Abstract
There is a direct correlation between protein levels and disease states in human serum making it an attractive target for sensors and diagnostics. However this is made challenging because serum features more than 20,000 proteins with an overall protein content of greater than 1 mM. Here we report a hybrid synthetic-biomolecule based sensor that uses green fluorescent protein-nanoparticle arrays to detect proteins at biorelevant concentrations in both buffer and human serum. Distinct and reproducible fluorescence response patterns were obtained from five serum proteins (human serum albumin, immunoglobulin G, transferrin, fibrinogen and α-antitrypsin) in buffer and when spiked into human serum. Using linear discriminant analysis we identified these proteins with an identification accuracy of 100% in buffer and 97% in human serum. The arrays were also able to discriminate between different concentrations of the same protein as well as a mixture of different proteins in human serum.
The rapid and efficient identification of protein imbalances in serum (the clear yellowish solution obtained after removal of blood cells and clotting factors from whole blood), is an important tool for disease diagnosis1, 2. It contains >20,000 different proteins ranging from 50 gL−1 (serum albumin)3, 4 to less than 1 ngL−1 (troponin)5, with an overall protein concentration of ~1 mM. The relative and absolute level of these proteins is directly related to specific disease states. Two different approaches have been employed for serum-based diagnostics: specific recognition of biomarkers and techniques that focus on the overall levels of serum proteins. Proteins present in small quantities are specifically detected by monoclonal antibodies. With this method each monoclonal antibody has to be developed and can detect only one specific protein6,7, and technical difficulties in regard to quantification are significant8. Alternatively, electrophoresis is the current tool of choice in clinics for overall serum analysis, despite the relative insensitivity, lack of resolution, and difficulty in quantification of this method9. Better resolution is provided by 2D-SDS-PAGE electrophoresis. However, quantification and slow analysis times remain an issue. Mass spectrometry (SELDI) likewise provides a potentially powerful tool10, 11, but the expensive instrumentation, low throughput and the limited dynamic range restrict its applicability. The indicator displacement assay (IDA) has also been used to detect the key biological targets (e.g. heparin,12 inorganic phosphate13) in serum. In spite of the convenience, sensitivity and promptness of these systems, the specificity of the sensor for particular analytes limits its applicability in multiple analyte detection in undiluted serum.
A “chemical nose/tongue” strategy14,15 provides an alternative strategy to the above methods for protein sensing. In the “nose” approach, differential interactions of analytes with a receptor array generate a pattern that is used for identification. A variety of scaffolds have been employed for array-based sensing of proteins, including oligopeptide-functionalized resins16, substituted porphyrins17, polymers18, 19 and synthetic polymer-nanoparticle systems20, 21. While highly effective at identifying proteins, these systems generally feature high limits of detection (generally 8–40 μM) and require a large number of detector elements relative to the number of proteins sensed. Moreover, these methods have not been applied to sensing in challenging matrices such as biofluids.
To provide a more effective system suitable for protein sensing in serum, we created hybrid synthetic-biomolecular sensor elements. In the sensing process an array of green fluorescent protein (GFP)-nanoparticle (NP) complexes generates a signature that can be employed to identify proteins in human serum. Compared to our previous sensor array using polymers, the biocompatibility of both the nanoparticles and GFP allows us to use this system without affecting the target protein conformation during their detection22,23. In addition, the GFP-NP conjugate mimics protein-protein surface interactions, which is instrumental in reaching much lower detection limits and thus enabling detection of biomedically relevant changes in protein concentration in undiluted human serum.
