Abstract
Purpose
Recent studies suggest that children <24 months with stage 1 favorable histology Wilms tumors <550g (Very Low Risk Wilms Tumors, VLRWT) have an excellent prognosis when treated with nephrectomy only, without adjuvant chemotherapy. The identification of risk categories within VLRWT may enable refinement of their definition and optimization of their therapy.
Experimental Design
To define biologically distinct subsets, global gene expression analysis was performed on 39 VLRWT that passed all quality control parameters and the clusters identified were validated on in independent set of 11 VLRWT. Validation of select differentially expressed genes was performed with immunohistochemistry on a tissue microarray from 20/39 tumors. Loss of heterozygosity for 11p15, 1p, and 16q was analyzed in 52 tumors using polymerase chain reaction.
Results
Two distinctive clusters were identified. One cluster included nine tumors with epithelial tubular differentiated histology, paucity of nephrogenic rests, lack of LOH for 1p, 16q and 11p, absence of relapse, and a unique gene expression profile consistent with arrest following mesenchymal-to-epithelial transition. The second cluster included 13 tumors with mixed histology, intralobar nephrogenic rests, and decreased expression of WT1. Three of six relapses occurred in this cluster. Of 43 informative tumors, 11p LOH was present in 5/5 relapses and in 11/38 non-relapses.
Conclusions
Two subsets comprising a total of 56% of VLRWT are identified that have pathogenetic and molecular differences and apparent differences in risk for relapse. If these predictors can be prospectively validated, this would enable the refinement of clinical stratification and less arbitrary definition of VLRWT.
Keywords: Wilms tumor, gene expression analysis, renal development, WT-1, kidney
Introduction
Modifications in treatment strategies have greatly improved the prognosis for children with Favorable Histology Wilms Tumor (FHWT). Recent efforts seek to reduce morbidity of treatment while maintaining overall survival. In particular, young patients with small, low stage FHWT have been targeted. In 1979 a review of 176 patients with Stage I FHWT suggested that nephrectomy only, without adjuvant chemotherapy, may be adequate for patients <24 months of age with tumors <550g, referred to here as Very Low Risk Wilms Tumors (VLRWT) (1). A retrospective analysis of children treated on the first three NWTS protocols supported this hypothesis by noting that changes in the NWTS treatment regimens over a period of more than 20 years have not improved on the excellent prognosis of this group of patients (2). The first prospective cooperative group evaluation of nephrectomy as the only treatment for VLRWT was initiated in 1995 within NWTS-5, which registered 75 eligible patients. Eight patients recurred and three developed metachronous contralateral tumors, resulting in a 2-year disease-free survival estimate of 86.5% (3). Due to pre-established stopping rules, this therapeutic arm was closed and patients previously registered were given the option of receiving chemotherapy late in their course. Following this closure, NWTS-5 patients meeting the criteria for VLRWT were provided treatment with vincristine and dactinomycin. Seven of the eight patients who initially did not receive chemotherapy and relapsed responded well to subsequent therapy and were alive at 2.84 years (3). The rate of successful salvage was greater than anticipated, and the current Children's Oncology Group (COG) protocols again include a treatment arm with no adjuvant chemotherapy for children with VLRWT. Approximately 10% of VLRWT will relapse without chemotherapy. The ability to identify subsets of patients with higher or lower risks of recurrence may enable more precise stratification of patients and refinement of the definition of VLRWT. To accomplish this, we investigated the patterns of global gene expression patterns and Loss of Heterozygosity (LOH) for 11p, 1p and 16q in VLRWT in order to define and characterize subsets within VLRWT.
Materials and Methods
Clinical samples
Patient samples analyzed were taken from a case:cohort prepared on 6/5/2002 as previously described (4). Briefly, a 30% random sampling of all 1495 eligible patients from NWTS-5 was taken, and to this all relapses from the NWTS-5 were added, resulting in 600 cases enriched for relapse. This set includes 52 patients who meet the criteria for VLRWT (tumor size <550g, age <24 months, and stage 1 FHWT as determined by central pathology review). Patients who both did and did not receive adjuvant chemotherapy are included (as reviewed in the introduction). To validate the findings identified in the above tumors, an additional 11 tumors meeting the criteria for VLRWT from outside the case:cohort were randomly selected and also analyzed. Lastly, 7 tumors within the case:cohort that did not meet the criteria for VLRWT, but which demonstrated epithelial differentiated histology were identified and analyzed (vida infra). Institutional review board approval and informed consent were obtained for all tumor specimens. Specimens were obtained from the initial nephrectomy, prior to the initiation of chemotherapy. Samples were snap-frozen and stored at −80°C (5). Frozen section confirmed at least 80% viable tumor.
Gene Expression Analysis
RNA was extracted and hybridized to Affymetrix U133A arrays, scanned, and subjected to quality control parameters according to the previously described protocol (6). The microarray data consists of 22,215 probe sets from Affymetrix HG-U133A chip.9 Data were normalized using robust multi-array average (RMA) method (7). To detect native similarities and differences within the gene expression data, average-linkage clustering was performed using CLUSTER and the results were displayed using TREEVIEW (8).10 Leave-one-out cross validation was performed by leaving one tumor out and identifying the most variable genes (k=1000–9000 genes) from the remaining tumors. The tumor left out was then placed in a cluster using K-nearest neighbors (K=3). This was performed iteratively until all tumors were clustered. For each iteration, the top genes were re-determined.
Immunohistochemistry
A tissue microarray was created from formalin-fixed, paraffin-embedded tissue of 20/39 VLRWT analyzed for gene expression. The following monoclonal antibodies were tested: WT1 (DAKO, Carpinteria, CA, dilution 1:75), PAX8 (ProteinTech Group, Chicago, IL, dilution 1:100), PDGFRa (Labvision, Freemont, CA, dilution 1:50), and HMGA2 (R&D Systems, Minneapolis, MN, dilution 1:750). Staining was visualized by streptavidin-biotin (Vectastain Elite ABC Kit, Vector Laboratories) followed by ImmunoPure Metal Enhanced DAB Substrate (Therm Scientific, Rockford, IL) and counter-stained with hematoxylin (Richard-Allen Scientific, Kalamazoo, MI). Immunohistochemical staining was graded based on percentage of cells showing nuclear positivity for HMGA2, PAX8, and WT-1, and cytoplasmic positivity for PDGFα (Grade 0= 0 staining; Grade 1= <10% of cells; Grade 2= 10–20% of cells; Grade 3= >20% of cells.)
