Abstract
Escherichia coli mutants deficient in 2-keto-3-deoxy-d-manno-octulosonic acid (Kdo) biosynthesis are conditionally lethal, but their phenotypes are bypassed by certain suppressor mutations or by over-expression of MsbA, the inner membrane flippase for core-lipid A. These strains grow on broth with the tetra-acylated precursor lipid IVA replacing lipopolysaccharide (Meredith, T. C. et al. ACS Chem. Biol. 1, 33–42, 2006). Deletion of kdtA, which encodes the Kdo transferase, is possible under these conditions. We now show that lipid IVA reaches the outer surface of the outer membrane in these strains, as judged by its accessibility to the lipase PagL. On the assumption that MsbA is optimized to transport penta- or hexa-acylated lipid A, we over-expressed the lauroyl or the myristoyl transferase of lipid A biosynthesis, encoded by lpxL and lpxM respectively, and demonstrated that kdtA deletion mutants were also viable in this setting. Although E. coli LpxL is stimulated by the presence of the Kdo-disaccharide in its acceptor substrate, LpxL does slowly acylate lipid IVA. Over-expression of LpxL from a plasmid suppressed the lethality of kdtA deletions on nutrient broth at 30 or 37 °C without the need for MsbA over-production. These strains accumulated penta- and hexa-acylated free lipid A containing a secondary laurate chain, or a laurate and a myristate chain, respectively. Deletion of kdtA in strains over-expressing LpxM accumulated penta-acylated lipid A with a secondary myristate moiety. None of the strains lacking kdtA grew in the presence of bile salts at any temperature or on nutrient broth at 42 °C. Our findings show that the main function of Kdo is to provide the right substrates for the acyltransferases LpxL and LpxM, resulting in the synthesis of penta- and hexa-acylated lipid A, which is optimal for the MsbA flippase.
Mutants of Escherichia coli or Salmonella defective in the biosynthesis of ADP-heptose or in the transfer of heptose to the inner2-keto-3-deoxy-d-manno-octulosonic acid (Kdo) residue of nascent lipopolysaccharide (LPS) accumulate Kdo2-lipid A (Figure 1), which is transported to their outer membranes (1–3). The Kdo2-lipid A substructure of LPS (4) is sufficient to support bacterial growth on nutrient broth at 30 or 42 °C, albeit with alterations in outer membrane permeability (5). Kdo2-lipid A is the most conserved portion of LPS in all Gram-negative bacteria (2, 3) and is a potent agonist against the Toll-like receptor 4 (TLR4)/MD2 complex of the innate immune system (6–8), which detects LPS as foreign and triggers inflammation.
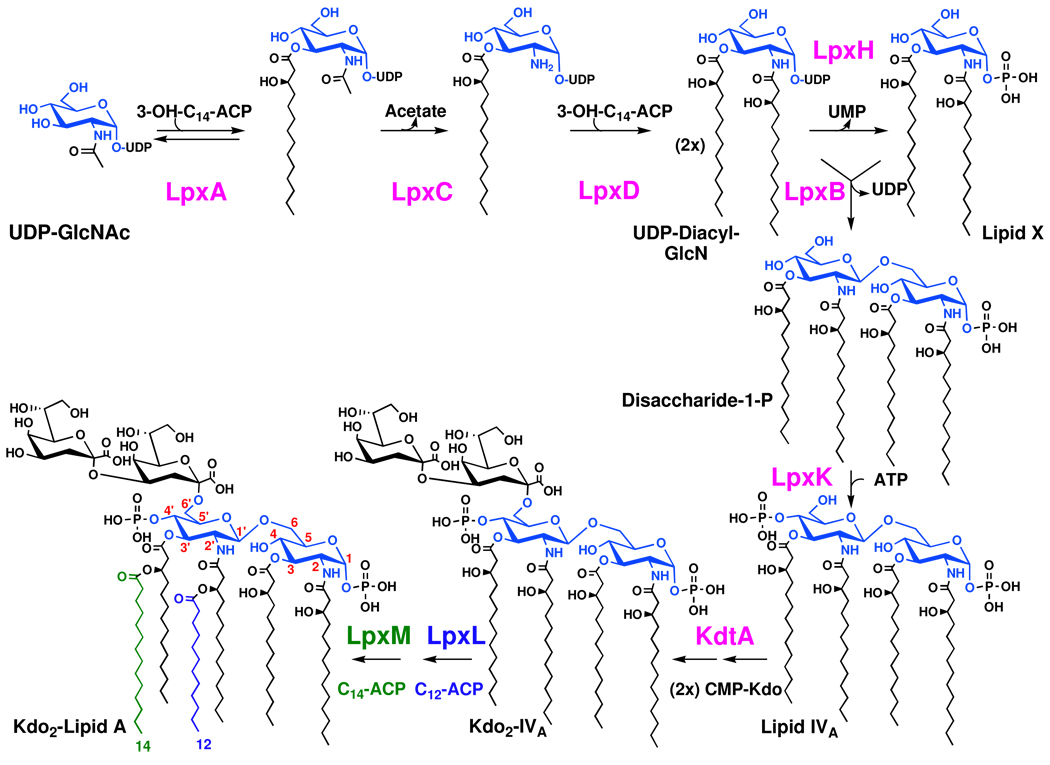
Figure 1. Biosynthesis of lipid A in wild-type E. coli K-12.
The first three enzymes are cytoplasmic, whereas the others are peripheral or integral inner membrane proteins that face the cytoplasm (2, 3). LpxL is stimulated ~1000 fold in vitro by the presence of the Kdo disaccharide in its substrate, but its residual activity with lipid IVA is significant (38). The Kdo dependency of LpxM has not been studied in vitro. Mutants of E. coli or Salmonella defective in Kdo biosynthesis or transfer accumulate mainly tetra-acylated lipid IVA and some of its modified derivatives (not shown) (11, 12). No free hexa-acylated lipid A accumulates in Kdo-deficient mutants of cells grown at 30 °C or above.
E. coli or Salmonella mutants defective in Kdo biosynthesis, or lacking Kdo-transferase, are not viable on nutrient broth, and conditional mutants accumulate lipid IVA (Figure 1) in their inner membranes (9–14). However, Kdo biosynthesis genes can be deleted in the presence of certain point mutations in MsbA (15, 16), an essential ABC transporter that normally functions as the inner membrane flippase for nascent LPS (Figure 2) (17–20), or in the non-essential inner membrane of protein YhdJ (15), the function of which is unknown. Alternatively, Kdo biosynthesis genes can be deleted if MsbA is overexpressed (16), and the gene encoding Kdo transferase can be deleted if cells are grown very slowly on minimal medium at 21 °C (21). In all these situations, cells replace their LPS with the recursor lipid IVA (Figure 1), which accumulates to high levels (16, 21).
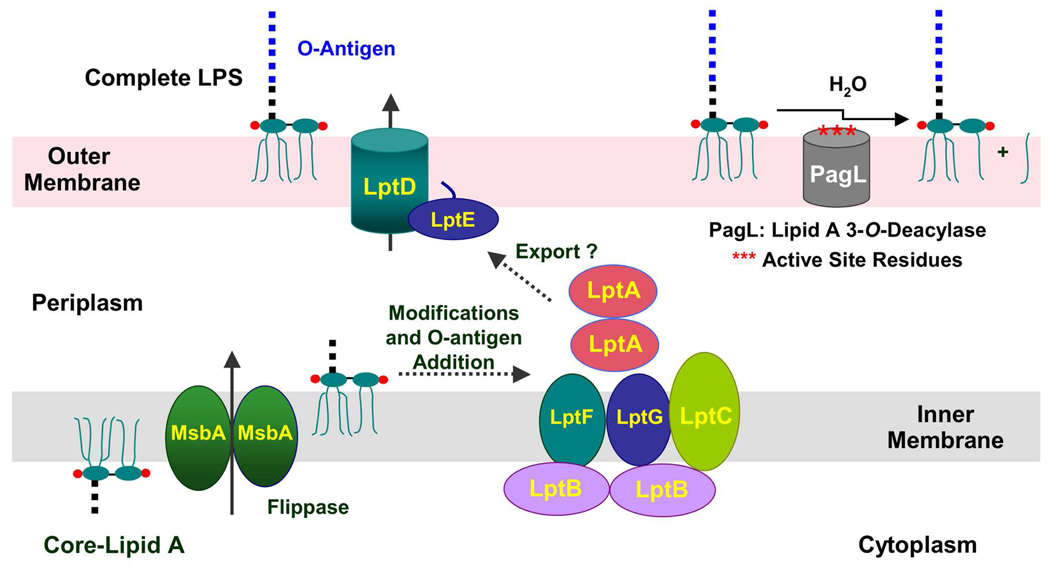
Figure 2. Proteins involved in the export of nascent LPS to the outer membrane.
The main steps of core-lipid A biosynthesis occur on the inner surface of the inner membrane, whereupon the essential ABC transporter MsbA flips the nascent LPS to the outer surface of the inner membrane (2, 3). In wild-type strains (though not E. coli K-12), O-antigen is polymerized and transferred to the nascent LPS on the outer surface of the inner membrane (2, 3). The downstream ABC transporter, consisting of the LptA, LptB, LptC, LptG and LptF proteins, is thought to deliver LPS to the outer membrane LptD/E complex, which is responsible for the proper integration of LPS into the outer surface of the outer membrane (22–27). Modification of lipid A with pEtN or l-Ara4N units in polymyxin-resistant mutants occurs on the outer surface of the inner membrane (not shown), whereas PagL catalyzed removal of the 3-O-acyl chain or PagP-catalyzed addition of palmitate (not shown) occurs in the outer surface of the outer membrane (3).
Following several covalent modifications of core-lipid A and attachment of O-antigen on the outer surface of the inner membrane (2, 3), nascent LPS is transported to the outer surface of the outer membrane by the Lpt proteins (Figure 2) (22–27). The inner membrane proteins LptB, LptF, and LptG, in conjunction with the accessory protein LptC, constitute a second essential ABC transporter (26), apparently required for the ejection of LPS from the outer surface of the inner membrane (Figure 2). The soluble periplasmic protein LptA may bind to lipid A directly (28) and deliver it to the LptD/E complex, or it may function to appose the inner and outer membranes (27, 29) so that the LptD and LptE proteins can incorporate nascent LPS into the outer surface of the outer membrane (Figure 2). The biochemical mechanisms by which these transport proteins accomplish their tasks have not been elucidated.