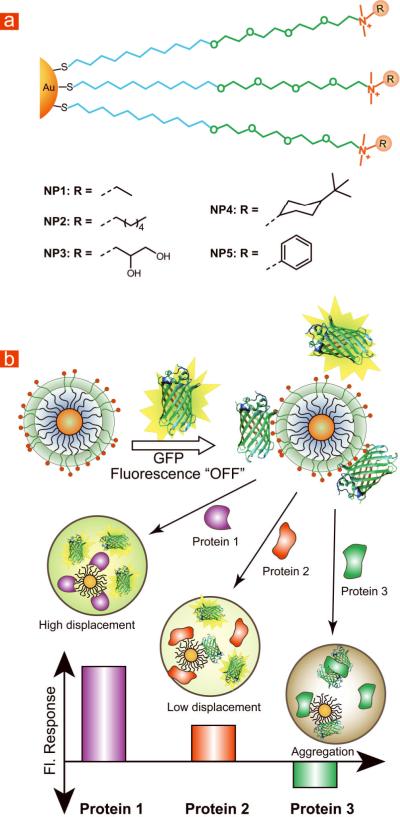
Our sensing strategy relies on the electrostatic complementarity between GFP and the NPs. GFP is a beta barrel shaped marker protein that is negatively charged at physiological conditions (3.0 diameter × 4.0 nm length, MW = 27 KDa, pH 7.4, pI = 5.92) 24, 25, with an excitation peak at 490 nm and emission peak at 510 nm. Due to their positive charges, the gold NPs complex the anionic GFP, resulting in fluorescence quenching. We hypothesized that in the presence of analyte proteins the binding equilibrium between GFP and NP would be altered due to competitive binding, thus modulating the fluorescence response (Figure 1b). The fluorescence response can be positive or negative depending on the binding affinity of analyte proteins towards NPs and GFP. A higher affinity of the protein to NPs produces positive response, while a higher affinity to GFP generates a negative response as a result of analyte protein-GFP aggregation (Figure S17). To confirm this hypothesis, five cationic gold NPs (NP1–NP5) were fabricated as sensor elements. In addition to their cationic charges, the ligand shells of these NPs differ in hydrophobicity, aromaticity, and hydrogen bonding ability (Figure 1a).
Figure 1. Structural features of nanoparticles (NPs) and modes of sensor response.
(a) Chemical structures of cationic gold NPs. NP1 features primarily cationic charge, NP2 and NP4 feature groups capable of hydrophobic interaction, NP3 features groups capable of hydrogen bonding and NP5 has aromatic recognition unit capable of π-π interaction. (b) Schematic illustration of the competitive binding between protein and quenched GFP-NP complexes and protein aggregation leading to the fluorescence light-up or further quenching.
RESULT AND DISCUSSION
Roughly 20 serum proteins with different charge and molecular weights constitute 99% by mass of the serum protein content26. We choose five of the most abundant serum proteins for our studies: albumin, immunoglobulin G (IgG), transferrin, fibrinogen and α-antitrypsin (Table 1).
Table 1.
Molecular weight, isoelectric point and concentration of five analyte serum proteins26 in human serum.
| Protein | μML−1 | Mwa | pIb | %c |
|---|---|---|---|---|
| Albumin | 769 | 65 | 5.2 | 70 |
| IgG | 66.7 | 150 | 7.5–7.8 | 14 |
| Transferrin | 25–50 | 80 | 5.6 | 5.7 |
| Fibrinogen | 5.9 | 340 | 5.6 | 2.8 |
| α-Antitrypsin | 9.6 | 52 | 5.4 | 0.7 |
) molecular weight in kDa
) isoelectric point
) weight percent in human serum.
Preliminary studies were performed in buffer. We optimized the binding ratio between GFP and NPs (NP1–NP5) through fluorescence titration. The fluorescence of GFP was significantly quenched for all NPs and the change of fluorescence intensity against increasing NP concentrations was plotted (Figure S2). The complex stability constants (KS) and association stoichiometries (n) were obtained through nonlinear least-squares curve-fitting analysis (Table S1)27. The variation in complex stabilities and the binding stoichiometry demonstrate the significant effect of head groups in NP-protein affinity.