Loss of heterozygosity (LOH)
Samples were analyzed using polymerase chain reaction (PCR) as previously described (5). For chromosome 16q the following loci were examined: D16S7 or D16S2621; D16S422 and D16S402 only if D16S422 was noninformative (NI); D16S518 and D16S3101 only if D16S518 was NI; D16S421; and D16S400. For chromosome 1p the following loci were examined: D1S80 and D1S243 only if D1S80 was NI, and D1S468 only if D1S243 was NI; D1S214 and D1S244 and D1S1612 only if both D1S214 and D1S244 were not informative. For chromosome 11p the following loci were examined: INS; TH and D11S1984. LOH was considered to be present if one of the two alleles in the constitutional DNA was absent or definitely reduced in the tumor DNA as determined by visual inspection of the ethidium stained gel.
Results
Gene expression analysis
52 tumors from the original case:cohort of 600 tumors met the criteria for VLRWT. Thirteen tumors were excluded from gene expression analysis for quality control reasons, resulting in 39 tumors for analysis. Hierarchical clustering using the genes with the highest coefficient of variation (CV) was performed in order to identify native differences between subsets of tumors. The top 1000–5000 genes demonstrated three clusters that were stable in each analysis, identifying the same samples within each cluster (Figure 1A). The demographic and histologic data within each cluster are shown in Table 1. To further evaluate the stability and robustness of these clusters, leave-one-out cross validation was performed using 1000–9000 genes and K-nearest neighbors. Each tumor was correctly identified into the appropriate cluster in each analysis. The top differentially expressed genes were identified by first filtering out probesets with a maximum expression of less than 6.5 in all 39 tumors. Using the remaining 14,452 probesets, the expression of each cluster was compared with the expression in the remaining tumors, and those genes with a p-value <0.00001 and positive or negative fold-change (FC) of greater than 2.5 were identified. This resulted in 161 probesets from the comparison of C1 with C2+C3, 43 probesets from the comparison of C2 with C1+C3, and 71 probesets from the comparison of C3 with C1+C2. The false discovery rates are 0.02%, 0.06% and 0.05% for C1, C2, and C3, respectively. Because of the nature of these comparisons, the genes in these lists are not mutually exclusive (a gene significantly up-regulated in one cluster may be down-regulated in another). In order to obtain the most accurate representation of expression across all clusters, these three lists were combined. The resulting 239 probesets (161 genes) underwent hierarchical analysis, resulting in discrete patterns of over- and under expression characteristic of each cluster (Figure 1B). These probesets are provided in Supplemental Table 1 within the order of their clustering in Figure 1B. These genes were compared with the comprehensive gene expression information recently compiled by Brunskill et al. for the different regions and different stages within the developing mouse kidney. The different Brunskill groups are provided in supplemental Table 1 (9). The genes were likewise compared with gene expression patterns of Wilms tumors in the literature (6, 10, 11). Top genes by p-value and fold change for each cluster are provided in Table 2 according to overall categories established by their Brunskill groups. Those that were not identified as expressed during mouse renal development are classified as Miscellaneous. The entire gene expression data and description of the experiment using the MIAME format can be found on the GEO website. 11
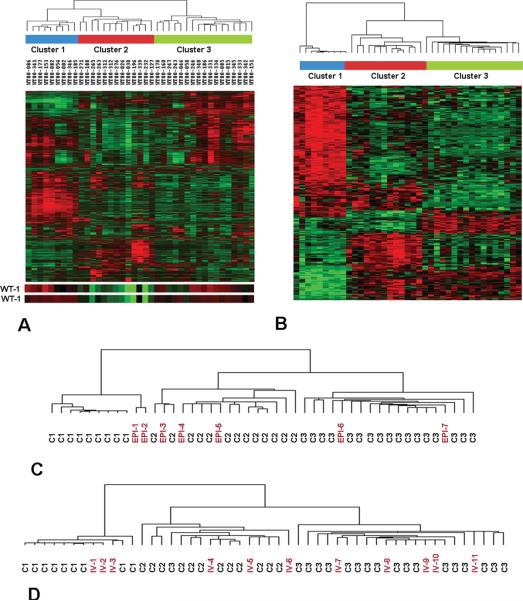
Figure 1. Unsupervised Hierarchical Clustering of 39 VLRWT.
A. The 4000 probesets with the highest coefficient of variation were utilized for unsupervised analysis using hierarchical clustering. Three primary clusters can be discriminated, as shown. Expression of genes, clustered on the Y-axis, is shown with levels ranging from high (red) to low (green). The expression of the two WT1 probesets are located at the bottom, illustrating decreased WT1 expression in Cluster 2 tumors.
B. Heirarchical analysis of the top 239 genes with fold change >2.5 and p value <0.00001, showing distinctive grouping of genes within the clusters (genes are identified within Supplemental Table 1 in the order of their appearance on this heat map).
C. Heirarchical analysis was performed using the 239 genes in Supplemental Table 1 to analyze the above 39 tumors plus 7 epithelial differentiated tumors identified from older patients. The dendogram demonstrates that these tumors do not cluster with C1 tumors.
D. Hierarchical clustering was performed using the 239 genes in Supplemental Table 1 to analyze the above 39 tumors plus an independent validation set of 11 tumors. The dendogram reveals tight clustering of the validation set within these clusters, with preserved clinical and pathologic associations within these clusters (Table 1).
TABLE 1.