Enzymatic modifications to the lipid A moiety of LPS occur on the outer surface of the inner membrane or in the outer membrane (3). For instance, in polymyxin-resistant mutants, phosphoethanolamine (pEtN) and 4-amino-4-deoxy-l-arabinose (l-Ara4N) units are attached to the phosphate groups of lipid A on the outer surface of the inner membrane (18, 30). After incorporation into the outer surface of the outer membrane, the lipase PagL from Salmonella (Figure 2) can remove the R-3-hydroxyacyl chain from the 3-position of lipid A (31, 32). Alternatively, the outer membrane acyltransferase PagP (not shown) can add a secondary palmitate chain to the R-3-hydroxymyristate unit at the 2-position of lipid A (33–35). These modification enzymes are useful as reporters for the trafficking of lipid A from the inside of the cell to the outer leaflet of the outer membrane.
We now report several new E. coli constructs in which the kdtA gene (Figure 1) can be deleted. Over-expression of MsbA from the plasmid pWSK29 (36) permits the deletion of kdtA in E. coli cells grown on nutrient broth at 30 or 37 °C, accompanied by the accumulation of lipid IVA, but these strains do not grow at 42 °C or on MacConkey agar (37), which contains bile salts. The kdtA gene can also be deleted when LpxL or LpxM (38–41) (Figure 1) are over-expressed from pWSK29. When LpxL is over-expressed in strains lacking kdtA, large amounts of penta- and hexa-acylated free lipid A species are produced. The penta-acylated species contains laurate as its secondary acyl chain, whereas the hexa-acylated material contains laurate and myristate, as does the lipid A moiety of wild-type LPS (3). When LpxM is over-expressed in strains lacking kdtA, penta-acylated free lipid A bearing a secondary myristate chain accumulates. These results support the view that penta- and hexa-acylated lipid A are better substrates for the MsbA flippase than is the tetra-acylated precursor lipid IVA (17, 20); consequently over-expression of MsbA is not necessary to support growth in these constructs. Finally, kdtA can also be deleted in cells over-expressing LptA, LptB and LptC (Figure 2), but these cells only grow slowly at 30 °C.
Experimental Procedures
Materials
Chloroform, methanol, and silica gel 60 (0.25 mm) thin layer chromatography plates were from EMD Chemicals Inc. (Gibbstown, NJ). Tryptone, yeast extract, and agar were from Becton, Dickinson and Co. (Franklin Lakes, NJ). Isopropyl 1-thio-β-d-galactopyranoside (IPTG) was from Invitrogen Corp. (Carlsbad, CA). 32Pi (10 µCi/µL) was from PerkinElmer Life and Analytical Sciences, Inc. (Waltham, MA). All other chemicals were reagent grade and were purchased from either Sigma-Aldrich (St. Louis, MO) or Mallinckrodt Baker, Inc. (Phillipsburg, NJ).
Bacterial strains
Bacterial strains used in this study are described in Table 1. They are all derivatives of E. coli K-12, which makes a complete LPS core but no O-antigen (2). Typically, bacteria were grown in LB medium, a nutrient broth that contains 10 g tryptone, 5 g yeast extract and 10 g NaCl per liter (37), and tested for viability on MacConkey medium (37). When required for the selection of plasmids, cells were grown in the presence of 100 µg/mL ampicillin, 25 µg/mL chloramphenicol, 12.5 µg/mL tetracycline, and/or 20 µg/mL kanamycin.
Table 1.
Bacterial strains and plasmids.
| E. coli Strain | Relevant genotype | Source or References |
|---|---|---|
| W3110 | Wild type, F−, λ− |
E. coli Genetic Stock center, Yale University |
| DY330 | W3110 ΔlacU169 gal490 λcI857 Δ (cro-bioA) |
(44) |
| XL1-Blue | ΔmcrABC recA1 endA1 gyrA96 relA1 supE44 thi-1 lac |
Stratagene |
| WBB06 | W3110 mtl, (ΔwaaC-waaF)::tet6, heptose-deficient |
(1) |
| MST100* | DY330 PmR | (55) |
| CMR100 | DY330 (kdtA::kan) pWMsbA | This work |
| CMR101 | DY330 (kdtA::kan) pWLpxL | This work |
| CMR102 | DY330 (kdtA::kan) pWLpxM | This work |
| CMR103 | DY330 (kdtA::kan) pLptCAB2 | This work |
| CMR200 | MST100 (kdtA::kan) pWMsbA | This work |
| CMR300 | W3110 (kdtA::kan) pWMsbA | This work |
| CMR301 | W3110 (kdtA::kan) pWLpxL | This work |
| CMR302 | W3110 (kdtA::kan) pWLpxM | This work |
| Plasmids | ||
| pWSK29 | Low copy vector, lac promoter, AmpR | (36) |
| pWSK130 | Low copy vector, lac promoter, KanR | (36) |
| pACYC184 | Medium copy vector, TetR, CamR | New England Biolabs |
| pBAD33 | Medium copy vector, CamR | (62) |
| pBAD-pagL | pBAD33 harboring pagL | (27) |
| pWTD1 | pET28b harboring msbA | (19) |
| pWTD2 | pET23a(+) harboring msbA | This work |
| pACYC-KdtA | pACYC184 harboring kdtA | This work |
| pWMsbA | pWSK29 harboring msbA | This work |
| pWLpxL | pWSK29 harboring lpxL | (38)** |
| pMsbB | pET21a(+) harboring lpxM | (39) |
| pWLpxM | pWSK29 harboring lpxM | This work |
| pLptCAB2 | pWSK29 harboring lptC, lptA, lptB | (27) |
PmR: polymyxin resistant
Same as pWSK-LpxL (38)
Molecular biology applications
Protocols for handling of DNA samples and preparing E. coli for electroporation were those of Sambrook and Russell (42). Transformation-competent cells of E. coli were prepared by the method of Inoue et al. (43). Plasmids were isolated from cell cultures using the QIAprep Miniprep kit or extracted from agarose gel with the QIAquick gel extraction kit (Qiagen, Valencia, CA). Genomic DNA was isolated using the protocol for bacterial cultures in the Easy-DNA kit (Invitrogen, Carlsbad, CA). T4 DNA ligase (Invitrogen), restriction endonucleases (New England Biolabs, Ipswich, MA), and shrimp alkaline phosphatase (USB, Cleveland, OH) were used according to the manufacturers’ instructions. Double-stranded DNA sequencing was performed with an ABI Prism 377 instrument at the Duke University DNA Analysis Facility. Primers were purchased from MWG-Biotech (Huntsville, AL).
Construction of MsbA, LpxM, and KdtA expression vectors
The msbA gene was excised from pWTD1 (19) by digesting the plasmid with NdeI and BamHI. The msbA gene was then ligated into the corresponding restriction sites of pET23a(+) to create pWTD2. Next, the XbaI/BamHI-digested fragment, consisting of the msbA gene as well as the pET23a(+)-derived ribosome binding site, was ligated to the corresponding sites of pWSK29, a lactose-inducible, low-copy expression vector (36). This plasmid, designated pWMsbA, was transformed into competent cells of E. coli WBB06, DY330, W3110, or MST100 (Table 1).
The lpxM gene was excised from pMsbB (39) using the XbaI and HindIII sites. The XbaI/HindIII-digested fragment, consisting of the lpxM gene as well as the pET21a(+)-derived ribosome binding site, was ligated to the corresponding sites of pWSK29. This plasmid, designated pWLpxM, was transformed into competent cells of E. coli DY330 or W3110.
The kdtA gene of E. coli W3110 was cloned into pACYC184. The coding region for kdtA was amplified by PCR from E. coli W3110 genomic DNA, using the primers KdtAXbaI and KdtABamHI (Table 2). The PCR reaction was carried out using Pfu DNA polymerase (Stratagene, La Jolla, CA), according to the manufacturer’s instructions. The PCR product and the vector were digested with both XbaI and BamHI, ligated together, and transformed into XL1-Blue cells (Table 1) for propagation of the desired plasmid. This plasmid, designated pACYC-KdtA, was transformed into CMR100 (see below).
Table 2.
Oligonucleotide primers used in this work.
| Name | Sequence |
|---|---|
| KdtAKOFOR | 5′- CATAGAATCCCCAGCACATCCATAAGTCAGCTATTTACTAA GGAGATATAATGAGCCATATTCAACGGGAA-3′* |
| KdtAKOREV | 5′- GAAAGTACCCGGATAAATCGCCCGTTTTTGCATAACAACCT TAGAAAAACTCATCGAGCAT-3′* |
| KdtACFor | 5′-CCAAACTGAAGCTATTTAAGTC-3′ |
| KdtACRev | 5′-GTGATCGAACATCTGCGTCGTG-3′ |
| KdtAXbaI | 5′- GCGCGCTCTAGAAAGGAGATATAATGCTCGAATTGCTTTAC ACC-3′ ** |
| KdtABamHI | 5′-GCGCGCGGATCCTCAATGCGTTTTCGGTGGCAG-3′ ** |
The letters in italics are part of the kan gene plus the ribosome binding site. The kdtA flanking genomic sequence is not italicized.
The underlined letters are the recognition sites for the restriction enzymes.
Construction of kdtA deletions in E. coli DY330 derivatives
PCR was used to construct a linear piece of DNA containing the kanamycin resistance cassette (kan), flanked on the 5′ end by a ribosome binding site plus 39 bp of chromosomal DNA upstream of kdtA, and flanked on the 3′ end by 40 bp of chromosomal DNA located downstream of kdtA. The sequences of the forward (KdtAKOFOR) and reverse (KdtAKOREV) primers used to construct this PCR product are presented in Table 2. The kanamycin resistance gene (Tn903) from plasmid pWSK130 (36) served as the template. The PCR reaction was carried out using Pfu DNA polymerase (Stratagene, La Jolla, CA), according to the manufacturer’s instructions. The PCR product was resolved on a 1% agarose gel and purified with the QIAquick gel kit. The product was electroporated into DY330/pWMsbA, DY330/pWLpxL, DY330/pWLpxM, DY330/pLptCAB, and MST100/pWMsbA (Table 1), which had been grown at 30 °C in the presence of 100 µg/mL ampicillin and 1 mM IPTG until the A600 reached ≈0.4. Just prior to electroporation, each culture was shifted to 42 °C for 15 min to activate the λ–RED genes (44), and the cells were washed twice with 30 mL ice cold distilled water. The cells were resuspended in cold distilled water and then electroporated with 100–250 ng of the PCR product. After growth for 2 h at 30 °C in LB medium with 100 µg/mL ampicillin and 1 mM IPTG, the cells were plated on LB agar, containing kanamycin, ampicillin and IPTG. The plates were incubated at 30 °C, and the resulting colonies were re-purified on LB-kanamycin, ampicillin and IPTG plates at 30 °C. Colonies of all the strains remained viable for only 1 to 2 days on LB agar plates. Liquid cultures lost viability in stationary phase, and therefore they were only grown to A600 = 0.3 to 0.4, except as indicated. All the strains remained viable at −80 °C in glycerol stocks. The kdtA::kan replacement on the chromosome of DY330 harboring the various covering plasmids was verified by PCR using external primers KdtACFor and KdtACRev (Table 2). The PCR products were resolved on a 1% agarose gel, purified with the QIAquick gel extraction kit, and sequenced to confirm the replacement. The kdtA::kan derivatives of DY330/pWMsbA, DY330/pWLpxL, DY330/pWLpxM, DY330/pLptCAB2, and MST100/pWMsbA were designated as CMR100, CMR101, CMR102, CMR103, and CMR200, respectively.
