Clonality of prostate-infiltrating CD8+ T cells as evidenced by TCR Vβ CDR3 repertoire sequencing. A) RNA from positively isolated peripheral blood and prostate CD8+ T cells was subject to 5′ RACE analysis using a TCR Cβ constant region primer. PCR products were cloned and sequenced. Identical sequences found either five or more times in one anatomic site (blue) or ≥ 5 times in both right and left peripheral prostate (red) are shown. Identical sequences (< 5) were also identified in both right and left peripheral zone of the prostate of Patients 1,2,3,4 and 6. PB = peripheral blood, RP = right peripheral prostate, LP = left peripheral prostate. Number in parentheses is number of clones sequenced. B) Amino acid sequence of junctional regions observed from all sequences belonging to Vβ family 13.6 derived from Patient 1 and Vβ family 2.1 derived from Patient 3. * = Identical sequences observed in both right and left peripheral prostate. C) The same RNA samples used for Patient 1 5′ RACE and sequence analysis were reverse transcribed using an oligo d(T) primer. cDNA was then amplified using a Vβ13.6 family-specific forward primer and a 6-FAM labeled TCR Cβ constant region reverse primer. Spectratyping analysis of the PCR products reveals marked skewing towards one CDR3 region length for both right and left peripheral prostate, indicative of local clonal expansion. These results are in concordance with the nucleotide sequence analyses.
HLA typing
To test whether CDR3 clonality (Fig. 3) or Vβ skewing (Figs. 1,2) of CD8+ T cells correlates with particular HLA alleles, we performed Class I HLA typing of the patients studied (Table I). This limited analysis did not reveal any particular association between HLA allelic expression and clonality – however the tremendous diversity at the HLA locus would suggest that a large number of patient samples would most likely need to be analyzed to demonstrate such an association.
Table I.
Class I HLA Genotypes of Patients Analyzed for Vβ Repertoire
| CDR3 Sequencing | Skewed family | HLA A | HLA Cw | HLA B | HLA Bw |
|---|---|---|---|---|---|
| Pt # | |||||
| 1 | 13.6 | 1,32 | 5,7 | 8,44 | 4,6 |
| 2 | 5.1 | 23,68 | 6,7 | 49,58 | 4 |
| 3 | 2.1, 8.2, 13.3, 14.1, 21.3 | 1,2 | 2,7 | 8,61 | 6 |
| 4 | 2.1, 8.2, 22.1, 24.1 | 1,2 | 6,15 | 7,57 | 4,6 |
| 5 | 6.5, 7.1, 12.1, 13.2, 21.1 | 1,2 | 5,7 | 8,61 | 6 |
| 6 | 13.6 | 2,68 | 4,5 | 35,44 | 4,6 |
| 7 | 2.1, 7.1, 13.3 | 3,24 | 1,12 | 27,35 | 4,6 |
| Flow Cytometry * | |||||
| Pt # | |||||
| 3 | 13.6 | 2,11 | 7,10 | 7,60 | 6 |
| 5 | 8 | 32 | 8 | 64 | 6 |
| 12 | 14 | 3,30 | 4,15 | 35,51 | 4,6 |
| 16 | 13.6 | 2,3 | 6,7 | 7,50 | 6 |
| 28 | 14 | 1,30 | 6 | 13,50 | 4,6 |
| 29 | 8 | 26,68 | 4,8 | 35,65 | 6 |
| 32 | 13.6,14 | 24 | 2,9 | 61,62 | 6 |
Only patients with significant Vβ skewing in prostate are shown.
