Abstract
Ras proteins have become paradigms for isoform- and compartment-specific signaling. Recent work has shown that Ras isoforms are differentially distributed within cell surface signaling nanoclusters and on endomembranous compartments. The critical feature regulating Ras protein localization and isoform-specific functions is the C-terminal hypervariable region (HVR). In this review we discuss the differential post-translational modifications and reversible targeting functions of Ras isoform HVR motifs. We describe how compartmentalized Ras signaling has specific functional consequences and how cell surface signaling nanoclusters generate precise signaling outputs.
Keywords: Palmitoylation, GTPase, isoforms, plasma membrane, nanoclusters
Introduction
Ras proteins are small GTPase molecular switches that regulate cell proliferation, differentiation, migration and apoptosis. They sit on membranes and following activation by cell surface receptors act as adaptors that recruit and facilitate activation of a wide variety of effectors. Mutations that generate aberrant, hyper-active Ras promote cancer and developmental defects [1]. Interestingly, there are three major isoforms, H-, K- and N-Ras that are ubiquitously expressed and share >90% sequence homology but are not functionally redundant. For example, only K-Ras is essential for normal mouse development [2], while K-Ras is the most frequently mutated isoform associated with human cancers, possibly due to an essential role for K-Ras but not N- and H-Ras in endodermal stem cell expansion [3,4]. Differences in the membrane interacting motifs and consequent trafficking and localization of Ras isoforms are believed to underlie their biological differences. Whilst the majority of Ras activity is associated with the plasma membrane, each isoform also resides on intracellular organelles to differing extents. As discussed later, both the cell surface and intracellular organelles regulate access to different pools of signaling complexes, allowing diversification of Ras output and potentially reducing cross-talk between isoforms.
Ras post-translational modifications and trafficking
Stabilizing membrane interactions
The first 185 amino acids of Ras proteins exhibit a high degree of homology between isoforms and contain all of the nucleotide and effector interacting domains required for Ras function. The final 23/24 amino acids comprise the hypervariable region (HVR) that defines the isoform and contains the membrane interacting and targeting sequences (Figure 1). After synthesis on cytosolic ribosomes, all three major Ras isoforms undergo sequential post-translational modifications of the HVR to enable them to more stably interact with membranes. The cysteine of the C-terminal CAAX motif is farnesylated in the cytosol by farnesyl protein transferase. This enables Ras to interact with the endoplasmic reticulum (ER) for subsequent AAX proteolysis of the farnesylated CAAX by Rce1 (homologous to Afc1p/Ste24p and Rce1p in yeast), followed by carboxymethylation of the farnesylated cysteine by Icmt (homologous to Ste14p in yeast).
Figure 1.
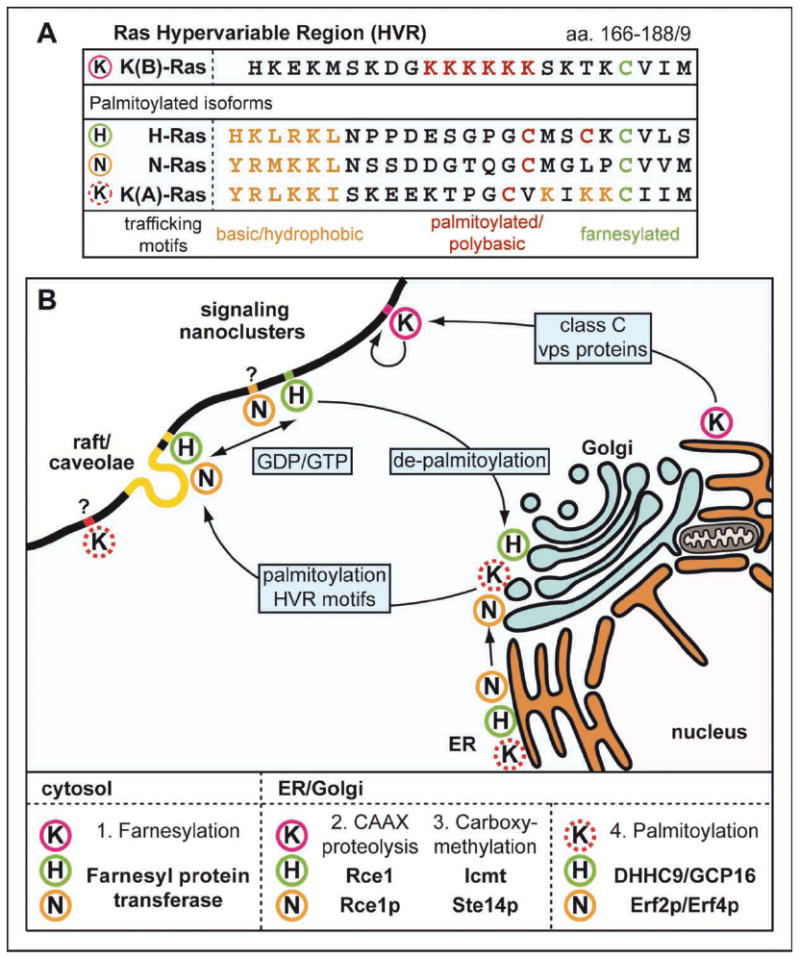
The Ras Hypervariable Region (HVR). (A) Ras isoform C-terminal HVRs have different combinations of post-translational lipid modifications and membrane interacting polybasic motifs that specify differential trafficking and localization. (B) Ras isoform localization is dynamic; changes in H- and N-Ras activation state or palmitoylation alter the association with cell surface subdomains/clusters and endomembranous compartments.
The weak membrane binding allowed by the farnesylated cysteine is supported by a second motif in the HVR that strengthens membrane interactions. This motif varies amongst Ras isoforms. For H-, N-and the 4A splice variant of K-Ras (K(A)-Ras), the second signal consists of one or two palmitoyl groups (Figure 1A). In the 4B splice variant of K-Ras (K(B)-Ras, referred to from here on as K-Ras), a hexa-lysine polybasic sequence electrostatically interacts with acidic lipid headgroups enriched in the inner leaflet of the plasma membrane. These motifs enable plasma membrane localization of the Ras isoforms and minimal sequences consisting of the H- or K-Ras CAAX motif plus second motif (dipalmitoyl or polybasic groups) recapitulate the localization of the full length proteins [5,6]. For the mono-palmitoylated K(A)-Ras and N-Ras isoforms, a third HVR motif consisting of basic/hydrophobic amino acids is necessary for plasma membrane localization (Figure 1A; [7]). These second signal motifs also specify the trafficking routes: H- and N-Ras traffic via the conventional secretory pathway, whilst preliminary data from yeast indicates that K-Ras transits via a poorly understood Golgi-independent route that requires mitochondrial function and class C vps proteins [5,8,9]. Once at the cell surface, the HVR motifs regulate interactions with different microdomains within the plasma membrane (discussed later). Importantly, the reversibility of palmitoylation and electrostatic interactions is critical for ensuring correct and dynamic localization of each isoform.
Ras acylation and reversible membrane interactions
Metazoan Ras palmitoylation is catalyzed by an ER/Golgi-localized heterodimeric complex consisting of DHHC9 (Erf2p in yeast) and GCP16 (Erf4p in yeast) [10-13]. DHHC9 is a member of a large family of 23 DHHC-motif containing mammalian protein S-acyltransferases (PATs; [14]). GCP16 was identified as a dually palmitoylated golgin (GCP170)-interacting protein [15]. The precise function of GCP16 is unclear; however it is required for DHHC9 ER/Golgi localization and function and in the absence of GCP16 DHHC9 suffers extensive proteolysis [10].
Although H- and N-Ras are ubiquitously expressed, DHHC9 is not expressed in thymus, skeletal muscle, spleen and leukocytes, indicating that other DHHC family members must also palmitoylate Ras proteins [10,13]. The extent of redundancy amongst PATs for Ras palmitoylation has not been extensively investigated. All 22 human DHHC proteins have now been cloned [13,16] and the subfamily of six Erf2-like DHHC proteins (DHHCs: 5, 8, 9, 14, 18 and 19) represent the best candidates for alternative Ras palmitoylation [14]. All are ER/Golgi localized except for DHHC5 (found exclusively on the plasma membrane) and all except DHHC9 and DHHC19 are ubiquitously expressed [13]. Preliminary support for the idea that other members of the Erf2 family of PATs can act as Ras palmitoylators comes from a study that overexpressed DHHC18 and H-Ras in HEK293 cells, resulting in increased palmitoylation of H-Ras [17].