Construction of kdtA deletions in E. coli W3110
DY330 is inherently temperature-sensitive (44); thus, it was necessary to transduce the kdtA::kan marker into W3110 harboring the various pWSK29-derived multi-copy suppressor constructs (Table 1) in order to evaluate the viability of the kdtA::kan strains at higher temperatures. To accomplish this, we first transformed CMR100 with pACYC-KdtA to restore LPS, which is needed for P1vir infection. Next, a P1vir bacteriophage lysate of donor strain CMR100/pACYC-KdtA was made (37) and used to transduce stationary cells of W3110/pWMsbA, W3110/pWLpxL, and W3110/pWLpxM, which had been grown at 30 °C in the presence of 100 µg/mL ampicillin and 1 mM IPTG. The transduction mixtures were plated onto LB agar, containing 20 µg/mL kanamycin, 100 µg/mL ampicillin, 1 mM IPTG, and 4 mM citrate. Colonies were re-purified with the same selection, and the kdtA::kan replacements on the chromosome of each of the constructs were verified by PCR, using external primers KdtACFor and KdtACRev (Table 2). The PCR products were resolved on a 1% agarose gel, purified with the QIAquick gel extraction kit, and sequenced to confirm the replacement. The kdtA::kan derivatives of W3110/pWMsbA, W3110/pWLpxL, and W3110/pWLpxM were designated as CMR300, CMR301, and CMR302, respectively. As in the DY330 background, colonies on LB agar remained viable for 1 or 2 days, and in liquid culture the strains lost viability in stationary phase.
Growth phenotypes of kdtA deletion mutants
Strains were grown overnight on LB broth containing 1 mM IPTG and 100 µg/mL ampicillin at 30 °C, and used to inoculate 50 mL of pre-warmed LB broth containing 1 mM IPTG and 100 µg/mL ampicillin to A600 = 0.02. Growth was allowed to continue with shaking at 220 rpm. Whenever the A600 reached 0.3–0.4, the cultures were diluted 10-fold into 50 mL of pre-warmed LB broth containing 1 mM IPTG and 100 µg/mL ampicillin in order to keep the cells in mid log-phase.
Lipid isolation from kdtA deletion mutants
Typically, strains were grown from single colonies to A600 = 0.3 on LB broth containing 1 mM IPTG and 100 µg/mL ampicillin at 30 °C, and used to inoculate 50 mL of LB broth containing 1 mM IPTG and 100 µg/mL ampicillin to A600 = 0.02. Growth was allowed to continue with shaking at 220 rpm. When the A600 reached 1.0, the cultures were harvested by centrifuging at 4000 × g for 20 min. Cells were then washed once with 30 mL of phosphate-buffered saline (PBS) (45). The pellet was then resuspended in 8 mL of PBS. Chloroform (10 mL) and methanol (20 mL) were added to make a one-phase Bligh-Dyer system (46). The mixture was incubated at room temperature for 1 h with occasional mixing and then centrifuged at 4000 × g for 20 min. The supernatant was converted to a two-phase Bligh and Dyer mixture by the addition of chloroform (10 mL) and PBS (10 mL). The mixture was vortexed and centrifuged at 4000 × g for 20 min. The lower phase was removed, and the upper aqueous-methanol phase was extracted a second time by the addition of pre-equilibrated lower phase. The lower phases were pooled and dried by rotary evaporation. The lipids were stored at −80 °C.
Electrospray ionization mass spectrometry of lipids
Spectra were acquired on an ABI QSTAR XL quadropole time-of-flight tandem mass spectrometer (ABI/MDS-Sciex, Toronto, Canada), equipped with an electrospray ionization (ESI) source. Spectra were acquired in the negative-ion mode and typically were the accumulation of 60 scans collected from m/z 200–2000. Typically, one-third of the total lipids extracted from a 50 mL culture were dissolved in 200 µL chloroform/methanol (2:1, v/v), supplemented with 1% piperidine, and immediately infused into the ion source at 5–10 µL/min. The negative ion ESI was carried out at −4200 V. Data acquisition and analysis were performed using the Analyst QS software (ABI/MDS-Sciex, Toronto, Canada).
Results
Meredith et al. first reported that E. coli mutants defective in Kdo biosynthesis, which are conditionally lethal in a wild-type background, were rendered viable by point mutations in either MsbA (Fig. 2) or in YhdJ, a non-essential, inner-membrane protein of unknown function (15, 16). Their constructs lack LPS, but still synthesize the precursor lipid IVA (Fig. 1) and transport it to the outer membrane (16). Meredith et al. did not determine the orientation of the lipid IVA within the outer membrane.
Phenotypes of kdtA deletions in cells over-expressing msbA
We constructed a new strain of E. coli in which the kdtA gene, encoding Kdo transferase (Fig. 1), could be deleted without loss of viability. The MsbA-expressing plasmid pWMsbA was introduced into DY330 (Table 1), a strain that makes the λ-red recombinase to permit homologous recombination between exogenous linear DNA molecules and their corresponding chromosomal sequences (44). In this setting it was possible to obtain kanamycin-resistant recombinants of DY330 on nutrient broth at 30 °C. The kan cassette in DY330 kdtA::kan(pWMsbA) replaced the entire kdtA gene; the strain was designated CMR100. When lipids of CMR100 were analyzed by ESI/MS in the negative ion mode, peaks consistent with accumulation of lipid IVA were observed (Supporting Fig. 1).
The presence of the heat-inducible λ-red recombinase in DY330 renders this strain temperature-sensitive for growth (44). To evaluate the growth phenotype of kdtA deletion mutations in the absence of the λ-red recombinase, the kan cassette of DY330 kdtA::kan(pWMsbA) was transferred into E. coli W3110(pWMsbA) by P1vir transduction and selection for kanamycin-resistant transductants on LB broth at 30 °C. W3110 kdtA::kan(pWMsbA), renamed CMR300 (Table 1), grew normally on broth at 30 and 37 °C (Fig. 3A), but stopped growing after ~2 hours at 42 °C (Fig. 3D). CMR300 failed to grow at any temperature on MacConkey agar (not shown), possibly because of the presence of bile salts in this medium. CMR300 remained very sensitive to the selective LpxC inhibitor CHIR-090 (47) (data not shown); apparently, the lipid IVA that replaces LPS in CMR300 is still essential for some aspect of cell physiology or outer membrane biogenesis. All kdtA deletion mutants lose viability if A600 exceeds 1 (data not shown).
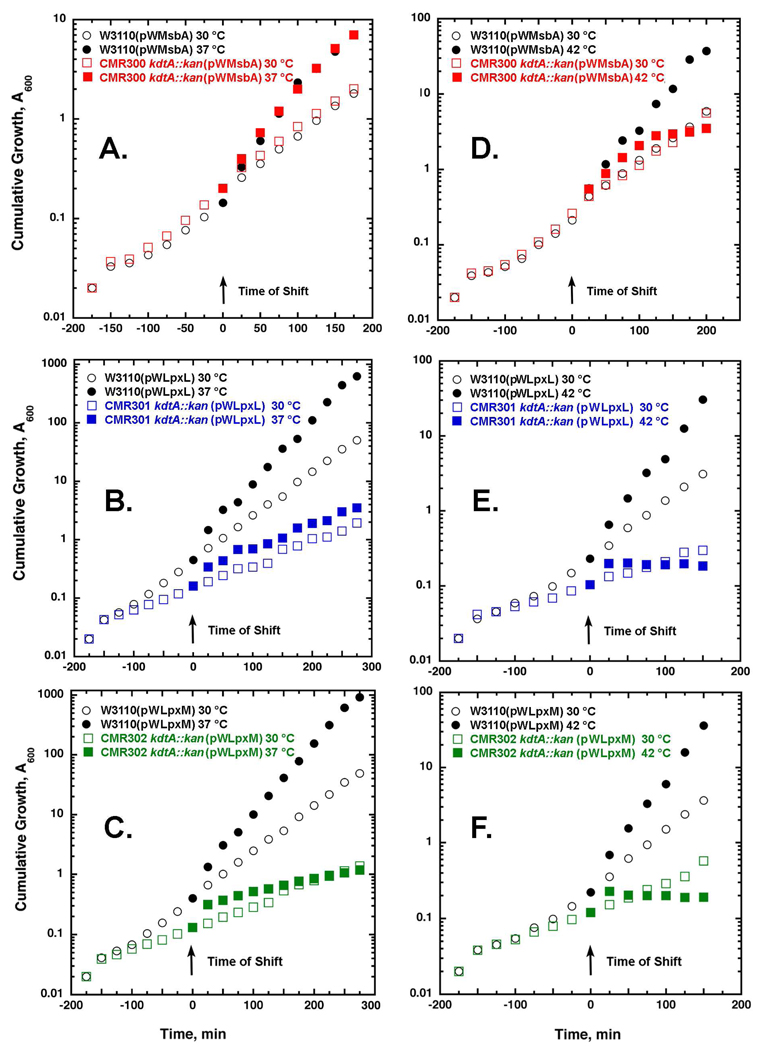
Figure 3. Temperature-sensitive growth of CMR300, CMR301, and CMR302 on LB broth.
Cells were grown on LB broth with 100 µg/mL ampicillin and 1 mM IPTG from A600 0.02 to 0.2 at 30 °C; then half of each culture was shifted to 37 or 42 °C, as indicated. Cultures were diluted 10-fold whenever A600 reached 0.3–0.4. The cumulative A600 was corrected for the dilutions. Panels A, B and C show the temperature shifts of CMR300, CMR301 and CMR302, respectively, from 30 to 37 °C. CMR300 nearly matches the W3110(pWMsbA) control at both temperatures. Reduced growth rates, albeit with good viability, were observed for CMR301 and CMR302 at both 30 and 37 °C versus their matched vector control strains. Panels D, E and F show the temperature shifts of exponentially growing CMR300, CMR301 and CMR302, respectively, from 30 to 42 °C in LB broth, demonstrating the rapid growth cessation of all three kdtA deletion constructs.