Prostate-infiltrating CD8+ T cells express PD-1, a marker of CD8+ T cell exhaustion
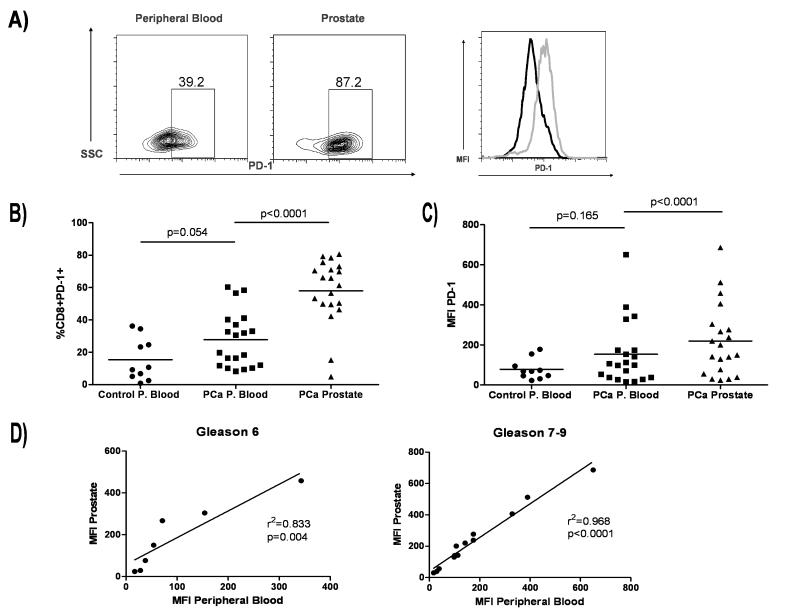
In view of the CDR3 clonality we observed in CD8+ T cells that infiltrate the prostates of PCa patients, we examined the cell surface expression of Programmed cell death 1 (PD-1), which has been demonstrated to mediate an inhibitory role on CD8+ T cells in models of infectious disease, autoimmunity, and tolerance [28]. As shown in Fig. 4, PD-1 was relatively up-regulated on CD8+ T cells infiltrating the prostate gland in men with cancer (Fig. 4a-c). In some patients, almost 90% of prostate-infiltrating CD8+ T cells were PD-1+, strongly suggesting an “exhausted” phenotype. Interestingly, in patients with high grade disease (Gleason score ≥ 7), we also observed a strong correlation between PD-1 mean fluorescence intensity (MFI) between peripheral blood and prostate-infiltrating CD8+ T cells (Fig. 4d), suggesting the possibility that systemic cytokine levels may be regulating PD-1 expression in some manner.
Figure 4.
Prostate-infiltrating CD8+ T cells exhibit an exhausted phenotype. A) FACS analysis of prostate-infiltrating CD8+ T cells. Plots are gated on CD8+ T cells and gates set using appropriate isotype controls. Far right, mean fluorescence intensity (MFI) comparison of peripheral blood CD8+PD-1+ T cells (black histogram) and prostate CD8+PD-1+ T cells (gray histogram). B) Summary of data above. % CD8+ T cells positive for PD-1 are shown. Bar = mean. C) Summary of data above. PD-1 MFI is shown. Bar = mean. P values were calculated for control peripheral blood (P. blood) versus PCa P. blood by unpaired Student’s t test, two sided and for PCa P. blood versus prostate by paired Student’s t test, two sided. D) Correlative analysis for PD-1 MFI for PCa P. blood versus prostate for low grade (Gleason 6) versus high grade (Gleason 7-9) disease.
Discussion
The results of the present study provide the first reported evidence of a substantial degree of prostate-specific CD8+ T cell clonality in prostate cancer patients as determined by TCR Vβ CDR3 sequence analysis, as well as by CDR3 spectratyping and Vβ family-specific flow cytometry. As has been previously demonstrated in other disorders, these results are potentially consistent with the recruitment of CD8+ T cells into the prostate after stimulation (or chronic stimulation) by persistent infection [4], autoimmunity [3], or the emergence of cancerous cells within the prostate. Evidence for the latter stimulus is supported by a recent study by Savage et al., which demonstrated in the TRAMP mouse model a naturally arising CD8+ T cell response reactive to a peptide derived from histone H4 [8]. Interestingly, T cell recognition of this self-peptide was specifically associated with the presence of prostate cancer. Whereas the abundance of these histone H4-reactive cells was relatively low (with a median frequency of 0.15% of prostate-infiltrating CD8+ T cells), the results of that study demonstrated for the first time a prostate cancer-specific immune response. Our results are broadly consistent with those data, supporting the notion that the CD8+ T cells in the prostate gland may be responding to a specific antigen.
