Summary
Previous studies have suggested that the HIF transcription factors can both activate and inhibit gene expression. Here we show that HIF1 regulates the expression of mir-210 in a variety of tumor types through a hypoxia responsive element. Expression analysis in primary head & neck tumor samples indicates that mir-210 may serve as an in vivo marker for tumor hypoxia. By Argonaute protein immunoprecipitation, we identified 50 potential mir-210 targets and validated randomly selected ones. The majority of these 50 genes are not classical hypoxia inducible genes, suggesting mir-210 represses genes expressed under normoxia that are no longer necessary to adapt and survive in a hypoxic environment. When human head and neck or pancreatic tumor cells ectopically expressing mir-210 were implanted into immunodeficient mice, mir-210 repressed initiation of tumor growth. Taken together, these data implicate an important role for mir-210 in regulating the hypoxic response of tumor cells and tumor growth.
Keywords: Hypoxia, microRNA, mir-210, gene expression, tumor initiation
Introduction
Hypoxia, the condition of insufficient oxygen supply to tissues, results from a reduction in oxygen availability, inadequate oxygen transport, or the inability of the tissues to utilize oxygen. In normal tissues, high attitude exposure, anemia, and drugs can induce hypoxia responsive pathways in the body. However, hypoxia is also a hallmark of cancer, where cancer cells shift their energy production from the TCA cycle to glycolysis (Kim and Dang, 2006). The hypoxia-inducible factors (HIFs) are a family of transcription factors that have been identified as important regulators of the cellular response to hypoxia (Semenza, 1998). Under normoxic conditions, the alpha subunit of HIF1 is hydoxylated at proline 402 and 564 by prolyl-4-hydroxylases (PHDs), targeting HIF1α for proteasome destruction mediated by the von Hippel Lindau (VHL) protein, an E3 ubiquitin ligase (Chan et al., 2005; Ivan et al., 2001; Jaakkola et al., 2001). Under hypoxic conditions, the activity of PHDs decreases, HIF1α is stabilized and it transcriptionally regulates a large number of target genes involved in adaptation and protection against low oxygen conditions (Chi et al., 2006).
HIFs regulate an ever increasing number of genes involved in glycolytic metabolism, angiogenesis, erythropoiesis, and metastasis (Chan and Giaccia, 2007; Semenza, 1998; Sullivan and Graham, 2007). Recently, microRNAs (miRNA) have emerged as a new class of non-coding genes involved in regulating cell proliferation, differentiation, and viability (Bartel, 2004; Stefani and Slack, 2008). MiRNAs are single-stranded small RNA molecules that are approximately 22 nucleotides in length. MiRNAs primarily regulate gene expression through inhibition of RNA translation by base-pairing of their “seed region”, nucleotides 2–8, to their target genes’ 3′ UTR (Nilsen, 2007). MiRNAs can also facilitate targeting of specific mRNAs for cleavage, resulting in the down-regulation of target mRNAs (Jackson and Standart, 2007; Lim et al., 2005). It has been shown that miRNA expression is regulated by certain physiological stimuli (van Rooij et al., 2006). We hypothesized that some miRNAs are regulated by hypoxia given the critical roles oxygen homeostasis plays in cellular physiology and the broad spectrum of genes hypoxia regulates. Several recent studies have shown that mir-210 is induced by hypoxia and appears to be a HIF target gene, although no identification of functional HREs have been demonstrated (Camps et al., 2008; Giannakakis et al., 2007; Kulshreshtha et al., 2007).
In this report, we identified several hypoxia regulated miRNAs through miRNA microarray analysis. We present evidence showing that one of the miRNAs, mir-210, is the predominant miRNA gene induced under hypoxic conditions in a broad spectrum of cancer types, and its transcriptional induction is HIF1α-dependent. By immunoprecipitating the major functional component of the miRNA pathway, Argonaute 2, we identified a list of fifty genes as potential mir-210 targets, of which the majority are not known to be induced under hypoxia. The regulation of mir-210 by HIF can explain how HIF activation leads to inhibition of gene expression. Functionally, we show that ectopic expression of mir-210 represses tumor growth, linking HIF regulation to inhibition of tumor growth through mir-210 regulation.
Results
Mir-210 is the predominant hypoxia-inducible microRNA
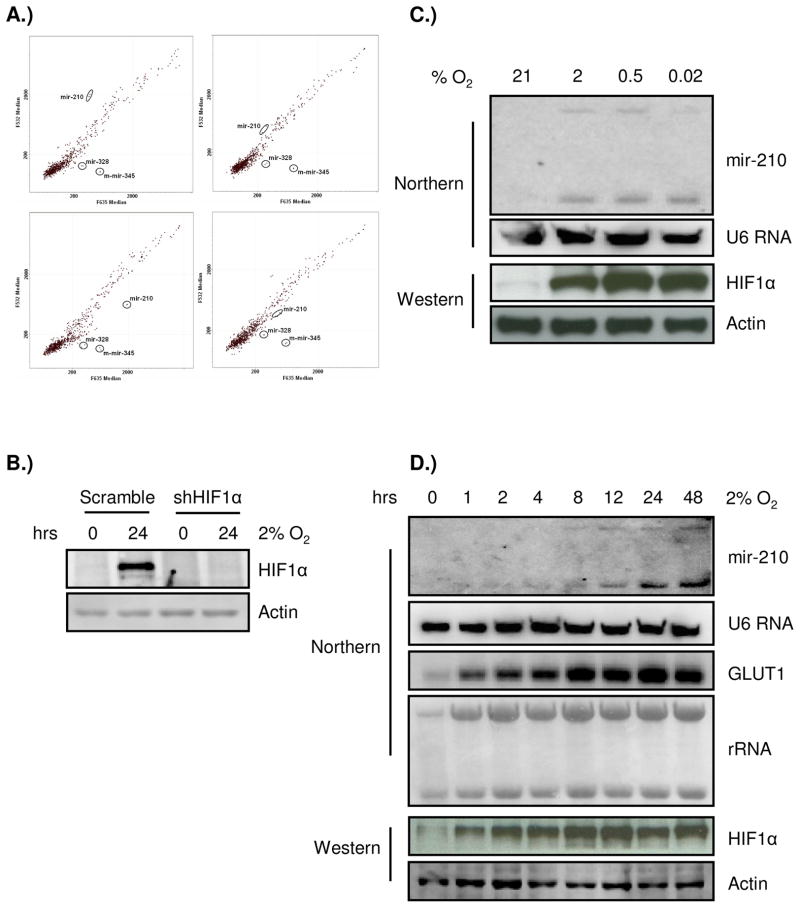
To identify miRNA genes regulated by hypoxia, we analyzed the expression profiles of the human pancreatic cancer cell lines SU86.86 and PANC1 using a miRNA microarray containing 314, 49, and 14 probes corresponding to mature forms of human, mouse, and rat miRNAs, respectively. After 16 hours under 2% oxygen, paired nomoxia/hypoxia miRNA samples were labeled with either cy3 or cy5. In order to detect non-specific labeling and hybridization, dye-swapped experiments were performed in parallel. Several miRNAs appeared to be robustly induced by hypoxia (Fig. 1A), such as has-mir-328, mmu-mir-345, and hsa-mir-210. However, in the dye swap experiment only the expression of hsa-mir-210 changed reciprocally (Fig. 1A). Because HIF1α is a major regulator of the cellular response to hypoxia, we established a SU86.86 cell line that expresses a small hairpin RNA (shRNA) that reduces HIF1α expression to less than 10% of the wild-type controls (Fig. 1B). After stable knock-down of HIF1α, the induction of mir-210 under hypoxia was undetectable (Fig. 1A), which suggests that mir-210 is a HIF1α regulated gene. Subsequent miRNA Northern verification of the miRNA microarrays indicated that several other miRNA genes were also induced by hypoxia, although their induction was not as pronounced as mir-210 (supplementary Figure 1). For these reasons, mir-210 was chosen for further analysis in the present study.