ESI/MS and TLC of lipids from W3110(pWMsbA) and CMR300
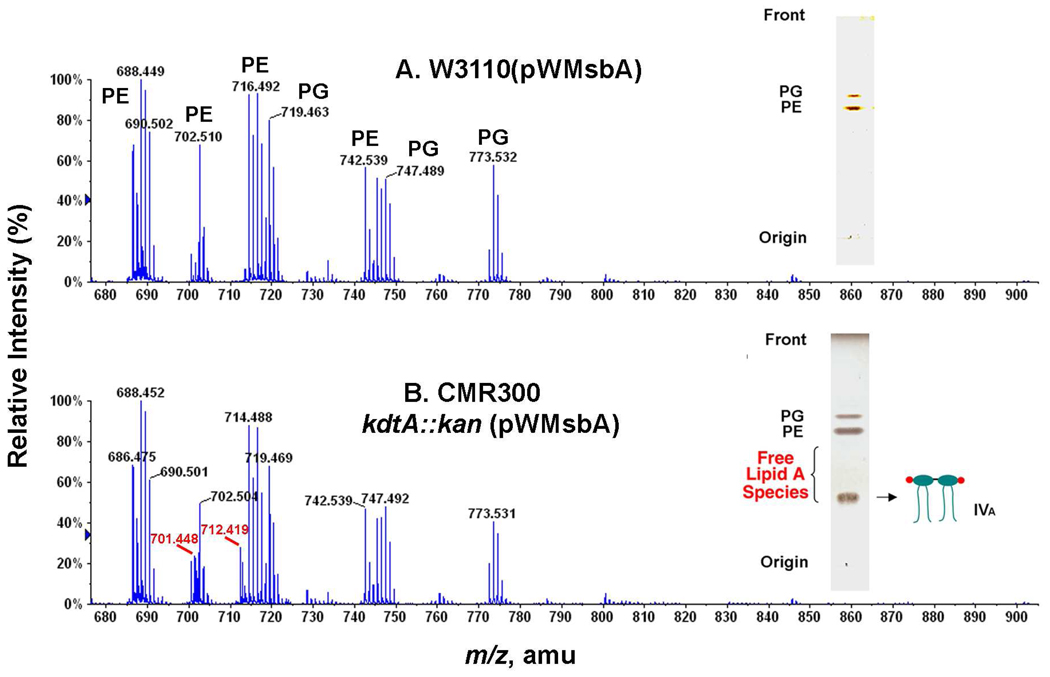
Like DY330 kdtA::kan(pWMsbA), CMR300 cells accumulate high levels of lipid IVA when grown at 30 °C on LB broth, representing between 15 and 20% of the total chloroform-soluble material, as quantified by 32Pi-labeling (see below). ESI/MS of the total cellular lipids in the negative ion mode showed new peaks interpreted as the [M-2H]2− and [M-3H+Na]2− ions of lipid IVA at m/z 701.448 and 712.419 in CMR300, but not in W3110(pWMsbA) (Figs. 4B and 4A, respectively). These values are in reasonable agreement with the predicted m/z of 701.420 and 712.411 for the [M-2H]2− and the [M-3H+Na]2− ions of lipid IVA, which ionizes predominantly as a dianion during ESI/MS. The slight discrepancy between the observed and predicted values for the [M-2H]2− ion may reflect the presence of overlapping minor glycerophospholipids in this region of the spectrum (Fig. 4B). The overall distribution of phosphatidylethanolamine (PE) and phosphatidylglycerol (PG) molecular species was very similar in both strains (Figs. 4A and 4B). The accumulation of lipid IVA in CMR300 versus W3110(pWMsbA) was confirmed by TLC and charring with 10% sulfuric acid in ethanol (Fig. 4, insets).
Figure 4. ESI/MS and TLC demonstrating accumulation of lipid IVA in CMR300.
Exponentially-growing cells in 50 mL LB broth supplemented with 1 mM IPTG and 100 µg/mL ampicillin were harvested in late log phase. The lipids were extracted with a two-phase neutral Bligh-Dyer system (46), re-dissolved in chloroform/methanol/piperidine (2:1:0.03, v/v/v), and immediately analyzed in the negative ion mode by direct infusion ESI/MS, using an ABI QSTAR XL quadrupole time-of-flight mass spectrometer. Panel A. Major glycerophospholipid ions of the control strain W3110(pWMsbA) between m/z 680 and 900 consist mainly of molecular species of PE and PG, as indicated. Panel B. The kdtA deletion mutant CMR300 contains similar glycerophospholipids, but accumulates additional peaks (red), which are interpreted as the [M-2H]2− and [M-3H+Na]2− ions of lipid IVA. The accumulation of lipid IVA was confirmed by TLC (insets).
Presence of lipid IVA on the outer surface of the outer membrane in CMR300
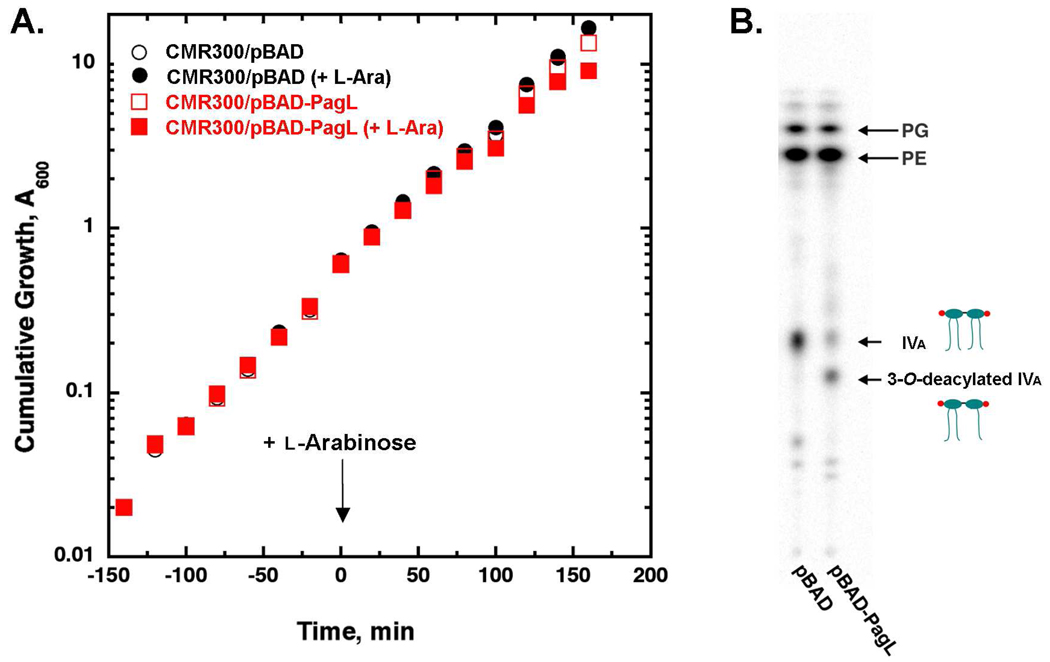
In order to demonstrate that lipid IVA in CMR300 is transported to the outer surface of the outer membrane, a second compatible hybrid plasmid (pBAD-PagL) (27) was transformed into CMR300 (Table 1). In parallel, the pBAD vector without insert was introduced into CMR300. The resulting constructs were grown at 37 ° C in LB broth and induced with 0.2% l-arabinose when A600 reached 0.3 (Fig. 5A). Concomitantly, the cells were labeled with 32Pi and grown for three more hours with occasional 10-fold back-dilution when A600 reached 1.0 to maintain exponential growth. As shown in Fig. 5B, about two-thirds of the lipid IVA was deacylated when PagL was present, supporting the view that much of the lipid IVA reaches the outer surface of the outer membrane in CMR300. The loss of one of the four acyl chains of lipid IVA (31, 32) does not further compromise the viability of CMR300 under these conditions.
Figure 5. Efficient transport of lipid IVA to the outer surface of the outer membrane in CMR300.
Panel A. Cultures of CMR300 harboring either pBAD33 or pBAD-pagL were grown at 37 °C in the presence of 100 µg/mL ampicillin, 25 µg/mL chloramphenicol from an initial A600 of 0.02. When the A600 reached 0.3, 0.2% l-arabinose and 5 µCi/mL 32Pi were added, and growth was allowed to continue for three more hours with the cultures back-diluted as needed. The cells were harvested when the cumulative A600 reached 10 and washed once with PBS. Panel B. A single-phase neutral Bligh-Dyer mixture (46) was used to extract the lipids, followed by conversion to two phases and drying down of the lower phase that contains the lipids, as described previously (17, 48). The lipids were dissolved in chloroform/methanol (2:1, v/v) and spotted onto a silica gel 60 TLC plate. The plate was developed with a chloroform:pyridine:88% formic acid:water (50:50:16:5, v/v/v/v) system and then analyzed with a PhosphorImager.
Phosphoethanolamine addition to lipid IVA in polymyxin-resistant DY330
In polymyxin-resistant mutants of E. coli, the lipid A moiety of LPS is modified with pEtN and l-Ara4N units (3), which reduce its affinity for polymyxin and other cationic antimicrobial peptides. The pEtN and l-Ara4N moieties are added to lipid A on the outer surface of the inner membrane (18). In the presence of the Kdo disaccharide, pEtN goes mainly to position-1, whereas l-Ara4N is targeted to position-4′ (48). In temperature-sensitive Salmonella mutants defective in Kdo biosynthesis, pEtN is incorporated exclusively at position-4′ and a small amount of l-Ara4N is added to position-1 (12, 48, 49).