Clinical, Pathologic, and Molecular Characteristics of VLRWT
| Sample | 1p LOH | 16q LOH | 11p LOH | Histology | nephrogenic rests | age at dx (m) | Weight (grams) | Relapse (days) |
|---|---|---|---|---|---|---|---|---|
| CLUSTER 1 | ||||||||
| WT00-086 | NO LOH | NO LOH | NI | Epithelial Diff. Tubular | inconclusive | 6 | 230 | No |
| WT00-341 | NO LOH | NO LOH | NO LOH | Epithelial Diff. Tubular | none | 11 | 225 | No |
| WT00-177 | NO LOH | NO LOH | NO LOH | Epithelial Diff. Tubular | none | 13 | 337 | No |
| WT00-153 | NO LOH | NO LOH | NO LOH | Epithelial Diff. Tubular | none | 15 | 485 | No |
| WT00-082 | NO LOH | NO LOH | NO LOH | Epithelial Diff. Tubular | none | 8 | 390 | No |
| WT00-054 | NO LOH | NO LOH | NO LOH | Epithelial Diff. Tubular | none | 17 | 207 | No |
| WT00-002* | NA | NA | NA | Epithelial Diff. Tubular | none | 18 | 312 | No |
| WT00-185 | NO LOH | NO LOH | NO LOH | Epithelial Diff. Tubular | inconclusive | 7 | 315 | No |
| WT00-346 | NO LOH | NO LOH | NO LOH | Epithelial Diff. Tubular | none | 3 | 430 | No |
| CLUSTER 2 | ||||||||
| WT00-188 | NA | NA | NA | Mixed | ILNR | 21 | 249 | No |
| WT00-271 | NO LOH | NO LOH | NO LOH | Mixed | ILNR | 17 | 462 | No |
| WT00-276 | NO LOH | NO LOH | LOH | Mixed | ILNR | 20 | 331 | No |
| WT00-311* | NA | NA | NA | Mixed | none | 11 | 526 | Y (140d) |
| WT00-312* | NO LOH | LOH | LOH | Mixed | ILNR | 22 | 545 | Y (154d) |
| WT00-038 | NO LOH | NO LOH | NO LOH | Mixed | ILNR | 21 | 444 | No |
| WT00-026* | NO LOH | NO LOH | LOH | Mixed | ILNR | 8 | 260 | Y (182d) |
| WT00-339 | NA | NA | NA | Mixed | ILNR | 19 | 215 | No |
| WT00-196 | NO LOH | NO LOH | NO LOH | Mixed | ILNR | 3 | 191 | No |
| WT00-232 | NO LOH | NO LOH | NO LOH | Mixed | ILNR | 12 | 360 | No |
| WT00-127 | NO LOH | NO LOH | NO LOH | Mixed | ILNR | 17 | 360 | No |
| WT00-265* | NO LOH | NO LOH | NO LOH | Mixed | ILNR | 18 | 110 | No |
| WT00-263 | NO LOH | NO LOH | NO LOH | Mixed | ILNR | 12 | 210 | No |
| CLUSTER 3 | ||||||||
| WT00-247 | NO LOH | NO LOH | LOH | Mixed | none | 7 | 496 | Y (383d) |
| WT00-243 | NO LOH | NO LOH | LOH | Blastemal | ILNR | 24 | 94 | No |
| WT00-046 | NO LOH | NO LOH | NO LOH | Blastemal | PLNR | 4 | 160 | No |
| WT00-178 | NA | NA | NA | Blastemal | ILNR | 6 | 350 | No |
| WT00-160 | NO LOH | NO LOH | NO LOH | Mixed | ILNR | 9 | 198 | No |
| WT00-186* | NO LOH | NO LOH | LOH | Mixed | ILNR | 20 | 388 | Y (202d) |
| WT00-058* | NO LOH | NO LOH | LOH | Mixed | inconclusive | 2 | 405 | Y (164d) |
| WT00-246 | LOH | NO LOH | NO LOH | Mixed | ILNR+PLNR | 13 | 472 | No |
| WT00-345 | LOH | NO LOH | NO LOH | Mixed | none | 7 | 175 | No |
| WT00-333 | NO LOH | NO LOH | LOH | Mixed | ILNR | 21 | 91 | No |
| WT00-342 | LOH | LOH | LOH | Blastemal | none | 2 | 157 | No |
| WT00-151 | LOH | NO LOH | NI | Epithelial Undiff. | inconclusive | 15 | 154 | No |
| WT00-340 | NO LOH | NO LOH | LOH | Mixed | ILNR | 5 | 442 | No |
| WT00-334 | NO LOH | NO LOH | NI | Mixed | none | 20 | 130 | No |
| WT00-331 | NO LOH | NO LOH | NO LOH | Mixed | ILNR | 9 | 163 | No |
| WT00-085 | NO LOH | NO LOH | LOH | Mixed | inconclusive | 10 | 180 | No |
| WT00-015* | NO LOH | NO LOH | NO LOH | Blastemal | none | 3 | 380 | No |
| No Gene Expression Available | ||||||||
| WT00-298 | NO LOH | NO LOH | NO LOH | Epithelial Diff. Tubular | none | 6 | 139 | No |
| WT00-303 | NO LOH | NO LOH | NO LOH | Mixed | none | 12 | 514 | No |
| WT00-304 | NO LOH | NO LOH | NO LOH | Epithelial Diff. Tubular | none | 12 | 528 | No |
| WT00-305 | NO LOH | NO LOH | LOH | Mixed | ILNR+PLNR | 15 | 375 | No |
| WT00-056 | NO LOH | NO LOH | NO LOH | Mixed | none | 5 | 328 | No |
| WT00-335 | NO LOH | NO LOH | LOH | Mixed | ILNR | 19 | 95 | No |
| WT00-336 | NO LOH | NO LOH | LOH | Mixed | ILNR | 15 | 544 | No |
| WT00-337 | NO LOH | NO LOH | NO LOH | Mixed | ILNR | 13 | 532 | No |
| WT00-343 | NO LOH | NO LOH | NO LOH | Rhabdomyomatous | none | 20 | 275 | No |
| WT00-344 | LOH | LOH | LOH | Mixed | ILNR | 19 | 333 | No |
| WT00-330 | NO LOH | NO LOH | NO LOH | Epithelial Undiff. | ILNR+PLNR | 6 | 168 | No |
| WT00-332 | NO LOH | NO LOH | LOH | Mixed | ILNR+PLNR | 14 | 423 | No |
| WT00-338 | NO LOH | NO LOH | NI | Mixed | ILNR+PLNR | 4 | 253 | No |
| Independent Validation Set | ||||||||
| IV-1 | Epithelial Diff. Tubular | none | 10 | 178 | No | |||
| IV-2 | Epithelial Diff. Tubular | none | 12 | 243 | No | |||
| IV-3 | Epithelial Diff. Tubular | inconclusive | 2 | 105 | No | |||
| IV-4 | Mixed central | ILNR | 20 | 380 | No | |||
| IV-5 | Blastemal | none | 12 | 549 | No | |||
| IV-6 | Mixed | ILNR | 6 | 412 | No | |||
| IV-7 | Epithelial Undiff. | none | 4 | 105 | No | |||
| IV-8 | Blastemal | none | 7 | 210 | No | |||
| IV-9 | Mixed | ILNR | 15 | 272 | No | |||
| IV-10 | Blastema | PLNR | 17 | 211 | No | |||
| IV-11 | Epithelial Undiff. | none | 11 | 523 | No | |||
Patients who did not receive adjuvant chemotherapy; ILNR=intralobar nephrogenic rest; PLNR=perilobar nephrogenic rest
TABLE II.