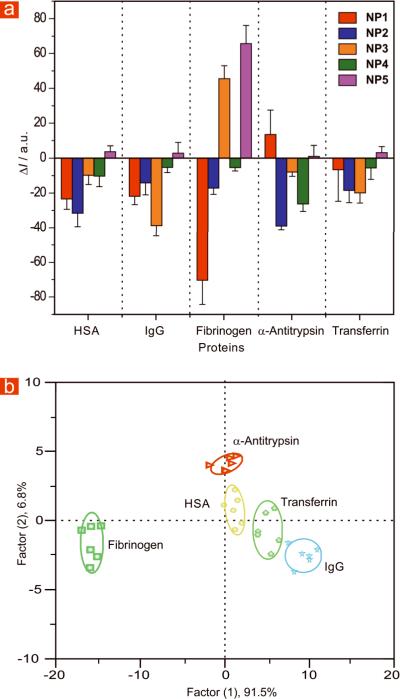
Once the binding ratio that provided maximum quenching of fluorescence was determined, we tested the ability of our sensor to detect serum proteins in 5 mM sodium phosphate buffer using a solution of 100 nM of both NPs and GFP. Testing at varying protein concentrations demonstrated that complete differentiation of the five analyte proteins was obtained at 25 nM (Figure 2a).
Figure 2. Array based sensing of five serum proteins in 5 mM sodium phosphate buffer (pH 7.40).
(a) Fluorescence response (ΔI) patterns of five GFP-NP adducts in the presence of five high abundant serum proteins at 25 nM concentration (responses are averages of six measurements and error bars are standard deviations). (b) Canonical score plot for the fluorescence patterns as obtained from LDA against five protein analytes at fixed concentration of 25 nM, with 95% confidence ellipses.
Linear discriminant analysis (LDA) was used to quantitatively differentiate the fluorescence response patterns of the GFP-NP conjugates with the serum proteins28. LDA is a statistical technique that maximizes the ratio between-class variance to the within-class variance, allowing the differentiation of response patterns. We generated the fluorescence responses six times for each protein against the five GFP-NP conjugates for this purpose. After the analysis, four canonical factors were generated (91.5%, 6.8%, 1.2% and 0.5%) that represent linear combinations of the response matrices obtained from the fluorescence response patterns (5 GFP-NP conjugates × 5 proteins × 6 replicates). The 30 training cases (5 proteins × 6 replicates) are separated in five respective groups with 100% accuracy according to the jackknifed classification matrix derived from analysis of subsets of the datasets, and the most significant two factors are plotted in 2D (Figure 2b). This detection efficiency was validated through identification of unknowns from our training set, where a randomized set of the five proteins from the training set were identified with 97% accuracy (Table S6).
Protein sensing in human serum provides a far more demanding testbed than pure proteins in buffer solution. The high overall protein content (~1 mM, 71 mg/mL) and multianalyte nature of human serum generates a complex matrix that is challenging for sensor design. To provide a controlled model for testing our methodology, we spiked physiologically relevant concentrations of the above five proteins into commercially available human serum. We first optimized the concentration and ratio of GFP-NP conjugates. Because of the optical density of pure human serum, a higher concentration of GFP (250 nM) was required to get sufficient response. The titration data (Figure 3) indicates that saturation in fluorescence quenching was not observed even at a 8:1 NP/GFP ratio due to the presence of the other proteins in human serum that compete with GFP for NP binding. To provide reproducible quenching and a high level of sensitivity, we used a particle concentration of 500 nM, corresponding to the inflexion point observed in the titration curves (Figure 3).
Figure 3. Determination of the optimum ratio of fluorophore to NP.

Change in fluorescence intensities of GFP (250 nM) at 510 nm were measured after the addition of cationic NPs (0–2 μM) with an excitation wavelength of 475 nm. The absorption effect from gold core was subtracted by using non-interacting tetra(ethylene glycol)-functionalized gold NPs. Inset shows the change of Fluorescence intensity of GFP (100 nM) at 510 nm upon addition of cationic NP3 in 5 mM sodium phosphate buffer. The red solid lines represent the best curve fitting using the model of single set of identical binding sites and the arrow indicates the optimum binding ratio used for our study.