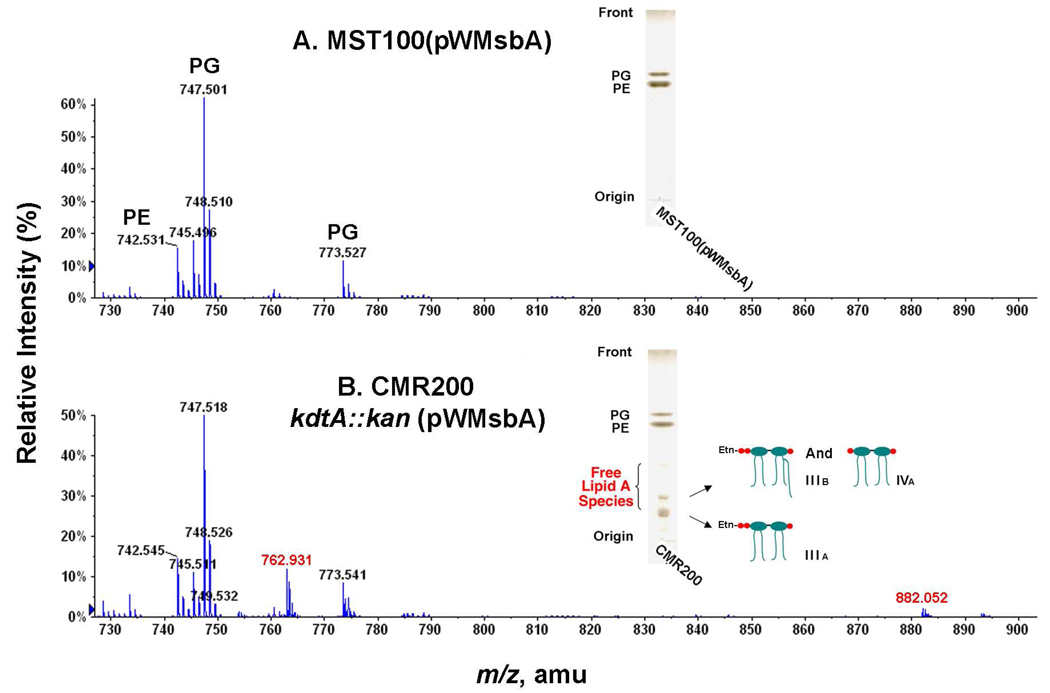
To demonstrate that lipid IVA reached the outer surface of the inner membrane (Fig. 2), we deleted kdtA in MST100(pWMsbA), a polymyxin-resistant derivative of DY330 (Table 1). In the kdtA deletion strain of MST100 (designated CMR200 in Table 1), relatively little lipid IVA accumulated, but a new compound migrating more slowly was observed (Fig. 6, insets). ESI/MS in the negative ion mode of the lipids from CMR200 revealed new peaks at m/z 762.931 and 882.052, interpreted as the [M-2H]2− ions of lipid IIIA (i.e. lipid IVA derivatized with one pEtN moiety)(12) and lipid IIIB (i.e. lipid IVA derivatized with one pEtN and one palmitoyl moiety) (12), respectively (Fig. 6B). The predicted [M-2H]2− ions for these species are m/z 762.923 and m/z 882.088, in reasonable agreement with the observed values. The proposed attachment of the pEtN moiety to the 4′-phosphate group (Fig. 6B, inset) was confirmed by 31P-NMR analysis (not shown) (12). The presence of some lipid IIIB (Fig. 6B) provides additional confirmation that lipid IIIA reaches the outer surface of the outer membrane in this construct. The palmitate moiety of IIIB is incorporated by the outer membrane acyltransferase PagP (33, 34). No peaks corresponding to l-Ara4N-modified lipid IVA were observed in CMR200 (Fig. 6B), presumably reflecting the requirement for the Kdo moiety and/or the secondary myristoyl chain of lipid A for l-Ara4N transferase activity (49, 50). CMR200 is polymyxin sensitive when compared to MST100, consistent with the loss of the l-Ara4N modification (data not shown).
Figure 6. Phosphoethanolamine-modified lipid IVA in CMR200, a PmrA-constitutive kdtA deletion mutant.
Cells were grown at 30 °C and lipids were extracted as in Fig. 4. Panel A. ESI/MS in the negative ion mode of the major lipid species in the control strain MST100(pWMsbA). Panel B. Accumulation of the pEtN-modified lipids IIIA and IIIB (11, 12) in CMR200, indicated by the red numbers. The accumulation of lipids IIIA ([M-2H]2− at m/z 762.931) and IIIB ([M-2H]2− at m/z 882.052) was confirmed by TLC (insets).
Viability of kdtA deletions in cells over-expressing lpxL or lpxM
Over-expression of the msbA gene suppresses the temperature-sensitive phenotype of deletions in lpxL(htrB), which encodes in the lauroyl transferase of lipid A biosynthesis (Fig. 1) (17, 51, 52). LpxL-deficient strains accumulate LPS containing a tetra-acylated lipid A moiety at 42 °C (17, 39). The requirement for MsbA over-expression to permit growth of lpxL mutants at 42 °C suggests that tetra-acylated lipid A is a relatively poor substrate for MsbA (39).
LpxL activity is stimulated ~1000 fold by the presence of the Kdo disaccharide in its acceptor substrate (Fig. 1), but the residual activity is significant (38). To determine whether or not hexa-acylated lipid A by itself is the preferred substrate for MsbA, we constructed W3110 kdtA::kan(pWLpxL), renamed CMR301, which grew more slowly on LB broth at 30 and 37 °C than did CMR300 (Fig. 3B), but formed single colonies on LB agar (not shown). Like CMR300, CMR301 failed to grow at 42 °C (Fig. 3D) on LB agar or on MacConkey agar at any temperature.
The same strategy for LpxL was used to delete the kdtA gene of W3110 in the presence of pWLpxM, which directs the over-expression of the myristoyl transferase of lipid A biosynthesis (Fig. 1) (40). The growth phenotypes of W3110 kdtA::kan(pWLpxM), renamed CMR302 (Figs. 3C and 3F), were very similar to those of CMR301 (Figs. 3B and 3E). No growth of CMR302 was observed at 42 °C on LB broth or LB agar, or on MacConkey agar at any temperature.
Penta- and hexa-acylated free lipid A in CMR301 and CMR302
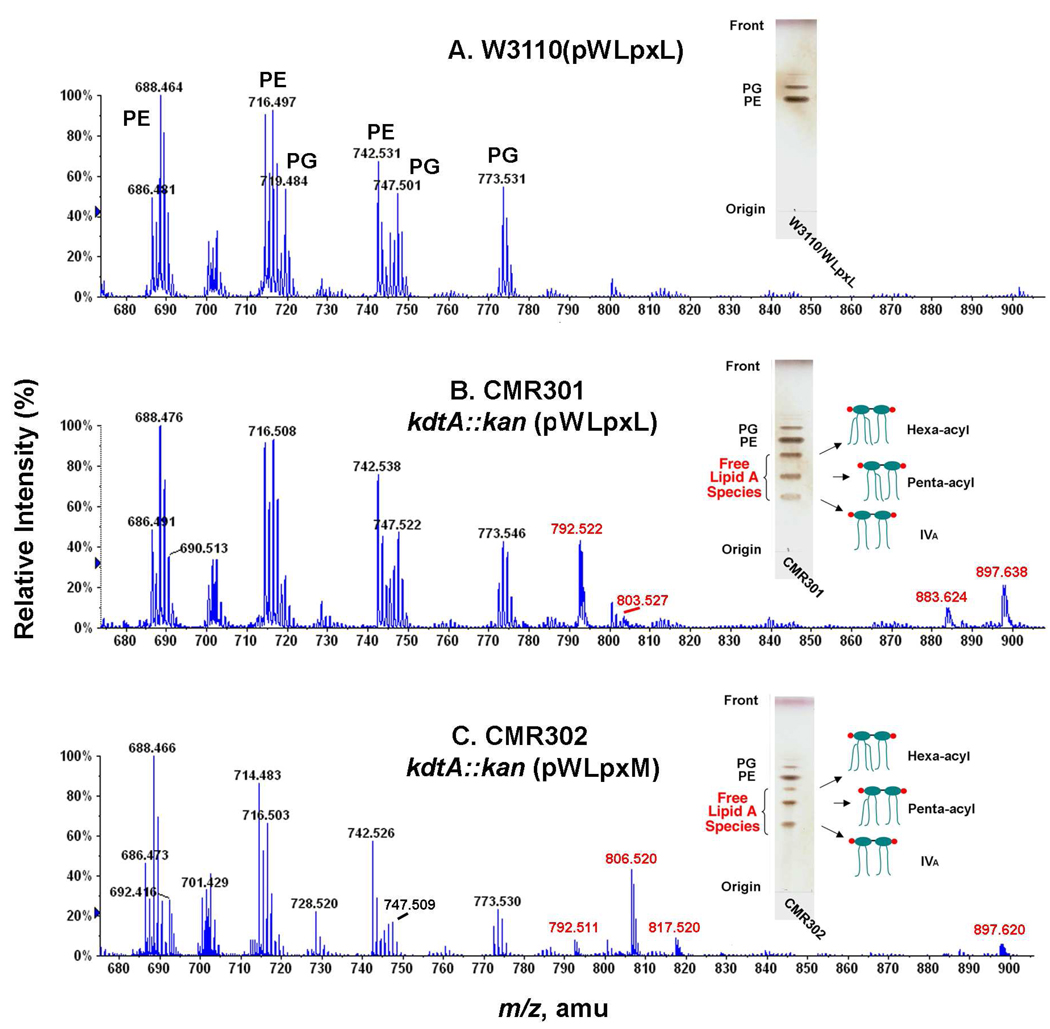
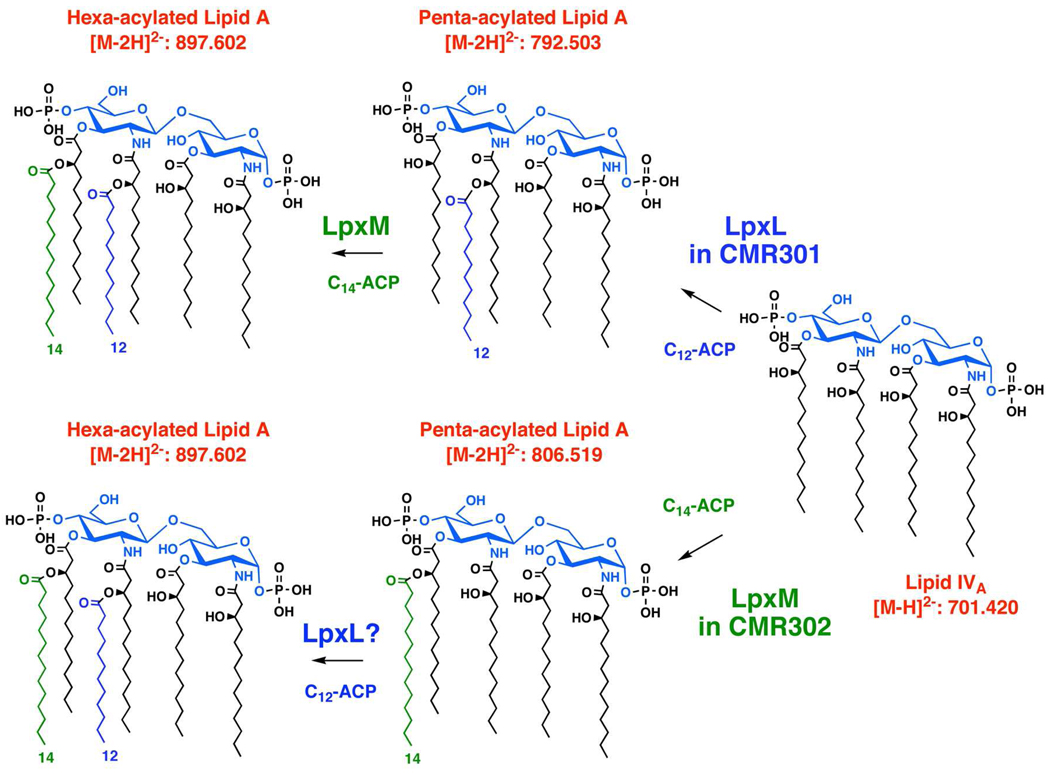
TLC and ESI/MS were used to evaluate the lipid compositions of CMR301 and CMR302 grown at 30 °C on LB broth with 1 mM IPTG and 100 µg/mL ampicillin. The control strain W3110(pWLpxL) contained no free lipid A, and its major PE and PG molecular species (Fig. 7A) were essentially the same as those in W3110(pWMsbA) (Fig. 4A) or W3110(pWLpxM) (not shown). Both CMR301 kdtA::kan(pWLpxL) and CMR302 kdtA::kan(pWLpxM) accumulated several free lipid A species, two of which migrated faster than lipid IVA (Figs. 7B and 7C, insets). ESI/MS in the negative ion mode showed a major new peak in the lipids of CM301 at m/z 792.522. This is interpreted as the [M-2H]2− ion of a penta-acylated, free lipid A species, bearing a secondary laurate chain at the 2′-position (calculated m/z of 792.503), consistent with the selectivity of LpxL (Fig. 8) (38, 39). The additional peak at m/z 897.638 (Fig. 7B) is interpreted as the [M-2H]2− ion of a hexa-acylated, free lipid A species, bearing one secondary laurate and one secondary myristate chain (calculated m/z of 897.602), as in the lipid A moiety of wild-type E. coli LPS (3). The small peak at m/z 803.527 represents the monosodium adduct [M-3H+Na]2− of the species at m/z 792.522 (Fig. 7B), whereas the peak at m/z 883.624 (Fig. 7B) likely arises from a hexa-acylated lipid A in which both secondary acyl chains are laurate because of the lack of absolute selectivity of LpxM (4, 40). The dependency of LpxM on the Kdo domain has not been studied, but the results of Fig. 7B suggest that it functions fairly well in its absence. The proposed pathway for the formation of these species in CMR301 is shown in the upper part of Fig. 8.