Another important finding in the present study is the high degree of expression of PD-1 on prostate-infiltrating CD8+ T cells. This finding is consistent with a model of persistent antigen stimulation, as has been shown for chronic viral infections [20,29]. Interestingly, a recent study utilizing a semi-quantitative immunohistochemial assay also identified cells expressing both PD-1 and its ligand B7-H1 in lymphocyte clusters surrounding prostate cancer lesions [30]. Importantly, this study also found that PD-1+ and B7-H1+ cells were rare in both benign prostatic hyperplasia (BPH) tissues and healthy prostate tissues obtained from autopsies, indicating that the presence of PD-1+ and B7-H1+ cells are specific to prostate cancer lesions. The findings of this study and our present study are of particular importance in the design of prostate cancer immunotherapy strategies. If the majority of CD8+ T cells present within the prostate tumor microenvironment are phenotypically exhausted, therapeutic strategies which include blockade of PD-1 (for example, using anti-PD-1 neutralizing monoclonal antibodies) could potentially reverse this exhaustion and enhance therapeutic efficacy.
A logical extension of these studies would be an attempt to identify the antigen(s) to which clonal cells identified in the prostate in this study responded. This line of study represents a challenging prospect as the current techniques available for antigen identification - such as screening cDNA expression libraries with tumor-infiltrating lymphocytes (TIL) or the generation of a transgenic mouse expressing specific TCR Vα and Vβ chains - are challenging. Likewise, several possibilities could account for prostate-specific T cell clonality, including a tumor antigen, self-antigen, or perhaps even an infectious agent-associated antigen. Precedent exists for each of these possibilities [3,4,8,9,31]. Despite the challenges, identification of an antigen capable of eliciting a prostate-specific CD8+ T cell response would be tremendously important, as it could lead to more specific immunotherapy for men with prostate cancer.
Conclusions
Our results suggest that CD8+ PIL in cancer patients have undergone clonal expansion in response to an as yet unidentified antigen. Due to the high expression of PD-1, these cells are likely incapable of mounting an effective anti-tumor immune response – a result which is particularly relevant in regards to the rational design of prostate cancer immunotherapy treatments. Our study provides an important basis for further efforts aimed at the identification of specific antigens responsible for prostatic inflammation.
Acknowledgements
We would like to acknowledge and thank Kathleen Wiley and Sally Isaacs for assistance with patient recruitment and coordination, Helen Fedor and Jurga Sauvageot for assistance with sample collection, and Laura Kasch of the Johns Hopkins Fragment Analysis Facility for assistance with spectratyping analyses. C.G.D. is a Damon Runyon Clinical Fellow. These studies were supported by National Institutes of Health (NIH) R01 CA127153 (C.G.D.), K08 CA096948 (C.G.D.), and the Patrick C. Walsh Fund. This publication was made possible by Grant Number 2T32DK007552 from NIH-NIDDK to K.S.S. and its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH-NIDDK.