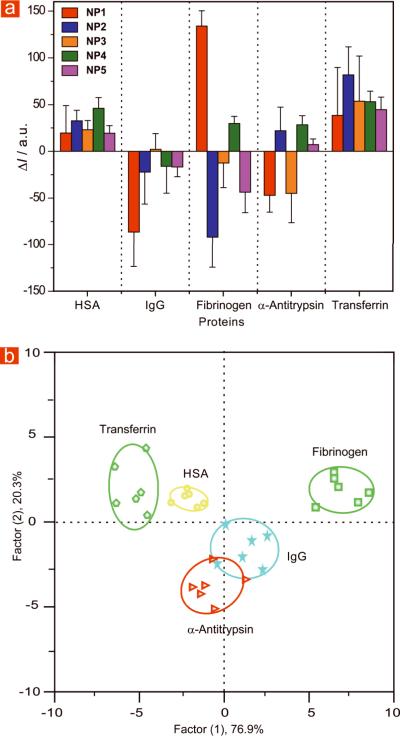
Using the conditions optimized above, 500 nM was the minimum amount of spiked analyte required for reproducible differentiation of the target proteins. We created a training matrix (5 GFP-NP conjugates × 5 proteins × 6 replicates) with GFP-NP conjugates and each of the proteins. Similar to the buffer studies, each of the proteins generated distinct fluorescence response. As before, these patterns were further subjected to LDA analysis, providing a 97% identification accuracy, as α-antitrypsin slightly overlaps with IgG. The four canonical factors are 76.9%, 20.3%, 2.7%, 0.1% and the plot of the first two factors with 95% confidence ellipses is presented in Figure 4b. We next tested the system against unknowns taken from the training set. Out of 30 samples 28 samples were correctly identified affording a 93% identification accuracy (see Table S7 for raw data). We identified target proteins in the complex serum matrix at physiologically relevant submicromolar concentrations. As a point of reference, we were able to detect and identify the analyte proteins between 0.06 to 8.4% (by molarity) of total serum protein concentration (Table 2).
Figure 4. Array based sensing of five serum proteins in human serum.
(a) Fluorescence response (ΔI) pattern of five GFP-NP adducts in the presence of serum proteins spiked in human serum at 500 nM concentration (responses are average of six measurements and error bars are standard deviations of the measurements). (b) Canonical score plot for the fluorescence patterns as obtained from LDA against five protein analytes at fixed concentration (500 nM) with 95% confidence ellipses.
Table 2.
Change of protein concentration as a percentage of its typical concentration in normal serum detected by the sensor array.
| Protein | μML−1 | %a |
|---|---|---|
| Albumin | 769 | 0.06 |
| IgG | 66.7 | 0.75 |
| Transferrin | 25–50 | 1–2 |
| Fibrinogen | 5.9 | 8.4 |
| α-Antitrypsin | 9.6 | 5.2 |
Change in protein concentrations in molarity.
After successful detection of serum proteins in human serum, the next challenge arises from the detection of a protein at variable concentration level and in mixture with other proteins. One of the limitations of immunosensing as well as other approaches is the differentiation between the variable concentration levels of a single protein. To probe the ability of our system in this dimension, we performed spiking experiments with HSA and IgG at different concentrations (500 nM, 1 μM and 2 μM). We observed that the LDA plots for various concentrations are not random, but rather follow certain patterns and can be differentiated from each other for many concentrations (Figure 5a). Essentially, the responses from different concentrations form clusters around a common center which are distinct for each protein (Figure 5b).
Figure 5. Discrimination of HSA and IgG at different concentrations and mixture of proteins.
(a) Canonical score plot for the fluorescence patterns as obtained from LDA for human serum albumin (HSA) and immunoglobulin G (IgG) at different concentrations (500 nM, 1 μM and 2 μM) with 95% confidence ellipses. (b) Clustering of all the data for the three concentrations mentioned for each protein as obtained from LDA analysis. (c) HSA and IgG were mixed at 1:1 molar ratio with 250 nM each and 500 nM each and added to five GFP-NP complexes. The canonical score plots obtained from LDA analysis were compared with that for the 500 nM of the individual proteins.