It has been known for a while that palmitoylation is labile, and that H- and N-Ras activation significantly decreases the half-life of the attachment of their palmitoyl groups from hours to minutes [18,19]. Ras depalmitoylation is important for correct localization, because when non-hydrolysable acyl groups are attached to H-Ras, it partitions non-specifically into the entire endomembrane system [20]. Depalmitoylation results in H- and N-Ras translocation to the cytosol before re-palmitoylation at the Golgi allows another cycle of trafficking back to the cell surface. Mono-palmitoylated N-Ras is more susceptible to loss of plasma membrane anchorage than dually palmitoylated H-Ras [20]; this mechanism may explain the more pronounced Golgi localization of N-Ras in many cell types.
In addition to regulating Ras macro-localization in the cell, palmitoylation also specifies localization within cell surface subdomains. These are discussed in detail in a following section; briefly, it should be noted that palmitoylation enables access to cholesterol-sensitive nanodomains or clusters, whereas non-palmitoylated K-Ras is excluded from these domains [21,22]. Interestingly, the positioning of the palmitoyl group relative the farnesylated C-terminal cysteine is important for both trafficking and eventual subdomain localization within the plasma membrane. Palmitoylated Cys181, shared by both N-Ras and H-Ras, supports trafficking to cell surface cholesterol-dependent domains/clusters, whereas mutant H-Ras monopalmitoylated on Cys184 remains confined in the Golgi [23].
In summary, the dynamic interactions of the Ras palmitoyl or polybasic HVR targeting motifs with membranes modulate the targeting of Ras isoforms to cell surface and intracellular organelles.
Ras antagonists that perturb Ras processing
Since membrane targeting is required for Ras function, drugs targeting Ras post-translational processing have been developed as potential anti-cancer agents with mixed results. For example, farnesyl transferase inhibitors (FTIs) that in the main mimic the Ras CAAX motif and compete for farnesyl transferase binding have two potential problems. Firstly, K-Ras and N-Ras, the isoforms most frequently mutated in cancer, can bypass farnesyl transferase inhibition by being alternatively prenylated by geranylgeranyl transferase in the presence of FTIs [24]. Secondly, FTIs are not specific for Ras but also inhibit the function of other prenylated proteins such as Rho family members and Rheb [25,26]. Work is ongoing to characterize FTI targets in FTI-sensitive tumors where oncogenic Ras is not present.
Inhibition of Icmt or Rce1 function appears to be more promising, as it has in vivo anti-transformation and anti-cancer efficacy [27-29]. Whilst these proteins are also required for the processing of Rho GTPases, their effects on proteins other than Ras are fortuitously limited, as demonstrated by the finding that knocking out Icmt and Rce1 does not perturb Rho localization or function [30]. Finally, the Ras PAT DHHC9 is also a potential drug target. However, the non-palmitoylated K-Ras, which is the most frequently mutated isoform in cancer, is not susceptible to this enzyme. Furthermore, as discussed above, the PAT redundancy for Ras palmitoylation complicates the usefulness of PAT as a target for oncogenic Ras inhibition.
Targeting of Ras proteins to membrane lateral domains/nanoclusters
Lateral domains and nanoclusters in the plasma membrane
The cellular plasma membrane is laterally heterogeneous. It is comprised of a large array of subdomains, evidenced in both spatial and temporal segregation of lipids, proteins and membrane-associated scaffolds [21,31-39]. The interactions between distinct lipids and proteins in these subdomains confer the formation of nanoscale domains or clusters, including ‘lipid rafts’ (defined below) as well as other types of nanoclusters.
Lipid rafts were originally viewed as liquid-ordered cholesterol- and sphingolipid-rich membrane regions into which specific proteins partition preferentially [35,40-42]. However, more recent models treat rafts as transient nanoscale cholesterol-dependent assemblies of specific lipids and proteins, where interactions with specific proteins, scaffolds and membrane lipids influence the formation, stability and size of the cluster [38,39,43]. The potential of domain-specific interactions to regulate signaling and cellular trafficking by selective segregation of multiple interacting proteins has led to a large number of studies on the involvement of rafts in cellular processes [37,44-50]. Clear evidence for the existence of such domains in artificial lipid bilayers [38,41,51-54] led to the suggestion that they exist also in cell membranes [35,38-40,55]. However, large-scale laterally segregated cholesterol-dependent domains were not detected in cell membranes, implying that their size in cells (if they exist) must be below the resolution of light microscopy, and casting doubts on their very existence in cell membranes [56-60]. Yet, some raft-related structures were undoubtedly identified in cells; these are caveolae, assembled around caveolin as a principal structural protein, which may be considered a subtype of lipid rafts [55,61,62].
Enrichment of specific proteins in detergent-resistant membranes (DRMs) floated on density gradients, commonly used to evaluate raft association of proteins in cell extracts, is not a sufficient criterion for raft association due to the potential complex effects of detergents on nanoscale membrane domains [38,63]. Therefore, biophysical and structural studies were initiated to explore the existence and properties of cholesterol-sensitive clusters in cells. Early fluorescence resonance energy transfer (FRET) and electron paramagnetic resonance studies indicated that such clusters are small and unstable, and/or that they can rapidly exchange proteins with the surrounding membrane [64,65]. FRET studies of acylated peptides tagged with green fluorescent protein (GFP) variants anchored to the cytoplasmic plasma membrane leaflet suggested that they form clusters [66]. More recently, FRET studies combined with mathematical modeling demonstrated that a fraction (20–40%) of glycosylpho-sphatidylinositol-anchored proteins (GPI-AP) reside in ~5 nm cholesterol-dependent clusters containing 3–4 GPI-APs [43,67]. Single particle tracking and single fluorophore video tracking (SFVT) have indicated that GPI-AP clusters are small (<10 nm), dynamic, and can be stabilized by crosslinking with antibody-coated gold particles, leading to transient cholesterol-dependent recruitment (0.1– 0.2 s lifetimes) of Lyn or Gαi2 proteins [50,68]. A rather similar distribution and size was found for the lipid anchor of H-Ras (amino acids 180–189) fused to GFP (GFP-tH), which is considered an inner-leaflet marker of cholesterol-dependent clusters [21,37]. Using electron microscopy (EM) spatial analysis of immunogold point patterns in plasma membrane sheets, ~40% of GFP-tH was found in 12–20 nm cholesterol-dependent clusters comprised of ~6 proteins, which were also sensitive to the integrity of the actin cytoskeleton [21,69]. The extent of clustering remained unaltered (~40% in clusters) over a wide cell-surface density, excluding the possibility that tH partitions into pre-existing raft domains and suggesting that the H-Ras lipid anchor itself drives the formation of cholesterol-dependent nanoclusters [38,69]. Importantly, the tH clusters, similar to clusters of full-length H- and K-Ras, are dynamic (Figure 2), with lifetimes between 0.1–0.5 s as deduced from SFVT [33,36,38,50]. Thus, cholesterol-dependent dynamic proteolipid nanoclusters, such as those observed for GPI-APs and for tH, can be regarded as a modern equivalent of rafts. It is important to note that these are not the only nanoclusters associated with Ras proteins, in view of the identification of non-overlapping cholesterol-independent clusters of K-Ras (both GDP- and GTP-loaded), H-Ras-GTP and N-Ras-GDP [21,22,38,69-71], and the laterally segregated assemblies suggested by the distinct FRET pair vectors measured between a set of domain markers and tH, tK (the lipid anchor of K-Ras) or their complete HVR counterparts ([72]; see also Figure 2). The interactions of the different Ras isoforms with these distinct nanoclusters are discussed below.
Figure 2.