Figure 1.
Mir-210 is the predominant hypoxia-responsive miRNA. A) Microarray analysis of miRNAs induced during the cellular response to hypoxia. SU86.86 and SU86.86/shHIF1α cells were split into two plates 24 hours before the hypoxic treatment, respectively. One plate stayed in normoxia and the other was exposed to 2% O2 for 16 hours. Then RNAs were harvested at the same time and used to conduct microarray analysis. Data are presented on a scatter plot showing log10-transformed signal intensities for each probe on both channels for the Cy3-labeled normoxic control and for the Cy5-labeled hypoxic sample (top panel). In the parallel dye swap experiment, normoxic sample was labeled with cy5 and hypoxic sample was labeled with cy3 (bottom panel). mir-210 is identified as the most robustly induced miRNA and its induction is dependent on HIF1α. B) Western blot confirms efficient HIF1α knock-down by the shRNA construct in SU86.86 cells used in the microarray experiment; C) mir-210 is induced under different hypoxic stringencies by Northern blot. SU86.86 cells were split into four plates 24 hours before treatment. Then each plate was exposed to normoxia, 2%, 0.5%, or < 0.02% (anoxia) O2 for 24 hours. Small nuclear RNA U6 was used as a loading control; D) The kinetics of mir-210 induction under 2% O2 in SU86.86 cells. mir-210 expression reaches the plateau after 24 hours of hypoxia and stayed high till 48 hours (top panel), which is consistent with the control, GLUT1, a classic HIF regulated gene (middle panel). Western blot of HIF1α was shown to indicate hypoxia condition (bottom panel).
Since it has been known that certain genes/pathways are activated under specific oxygen tensions, such as the unfolded protein response pathway (UPR), we investigated the inducibility of mir-210 under more stringent hypoxic conditions. As shown in Fig. 1C, mir-210 is induced by mild as well as stringent oxygen tensions. Furthermore, the kinetics of mir-210 induction under 2% oxygen is similar to a classic hypoxia regulated gene, GLUT1 (Fig. 1D). Taken together, our data suggest that mir-210 is highly responsive to changes in oxygen.
Mir-210 is broadly expressed and is HIF1α-dependent
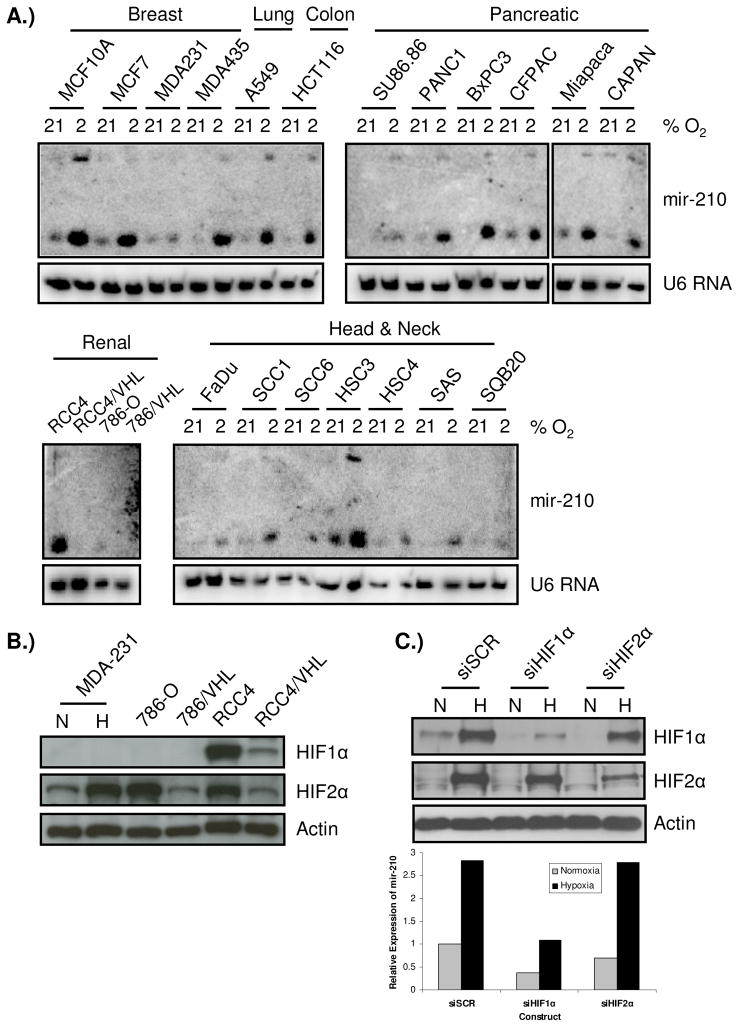
To investigate whether induction of mir-210 under hypoxia is tissue or cell line specific, we examined the induction of mir-210 in pancreatic, breast, head and neck, lung, colon, and renal cell lines exposed to 2% oxygen for 24 hours by Northern blotting. Mir-210 was almost universally induced in all cell lines across different tissue origins (Fig. 2A). Generally, mir-210 expression was low under normoxic conditions and its expression was greatly induced after exposure to hypoxia.
Figure 2.
Mir-210 is broadly expressed and is a HIF1α-specific gene. A) Up-regulation of mir-210 by hypoxia is detected in all six different tumor types examined. Each cell line was split into two identical plates 24 hours prior to treatment. One was exposed to 2% O2 for 24 hours while the other was kept under normoxia and then, RNAs were harvested at the same time and used for Northern blotting. RCC4/VHL and 786/VHL are RCC4 and 786-O cell lines with stably reconstituted wild-type VHL gene. mir-210 is well induced by hypoxia in almost all cell lines examined, with highest induction in HSC3, MCF10A, and MCF7 cells. However, up-regulation by hypoxia is barely seen in MDA231, SQB20, and 786-O cells. U6 RNA was used as a loading control; B) Western blot of HIF1α and HIF2α in MDA-MB-231 and paired RCC4, 786-O cells. No HIF1α could be detected in 786-O and MDA-231 cells under 2% O2 for 24 hr. However, HIF2α could be readily detected in all cell lines. C) mir-210 expression was assayed by qPCR in RNAs harvested from RCC4/VHL cells with siRNA knock-down of HIF1α, HIF2α, or a scramble control siRNA with or without exposing to 2% O2 for 24 hours (lower panel). RNA input was normalized on small nuclear RNA RNU48. Upper panel shows the hypoxia induction and siRNA knock-down of HIF1α and HIF2α in these cells.
Interestingly, in two pairs of renal cancer cell lines, RCC4/RCC4-VHL and 786-O/786-VHL, mir-210 expression is clearly different (Fig. 2A). It is well-known that both RCC4 and 786-O harbor VHL gene mutations that in turn cause stabilization of HIF transcriptional factors. In RCC4 cells both HIF1α and HIF2α are expressed, but in 786-O only HIF2α is expressed (Fig. 2B). In addition, we possess a variant of the MDA-231 breast cancer cell line that has lost HIF1α expression (Fig. 2B). Since both MDA-231 and 786-O cells exhibited poor mir-210 induction by hypoxia, this prompted us to determine whether mir-210 expression is HIF1-specific. In order to address the role of HIF1α versus HIF2α in mir-210 expression, siRNAs against HIF1α, HIF2α, or scramble control siRNAs were transfected into RCC4/VHL cell line. The cells transfected with siRNA scramble controls and siRNAs against HIF2α had robust induction of mir-210 expression under 2% oxygen as detected by quantitative RT-PCR (qPCR). However, this induction was abolished in RCC4/VHL cells that were transfected with siRNAs against HIF1α (Fig. 2C). Interestingly, the baseline expression of mir-210 was also reduced even under normoxic conditions, indicating that HIF1α also regulates the transcriptional expression of mir-210 under normoxia.