Genes Differentially Expressed in Clusters 1, 2, and 3
| Probes et | Gene Name | FC cluster 1 vs all | FC cluster 2 vs all | FC cluster 3 vs all | GO Molecular Function |
|---|---|---|---|---|---|
| Renal Development: Metanephric mesenchyme | |||||
| 204069_at | MEIS1 | −4.0 | ns | 2.3 | DNA binding, transcription factor activity |
| 207480_s_at | MEIS2 | −13.4 | ns | 3.0 | DNA binding, transcription factor activity |
| 206510_at | SIX2 | −6.5 | ns | 2.1 | DNA binding, transcription factor activity |
| 206051_at | ELAVL4 | −3.1 | ns | ns | RNA binding, mRNA 3'-UTR binding |
| 204672_s_at | ANKRD6 | ns | ns | 2.6 | protein binding |
| 214265_at | ITGA8 | ns | ns | 3.2 | receptor activity,calcium ion binding |
| Renal Development: Post-induction renal vesicle, S-shaped body | |||||
| 209552_at | PAX8* | 3.1 | −3.00 | ns | DNA binding, transcription factor activity |
| 206230_at | LHX1 | 5.1 | ns | ns | DNA binding, transcription factor activity |
| 208712_at | CCND1 | 5.3 | ns | −8.8 | protein kinase activity |
| 212226_s_at | PPAP2B (inhibitory) | 2.7 | ns | −2.9 | phosphoprotein phosphatase activity |
| 208791_at | CLU | 6.0 | ns | ns | protein binding |
| 203910_at | PARG1 (ARHGAP29) | 4 | ns | −4.5 | GTPase activator activity |
| 202157_s_at | CUGBP2 | 7.2 | ns | −7.0 | nucleotide binding |
| 213348_at | CDKN1C | 2.7 | ns | −6.0 | protein kinase inhibitor activity |
| 202620_s_at | PLOD2 | 6.1 | ns | ns | procollagen-lysine 5-dioxygenase activity |
| 210095_s_at | IGFBP3 | ns | ns | −3.8 | insulin-like growth factor binding |
| 201860_s_at | PLAT | ns | ns | −3.5 | serine-type endopeptidase activity |
| 201131_s_at | CDH1 | ns | ns | −3.9 | calcium ion binding, beta-catenin binding |
| 206067_s_at | WT1* | ns | −3.2 | ns | nucleic acid binding, transcription factor activity |
| Renal Development: terminal epithelial differentiation | |||||
| 206522_at | MGAM | 9.7 | −11.3 | ns | alpha-glucosidase activity |
| 216268_s_at | JAG1 | 5.2 | ns | −4.4 | Notch binding, calcium ion binding |
| 202669_s_at | EFNB2 | 3.7 | ns | −3.5 | protein binding, ephrin receptor binding |
| 209101_at | CTGF | 6.2 | ns | ns | integrin binding, insulin-like growth factor binding |
| 201150_s_at | TIMP3 | 3.7 | ns | ns | enzyme inhibitor activity |
| 203509_at | SORL1 | 7.7 | ns | ns | transmembrane receptor activity |
| 203817_at | GUCY1B3 | 10.0 | ns | −7.0 | guanylate cyclase activity |
| 202948_at | IL1R1 | 17.5 | ns | −10.0 | signal transducer activity |
| 203710_at | ITPR1* | ns | −2.80 | 2.6 | receptor activity, calcium channel activity |
| 208228_s_at | FGFR2* | ns | −2.2 | 2.8 | nucleotide binding. protein kinase activity |
| 209292_at | ID4* | ns | −2.7 | ns | transcription corepressor activity |
| 206558_at | SIM2* | ns | 4.40 | 3.2 | DNA binding, transcription factor activity |
| 204463_s_at | EDNRA* | ns | 6.7 | −4.7 | endothelin-A receptor activity |
| 203131_at | PDGFRA* | ns | 5.7 | ns | nucleotide binding,protein kinase activity |
| 202465_at | PCOLCE* | ns | 3.7 | ns | protein binding |
| 201983_s_at | EGFR* | ns | 3.7 | ns | nucleotide binding, protein kinase activity |
| 202283_at | SERPINF1*(PEDF) | −2.8 | 3.8 | ns | serine-type endopeptidase inhibitor activity |
| Wilms tumor-associated | |||||
| 202575_at | CRABP2** | ns | ns | 2.5 | retinoic acid binding |
| 204042_at | WASF3** | ns | ns | 2.9 | actin binding |
| 206432_at | HAS2*,** | ns | −2.9 | 2.9 | transferase activity |
| 208025_s_at | HMGA2** | −7.0 | ns | ns | DNA binding, transcription factor activity |
| 206002_at | GPR64** | ns | −4.1 | 4.0 | signal transducer activity |
| 205818_at | DBC1**(DBCCR1) | −2.8 | ns | 2.9 | protein binding |
| Miscellaneous | |||||
| 217478_s_at | HLA-DMA | 20.3 | ns | ns | MHC class II receptor activity |
| 203932_at | HLA-DMB | 4.8 | ns | −3.2 | MHC class I protein binding |
| 201508_at | IGFBP4 | 3.3 | ns | ns | insulin-like growth factor binding /// growth factor binding |
| 205372_at | PLAGL1 | −3.9 | ns | ns | nucleic acid binding, transcription factor activity |
| 40093_at | BCAM | 3.9 | −2.6 | ns | receptor activity |
| 202934_at | HK2 | 3.1 | ns | ns | nucleotide binding, hexokinase activity |
| 204733_at | KLK6* | 2.6 | −4.00 | ns | catalytic activity,serine-type endopeptidase activity |
| 203819_s_at | KOC1 (IGF2BP3) | −7.7 | ns | ns | RNA binding, mRNA 5'-UTR binding |
| 209160_at | AKR1C3 | 9.0 | ns | −7.0 | aldo-keto reductase activity |
| 206118_at | STAT4 | ns | −6 | ns | DNA binding, transcription factor activity |
| 220679_s_at | CDH7 | −4 | ns | ns | calcium ion binding,protein binding |
| 206398_s_at | CD19 | 3.9 | −2.7 | ns | receptor signaling protein activity |
| 203886_s_at | FBLN2 | 14.1 | ns | ns | extracellular matrix structural constituent |
| 211896_s_at | DCN | −10.8 | 8.1 | ns | protein binding |
Cluster 1 contains nine tumors (23% of total) that were exclusively of epithelial differentiated tubular (EDT) histology, none of which are associated with conclusive nephrogenic rests, and none of which relapsed (Table I). Striking differential expression of genes involved with renal development is present. Of the 161 genes in Supplemental Table I that were differentially regulated in Cluster 1, 83 have been shown to be up-regulated within the developing kidney (9). Cluster 1 tumors showed decreased levels of genes expressed in the pre-induction metanephric mesenchyme, and increased levels of genes expressed in post-induction epithelial differentiation. (The exceptions are those genes that are strikingly upregulated in Cluster 2). Key genes are shown in Table II, and the expression pattern of selected genes is shown in Figure 2. Immunohistochemistry confirms low expression of HMGA2 and PDGFRa, and high expression of WT-1 and PAX8 proteins in Cluster 1 tumors (Figure 3, Supplemental Table 2).