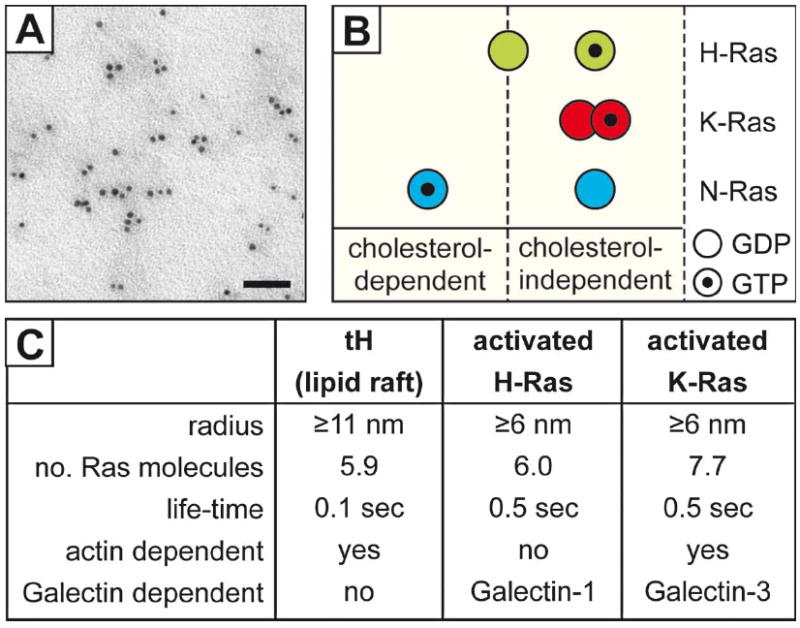
Plasma membrane Ras nanocluster parameters. EM imaging of immunogold-labeled H-Ras molecules on 2-D plasma membrane sheets (A); bar =50 nm. Ras isoforms dynamically localize to distinct signaling nanoclusters with differential cholesterol dependence (B). Other characteristics of Ras nanoclusters obtained from EM and advanced light microscopy studies are summarized in (C). This Figure is reproduced in color in Molecular Membrane Biology online.
Localization of Ras proteins to nanoclusters/assemblies and its roles in signaling
Recent data suggest that Ras proteins at the plasma membrane reside in distinct assemblies/nanodomains that depend on the Ras isoform and its GDP/GTP loading (reviewed in [37,38,73]). Initial evidence that H- and K-Ras differ in their association with cholesterol-dependent assemblies came from biochemical fractionation experiments [22,74]. These were followed by EM spatial immuno-gold point-pattern analysis and biophysical studies that measured the clustering, lateral diffusion and interactions of Ras proteins in cell membranes. These studies yielded compelling evidence that Hand K-Ras in the plasma membrane are targeted to largely non-overlapping nanoclusters. Their distributions are modulated by GDP/GTP exchange; unactivated wild-type H-Ras (H-Ras-GDP) exhibits the highest affinity to cholesterol-sensitive clusters, H-Ras-GTP (e.g. constitutively active H-RasG12V) has a preference for cholesterol-independent domains/clusters, while either wild-type K-Ras (K-Ras-GDP) or K-RasG12V (K-Ras-GTP) interact with cholesterol-independent clusters ([21,22,32,70,75,76]; see Figure 2). Although both are cholesterol-independent, the H-Ras-GTP and K-Ras-GTP nanoclusters are physically distinct [21,32,69], exhibit different dependencies on the actin cytoskeleton (only K-Ras-GTP clusters and signaling are actin-dependent; [69]), and differ in their ability to recruit Raf-1 [70]. The selective association of H-Ras-GDP (as opposed to K-Ras) with cholesterol-dependent clusters is in accord with the demonstration that prenylation alone (e.g. K-Ras) targets proteins to non-raft (non-DRM) domains, while dual acylation (S-palmitoyl and/or N-myristoyl residues) enhances their association with DRMs [77,78]. Interestingly, a dependence of the balance between cholesterol-dependent and independent clusters on the GDP/GTP loading state was also observed for N-Ras (Figure 2), although in the latter case it is N-Ras-GTP which is preferentially localized in cholesterol-sensitive clusters [23]. Apart from the important roles of differential targeting to nanoclusters in Ras signaling (see next section), it may also affect the susceptibility of distinct Ras isoforms to specific modifications. One such example is the selective ubiquitination of the G-domain of H-Ras (and N-Ras, but not K-Ras), which was shown to require both the CAAX box and the palmitoylation sites on H-Ras, and results in its transport to endosomes [79]. It should be noted that some studies reported that the lateral diffusion of wild-type H-Ras is insensitive to cholesterol depletion [33,80], seemingly at odds with its preferential localization to cholesterol-dependent clusters. However, this discrepancy is likely due to the use of methyl–cyclodextrin to deplete cholesterol, a treatment that has additional effects on the plasma membrane that are unrelated to cholesterol depletion [81,82]. An alternative method (metabolic inhibition of cholesterol synthesis using statins) did not lead to such artifacts and increased (~2-fold) the lateral diffusion rate of wild-type H-Ras without affecting K-Ras diffusion [32,81,82].
The lateral segregation of Ras proteins depends not only on their lipid anchors, but also on additional features in the HVR regions (depicted in Figure 1). This was most thoroughly investigated for H-Ras. Combining fluorescence recovery after photobleaching (FRAP) beam-size analysis (a FRAP variation that measures the relative contribution of membrane-cytoplasmic exchange and lateral diffusion to the FRAP recovery kinetics; [83]) with EM spatial pattern analysis, it was shown that the lipid anchor and the HVR linker region of H-Ras (residues 166–179; Figure 1) exert distinct and significant attractive forces targeted to specific membrane domains [75]. The lipid anchor, especially the palmitoyl at Cys181, favored association with cholesterol-sensitive assemblies, while the HVR linker region interacted preferentially with non-raft domains and clusters [23,75]. The G-domain (residues 1–165) had a negative contribution, the extent of which was modulated by GDP/GTP exchange (weaker membrane association in the GTP-bound form). Thus, apart from its role in the regulation of H-Ras segregation between cholesterol-dependent and independent domains/clusters, the HVR linker also contributes to the stabilization of H-Ras association with the plasma membrane, and both effects are modulated by the GDP/GTP loading state [37,75]. Insight into the mechanism underlying the guanyl nucleotide-dependent changes in H-Ras membrane association and lateral segregation was recently provided by applying molecular dynamics simulations [84] to NMR data on the farnesylated full-length H-Ras in DMPC bilayers [85], and by molecular dynamics simulations combined with FRET studies and mutational analysis of H-Ras with respect to a set of nanodomain markers [72]. These studies demonstrated different interactions of H-Ras with the membrane depending on GTP/GDP exchange, with different orientation of the G-domain and HVR region relative to the lipid bilayer due to different contributions of basic residues in the HVR linker (in H-Ras-GDP) and in helix α4 (H-Ras-GTP) to membrane binding (Figure 3). Notably, the extended conformation of the palmitoyl moieties for membrane-bound H-Ras-GDP results in deeper insertion into the bilayer than in the GTP-bound conformation ([72,84]; see Figure 3). This may increase the availability of the GTP-loaded conformation for interaction with the scaffold protein galectin-1, explaining its preferential binding to the GTP-loaded conformation of H-Ras [21,86-88].
Figure 3.
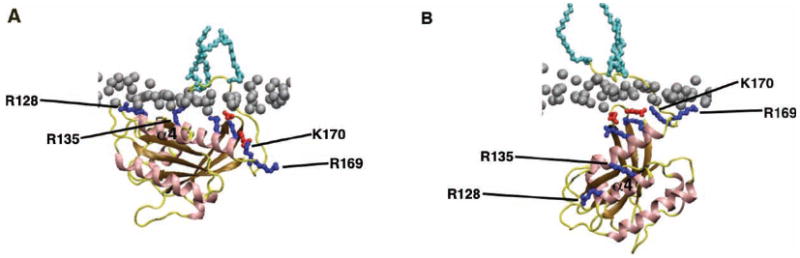
Molecular Dynamics simulations of H-Ras HVR interactions with the plasma membrane. The H-Ras G-domain and HVR lipid moieties adopt different orientations with respect to the plane of the membrane when GTP-bound (A) and GDP-bound (B). The GTP-bound conformation is stabilized by membrane contacts with basic residues (R128 and R135) on helix α4. These contacts are lost in GDP-H-Ras, which is stabilized by an alternative set of basic residues within the HVR. Note also that the palmitoyl groups exhibit a more extended conformation when H-Ras is GDP-bound. Phosphorous atoms of lipid head groups of the inner membrane leaflet are shown in grey and H-Ras lipid anchors are in light blue. Important basic residues in H-Ras are shown in dark blue and acidic residues in red. Reproduced with permission from [72].