Characterization of mir-210 promoter
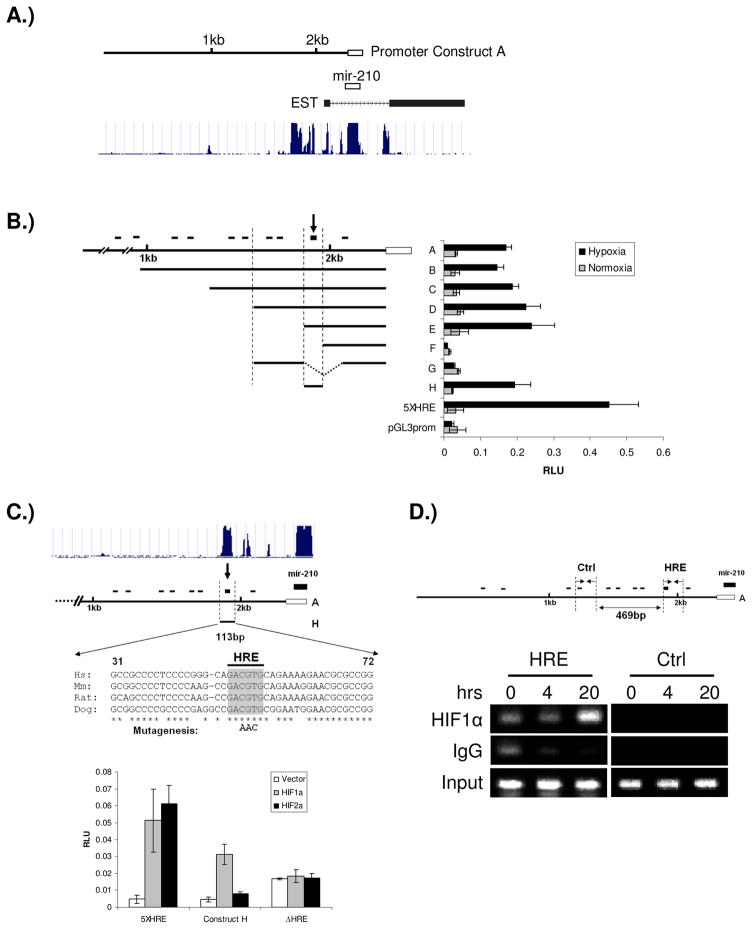
HIF1α regulates target gene activity by binding to a hypoxia responsive element (HRE) on genomic DNA. Since our data indicates that mir-210 is a HIF1α regulated gene, we sought to determine how HIF1α regulates it. A 2.3kb promoter sequence immediately upstream of mir-210 stem loop structure was cloned into a pGL3-Basic vector (Fig. 3A). By searching the 2.3kb promoter sequence, ten potential HRE sites were identified according to the consensus sequence (A/G)CGTG. In order to pinpoint the functional HRE, we made seven serial deletion constructs and performed reporter assays to determine the effect of the deletions on reporter gene activity (Fig. 3B). We found a 113bp fragment that harbors a HRE element is responsible for the robust induction of mir-210 promoter under hypoxia. Mutation of this HRE site completely abolished the responsiveness of the promoter to HIF1α (Fig. 3C). The promoter assay also confirmed that mir-210 is a HIF1α-dependent gene, and is not HIF2α responsive. Interestingly, when the human, mouse, rat, and dog mir-210 genomic sequences were compared, the mir-210 coding sequence and this HRE site are the only highly conserved sequences across species (Fig. 3C), indicating the functional importance of the HRE element.
Figure 3.
Identification of the functional HRE in mir-210 promoter. A) The schematic view of mir-210 genomic structure. The location of the cloned promoter related to mir-210 coding region was shown in top panel. Open box indicates the coding region of mir-210 and solid line indicates the 2.3kb promoter cloned. The conservation of mir-210 genomic sequences across species was shown in the bottom panel, which is from the UCSC database (http://genome.ucsc.edu). B) mir-210 promoter analysis. Promoter fragments were cloned into the pGL3-Basic vector; a 5XHRE construct, including five copies of the human VEGF HRE, serves as a positive control. Serial deletion constructs (identified by the letters A–H in the middle of the figure) are schematically represented in the bar diagram on the left, while their relative promoter activities in normoxia (gray bars) and 2% oxygen (black bars) are displayed on the right of the figure. The small black bars on the top represent the relative location of the ten potential HREs. The arrow at the top of the bar diagram indicates this HRE is essential for up-regulation of promoter activity under hypoxia. The open box represents mir-210 coding sequence; C) Mutation of the HRE sequence abolishes mir-210 promoter’s responsiveness to HIF1α. Constructs A and H are shown. Upper panel shows that the sequence surrounding the HRE site is the only region highly conserved across species in mir-210 promoter according to the UCSC database. The conserved nucleotides from 31–72 in construct H are indicated by asterisks. Three nucleotide mutations in the core of the HRE are in bold letters. Luciferase assays of construct H with/without HRE mutation are shown in the lower panel. Error bars indicate standard deviation. D) ChIP analysis of the mir-210 promoter. Upper panel, the relative locations of ChIP primers on mir-210 promoter. Lower panel, ChIP results. HSC3 cells were cultured under normoxia or 0.5% oxygen for 4 and 20 hours, respectively. PCR was performed with primers specific to the functional mir-210 HRE (HRE) and a region that is ~500bp upstream of the HRE site serving as a control (Ctrl). IgG refers to samples derived for the IP-negative control, HIF1α refers to DNA immunoprecipitated with a HIF1α antibody, and Input refers to the DNA derived from samples prior to immunoprecipitation.
We next wanted to determine if HIF1α is capable of binding to the mir-210 promoter directly in vivo. We designed two pairs of primers for chromatin immunoprecipitation (ChIP). The first pair flanks the functional HRE element identified through serial deletion while the other is approximately 500bp upstream of this HRE as a control. We found that HIF1α specifically binds to the functional HRE element, but not the control promoter sequence (Fig. 3D).
Mir-210 is hypoxia-regulated in vivo
We hypothesized that if mir-210 is such a robust hypoxia-regulated gene in cell lines in vitro, we should be able to detect its increased expression in primary tumors that exhibit significant levels of hypoxia. We took advantage of the microarray profiling data in 65 primary head & neck tumor samples developed at the M.D. Anderson Cancer Center. First, we performed a whole genome unsupervised clustering analysis based on the gene expression profiles and mir-210 expression in these samples. We found that expression of mir-210 is highly correlated with the expression of some well-known hypoxia inducible genes, with the strongest correlation with NDRG1 and SLC2A1 (GLUT1) (Supplementary Figure 2A & 2C). Interestingly, two genes in the list, HRAS and RASSF7, are not known for to be hypoxia inducible genes. However, they are both located on chromosome 11p15.5 and are proximally adjacent to mir-210 (Supplementary Figure 2D), suggesting that their expression may be coordinated due to genomic proximity. Next, we compared mir-210 expression with a list of 10 overlapping genes from two published studies describing a hypoxic gene expression signature in these 65 primary H&N tumor samples (Chi et al., 2006; Winter et al., 2007). We found that expression of mir-210 is closely correlated with that of these 10 genes (Supplementary Figure 2B). Since these 10 genes are obtained from two independent studies, they may represent a core gene set regulated by hypoxia in general and therefore, the close correlation of mir-210 expression to their expression strongly supports that mir-210 is a hypoxia-regulated gene in vivo.