After obtaining differentiation at varying level of protein concentration we continued our investigation towards the detection of mixture of proteins in serum. We mixed HSA and IgG in serum at 1:1 molar ratio with 250 nM each as well as 500 nM each and compared with 500 nM of individual proteins. When subjected to the LDA analysis of the fluorescence responses, it shows that the canonical score plots revolve around the HSA and IgG plots and are clearly distinct (Figure 5c). Although we can discriminate the mixture of proteins at different concentration from each other, no correlation between the plots could be drawn since the complex equillibria among the serum proteins, GFP and NP make the system behave differently. However, these studies indicate that profiles of mixtures of proteins can be generated that could enable detection of disease states featuring altered levels of proteins.
In conclusion, a GFP-NP array combining synthetic and biological elements provides a highly sensitive array-based sensor system. Significantly, this approach allowed identification of proteins in serum at physiologically relevant concentrations using only five NP/GFP constructs. Furthermore, the sensor has potential in discriminating different concentrations of same protein and a mixture of proteins. Current studies are exploring the use of approach to the profiling of serum samples for the diagnosis of disease states.
METHODS
Green fluorescence protein (GFP) was expressed according to known procedure29. In brief, starter cultures from a glycerol stock of GFP in BL21(DE3) was grown overnight in 50 ml culture media at 37 °C. The following day, 5 ml of the starter cultures was added to a Fernbach flask containing 1 L culture media and shaken until the OD600 = 0.6 – 0.7. The culture was then induced by adding isopropyl-β-D-thiogalactopyranoside (IPTG) (1 mM final concentration) and shaken at 28 °C. After three hours, the cells were harvested by centrifugation and the pellet was then resuspended in lysis buffer. Once lysed, the solution was pelleted and the supernatant was further purified using HisPur Cobalt columns. The analyte proteins, serum albumin (HSA), immunoglobulins (IgG), transferrin, fibrinogen and α-antitrypsin all from human serum were purchased from Sigma-Aldrich and used as received. Cationic NPs NP1, NP2 were synthesized according to the reported procedure and NP3 – NP5 were prepared following the similar procedure which is elaborately described in supporting information. 5 mM sodium phosphate buffer, pH 7.4 was used as a solvent for the experiment in buffer solution. The commercial human serum from untransfused male donors was purchased from MP Biomedicals, LLC and used without further treatment.
In the fluorescence titration between NPs and GFP, the change of fluorescence intensity at 510 nm was measured with an excitation wavelength of 475 nm at various concentrations of nanoparticles from 0 to 100 nM on a Molecular Devices SpectaMax M5 microplate reader at 25 °C in 5 mM sodium phosphate buffer. The change of fluorescence intensity against increasing NP concentrations was plotted (Figure S2), using a non-interacting gold NP (e.g. PEG-NP) as a control to compensate for particle absorption. Nonlinear least-squares curve-fitting analysis was employed to estimate the binding constant (Ks) and association stoichiometry (n) using the model in which the nanoparticle is assumed to possess n equivalent of independent binding sites. In case of human serum, similar procedure was followed, but the only modification was the concentration of GFP (250 nM) and the nanoparticles (0–2 μM).
To create the training matrix, GFP and NPs are mixed in the ratio obtained from fluorescence titration. In case of 5 mM sodium phosphate buffer solution the final concentration of NP and GFP were 100 nM each. On the other hand, in serum studies the final concentration of NP and GFP were 500 nM and 250 nM respectively. After 30 min of incubation 200 μL of each solution was loaded into a well on a 96-well plate (300 μL Whatman black bottom micropalte) and the fluorescence intensity at 510 nm recorded using fluorescence microplate reader (Molecular Devices SpectraMax M5). Subsequently, 10 μL of protein solution, 0.525 μM for buffer solution and 10.5 μM for serum solution, was added so that the final concentrations were 25 nM and 500 nM in buffer and serum respectively. After incubation for 30 min the fluorescence intensity at 510 nm was recorded again. The difference between the two intensities before and after addition of proteins was considered as the fluorescence response (Table S1 and S3). This process was repeated for five serum proteins with five selective cationic NP in six replicates. This data was used to generate the 6 × 5 × 5 (6 replicates × 5 proteins × 5 NPs) training matrix. This training matrix was used for classical linear discriminant analysis (LDA) in SYSTAT (version 11.0).