The K-Ras HVR region also plays a crucial role in K-Ras membrane association and recruitment to nanoclusters. In accord with the absence of palmitoylation sites in K-Ras, it is not targeted to raft-like domains/clusters [21,22,32,76,81]. Rather, the high positive charge (8+, including a 6-lysine basic cluster) at the K-Ras HVR region binds to negatively-charged lipids in the inner leaflet of the plasma membrane [89-91]. It should be noted that another small GTPase, Rac1, was reported to interact with raft-like domains [92], although its C-terminus resembles that of K-Ras [93,94]. This difference may be due to more subtle but still distinct differences between the C-terminal regions of K-Ras and Rac1, which may result in association with different scaffold and adaptor proteins that affect their targeting (see following section). Thus, while K-Ras is farnesylated, Rac1 is geranylgeranylated, and although it has a C-terminal polybasic cluster resembling K-Ras (but containing also Arg residues), it has an adjacent proline-rich region absent in K-Ras [93-95]. These distinct differences can lead to specific interactions with adaptor proteins such as Crk and to integrin regulation of Rac1 membrane binding sites [94,96].
Recent reports demonstrate that negatively charged phosphatidylinositol-4,5-bisphosphate and phosphatidylinositol-3,4,5-trisphosphate [97] are required for the charge-based membrane interactions of K-Ras, and phosphatidylserine also has a role in balancing K-Ras between the plasma membrane and endocytic compartments [98]. Accordingly, interference with these electrostatic interactions by a cationic amphiphilic drug (chlorpromazine) reduced the association of K-RasG12V (but not H-Ras) with the plasma membrane, leading to its accumulation in endosomal or mitochondrial membranes with corresponding effects of growth inhibition or apoptosis [99]. This is in line with the demonstration that the anesthetic dibucaine, which induces flipping of phosphatidylserine and is positively charged, can displace polybasic peptide probes resembling the positively charged C-terminus of K-Ras from the plasma membrane [100]. The important role of the HVR electrostatic interactions is further demonstrated by the partial translocation of K-Ras from the plasma membrane to internal membranes following phosphorylation of Ser181 [101]. Recent studies employing EM spatial mapping and FRET have shown that phosphorylation of Ser181 reduces K-Ras-GTP nanoclustering and modulates its signaling, suggesting that electrostatic interactions of the HVR region are involved in K-Ras clustering [70]. Interactions of K-Ras-GTP with the scaffold protein galectin-3 [102], which enhance K-Ras-GTP localization to nanoclusters [103], were insensitive to Ser181 phosphorylation, suggesting that they are induced by an independent mechanism [70] and raising the possibility that K-Ras-GTP may interact with more than one type of clusters. It is tempting to suggest that in essence both K-Ras-GTP and H-Ras-GTP can participate in a spectrum of nonidentical nanoclusters, which differ from each other in lipid composition and/or in the participating scaffold proteins, such as galectins, Sur-8 and perhaps others [88,103,104]. This, in turn, would enable preferential activation of specific signaling patterns depending on the nanoclusters subtype.
Cumulative evidence from biochemical, EM and biophysical studies (FRAP and SFVT) shows that the interactions of Ras proteins with nanoclusters are dynamic [22,32,33,37]. The transient, dynamic nature of the interactions was shown to be crucial for effective H-Ras signaling [21,22,32,76]. Biochemical and EM studies suggested that although H-Ras-GDP is preferentially localized to cholesterol-dependent nanodomains, activated H-Ras-GTP has to exit these domains to effectively signal via Raf [21,22]. Combining antibody-mediated clustering of a GPI-AP with FRAP studies on the lateral diffusion of wild type H-Ras and H-RasG12V, it was shown that GPI-AP clustering stabilizes the association of H-Ras-GDP with raft-like clusters, enhancing the step of GDP/GTP exchange but retarding the exit of H-Ras-GTP from the GPI-AP clusters and the ensuing activation of Raf [76]. Thus, the requirement for dynamic interactions of H-Ras with cluster sites flows from the need to shift from association with one type of clusters (cholesterol-dependent, preferred by H-Ras-GDP) to a different set of signaling clusters (non-raft, preferred by H-Ras-GTP).
Compartmentalized signaling
Cell surface nanoclusters: coupling analog and digital signaling
The contribution of nanoclusters to transmembrane signal transduction has recently been evaluated in the context of Ras-dependent activation of the Raf/MEK/ERK cascade. K-Ras-GTP nanoclusters operate as highly sensitive digital switches, that is a nanocluster dumps a fixed output of ERKpp into the cytosol for a wide range of Raf kinase inputs [71,105]. Thus even low levels of Raf kinase activity when scaffolded in a Ras nanocluster together with KSR/MEK and ERK generate the same ERKpp output as high levels of Raf activity. The number of K-Ras-GTP nanoclusters generated in response to non-saturating doses of EGF is a linear function of agonist concentration [71]. In combination these two characteristics allow the plasma membrane/nanocluster system to operate as a high fidelity analog-digital-analog relay that accurately reproduces an ERKpp output in the cytosol that precisely matches the EGF input signal that was delivered to the outer plasma membrane (Figure 4; [71]).
Figure 4.
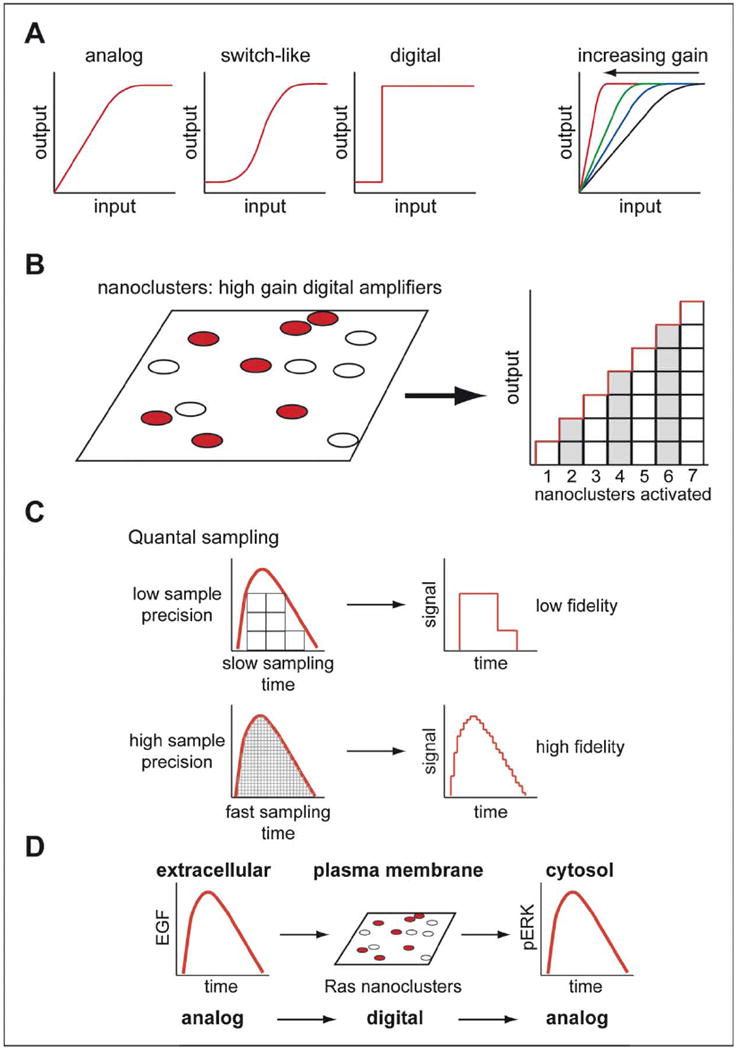
Analog-digital coupling involving Ras signaling nanoclusters. (A) Analog signals allow graded responses proportional to input whereas digital or high gain switch-like signaling results in maximal output from a wide range of inputs – amplifying weak inputs into maximum outputs. (B) Ras nanoclusters operate as digital amplifiers. (C) Digital quantal sampling of analog inputs can generate high fidelity analog-like outputs if sampling time (Ras activation/de-activation and nanocluster lifetime) is very fast and sample precision (number of nanoclusters) is high. (D) High fidelity digital Ras nanocluster signaling ensures that the analog extracellular signal is converted into a graded cytosolic signaling response. This Figure is reproduced in color in Molecular Membrane Biology online.