Direct identification of mir-210 target genes through Argonaute 2 (AGO2) ribonucleoprotein immunoprecipitation (miRNP-IP)
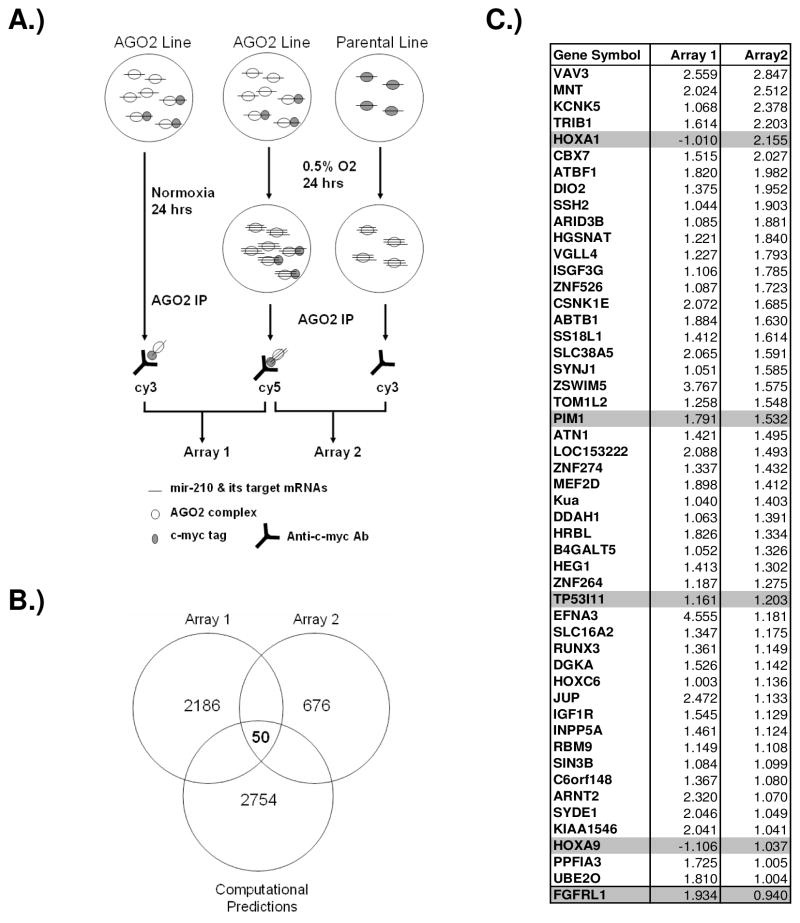
It has been known that miRNA could regulate its target genes by inhibiting protein translation and/or degrading messenger RNAs (Jackson and Standart, 2007; Nilsen, 2007). Since miRNA regulates its target genes through an imperfect match in their 3′ UTR region, and only a six- to seven-nucleotide perfect match in the “seed” region is required, each miRNA could potentially regulate hundreds of target genes as predicted by computational programs. However, many of them may just be artifacts. Therefore, currently it is still a major challenge to directly identify miRNA target genes. We hypothesized that miRNA functions through the argonaute protein complex and immunoprecipitation of the major functional component of this complex, AGO2, would be useful to identify mRNAs associated with AGO2 using microarrays. Thus, the combination of experimentally identified genes with computational predictions would be a powerful approach to identify mir-210 targets. Our experimental design is illustrated in Fig. 4A. The N-terminal of the AGO2 protein was labeled with a c-myc tag and a stable cell line with tagged AGO2 was established in MCF10A cells because of its robust mir-210 induction under hypoxia (Fig. 2A and supplementary Figure 3).
Figure 4.
Identification of mir-210 target genes by miRNP-IP. A) A schematic view of the strategy to identify mir-210 targets. B) Venn diagram shows potential mir-210 target genes predicted by four computational programs, TargetSacn, PicTar, miRanda, and PITA and genes enriched in miRNP-IP experiments. C) Fifty potential mir-210 target genes were identified. The four genes used in validation were highlighted in gray. The FGFRL1 gene with a little less than 2-fold enrichment in array 2 was also included at the bottom of the list. The values shown are normalized log2(R/G).
After miRNP-IP and microarray experiments, 246 genes with a higher than two fold enrichment in both array 1 and array 2 were selected. We then searched TargetScan, PicTar, mirRanda, and PITA programs compiling a list of 2754 potential mir-210 target genes (John et al., 2004; Kertesz et al., 2007; Krek et al., 2005; Lewis et al., 2005). Fifty of the 246 genes (20.3%) are computationally predicted to be mir-210 targets (Fig. 4B & C). Therefore, the 3′ UTR mir-210 target sites were greatly enriched in the genes identified through the miRNP-IP approach. Interestingly, except EFNA3 all other identified genes are not known to be induced by hypoxia. On the basis of functional annotation, genes involved in regulating gene expression and cellular metabolic process were strongly overrepresented among this set of 50 genes (p<7.2 × 10−5 and p<1.2 × 10−4, respectively) (supplementary Table 1), suggesting that mir-210 may down regulate genes expressed under normoxia when induced by HIF to better prepare cells to adapt and survive the hypoxic stress.
Validation of mir-210 target genes identified through miRNP-IP
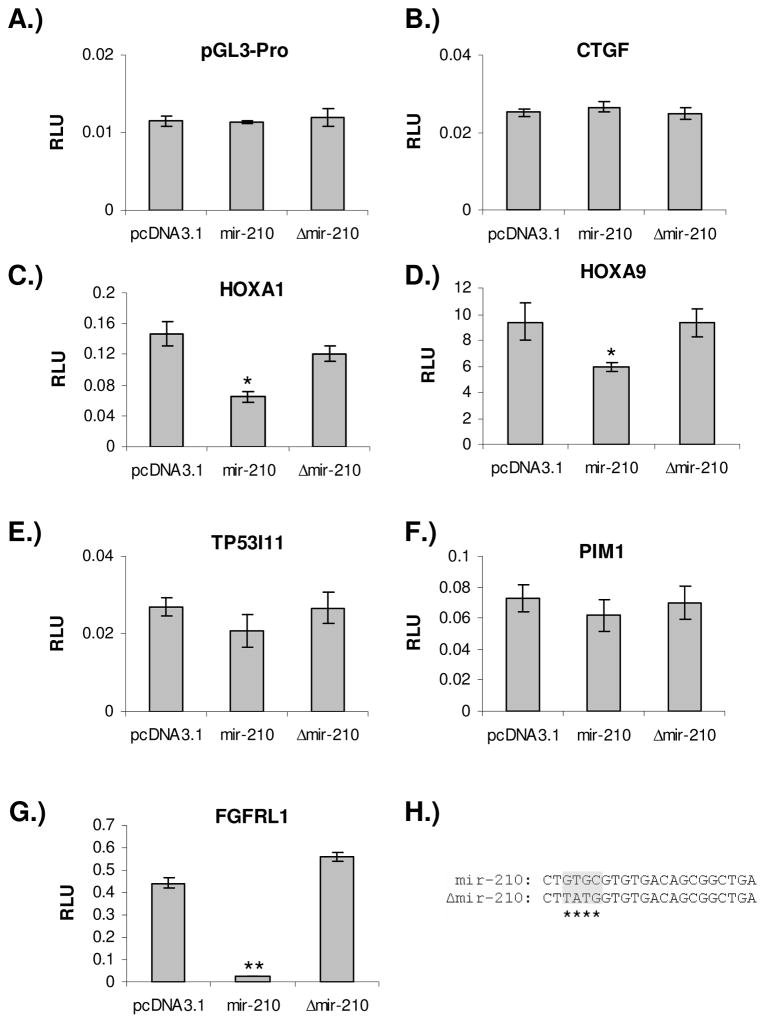
In order to validate mir-210 target genes identified through the RNA-IP approach, we cloned the 3′ UTRs of four randomly selected genes with different degrees of enrichment into pGL3 vector to perform reporter assays. When the mir-210 expression plasmid was co-transfected into the cell with these reporter constructs, luciferase activity was repressed more than 50% in HOXA1 construct, 35% in HOXA9, and 15% in PIM1, and 17% in TP53I11 constructs compared to co-transfection with pcDNA3.1 control plasmid (Fig. 5C – F). However, when a pGL3-Promoter control luciferase reporter construct and a CTGF 3′ UTR construct without mir-210 target sequences were co-transfected, there was no repression of luciferase activity (Fig. 5A & B). When a mir-210 expressing plasmid with mutated seed sequence was co-transfected, the repression of luciferase activity was completely abolished (Fig. 5C & D).