Figure 7. Penta- and hexa-acylated free lipid A molecules in a kdtA deletion mutants over-expressing LpxL or LpxM.
Exponentially-growing cells in 50 mL LB broth with 100 µg/mL ampicillin and 1 mM IPTG at 30 °C were harvested in late log phase. The lipids were extracted and analyzed by direct infusion ESI/MS in the negative ion mode, using an ABI QSTAR XL quadruplole time-of-flight mass spectrometer, as in Fig. 4. Panel A. Major glycerophospholipid ions of the control strain W3110(pWLpxL) between m/z 680 and 900 consist mainly of molecular species of PE and PG, as indicated. Panel B. The kdtA deletion mutant CMR301, which over-expresses LpxL, contains similar glycerophospholipids, but accumulates additional peaks (red), which are interpreted as the [M-2H]2− and [M-3H+Na]2− ions of penta-acylated and hexa-acylated free lipid A (see also Fig. 8), in addition to some lipid IVA (not labeled). The penta-acylated lipid A contains laurate as its secondary acyl chain, but no myristate. A significant amount of hexa-acylated lipid A with a wild-type [M-2H]2− is also generated, suggesting that LpxM may not be as Kdo-dependent as LpxL. Panel C. The kdtA deletion mutant CMR302, which over-expresses LpxM, accumulates a different penta-acylated lipid A than does CMR301 (see Fig. 8). In this case the predominant secondary acyl chain is myristate, although some laurate is also present because of the relative lack of selectivity of LpxM (40). The accumulation of these free lipid A species was confirmed by TLC (insets).
Figure 8. Origin of free lipid A molecules in kdtA deletion mutants over-expressing LpxL or LpxM.
High levels of LpxL or LpxM bypass the need for the Kdo disaccharide in the acceptor substrate. Consequently, CMR301 and CMR302 are able to synthesize significant quantities of penta- and hexa-acylated lipid A, which is exported at a sufficient rate by wild-type levels of MsbA to support cell growth. The locations of the secondary acyl chains are proposed based on their normal locations in wild-type lipid A (3) and have not been validated by independent methods.
In the ESI/MS analysis of the CMR302 lipids, a prominent new peak is seen at m/z 806.520 (Fig. 7C), which is not detectable in CMR301 (Fig. 7B). This species is consistent with the [M-2H]2− ion of a penta-acylated lipid A, bearing a secondary myristate chain (calculated m/z of 806.519). The myristate chain is presumably attached at the 3′-position, assuming the usual regioselectivity of LpxM (lower half of Fig. 8). The small peak at m/z 817.520 is interpreted the [M-3H+Na]2− adduct ion of the same penta-acylated species. The small peak at m/z 792.511 likely reflects the relaxed selectivity of LpxM for myristate versus laurate (Fig. 7C) (40), which also accounts for the minor hexa-acylated species at m/z 883.624 in CMR301 (Fig. 7B). The small peak at m/z 897.620 in CMR302 is interpreted as the [M-2H]2− ion of a hexa-acylated lipid A, bearing one secondary laurate and one secondary myristate chain, perhaps arising as shown in the lower half of Fig. 8 with LpxL functioning after LpxM.
Taken together, our results demonstrate that chromosomal levels of wild-type MsbA are sufficient to support E. coli growth in the absence of Kdo transfer to lipid IVA, provided that an alternative mechanism is available for the generation of sufficient amounts of penta- and hexa-acylated lipid A (Fig. 1 versus Fig. 8). The primary function of Kdo in E. coli therefore appears to be the enhancement of LpxL and possibly LpxM activity with the generation of the optimal penta- and hexa-acylated lipid A substrates for the flippase, MsbA (Figure 2).
Discussion
Mutants lacking the lauroyl transferase of lipid A biosynthesis, encoded by lpxL (Figure 1), do not grow on nutrient broth at 42 °C (41, 53) and concurrently accumulate LPS bearing a tetra-acylated lipid A moiety (39). Over-expression of the essential ABC transporter MsbA (51) suppresses this phenotype, permitting growth with tetra-acylated LPS (17). These findings led to the proposal that MsbA is the inner membrane flippase for nascent LPS (Figure 2) and that MsbA might preferentially transport hexa-acylated lipid A species (17, 20) Subsequent genetic, biochemical and structural studies have supported this hypothesis (18, 19, 39, 54). However, in vitro assays showing that purified MsbA can flip hexa-acylated LPS across a lipid bilayer have not yet appeared.
Mutants defective in Kdo biosynthesis because of a mutation in d-arabinose 5-phosphate isomerase (kdsD/gutQ) require the addition of d-arabinose 5-phosphate to the growth medium to support growth and LPS assembly (16). Meredith et al. also discovered that certain point mutations in MsbA or in the inner membrane protein YhdJ bypass this arabinose-5-phosphate auxotrophy, resulting in strains that can grow with lipid IVA as their sole LPS substructure (15, 16). The physiological function of YhdJ is unknown, but it is not normally required for growth (15). Presumably, certain point mutations in MsbA and YhdJ (15) somehow enhance the export of lipid IVA, which otherwise tends to accumulate in the inner membrane of mutants defective in Kdo biosynthesis or Kdo transfer to lipid IVA. Very recently, Klein et al. have shown that Kdo-transferase can be deleted in a wild-type background if the cells are grown very slowly on a minimal medium at 21 °C or below with lipid IVA as the only remaining LPS substructure (21).
We have now discovered several additional multi-copy suppressors that restore the growth of E. coli on nutrient broth when the kdtA gene is deleted. As suggested by the work of Meredith et al. (16), MsbA over-expression from a multi-copy plasmid permits the deletion the kdtA gene in E. coli cells at 30 or 37 °C with the accumulation of lipid IVA (Figure 3 and Figure 4). Furthermore, this lipid IVA reaches the outer surface of the outer membrane (Figure 2), as judged by its accessibility to cleavage by the outer membrane lipase PagL (Figure 5), the crystal structure of which is known (31). When kdtA is deleted in the polymyxin-resistant strain MST100 (55), the lipid IVA is modified with a pEtN unit (Figure 6), demonstrating that it may transiently reside on the outer surface of the inner membrane (18). In the absence of the Kdo disaccharide, the pEtN unit is attached to the 4′-phosphate group of lipid IVA (Figure 6) (12). The additional modification with palmitate by PagP demonstrates the further movement of the pEtN-modified product to the outer leaflet of the outer membrane (35). The absence of l-Ara4N-modified lipid IVA in this setting is explained by the specificity of the E. coli aminoarabinose transferase ArnT for hexa-acylated lipid A substrates (50) (not shown).
Given that MsbA may be optimized to transport hexa-acylated lipid A, we reasoned that over-expression of LpxL or LpxM (Figure 1) might also permit the deletion of kdtA under these conditions. Wild-type LpxL greatly prefers Kdo2-lipid IVA as its acceptor substrate, but it does have measurable activity with lipid IVA in vitro (38). Accordingly, the over-expression of LpxL from a multi-copy plasmid might enable the formation of penta- and/or hexa-acylated lipid A in the absence of KdtA (Figure 8). The preference of LpxM for the Kdo disaccharide has not been analyzed in vitro. Although LpxM prefers penta-acylated acceptor substrates containing a secondary laurate chain (40), it does function slowly when over-expressed in cells lacking LpxL (39).
Strains CMR301 and CMR302, which over-express LpxL or LpxM respectively, can grow when their kdtA gene is deleted (Figure 3), albeit more slowly than CMR300, the matched MsbA over-expressing construct (Table 1). As anticipated from the in vitro selectivity of LpxL and LpxM, CMR301 and CMR302 accumulate penta- and hexa-acylated free lipid A, and relatively little lipid IVA (Figure 7). The penta-acylated lipid A in CMR301 contains a secondary laurate chain, as judged by the ESI/MS analysis (Figure 7B and 7C), whereas the penta-acylated lipid A of CMR302 mainly contains a secondary myristate unit. CMR301 also accumulates hexa-acylated material (Figure 7B) with the molecular weight expected for wild-type lipid A (3, 4). Taken together, these phenotypes demonstrate that MsbA prefers penta- or hexa-acylated substrates to tetra-acylated molecules in the absence of core sugars. A primary function of the Kdo disaccharide in E. coli therefore appears to be the production of the optimal substrate for LpxL. Chromosomal levels of LpxM show significant activity with penta-acylated free lipid A generated by LpxL in the absence of the Kdo disaccharide (Figure 7B and Figure 8).
The idea that MsbA preferentially transports hexa-acylated lipid A gains further support from the effects of over-expressing msbA in the heptose-deficient mutant WBB06. This well-characterized strain synthesizes mostly hexa-acylated Kdo2-lipid A when grown on LB broth (4). However, when pWMsbA is introduced into WBB06, the pattern of lipid A species becomes more complex, and the levels of Kdo2-lipid A are reduced (Figure 10A). Analysis by ESI/MS demonstrates the accumulation of the precursors lipid IVA, Kdo2-lipid IVA and Kdo2-(lauroyl)-lipid IVA, as in the de-convoluted negative ion mode spectrum (Figure 10B). When MsbA flippase function is enhanced by over-expression in WBB06, we suggest that the rate of lipid IVA export may become comparable to the rates of Kdo and secondary acyl chain addition to lipid IVA, consistent with the ability of msbA over-expression to suppress the lethality of kdtA deletions. In the wild-type W3110, which generates a complete core, over-production of MsbA does not have a dramatic effect on the acyl chain composition of the lipid A moiety (not shown). The reasons for the different responses of WBB06 and W3110 to MsbA over-expression are unclear and require further investigation.