High fidelity signaling is achieved by the Ras nanocluster analog-digital-analog relay because of the high sampling rate and large number of quantization levels available to the system (Figure 4C; [71,106,107]). The high sampling rate is reflected in the short lifetime of a nanocluster (~0.4 s) that effectively samples the signal input ~150 × per min. The number of quantization levels reflects the total number of nanoclusters that can be assembled from the available Ras-GTP monomers (tens of thousands; [106,107]). The unique spatio-temporal properties of Ras organization on the plasma membrane therefore deliver critically important emergent signaling characteristics.
The original studies on signal output from Ras nanoclusters focused on plasma membrane localized K-Ras-GTP [71,105]; very recent work has now shown that N-Ras-GTP and H-Ras-GTP nanoclusters also operate digitally with respect to the Raf/MEK/ERK cascade [108]. Interestingly, however activation of the Raf/MEK/ERK cascade from Golgi Ras platforms is analog and not digital, leading to a delayed low fidelity signal response to EGF stimulation (see also below; [108]). H- and N-Ras nanoclusters occupied by GDP-loaded Ras do not support Raf activation and therefore do not signal, prompting the question as to whether these clusters serve any specific function. One possible role flows from a recent analysis of Sos activation on model membranes showing that allosteric activation of Sos exchange activity is regulated by cell surface Ras proteins [109]. The minimum surface density of Ras required to activate Sos is delivered on the nanoscale in Ras nanoclusters, but would not be achieved if Ras proteins were randomly distributed as monomers over the cell surface [109].
Ras signaling from intracellular organelles
As a result of differences in trafficking, internalization and localization within cell surface subdomains, the Ras isoforms have partially overlapping but distinct subcellular distributions [110]. Most noticeable is the different degree of association (N ≥H > K-Ras in most cell lines) with endomembranes (ER, Golgi, endosomes, mitochondria). Combined with differences in the relative abundance of each isoform across a wide range of cell lines (K ≥N ≫H-Ras; [111]), a simple conclusion would be that K(B)-Ras represents the pre-eminent cell surface Ras, whilst N-Ras dominates endomembranous Ras signaling. In addition to differential Ras distributions, Ras pathway scaffolds, activators and effectors have been localized to a variety of subcellular platforms [110]. For example, the MAP kinase scaffolds Sef, p14-MP1 and KSR are located on the Golgi, late endosome and plasma membrane respectively [112-116], whilst the Akt scaffold Appl1 localizes to a sub-population of early endosomes [117,118].
In recent years a series of studies have established that Ras signaling from intracellular organelles occurs and has specific phenotypic consequences. This appears to be an evolutionarily ancient phenomenon, because in yeast endomembranous Ras signaling controls morphology, whereas plasma membrane Ras regulates mating [119]. In mammalian cells, ectopically expressed Golgi-Ras promotes cell proliferation [120,121], and Ras activation in this organelle is delayed but prolonged (onset within 10 minutes, duration of 60 minutes; [122]). In a more physiological context, positive thymocyte selection requires endogenous Golgi Ras signaling whereas acute activation of cell surface Ras supports negative selection [123]. Interestingly, whilst both cell surface and Golgi Ras can be stimulated by the Ras activator RasGRP1 in T cells, the location of RasGRP1 stimulation can be precisely regulated by different extracellular ligands and second messengers. T cell receptor (TCR) induces Golgi RasGRP1 stimulation of Ras activation whilst co-stimulation of the TCR and the integrin LFA-1 generates plasma membrane localized diacylglycerol (DAG) and phosphatidic acid pools that recruit RasGRP1 to the cell surface for Ras activation [124].
An alternative location for Ras signaling is the mitochondria. N-Ras is required for normal mitochondrial morphology and from this location generates retrograde signaling to the nucleus [125]. As discussed earlier, K-Ras can translocate to the mitochondria when Ser181 is phosphorylated by protein kinase C (PKC), resulting in the initiation of apoptotic cascades [101]. The endosome is also a site for Ras signaling following endocytosis of growth factor receptors and Ras proteins [126]. Thus, inhibition of endocytosis selectively impairs H- and N-Ras signaling [111], and endosomes support sustained MAP kinase activation [127,128]. Interestingly, signaling divergence also occurs here: a subset of endosomes selectively supports Akt signaling via the scaffold Appl1 [117]. This is critical for zebra fish development by promoting cell survival in tissues where Appl1 is expressed.
Whilst the majority of Ras signaling emanates from the cell surface, the functionality of the alternative intracellular Ras pools, particularly at later time points following stimulation, indicates that they make a meaningful contribution to cell signaling.
Conclusions
Ras HVR motifs play a critical role in the correct positioning of Ras isoforms within the cell. Ras localization is dynamic and influenced by the Ras activation status and interacting proteins, allowing contact with different pools of regulators and effectors. This mechanism is likely to underlie isoform-specific Ras signaling. Further work is needed to identify the mechanisms of Ras regulation on each organelle and the precise signaling complement engaged in these locations. Most work to date has been performed with ectopically expressed Ras; however a variety of pioneering studies have been able to examine endogenous compartmentalized Ras outputs. Although challenging, this represents the optimal model for future studies, because of the potentially perturbing effects of over-expressing a protein within a signaling network. Insights from the studies reviewed here have widespread implications because compartment- and isoform-specific signaling is likely to occur in every signaling system.
Acknowledgments
Y.I.H. is an incumbent of the Zalman Weinberg Chair in Cell Biology. I.A.P is a Royal Society University Research Fellow. Work in the laboratory of J.F.H is supported by the NIH (GM066717).
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esteban LM, Vicario-Abejon C, Fernandez-Salguero P, Fernandez-Medarde A, Swaminathan N, Yienger K, Lopez E, Malumbres M, McKay R, Ward JM, Pellicer A, Santos E. Targeted genomic disruption of H-ras and N-ras, individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol Cell Biol. 2001;21:1444–1452. doi: 10.1128/MCB.21.5.1444-1452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quinlan MP, Quatela SE, Philips MR, Settleman J. Activated Kras, but not Hras or Nras, may initiate tumors of endodermal origin via stem cell expansion. Mol Cell Biol. 2008;28:2659–2674. doi: 10.1128/MCB.01661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The data was obtained from the Wellcome Trust Sanger Institute Cancer Genome Project web site. http://www.sanger.ac.uk/genetics/CGP.