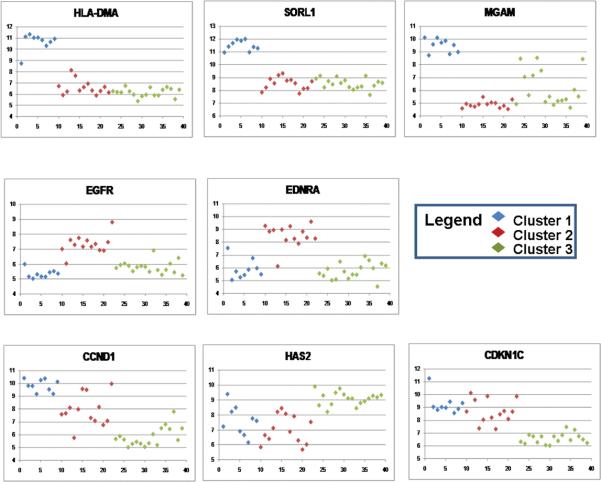
Figure 2. Gene expression patterns within the clusters of selected genes.
The expression levels (low to high) are plotted on the Y-axis. The X axis reflects an arbitrary tumor number, grouping the different tumor types starting with Cluster 1 tumors in light blue, Cluster 2 tumors in red, Cluster 3 tumors in green).
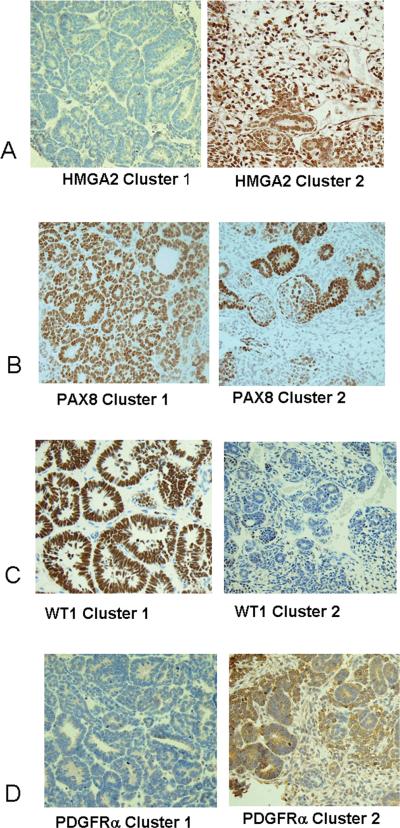
Figure 3. Immunohistochemistochemical validation of protein expression in VLRWT.
A. Immunohistochemical analysis for HMGA2: All four cluster 1 tumors were negative (left); all seven tumors within clusters 2 were immunoreactive for HMGA2 in greater than 10% of the cells.
B. Immunohistochemical analysis for PDGFRa: All four cluster 1 tumors showed immunoreactivity in fewer than 10% of cells (left); of seven cluster 2 tumors, fewer than 10% of the cells were immunoreactive in three tumors, and greater than 10% of the cells were immunoreactive in four tumors (right).
C. Immunohistochemical analysis for WT-1: All four cluster 1 tumors were immunoreactive for WT1 in greater than 10% of the cells (left); of seven cluster 2 tumors, five showed fewer than 10% of the cells to be positive and two showed greater than 10% of the cells positive (right).
D. Immunohistochemical analysis for PAX8: All four cluster 1 tumors showed greater than 80% of the cells to be immunoreactive for PAX8; of six evaluable cluster 2 tumors, all showed greater than 10% of the cells to be immunoreactive (right).
Epithelial differentiated Wilms tumors may be found within all age groups of Wilms tumors, although they are relatively infrequent (12). To determine if the expression pattern found in Cluster 1 tumors is simply a reflection of epithelial differentiation, we identified all epithelial predominant (>90% epithelial) Wilms tumors within our case:cohort that did not meet the criteria for VLRWT and performed global gene expression analysis. Seven cases were identified, ages 48–100 months, stages I (one case), II (two cases) and III (four cases); 4/7 tumors relapsed and two were associated with nephrogenic rests (one each ILNR and PLNR). Hierarchical analysis was performed using the expression of the top 239 genes within the 39 original tumors combined with the seven epithelial differentiated tumors. Five of seven tumors clustered with C2 or C3 tumors and two clustered adjacent to but outside of C1 (Figure 1C). Therefore, the gene expression pattern of C1 is not determined simply by its pattern of differentiation.
Cluster 2 includes 13 triphasic tumors (33% of total); 12 are associated with intralobar nephrogenic rests (ILNR). As can be seen in Figure 1A, the gene expression pattern is somewhat heterogeneous. Of the 43 genes significantly up- or down-regulated in C2, 21 are known to be involved in renal development (9). The most noteworthy is the down-regulation of WT1 (Figure 1A, bottom), and the coordinate expression of genes previously shown to be differentially expressed in Wilms tumors with WT1 mutation (genes with * in Table 2) (13). Three of six tumors that relapsed are in Cluster 2. The expression patterns of representative genes are illustrated in Figure 2. Immunohistochemistry demonstrates low protein expression of WT-1 in 6/7 tumors, and high expression of HMGA2 in all tumors. While the PAX8 RNA levels are decreased in Cluster 2 compared with the remaining tumors, consistently high protein expression is present (Figure 3, Supplemental Table 2.)
Cluster 3 contains 17 tumors with multiple different histologic subtypes; nine tumors demonstrate conclusive nephrogenic rests, the majority of the intralobar subtype. Of the 71 genes significantly differentially expressed, 50 have been demonstrated to be involved in renal development (9). This cluster shows increased levels of genes highly expressed by the pre-induction metanephric mesenchyme, and down-regulation of genes expressed later in development (with the exception of those down-regulated in Cluster 2). Increased levels of genes previously demonstrated to be highly expressed in the majority of Wilms tumors compared with fetal kidney was identified in Cluster 3 (10) (Table 2, Figure 2). The exception was HMGA2, which was strikingly downregulated in C1. Immunohistochemistry shows variable expression of WT-1, and high expression of HMGA2 and PAX8 in most Cluster 3 tumors (Supplemental Table 2).
Validation of gene expression in an independent set of VLRWT
To validate the above clusters within an independent set of patients, an additional 11 tumors that met the criteria for VLRWT were identified outside the case:cohort and gene expression analysis was performed. Heirarchical clustering was performed on the original 39 tumors and the additional 11 tumors using the 293 probesets within Supplemental Table 1. As shown in Figure 1D, 3/11 tumors clustered with C1 tumors, and all three showed epithelial differentiated tubular histology; 3/11 tumors clustered with C2, two with mixed histology and associated with ILNRs. The remaining 6/11 tumors clustered with C3 tumors and showed a variety of histologic patterns.