Figure 10. Accumulation of Kdo2-lipid A precursors in a heptose-deficient mutant over-expressing MsbA.
Strain WBB06 harboring pWMsbA was grown in the presence of 32Pi to late log phase on LB medium at 30 °C. Labeled lipids were extracted and analyzed by TLC (17) as in Fig. 5. Panel A. Accumulation of additional lipids in WBB06/pWMsbA versus its vector control. Panel B. The lipids from WBB06/pWMsbA were fractionated by DEAE cellulose chromatography (17, 48) and analyzed by ESI/MS in the negative ion mode. The de-convoluted spectrum of the lipids contained in the 480 mM ammonium acetate fraction (17, 48) revealed the measured exact masses of the peaks arising from Kdo2-lipid A and its precursors bearing four or five acyl chains.
Over-expression of MsbA does not enable the deletion of lpxK (Figure 2) (data not shown), suggesting that MsbA has a strong preference for substrates with two monophosphate groups. Expression of the Francisella lipid A 4′-phosphatase (56), the active site of which resides on the outer surface of the inner membrane, is toxic in CMR300, suggesting that the 4′-phosphate group may have additional functions in E. coli following the export of lipid IVA by MsbA.
The first six enzymes of the lipid A pathway are excellent targets for new antibiotic development, as illustrated by CHIR-090 (47, 57), a potent, selective inhibitor of LpxC (Figure 2). Kdo-deficient strains like CMR300 remain sensitive to killing by CHIR-090 under all conditions examined to date, demonstrating that CMR300 cannot grow without lipid IVA. Nevertheless, it may yet be possible to delete the E. coli lipid A pathway entirely, as in the case of Neisseria meningitidis (58, 59), by introducing additional, as yet unidentified suppressor mutations. However, most other Gram-negative bacteria are like E. coli in that lipid A is essential for growth.
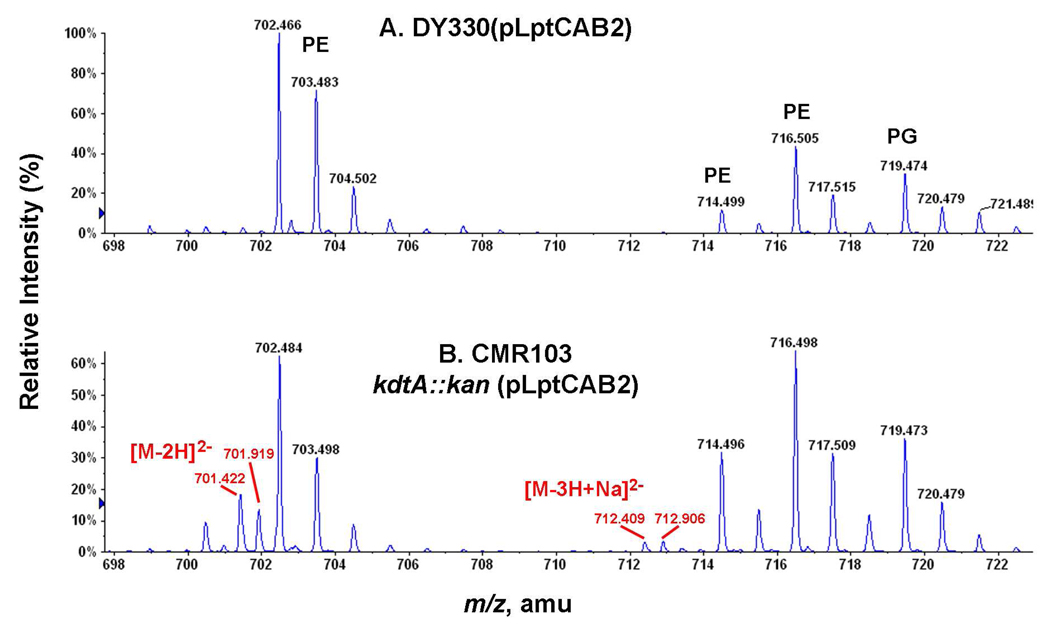
The recent discovery of the Lpt transport proteins (Figure 2) has provided the first molecular insights into the trafficking of nascent LPS from the outer surface of the inner membrane to the outer membrane (22–27). Interestingly, concurrent over-expression of LptA, LptB and LptC (Figure 2), which are encoded in an operon (22), partially suppresses the lethality of kdtA deletion at 30 °C with accumulation of lipid IVA (Figure 9). These constructs grew very slowly compared to the others described above (not shown). The partial suppression of kdtA deletions by LptABC over-expression suggests that rapid removal of lipid IVA from the outer surface of the inner membrane might increase the efficiency of MsbA (Figure 2), perhaps by mass action. Over-expression of the entire Lpt complex (Figure 2) might improve MsbA efficiency even further.
Figure 9. Accumulation of lipid IVA in CMR103 (kdtA::kan) over-expressing LptC, LptA and LptB.
Cells were grown at 30 °C on LB broth, and lipids were extracted as in Fig. 4. Panel A. ESI/MS in the negative ion mode of the major lipid species in the control strain DY330(pLptCAB2). Panel B. Accumulation of lipid IVA in CMR103, as indicated by the red numbers for its [M-2H]2− and [M-3H+Na]2− ions.
The ability of E. coli cells over-expressing LpxL or LpxM to grow with free lipid A replacing their LPS resembles the normal situation in Francisella novicida, in which most of the lipid A is not covalently attached to LPS (60, 61). In fact, kdtA deletion in Francisella does not inhibit cell growth (J. Zhao and C. R. H. Raetz, in preparation). One of the two LpxL orthologs present in F. novicida may be Kdo-independent (D. A. Six, W. Chen and C. R. H. Raetz, in preparation).
The growth phenotypes associated with kdtA deletions in CMR300, CMR301 and CMR302 (Figure 3) should facilitate the selection of second-site suppressor mutations that can grow without LPS at 42 °C or in the presence of bile salts. This genetic approach might reveal the presence of additional protein components required for lipid A trafficking or for the proper assembly of a functional outer membrane. If the strategy of replacing LPS with free lipid A is applicable to pathogens, like Salmonella or Shigella, it may be possible to attenuate these organisms and to use them for the development of novel vaccines.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. William Doerrler for constructing pWTD2, Dr. M. Stephen Trent for MST100, Dr. David Six for pWLpxL and pWLpxM, and Dr. Middleton Boon-Hinckley for pWMsbA. We are especially grateful to Drs. David Six, Jinshi Zhao, Louis Metzger, Hak Suk Chung and Ziqiang Guan for their critical reading of the manuscript.
This research was supported by NIH Grant GM-51310 to C. R. H. Raetz and the LIPID MAPS Large Scale Collaborative Grant GM-069338.
The abbreviations are
- l-Ara4N
4-amino-4-deoxy-l-arabinose
- ESI/MS
electrospray-ionization mass spectrometry
- IPTG
isopropyl-β-d-thiogalactoside
- Kdo
2-keto-3-deoxy-d-manno-octulosonic acid
- LPS
lipopolysaccharide
- PBS
phosphate-buffered saline
- PE
phosphatidylethanolamine
- pEtN
phosphoethanolamine
- PG
phosphatidylglycerol
- TLR4
Toll-like receptor 4
Footnotes
Supporting information available
The negative ion ESI/MS spectrum of the lipids of strain CMR100 is shown in Supporting Fig. 1, demonstrating the accumulation of lipid IVA when compared to a wild-type control. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Brabetz W, Muller-Loennies S, Holst O, Brade H. Deletion of the heptosyltransferase genes rfaC and rfaF in Escherichia coli K-12 results in an Re-type lipopolysaccharide with a high degree of 2-aminoethanol phosphate substitution. Eur. J. Biochem. 1997;247:716–724. doi: 10.1111/j.1432-1033.1997.00716.x. [DOI] [PubMed] [Google Scholar]
- 2.Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raetz CRH, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raetz CRH, Garrett TA, Reynolds CM, Shaw WA, Moore JD, Smith DC, Jr, Ribeiro AA, Murphy RC, Ulevitch RJ, Fearns C, Reichart D, Glass CK, Benner C, Subramaniam S, Harkewicz R, Bowers-Gentry RC, Buczynski MW, Cooper JA, Deems RA, Dennis EA. (Kdo)2-lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J. Lipid Res. 2006;47:1097–1111. doi: 10.1194/jlr.M600027-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S, Du X, Hoebe K. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu. Rev. Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- 7.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 8.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 9.Rick PD, Fung LW-M, Ho C, Osborn MJ. Lipid A mutants of Salmonella typhimurium. Purification and characterization of a lipid A precursor produced by a mutant in 3-deoxy-D-mannooctulosonate-8-phosphate synthetase. J. Biol. Chem. 1977;252:4904–4912. [PubMed] [Google Scholar]
- 10.Rick PD, Osborn MJ. Lipid A mutants of Salmonella typhimurium. Characterization of conditional lethal mutants in 3-deoxy-D-mannooctulosonate-8-phosphate synthetase. J. Biol. Chem. 1977;252:4895–4903. [PubMed] [Google Scholar]
- 11.Raetz CRH, Purcell S, Meyer MV, Qureshi N, Takayama K. Isolation and characterization of eight lipid A precursors from a 3-deoxy-D-manno-octulosonic acid-deficient mutant of Salmonella typhimurium. J. Biol. Chem. 1985;260:16080–16088. [PubMed] [Google Scholar]
- 12.Strain SM, Armitage IM, Anderson L, Takayama K, Qureshi N, Raetz CRH. Location of polar substituents and fatty acyl chains on lipid A precursors from a 3-deoxy-D-manno-octulosonic acid-deficient mutant of Salmonella typhimurium: Studies by 1H, 13C and 31P nuclear magnetic resonance. J. Biol. Chem. 1985;260:16089–16098. [PubMed] [Google Scholar]
- 13.Belunis CJ, Clementz T, Carty SM, Raetz CRH. Inhibition of lipopolysaccharide biosynthesis and cell growth following inactivation of the kdtA gene in Escherichia coli. J. Biol. Chem. 1995;270:27646–27652. doi: 10.1074/jbc.270.46.27646. [DOI] [PubMed] [Google Scholar]
- 14.Meredith TC, Woodard RW. Escherichia coli YrbH is a D-arabinose 5-phosphate isomerase. J Biol Chem. 2003;278:32771–32777. doi: 10.1074/jbc.M303661200. [DOI] [PubMed] [Google Scholar]
- 15.Mamat U, Meredith TC, Aggarwal P, Kuhl A, Kirchhoff P, Lindner B, Hanuszkiewicz A, Sun J, Holst O, Woodard RW. Single amino acid substitutions in either YhjD or MsbA confer viability to 3-deoxy-d-manno-oct-2-ulosonic acid-depleted Escherichia coli. Mol. Microbiol. 2008;67:633–648. doi: 10.1111/j.1365-2958.2007.06074.x. [DOI] [PubMed] [Google Scholar]
- 16.Meredith TC, Aggarwal P, Mamat U, Lindner B, Woodard RW. Redefining the requisite lipopolysaccharide structure in Escherichia coli. ACS Chem. Biol. 2006;1:33–42. doi: 10.1021/cb0500015. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Z, White KA, Polissi A, Georgopoulos C, Raetz CRH. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J. Biol. Chem. 1998;273:12466–12475. doi: 10.1074/jbc.273.20.12466. [DOI] [PubMed] [Google Scholar]
- 18.Doerrler WT, Gibbons HS, Raetz CRH. MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J. Biol. Chem. 2004;279:45102–45109. doi: 10.1074/jbc.M408106200. [DOI] [PubMed] [Google Scholar]
- 19.Doerrler WT, Raetz CRH. ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 2002;277:36697–36705. doi: 10.1074/jbc.M205857200. [DOI] [PubMed] [Google Scholar]
- 20.Doerrler WT, Reedy MC, Raetz CRH. An Escherichia coli mutant defective in lipid export. J. Biol. Chem. 2001;276:11461–11464. doi: 10.1074/jbc.C100091200. [DOI] [PubMed] [Google Scholar]