- 5.Apolloni A, Prior IA, Lindsay M, Parton RG, Hancock JF. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol Cell Biol. 2000;20:2475–2487. doi: 10.1128/mcb.20.7.2475-2487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 7.Laude AJ, Prior IA. Palmitoylation and localisation of RAS isoforms are modulated by the hypervariable linker domain. J Cell Sci. 2008;121:421–427. doi: 10.1242/jcs.020107. [DOI] [PubMed] [Google Scholar]
- 8.Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR. Endomembrane trafficking of Ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- 9.Wang G, Deschenes RJ. Plasma membrane localization of Ras requires class C vps proteins and functional mitochondria in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:3243–3255. doi: 10.1128/MCB.26.8.3243-3255.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem. 2005;280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- 11.Lobo S, Greentree WK, Linder ME, Deschenes RJ. Identification of a ras palmitoyltransferase in saccharomyces cerevisiae. J Biol Chem. 2002;277:41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- 12.Bartels DJ, Mitchell DA, Dong X, Deschenes RJ. Erf2, a novel gene product that affects the localization and palmitoylation of Ras2 in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6775–6787. doi: 10.1128/mcb.19.10.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohno Y, Kihara A, Sano T, Igarashi Y. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim Biophys Acta. 2006;1761:474–483. doi: 10.1016/j.bbalip.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res. 2006;47:1118–1127. doi: 10.1194/jlr.R600007-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Ohta E, Misumi Y, Sohda M, Fujiwara T, Yano A, Ikehara Y. Identification and characterization of GCP16, a novel acylated Golgi protein that interacts with GCP170. J Biol Chem. 2003;278:51957–51967. doi: 10.1074/jbc.M310014200. [DOI] [PubMed] [Google Scholar]
- 16.Fukata Y, Iwanaga T, Fukata M. Systematic screening for palmitoyl transferase activity of the DHHC protein family in mammalian cells. Methods. 2006;40:177–182. doi: 10.1016/j.ymeth.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Fukata M, Fukata Y, Adesnik H, Nicoll RA, Bredt DS. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44:987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Magee AI, Gutierrez L, McKay IA, Marshall CJ, Hall A. Dynamic fatty acylation of p21N-ras. EMBO J. 1987;6:3353–3357. doi: 10.1002/j.1460-2075.1987.tb02656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker TL, Zheng H, Walker J, Coloff JL, Buss JE. Distinct rates of palmitate turnover on membrane bound cellular and oncogenic H-Ras. J Biol Chem. 2003;278:19292–19300. doi: 10.1074/jbc.M206956200. [DOI] [PubMed] [Google Scholar]
- 20.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 21.Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prior IA, Harding A, Yan J, Sluimer J, Parton RG, Hancock JF. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat Cell Biol. 2001;3:368–375. doi: 10.1038/35070050. [DOI] [PubMed] [Google Scholar]
- 23.Roy S, Plowman S, Rotblat B, Prior IA, Muncke C, Grainger S, Parton RG, Henis YI, Kloog Y, Hancock JF. Individual palmitoyl residues serve distinct roles in H-ras trafficking, microlocalization, and signaling. Mol Cell Biol. 2005;25:6722–6733. doi: 10.1128/MCB.25.15.6722-6733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, Bishop WR, Pai JK. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem. 1997;272:14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 25.Basso AD, Mirza A, Liu G, Long BJ, Bishop WR, Kirschmeier P. The farnesyl transferase inhibitor (FTI) SCH66336 (lonafarnib) inhibits Rheb farnesylation and mTOR signaling. Role in FTI enhancement of taxane and tamoxifen anti-tumor activity. J Biol Chem. 2005;280:31101–31108. doi: 10.1074/jbc.M503763200. [DOI] [PubMed] [Google Scholar]
- 26.Lebowitz PF, Casey PJ, Prendergast GC, Thissen JA. Farnesyltransferase inhibitors alter the prenylation and growth-stimulating function of RhoB. J Biol Chem. 1997;272:15591–15594. doi: 10.1074/jbc.272.25.15591. [DOI] [PubMed] [Google Scholar]
- 27.Bergo MO, Ambroziak P, Gregory C, George A, Otto JC, Kim E, Nagase H, Casey PJ, Balmain A, Young SG. Absence of the CAAX endoprotease Rce1: effects on cell growth and transformation. Mol Cell Biol. 2002;22:171–181. doi: 10.1128/MCB.22.1.171-181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergo MO, Gavino BJ, Hong C, Beigneux AP, McMahon M, Casey PJ, Young SG. Inactivation of Icmt inhibits transformation by oncogenic K-Ras and B-Raf. J Clin Invest. 2004;113:539–550. doi: 10.1172/JCI18829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winter-Vann AM, Baron RA, Wong W, dela Cruz J, York JD, Gooden DM, Bergo MO, Young SG, Toone EJ, Casey PJ. A small-molecule inhibitor of isoprenylcysteine carboxyl methyltransferase with antitumor activity in cancer cells. Proc Natl Acad Sci USA. 2005;102:4336–4341. doi: 10.1073/pnas.0408107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michaelson D, Ali W, Chiu VK, Bergo M, Silletti J, Wright L, Young SG, Philips M. Postprenylation CAAX processing is required for proper localization of Ras but not Rho GTPases. Mol Biol Cell. 2005;16:1606–1616. doi: 10.1091/mbc.E04-11-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J Cell Biol. 2002;157:1071–1081. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niv H, Gutman O, Kloog Y, Henis YI. Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J Cell Biol. 2002;157:865–872. doi: 10.1083/jcb.200202009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murakoshi H, Iino R, Kobayashi T, Fujiwara T, Ohshima C, Yoshimura A, Kusumi A. Single-molecule imaging analysis of Ras activation in living cells. Proc Natl Acad Sci USA. 2004;101:7317–7322. doi: 10.1073/pnas.0401354101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edidin M. Lipids on the frontier: a century of cell-membrane bilayers. Nat Rev Mol Cell Biol. 2003;4:414–418. doi: 10.1038/nrm1102. [DOI] [PubMed] [Google Scholar]
- 35.Simons K, Vaz WL. Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 36.Kusumi A, Ike H, Nakada C, Murase K, Fujiwara T. Single-molecule tracking of membrane molecules: plasma membrane compartmentalization and dynamic assembly of raft-philic signaling molecules. Semin Immunol. 2005;17:3–21. doi: 10.1016/j.smim.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 37.Hancock JF, Parton RG. Ras plasma membrane signalling platforms. Biochem J. 2005;389:1–11. doi: 10.1042/BJ20050231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hancock JF. Lipid rafts: contentious only from simplistic standpoints. Nat Rev Mol Cell Biol. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobson K, Mouritsen OG, Anderson RG. Lipid rafts: at a crossroad between cell biology and physics. Nat Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- 40.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 41.Edidin M. The state of lipid rafts: from model membranes to cells. Annu Rev Biophys Biomol Struct. 2003;32:257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- 42.Brown DA, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- 43.Sharma P, Varma R, Sarasij RC, Gousset IK, Krishnamoorthy G, Rao M, Mayor S. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–590. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- 44.van Meer G, Simons K. Viruses budding from either the apical or the basolateral plasma membrane domain of MDCK cells have unique phospholipid compositions. EMBO J. 1982;1:847–852. doi: 10.1002/j.1460-2075.1982.tb01258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheiffele P, Rietveld A, Wilk T, Simons K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J Biol Chem. 1999;274:2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- 46.Ono A, Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci USA. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin S, Naim HY, Rodriguez AC, Roth MG. Mutations in the middle of the transmembrane domain reverse the polarity of transport of the influenza virus hemagglutinin in MDCK epithelial cells. J Cell Biol. 1998;142:51–57. doi: 10.1083/jcb.142.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurzchalia TV, Parton RG. Membrane microdomains and caveolae. Curr Opin Cell Biol. 1999;11:424–431. doi: 10.1016/s0955-0674(99)80061-1. [DOI] [PubMed] [Google Scholar]
- 49.Taner SB, Onfelt B, Pirinen NJ, McCann FE, Magee AI, Davis DM. Control of immune responses by trafficking cell surface proteins, vesicles and lipid rafts to and from the immunological synapse. Traffic. 2004;5:651–661. doi: 10.1111/j.1600-0854.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- 50.Kusumi A, Koyama-Honda I, Suzuki K. Molecular dynamics and interactions for creation of stimulation-induced stabilized rafts from small unstable steady-state rafts. Traffic. 2004;5:213–230. doi: 10.1111/j.1600-0854.2004.0178.x. [DOI] [PubMed] [Google Scholar]