Figure 5.
Validation of randomly selected mir-210 target genes identified by miRNP-IP. A) pGL3-promoter vector and B) CTGF 3′ UTR construct were used as negative controls. They are not responsive to expression of either wild type or mutant mir-210. C-G) Reporter luciferase activity are repressed or relieved by co-transfecting mir-210 wild type or mutant expression vector with HOXA1, HOXA9, TP53I11, PIM1, and FGFRL1 3′ UTR constructs. H) The mutations in mir-210 expression construct. The four-nucleotide mutation was introduced in the seed region of mir-210. Each reporter assay was repeated at least three times. Error bar indicates standard deviation. Student’s t-test was performed for statistical analysis, ** p< 0.001, * p<0.05.
In addition to these four genes we selected from the fifty-gene list, one gene, FGFRL1 also caught our attention because of the presence of multiple mir-210 target sites in its 3′UTR region. It has been reported that closely located multiple miRNA target sites in a gene’s 3′ UTR region may function synergistically in mediating miRNA translation repression (Grimson et al., 2007; Rajewsky, 2006). There are seven potential mir-210 target sites in the 3′ UTR of FGFRL1, the highest number of predicted mir-210 target sites in a gene’s 3′ UTR although none of them are well conserved across species (supplementary Figure 4). We cloned a 1kb fragment of the 3′ UTR of FGFRL1 that contains all seven predicted mir-210 target sequences into a pGL3-Promoter vector and performed reporter assays. When the mir-210 expression plasmid was co-transfected into the cell with FGFRL1 3′ UTR luciferase reporter construct, reporter luciferase activity was repressed more than 90% compared to co-transfection with pcDNA3.1 control plasmid (Fig. 5G). However, the repression was completely abolished when a mutant form of mir-210 expressing plasmid was co-transfected.
Taken together, our data show that HOXA1, FGFRL1, and HOXA9 are robust mir-210 target genes, suggesting that approximately 60% of the targets we identified through miRNP-IP experiments are real mir-210 targets, a number of which is consistent with the accuracy of miRNP-IP approach in identifying miRNA targets reported elsewhere (Karginov et al., 2007). Therefore, when combined with computational prediction, miRNP-IP is an efficient means to identify miRNA targets.
Expression of mir-210 represses tumor xenograft growth
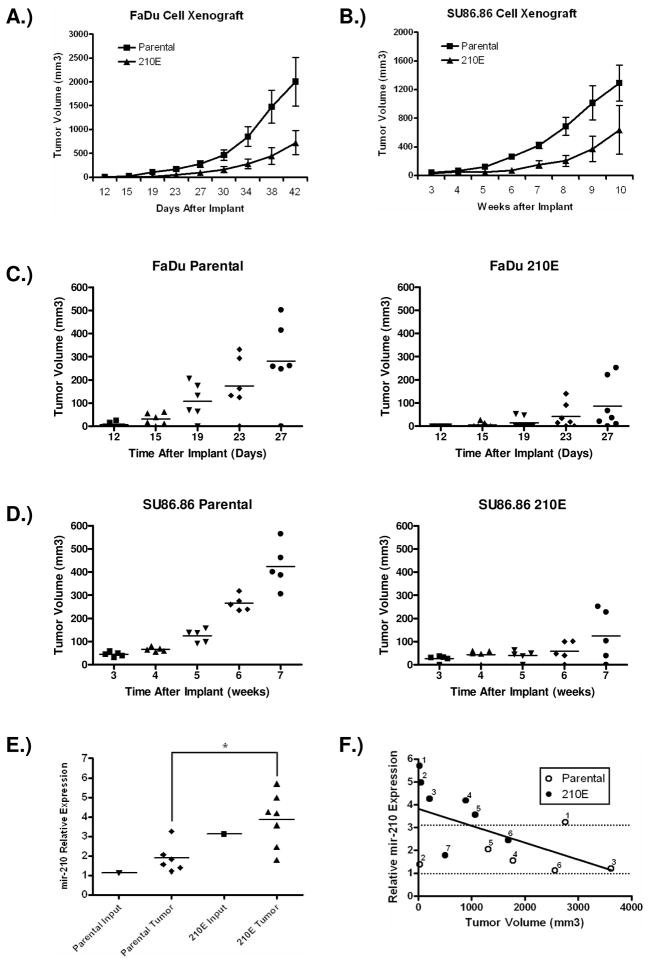
In order to elucidate the biological function of mir-210, we stably expressed mir-210 in a head & neck cancer cell line, FaDu and a pancreatic cancer cell line, SU86.86 by retroviral transduction. The ectopic expression of mir-210 is comparable to that found under hypoxic conditions (Supplementary Figure 5A). Although when exposed under 2% oxygen the cells ectopically expressing mir-210 had an even higher mir-210 level than that of parental cells under the same condition, their mir-210 level is still comparable to the hypoxic mir-210 expression in some other cell lines in the same type of cancer (Supplementary Figure 5B). Expression of mir-210 did not affect cell growth under normoxia or 2% oxygen conditions (supplementary Figure 6). However, when these two cell lines that stably express mir-210 were subcutaneously implanted into nude mice, tumor growth was significantly delayed (Fig. 6A & B). Sections of individual tumors were stained with antibodies to CD31, Ki67, and TUNEL to examine angiogenesis, proliferation, and apoptosis, respectively. Interestingly, it appeared that mir-210 expression had little effect on any of the processes, in agreement of our in vitro assay. However, in mice implanted with FaDu cells ectopically expressing mir-210, five out of seven mice did not have measurable tumors until week 4 compared to one out of six in the control group (Fig. 6C). In mice implanted with SU86.86 cells ectopically expressing mir-210, three out of five mice did not have measurable tumors until week 6 compared to none in the control group (Fig. 6D).
Figure 6.
Mir-210 represses tumor growth in a mouse xenograft model. A) FaDu cells with ectopic expression of mir-210 and parental cells were injected s.c in nude mice at a density of 106 cells. N=6 for parental group and N=7 for 210E group. B) SU86.86 cells with ectopic expression of mir-210 and parental cells were injected s.c in nude mice at a density of 107 cells. N=5 for the each group. Tumor volumes are represented as average ± SEM. C) Tumor growth in the first four weeks of FaDu cell xenograft implants. D) Tumor growth in the first seven weeks of SU86.86 cell xenograft implants. Left panel, cells without ectopic mir-210 expression; right panel, cells with ectopic mir-210 expression. E) Expression of mir-210 was measured by TaqMan RT-PCR from xenograft tumor samples with/without ectopic expression of mir-210. Expression of mir-210 was also measured from the same tumor cells left after tumor injection. RNA input was normalized on small nuclear RNA RNU48. There is a significant difference of mir-210 expression between the parental and 210E groups (student’s t-test, p=0.025). F) Negative correlation between the size of xenograft tumors and corresponding mir-210 expression level. The bottom and top dotted lines indicate mir-210 expression level in parental cells and in cells ectopically expressing mir-210, respectively.