LOH analysis
LOH analysis was informative for 1p and 16q in 47/52 tumors each, and for 11p in 43/52 tumors. Overall, 1pLOH was seen in 4/47 (8.5 %), 16q LOH in 3/47 (6.4 %), and 11p LOH in 15/42 (36 %) of tumors (Table 1). This compares with an overall published prevalence within FHWT of 11.3%, 17.4%, and 33%, respectively (5). Cluster 1 lacked LOH at any of these loci; Cluster 2 lacked LOH for 1p and 16q while LOH for 11p was present in 3/10(33%); Cluster 4 demonstrated LOH for 1p, 16q, and 11p of 4/12 (33%), 1/12 (8%) and 6/10 (66%), respectively. All five informative tumors that relapsed demonstrated LOH for 11p.
DISCUSSION
Over the past several decades the prognosis for children with FHWT has improved dramatically, largely due to improved chemotherapeutic regimens. However, combination chemotherapy with vincristine and dactinomycin may cause serious myelosupression and hepatic toxicity, particularly in very young patients. Studies suggest that children with VLRWT have an excellent prognosis when treated with nephrectomy only (1–3, 14). Further, chemotherapy in VLRWT may be associated with overall financial, quality of life, and outcome costs that are greater than its benefit (3). We evaluate global gene expression patterns and LOH for 1p, 11p, and 16q in VLRWT with the hope of identifying subgroups of patients that have different clinical characteristics.
While several groups have investigated gene expression patterns in FHWT in order to define differences associated with relapse or with mutation status, few studies have addressed the overall expression patterns characteristic of Wilms tumors as a group. Li et al compared FHWT with fetal kidney samples using an algorithm taking into account the patterns of gene expression during renal development (10). They demonstrated increased expression of genes corresponding to the earliest stage of metanephric development, including PAX2, SIX1, SIX2, EYA1, SALL2, and HOXA11, and decreased expression of genes corresponding to later stages of renal development. Additionally, genes such as GPR64, WASF3, CRABP2, HAS2, and DBC1 were over-expressed. They proposed that Wilms tumors arise in cells at least partially arrested very early in renal development (10). In another comparison between differentially expressed genes in Wilms tumor with renal developmental databases, the observation that genes over-expressed in most Wilms tumors tend to be those that are expressed within the metanephric mesenchyme was confirmed (11). Huang et al compared the expression of FHWT with other pediatric renal tumors and likewise demonstrated strong up-regulation of EYA1, PAX2, GPR64, and WASF3(6). The current data distinguishes three clusters within VLRWT; Cluster 3 demonstrates considerable overlap in gene expression with the three previously cited studies (6, 10, 11). Of the 27 genes over-expressed in the “signature” proposed by Li et al, many were also over-expressed in tumors within Cluster 3 relative to the other clusters (as indicated in Table II by **).(10) Therefore, Cluster 3 appears to represent the lower age range of the most common type/s of FHWT which show arrest early in renal development. By comparison, this allows us to define two unique subsets, discussed below.
VLRWTs include a distinctive subgroup of epithelial differentiated tumors
Cluster 1 comprises approximately 25% of VLRWT and is composed of tumors with a distinctive epithelial tubular differentiated (EDT) histology, as illustrated in Figure 4A. Beckwith et al reviewed the historical experience with Wilms tumors showing a dominance of epithelial differentiation. He noted that historically survival rates for epithelial predominant tumors were much better than all other patterns combined and the patients tended to be younger (12). A careful analysis of the NWTs data demonstrated that 81% of epithelial Wilms tumors were stage I at presentation, and comprised 13% of all unilateral stage I patients. Such patients had an excellent survival. However, approximately 6% of epithelial Wilms tumor were stage III or IV at presentation, presented at an older age, and had a 21% four year relapse free survival (compared with 79% for diffuse blastemal tumors of the same stage) (12). The European Society of Pediatric Oncology (SIOP) experience supports an excellent outcome for the majority of patients with epithelial Wilms tumors, although such tumors often show poor response to therapy.(15) These findings suggest heterogeneity may exist within Wilms tumors that are epithelial predominant.
Figure 4. Epithelial Differentiated Tubular VLRWT.
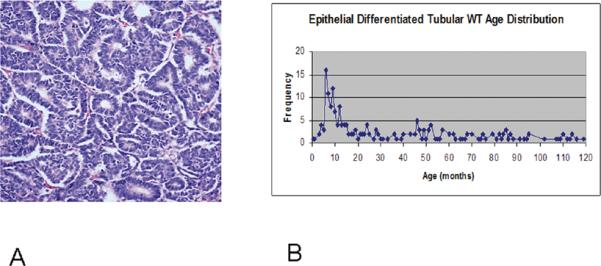
A. The histologic appearance of epithelial tubular differentiated Wilms tumors consists of epithelial structures that may show a range of differentiation throughout the tumor.
B. Age distribution of 214 patients with epithelial tubular FHWT submitted to the Renal Tumor Pathology Center from 1984—2008. Mean 36 months, median 22 months, mode 6 months.
Green et al reported that 23/75 VLRWT registered during NWTS-5 demonstrated EDT histology (3). By definition, VLRWT occur before 24 months of age. To further investigate the association between age and the diagnosis of EDT FHWT, all patients evaluated by the Renal Tumor Pathology Center from 1984–2008 and classified as EDT (215 patients) were evaluated. This demonstrates a distinct peak during infancy, and suggests that one source of heterogeneity with epithelial Wilms tumor may be reflected by the age at presentation (Figure 4B). The current study provides evidence to suggest that these distinctive Cluster 1-EDT tumors of infants may have a different pathogenesis. First, unlike the remaining VLRWT, most C1-EDTs do not arise in the setting of nephrogenic rests. Second, all evaluable C1-EDTs lacked LOH for 1p, 16q and 11p. In particular, the absence of LOH for 11p in Cluster 1 compares with an incidence in the remaining clusters of 46% (11/24). Lastly, C1-EDTs were characterized by intrinsic differences in gene expression, even when compared with histologically similar tumors at older ages. While the majority of Wilms tumors (as exemplified in Cluster 3) show arrest early in renal development (with over-expression of genes expressed in the early metanephric mesenchyme), the gene expression pattern in C1 is consistent with completion of the mesenchymal-to-epithelial transition and onset of terminal epithelial differentiation. This includes upregulation of PAX8, CCND1, CDH1, WT1, and down-regulation of SIX2 (16–18). Further, differentiation into specific epithelial types is evidenced by increased expression of genes such as WT1 and, MAFB in glomeruli, and JAG1 in proximal tubules.
Cluster 1 also demonstrates differential expression of genes whose role is to repress IGF signaling and growth. The availability and activity of IGFs depends greatly on IGF binding proteins, which may increase or decrease IGF actions. The undifferentiated metanephric mesenchyme shows predominately IGFBP-5 expression. Following induction, increasing IGFBP4 occurs with increasing differentiation (19). Therefore, upregulation of IGFBP4 seen in Cluster 1 tumors is expected and is present (Table 2). Unlike other IGFBPs, IGFBP4 inhibits IGF action (20). PPAP2B, which is upregulated in Cluster 1 tumors, also cleaves IGFBP5, further inhibiting IGF signaling and growth (21). Lastly, PLAGL1 (downregulated in Cluster 1 tumors) is a proto-oncogene that upregulates IGF2 and Wnt signaling (22). This reveals a coordinated repression of IGF signaling.