- 51.Feigenson GW, Buboltz JT. Ternary phase diagram of dipalmitoyl-PC/dilauroyl-PC/cholesterol: nanoscopic domain formation driven by cholesterol. Biophys J. 2001;80:2775–2788. doi: 10.1016/S0006-3495(01)76245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Almeida RF, Fedorov A, Prieto M. Sphingomyelin/phosphatidylcholine/cholesterol phase diagram: boundaries and composition of lipid rafts. Biophys J. 2003;85:2406–2416. doi: 10.1016/s0006-3495(03)74664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Almeida RF, Loura LM, Fedorov A, Prieto M. Lipid rafts have different sizes depending on membrane composition: a time-resolved fluorescence resonance energy transfer study. J Mol Biol. 2005;346:1109–1120. doi: 10.1016/j.jmb.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 54.Zhao J, Wu J, Heberle FA, Mills TT, Klawitter P, Huang G, Costanza G, Feigenson GW. Phase studies of model biomembranes: complex behavior of DSPC/DOPC/cholesterol. Biochim Biophys Acta. 2007;1768:2764–2776. doi: 10.1016/j.bbamem.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson RG, Jacobson K. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science. 2002;296:1821–1825. doi: 10.1126/science.1068886. [DOI] [PubMed] [Google Scholar]
- 56.Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang TY, Leventis R, Silvius JR. Artificially lipid-anchored proteins can elicit clustering-induced intracellular signaling events in Jurkat T-lymphocytes independent of lipid raft association. J Biol Chem. 2005;280:22839–22846. doi: 10.1074/jbc.M502920200. [DOI] [PubMed] [Google Scholar]
- 58.Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 59.Nichols B. Cell biology: without a raft. Nature. 2005;436:638–639. doi: 10.1038/436638a. [DOI] [PubMed] [Google Scholar]
- 60.Shaw AS. Lipid rafts: now you see them, now you don’t. Nat Immunol. 2006;7:1139–1142. doi: 10.1038/ni1405. [DOI] [PubMed] [Google Scholar]
- 61.Parton RG, Hanzal-Bayer M, Hancock JF. Biogenesis of caveolae: a structural model for caveolin-induced domain formation. J Cell Sci. 2006;119:787–796. doi: 10.1242/jcs.02853. [DOI] [PubMed] [Google Scholar]
- 62.Parton RG, Simons K. The multiple faces of caveolae. Nat Rev Mol Cell Biol. 2007;8:185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- 63.Lichtenberg D, Goni FM, Heerklotz H. Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem Sci. 2005;30:430–436. doi: 10.1016/j.tibs.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 64.Kenworthy AK, Petranova N, Edidin M. High-resolution FRET microscopy of cholera toxin B-subunit and GPI- anchored proteins in cell plasma membranes. Mol Biol Cell. 2000;11:1645–1655. doi: 10.1091/mbc.11.5.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kawasaki K, Yin JJ, Subczynski WK, Hyde JS, Kusumi A. Pulse EPR detection of lipid exchange between protein-rich raft and bulk domains in the membrane: methodology development and its application to studies of influenza viral membrane. Biophys J. 2001;80:738–748. doi: 10.1016/S0006-3495(01)76053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 67.Mayor S, Rao M. Rafts: scale-dependent, active lipid organization at the cell surface. Traffic. 2004;5:231–240. doi: 10.1111/j.1600-0854.2004.00172.x. [DOI] [PubMed] [Google Scholar]
- 68.Suzuki KG, Fujiwara TK, Sanematsu F, Iino R, Edidin M, Kusumi A. GPI-anchored receptor clusters transiently recruit Lyn and Ga for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1. J Cell Biol. 2007;177:717–730. doi: 10.1083/jcb.200609174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Plowman SJ, Muncke C, Parton RG, Hancock JF. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc Natl Acad Sci USA. 2005;102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Plowman SJ, Ariotti N, Goodall A, Parton RG, Hancock JF. Electrostatic interactions positively regulate K-Ras nanocluster formation and function. Mol Cell Biol. 2008;28:4377–4385. doi: 10.1128/MCB.00050-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tian T, Harding A, Inder K, Plowman S, Parton RG, Hancock JF. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol. 2007;9:905–914. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- 72.Abankwa D, Hanzal-Bayer M, Ariotti N, Plowman SJ, Gorfe AA, Parton RG, McCammon JA, Hancock JF. A novel switch region regulates H-ras membrane orientation and signal output. EMBO J. 2008;27:727–735. doi: 10.1038/emboj.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abankwa D, Gorfe AA, Hancock JF. Ras nanoclusters: molecular structure and assembly. Semin Cell Dev Biol. 2007;18:599–607. doi: 10.1016/j.semcdb.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roy S, Luetterforst R, Harding A, Apolloni A, Etheridge M, Stang E, Rolls B, Hancock JF, Parton RG. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat Cell Biol. 1999;1:98–105. doi: 10.1038/10067. [DOI] [PubMed] [Google Scholar]
- 75.Rotblat B, Prior IA, Muncke C, Parton RG, Kloog Y, Henis YI, Hancock JF. Three separable domains regulate GTP-dependent association of H-ras with the plasma membrane. Mol Cell Biol. 2004;24:6799–6810. doi: 10.1128/MCB.24.15.6799-6810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eisenberg S, Shvartsman DE, Ehrlich M, Henis YI. Clustering of raft-associated proteins in the external membrane leaflet modulates internal leaflet H-Ras diffusion and signaling. Mol Cell Biol. 2006;26:7190–7200. doi: 10.1128/MCB.01059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Melkonian KA, Ostermeyer AG, Chen JZ, Roth MG, Brown DA. Role of lipid modifications in targeting proteins to detergent- resistant membrane rafts. Many raft proteins are acylated, while few are prenylated. J Biol Chem. 1999;274:3910–3917. doi: 10.1074/jbc.274.6.3910. [DOI] [PubMed] [Google Scholar]
- 78.Wang TY, Leventis R, Silvius JR. Partitioning of lipidated peptide sequences into liquid-ordered lipid domains in model and biological membranes. Biochemistry. 2001;40:13031–13040. doi: 10.1021/bi0112311. [DOI] [PubMed] [Google Scholar]
- 79.Jura N, Scotto-Lavino E, Sobczyk A, Bar-Sagi D. Differential modification of Ras proteins by ubiquitination. Mol Cell. 2006;21:679–687. doi: 10.1016/j.molcel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 80.Kenworthy AK, Nichols BJ, Remmert CL, Hendrix GM, Kumar M, Zimmerberg J, Lippincott-Schwartz J. Dynamics of putative raft-associated proteins at the cell surface. J Cell Biol. 2004;165:735–746. doi: 10.1083/jcb.200312170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shvartsman DE, Gutman O, Tietz A, Henis YI. Cyclodextrins but not compactin inhibit the lateral diffusion of membrane proteins independent of cholesterol. Traffic. 2006;7:917–926. doi: 10.1111/j.1600-0854.2006.00437.x. [DOI] [PubMed] [Google Scholar]
- 82.Goodwin JS, Drake KR, Remmert CL, Kenworthy AK. Ras diffusion is sensitive to plasma membrane viscosity. Biophys J. 2005;89:1398–1410. doi: 10.1529/biophysj.104.055640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Henis YI, Rotblat B, Kloog Y. FRAP beam-size analysis to measure palmitoylation-dependent membrane association dynamics and microdomain partitioning of Ras proteins. Methods. 2006;40:183–190. doi: 10.1016/j.ymeth.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 84.Gorfe AA, Hanzal-Bayer M, Abankwa D, Hancock JF, McCammon JA. Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoyl-glycero-3-phosphocholine bilayer. J Med Chem. 2007;50:674–684. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
- 85.Thapar R, Williams JG, Campbell SL. NMR characterization of full-length farnesylated and non-farnesylated H-Ras and its implications for Raf activation. J Mol Biol. 2004;343:1391–1408. doi: 10.1016/j.jmb.2004.08.106. [DOI] [PubMed] [Google Scholar]
- 86.Paz A, Haklai R, Elad-Sfadia G, Ballan E, Kloog Y. Galectin-1 binds oncogenic H-Ras to mediate Ras membrane anchorage and cell transformation. Oncogene. 2001;20:7486–7493. doi: 10.1038/sj.onc.1204950. [DOI] [PubMed] [Google Scholar]
- 87.Elad-Sfadia G, Haklai R, Ballan E, Gabius HJ, Kloog Y. Galectin-1 augments Ras activation and diverts Ras signals to Raf-1 at the expense of phosphoinositide 3-kinase. J Biol Chem. 2002;277:37169–37175. doi: 10.1074/jbc.M205698200. [DOI] [PubMed] [Google Scholar]
- 88.Belanis L, Plowman SJ, Rotblat B, Hancock JF, Kloog Y. Galectin-1 Is a Novel Structural Component and a Major Regulator of H-Ras Nanoclusters. Mol Biol Cell. 2008;19:1404–1414. doi: 10.1091/mbc.E07-10-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cadwallader KA, Paterson H, Macdonald SG, Hancock JF. N-terminally myristoylated Ras proteins require palmitoylation or a polybasic domain for plasma membrane localization. Mol Cell Biol. 1994;14:4722–4730. doi: 10.1128/mcb.14.7.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ghomashchi F, Zhang X, Liu L, Gelb MH. Binding of prenylated and polybasic peptides to membranes: affinities and intervesicle exchange. Biochemistry. 1995;34:11910–11918. doi: 10.1021/bi00037a032. [DOI] [PubMed] [Google Scholar]