For the unknown detection, we prepared the protein solutions (0.525 μM or 10.5 μM) out of the five serum proteins according to buffer or serum study. From this prepared solution we randomly choose 30 samples for each system (buffer or serum) and the same method was followed using the GFP-NP motif. We replicated each unknown samples three times instead of six for preparing training matrix. We considered the average response of three replicates for a single unknown sample and analyzed with five known proteins in LDA analysis.
Supplementary Material
Acknowledgements
This work was supported by the National Science Foundation (NSF) Center for Hierarchical Manufacturing at the University of Massachusetts (NSEC, DMI-0531171) and the NIH (GM077173).
Footnotes
Supplementary Information Synthesis of ligands and nanoparticles, fluorescence titration, analysis of fluorescence response patterns and unknown identification studies.
Competing financial interests None
References
- 1.Hardy J, Selkoe DJ. Medicine - The amyloid hypothesis of Alzheimer's disease: Progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Pulido R, van Huijsduijnen RH. Protein tyrosine phosphatases: dual-specificity phosphatases in health and disease. Febs Journal. 2008;275:848–866. doi: 10.1111/j.1742-4658.2008.06250.x. [DOI] [PubMed] [Google Scholar]
- 3.Adkins JN, et al. Toward a human blood serum proteome - Analysis by multidimensional separation coupled with mass spectrometry. Molecular & Cellular Proteomics. 2002;1:947–955. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- 4.Pieper R, et al. The human serum proteome: Display of nearly 3700 chromatographically separated protein spots on two-dimensional electrophoresis gels and identification of 325 distinct proteins. Proteomics. 2003;3:1345–1364. doi: 10.1002/pmic.200300449. [DOI] [PubMed] [Google Scholar]
- 5.Antman EM, et al. Cardiac-specific troponin I levels to predict the risk of mortality in patients with acute coronary syndromes. N. Engl. J. Med. 1996;335:1342–1349. doi: 10.1056/NEJM199610313351802. [DOI] [PubMed] [Google Scholar]
- 6.Anderson GP, et al. Quantifying serum antiplague antibody with a fiber-optic biosensor. Clinical and Diagnostic Laboratory Immunology. 1998;5:609–612. doi: 10.1128/cdli.5.5.609-612.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Badgwell D, Bast RC. Early detection of ovarian cancer. Dis. Markers. 2007;23:397–410. doi: 10.1155/2007/309382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy GP, Elgamal AAA, Su SL, Bostwick DG, Holmes EH. Current evaluation of the tissue localization and diagnostic utility of prostate specific membrane antigen. Cancer. 1998;83:2259–2269. [PubMed] [Google Scholar]
- 9.McPherson RA, Pincus MR. Henry's Clinical Diagnosis and Manangement by Laboratory Methods. Saunders-Elsevier; 2007. Ch 19. [Google Scholar]
- 10.Li JN, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clinical Chemistry. 2002;48:1296–1304. [PubMed] [Google Scholar]
- 11.Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics. 2004;20:777–U710. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- 12.Wright AT, Zhong Z, Anslyn EV. A Functional Assay for Heparin in Serum Using a Designed Synthetic Receptor. Angew. Chem., Int. Ed. 2005;44:5679–5682. doi: 10.1002/anie.200501437. [DOI] [PubMed] [Google Scholar]
- 13.Tobey SL, Anslyn EV. Determination of Inorganic Phosphate in Serum and Saliva Using a Synthetic Receptor. Org. Lett. 2003;5:2029–2031. doi: 10.1021/ol034427x. [DOI] [PubMed] [Google Scholar]