In fact, many of the genes differentially expressed in Cluster 1 largely act to promote differentiation, decrease proliferation, and increase growth suppression. The underlying explanation for the fact that these are tumors that show a proliferative advantage remains limited. One exception is the striking up-regulation of CUGBP2 (Etr-3, NAPOR) an RNA binding protein implicated in the regulation of RNA splicing, editing, stability and translation. The splicing of insulin receptor is regulated by CUGBP2, resulting in a switch to the IR-A isoform, an isoform particularly over-expressed in cancer (23, 24). IR-A, unlike IR-B, binds not only insulin but also IGF-II, resulting in mitogenic effects. Another exception is DBC1, a tumor suppressor gene that promotes apoptosis (25). DBC1 expression is frequently lost in several tumor types either by genetic loss or by hypermethylation (26). It is characteristically up-regulated in most FHWT (10) and down-regulated in Cluster 1 tumors.
In summary, Cluster 1 represents a subset of VLRWT with a characteristic histology and several molecular differences from other WT, all pointing toward a different pathogenesis. Of greatest clinical interest is the absence of relapse in this group of patients, which was also reported by Green et al (3). If this group can be adequately defined using features other than histology (which are not reliable), the age and tumor size restriction for eligibility for reduction in therapy, which are currently arbitrary, may be broadened. This is critically important because it has been shown that EDT tumors of older patients are pathogenetically different and are capable of metastasizing and relapsing, and when they do so they are often refractory to therapy (12).
VLRWT include a subgroup of tumors with decreased WT-1 expression
Cluster 2 comprises 13/39 (33%) of VLRWT. These demonstrate mixed histology and a strong association with intralobar nephrogenic rests. A key gene downregulated in Cluster 2 was WT1, a gene critical to normal renal development whose inactivation through genetic alteration is responsible for up to 20% of Wilms tumors (27). It has been previously shown that patients with WT1-mutant tumors present at an earlier age and are associated with intralobar nephrogenic rests (28, 29). In addition, Cluster 2 tumors demonstrate a gene expression pattern that is quite similar to that seen in previously reported studies comparing FHWTs with WT1 mutations to those with wild-type WT1 (Table 2, genes designated *) (13). Genes recognized to be targets of WT1 (PAX8, FGFR2) were down-regulated and EGFR, previously shown to be repressed by WT1, was upregulated in Cluster 2 tumors (30, 31).
Cluster 2 tumors demonstrate LOH for 11p in 3/10 (33%) of the evaluable tumors. This is consistent with previous reports showing that approximately half of tumors with abnormalities of WT1 also show uniparental isodisomy at 11p15 (32). While this group of tumors cannot be evaluated for outcome due to the fact that it includes patients that both did and did not receive adjuvant chemotherapy, it is of considerable interest that 3/6 relapses within this group of VLRWT occurred within Cluster 2, and all informative relapses demonstrated LOH for 11p. No relationship has been reported between relapse risk and either WT1 mutation or 11p LOH in patients with FHWT that receive chemotherapy. However, our study raises a question regarding whether there may be an association between relapse and WT1 mutation and/or 11p LOH in patients who do NOT receive chemotherapy. This needs to be further addressed by performing 11p LOH and WT1 mutation analyses in a larger group of prospectively identified patients who did not receive chemotherapy.
In summary, two subsets of VLRWT are identified that may have pathogenetic differences and different risks of relapse. If these findings can be validated within the ongoing clinical protocols, it may be possible to identify tumors with an extremely good outcome after nephrectomy only. Additional patients older than 2 years, or with a tumor weight > 550g may also show the same excellent outcome. Similarly, a second subset of patients may be defined that has an increased risk of relapse, and decisions will need to be carefully considered that balance the toxicity of therapy with the risk of relapse.
Statement of Translational Relevance.
VLRWT are defined by somewhat arbitrary stage, age and tumor weight criteria. Currently, eligible VLRWT enrolled in the current Children's Oncology Group clinical protocols are treated by nephrectomy alone, without adjuvant chemotherapy. This study provides evidence for two biologically distinctive subsets within VLRWT, and suggests that these two subsets are associated with differences in relapse. Cluster 1 (23% of tumors) is characterized by absence of relapse. Cluster 2 (33% of tumors) is characterized by an increased risk of relapse. If confirmed in prospective validation studies, these results may enable clinicians to broaden the definition of VLRWT by including tumors that have the characteristics of Cluster 1 tumors, yet fall outside of the stage, age, and tumor weight parameters that currently define VLRWT. Conversely, the additional risk for relapse of Cluster 2 tumors will require careful consideration to determine if such patients should receive adjuvant chemotherapy.
Acknowledgments
Grant Support: NIH U10CA42326 (NB, DMG, EJP); U10CA98543 (JSD, PEG, EJP); UO1CA88131 (EJP, CCH).
Footnotes
None of the authors has a conflict of interest with regard to this manuscript.