- 21.Klein G, Lindner B, Brabetz W, Brade H, Raina S. Escherichia coli K-12 suppressor-free mutants lacking early glycosyltransferases and late acyltransferases: minimal lipopolysaccharide structure and induction of envelope stress response. J. Biol. Chem. 2009;284:15369–15389. doi: 10.1074/jbc.M900490200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sperandeo P, Cescutti R, Villa R, Di Benedetto C, Candia D, Deho G, Polissi A. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J. Bacteriol. 2007;189:244–253. doi: 10.1128/JB.01126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sperandeo P, Pozzi C, Deho G, Polissi A. Non-essential Kdo biosynthesis and new essential cell envelope biogenesis genes in the Escherichia coli yrbG-yhbG locus. Res. Microbiol. 2006;157:547–558. doi: 10.1016/j.resmic.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 24.Wu T, McCandlish AC, Gronenberg LS, Chng SS, Silhavy TJ, Kahne D. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U S A. 2006;103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U S A. 2008;105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sperandeo P, Lau FK, Carpentieri A, De Castro C, Molinaro A, Deho G, Silhavy TJ, Polissi A. Functional analysis of the protein machinery required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. J. Bacteriol. 2008 doi: 10.1128/JB.00270-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma B, Reynolds CM, Raetz CR. Periplasmic orientation of nascent lipid A in the inner membrane of an Escherichia coli LptA mutant. Proc. Natl. Acad. Sci. U S A. 2008;105:13823–13828. doi: 10.1073/pnas.0807028105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran AX, Trent MS, Whitfield C. The LptA protein of Escherichia coli is a periplasmic lipid A binding protein involved in the lipopolysaccharide export pathway. J. Biol. Chem. 2008 doi: 10.1074/jbc.M802503200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suits MD, Sperandeo P, Deho G, Polissi A, Jia Z. Novel structure of the conserved gram-negative lipopolysaccharide transport protein A and mutagenesis analysis. J. Mol. Biol. 2008;380:476–488. doi: 10.1016/j.jmb.2008.04.045. [DOI] [PubMed] [Google Scholar]
- 30.Trent MS, Ribeiro AA, Doerrler WT, Lin S, Cotter RJ, Raetz CRH. Accumulation of a polyisoprene-linked amino sugar in polymyxin resistant mutants in Salmonella typhimurium and Escherichia coli. Structural characterization and possible transfer to lipid A in the periplasm. J. Biol. Chem. 2001;276:43132–43144. doi: 10.1074/jbc.M106962200. [DOI] [PubMed] [Google Scholar]
- 31.Rutten L, Geurtsen J, Lambert W, Smolenaers JJ, Bonvin AM, de Haan A, van der Ley P, Egmond MR, Gros P, Tommassen J. Crystal structure and catalytic mechanism of the LPS 3-O-deacylase PagL from Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U S A. 2006;103:7071–7076. doi: 10.1073/pnas.0509392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trent MS, Pabich W, Raetz CRH, Miller SI. A PhoP/PhoQ-induced lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J. Biol. Chem. 2001;276:9083–9092. doi: 10.1074/jbc.M010730200. [DOI] [PubMed] [Google Scholar]
- 33.Bishop RE. The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis. Mol. Microbiol. 2005;57:900–912. doi: 10.1111/j.1365-2958.2005.04711.x. [DOI] [PubMed] [Google Scholar]
- 34.Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CRH. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. Embo J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang PM, Choy WY, Lo EI, Chen L, Forman-Kay JD, Raetz CRH, Prive GG, Bishop RE, Kay LE. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc. Natl. Acad. Sci. U S A. 2002;99:13560–13565. doi: 10.1073/pnas.212344499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 37.Miller JR. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 38.Six DA, Carty SM, Guan Z, Raetz CRH. Purification and mutagenesis of LpxL, the Lauroyltransferase of Escherichia coli lipid A biosynthesis. Biochemistry. 2008;47:8623–8637. doi: 10.1021/bi800873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vorachek-Warren MK, Ramirez S, Cotter RJ, Raetz CRH. A triple mutant of Escherichia coli lacking secondary acyl chains on lipid A. J. Biol. Chem. 2002;277:14194–14205. doi: 10.1074/jbc.M200409200. [DOI] [PubMed] [Google Scholar]
- 40.Clementz T, Zhou Z, Raetz CRH. Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A; acylation by MsbB follows laurate incorporation by HtrB. J. Biol. Chem. 1997;272:10353–10360. doi: 10.1074/jbc.272.16.10353. [DOI] [PubMed] [Google Scholar]
- 41.Clementz T, Bednarski JJ, Raetz CRH. Function of the htrB high temperature requirement gene of Escherichia coli in the acylation of lipid A. HtrB catalyzed incorporation of laurate. J. Biol. Chem. 1996;271:12095–12202. doi: 10.1074/jbc.271.20.12095. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook JG, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor; 2001. [Google Scholar]
- 43.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 44.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dulbecco R, Vogt M. Plaque formation and isolation of pure lines with poliomyelitis viruses. J. Exp. Med. 1954;99:167–182. doi: 10.1084/jem.99.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bligh EG, Dyer JJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 47.McClerren AL, Endsley S, Bowman JL, Andersen NH, Guan Z, Rudolph J, Raetz CRH. A slow, tight-binding inhibitor of the zinc-dependent deacetylase LpxC of lipid A biosynthesis with antibiotic activity comparable to ciprofloxacin. Biochemistry. 2005;44:16574–16583. doi: 10.1021/bi0518186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Z, Ribeiro AA, Raetz CRH. High-resolution NMR spectroscopy of lipid A molecules containing 4-amino-4-deoxy-L-arabinose and phosphoethanolamine substituents. Different attachment sites on lipid A molecules from NH4VO3-treated Escherichia coli versus kdsA mutants of Salmonella typhimurium. J. Biol. Chem. 2000;275:13542–13551. doi: 10.1074/jbc.275.18.13542. [DOI] [PubMed] [Google Scholar]
- 49.Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CRH. An inner membrane enzyme in Salmonella typhimurium and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A. Induction in polymyxin resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 2001;276:43122–43131. doi: 10.1074/jbc.M106961200. [DOI] [PubMed] [Google Scholar]
- 50.Tran AX, Lester ME, Stead CM, Raetz CRH, Maskell DJ, McGrath SC, Cotter RJ, Trent MS. Resistance to the antimicrobial peptide polymyxin requires myristoylation of Escherichia coli and Salmonella typhimurium lipid A. J. Biol. Chem. 2005;280:28186–28194. doi: 10.1074/jbc.M505020200. [DOI] [PubMed] [Google Scholar]
- 51.Karow M, Georgopoulos C. The essential Escherichia coli msbA gene, a multicopy suppressor of null mutations in the htrB gene, is related to the universally conserved family of ATP-dependent translocators. Mol. Microbiol. 1993;7:69–79. doi: 10.1111/j.1365-2958.1993.tb01098.x. [DOI] [PubMed] [Google Scholar]
- 52.Polissi A, Georgopoulos C. Mutational analysis and properties of the msbA gene of Escherichia coli, coding for an essential ABC family transporter. Mol. Microbiol. 1996;20:1221–1233. doi: 10.1111/j.1365-2958.1996.tb02642.x. [DOI] [PubMed] [Google Scholar]
- 53.Karow M, Fayet O, Cegielska A, Ziegelhoffer T, Georgopoulos C. Isolation and characterization of the Escherichia coli htrB gene, whose product is essential for bacterial viability above 33°C in rich media. J. Bacteriol. 1991;173:741–750. doi: 10.1128/jb.173.2.741-750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl. Acad. Sci. U S A. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Breazeale SD, Ribeiro AA, McClerren AL, Raetz CRH. A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-amino-4-deoxy-L-arabinose. Identification and function of UDP-4-deoxy-4-formamido-L-arabinose. J. Biol. Chem. 2005;280:14154–14167. doi: 10.1074/jbc.M414265200. [DOI] [PubMed] [Google Scholar]
- 56.Wang X, McGrath SC, Cotter RJ, Raetz CRH. Expression cloning and periplasmic orientation of the Francisella novicida lipid A 4'-phosphatase LpxF. J. Biol. Chem. 2006;281:9321–9330. doi: 10.1074/jbc.M600435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barb AW, McClerren AL, Snehelatha K, Reynolds CM, Zhou P, Raetz CRH. Inhibition of lipid A biosynthesis as the primary mechanism of CHIR-090 antibiotic activity in Escherichia coli. Biochemistry. 2007;46:3793–3802. doi: 10.1021/bi6025165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steeghs L, de Cock H, Evers E, Zomer B, Tommassen J, van der Ley P. Outer membrane composition of a lipopolysaccharide-deficient Neisseria meningitidis mutant. Embo J. 2001;20:6937–6945. doi: 10.1093/emboj/20.24.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- 60.Wang X, Ribeiro AA, Guan Z, McGrath S, Cotter R, Raetz CRH. Structure and biosynthesis of free lipid A molecules that replace lipopolysaccharide in Francisella tularensis subsp. novicida. Biochemistry. 2006;45:14427–14440. doi: 10.1021/bi061767s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, Ribeiro AA, Guan Z, Abraham SN, Raetz CRH. Attenuated virulence of a Francisella mutant lacking the lipid A 4'-phosphatase. Proc. Natl. Acad. Sci. U S A. 2007;104:4136–4141. doi: 10.1073/pnas.0611606104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.