References
- 1.Bronte V, Kasic T, Gri G, Gallana K, Borsellino G, Marigo I, Battistini L, Iafrate M, Prayer-Galetti T, Pagano F, Viola A. Boosting antitumor responses of T lymphocytes infiltrating human prostate cancers. J Exp Med. 2005;201(8):1257–1268. doi: 10.1084/jem.20042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karja V, Aaltomaa S, Lipponen P, Isotalo T, Talja M, Mokka R. Tumour-infiltrating lymphocytes: A prognostic factor of PSA-free survival in patients with local prostate carcinoma treated by radical prostatectomy. Anticancer Res. 2005;25(6C):4435–4438. [PubMed] [Google Scholar]
- 3.Striebich CC, Falta MT, Wang Y, Bill J, Kotzin BL. Selective accumulation of related CD4+ T cell clones in the synovial fluid of patients with rheumatoid arthritis. J Immunol. 1998;161(8):4428–4436. [PubMed] [Google Scholar]
- 4.Umemura T, Yoshizawa K, Ota M, Katsuyama Y, Inada H, Tanaka E, Kiyosawa K. Analysis of T cell repertoire in the liver of patients with chronic hepatitis C. Clin Exp Immunol. 2000;121(1):120–126. doi: 10.1046/j.1365-2249.2000.01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sfanos KS, Sauvageot J, Fedor HL, Dick JD, De Marzo AM, Isaacs WB. A molecular analysis of prokaryotic and viral DNA sequences in prostate tissue from patients with prostate cancer indicates the presence of multiple and diverse microorganisms. Prostate. 2008;68(3):306–320. doi: 10.1002/pros.20680. [DOI] [PubMed] [Google Scholar]
- 6.Palapattu GS, Sutcliffe S, Bastian PJ, Platz EA, De Marzo AM, Isaacs WB, Nelson WG. Prostate carcinogenesis and inflammation: emerging insights. Carcinogenesis. 2005;26(7):1170–1181. doi: 10.1093/carcin/bgh317. [DOI] [PubMed] [Google Scholar]
- 7.De Marzo AM, Platz EA, Sutcliffe S, Xu J, Gronberg H, Drake CG, Nakai Y, Isaacs WB, Nelson WG. Inflammation in prostate carcinogenesis. Nat Rev Cancer. 2007;7(4):256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savage PA, Vosseller K, Kang C, Larimore K, Riedel E, Wojnoonski K, Jungbluth AA, Allison JP. Recognition of a ubiquitous self antigen by prostate cancer-infiltrating CD8+ T lymphocytes. Science. 2008;319(5860):215–220. doi: 10.1126/science.1148886. [DOI] [PubMed] [Google Scholar]
- 9.Fasso M, Waitz R, Hou Y, Rim T, Greenberg NM, Shastri N, Fong L, Allison JP. SPAS-1 (stimulator of prostatic adenocarcinoma-specific T cells)/SH3GLB2: A prostate tumor antigen identified by CTLA-4 blockade. Proceedings of the National Academy of Sciences. 2008;105(9):3509–3514. doi: 10.1073/pnas.0712269105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klyushnenkova EN, Ponniah S, Rodriguez A, Kodak J, Mann DL, Langerman A, Nishimura MI, Alexander RB. CD4 and CD8 T-lymphocyte recognition of prostate specific antigen in granulomatous prostatitis. J Immunother. 2004;27(2):136–146. doi: 10.1097/00002371-200403000-00007. [DOI] [PubMed] [Google Scholar]
- 11.Elkord E, Rowbottom AW, Kynaston H, Williams PE. Correlation between CD8+ T cells specific for prostate-specific antigen and level of disease in patients with prostate cancer. Clin Immunol. 2006;120(1):91–98. doi: 10.1016/j.clim.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Kiessling A, Schmitz M, Stevanovic S, Weigle B, Holig K, Fussel M, Fussel S, Meye A, Wirth MP, Rieber EP. Prostate stem cell antigen: Identification of immunogenic peptides and assessment of reactive CD8+ T cells in prostate cancer patients. Int J Cancer. 2002;102(4):390–397. doi: 10.1002/ijc.10713. [DOI] [PubMed] [Google Scholar]
- 13.Peshwa MV, Shi JD, Ruegg C, Laus R, van Schooten WC. Induction of prostate tumor-specific CD8+ cytotoxic T-lymphocytes in vitro using antigen-presenting cells pulsed with prostatic acid phosphatase peptide. Prostate. 1998;36(2):129–138. doi: 10.1002/(sici)1097-0045(19980701)36:2<129::aid-pros8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 14.Brian M, Olson DGM. Antibody and T-cell responses specific for the androgen receptor in patients with prostate cancer. The Prostate. 2007;67(16):1729–1739. doi: 10.1002/pros.20652. [DOI] [PubMed] [Google Scholar]