Interestingly, although the initiation of xenograft growth was severely delayed in mice implanted with cells ectopically expressing mir-210, these tumors eventually overcome this inhibitory effect and start to grow. It is possible that the growth of these tumors may be due to lost of mir-210 expression or utilization of alternative pathways to circumvent mir-210’s inhibitory effect. To distinguish these two possibilities, we measured mir-210 expression in harvested FaDu tumor samples (Fig. 6E). Although generally mir-210 expression still remains significantly higher in tumors ectopically expressing mir-210 compared to that of in tumors from parental cells, two of the tumors have less mir-210 expression compared to the original input cells, suggesting that the tumor cells utilized both means to overcome mir-210’s inhibition on tumor growth initiation. Since mir-210 showed a clear inhibitory effect on tumor growth initiation, we analyzed the sizes of harvested xenograft samples and their corresponding mir-210 expression levels. As shown in Fig 6F, there is a statistically significant negative correlation between FaDu xenograft size and mir-210 expression level (r=−0.58, p=0.0367), indicating mir-210 expression is growth inhibitory.
Taken together, our data indicate that ectopic expression of mir-210 is primarily inhibiting initiation of tumor growth since a significantly higher mir-210 expression still remains in these growing tumors whose growth was inhibited initially.
HOXA1 is a mir-210 target in vivo
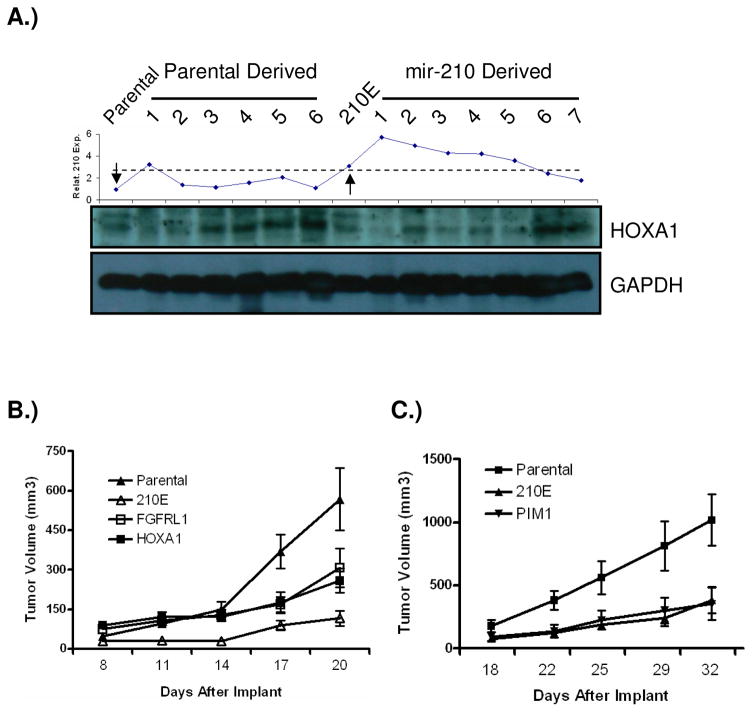
In an effort to investigate if the mir-210 targets we identified are also regulated by mir-210 in vivo, we examined HOXA1 protein level in harvested tumor xenograft samples (Fig. 7A). HOXA1 protein level is generally higher in xenograft samples with lower mir-210 expression in each group. The two samples (#6 & 7) with lowest mir-210 expression in mir-210 derived xenograft tumors also had the highest expression of HOXA1 in this group and in the parental group the sample (#1) with highest mir-210 expression had the lowest expression of HOXA1, suggesting that HOXA1 is a mir-210 target in vivo.
Figure 7.
HOXA1 is regulated by mir-210 in vivo. A) HOXA1 protein in harvested tumor xenograft samples. Top panel, mir-210 expression in each sample; bottom panel, HOXA1 protein level in corresponding samples. The left and right arrows in the top panel indicate the input parental and input mir-210 expressing cells, respectively. Dotted line indicates two groups of samples with inversed profiles of mir-210 expression and HOXA1 protein level. B) The inhibitory effect of mir-210 on tumor growth initiation was partially rescued by expressing HOXA1 or FGFRL1 coding sequence without a 3′ UTR region. Each group of FaDu cells was injected s.c in nude mice at a density of 2.5x106 cells. N=7 for the each group. C) Expressing PIM1 coding sequence without a 3′ UTR region has no effect on the inhibitory effect of mir-210 on tumor growth initiation. Each group of FaDu cells was injected s.c in nude mice at a density of 3x106 cells. N=7 for the each group.
Rescuing mir-210 phenotype by HOXA1 and FGFRL1 in tumor xenograft
Because miRNA binds to the 3′ UTR region of its target gene to repress protein translation or cause degradation of mRNA, we tested if the protein coding regions of two top candidate genes, HOXA1 and FGFRL1 that are identified by miRNA-IP approach and validated by reporter assays, could rescue the phenotype caused by ectopic expression of mir-210. We also included a gene, PIM1 that is not a mir-210 target as a control. The protein coding regions of HOXA1, FGFRL1, and PIM1 were cloned and stably expressed in FaDu cells expressing mir-210 (Supplementary Figure 7). The resulting cells were subcutaneously implanted into nude mice, and the growth of tumors derived from these cells was compared to parental cells and cells ectopically expressing mir-210. We found that the inhibitory effect elicited by mir-210 on tumor growth initiation was rescued in part by either expressing HOXA1 or FGFRL1, but not by PIM1 (Fig. 7B & C). However, neither gene could completely rescue the phenotype, suggesting that more than one target gene is involved in inhibiting tumor growth initiation by mir-210. This is not surprising given that miRNAs regulate the expression of groups of genes, and that we identified other potential mir-210 targets as well in this study.
Discussion
Since the discovery of first miRNA in 1993, the role miRNAs play in gene regulation has increasingly been appreciated, especially in recent years (Bartel 2004; Lee et al., 1993). In this study, we utilized microarray technology to identify mir-210 as the predominant miRNA induced by hypoxia in agreement with what other groups have reported recently (Camps et al., 2008; Giannakakis et al., 2007; Kulshreshtha et al., 2007). However, in these previously published reports the structure of mir-210 promoter has not been analyzed and the HRE responsible for the robust mir-210 hypoxia induction has not been identified. By constructing a series of mir-210 promoter deletion constructs, we identified a functional HRE and demonstrated that its hypoxia induction is HIF1α specific and is not regulated by HIF2α. Interestingly, the HRE we identified and the mir-210 coding sequence are the only loci in the mir-210 genomic region highly conserved across species, which strongly suggests the functional importance of this HRE and the unique and critical role of hypoxia plays in regulating mir-210 expression.
Recently, it has been reported that HIF1α binds to two predicted HRE sites up- stream of the one we identified by ChIP assay (Kulshreshtha et al., 2007). Since in our study a different HRE that is functional was identified through serial deletions of mir-210 promoter, it is possible that HIF1α binds to HRE sequences whenever the binding sites are physically available, but binding of HIF1α to HRE does not necessarily results in active transcription of its target genes. Other co-factors need to be recruited to cooperate with HIF1α to initiate transcription, which may be determined by surrounding genomic sequences that provide binding sequence for additional co-factors.