- 91.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 92.Del Pozo MA, Schwartz MA. Rac, membrane heterogeneity, caveolin and regulation of growth by integrins. Trends Cell Biol. 2007;17:246–250. doi: 10.1016/j.tcb.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 93.Wennerberg K, Der CJ. Rho-family GTPases: it’s not only Rac and Rho (and I like it) J Cell Sci. 2004;117:1301–1312. doi: 10.1242/jcs.01118. [DOI] [PubMed] [Google Scholar]
- 94.van Hennik PB, ten Klooster JP, Halstead JR, Voermans C, Anthony EC, Divecha N, Hordijk PL. The C-terminal domain of Rac1 contains two motifs that control targeting and signaling specificity. J Biol Chem. 2003;278:39166–39175. doi: 10.1074/jbc.M307001200. [DOI] [PubMed] [Google Scholar]
- 95.Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 96.del Pozo MA, Alderson NB, Kiosses WB, Chiang H-H, Anderson RGW, Schwartz MA. Integrins regulate rac targeting by internalization of membrane domains. Science. 2004;303:839–842. doi: 10.1126/science.1092571. [DOI] [PubMed] [Google Scholar]
- 97.Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 99.Eisenberg S, Giehl K, Henis YI, Ehrlich M. Differential interference of chlorpromazine with the membrane interactions of oncogenic K-Ras and its effects on cell growth. J Biol Chem. 2008;283:27279–27288. doi: 10.1074/jbc.M804589200. [DOI] [PubMed] [Google Scholar]
- 100.Yeung T, Terebiznik M, Yu L, Silvius J, Abidi WM, Philips M, Levine T, Kapus A, Grinstein S. Receptor activation alters inner surface potential during phagocytosis. Science. 2006;313:347–351. doi: 10.1126/science.1129551. [DOI] [PubMed] [Google Scholar]
- 101.Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, Yim D, Fein A, Perez de Castro I, Li C, Thompson CB, Cox AD, Philips MR. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21:481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 102.Elad-Sfadia G, Haklai R, Balan E, Kloog Y. Galectin-3 augments K-Ras activation and triggers a Ras signal that attenuates ERK but not phosphoinositide 3-kinase activity. J Biol Chem. 2004;279:34922–34930. doi: 10.1074/jbc.M312697200. [DOI] [PubMed] [Google Scholar]
- 103.Shalom-Feuerstein R, Plowman SJ, Rotblat B, Ariotti N, Tian T, Hancock JF, Kloog Y. K-ras nanoclustering is subverted by overexpression of the scaffold protein galectin-3. Cancer Res. 2008;68:6608–6616. doi: 10.1158/0008-5472.CAN-08-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sieburth DS, Sun Q, Han M. SUR-8, a conserved Ras-binding protein with leucine-rich repeats, positively regulates Ras-mediated signaling in C. elegans. Cell. 1998;94:119–130. doi: 10.1016/s0092-8674(00)81227-1. [DOI] [PubMed] [Google Scholar]
- 105.Harding A, Tian T, Westbury E, Frische E, Hancock JF. Subcellular localization determines MAP kinase signal output. Curr Biol. 2005;15:869–873. doi: 10.1016/j.cub.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 106.Harding AS, Hancock JF. Using plasma membrane nanoclusters to build better signaling circuits. Trends Cell Biol. 2008;18:364–371. doi: 10.1016/j.tcb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Harding A, Hancock JF. Ras nanoclusters: combining digital and analog signaling. Cell Cycle. 2008;7:127–134. doi: 10.4161/cc.7.2.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Inder K, Harding A, Plowman SJ, Philips MR, Parton RG, Hancock JF. Activation of the MAPK module from different spatial locations generates distinct system outputs. Mol Biol Cell. 2008;19:4776–4784. doi: 10.1091/mbc.E08-04-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gureasko J, Galush WJ, Boykevisch S, Sondermann H, Bar-Sagi D, Groves JT, Kuriyan J. Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat Struct Mol Biol. 2008;15:452–461. doi: 10.1038/nsmb.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Omerovic J, Laude AJ, Prior IA. Ras proteins: paradigms for compartmentalised and isoform-specific signalling. Cell Mol Life Sci. 2007;64:2575–2589. doi: 10.1007/s00018-007-7133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Omerovic J, Hammond DE, Clague MJ, Prior IA. Ras isoform abundance and signalling in human cancer cell lines. Oncogene. 2008;27:2754–2762. doi: 10.1038/sj.onc.1210925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Luttrell LM, Roudabush FL, Choy EW, Miller WE, Field ME, Pierce KL, Lefkowitz RJ. Activation and targeting of extracellular signal-regulated kinases by beta-arrestin scaffolds. Proc Natl Acad Sci USA. 2001;98:2449–2454. doi: 10.1073/pnas.041604898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Teis D, Wunderlich W, Huber LA. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev Cell. 2002;3:803–814. doi: 10.1016/s1534-5807(02)00364-7. [DOI] [PubMed] [Google Scholar]
- 114.Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- 115.Torii S, Kusakabe M, Yamamoto T, Maekawa M, Nishida E. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev Cell. 2004;7:33–44. doi: 10.1016/j.devcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 116.Wunderlich W, Fialka I, Teis D, Alpi A, Pfeifer A, Parton RG, Lottspeich F, Huber LA. A novel 14-kilodalton protein interacts with the mitogen-activated protein kinase scaffold mp1 on a late endosomal/lysosomal compartment. J Cell Biol. 2001;152:765–776. doi: 10.1083/jcb.152.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schenck A, Goto-Silva L, Collinet C, Rhinn M, Giner A, Habermann B, Brand M, Zerial M. The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell. 2008;133:486–497. doi: 10.1016/j.cell.2008.02.044. [DOI] [PubMed] [Google Scholar]
- 118.Miaczynska M, Christoforidis S, Giner A, Shevchenko A, Uttenweiler-Joseph S, Habermann B, Wilm M, Parton RG, Zerial M. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell. 2004;116:445–456. doi: 10.1016/s0092-8674(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 119.Onken B, Wiener H, Philips MR, Chang EC. Compartmentalized signaling of Ras in fission yeast. Proc Natl Acad Sci USA. 2006;103:9045–9050. doi: 10.1073/pnas.0603318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Matallanas D, Sanz-Moreno V, Arozarena I, Calvo F, Agudo-Ibanez L, Santos E, Berciano MT, Crespo P. Distinct utilization of effectors and biological outcomes resulting from site-specific Ras activation: Ras functions in lipid rafts and Golgi complex are dispensable for proliferation and transformation. Mol Cell Biol. 2006;26:100–116. doi: 10.1128/MCB.26.1.100-116.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bivona TG, Perez De Castro I, Ahearn IM, Grana TM, Chiu VK, Lockyer PJ, Cullen PJ, Pellicer A, Cox AD, Philips MR. Phospholipase Cg activates Ras on the Golgi apparatus by means of RasGRP1. Nature. 2003;424:694–698. doi: 10.1038/nature01806. [DOI] [PubMed] [Google Scholar]
- 122.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, 2nd, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 123.Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, Werlen G, Hollander GA, Gascoigne NR, Palmer E. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 124.Mor A, Campi G, Du G, Zheng Y, Foster DA, Dustin ML, Philips MR. The lymphocyte function-associated antigen-1 receptor costimulates plasma membrane Ras via phospholipase D2. Nat Cell Biol. 2007;9:713–719. doi: 10.1038/ncb1592. [DOI] [PubMed] [Google Scholar]
- 125.Wolfman JC, Planchon SM, Liao J, Wolfman A. Structural and functional consequences of c-N-Ras constitutively associated with intact mitochondria. Biochim Biophys Acta. 2006;1763:1108–1124. doi: 10.1016/j.bbamcr.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 126.Pol A, Calvo M, Enrich C. Isolated endosomes from quiescent rat liver contain the signal transduction machinery. Differential distribution of activated Raf-1 and Mek in the endocytic compartment. FEBS Lett. 1998;441:34–38. doi: 10.1016/s0014-5793(98)01517-8. [DOI] [PubMed] [Google Scholar]
- 127.Taub N, Teis D, Ebner HL, Hess MW, Huber LA. Late endosomal traffic of the epidermal growth factor receptor ensures spatial and temporal fidelity of mitogen-activated protein kinase signaling. Mol Biol Cell. 2007;18:4698–4710. doi: 10.1091/mbc.E07-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oksvold MP, Skarpen E, Wierod L, Paulsen RE, Huitfeldt HS. Re-localization of activated EGF receptor and its signal transducers to multivesicular compartments downstream of early endosomes in response to EGF. Eur J Cell Biol. 2001;80:285–294. doi: 10.1078/0171-9335-00160. [DOI] [PubMed] [Google Scholar]