Reference List
- [1].Green DM, Jaffe N. The role of chemotherapy in the treatment of Wilms' tumor. Cancer. 1979;44:52–7. doi: 10.1002/1097-0142(197907)44:1<52::aid-cncr2820440110>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- [2].Green DM, Breslow NE, Beckwith JB, Takashima J, Kelalis P, D'Angio GJ. Treatment outcomes in patients less than 2 years of age with small, stage I, favorable-histology Wilms' tumors: a report from the National Wilms' Tumor Study. J Clin Oncol. 1993;11:91–5. doi: 10.1200/JCO.1993.11.1.91. [DOI] [PubMed] [Google Scholar]
- [3].Green DM, Breslow NE, Beckwith JB, et al. Treatment with nephrectomy only for small, stage I/favorable histology Wilms' tumor: a report from the National Wilms' Tumor Study Group. J Clin Oncol. 2001;19:3719–24. doi: 10.1200/JCO.2001.19.17.3719. [DOI] [PubMed] [Google Scholar]
- [4].Huang CC, Gadd S, Breslow NB, et al. Predicting relapse in favorable histology Wilms tumor using gene expression analysis. A report from the renal tumor committee of the Children's Oncology Group. Clin Cancer Res. 2009;15:1770–8. doi: 10.1158/1078-0432.CCR-08-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Grundy PE, Breslow NE, Li S, et al. Loss of heterozygosity for chromosomes 1p and 16q is an adverse prognostic factor in favorable-histology Wilms tumor: a report from the National Wilms Tumor Study Group. J Clin Oncol. 2005;23:7312–21. doi: 10.1200/JCO.2005.01.2799. [DOI] [PubMed] [Google Scholar]
- [6].Huang CC, Cutcliffe C, Coffin C, Sorensen PH, Beckwith JB, Perlman EJ. Classification of malignant pediatric renal tumors by gene expression. Pediatr Blood Cancer. 2006;46:728–38. doi: 10.1002/pbc.20773. [DOI] [PubMed] [Google Scholar]
- [7].Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- [8].Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Brunskill EW, Aronow BJ, Georgas K, et al. Atlas of gene expression in the developing kidney at microanatomic resolution. Dev Cell. 2008;15:781–91. doi: 10.1016/j.devcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Li CM, Guo M, Borczuk A, et al. Gene expression in Wilms' tumor mimics the earliest committed stage in the metanephric mesenchymal-epithelial transition. Am J Pathol. 2002;160:2181–90. doi: 10.1016/S0002-9440(10)61166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Li W, Kessler P, Yeger H, et al. A gene expression signature for relapse of primary wilms tumors. Cancer Res. 2005;65:2592–601. doi: 10.1158/0008-5472.CAN-04-1532. [DOI] [PubMed] [Google Scholar]
- [12].Beckwith JB, Zuppan CE, Browning NG, Moksness J, Breslow NE. Histological analysis of aggressiveness and responsiveness in Wilms' tumor. Med Pediatr Oncol. 1996;27:422–8. doi: 10.1002/(SICI)1096-911X(199611)27:5<422::AID-MPO6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- [13].Li CM, Kim CE, Margolin AA, et al. CTNNB1 mutations and overexpression of Wnt/beta-catenin target genes in WT1-mutant Wilms' tumors. Am J Pathol. 2004;165:1943–53. doi: 10.1016/s0002-9440(10)63246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Folgueira MA, Carraro DM, Brentani H, et al. Gene expression profile associated with response to doxorubicin-based therapy in breast cancer. Clin Cancer Res. 2005;11:7434–43. doi: 10.1158/1078-0432.CCR-04-0548. [DOI] [PubMed] [Google Scholar]
- [15].Weirich A, Leuschner I, Harms D, et al. Clinical impact of histologic subtypes in localized non-anaplastic nephroblastoma treated according to the trial and study SIOP-9/GPOH. Ann Oncol. 2001;12:311–9. doi: 10.1023/a:1011167924230. [DOI] [PubMed] [Google Scholar]
- [16].Schwab K, Patterson LT, Aronow BJ, Luckas R, Liang HC, Potter SS. A catalogue of gene expression in the developing kidney. Kidney Int. 2003;64:1588–604. doi: 10.1046/j.1523-1755.2003.00276.x. [DOI] [PubMed] [Google Scholar]
- [17].Schmidt-Ott KM, Masckauchan TN, Chen X, et al. beta-catenin/TCF/Lef controls a differentiation-associated transcriptional program in renal epithelial progenitors. Development. 2007;134:3177–90. doi: 10.1242/dev.006544. [DOI] [PubMed] [Google Scholar]
- [18].Self M, Lagutin OV, Bowling B, et al. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 2006;25:5214–28. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Matsell DG, Delhanty PJ, Stepaniuk O, Goodyear C, Han VK. Expression of insulin-like growth factor and binding protein genes during nephrogenesis. Kidney Int. 1994;46:1031–42. doi: 10.1038/ki.1994.364. [DOI] [PubMed] [Google Scholar]
- [20].Zhou R, Diehl D, Hoeflich A, Lahm H, Wolf E. IGF-binding protein-4: biochemical characteristics and functional consequences. J Endocrinol. 2003;178:177–93. doi: 10.1677/joe.0.1780177. [DOI] [PubMed] [Google Scholar]
- [21].Christians JK, Hoeflich A, Keightley PD. PAPPA2, an enzyme that cleaves an insulin-like growth-factor-binding protein, is a candidate gene for a quantitative trait locus affecting body size in mice. Genetics. 2006;173:1547–53. doi: 10.1534/genetics.106.057513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Van Dyck F, Declercq J, Braem CV, Van de Ven WJ. PLAG1, the prototype of the PLAG gene family: versatility in tumour development (review) Int J Oncol. 2007;30:765–74. [PubMed] [Google Scholar]
- [23].Belfiore A. The role of insulin receptor isoforms and hybrid insulin/IGF-I receptors in human cancer. Curr Pharm Des. 2007;13:671–86. doi: 10.2174/138161207780249173. [DOI] [PubMed] [Google Scholar]
- [24].Frasca F, Pandini G, Sciacca L, et al. The role of insulin receptors and IGF-I receptors in cancer and other diseases. Arch Physiol Biochem. 2008;114:23–37. doi: 10.1080/13813450801969715. [DOI] [PubMed] [Google Scholar]
- [25].Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–6. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- [26].Izumi H, Inoue J, Yokoi S, et al. Frequent silencing of DBC1 is by genetic or epigenetic mechanisms in non-small cell lung cancers. Hum Mol Genet. 2005;14:997–1007. doi: 10.1093/hmg/ddi092. [DOI] [PubMed] [Google Scholar]
- [27].Huff V. Wilms tumor genetics. Am J Med Genet. 1998;79:260–7. doi: 10.1002/(sici)1096-8628(19981002)79:4<260::aid-ajmg6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- [28].Schumacher V, Schneider S, Figge A, et al. Correlation of germ-line mutations and two-hit inactivation of the WT1 gene with Wilms tumors of stromal-predominant histology. Proc Natl Acad Sci U S A. 1997;94:3972–7. doi: 10.1073/pnas.94.8.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Beckwith JB, Kiviat NB, Bonadio JF. Nephrogenic rests, nephroblastomatosis, and the pathogenesis of Wilms' tumor. Pediatr Pathol. 1990;10:1–36. doi: 10.3109/15513819009067094. [DOI] [PubMed] [Google Scholar]
- [30].Englert C, Hou X, Maheswaran S, et al. WT1 suppresses synthesis of the epidermal growth factor receptor and induces apoptosis. EMBO J. 1995;14:4662–75. doi: 10.1002/j.1460-2075.1995.tb00148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Scharnhorst V, van der Eb AJ, Jochemsen AG. WT1 proteins: functions in growth and differentiation. Gene. 2001;273:141–61. doi: 10.1016/s0378-1119(01)00593-5. [DOI] [PubMed] [Google Scholar]
- [32].D'Angio GJ, Rosenberg H, Sharples K, Kelalis P, Breslow N, Green DM. Position paper: imaging methods for primary renal tumors of childhood: costs versus benefits. Med Pediatr Oncol. 1993;21:205–12. doi: 10.1002/mpo.2950210310. [DOI] [PubMed] [Google Scholar]