Because mir-210 is such a robustly induced gene under hypoxia in vitro, we took an unbiased approach to assess whether expression of mir-210 could reflect tumor hypoxia in vivo. Using an unsupervised clustering analysis, we found that mir-210 expression highly correlated with well characterized hypoxia-regulated genes in primary head & neck tumor samples. In addition to the four well-known hypoxia inducible genes highlighted in Supplementary Figure 4C, the other two genes, AK3L1 and ITGB4 have also been reported as hypoxia- regulated (Elvidge et al., 2006; Martin-Rendon et al., 2007). This indicates that expression of mir-210 could potentially serve as a surrogate marker for tumor hypoxia in vivo. Interestingly, mir-210 has been found overexpressed in several tumor types (Volinia et al., 2006). Overexpression of mir-210 has been detected in breast cancer patients recently and was correlated with worse prognosis and was also found correlated to “VEGF, hypoxia and angiogenesis” (Camps et al., 2008; Foekens et al., 2008; Zhang et al., 2009). In these reports, the expression of mir-210 may actually reflect the hypoxic status of those tumor samples instead of its biological function in tumorigenesis. Therefore, overexpression of mir-210 could predict poor prognosis in these cases may be because mir-210 is a surrogate marker for tumor hypoxia, which is a well known independent indicator of poor patient outcome (Vaupel and Mayer, 2007).
In an effort to experimentally identify mir-210 target genes, we used a miRNP-IP approach that has been successfully used to identify miRNA targets (Beitzinger et al., 2007; Easow et al., 2007; Karginov et al., 2007). MiRNP-IP is particularly suited to identify mir-210 targets because induction of mir-210 expression and enrichment of its target mRNAs in the argonaute complex are all under physiological conditions and therefore, eliminate the requirement to transfect miRNA into cells that may cause experimental artifacts. Fifty genes with at least two-fold enrichment were identified to be potential mir-210 targets, consistent with recent discoveries that miRNAs regulate the protein output of a large number of genes and that even one single miRNA could affect the expression of hundreds of genes (Baek et al., 2008; Selbach et al., 2008). Since several other miRNAs are also hypoxia inducible (Kulshreshtha et al., 2007) (supplementary Figure 1) the 50 genes in the list may include their targets as well. This may explain why the PIM1 and TP53I11 genes are enriched by microarray analysis but the 3′ UTR reporter assays only demonstrate a weak repression by mir-210. Alternatively, they may represent the intrinsic limitation of this approach in identifying miRNA targets because the similar efficiency (70% vs. 60% in the current study) was reported in an independent study (Karginov et al., 2007).
Interestingly, we observed that the majority of the 50 genes are not known to be hypoxia inducible. This suggests activation of two parallel pathways by HIF1α when cells are under hypoxic stress. The first pathway is well-studied and is directly regulated by HIF1α transcriptionally, and includes genes involved in glycolysis and angiogenesis. Genes induced in this pathway are necessary for cells to survive or adapt to hypoxic stress. The second pathway is indirectly regulated by HIF1α through the induction of mir-210. We hypothesize that steady expression of the genes in this second pathway are necessary for cells under a normoxic environment, but must be inhibited under hypoxic conditions. This is supported by gene ontology (GO) analysis that 19 of the 50 mir-210 target genes are involved in regulation of cellular metabolic processes and gene expression. Thus, the mir-210 miRNA pathway functions as a rheostat, allowing cells to quickly and efficiently turn off or tune down the protein output of dozens of genes under hypoxia. One exception is the EFNA3 gene identified in our miRNP-IP experiment, whose expression is highly induced under hypoxia. Cells may utilize mir-210 to fine-tune EFNA3 expression as EFNA3 has also been reported as a mir-210 target by other groups recently (Fasanaro et al., 2008; Pulkkinen et al., 2008).
Interestingly, ectopic expression of mir-210 delayed subcutaneous tumor growth and the expression level of mir-210 in harvested tumor xenograft samples negatively correlated with tumor size. This was a surprising result because HIF1α is generally considered a promoter of tumorigenesis, although some published data suggests it can also inhibit tumor growth (Carmeliet et al., 1998; Maranchie et al., 2002; Raval et al., 2005). HIF1α has been well known to regulate a variety of downstream target genes, including those involved in angiogenesis, glycolysis and glucose transport, such as VEGF, PGK and GLUT1. However, HIF1α also induces genes leading to cell cycle arrest, and cell death, such as p21, p27 and NOXA (Gardner et al., 2001; Goda et al., 2003; Green et al., 2001; Kim et al., 2004). The functional balance between these pro- and anti-tumorigenesis genes regulated by HIF1α may determine the tumor-promoting or tumor-suppressing phenotypes observed previously under different circumstances. To tease out how each HIF1α target gene contributes to this overall phenotype, individual genes need to be studied.
Analyzing mir-210 expression in harvested tumor samples suggests that ectopic expression of mir-210 only affects the initiation phase of tumor growth since a significantly higher mir-210 expression still remains in these growing tumors whose growth was inhibited initially. In tumors with ectopic mir-210 expression, the three smallest tumors also have the highest mir-210 expression, further demonstrating the ectopic expression of mir-210 accentuates the inhibitory function of this microRNA on tumor initiation. However, once tumor cells transit through the initial inhibitory effect of mir-210, they start to grow in a similar manner as their parental cells. In agreement with our findings, the mir-210 genomic region has been shown to be deleted and its expression lost at a high frequency in ovarian cancer (Giannakakis et al., 2008), suggesting a potential tumor suppressor function of mir-210. Interestingly, mir-210 was recently shown to promote cell cycle progression by activating c-MYC through inhibiting MNT, an antagonist of c-MYC and a gene ranked second in our fifty-gene list (Zhang et al., 2009). However, in our systems we did not observe an in vitro growth advantage of cells ectopically expressing mir-210, and in vivo mir-210 actually repressed tumor growth initiation.
In our miRNP-IP experiment several oncogenes were identified as potential mir-210 targets, such as HOXA1 (Zhang et al., 2003). It is plausible that by down-regulating oncogenes through ectopic expression of mir-210, the initiation of tumor growth may be temporarily impaired, providing tumor cells time to adapt to a hypoxic environment. Our rescue experiment by implanting cells expressing HOXA1 coding region on a mir-210 expressing background provides evidence to this hypothesis. When HOXA1 protein levels were examined in the harvested xenograft tumor samples, they had the opposite profile of mir-210 expression, confirming that HOXA1 is a mir-210 target in vivo. However, HOXA1 protein was also up-regulated in parental tumors (#3–6), although they all have slightly increased mir-210 expression (1.2, 1.6, 2.1, and 1.1, respectively) compared to parental input mir-210 expression, suggesting factors other than mir-210 controlling HOXA1 expression and its importance in tumorigenesis.
In summary, our work identified mir-210 as a robust hypoxia-inducible miRNA regulating a number of genes that are not induced under hypoxia. Our finding reveals a new mechanism of gene regulation by HIF1α through a miRNA pathway and therefore sheds new light on the complexity of hypoxia gene regulation. More importantly, ectopic expression of mir-210 repressed tumor growth initiation and thus, mir-210 may account for at least part of the repressive effect of HIF1α on tumor growth previously observed.
Experimental Procedures
Reporter assay
All transfections were carried out using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. Luciferase activity was measured using the Dual Luciferase assay system (Promega) according to the manufacturer’s instructions.
Quantitative RT-PCR (QRT-PCR)
Hsa-mir-210 and endogenous control RNU48 TaqMan QRT-PCR kits were purchased from Applied Biosystems (Foster City, CA). The RT and TaqMan quantitative PCR were performed according to the manufacturer’s instruction.
Expression level of FGFRL1 and PIM1 was measured by quantitative RT-PCR using a SYBR green PCR master mix (Applied Biosystems). Primer sequences are available upon request.