- 14.Wright AT, Anslyn EV. Differential receptor arrays and assays for solution-based molecular recognition. Chem. Soc. Rev. 2006;35:14–28. doi: 10.1039/b505518k. [DOI] [PubMed] [Google Scholar]
- 15.Lavigne JJ, Anslyn EV. Sensing a paradigm shift in the field of molecular recognition: From selective to differential receptors. Angew. Chem., Int. Ed. 2001;40:3119–3130. doi: 10.1002/1521-3773(20010903)40:17<3118::AID-ANIE3118>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 16.Wright AT, Edwards NY, Anslyn EV, McDevitt JT. The discriminatory power of differential receptor Arrays is improved by prescreening - A demonstration in the analysis of Tachykinins and similar peptides. Angew. Chem., Int. Ed. 2007;46:8212–8215. doi: 10.1002/anie.200701236. [DOI] [PubMed] [Google Scholar]
- 17.Zhou HC, Baldini L, Hong J, Wilson AJ, Hamilton AD. Pattern recognition of proteins based on an array of functionalized porphyrins. J. Am. Chem. Soc. 2006;128:2421–2425. doi: 10.1021/ja056833c. [DOI] [PubMed] [Google Scholar]
- 18.Miranda OR, et al. Array-based sensing of proteins using conjugated polymers. J. Am. Chem. Soc. 2007;129:9856–9857. doi: 10.1021/ja0737927. [DOI] [PubMed] [Google Scholar]
- 19.Stephenson CJ, Shimizu KD. Colorimetric and fluorometric molecularly imprinted polymer sensors and binding assays. Polym. Int. 2007;56:482–488. [Google Scholar]
- 20.You CC, et al. Detection and identification of proteins using nanoparticle-fluorescent polymer `chemical nose' sensors. Nature Nanotechnology. 2007;2:318–323. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]
- 21.Phillips RL, Miranda OR, You CC, Rotello VM, Bunz UHF. Rapid and efficient identification of bacteria using gold-nanoparticle - Poly(para-phenyleneethynylene) constructs. Angew. Chem., Int. Ed. 2008;47:2590–2594. doi: 10.1002/anie.200703369. [DOI] [PubMed] [Google Scholar]
- 22.De M, You CC, Srivastava S, Rotello VM. Biomimetic interactions of proteins with functionalized nanoparticies: A thermodynamic study. J. Am. Chem. Soc. 2007;129:10747–10753. doi: 10.1021/ja071642q. [DOI] [PubMed] [Google Scholar]
- 23.Hong R, et al. Control of protein structure and function through surface recognition by tailored nanoparticle scaffolds. J. Am. Chem. Soc. 2004;126:739–743. doi: 10.1021/ja037470o. [DOI] [PubMed] [Google Scholar]
- 24.Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 25.Chalfie M, Kain SR. Green Fluorescent Protein: Properties, Applications, and Protocols. Wiley-Interscience; Hoboken, N.J.: 2006. p. 69. [Google Scholar]
- 26.Anderson NL, Anderson NG. The human plasma proteome - History, character, and diagnostic prospects. Molecular & Cellular Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 27.You CC, De M, Han G, Rotello VM. Tunable inhibition and denaturation of alpha-chymotrypsin with amino acid-functionalized gold nanoparticles. J. Am. Chem. Soc. 2005;127:12873–12881. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- 28.Jurs PC, Bakken GA, McClelland HE. Computational methods for the analysis of chemical sensor array data from volatile analytes. Chem. Rev. 2000;100:2649–2678. doi: 10.1021/cr9800964. [DOI] [PubMed] [Google Scholar]
- 29.De M, Rana S, Rotello VM. Nickel-Ion-Mediated Control of the Stoichiometry of His-Tagged Protein/Nanoparticle Interactions. Macromol. Biosci. 2009;9:174–178. doi: 10.1002/mabi.200800289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.