- 15.Klyushnenkova EN, Kouiavskaia DV, Berard CA, Alexander RB. Cutting edge: Permissive MHC Class II allele changes the pattern of antitumor immune response resulting in failure of tumor rejection. J Immunol. 2009;182(3):1242–1246. doi: 10.4049/jimmunol.182.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, Costagliola D, Rouzioux C, Agut H, Marcelin A-G, Douek D, Autran B, Appay V. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204(10):2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole DJ, Wilson MC, Rivoltini L, Custer M, Nishimura MI. T-cell receptor repertoire in matched MART-1 peptide-stimulated peripheral blood lymphocytes and tumor-infiltrating lymphocytes. Cancer Res. 1997;57(23):5320–5327. [PubMed] [Google Scholar]
- 18.Mercader M, Bodner BK, Moser MT, Kwon PS, Park ESY, Manecke RG, Ellis TM, Wojcik EM, Yang D, Flanigan RC, Waters WB, Kast WM, Kwon ED. T cell infiltration of the prostate induced by androgen withdrawal in patients with prostate cancer. Proceedings of the National Academy of Sciences. 2001;98(25):14565–14570. doi: 10.1073/pnas.251140998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebelt K, Babaryka G, Figel AM, Pohla H, Buchner A, Stief CG, Eisenmenger W, Kirchner T, Schendel DJ, Noessner E. Dominance of CD4+ lymphocytic infiltrates with disturbed effector cell characteristics in the tumor microenvironment of prostate carcinoma. Prostate. 2008;68(1):1–10. doi: 10.1002/pros.20661. [DOI] [PubMed] [Google Scholar]
- 20.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439(7077):682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 21.Chen L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat Rev Immunol. 2004;4(5):336–347. doi: 10.1038/nri1349. [DOI] [PubMed] [Google Scholar]
- 22.Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, Horton HF, Fouser L, Carter L, Ling V, Bowman MR, Carreno BM, Collins M, Wood CR, Honjo T. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192(7):1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sfanos KS, Bruno TC, Maris CH, Xu L, Thoburn CJ, DeMarzo AM, Meeker AK, Isaacs WB, Drake CG. Phenotypic analysis of prostate-infiltrating lymphocytes reveals TH17 and Treg skewing. Clin Cancer Res. 2008;14(11):3254–3261. doi: 10.1158/1078-0432.CCR-07-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefranc M-P, Giudicelli V, Kaas Q, Duprat E, Jabado-Michaloud J, Scaviner D, Ginestoux C, Clement O, Chaume D, Lefranc G. IMGT, the international ImMunoGeneTics information system(R) Nucl Acids Res. 2005;33(suppl_1):D593–597. doi: 10.1093/nar/gki065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arden B, Clark SP, Kabelitz D, Mak TW. Human T-cell receptor variable gene segment families. Immunogenetics. 1995;42(6):455–500. doi: 10.1007/BF00172176. [DOI] [PubMed] [Google Scholar]
- 26.Arstila TP, Casrouge A, Baron V, Even J, Kanellopoulos J, Kourilsky P. A direct estimate of the human T cell receptor diversity. Science. 1999;286(5441):958–961. doi: 10.1126/science.286.5441.958. eacute, ronique. [DOI] [PubMed] [Google Scholar]
- 27.Khan N, Shariff N, Cobbold M, Bruton R, Ainsworth JA, Sinclair AJ, Nayak L, Moss PAH. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J Immunol. 2002;169(4):1984–1992. doi: 10.4049/jimmunol.169.4.1984. [DOI] [PubMed] [Google Scholar]
- 28.Keir ME, Francisco LM, Sharpe AH. PD-1 and its ligands in T-cell immunity. Curr Opin Immunol. 2007;19(3):309–314. doi: 10.1016/j.coi.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 29.Urbani S, Amadei B, Tola D, Massari M, Schivazappa S, Missale G, Ferrari C. PD-1 expression in acute hepatitis C virus (HCV) infection is associated with HCV-specific CD8 exhaustion. J Virol. 2006;80(22):11398–11403. doi: 10.1128/JVI.01177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebelt K, Babaryka G, Frankenberger B, Stief CG, Eisenmenger W, Kirchner T, Schendel DJ, Noessner E. Prostate cancer lesions are surrounded by FOXP3(+), PD-1(+) and B7-H1(+) lymphocyte clusters. Eur J Cancer. 2009 doi: 10.1016/j.ejca.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 31.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254(5038):1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]