In vivo experiments
Five- to six-week-old male nude mice supplied by Charles Rivers were housed in the AALAC-approved Stanford University Animal Facility with 12 hr light cycles. Food and water were provided ad libitum. All experimental protocols were APLAC approved. Cells were injected subcutaneously (s.c.) on the back of mice. Tumors were measured every three days, and tumor volume was calculated by the following formula: (W2 × L) × 0.52 (W, width; L, length) (Arbiser et al., 1997). Tumors were frozen in OCT, and 5 μm sections were cut.
Supplementary Material
Acknowledgments
We thank Drs. Scott Welford, Denise Chan, and Elizabeth Finger for help on collecting mouse data, Dr. Adam Krieg for technical assistance on ChIP assay, and Dr. Denise Chan for helpful discussions and critical reading of the manuscript. We also would like to thank Dr. Anindya Dutta at the University of Virginia for the AGO2 antibody, Dr. Jiahuai Han at the Scripps Research Institute for c-myc tagged AGO2 plasmid, and Dr. Yoel Sadovsky at MWRI for support on Northern blot. This work was supported by the following grants from NIH: PO1-CA67166 (AJG, KB, XH, QTL) and 1R01-CA118582 (QTL, AJG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biology. 2007;4:76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 Is Induced by Hypoxia and Is an Independent Prognostic Factor in Breast Cancer. Clin Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, et al. Role of HIF- 1[alpha] in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- Chan DA, Giaccia A. Hypoxia, gene expression, and metastasis. Cancer and Metastasis Reviews. 2007;26:333–339. doi: 10.1007/s10555-007-9063-1. [DOI] [PubMed] [Google Scholar]
- Chan DA, Sutphin PD, Yen S-E, Giaccia AJ. Coordinate Regulation of the Oxygen-Dependent Degradation Domains of Hypoxia-Inducible Factor 1{alpha} Mol Cell Biol. 2005;25:6415–6426. doi: 10.1128/MCB.25.15.6415-6426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi JT, Wang Z, Nuyten DSA, Rodriguez EH, Schaner ME, Salim A, Wang Y, Kristensen GB, Helland slaug, et al. Gene Expression Programs in Response to Hypoxia: Cell Type Specificity and Prognostic Significance in Human Cancers. PLoS Medicine. 2006;3:e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198–1204. doi: 10.1261/rna.563707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvidge GP, Glenny L, Appelhoff RJ, Ratcliffe PJ, Ragoussis J, Gleadle JM. Concordant Regulation of Gene Expression by Hypoxia and 2-Oxoglutarate-dependent Dioxygenase Inhibition: THE ROLE OF HIF-1alpha, HIF-2alpha, AND OTHER PATHWAYS. J Biol Chem. 2006;281:15215–15226. doi: 10.1074/jbc.M511408200. [DOI] [PubMed] [Google Scholar]
- Fasanaro P, D’Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 Modulates Endothelial Cell Response to Hypoxia and Inhibits the Receptor Tyrosine Kinase Ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AWM, Klijn JGM, Wiemer EAC, Martens JWM. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proceedings of the National Academy of Sciences. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner LB, Li Q, Park MS, Flanagan WM, Semenza GL, Dang CV. Hypoxia Inhibits G1/S Transition through Regulation of p27 Expression. J Biol Chem. 2001;276:7919–7926. doi: 10.1074/jbc.M010189200. [DOI] [PubMed] [Google Scholar]
- Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, Li C, O’Brien-Jenkins A, Katsaros D, Weber BL, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biology & Therapy. 2008;7:255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda N, Ryan HE, Khadivi B, McNulty W, Rickert RC, Johnson RS. Hypoxia-Inducible Factor 1{alpha} Is Essential for Cell Cycle Arrest during Hypoxia. Mol Cell Biol. 2003;23:359–369. doi: 10.1128/MCB.23.1.359-369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SL, Freiberg RA, Giaccia AJ. p21Cip1 and p27Kip1 Regulate Cell Cycle Reentry after Hypoxic Stress but Are Not Necessary for Hypoxia-Induced Arrest. Mol Cell Biol. 2001;21:1196–1206. doi: 10.1128/MCB.21.4.1196-1206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KKH, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha Targeted for VHL-Mediated estruction by Proline Hydroxylation: Implications for O2 Sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, et al. Targeting of HIF-alpha to the von Hippel-Lindau Ubiquitylation Complex by O2-Regulated Prolyl Hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Standart N. How Do MicroRNAs Regulate Gene Expression? Sci STKE 2007. 2007:re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA Targets. PLoS Biology. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proceedings of the National Academy of Sciences. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Kim J-w, Dang CV. Cancer’s Molecular Sweet Tooth and the Warburg Effect. Cancer Res. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- Kim JY, Ahn HJ, Ryu JH, Suk K, Park JH. BH3-only Protein Noxa Is a Mediator of Hypoxic Cell Death Induced by Hypoxia-inducible Factor 1{alpha} J Exp Med. 2004;199:113–124. doi: 10.1084/jem.20030613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krieg AJ, Hammond EM, Giaccia AJ. Functional Analysis of p53 Binding under Differential Stresses. Mol Cell Biol. 2006;26:7030–7045. doi: 10.1128/MCB.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, et al. A MicroRNA Signature of Hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Maranchie JK, Vasselli JR, Riss J, Bonifacino JS, Linehan WM, Klausner RD. The contribution of VHL substrate binding and HIF1-[alpha] to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell. 2002;1:247–255. doi: 10.1016/s1535-6108(02)00044-2. [DOI] [PubMed] [Google Scholar]
- Martin-Rendon E, Hale SJM, Ryan D, Baban D, et al. Transcriptional Profiling of Human Cord Blood CD133+ and Cultured Bone Marrow Mesenchymal Stem Cells in Response to Hypoxia. Stem Cells. 2007;25:1003–1012. doi: 10.1634/stemcells.2006-0398. [DOI] [PubMed] [Google Scholar]
- Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends in Genetics. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Pulkkinen K, Malm T, Turunen M, Koistinaho J, Ylä-Herttuala S. Hypoxia induces microRNA miR-210 in vitro and in vivo: Ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Letters. 2008;582:2397–2401. doi: 10.1016/j.febslet.2008.05.048. [DOI] [PubMed] [Google Scholar]
- Rajewsky N. microRNA target predictions in animals. Nature Genetics. 2006;38(Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Raval RR, Lau KW, Tran MGB, Sowter HM, Mandriota SJ, Li J-L, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. Contrasting Properties of Hypoxia-Inducible Factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-Associated Renal Cell Carcinoma. Mol Cell Biol. 2005;25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Current Opinion in Genetics & Development. 1998;8:588–594. doi: 10.1016/s0959-437x(98)80016-6. [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Sullivan R, Graham C. Hypoxia-driven selection of the metastatic phenotype. Cancer and Metastasis Reviews. 2007;26:319–331. doi: 10.1007/s10555-007-9062-2. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proceedings of the National Academy of Sciences. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer and Metastasis Reviews. 2007;26:225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SC, Buffa FM, Silva P, et al. Relation of a Hypoxia Metagene Derived from Head and Neck Cancer to Prognosis of Multiple Cancers. Cancer Res. 2007;67:3441–3449. doi: 10.1158/0008-5472.CAN-06-3322. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhu T, Chen Y, Mertani HC, Lee K-O, Lobie PE. Human Growth Hormone-regulated HOXA1 Is a Human Mammary Epithelial Oncogene. J Biol Chem. 2003;278:7580–7590. doi: 10.1074/jbc.M212050200. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Sun H, Dai H, Walsh RM, Imakura M, Schelter J, Burchard J, Dai X, Chang AN, Diaz RL, et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:1–13. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.